Abstract
Steady-state RNA levels are controlled by the balance between RNA synthesis and RNA turnover. A selective RNA turnover mechanism that has received recent attention in neurons is nonsense-mediated RNA decay (NMD). NMD has been shown to influence neural development, neural stem cell differentiation decisions, axon guidance and synaptic plasticity. In humans, NMD factor gene mutations cause some forms of intellectual disability and are associated with neurodevelopmental disorders, including schizophrenia and autism spectrum disorder. Impairments in NMD are linked to neurodegenerative disorders, including amyotrophic lateral sclerosis. We discuss these findings, their clinical implications, and challenges for the future.
Introduction
Neurons rely extensively on post-transcriptional mechanisms to control the location and amounts of specific proteins1. A post-transcriptional mechanism that recently has been found to have major roles in neurons is nonsense-mediated RNA decay (NMD), a RNA turnover pathway conserved from yeast to man2–4. NMD was originally discovered as a RNA surveillance mechanism that degrades aberrant RNAs harboring premature translation termination codons (PTCs)5. This quality control function of NMD has medical implications, as the truncated proteins translated from PTC-bearing mRNAs can sometimes act as dominant-negative proteins that cause disease. Indeed, there is considerable evidence that NMD reduces genetic disease symptoms by decreasing the levels of such potentially deleterious proteins3,6,7.
NMD degrades subsets of normal mRNAs.
In recent years, it has become clear that NMD is more than merely a RNA surveillance pathway to protect cells from transcripts that contain genetic mutations. Genome-wide studies—first conducted in yeast and later in higher eukaryotes—have demonstrated that a wide variety of normal mRNAs are subject to rapid decay by NMD4,8–11. These studies showed that knockout or depletion of NMD factors causes a substantial subset (~5 to 10%) of normal mRNAs to be upregulated. Although it has remained unclear what proportion of these upregulated mRNAs are directly targeted by NMD, a wealth of studies have identified likely NMD target transcripts using a battery of different approaches, including RNA half-life analysis and immunoprecipitation analysis of mRNAs bound by the NMD factor, UPF14,10,12,13.
Why does NMD degrade subsets of normal mRNAs? As we argue below, there is increasing evidence that NMD serves as a regulatory mechanism to control the steady-state levels of such mRNAs in different biological contexts. This follows from the fact that NMD itself is a highly regulated pathway14,15. Thus, rather than being simply “on” or “off,” NMD efficiency can be regulated, resulting in differential degradation of NMD targets. For example, decreased NMD efficiency at a specific developmental stage leads to stabilization of NMD target RNAs at that developmental stage. Conversely, increased NMD magnitude will destabilize NMD target RNAs. Such shift in the levels of NMD target RNAs has the potential to drive and shape biological processes; indeed, NMD has been shown to influence many biological processes, including differentiation, cell survival, and stress responses4,16–20.
NMD factors and NMD-inducing features.
NMD is a complex pathway involving numerous factors (Fig. 1). Some of these factors are involved in the “recognition phase” of NMD, which establishes which transcripts are NMD targets. This recognition phase requires several NMD factors, including upframeshift protein 1 (UPF1), UPF2, and UPF3B (also called UPF3X). The “decay phase” of NMD is driven by other factors, including SMG6 (suppressor with morphological effect on genitalia 6), an endonuclease that cleaves NMD target mRNAs near the stop codon terwminating the main open reading frame (ORF)4.
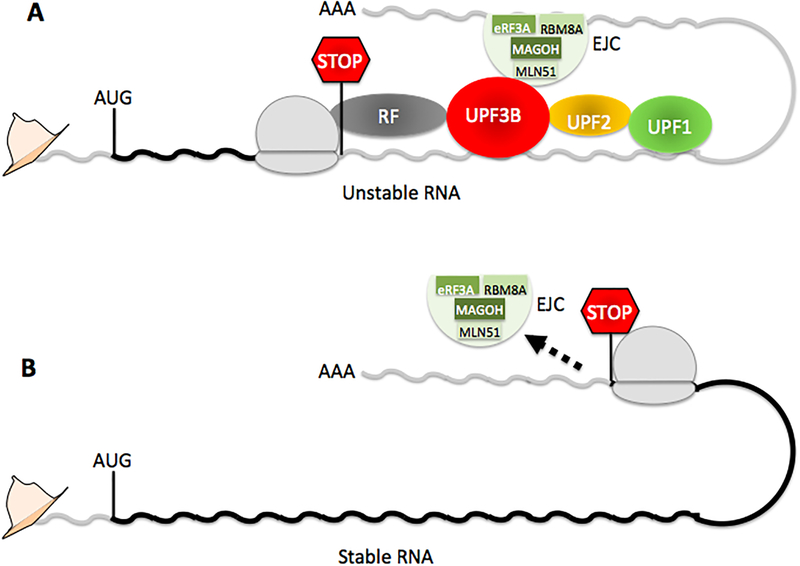
Figure 1. Nonsense-mediated RNA decay (NMD).
(A) mRNAs with at least one exon-exon junction downstream of the stop codon terminating the main ORF are degraded by NMD through the protein–protein interactions shown. A key interaction is between the RNA-binding protein upframeshift 3B (UPF3B) and the exon-junction complex (EJC), the latter of which is recruited just upstream of exon–exon junctions after RNA splicing. UPF3B is a scaffolding factor that also directly interacts with UPF1, UPF2, eukaryotic release factor 1 (eRF1), and eRF3A4,24. (B) mRNAs with all exon-exon junctions upstream of the stop codon fail to be degraded by EJC-dependent NMD because all EJCs are displaced by ribosomes prior to translation termination. Untranslated and coding regions are shown in grey and black, respectively.
NMD is triggered by so-called “NMD-inducing features,” all of which revolve around translation termination. In most transcripts, the stop codon terminating the main ORF is in the last exon, a context that typically does not trigger NMD. In contrast, transcripts harboring the stop codon in a middle exon are targeted for decay by NMD by virtue of the exon-exon junctions downstream of the stop codon (for the reasons described below; see Figs. 1, 2a, and 2b). Although originally considered to be rare21, mRNAs with middle-exon stop codons have since been shown to be relatively common, probably for regulatory purposes, as described below.
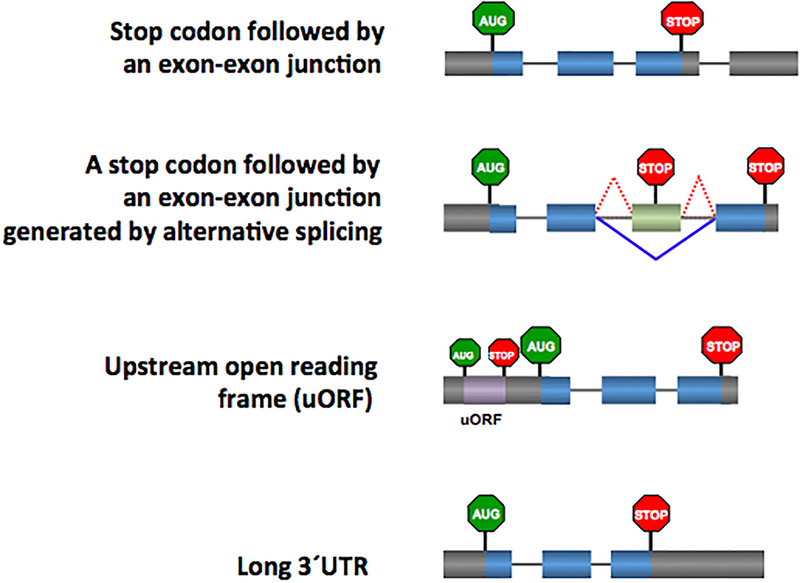
Figure 2. NMD-inducing features.
Many normal genes have a stop codon in a context that triggers NMD, including an exon–exon junction downstream of the stop codon, as shown in Fig. 1. This context is present in some normally spliced RNAs (panel a). b. NMD can also be triggered by alternative splicing, such as by the splicing pattern marked with red dotted lines. By contrast, an alternative splicing pattern that excises the green exon harboring the stop codon (grey line) will not trigger NMD (see also Fig. 3).
c. and d. Short reading frames upstream of the main reading frame (uORFs) and long 3’ UTRs (>1 kb) can also elicit NMD. However, the presence of a uORF or a long 3’ UTR do not necessarily trigger NMD and thus these NMD-inducing features only act in specific contexts111.
In some cases, transcripts with a middle-exon stop codon are generated by alternative RNA splicing. For example, this may involve inclusion of an alternative middle exon that includes an in-frame stop codon, sometimes called a “poison exon.” As a result, this transcript becomes an NMD target (Figs. 2 and 3). Alternative splice donors or acceptors that lead to a shift in the reading frame or inclusion of a sequence with an in-frame stop codon also lead to a transcript that is targeted by NMD (Fig. 3). Thus, alternative splicing can suppress the “functional output” of a gene by causing its encoded transcripts to be rapidly degraded.
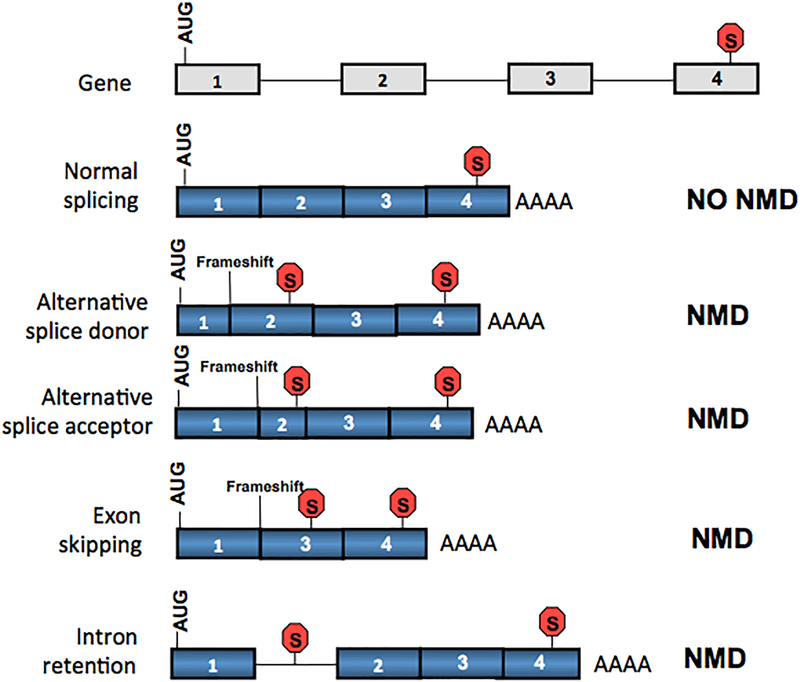
Figure 3. Alternatively spliced mRNAs are often targeted by NMD.
