Abstract
STUDY QUESTION
Do mitochondrial DNA (mtDNA) copy number and heteroplasmy in human embryos affect the ongoing pregnancy rate?
SUMMARY ANSWER
Our study suggests that mtDNA copy number above a specific threshold is associated with the ongoing pregnancy rate.
WHAT IS KNOWN ALREADY
Mitochondria play a vital role in cell function. Recently, there has been increasing research on mtDNA as a biomarker of embryo implantation. Although reports showed that high levels of mtDNA in the blastocyst are associated with low implantation potential, other publications were unable to confirm this. Confounding factors may influence the mtDNA copy number in euploid embryos. On the other hand it has been speculated that both mtDNA heteroplasmy and copy number contribute to mitochondrial function. Next generation sequencing (NGS) allows us to study in depth mtDNA heteroplasmy and copy number simultaneously.
STUDY DESIGN, SIZE, DURATION
A prospective non-selection study was performed. We included 159 blastocyst biopsies from 142 couples who attended our clinic for preimplantation genetic testing for aneuploidies (PGT-A), from January 2017 to December 2017. All embryos were biopsied on Day 5 or Day 6. The aneuploid testing was performed by NGS. All blastocysts were diagnosed as euploid non-mosaic and were transferred. The mtDNA analysis was performed once the embryo diagnosis was known.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Sequencing reads mapping to the mtDNA genome were extracted from indexed bam files to identify copy number and heteroplasmy. The relative measure of mtDNA copy number was calculated by dividing the mtDNA reads by the nuclear DNA value to normalize for technical variants and the number of cells collected at the biopsy. All the results were subjected to a mathematical correction factor according to the embryo genome. Heteroplasmy was assigned by MitoSeek.
MAIN RESULTS AND THE ROLE OF CHANCE
The mean average copy number and SD of mtDNA per genome was 0.0016 ± 0.0012. Regarding heteroplasmy, 40 embryos were heteroplasmy carriers (26.32%). MtDNA variants were detected in coding and non-coding regions and the highest number of variants in an embryo was eight. With respect to IVF outcome for mtDNA copy number analysis, we set a threshold of 0.003 for the following analysis. The vast majority of the embryos were below the threshold (142/159, 89.31%) and 17 embryos were classified as having higher mtDNA levels. We showed a reduction in ongoing pregnancy rate associated with elevated mtDNA copy number (42.96% versus 17.65%, P < 0.05). This result was independent of maternal age and day of the biopsy: these factors were included as confounding factors because mtDNA copy number was negatively correlated with female age (25 –30 y: 0.0017 ± 0.0011, 30 –35 y: 0.0012 ± 0.0007, 35 –40 y: 0.0016 ± 0.0009, over 40 y: 0.0024 + 0.0017, P < 0.05). Embryos biopsied on Day 5 were more likely to have higher quantities of mtDNA compared with those biopsied on Day 6 (0.0017 versus 0.0009, P < 0.001). According to IVF outcome and heteroplasmy, a lower ongoing pregnancy rate was reported for embryos that carried more than two variants. However, this did not reach statistical significance when we compared embryos with a number of variants lower or higher than two (39.15 versus 20.0, P = 0.188). Finally, a clear positive association between the mtDNA variants and copy number was reported when we compare embryos with or without heteroplasmy (0.0013 ± 0.0009 versus 0.0025 ± 0.0014, P < 0.001) and among different numbers of variants (0:0.0013 ± 0.0009, 1–2:0.0023 ± 0.0012, >2:0.0043 ± 0.0014, P < 0.05).
LIMITATIONS, REASONS FOR CAUTION
A limitation may be the size of the sample and the high-throughput sequencing technology that might not have detected heteroplasmy levels below 2% which requires high sequence depth A clinical randomized trial comparing the clinical outcome after the transfer of embryos selected according to mtDNA levels or only by morphological evaluation will be necessary. More research into the impact of mtDNA heteroplasmy and copy number on IVF outcome is needed.
WIDER IMPLICATIONS OF THE FINDINGS
Our results demonstrate that embryos with elevated mtDNA copy number have a lower chance of producing an ongoing pregnancy. MtDNA copy number is higher in older women and is dependent upon the number of cell divisions that preceded biopsy. Moreover, our data suggest that mitochondrial activity could be a balance between functional capacity and relative mtDNA copy number.
STUDY FUNDING/COMPETING INTEREST(S)
There are no conflicts of interest or sources of funding to declare.
Trial registration number
Not applicable.
Keywords: mtDNA, ongoing pregnancy, heteroplasmy, PGT-A, implantation rate
Introduction
Mitochondria constitute the powerhouses of cells, producing the energy required for all cellular functions. In addition, mitochondria have important roles in calcium homoeostasis, fatty acid oxidation and apoptosis. Maternally inherited, mitochondria are highly dynamic organelles that continuously move, fuse and divide in response to variations in cellular energy demands. Mitochondria differ from all other organelles in animals in having their own genome in the form of mitochondrial DNA (mtDNA), a double-stranded circular 16569 bp DNA molecule in humans (Anderson et al., 1981), which contains 37 genes: 22 tRNAs, two rRNAs and 13 protein subunits of the electron transport chain. The mtDNA content of each human cell type is highly variable, with the extremes being the spermatozoa that contain only a few copies (May-Panloup et al., 2003) and mature oocytes with up to several hundred thousand mtDNA copies (Reynier et al., 2001). The mtDNA copy number is strictly regulated ensuring that mitochondria can generate appropriate levels of energy. Altered mtDNA copy number has been shown to be involved in cancer (Reznik et al., 2016), neurodegeneration disorders (Schon and Manfredi, 2003), ageing (Goldberg et al., 2018) and diabetes (Kwak et al., 2010).
WHAT DOES THIS MEAN FOR PATIENTS?
This research looks at whether the DNA found in cells called the mitochondria is important in the outcome of IVF treatment. Some previous reports have found that high levels of this mitochondrial DNA are linked with lower chances of an embryo implanting, but this has not been confirmed. Other studies suggest that the structure and whether there are different types of genetic material in the mitochondrial DNA may be relevant.
This research team looked at mitochondrial DNA samples from 159 blastocysts, which were then transferred. They analysed both the quantity and quality of the samples. Their research showed that embryos from women who were over 40 years old had higher levels of mitochondrial DNA than those from younger women, and that embryos with higher levels of mitochondrial DNA have a lower chance of producing an ongoing pregnancy. They say that more research in this field is needed to understand more about the role of mitochondrial DNA in the growing embryo.
Usually, mtDNA molecules of all the cells of an individual are homoplasmic, carrying only one genome with a given nucleotide sequence but due to the proximity of mtDNA with the respiratory chain, and the lack of protective histones and efficient repair mechanisms, the mtDNA mutation rate is almost 25 times higher than that of nuclear DNA (Lynch et al., 2006). Thus, a large number of mutations (>200) and polymorphisms have been described for mtDNA in its heteroplasmic forms (Brandon et al., 2005). In particular, recent studies have shown that mtDNA heteroplasmy is common in the general population (Payne et al., 2013; Ye et al., 2014).
Beyond a certain percentage of heteroplasmy the synchronization between the nuclear and mitochondrial genes may break down, particularly in terms of energy production (Reinhardt et al., 2013). In fact, during a bottleneck, genetic drift occurs together with mutation-specific negative and positive selection mechanisms at the level of oxidative phosphorylation (Otten et al., 2018).
Infertility is a common disease affecting up to 10% of all couples (Mascarenhas et al., 2012) Identifying the genetic basis of infertility is important in order to develop treatments and potentially improve the outcomes of ART. The functional role of mitochondria in infertility is becoming an increasingly important consideration. The mtDNA in oocytes or embryos have been shown to be involved in some causes of infertility: ovarian insufficiency (May-Panloup et al., 2016), endometriosis (Xu et al., 2015), female age (Fragouli et al., 2015) and aneuploidy of embryos (Diez-Juan et al., 2015). Mitochondrial function has also been proposed as a biomarker for embryo implantation (Seli, 2016).
About one-third to one-quarter of the morphologically and chromosomally normal embryos fail to implant. Research has been focused on new technologies, such as metabolomics (Marhuenda-Egea et al., 2010), that would assess embryo viability and ultimately allow transfer of a single competent embryo. One important factor in embryo implantation potential could be an adequate energy supply (Leese, 2012). It has been speculated that both mtDNA heteroplasmy (Shamsi et al., 2013) and copy number (Diez-Juan et al., 2015; Fragouli et al., 2015, 2017; Ravichandran et al., 2017) may contribute to embryo implantation potential. However, conflicting results in this area have already challenged the potential significance of mtDNA in embryo implantation (Treff et al., 2017; Victor et al., 2017) and further studies are needed in order to provide clarity. Moreover, the effects of mtDNA heteroplasmy and copy number were only discussed separately, with the results thus remaining inconclusive. Next generation sequencing (NGS) offers the great sensitivity required for the detection of low levels of heteroplasmy. Thus, NGS has provided the opportunity to conduct an in-depth analysis and allowed the study of mtDNA heteroplasmy and copy number simultaneously.
The aim of this study was to examine whether maternal and embryo factors influence mtDNA quantity and to investigate what influence mtDNA copy number and heteroplasmy in euploid non-mosaic embryos has on IVF outcome.
Materials and Methods
Study design
This study is a prospective non-selection study. We included blastocyst biopsies from couples who attended the Instituto Bernabeu for preimplantation genetic testing for aneuploidies (PGT-A). All blastocysts were diagnosed as euploid non-mosaic and were transferred. The mtDNA analysis was performed once the embryo diagnosis was known. We included 159 blastocysts biopsies from 142 couples from January 2017 to December 2017. Patients signed an informed consent form. This study was approved by the Instituto Bernabeu Institutional Review Board.
IVF procedures, embryo biopsy and aneuploidy testing
Briefly, all patients started controlled ovarian stimulation on Day 2–4 of their menstrual cycle with an initial dose of 150–225 UI/day of recombinant FSH alone (Gonal F. Merk-Serono, Madrid, Spain) or with HMG (Menopur. Ferring, Madrid, Spain) according to ovarian reserve parameters and BMI. When the lead follicle reached 13–14 mm a GnRH antagonist (Cetrotide®, Merck-Serono, Madrid, Spain) was administered daily (0.25 mg/day). Finally a GnRH agonist (Triptoreline, 0.2 mg, Decapeptyl®, Ipsen Pharma, Barcelona, Spain) was used for final oocyte maturation when follicles were >18 mm in diameter. Oocyte aspiration was performed 36 h after GnRH agonist injection s.c. by transvaginal ultrasound-guided needle-aspiration. Oocyte manipulation and ICSI were performed according to our own IVF laboratory guidelines. Fertilization was checked 16–18 h post ICSI. Zygotes were cultured in bench-top incubators (MINC-Cook and Planer-Origio, Barcelona, Spain) with low oxygen tension (5%) and a single-phase culture medium (Global Total®: LifeGlobal, Canada) was used for embryo development. Two or three laser shots at a pulse of 0.536 ms were used to breach the zona pelluzida on Day 3. Blastocysts graded ≥3 (Gardner et al., 2000) with herniating cells were biopsied with the same laser pulses on Day 5 or 6. A Saturn Active laser from Research Instruments (RI, Germany) was used for this purpose. Three to six trophoectoderm cells were collected from the blastocysts and tubed for aneuploidy testing. Whole genome amplification on each biopsy was performed using the Sureplex method and followed by NGS using the Veriseq protocol (Illumina®) with the MiSeq Sequencer (Illumina®). Analyses for aneuploidy testing were performed using Bluefuse Multi Software (Illumina). All embryos were vitrified after biopsy. Euploid blastocysts were subsequently thawed after preparation and synchronization of the endometrium. For vitrification and warming we used the Irvine protocol (IrvineScientific, CA, USA). A single euploid non-mosaic blastocyst was transferred to each patient. For the embryo transfer patients received hormonal replacement therapy with oral or transdermal oestrogens, adding vaginal pessaries of progesterone, once the endometrial thickness was above 7 mm and showed a trilaminar pattern. All the procedures were performed by senior gynecologists.