A generic gene with a stop codon in the final exon in frame with the start codon (AUG) is shown at the top. Five alternative RNAs expressed from this gene are shown below. The normally spliced RNAs is not targeted for decay by NMD because its stop codon is not followed by an exon-exon junction and thus it avoids EJC-dependent NMD (see Figs. 1 and 2). By contrast, the four mRNAs depicted below the normally spliced mRNA have a context that triggers NMD, as they all acquire a premature stop codon, also called a premature termination codon (PTC), upstream of the final exon–exon junction as a result of alternative splicing. In the first 3 alternatively spliced transcripts shown, a PTC is created by a frameshift, which occurs by chance in 2 of 3 instances of alternative splicing. Intron retained RNAs are almost always degraded by NMD, as they typically contain multiple stop codons in the retained intron (the occurrence of stop codons in random sequences is ~1:20).
Exon-exon junctions downstream of stop codons elicit NMD by acting as nucleation points that recruit a set of NMD-promoting proteins collectively called the exon junction complex (EJC)22,23. In a current consensus model, RNA decay is triggered in an EJC-dependent manner through a series of steps. First, when the ribosome reaches a stop codon defining the end of the main ORF, this leads to the formation of a termination complex (Fig. 1). The stop codon is recognized by eukaryotic release factors (eRFs), which are key proteins that physically connect the termination complex to the EJC, and thereby trigger NMD. A key protein in this coupling event is UPF3B, which binds both the EJC and eRFs, thereby acting as a bridge that forms a NMD-promoting complex24,25. Also present in this NMD-promoting complex are the RNA helicase, UPF1, and the adaptor protein UPF225, the latter of which may also directly interact with eRFs26 (Fig. 3). The formation of this NMD-promoting complex enables UPF1 to be phosphorylated by SMG1, leading to recruitment of proteins critical for the effector phase of NMD27.
Notably, the formation of the complex comprising the stop codon-bound ribosome and the EJC, is limited to EJCs downstream of the main ORF stop codon, as upstream EJCs are ejected during the pioneer round of translation28. As a consequence, only stop codons in middle exons, not the final exon, elicit EJC-dependent NMD. This provides a simple mechanism by which PTCs are distinguished from normal stop codons.
Of note, NMD is typically only triggered by stop codons at least ~50-nt upstream of the last exon-exon junction21. This “−50 boundary rule” most likely derives from the ability of the ribosome to displace the EJC from an mRNA28–30. Stop codons closer than ~50 nt to the final exon-exon junction would be expected to elicit ribosome-mediated displacement of the most 3’ EJC prior to translation termination, as the EJC is typically centered ~24 nt upstream of the exon-exon junction23 and the ribosome footprint is ~20 nt on either side. Without at least one retained EJC, NMD is not elicited.
In addition to downstream exon–exon junctions, other NMD-inducing features have been identified, including long 3’ untranslated regions (UTRs) and short upstream (u) ORFs4 (Figs. 2c and 2d).
NMD and neural disease
UPF3B and intellectual disability.
In 2007, Gecz and colleagues reported that mutations in the X-linked NMD gene, UPF3B, cause intellectual disability (ID) in humans31 (Table 1). Through sequence analysis of X chromosome genes from 250 families with X-linked ID, they identified UPF3B mutations in 4 families. Later pedigree studies identified several other families with UPF3B mutations; in each case, males with such mutations had ID32–35. Intriguingly, several of these ID individuals also have schizophrenia (SCZ) or autism spectrum disorder (ASD), raising the possibility that NMD dysfunction also contributes to these neuro-developmental disorders.
Table 1. NMD and Neural Disease.
Pedigree analysis of UPF3B mutations indicates causality in intellectual disability. The evidence for other NMD genes being involved in neural disease is less strong. Only statistically significant associations of CNVs with neural disease are shown, but note that the CNVs often amplify or delete other genes in addition to the NMD and EJC genes indicated.
| Gene | Disease | Evidence | Reference |
|---|---|---|---|
| EIF4A3 | Neuro-development disorders | CNV gain | 43 Coe 14 |
| EIF4A3 | RCP Syndrome | 5’ UTR repeat | Favora et al.110 |
| RBM8A | Neuro-development disorders | CNV gain or loss | 43, Coe 14 |
| RBM8A | ID, epilepsy, ASD, SCZ | 1q21.1 del | Various45–47 |
| RBM8A | TAR Syndrome | 1q21.1 del + RBM8A mtn | Albers et al.112 |
| RNPS1 | Neuro-development disorders | CNV gain | 43, Coe 14 |
| SMG6 | Neuro-development disorders | CNV gain | 43, Coe 14 |
| UPF2 | Neuro-development disorders | CNV gain or loss | Various43,44 |
| UPF3A | Neuro-development disorders | CNV loss | 43, Coe 14 |
| UPF3B | ID | Pedigree analysis | Various31–35 |
| UPF3B | ASD, SCZ, ADHD | Various mutations | Various31–35 |
As one approach to understand the underlying mechanism, Upf3b-null mice have been generated36. These NMD-deficient mice are viable and are not significantly different from wild-type controls in most respects. However, they exhibit a profound defect in pre-pulse inhibition (PPI), a measure of sensorimotor gating commonly deficient in individuals with SCZ and other brain disorders. They also have cued and contextual fear learning deficits, but no significant defect in spatial memory tests (Y maze and Barnes maze). This selective learning defect is intriguing and suggests a role for UPF3B in the generation and/or function of neural circuits specifically important for fear conditioned learning. Consistent with both their learning and PPI deficits, Upf3b-null mice display deficient dendritic spine maturation in their cortical pyramidal neurons. The learning and PPI deficits in Upf3b-null mice may serve as a model for understanding the behavioral and learning abnormalities in humans with UPF3B mutations.
UPF3B is a critical adaptor protein that physically links the EJC with the core NMD factors, thereby triggering RNA decay. Although an important NMD factor, UPF3B is not universally required for NMD but instead evidence suggests it directs a specific branch of the NMD pathway (Fig. 4A). Thus, depletion of UPF3B from mammalian cell lines only leads to upregulation of some, but not all, NMD substrates, providing evidence for UPF3B-dependent and -independent branches of NMD17,37. This model was confirmed by analysis of Upf3b-null mice, which upregulate some NMD substrates but not others38. Further evidence that UPF3B functions in only a branch of the NMD pathway is the finding that Upf3b-null mice progress through embryogenesis and reach adulthood36, whereas mice lacking factors crucial for the entire NMD pathway (e.g., UPF1) suffer from early embryonic lethality18,39–42.
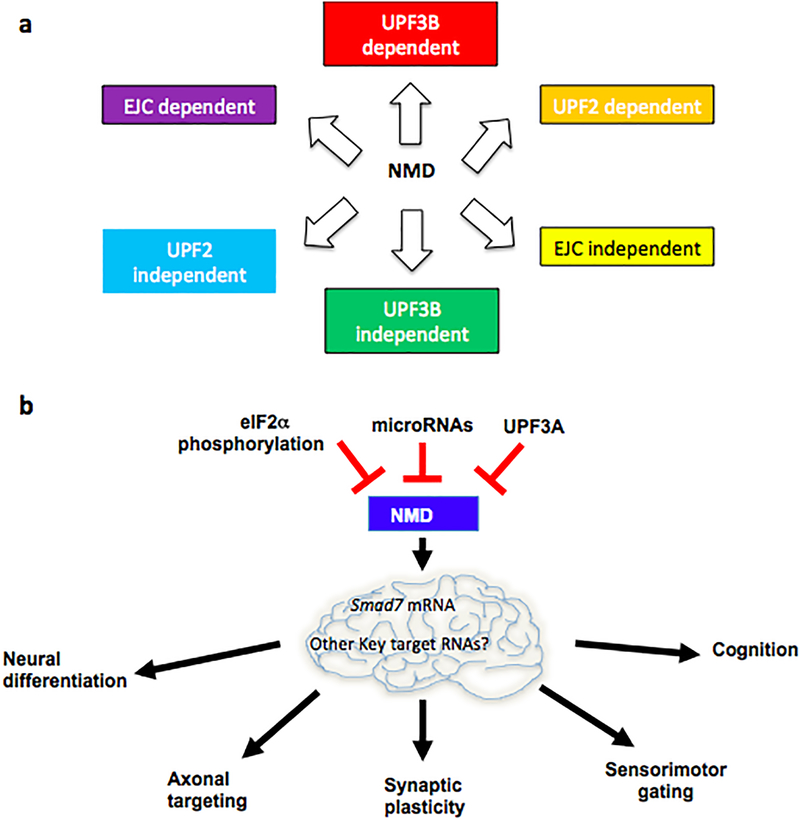
Figure 4. NMD branches, regulation, and neural functions.
a. NMD branches. NMD factor-depletion studies have suggested the existence of distinct NMD branches that require different factors and degrade different subsets of transcripts. The extent of overlap between the different branches is not known. The NMD targets degraded by a specific branch can vary in a cell type-specific manner.
b. Regulation and function of NMD in neurons. In order for NMD to modulate the levels of its target RNAs, it must be regulated. Several NMD regulatory factors, including those shown here, have been identified. One target RNA critical for the ability of NMD to dictate neural stem cell vs. differentiation decisions is Smad7 mRNA, which encodes a negative regulator of TGFβ signaling and promotes neuronal differentiation55. Many candidate RNAs that might function downstream of NMD in its other neuronal roles have been identified, including Robo3.2 mRNA, which encodes a protein involved in axon guidance73.
Together, these studies suggest that the UPF3B-dependent NMD branch is critical for normal behavior. As we discuss later, one possible explanation is that RNAs specifically degraded by this branch of the NMD pathway have critical roles in developing and mature neural cells.
NMD genes and neural diseases
In addition to UPF3B, other genes in the NMD pathway have also been linked with ID and neural disease. For example, heterozygous deletions of the NMD gene, UPF2, and adjacent genomic regions, have been shown to be associated with ID and neuro-developmental disorders43 (Table 1). Further support for the involvement of UPF2 in psychiatric disorders comes from the identification of a de novo missense mutation in UPF2 in a SCZ patient44.
Further evidence that NMD defects cause neurological disease comes from a study by Nguyen et al., who found that copy number variations (CNVs) encompassing several NMD genes were significantly associated with several types of neural dysfunction, including neurodevelopmental disorders (Table 1)43. Among genes exhibiting a significant copy number loss were UPF2 and UPF3A, the latter of which encodes a NMD repressor42, raising the possibility that not only too little NMD, but also too much NMD, can cause neural dysfunction. As further evidence for this “Goldilocks” principal, copy number gain of several NMD genes were shown to be significantly associated with ID, neuro-developmental disorders, and brain malformations, including macrocephaly (Table 1). Together, these human genetic studies raise the possibility that either too much or too little of NMD factors can cause neurological and psychiatric disorders.
Mutations in EJC genes are also associated with neurological disease (Table 1). For example, deletions in 1q21.1, a small region of human chromosome 1 that includes the gene encoding the EJC core component, RBM8A (also called Y14), is associated with increased incidence of ID, epilepsy, ASD, and SCZ45–47. In support of a causal role, Rmb8a-haploinsufficient mice have dramatic neural defects, including microcephaly48,49, a defect observed in many 1q21.1 deletion patients47. As further evidence for a role of Rbm8a in neural function, overexpression of this EJC gene in mice increases anxiety-like behavior, impairs social skills, decreases immobile time, and enhances the frequency of miniature excitatory postsynaptic currents50.
These data from EJC-deficient mice and humans raise the possibility that defects in NMD cause aberrant neuronal activation, neurodevelopmental disorders, microcephaly, and epilepsy. However, it should be noted that EJC factors not only bind to splice junctions51,52 but are involved in several molecular processes in addition to NMD22,23, and thus disruption of one or more of these other processes are likely to have a major role in many of the phenotypic defects in EJC-deficient mice.
NMD and neural development
The finding that mutations in NMD factor genes are strongly associated with neural disease suggests that NMD is critical for neural development and/or function. In this section, we consider the former. We first go over the evidence for the role of the central NMD factor, UPF1, in neural stem vs. differentiation decisions. We then discuss several studies conducted on the neural differentiation/maturation roles of UPF3B — the only NMD factor gene definitively shown to cause neural disease when mutated in humans31–35.
MicroRNA circuits controlling NMD.
The first hint that NMD had a role in neural development was the discovery—by Bruno et al.—that UPF1 is dramatically downregulated during mouse brain development53. Later studies showed that this downregulation also occurs during both mouse and human neural cell differentiation in vitro54–56. Other NMD factors were downregulated as well, raising the possibility that NMD efficiency is decreased during neural differentiation, a possibility directly demonstrated using two independent NMD reporters55.
Several lines of evidence indicated that this downregulated NMD-response is critical for neural stem cells to differentiate55. Lou et al. found that preventing UPF1 downregulation during neural differentiation (through forced expression of modest levels of UFP1) inhibited neural differentiation. Conversely, the high levels of NMD factors inherent in stem cells was found to be sufficient to maintain “stemness,” as depleting UPF1 triggered the differentiation of both mouse neural stem cells and neural precursor (P19) cells. Thus, the high expression level of NMD factors is essential to maintain neural precursor cells in a stem cell state.
NMD downregulation appears to be a general property of differentiation, as it has been shown to occur in several differentiation scenarios, including adipogenesis57, myogenesis58, and endoderm differentiation16. In the case of endoderm, the formation of this germ layer from human embryonic stem cells was found to critically depend on the NMD downregulatory response16.
What is responsible for differentiation-dependent NMD downregulation? One class of factors that have a role in NMD downregulation is microRNAs (miRNAs) (Fig. 4B). A particularly critical NMD-regulatory miRNA is the brain-enriched miRNA—miR-128—which was found to repress NMD by targeting UPF1, UPF3B, and the EJC core component MLN5153,55. miR-128 is dramatically induced during neural differentiation, and is at least partially responsible for the downregulation of NMD factors during neural differentiation, based on rescue experiments53,55. A miRNA-based bistable feedback circuit was defined by Lou et al. that stabilizes the neural stem cell state (when NMD is high) or promotes neural differentiation (when NMD is low)55. In addition to miR-128, other NMD-inhibitory miRNAs may participate in NMD down-modulation, including the brain-enriched miRNAs miR-9 and −12455. Another recently defined NMD-regulatory miRNA, miR-12559, has roles in synaptic function of adult born interneurons in the olfactory bulb60 and neural stem differentiation61.
How do NMD factor dynamics influence whether a cell is in a stem cell state or differentiate? Lou et al. found that the majority of well-established pro-neural differentiation proteins are encoded by high-confidence NMD target mRNAs, supporting the possibility that NMD promotes the stem cell state by degrading pro-differentiation mRNAs55. Using a rescue experiment approach, Lou et al. identified one particular neural differentiation factor—SMAD7—whose mRNA must be degraded by NMD to maintain the neural stem cell state55. This data supported a model in which Smad7 mRNA levels are low in neural stem cells because of high NMD magnitude, but upon receiving a neural induction signal, NMD efficiency is decreased, leading to stabilization of the Smad7 transcript and consequent neural differentiation. Lou et al. found that NMD also preferentially targets mRNAs encoding proliferation inhibitors, such as p21 and p27. The degradation of one or more of these mRNAs encoding proliferation inhibitors may explain how NMD promotes the proliferation of neural precursor cells55.
Another likely mechanism by which NMD acts is by regulating the stability of alternatively spliced mRNAs. Alternatively spliced transcripts predicted to be targeted by NMD in the developing mouse and brain cortex are enriched for those encoding transcriptional regulators and RNA-binding proteins62. Thus, the changes in NMD efficiency during neural differentiation could affect the stability and translation of these transcripts to influence brain development and function.
UPF3B impacts neural differentiation.
The studies described above demonstrate that high magnitude of NMD promotes the neural stem cell state and must be downregulated to initiate neural differentiation. However, Jolly et al. obtained evidence that NMD has the opposite role at later stages of neural development – it promotes neural differentiation. In their study, Jolly et al. used mouse neural progenitor cells (generated from the mouse E18.5 cortex) derived from older mouse embryos than the neural stem cells (derived from E14.5 brain) used by Lou et al. to show that NMD inhibits early stages of neural development55. Jolly et al. found that depletion of the NMD branch-specific factor UPF3B reduced the ability of mouse neural progenitor cells to differentiate54. This suggests that the UPF3B-regulated branch of NMD promotes the differentiation of already committed neural progenitor cells.
Further evidence that UPF3B promotes neuronal differentiation comes from a recent report from Huang et al.36, who found that neural stem cells (NSCs) from Upf3b-null mice exhibit hyper self-renewal and poorly differentiate when cultured under conditions that efficiently differentiate control mouse NSCs. Together with the experiments of Lou et al., these results supported a model in which NMD must be downregulated to allow uncommitted stem cells and neural stem cells to begin neural differentiation, but that once committed to the neural cell lineage, NMD is crucial for these cells to proceed through neural differentiation and exit from the cell cycle.
An alternative (but not mutually exclusive) explanation for the results of Jolly et al. and Huang et al. is the UPF3B-dependent branch of NMD has unique roles that differ from the NMD pathway as a whole. This follows from the fact that Jolly et al. and Huang et al. manipulated the level of the branch-specific factor, UPF3B54, whereas Bruno et al. and Lou et al. manipulated the level of the central NMD factor, UPF1, and thus it is likely that the entire NMD pathway was modulated in the latter gain- and loss-of-function experiments53,55. Although the UPF3B-dependent branch of NMD may exhibit unique effects in neural cells, it is worth noting that in another system—human embryonic stem cells—UPF1 and UPF3B manipulation (whether depletion or forced expression) had very similar effects on primary germ layer differentiation16.
A specific function of UPF3B may be to control the formation of dendritic arbors. Depletion of UPF3B in post-mitotic hippocampal neurons caused a modest decrease in neurite length and an increase in arborization (branching) of both axons and dendrites of hippocampal neurons54. This suggests that UPF3B promotes neurite growth but suppresses neurite arborization. Unexpectedly, another study showed that over-expression of dominant-negative UPF3B mutant proteins (encoded by ID patients with UPF3B mutations) led to dramatically decreased neurite arborization in a rat neural cell line. It is not clear why these two different conditions that presumably both suppress NMD have opposite effects on neurite arborization54,56. Regardless, the discovery that UPF3B impacts neurites is potentially clinically significant, as both mouse models and post-mortem human studies have linked neuronal branching defects to several neurodevelopmental disorders63.
A critical question for the future is to identify UPF3B target transcripts as a first step towards understanding how UPF3B influences neuronal differentiation and maturation. Towards this end, UPF3B-regulated transcripts have been identified in human lymphoblastoid cells64 and more recently UPF3B-regulated transcripts were defined in the frontal cortex36. Among the candidate UPF3B target transcripts identified in the frontal cortex (those upregulated in Upf3b-null mice and harboring NMD-inducing features) were several known to have roles in neural development, function and disease, including AKR1C14, BRCA2 CDH24, DSCAM, FBN2, KCNH4, PTCH1, and RMST.