Determination of mtDNA copy number and heteroplasmy
Sequencing reads mapping to the mtDNA genome were extracted from indexed bam files to identify copy number and heteroplasmy aligning to the mtDNA reference genome, as per the Genome Reference Consortium (GRC)h37. The relative measure of mtDNA copy number was calculated by dividing the mtDNA reads by the nuclear DNA value to normalize for technical batch to batch variation and the number of cells collected at the biopsy. All the results were subjected to a mathematical correction factor according to the embryo genome (Victor et al., 2017). For all euploid male embryos a multiplier of 0.9842 was included, because the male genome is 98.42% the length of the female genome. Regarding heteroplasmy, the NGS data for each sample was then remapped against the representative sequence in order that variant selection could be performed. Heteroplasmy was identified with the following criteria: a sequencing coverage >40× a minor allele frequency ≥2 and observed on both strands using the MitoSeek pipeline, i an open-source software tool. Only substitution variants were studied.
Statistics
All the data are represented using mean ± SD. For maternal and embryo factors, linear models and ANOVA were used to test for correlations with mtDNA copy number. Logistic regression models were utilized to test if relative mtDNA levels and heteroplasmy predict overall outcome among all embryos transferred. Maternal age, embryo quality and the day of the biopsy were included as confounding factors. Logistic regression was used to detect any correlation between mtDNA and heteroplasmy. A P-value <0.05 was considered to be statistically significant. The statistical analysis was performed in R Statistical Software version 3.4 (The R Foundation) and Statistical Package for the Social Sciences (SPSS) software (version 20.0, SPSS, Inc., Chicago, IL, USA). The ROCR package (Sing et al., 2005) was also used. ROCR is a package for evaluating and visualizing the performance of scoring classifiers using the statistical language R.
Results
Maternal and embryo factors
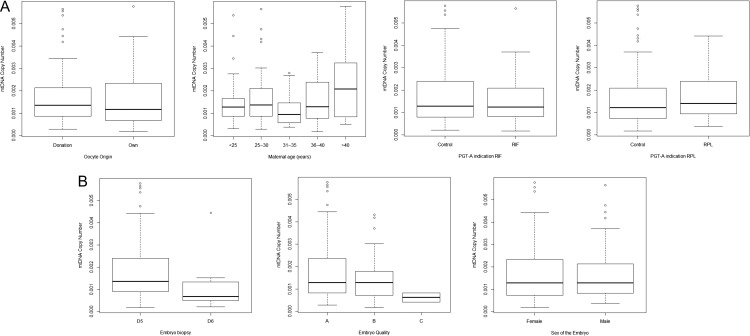
First we evaluated whether maternal and embryological parameters were associated with mtDNA copy number (Fig. 1). Concerning maternal factors, there are no statistically significant differences according to the egg origin, i.e. donated or own egg (0.0017 ± 0.0012 versus 0.0017 ± 0.0011, respectively; P = 0.423). Second, regarding age, mtDNA was negatively correlated with female age (ranges 25 –30 y: 0.0017 ± 0.0011, 31 –35 y: 0.0012 ± 0.0007, 36 –40 y: 0.0016 ± 0.0009, older than 40 y: 0.0024 ± 0.0017, P = 0.044). Post-hoc analysis reveals that embryos from women older than 40 y had higher mtDNA levels (copy number) when compared to embryos from younger patients (0.0024 ± 0.0017 versus 0.0015 ± 0.0011, respectively; P = 0048). Third, no significant differences in mtDNA copy number were reported according to the indication for PGT-A: recurrent implantation failure (control 0.0016 ± 0.0012 versus RIF patients 0.0015 ± 0.0010; P = 0.365) or repeated miscarriage (control 0.0016 ± 0.0012 versus RPL patients 0.0017 ± 0.0010; P = 0.523). With regards to embryo factors, embryos biopsied on Day 5 were more likely to have higher quantities of mtDNA compared with those biopsied on Day 6 (0.0017 ± 0.0011 versus 0.0009 ± 0.0008, respectively: P = 0.0014). The quality of the blastocyst, extrapolating Gardner’s criteria to the Spanish classification (Spanish Association for the Study of the Biology of Reproduction, ASEBIR, www.asebir.com) was not associated with mtDNA copy number (Grade A:0.0017 ± 0.0012, Grade B:0.0015 ± 0.0009, Grade C: 0.0006 ± 0.0003; P = 0.304). Finally, we also examined the possible relationship between the sex of the embryo and mtDNA copy number. We did not observe any significant differences in mtDNA levels between male and female embryos (0.0016 ± 0.0012 versus 0.0016 ± 0.0011, respectively: P = 0.851).
Figure 1.
Association between mitochondrial DNA copy number and maternal and embryo factors. (A) mitochondrial DNA (mtDNA) and maternal factors, (B) mtDNA and embryo factors. PGT-A, preimplantation genetic testing for aneuploidies; RIF, recurrent implantation failure; RPL, recurrent pregnancy loss.
mtDNA copy number and IVF outcome
We analysed the mtDNA from 159 non-mosaic euploid embryos corresponding to 142 couples who underwent PGT-A with frozen single embryo transfer. Euploid embryos were selected for transfer based on the standard morphological criteria. The average maternal age at time of oocyte retrieval was 30.76 ± 6.81 years (range: 22–45 y). Of these, 51.56% correspond to donated oocytes. According to IVF outcome, 41.5% of the embryos failed to result in a pregnancy and 42% resulted in an ongoing pregnancy. The overall implantation rate was 43.40% per embryo.