Another important future goal is to make use of model organisms in addition to mice to define conserved functions of NMD in neural development. In this regard, morpholino-mediated knockdown of NMD factors in zebrafish embryos has been shown to cause brain patterning and growth defects65. These NMD-deficient zebrafish embryos exhibited extensive CNS necrosis, aberrant eye development, and brain patterning defects, particularly at the midbrain-hindbrain boundary. The same set of defects was elicited by knockdown of any of several different NMD factors (UPF1, UPF2, SMG5, or SMG6), suggesting that deficient NMD was responsible.
EJC mouse mutants exhibit robust neurodevelopmental defects.
As described above, the EJC is recruited to splice junctions after RNA splicing, where it functions as a branch-specific NMD factor or NMD amplifier. Importantly, the EJC has other functions, including RNA splicing, nuclear-cytoplasmic RNA transport efficiency, translation, and cytoplasmic RNA localization22,23. Given this, EJC component-deficient mice would be predicted to exhibit more severe phenotypes than NMD factor-deficient mice. Consistent with this prediction, even modest reduction in EJC factor levels (as a result of haploinsufficiency) causes severe phenotypic defects. For example, mice with loss of one allele of the EJC factor gene, Magoh, suffer from microcephaly and several brain cortical defects, including: (i) decreased thickness of all cortical layers, (ii) few intermediate neural progenitors, (iii) premature generation of neurons, and (iv) extensive apoptosis66. Aberrant mitosis was found to be a likely culprit for at least some of these defects67. Deletion of one allele of two other core EJC factor genes—Rbm8a and Eif4a3—in neural progenitors cause essentially the same defects as observed in Magoh heterozygotes48,49, consistent with the idea that these phenotypes reflect impairment of EJC function in neural development.
Several indirect lines of evidence suggest that some of the neural defects caused by EJC factor haploinsufficiency result from perturbed NMD. First, NMD has been shown to be deficient in EJC factor-haploinsufficient neural progenitors49. Second, depletion of the EJC factor, RBM8A, promotes the proliferation and inhibits the differentiation of neural cells in vitro, a phenotype also exhibited by UPF1-deficient neural cells55,68. Finally, the defining feature of EJC factor insufficiency – microcephaly – is also observed in a subset of ID patients with mutations in the NMD factor gene UPF3B31,34. Although this constitutes some evidence that NMD deficiency has a role in the severe defects in EJC heterozygotes, it is important to note that little or no defects have been observed in mice heterozygous for NMD factor genes39–41, raising the possibility that EJC factors are more important for non-NMD functions than NMD functions.
However, an alternative explanation for the more severe phenotype of EJC mutants as compared to NMD factor mutants is that EJC factors are more limiting for NMD than NMD factors in neural cells. To tease out the independent contributions of the EJC’s various functions to specific phenotypes, it will be critical to identify EJC factor mutants with selective functional defects. This approach has been used to define the function of the NMD protein SMG6 in mouse ES cell differentiation. Thus, mutants of SMG6 that selectively perturb either its NMD or telomerase-promoting functions have allowed investigators to assign its NMD function as responsible for its ability to drive mouse ES cell differentiation18.
NMD and neuronal function
In addition to its roles in developing neurons, NMD has functions in fully differentiated neurons. In this section, we review the evidence for NMD’s roles in axon guidance and synaptic plasticity.
NMD in axon guidance.
Axon guidance is the process by which axons are guided and ultimately form connections with their synaptic targets. One of the best known axonal guidance mechanisms involves the SLIT proteins, which are secreted extracellular matrix proteins that are recognized by the cell-surface ROBO proteins located on the tips of elongating axons69–72. Two of the ROBO proteins—ROBO3.1 and ROBO3.2—are generated by alternatively RNA splicing from a single gene. Robo3.2 is an alternatively spliced isoform that encodes in a shortened form of ROBO with a novel C terminus (Fig. 5). The roles of ROBO3.1 and ROBO3.2 in axon guidance has been well studied in “commissural” axons, which cross midline structures in the brain. ROBO3.1 and ROBO3.2 protein exhibit highly specific spatial and temporal expression within commissural axons (Fig. 5) and the precise timing and location of these two proteins are known to be critical for the proper guidance of commissural axons in the spinal cord69. ROBO3.1 is the only isoform expressed in axons when they are initially attracted to the ventral midline69. After the commissural axons have crossed through the midline, the other isoform—ROBO3.2—is translationally activated. This induction of ROBO3.2 is critical, as this protein enhances the ability of ROBO1 and ROBO2 to sense SLIT, and, as a result, the axon becomes repelled from the SLIT-expressing midline69. Thus, the selective induction of ROBO3.2 after axons have crossed the midline ensures that the axon loses its initial attraction to the midline so that the axon can elongate away from the midline.
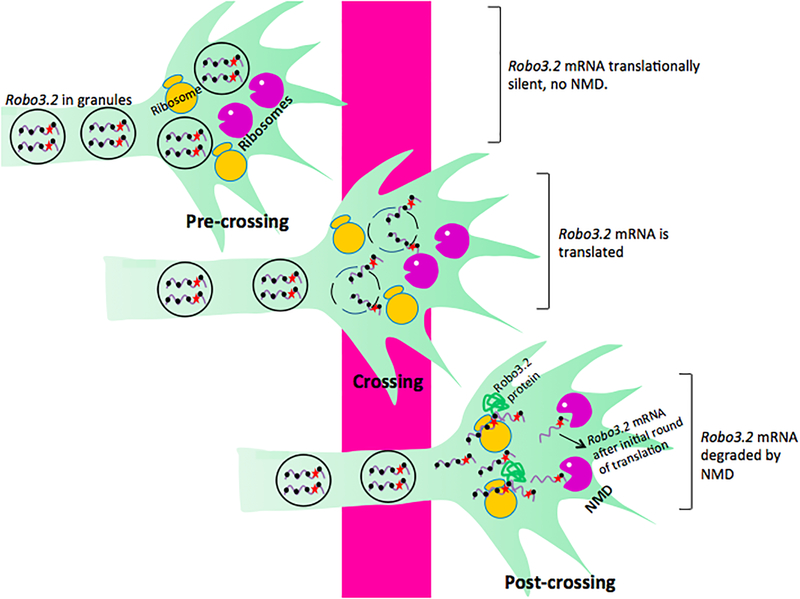
Figure 5. Control of protein synthesis and mRNA levels in axons by local NMD during axon outgrowth.
NMD proteins are localized in axons, where they serve to control mRNA levels and protein expression during axon guidance. This effect has been characterized in axons of commissural neurons73, a subset of neurons whose axons cross midline structures in the brain. In the case of spinal commissural neurons, the axons express Roundabout 3.2 (Robo3.2), an alternatively spliced transcript encoded by the Robo3 gene. ROBO3.2 protein enhances the activity other ROBO proteins such as ROBO1 and ROBO2, which detect and mediate the chemorepellant effect of axons toward SLIT proteins found in the midline. When axons are undergoing migration to the midline (pre-crossing, top image), Robo3.2 mRNA accumulates in RNA granules in a non-translated state. When the axon encounters the midline (middle image), midline-derived factors trigger the translation of Robo3.2 mRNA, presumably, in part, by triggering their release from granules. Translation of Robo3.2 mRNA also triggers its decay by NMD (post-crossing, bottom image), thereby allowing a short burst of ROBO3.2 protein production to enhance the function of ROBO1 and ROBO2 and enable the axon to be repelled by SLIT in the midline.
Robo3.2 mRNA only differs from Robo3.1 mRNA in possessing a retained intron. This shifts the reading frame of the main ORF, leading to the introduction of a stop codon upstream of exon–exon junctions and hence the potential to be degraded by NMD. Indeed, Colak et al. demonstrated—through several approaches—that Robo3.2 is a bona fide NMD target mRNA73. Notably, Robo3.2 mRNA is localized in axons and is in a translationally repressed state until commissural axons have crossed the spinal midline. This translationally repressed state allows Robo3.2 mRNA to escape NMD and thus accumulate in axons. Upon reaching the midline, ROBO3.2 translation is activated, which generates ROBO3.2 protein and sensitizes Robo3.2 mRNA to decay by NMD, leading to its eventual downregulation.
The finding that activation of translation triggers both protein production and RNA decay brings up an interesting conceptual conundrum. How can an RNA serve as a template for protein synthesis if it is degraded? This is particularly perplexing given the model that mRNAs are exclusively recognized as NMD targets during the first “pioneer” round of translation in mammalian cells74. If this was the case for Robo3.2 mRNA, this would allow for little or no ROBO3.2 protein to be produced. Instead, considerable ROBO3.2 protein is detectable in commissural axons73 and thus it seems likely that instead Robo3.2 mRNA is degraded after a lag period in post-pioneer rounds of translation.
How can NMD occur after the pioneer round of translation? In a widely recognized model, NMD is triggered when NMD factors that are normally recruited to the eRF-bound ribosome upon translation termination interact with EJCs27,75. However, this interaction would not occur if the ribosome is disassembled and released from the stop codon before it interacts with the EJC. Therefore, highly efficient ribosome disassembly at stop codons would prevent the stop codon-bound ribosome from detecting a downstream EJC. Alternatively, if the interaction with the EJC forms slowly, then ribosome release will occur before NMD is induced. Thus, a slow rate of ribosome disassembly would facilitate NMD occurring in the first round, whereas rapid ribosome disassembly may require multiple rounds of translation before the stop codon-stalled ribosome can interact with the EJC. The rate of ribosome disassembly could be regulated, for example, by levels of ribosome-release factors76. Regardless of the underlying mechanism, it is clear that mRNAs can—under certain circumstances—be recognized by NMD after the pioneer round of translation77,78.
Based on this model, NMD is intrinsically inefficient in commissural neuron axons and, thus, several rounds of ROBO3.2 protein synthesis typically occurs before Robo3.2 mRNA is degraded via the NMD pathway. Perhaps NMD is downregulated specifically in commissural neuron axons to allow for this outcome, a possibility supported by the evidence that NMD magnitude is highly regulated in developing neurons53,55,56. It is intriguing to also speculate that different types of neurons may have different NMD efficiencies; indeed, even individual axons may differ in NMD efficiency. As a result, specific axons would produce different amounts of ROBO3.2 and other proteins from NMD target mRNAs, which could be used as a strategy in commissural neuron axons to determine the precise degree of repulsion of the commissural axons from the spinal midline, thereby leading to targeting to different positions in the spinal cord.