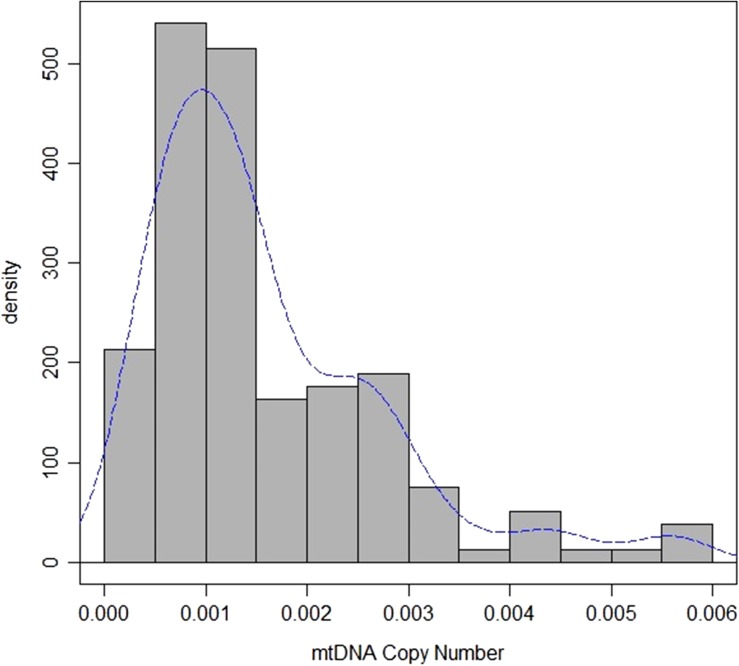
With respect to mtDNA copy number and IVF outcome, quantification of mtDNA was successful in all cases. The average copy number of mtDNA per embryo was 0.0016 ± 0.0012. The distribution of mtDNA quantity in the euploid blastocyst showed a positive asymmetry and a long kurtosis (Fig. 2). Due to the fact that mitochondrial dysfunction is only revealed when overall mitochondrial function drops below a threshold, we set a cut-off. In order to select the most extreme values we chose the 90 percentile of mtDNA values to set the threshold. The value of 0.003 was then used as a threshold for the following analysis. The vast majority of the embryos are below the threshold (142/159, 89.31%), while 17 embryos were classified as having high mtDNA levels. When we compared embryos with mtDNA below or above the threshold, respectively, no significant differences were reported for pregnancy rate (59.15% versus 47.10%, P = 0.33), biochemical pregnancy (13.38% versus 17.65%, P = 0.63) and implantation rate (45.77% versus 29.41% P = 0.19). However, we detected an increase in the miscarriage rate in embryos with high mtDNA (6.15% versus 40.00%, P < 0.05). Hence, a lower ongoing pregnancy rate is associated with elevated mtDNA (42.96% versus 17.65% P < 0.05) (Table I). This result was corrected for maternal age, day of the biopsy, egg origin (own or donated) and embryo morphology, as these factors were included as confounding factors.
Figure 2.
Distribution of mtDNA copy number in the trophoectoderm of human non-mosaic euploid embryos included in the study. The distribution of mtDNA quantity in the euploid blastocyst shows a positive asymmetry and a long kurtosis.
Table I.
Comparison of IVF cycle outcome between embryos with a mitochondrial DNA copy number lower or higher than the threshold value.
| mtDNA levels | LOW | HIGH | P |
|---|---|---|---|
| n | 142 | 17 | |
| Pregnancy rate n (%) | 59.15 (84) | 47.10 (8) | 0.33 |
| Biochemical miscarriage n (%) | 13.38 (19) | 17.65 (3) | 0.63 |
| Implantation rate n (%) | 45.77 (65) | 29.41 (5) | 0.19 |
| Miscarriage rate n (%) | 6.15 (4) | 40.0 (2) | 0.011* |
| Ongoing pregnancy rate n (%) | 42.96 (61) | 17.65 (3) | 0.048+ |
Statistical analysis was by logistic regression including confounding factors (maternal age, embryo quality and the day of the biopsy).
mtDNA, mitochondrial DNA.
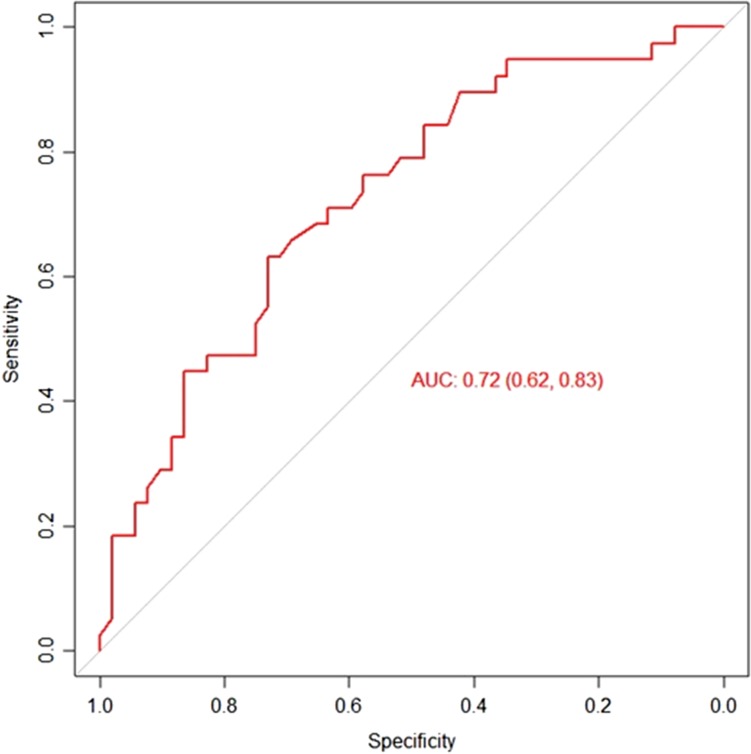
With the combination of all the factors with significance in mtDNA copy number, a predictive performance of the embryo implantation score was calculated using the mtDNA levels and the receiver operating characteristic (ROC) curve. The AUC was 0.72 with a specificity of 83% (Fig. 3).