The ability of translational activation coupled with NMD to both generate protein and trigger RNA decay is seemingly contradictory. We suggest that this “drive with both the gas-and-brake” strategy allows for a transient bolus of protein to be made in a highly regulated manner. In the case of ROBO3.2, it allows for transient production of this protein specifically when the axon reaches the midline, thus restricting its axonal repulsive action to precisely when it is needed. We also suggest that this coupled translation induction-RNA decay system is a general strategy that could be used in other settings. For example, coupled translation-RNA decay could provide a means to trigger the transient production of specific proteins in mature neurons to confer synaptic plasticity and thereby store memories. It will be important to identify RNAs in addition to Robo3.2 that undergo coupled translation and RNA decay in neurons. It will also be interesting to determine whether UPF3B contributes to the degradation of Robo3.2 since it could link this form of regulation to neurodevelopmental phenotypes seen in humans with UPF3B mutations31.
Local translation may serve as a trigger for NMD in neurites. A prime target for this type of local regulation is mRNAs that are not allowed to be translated when they first reach the cytoplasm from the nucleus. Such mRNAs become translationally active only after they are transported to dendrites or axons79. Since NMD requires translation80, such RNAs would not be predicted to be degraded NMD until they reached these distal sites. Support that this type of regulation may occur commonly is the finding that EJC proteins, such as eIF4A3, are abundant in RNA transport granules in dendrites81. Accumulation of EJC proteins would be expected if many mRNAs in such granules have not yet undergone translation and thus retain EJC proteins bound at exon-exon junctions.
Local NMD control may also be exerted in other ways and at other sites within neurons. A hint that this is the case is the finding that NMD factors are enriched in neurites56,73,81. Perhaps this allows NMD to be spatially localized, enabling mRNA degradation to occur at specific sites in neurons, such as distal axons.
NMD efficiency may also be regulated during development. In support, the NMD factor, UPF3B, exhibits a different nuclear-to-cytoplasmic distribution in developing neurons that depends on the stage of development. NPCs within the ventricular zone from E10.5 and E14.5 mice expressed predominantly cytoplasmic UPF3B, while NPCs at a later stage (E18.5) expressed mainly nuclear UPF3B54. Thus, NMD efficiency may be reduced at later stages in development due to lack of availability of UPF3B in the cytoplasm. It will be interesting to elucidate the molecular underpinnings behind this cytoplasmic-to-nuclear switch and to determine whether the efficiency of NMD is indeed reduced as a result of UPF3B relocalization.
NMD in synaptic plasticity.
NMD has also been implicated as having a role in synaptic plasticity, the process in which synaptic strength is altered in response to shifts in synaptic activity. There is considerable interest in this process, as synaptic plasticity is thought to underlie learning and memory. The first hint of that NMD might have a role in synaptic plasticity was the discovery that NMD degrades the mRNA encoding ARC, a cytoskeletal-associated protein enriched in dendrites that accumulates at synapses upon neural stimulation and functions in synaptic plasticity82,83. The Arc gene is unusual in that it responds to neural stimuli by undergoing not only rapid transcriptional activation84, but also rapid decay of its mRNA.
Several lines of evidence suggest that Arc mRNA instability is driven by NMD. First, Arc mRNA has two introns in its 3’ UTR and thus has two EJC-landing pads downstream of its stop codon, which would be predicted to trigger NMD (Figs. 1 and 2). The ability of these 2 introns to trigger NMD was recently directly shown in reporter experiments85. Second, Arc mRNA is bound by eIF4A381. Finally, knockdown of either UPF1 or the EJC factor, eIF4A3, triggers Arc mRNA upregulation81. As described below, the find that Arc mRNA is a NMD target has important implications for how it is regulated and how it functions in synaptic plasticity.
How does NMD influence the spatial and temporal pattern of Arc expression after synaptic stimulation? Arc mRNA instability by NMD allows ARC to achieve its highly transient induction in response in neuronal stimulation86. This follows from the known relationship of RNA synthesis and decay kinetics: (i) transcriptionally induced unstable mRNAs reach steady state more quickly than do stable mRNAs and (ii) unstable mRNAs decline more rapidly than do stable mRNA after transcriptional shut-off87. Thus, the instability of the Arc mRNA, coupled with its rapid transcriptional induction, allows ARC protein to be strongly and transiently expressed in activated neurons. Of note, it was recently shown that ARC translation is stimulated by brain-derived neurotrophic factor through a RNA splicing- and 3’UTR-dependent mechanism85. This provides another level of regulation (also acting through the 3’UTR) that drives the unique expression pattern of ARC in neurons.
With regard to Arc’s spatial regulation, this has been studied in depth by Farris et al.84. In their model system, a single electroconvulsive seizure in rats triggers rapid Arc mRNA induction in the dendrites of neurons in the dentate gyrus. Subsequent in vivo synaptic activation drives Arc mRNA to become specifically localized at the synapse and largely disappear from synaptically inactive dendritic domains. Farris et al. obtained several lines evidence that this pattern of expression is achieved by two events: (i) delivery of newly synthesized Arc mRNA to activated dendritic domains (e.g., near synapses) and (ii) Arc mRNA degradation throughout dendrites84. The net result of this two-fold mechanism is selective accumulation of both Arc mRNA and locally translated ARC protein at activated synapses.
NMD is likely to be responsible for the dendritic decay of Arc mRNA for three reasons. First, Arc mRNA is a well-established NMD substrate, as described above. Second, both UPF1 and EJC components are present all along dendritic processes (Giorgi et al., 2007). Third, Farris et al. found that local introduction of the translation inhibitor, cycloheximide (CHX), into the dentate gyrus region greatly increased Arc mRNA levels in dendrites84. Given that NMD absolutely depends on translation80, this provided evidence that Arc mRNA degradation in dendrites depends on NMD. As evidence for specificity, another mRNA localized to synaptic regions of dendrites—Camk2a mRNA—is not subject to translation-dependent mRNA downregulation in response to synaptic activation84.
Interestingly, Farris et al. found there was one dendritic site that did not exhibit a significant increase in Arc expression in response to CHX – the outer dendritic region. Might this region lack NMD? Perhaps there is little or no translation in this outer dendritic region. Or this site may have active NMD, but Arc mRNA is specifically immune to its effects in this region. Regardless of the underlying mechanism, the ability of local NMD to regulate the levels of its target transcripts in a temporally and spatially restricted manner in specific dendritic regions has potential implications for synaptic plasticity and other neuronal functions.
Arc mRNA also appears to be regulated by NMD in a stimulus-specific manner. Farris et al. found that electroconvulsive-seizure-induced Arc mRNA—which is broadly expressed across dendrites—is only modestly subject to translation-dependent downregulation84. By contrast, subsequent in vivo synaptic stimulation leads to strong translation-dependent downregulation of Arc mRNA in a dendritic region-specific manner, as described above. Together, these results support a model in which synaptic activation triggers a dramatic increase in Arc mRNA decay in a region-specific manner through the NMD pathway. It will be interesting to determine whether this reflects an Arc-specific regulation by NMD or a generalized shift in NMD magnitude. In support of synaptic stimulation regulating NMD magnitude, the NMD factor gene, UPF3B, has been shown to exhibit reduced expression in hippocampal neurons undergoing chronic depolarization54.
To more directly examine how NMD influence synaptic activity, Giorgi et al. evaluated the role of the NMD-promoting factor, eIF4A3, in neurons81. As described above, eIF4A3 is poised to play a role in synaptic functions, as this EJC factor is concentrated in neurites, where it is present in RNP complexes bound to NMD target mRNAs. eIF4A3 also associates with dendritic proteins such as FRMP81. Giorgi et al. found that eIF4A3 knockdown significantly increased excitatory miniature postsynaptic currents mediated by AMPA receptors in cortical neurons. In addition, AMPA receptor level was increased at putative synaptic sites. Together, these data suggest that eIF4A3 normally acts as a brake on synaptic strength.
Although the studies described above focused on Arc, many other high-confidence NMD substrates have been identified in the brain36,62,81 and potential NMD targets transcripts (e.g., those with retained introns) are localized in dendrites88,89. Thus, Arc is only one of many transcripts likely to be regulated by NMD in axons and dendrites. We suggest that NMD degrades diverse mRNAs at synapses to enable transient expression of their encoded proteins and thereby avoid overly prolonged synaptic translational activation. By shaping both the temporal and spatial expression pattern of the synaptically induced ARC and other proteins, NMD may be critical for “synaptic consolidation,” thereby explaining why defects in NMD cause intellectual disability in humans and mice31,36. In this regard, it is interesting to note that rats introduced to a novel learning environment downregulate Arc mRNA in a translation-dependent manner84, raising the possibility that NMD is critical for this form of learning.
Further evidence for a role of NMD in synaptic function comes from flies. Mutations in Smg1, a NMD gene, causes photoreceptor synaptic transmission defects, impaired synaptic structural architecture, decreased terminal arbor size, branching, and presynaptic terminal defects at neuromuscular junction synapses in Drosophila melanogaster90. In support of the notion that NMD is responsible for these functions, Long et al. found that mutations in two other NMD genes—Upf2 and Smg6—also caused similar impairments90. Together with the data described above from mammalian synapses, it appears that NMD serves as an evolutionarily conserved mechanism to influence synaptic events.
NMD drives a neural expression program.