Figure 3.
Receiver operating characteristic curve analysis to calculate the discriminative power of mtDNA to predict an ongoing pregnancy.
Heteroplasmy and IVF outcome
Regarding heteroplasmy, all the blastocyst biopsies were analysed for mtDNA mutations. The average depth for the mtDNA sequence reads was 205. According to our criteria described previously to detect heteroplasmic sites, we observed that 40 embryos were heteroplasmy carriers (26.32%). The number of variants per embryo ranges from one to eight. The vast majority of heteroplasmic embryos (70%) carried one mtDNA variant. Mutations in heteroplasmic form were detected in the coding and non-coding regions. Heteroplasmy was detected in 53 different positions across different regions of the mtDNA, and 46 (88.46%) of these sites were heteroplasmic in only one embryo. The majority of heteroplasmies were at low frequency, and the mean rate of heteroplasmy per embryo was 4.25%. Of the 53 detected heteroplasmies, 24 (45.28%) occurred in coding regions and only two out of 24 were synonymous. The highest number of coding heteroplasmic sites was located in the ND3 gene, encoding the ND3 protein. The ND3 protein is a subunit of NADH dehydrogenase, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain.
As for the relationship between the presence of heteroplasmy and the IVF outcome, when we compared the IVF outcome between embryos with different number of variants it appears that increased numbers were associated with a higher biochemical miscarriage rate (0 variants: 13.45%, 1–2 variants: 11.43%, >2 variants: 40.0% P = 0.22) and lower implantation (0:44.54%, 1–2:51.42%, >2:20.0; P = 0.39) and ongoing pregnancy rate (0:41.18%, 1–2:42.85%, >2:20.0%; P = 0.60) without reaching statistical significance (Table II). No significant difference was reported in pregnancy rate (0:57.98%, 1–2:62.9%, 60.0%; P = 0.87) and miscarriage (0:7.84%, 1–2:12.50%, >2:0%; P = 0.53) according to number of mtDNA variants (Table II).
Table II.
Comparison of IVF cycle outcome according to the number of heteroplasmic sites in mtDNA.
| Heteroplasmic sites | Not carrier | 1–2 | >2 | P |
|---|---|---|---|---|
| n | 119 | 35 | 5 | |
| Pregnancy rate (%) | 57.98 (69) | 62.9 (22) | 60.0 (3) | 0.87 |
| Biochemical miscarriage (%) | 13.45 (16) | 11.43 (4) | 40.0 (2) | 0.22 |
| Implantation rate (%) | 44.54 (53) | 51.42 (18) | 20.0 (1) | 0.39 |
| Miscarriage rate (%) | 7.84 (4) | 12.50 (3) | 0 (0) | 0.53 |
| Ongoing pregnancy rate (%) | 41.18 (49) | 42.85 (15) | 20.0(1) | 0.60 |
Statistical analysis was by logistic regression including confounding factors (maternal age, embryo quality and the day of the biopsy were included as confounding factors).
Heteroplasmy and mtDNA copy number
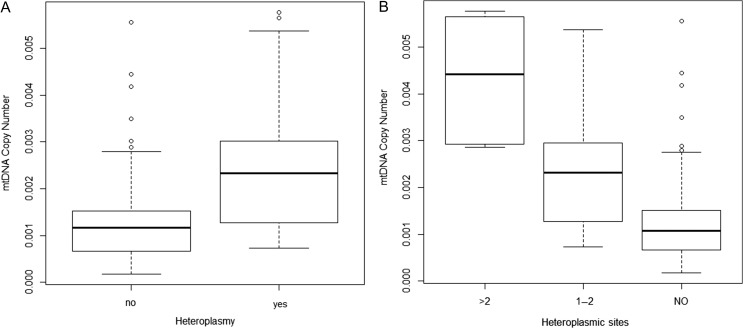
Finally, a clear association between the presence of heteroplasmy and mtDNA copy number was reported (0.0013 versus 0.0025, P < 0.001) (Fig. 4A). Moreover, when we compared the mtDNA copy number among the number of heteroplasmic positions it appears that there is a clear difference in terms of mitochondrial activities (Fig. 4B). Embryos with a higher number of heteroplasmic sites carried a higher mtDNA copy number (0 variants: 0.0013 ± 0.0009, 1–2 variants: 0.0023 ± 0.0011, >2 variants: 0.0043 ± 0.0014; P < 0.001). Thus, heteroplasmy was more prevalent in blastocysts with high mtDNA levels than in embryos with lower quantities of mtDNA.
Figure 4.
Association between mtDNA copy number and the presence of heteroplasmy and number of heteroplasmic sites.
Discussion
In this study, we evaluated the possible influence of maternal and embryonic factors on mtDNA levels in order to show if previous research discrepancies in the effect of mtDNA levels on embryo implantation are related to confounding factors. We focused not only on the quantitative (copy number) but also the qualitative (heteroplasmy) changes in mtDNA in human embryos. Our data suggested that an elevated mtDNA copy number in euploid non-mosaic embryos is indicative of lower embryo implantation potential. Although there were no statistically significant differences in pregnancy rate, it appears that high heteroplasmy numbers were associated with reduced pregnancy rate, however a higher sample size will corroborate this finding. In addition, heteroplasmy seems to affect mtDNA copy numbers.