NMD can be used to ensure that a gene exhibits a temporally regulated and cell type-specific pattern of gene expression. This was recently described for the gene encoding PSD95, a scaffold protein essential for synaptic maturation and plasticity that is highly expressed in mature neurons. Zheng et al. showed that whereas the Psd95 gene is transcribed broadly, PSD95 protein is selectively expressed in neurons through a post-transcriptional mechanism involving NMD91. In early embryonic neural development and in non-neuronal cell type, the polypyrimidine tract-binding proteins, PTBP1 and PTBP2, repress the splicing of PSD95 exon 18, leading to the generation of a PTC and decay by the NMD pathway92. PTBP1 and PTBP2 are downregulated later in brain development, allowing for exon 18 inclusion, consequent escape from NMD, and high level expression of PSD95. Thus, this study provides evidence that alternative splicing coupled with NMD—commonly referred to as “AS-NMD”—is an important means to establish neuron-specific gene expression. An important question for the future is whether the decay of the exon 18-skipped PSD95 mRNA by NMD is crucial for normal neural development. For example, this mRNA may be degraded by NMD because it encodes a dominant-negative protein that interferes with synaptic maturation.
NMD as a therapeutic target in disease
In this section, we advance the notion that “NMD therapy” may ameliorate the symptoms of patients with some neural diseases.
NMD and ALS.
The notion that neurodegenerative diseases are potential clinical target for NMD therapy was first suggested by studies on an inherited form of amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease. Two groups developed yeast ALS models expressing the human FUS/TLS gene. This resulted in cytotoxicity and cytoplasmic inclusion bodies, thereby replicating what is observed in ALS patient cells93,94. To screen for genes that suppress this toxicity, these groups introduced plasmids over-expressing most known yeast genes. These screens identified a small group of yeast genes, most of which encode RNA-metabolism proteins and RNA-binding proteins.
One of the yeast genes that suppressed the toxicity of the mutant FUS was ECM32, a RNA helicase related to human UPF1. This led one of the groups to test human UPF1, which they found also suppressed FUS-mediated toxicity in their yeast ALS model93. Two lines of evidence suggested that human UPF1 rescues FUS-mediated toxicity through the NMD pathway. First, overexpressed human UPF1 appeared to stimulate NMD, based on downregulated expression of one endogenous NMD substrate RNA. Second, overexpression of two other NMD factors—human UPF2 and UPF3B—also rescued yeast cells from the toxic effects of FUS.
To assess whether these findings in yeast apply to mammals, Jackson et al. used a rat model of TDP-43-induced motor paralysis95. In this model, TDP-43 is expressed in rat spinal cord, resulting in limb paralysis similar to that seen in human ALS. To simulate a therapeutic approach that might be used in humans, the authors over-expressed human UPF1 from an adeno-associated virus (AAV) vector. These authors observed that this UPF1-AAV therapy improve motor function, motor coordination, grip strength, and forelimb function in their ALS rat model.
Similar results were seen by Barmada et al., who tested whether enhanced NMD could protect from either TDP-43 and FUS/TLS-mediated neurotoxicity in rodent neurons96. They found that the decreased survival of primary rodent cortical neurons triggered by either FUS or TDP43 (wild-type or mutant) was rescued by expression of human UPF1. Of note, the protective effect of UPF proteins was specific for FUS- and TDP-induced ALS, as it was not observed in other models of neurotoxicity; i.e., a Huntington’s disease model and ALS disease triggered by mutant superoxide dismutase.
How does overexpressed UPF1 protect from FUS and TDP-induced toxicity? Barmada et al. obtained several lines of evidence that overexpressed UPF1 acts by elevating the magnitude of NMD. First, the chemical NMD inhibitor, NMDI, reversed the protective effect of UPF1. Second, another NMD factor—UPF2—also rescued toxicity. Third, even greater protection was seen upon forced co-expression of both UPF1 and UPF296. Finally, expression of a RNA helicase that has been shown to function in NMD—MOV1097—also rescued toxicity.
It is commonly assumed that the accumulation of cytoplasmic FUS- and TDP-43-containing bodies in neuronal cells from ALS patients are responsible for ALS symptoms. However, although UPF1 and UPF2 overexpression greatly reduced the toxicity of FUS and TDP-43 overexpression, this treatment does not reverse the formation of the cytosolic inclusion bodies in yeast93. This raises the possibility that the inclusion bodies present in ALS patient cells are not responsible for toxicity.
Why might overexpressed UPF1 and UPF2 suppress ALS-induced toxicity? One possibility is that these NMD factors are rate-limiting in neural cells and thus their overexpression increases NMD magnitude as a means to combat an increased need for NMD in ALS. This follows from the fact that FUS and TDP-43 are RNA splicing factors and thus when their function is impaired in ALS, this would be expected to lead to high levels of mis-spliced mRNAs, many of which are NMD substrates. In support of this model, mutant FUS/TDP-43 are largely lost from the nucleus of neuronal cells from ALS patients and instead accumulate in cytoplasmic granules98. Because RNA splicing occurs in the nucleus, mislocalization to the cytosol would be predicted to disrupt the nuclear functions of these RNA splicing factors, thereby leading to the accumulation of mis-spliced RNAs. The massive increase in such mis-spliced RNAs could overload the NMD machinery, thereby creating a need for more NMD factors (Fig. 6).
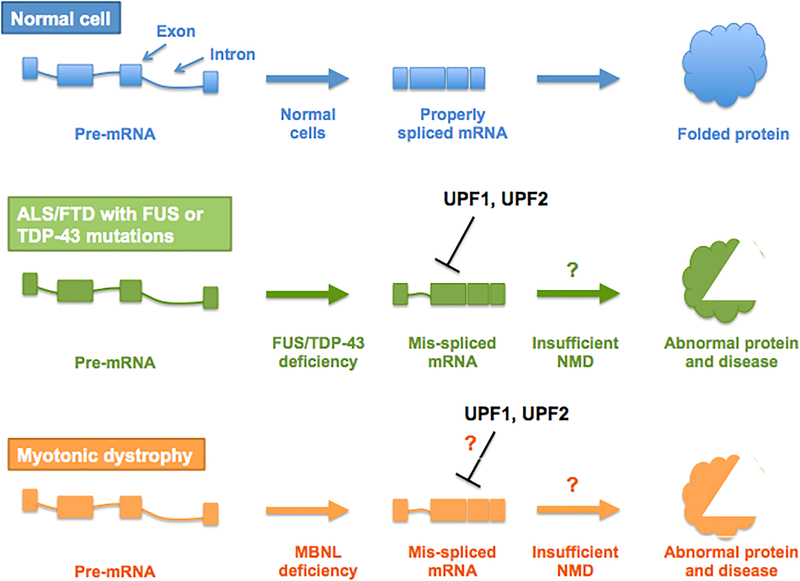
Figure 6. Potential roles of NMD in suppressing neurotoxic effects of mis-spliced mRNAs.
Upframeshift 1 (UPF1) and UPF2 overexpression reverses the neurotoxic effects of mutant Fused in sarcoma (FUS)- and TAR DNA-binding protein of 43 kDa (TDP-43) in amyotrophic lateral sclerosis (ALS) disease models96,102. This raises the possibility that these neurodegenerative diseases are mediated by insufficient NMD, which is restored by UPF1 or UPF2 overexpression. In normal cells (top), properly spliced mRNAs predominate, as NMD degrades most aberrantly spliced mRNAs. However, in cases where mutant TDP-43 and FUS is expressed (middle), these mutant proteins are mislocalized to the cytoplasm, preventing the ability of these proteins to regulate RNA splicing, thereby leading to abundant aberrant splicing99. NMD may not be sufficient to handle this increased load of aberrantly spliced transcripts, leading to the production of abnormal proteins that cause neurotoxicity. The protective effects of UPF1 and/or UPF2 overexpression may extend to other diseases where impaired RNA splicing is seen. In the case of myotonic dystrophy (bottom), the splicing factor, muscleblind-like splicing regulator 1 (MBNL), is sequestered in nuclear RNA granules, resulting in impaired MBNL-dependent splicing100. The toxicity of these mis-spliced mRNAs may also be mitigated by UPF1 and UPF2 overexpression (this remains speculative, indicated by the question mark).
In support of the idea that there is an increase in the load of mis-spliced RNA, studies have detected elevated levels of retained introns in transcripts in the spinal cord of mice expressing an ALS-associated FUS mutation99. It is well established that retained introns are NMD substrates, as most introns contain stop codons in all 3 reading frames and thus have PTCs, a key feature that triggers NMD.
How does an accumulation of mis-spliced mRNAs lead to ALS toxicity? If such mRNAs are not efficiently degraded by NMD, their translation would be expected to produce truncated proteins, some of which probably act as dominant-negative proteins. These proteins may themselves be toxic, or they may activate neuronal stress responses normally triggered by misfolded proteins.
Together, these studies suggest a model in which ALS and NMD are intimately intertwined, providing a potential rationale for therapeutic augmentation of NMD in the treatment of this disease. Insufficient levels of NMD may also be involved in other diseases that have been linked to sequestration of RNA splicing factors, such as trinucleotide repeat-expansion diseases100. Similarly, spinal muscular atrophy is associated with impaired RNA splicing due to inefficient assembly of spliceosome subunits101. As described below, we suggest that restoring normal NMD activity through “NMD therapy” could potentially restore normal function.
NMD activation therapy.
In order to reverse NMD deficiency in neurodegenerative diseases such as ALS, it is critical to devise therapeutic approaches to elevate the magnitude of NMD. How can this be achieved? One approach is to overexpress NMD factors such as UPF1 or UPF2 with viral vectors as done by Jackson et al. and Barmada et al.96,102. However, the efficiency of delivery, particularly in the brain, will likely be challenging. Another approach is to develop small molecule NMD activators. Although many small-molecule NMD inhibitors have been identified and applied to clinical scenarios6,103, no well-defined NMD activators have been discovered. One appealing strategy is to screen for inhibitors of UPF3A, which was recently shown to be a NMD repressor42, and thus UPF3A inhibitors would be predicted to activate NMD. Another strategy is to identify activators of SMG1, the kinase that phosphorylates UPF1 and is essential for NMD27,104,105.
Although NMD activators could be beneficial for neurodegenerative diseases associated with impaired function of RNA splicing factors or NMD proteins, it is possible that these activators could also have unwanted side effects. For example, NMD activators would be predicted to destabilize critical NMD target RNAs, such as Arc, which could impair synaptic plasticity and learning.
NMD inhibition therapy.