We want to identify what maternal and embryo factors may affect the mtDNA levels. With regards to maternal factors, our study showed a significant association between mtDNA and maternal age providing evidence that preimplantation embryos from older patients have higher levels of mtDNA than embryos from younger women. The association of age and mtDNA levels has been studied previously. Published results have not been entirely concordant, but most reported that mtDNA levels are increased in blastocysts from older women (Fragouli et al., 2017; Ravichandran et al., 2017). Our result agrees with those studies and suggests that blastocysts from older women may have more demanding energy requirements due to continuously declining organelle function over time (Fragouli et al., 2015), although the relationship between both factors is complicated to show because a single organelle may contain more than one copy of mtDNA. Concerning embryo factors, mtDNA copy number was significantly lower in embryos biopsed on Day 6 compared with those biopsed on Day 5. There was a dilution effect in relative mtDNA content in Day 6 trophoectoderm biopsies versus Day 5 trophoectoderm biopsies from euploid embryos. Thus, given that the mitochondrial content in a given cell of a preimplantation embryo is dependent upon the number of cell divisions that preceded the biopsy, it is useful to evaluate whether embryologic parameters also influence mtDNA quantity in embryo biopsy samples. It is possible that the higher mtDNA quantity observed in certain embryos in prior studies was a function of fewer cell divisions in that individual embryo prior to biopsy and thus less dilution of mitochondria in the analysed sample. Based on mouse models, it has been alleged that no additional mtDNA replication occurs between fertilization and the early post-implantation stage (Pikó and Taylor, 1987). As a result, the total amount of mtDNA must be split among cells during embryo division, thus by Day 6 of development each embryonic cell should contain very few copies of mtDNA. The discrepancy in the previous studies could represent the confounding influence of the day when the embryo was biopsied and the maternal age.
Mitochondrial numbers are quite variable in oocytes and embryos, suggesting that mitochondrial activity is not dictated solely by copy number (Chappel, 2013). Adequate amounts of mitochondrial activity are strictly regulated by nuclear signals, intracellular ion concentration and the availability of substrates (Dumollard et al., 2006, 2007). Since mitochondria contain multiple copies of DNA and cells may contain hundreds or thousands of mitochondria, a mutational load may be tolerated before a deficit in cellular function is apparent. An estimation of 18% mutation level has been associated with a 95% or higher chance of being unaffected (Hellebrekers et al., 2012). It suggests that mitochondrial dysfunction is only revealed when overall mitochondrial function drops below a threshold (Rossignol et al., 2003). A study using mice lacking the mitochondrial transcription factor A (TFAM), which is essential for the replication and maintenance of mitochondrial DNA, has demonstrated that oocytes and embryos must contain a threshold copy number of mtDNA to support the developmental competence of embryos to grow into viable foetuses (Wai et al., 2010). According to these previous works, we evaluated the effect of mtDNA copy number by setting a cut-off. Our observations suggest that embryos with mtDNA copy number above the threshold have a lower chance to achieve an ongoing pregnancy. Despite published data supporting our results (Diez-Juan et al., 2015; Spinella et al., 2016; Fragouli et al., 2017), not all studies are in agreement. Victor and colleagues (2017) failed to detect a relationship between mtDNA levels and embryo implantation suggesting that a correction factor calculation should be included in order to accurately evaluate the mtDNA levels. We had used the proposed outlined correction factor from Victor et al. (2017) to take the variability of embryonic genomes into account. Even so, we reported that mtDNA levels are a useful tool for pregnancy assessment. Another recent study (Treff et al., 2017) was also unable to show an association between mtDNA copy number and embryo viability, however technical limitations, such as storage of samples, could be a possible explanation. In our study, samples were not stored prior to analysis as they were analysed in the current PGT-A cycle. According to our results, the clinical application is the incorporation of mtDNA copy number analysis into the routine genetic analyses performed by NGS. The ROC curve obtained showed its value as a clinical tool to improve embryo selection. The ROC curve plays a central role in evaluating the diagnostic ability of tests. The derived summary measure of accuracy, such as the AUC, determines the inherent ability of the test to discriminate between the two possible results. The AUC is an effective and combined measure of sensitivity and specificity that describes the inherent validity of diagnostic tests. The reasonable AUC obtained in our model suggests that mtDNA levels have predictive value in discriminating between embryos that are able to achieve an ongoing pregnancy and those that are not. However, the overall percentage of embryos with mtDNA above the threshold is around 10% in most clinics (Wells, 2017) and thereby this limits the clinical value of assessing this feature. Nevertheless, considering that mtDNA quantification is inexpensive for embryos that have already undergone PGT-A, cost-effectiveness remains positive.
It is generally assumed that each individual is characterized by a single mtDNA type, but in fact even an isolated cell can harbour a population of distinct mtDNA genomes, termed heteroplasmy. A recent study reported clear evidence in favour of the existence of purifying selection from mtDNA mutations in human oocytes (De Fanti et al., 2017), however, little is known about the load of mutations in the mtDNA of human embryos and the possible effect of these mutations. To investigate this, we analysed the mtDNA sequence reads obtained from the NGS files for PGT-A. We adjusted the criteria for heteroplasmy identification according to the type of fragments that we obtained after the library preparation (Zhang et al., 2016). According to this criterion we found heteroplasmic changes over the entire mitochondrial genome in 26.32% of the embryos. This result agrees with previous research (Boucret et al., 2017), which means that probably ~1% of the embryos will carry a pathogenic point mutation. This would account for 1:10 000 de novo cases of mtDNA disease (Jacobs et al., 2007). We observed that the ongoing pregnancy rate, biochemical pregnancy and the embryo implantation rate differed among groups, without reaching statistical significance (1–2 variants, >2 variants and homoplasmic). The limited sample size could be an explanation for this result. More data are needed to determine the effect of mtDNA heteroplasmy on embryo developmental potential.
We next tested for a correlation between mtDNA copy number and heteroplasmy. We reported a clear association between heteroplasmy and mtDNA levels. Our data are also consistent with the idea that mtDNA copy number and heteroplasmy can influence each other (Zhang et al., 2017). In mitochondrial disease, a compensatory increase in mtDNA copy number via mitochondrial biogenesis may effectively compensate for heteroplasmic mtDNA mutations and mitochondrial dysfunction (Kauppila et al., 2017). An increase in the mtDNA content of human preimplantation embryos in response to mutation has previously been documented (Monnot et al., 2013) suggesting that human preimplantation embryos have the intrinsic ability to adapt their mtDNA content in response to impaired respiratory capacities triggered by high mtDNA mutant loads. Thus, mtDNA levels are a consequence of a compensatory mechanism in embryos carrying functionally deficient organelles caused by mutations of the mitochondrial genome. Another possible hypothesis that could explain why embryos with higher mtDNA have a lower chance to achieve an ongoing pregnancy is in line with the ‘quiet embryo hypothesis’ which postulates that under ideal circumstances embryos are engaged in low metabolic activity (Leese, 2002). Conditions that deviate from the steady state, such as aneuploidy, advance maternal age or chemically induced stress, tend to associate with higher mtDNA content (Victor et al., 2017). However, a relationship between cellular stress and more DNA molecules in mitochondria remains unclear. A confounding aspect is that a mitochondrial organelle can contain a broad range of mtDNA molecules. High mtDNA content in a cell does not mean a large number of mitochondria. Thus, elevated numbers of mitochondria do not necessarily mean enhanced mitochondrial function or metabolic activity (Viotti et al., 2017). We have demonstrated a link between mutation of the mitochondrial genome and the increased mtDNA levels associated with reduced implantation. An accumulation of mutations in mtDNA may limit energy production. As a result, the cell has decreased the capacity to support all cellular events and especially normal segregation during cell division (Chappel, 2013), therefore will negatively impact on the development of an embryo (Schatten et al., 2014).
In conclusion, to the best of our knowledge this is the first time that a comprehensive mtDNA study has been performed and associated with IVF outcome. Our study goes one step forward by not only analysing the quantity of mtDNA but also the quality. Our results have demonstrated that embryos with elevated mtDNA content have a lower chance to produce an ongoing pregnancy. Moreover, our data suggest a link between mutation of the mtDNA (heteroplasmy) and the increased mtDNA copy number. Mitochondrial activity is a balance between functional capacity (mutational load), absolute mtDNA copy number within the cell and motility of the organelle (Suzuki, Toyokawa, 2005). In addition, our data suggest that blastocysts from older women had a higher mtDNA content, thus oocyte ageing could affect ooplasmic factors. Moreover, our results suggest that the mtDNA in a given cell of an embryo is dependent upon the number of cell divisions that preceded biopsy because higher levels of mtDNA were found in Day 5 than Day 6 biopsied embryos. Finally, to better understand the involvement of mtDNA in embryo development and its clinical utility more research into mitochondrial function is needed and clinical randomized trials will be necessary.
Authors’ roles
B.L., J.A.O. and R.M.: study design. A.B, R.B. and J.LL.: patient recruitment. B.L., E.G. J.A.O. and R.M.: laboratory experiments. B.L. and J.A.O.: analysis. J.T., embryology laboratory data collection. B.L., manuscript drafting. B.L., J.A.O., R.M., J.LL, E.G. A.B., J.T and R.B.: critical discussion.
Funding
There was no external source of funding for this study.
Conflict of interest
There are none to declare.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F et al. Sequence and organization of the human mitochondrial genome. Nature 1981;290:457–465. [DOI] [PubMed] [Google Scholar]
- Boucret L, Bris C, Seegers V, Goudenège D, Desquiret-Dumas V, Domin-Bernhard M, Ferré-L’Hotellier V, Bouet PE, Descamps P, Reynier P et al. Deep sequencing shows that oocytes are not prone to accumulate mtDNA heteroplasmic mutations during ovarian ageing. Hum Reprod 2017;32:2101–2109. [DOI] [PubMed] [Google Scholar]
- Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC. MITOMAP: a human mitochondrial genome database—2004 update. Nucleic Acids Res 2005;33:D611–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappel S. The role of mitochondria from mature oocyte to viable blastocyst. Obstet Gynecol Int 2013;2013:183024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fanti S, Vicario S, Lang M, Simone D, Magli C, Luiselli D, Gianaroli L, Romeo G. Intra-individual purifying selection on mitochondrial DNA variants during human oogenesis. Hum Reprod 2017;32:1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, Díaz-Gimeno P, Valbuena D, Simón C. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril 2015;104:534–541. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol 2007;77:21–49. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Sardet C. Calcium signals and mitochondria at fertilisation. Semin Cell Dev Biol 2006;17:314–323. [DOI] [PubMed] [Google Scholar]
- Fragouli E, McCaffrey C, Ravichandran K, Spath K, Grifo JA, Munné S, Wells D. Clinical implications of mitochondrial DNA quantification on pregnancy outcomes: a blinded prospective non-selection study. Hum Reprod 2017;32:2340–2347. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. PLoS Genet 2015;11:e1005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril 2000;73:1155–1158. [DOI] [PubMed] [Google Scholar]
- Goldberg J, Currais A, Prior M, Fischer W, Chiruta C, Ratliff E, Daugherty D, Dargusch R, Finley K, Esparza-Moltó PB et al. The mitochondrial ATP synthase is a shared drug target for aging and dementia. Aging Cell 2018;17:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers DM, Wolfe R, Hendrickx AT, de Coo IF, de Die CE, Geraedts JP, Chinnery PF, Smeets HJ. PGD and heteroplasmic mitochondrial DNA point mutations: a systematic review estimating the chance of healthy offspring. Hum Reprod Update 2012;18:341–349. [DOI] [PubMed] [Google Scholar]