There has been considerable interest in identifying agents that inhibit the NMD pathway, as they would be expected to elevate the expression of mutant PTC-bearing genes that still encode a functional protein. For example, some types of muscular dystrophy are caused by point mutations that do not abolish protein function but cause the Dystrophin mRNA to be degraded by NMD106,107. By inhibiting NMD, mutant—but still functional—Dystrophin mRNA could be elevated in level, thereby potentially reducing disease symptoms. Likewise, NMD inhibitors have the potential to treat neural diseases caused by nonsense or frameshift mutations that do not completely ablate function. Several NMD inhibitors have already been identified, many with defined biochemical functions103,108,109, and thus the field is poised to investigate the clinical utility of such agents. Given that NMD inhibitors would be predicted to enhance ARC expression, another interesting potential future application is to determine whether such agents enhance synaptic plasticity after synaptic activation and thus improve learning and memory in neurodegenerative and neurodevelopmental diseases associated with impaired synaptic function.
Concluding remarks
NMD is emerging as a critical regulator of neuronal development and function, and more recently as a potential regulator of neuronal viability in disease contexts. Insufficient NMD may underlie neurodegenerative diseases, such as ALS, which are caused by sequestration of RNA splicing factors. NMD suppression may be valuable for enhancing the expression of synaptic proteins and synaptic function. It will be important for future studies to direct their efforts towards designing NMD modulators to treat neurodegenerative disease and other neural disorders.
Acknowledgements
This work was supported by NIH grants NS056306 (S.R.J.) and R01 GM111838 (M.F.W.)
Footnotes
Competing interests policy There is NO Competing Interest.
REFERENCES
- 1.Lennox AL, Mao H, Silver DL. RNA on the brain: emerging layers of post-transcriptional regulation in cerebral cortex development. Wiley Interdiscip Rev Dev Biol 2017; : e290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 2007; 76: 51–74. [DOI] [PubMed] [Google Scholar]
- 3.Popp MW-L, Maquat LE. Organizing Principles of Mammalian Nonsense-Mediated mRNA Decay. Annu Rev Genet 2013; 47: 139–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 2015; 16: 665–677. [DOI] [PubMed] [Google Scholar]
- 5.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A 1979; 76: 5134–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet 2004; 36: 801–8. [DOI] [PubMed] [Google Scholar]
- 7.Inoue K, Khajavi M, Ohyama T, Hirabayashi S, Wilson J, Reggin JD et al. Molecular mechanism for distinct neurological phenotypes conveyed by allelic truncating mutations. Nat Genet 2004; 36: 361–9. [DOI] [PubMed] [Google Scholar]
- 8.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol 1999; 19: 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5’ to 3’ mRNA decay pathways in yeast. Mol Cell 2003; 12: 1439–1452. [DOI] [PubMed] [Google Scholar]
- 10.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 2004; 36: 1073–8. [DOI] [PubMed] [Google Scholar]
- 11.Ge Y, Porse BT. The functional consequences of intron retention: Alternative splicing coupled to NMD as a regulator of gene expression. BioEssays 2014; 36: 236–243. [DOI] [PubMed] [Google Scholar]
- 12.Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 2012; 9: 1370–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muir VS. The Substrates of Nonsense-Mediated mRNA Decay in Caenorhabditis elegans. 2017; : 1–42. [DOI] [PMC free article] [PubMed]
- 14.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA 2012; 3: 807–828. [DOI] [PubMed] [Google Scholar]
- 15.Karam R, Wengrod J, Gardner LLB, Wilkinson MFM. Regulation of nonsense-mediated mRNA decay: implications for physiology and disease. Biochim Biophys Acta 2013; 1829: 624–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lou C-H, Dumdie J, Goetz A, Shum EYE, Brafman D, Liao XX et al. Nonsense-Mediated RNA Decay Influences Human Embryonic Stem Cell Fate. Stem Cell Reports 2016; 6: 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karam R, Lou C-H, Kroeger H, Huang L, Lin JH, Wilkinson MF. The unfolded protein response is shaped by the NMD pathway. EMBO Rep 2015; 16: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Shi Y, Wang P, Guachalla LM, Sun B, Joerss T et al. Smg6/Est1 licenses embryonic stem cell differentiation via nonsense-mediated mRNA decay. EMBO J 2015; 34: 1630–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson JO, Moore KA, Chapin A, Hollien J, Metzstein MM. Degradation of Gadd45 mRNA by nonsense-mediated decay is essential for viability. Elife 2016; 5. doi: 10.7554/eLife.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goetz AE, Wilkinson M. Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci 2017; 74: 3509–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998; 23: 198–9. [DOI] [PubMed] [Google Scholar]
- 22.Boehm V, Gehring NH. Exon Junction Complexes: Supervising the Gene Expression Assembly Line. Trends Genet 2016; 32: 724–735. [DOI] [PubMed] [Google Scholar]
- 23.Woodward LA, Mabin JW, Gangras P, Singh G. The exon junction complex: a lifelong guardian of mRNA fate. Wiley Interdiscip Rev RNA 2017; 8. doi: 10.1002/wrna.1411. [DOI] [PubMed] [Google Scholar]
- 24.Neu-Yilik G, Raimondeau E, Eliseev B, Yeramala L, Amthor B, Deniaud A et al. Dual function of UPF3B in early and late translation termination. EMBO J 2017; 36: e201797079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol 2008; 15: 85–93. [DOI] [PubMed] [Google Scholar]
- 26.López-Perrote A, Castaño R, Melero R, Zamarro T, Kurosawa H, Ohnishi T et al. Human nonsense-mediated mRNA decay factor UPF2 interacts directly with eRF3 and the SURF complex. Nucleic Acids Res 2015; 44: 1909–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 2006; 20: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 2002; 12: 1060–1067. [DOI] [PubMed] [Google Scholar]
- 29.Zünd D, Gruber AR, Zavolan M, Mühlemann O. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3’ UTRs. Nat Struct Mol Biol 2013; 20: 936–43. [DOI] [PubMed] [Google Scholar]
- 30.Hurt JA, Robertson AD, Burge CB. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 2013; 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarpey PS, Raymond FL, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 2007; 39: 1127–1133.This was the first report demonstrating that UPF3B mutations are likely to cause intellectual disability in humans. They also found that intellectual disability patients with UPF3B mutations sometimes have neuro-developmental disorders, such as schizophrenia.
- 32.Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry 2010; 15: 767–76. [DOI] [PubMed] [Google Scholar]
- 33.Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry 2011; 16: 238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch SA, Nguyen LS, Ng LY, Waldron M, McDonald D, Gecz J et al. Broadening the phenotype associated with mutations in UPF3B: two further cases with renal dysplasia and variable developmental delay. Eur J Med Genet 2012; 55: 476–9. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, Zhang L, Tong P, Xun G, Su W, Xiong Z et al. Exome sequencing identifies UPF3B as the causative gene for a Chinese non-syndrome mental retardation pedigree. Clin Genet 2012; 83: 560–4. [DOI] [PubMed] [Google Scholar]
- 36.Huang L, Shum EY, Jones SH, Lou C-H, Dumdie J, Kim H et al. A Upf3b-mutant mouse model with behavioral and neurogenesis defects. Mol Psychiatry 2017; : 1–14.This study showed that mice harboring loss-of-function mutation in Upf3b have specific behavioral defects and reduced dendritic spine maturation in specific brain regions. Cultured neural progenitors from these NMD-deficient mice exhibit perturbed neural differentation.
- 37.Chan W-K, Huang L, Gudikote JP, Chang Y-F, Imam JS, MacLean JA et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J 2007; 26: 1820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang L, Lou C-HH, Chan W, Shum EY, Shao A, Stone E et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell 2011; 43: 950–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 2001; 10: 99–105. [DOI] [PubMed] [Google Scholar]
- 40.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Mönch K, Thoren LA, Nielsen FC et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev 2008; 22: 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC et al. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci U S A 2010; 107: 12186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shum EY, Jones SH, Shao A, Dumdie J, Krause MD, Chan WK et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell 2016; 165: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen LS, Kim H-G, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y et al. Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet 2013; 22: 1816–25. [DOI] [PubMed] [Google Scholar]
- 44.Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 2013; 154: 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet 2008; 40: 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K et al. Recurrent Rearrangements of Chromosome 1q21.1 and Variable Pediatric Phenotypes. N Engl J Med 2008; 359: 1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenfeld JA, Traylor RN, Schaefer GB, McPherson EW, Ballif BC, Klopocki E et al. Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Hum Genet 2012; 20: 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao H, Pilaz L-J, McMahon JJ, Golzio C, Wu D, Shi L et al. Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly. J Neurosci 2015; 35: 7003–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mao H, McMahon JJ, Tsai Y-H, Wang Z, Silver DL. Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly. PLOS Genet 2016; 12: e1006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alachkar A, Jiang D, Harrison M, Zhou Y, Chen G, Mao Y. An EJC factor RBM8a regulates anxiety behaviors. Curr Mol Med 2013; 13: 887–99. [DOI] [PubMed] [Google Scholar]
- 51.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 2012; 151: 750–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol 2012; 19: 1124–1131. [DOI] [PubMed] [Google Scholar]
- 53.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 2011; 42: 500–510.This study identified the first miRNA that regulates NMD. This miRNA--miR-128--is induced during neural differentiation, where it functions to downregulates NMD.
- 54.Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet 2013; 22: 4673–4687.Using a knock-down approach, this study showed that Upf3b influences neural progenitor proliferation vs. differentiation decisions, as well as neurite outgrowth. This study also demonstrated that Upf3a and Upf3b exhibit interesting patterns of expression during brain development.
- 55.Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R et al. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep 2014; 6: 748–64.This study provided evidence for a NMD-based molecular circuit that controls neural differentiation. Neurally expressed miRNAs downregulate NMD, which, in turn, upregulates pro-neural and anti-proliferation proteins encoded by NMD target mRNAs.