- Jacobs L, Gerards M, Chinnery P, Dumoulin J, de Coo I, Geraedts J, Smeets H. mtDNA point mutations are present at various levels of heteroplasmy in human oocytes. Mol Hum Reprod 2007;13:149–154. [DOI] [PubMed] [Google Scholar]
- Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: an update. Cell Metab 2017;25:57–71. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Park KS, Lee KU, Lee HK. Mitochondrial metabolism and diabetes. J Diabetes Investig 2010;1:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. Bioessays 2002;24:845–849. [DOI] [PubMed] [Google Scholar]
- Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction 2012;143:417–427. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science 2006;311:1727–1730. [DOI] [PubMed] [Google Scholar]
- Marhuenda-Egea FC, Martínez-Sabater E, Gonsálvez-Alvarez R, Lledó B, Ten J, Bernabeu R. A crucial step in assisted reproduction technology: human embryo selection using metabolomic evaluation. Fertil Steril 2010;94:772–774. [DOI] [PubMed] [Google Scholar]
- Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S, Stevens GA. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med 2012;9:e1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Panloup P, Boucret L, Chao de la Barca JM, Desquiret-Dumas V, Ferré-L’Hotellier V, Morinière C, Descamps P, Procaccio V, Reynier P. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update 2016;22:725–743. [DOI] [PubMed] [Google Scholar]
- May-Panloup P, Chrétien MF, Savagner F, Vasseur C, Jean M, Malthièry Y, Reynier P. Increased sperm mitochondrial DNA content in male infertility. Hum Reprod 2003;18:550–556. [DOI] [PubMed] [Google Scholar]
- Monnot S, Samuels DC, Hesters L, Frydman N, Gigarel N, Burlet P, Kerbrat V, Lamazou F, Frydman R, Benachi A et al. Mutation dependance of the mitochondrial DNA copy number in the first stages of human embryogenesis. Hum Mol Genet 2013;22:1867–1872. [DOI] [PubMed] [Google Scholar]
- Otten ABC, Sallevelt SCEH, Carling PJ, Dreesen JCFM, Drüsedau M, Spierts S, Paulussen ADC, de Die-Smulders CEM, Herbert M, Chinnery PF et al. Mutation-specific effects in germline transmission of pathogenic mtDNA variants. Hum Reprod 2018;33:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet 2013;22:384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikó L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol 1987;123:364–374. [DOI] [PubMed] [Google Scholar]
- Ravichandran K, McCaffrey C, Grifo J, Morales A, Perloe M, Munne S, Wells D, Fragouli E. Mitochondrial DNA quantification as a tool for embryo viability assessment: retrospective analysis of data from single euploid blastocyst transfers. Hum Reprod 2017;32:1282–1292. [DOI] [PubMed] [Google Scholar]
- Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science 2013;341:1345–1346. [DOI] [PubMed] [Google Scholar]
- Reynier P, May-Panloup P, Chrétien MF, Morgan CJ, Jean M, Savagner F, Barrière P, Malthièry Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod 2001;7:425–429. [DOI] [PubMed] [Google Scholar]
- Reznik E, Miller ML, Şenbabaoğlu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016;5:e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J 2003;15:751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H, Sun Q, Prather R. The impact of mitochondrial function/dysfunction on IVF and new treatment possibilities for infertility. Reprod Biol Endocrinol 2014;12:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Manfredi G. Neuronal degeneration and mitochondrial dysfunction. J Clin Invest 2003;111:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seli E. Mitochondrial DNA as a biomarker for in-vitro fertilization outcome. Curr Opin Obstet Gynecol 2016;28:158–163. [DOI] [PubMed] [Google Scholar]
- Shamsi MB, Govindaraj P, Chawla L, Malhotra N, Singh N, Mittal S, Talwar P, Thangaraj K, Dada R. Mitochondrial DNA variations in ova and blastocyst: implications in assisted reproduction. Mitochondrion 2013;13:96–105. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:7881. [DOI] [PubMed] [Google Scholar]
- Spinella F, Cotroneo E, Bono S, Biricik A, Greco E, Minasi MG et al. Quantification of mitochondrial DNA in preimplantation embryos: a tool to predict implantation potential of chromosomally normal embryos. Hum Reprod 2016;31:i26. [Google Scholar]
- Suzuki S, Toyokawa K. Changes in distribution of active mitochondria during oocyte maturation and fertilization in the Hamster. J Mamm Ova Res 2005;22:163–169. [Google Scholar]
- Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, Morrison L, Morin SJ, Scott RT Jr.. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod 2017;32:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor AR, Brake AJ, Tyndall JC, Griffin DK, Zouves CG, Barnes FL, Viotti M. Accurate quantitation of mitochondrial DNA reveals uniform levels in human blastocysts irrespective of ploidy, age, or implantation potential. Fertil Steril 2017;107:34–42.e3. [DOI] [PubMed] [Google Scholar]
- Viotti M, Victor AR, Zouves CG, Barnes FL. Is mitochondrial DNA quantitation in blastocyst trophectoderm cells predictive of developmental competence and outcome in clinical IVF? J Assist Reprod Genet 2017;34:1581–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod 2010;83:52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D. Mitochondrial DNA quantity as a biomarker for blastocyst implantation potential. Fertil Steril 2017;108:742–747. [DOI] [PubMed] [Google Scholar]
- Xu B, Guo N, Zhang XM, Shi W, Tong XH, Iqbal F, Liu YS. Oocyte quality is decreased in women with minimal or mild endometriosis. Sci Rep 2015;5:10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci USA 2014;111:10654–10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Nakahira K, Guo X, Choi AM, Gu Z. Very short mitochondrial DNA fragments and heteroplasmy in human plasma. Sci Rep 2016;6:36097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wang Y, Ye K, Picard M, Gu Z. Independent impacts of aging on mitochondrial DNA quantity and quality in humans. BMC Genomics 2017;18:890. [DOI] [PMC free article] [PubMed] [Google Scholar]