- 56.Alrahbeni T, Sartor F, Anderson J, Miedzybrodzka Z, McCaig C, Müller B. Full UPF3B function is critical for neuronal differentiation of neural stem cells. Mol Brain 2015; 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho H, Kim KM, Han S, Choe J, Park SG, Choi SS et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol Cell 2012; 46: 495–506. [DOI] [PubMed] [Google Scholar]
- 58.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev 2009; 23: 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang G, Jiang B, Jia C, Chai B, Liang A. MicroRNA 125 represses nonsense-mediated mRNA decay by regulating SMG1 expression. Biochem Biophys Res Commun 2013; 435: 16–20. [DOI] [PubMed] [Google Scholar]
- 60.Akerblom M, Petri R, Sachdeva R, Klussendorf T, Mattsson B, Gentner B et al. microRNA-125 distinguishes developmentally generated and adult-born olfactory bulb interneurons. Development 2014; 141: 1580–1588. [DOI] [PubMed] [Google Scholar]
- 61.Lattanzi A, Gentner B, Corno D, Di Tomaso T, Mestdagh P, Speleman F et al. Dynamic Activity of miR-125b and miR-93 during Murine Neural Stem Cell Differentiation In Vitro and in the Subventricular Zone Neurogenic Niche. PLoS One 2013; 8. doi: 10.1371/journal.pone.0067411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan Q, Weyn-Vanhentenryck SM, Wu J, Sloan SA, Zhang Y, Chen K et al. Systematic discovery of regulated and conserved alternative exons in the mammalian brain reveals NMD modulating chromatin regulators. Proc Natl Acad Sci 2015; 112: 3445–3450.This study identified hundreds of alternatively spliced NMD-inducing exons expressed in the mouse brain cortex. These “NMD exons” are enriched in genes encoding RNA-binding proteins and chromatin regulators, suggesting the existance of a gene regulatory feedback loop operating in the cortex.
- 63.Krey JF, Pa??ca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci 2013; 16: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen LS, Jolly L, Shoubridge C, Chan WK, Huang L, Laumonnier F et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry 2012; 17: 1103–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wittkopp N, Huntzinger E, Weiler C, Saulière J, Schmidt S, Sonawane M et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol 2009; 29: 3517–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci 2010; 13: 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilaz L, Mcmahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E et al. Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain Article Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron 2016; 89: 83–99.Using mice haploid for the EJC factor gene, Magoh, this study found that Magoh is critical for developing radial glia, including their survival, the timing at which they initiate mitosis, and the number of cell divisions that they undergo.
- 68.Zou D, McSweeney C, Sebastian A, Reynolds DJ, Dong F, Zhou Y et al. A critical role of RBM8a in proliferation and differentiation of embryonic neural progenitors. Neural Dev 2015; 10: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M. Alternative Splicing of the Robo3 Axon Guidance Receptor Governs the Midline Switch from Attraction to Repulsion. Neuron 2008; 58: 325–332. [DOI] [PubMed] [Google Scholar]
- 70.Jaworski A, Long H, Tessier-Lavigne M. Collaborative and Specialized Functions of Robo1 and Robo2 in Spinal Commissural Axon Guidance. J Neurosci 2010; 30: 9445–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM et al. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 2004; 42: 213–223. [DOI] [PubMed] [Google Scholar]
- 72.Sabatier C, Plump AS, Ma L, Brose K, Tamada A, Murakami F et al. The divergent robo family protein Rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell 2004; 117: 157–169. [DOI] [PubMed] [Google Scholar]
- 73.Colak D, Ji S-J, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell 2013; 153: 1252–65.This study revealed the existence of “local” NMD occurring within axon terminals that controls the mRNA composition of pathfinding axons. NMD was found to target Robo3.2 mRNA, thereby controlling the expression this important axon guidance protein.
- 74.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: Features and functions. Cell 2010; 142: 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J 2008; 27: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mills EW, Wangen J, Green R, Ingolia NT. Dynamic Regulation of a Ribosome Rescue Pathway in Erythroid Cells and Platelets. Cell Rep 2016; 17: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rufener SC, Mühlemann O. EIF4E-bound mRNPs are substrates for nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol 2013; 20: 710–717. [DOI] [PubMed] [Google Scholar]
- 78.Durand S, Lykke-Andersen J. Nonsense-mediated mRNA decay occurs during eIF4F-dependent translation in human cells. Nat Struct Mol Biol 2013; 20: 702–709. [DOI] [PubMed] [Google Scholar]
- 79.Kiebler MA, Bassell GJ. Neuronal RNA Granules: Movers and Makers. Neuron 2006; 51: 685–690. [DOI] [PubMed] [Google Scholar]
- 80.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 1995; 270: 28995–9003. [DOI] [PubMed] [Google Scholar]
- 81.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 2007; 130: 179–191.This study linked synaptic activity to NMD-dependent degradation of the Arc mRNA, a natural NMD target which encodes a key synaptic regulator. This study showed that NMD factor components are present at synapses, providing the first hints that NMD can function at synaptic sites.
- 82.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG et al. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron 1995; 14: 433–445. [DOI] [PubMed] [Google Scholar]
- 83.Link WW, Konietzko UU, Kauselmann GG, Krug MM, Schwanke BB, Frey UU et al. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci U S A 1995; 92: 5734–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farris S, Lewandowski G, Cox CD, Steward O. Selective Localization of Arc mRNA in Dendrites Involves Activity- and Translation-Dependent mRNA Degradation. J Neurosci 2014; 34: 4481–4493.This study revealed evidence that NMD controls the levels of the Arc transcript at synapses, which encodes a key synaptic regulator.
- 85.Paolantoni C, Ricciardi S, De Paolis V, Okenwa C, Catalanotto C, Ciotti MT et al. Arc 3′ UTR Splicing Leads to Dual and Antagonistic Effects in Fine-Tuning Arc Expression Upon BDNF Signaling. Front Mol Neurosci 2018; 11: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr Opin Neurobiol 2005; 15: 599–606. [DOI] [PubMed] [Google Scholar]
- 87.Alonso CR. A complex ‘mRNA degradation code’ controls gene expression during animal development. Trends Genet 2012; 28: 78–88. [DOI] [PubMed] [Google Scholar]
- 88.Jaagura M, Taal K, Koppel I, Tuvikene J, Timmusk T, Tamme R. Rat NEURL1 3′UTR is alternatively spliced and targets mRNA to dendrites. Neurosci Lett 2016; 635: 71–76. [DOI] [PubMed] [Google Scholar]
- 89.Li Y, Bor Y -c., Fitzgerald MP, Lee KS, Rekosh D, Hammarskjold M-L. An NXF1 mRNA with a retained intron is expressed in hippocampal and neocortical neurons and is translated into a protein that functions as an Nxf1 cofactor. Mol Biol Cell 2016; 27: 3903–3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Long AA, Mahapatra CT, Woodruff EA, Rohrbough J, Leung H-TT, Shino S et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci 2010; 123: 3303–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng S, Gray EE, Chawla G, Porse BT, O’Dell TJ, Black DL. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci 2012; 15: 381–8, S1.This study provided evidence that NMD is critical for the regulation of a factor that mediates synaptic maturation, and plasticity.
- 92.Zheng S HHS Public Access. Int J Dev Neurosci 2016; 55: 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ju S, Tardiff DF, Han H, Divya K, Zhong Q, Maquat LE et al. A Yeast Model of FUS/TLS-Dependent Cytotoxicity. PLoS Biol 2011; 9. doi: 10.1371/journal.pbio.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the als disease protein fus/tls. PLoS Biol 2011; 9. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nickless A, Jackson E, Marasa J, Nugent P, Mercer RW, Piwnica-Worms D et al. Intracellular calcium regulates nonsense-mediated mRNA decay. Nat Med 2014; 20: 961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barmada SJ, Ju S, Arjun A, Batarse A, Archbold HC, Peisach D et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci 2015; 112: 7821–7826.This study showed that overexpression of the NMD factor, UPF1, reduces toxicicity in a neuronal model of amyotrophic lateral sclerosis.
- 97.Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S et al. MOV10 Is a 5’ to 3’ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3’ UTRs. Mol Cell 2014; 54: 573–585. [DOI] [PubMed] [Google Scholar]
- 98.Conlon EG, Manley JL. RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev 2017; 31: 1509–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiu H, Lee S, Shang Y, Wang WY, Au KF, Kamiya S et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest 2014; 124: 981–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miller JW. Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J 2000; 19: 4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lotti F, Imlach WL, Saieva L, Beck ES, Hao LT, Li DK et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell 2012; 151: 440–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jackson KL, Dayton RD, Orchard EA, Ju S, Ringe D, Petsko GA et al. Preservation of forelimb function by UPF1 gene therapy in a rat model of TDP-43-induced motor paralysis. Gene Ther 2015; 22: 20–28.Using a rat model of amyotrophic lateral sclerosis, these investigators showed that overexpression of the NMD protein, UPF1, can reduce symptoms of ALS, including motor paralysis.
- 103.Nickless A, Bailis JM, You Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci 2017; 7: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wilkinson MF. The Cycle of Nonsense. Mol Cell 2003; 12: 1059–1061. [DOI] [PubMed] [Google Scholar]
- 105.Melero R, Hug N, Lopez-Perrote A, Yamashita A, Caceres JF, Llorca O. The RNA helicase DHX34 functions as a scaffold for SMG1-mediated UPF1 phosphorylation. Nat Commun 2016; 7. doi: 10.1038/ncomms10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Phelps SF, Hauser MA, Cole NM, Rafael JA, Hinkle RT, Faulkner JA CJ. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet 1995; 4: 1251–1258. [DOI] [PubMed] [Google Scholar]
- 107.Yuasa K, Miyagoe Y, Yamamoto K, Nabeshima YI, Dickson G, Takeda S. Effective restoration of dystrophin-associated proteins in vivo by adenovirus-mediated transfer of truncated dystrophin cDNAs. FEBS Lett 1998; 425: 329–336. [DOI] [PubMed] [Google Scholar]
- 108.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen C-H, Grierson DS, Bertrand E et al. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol 2007; 178: 1145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martin L, Grigoryan A, Wang D, Wang J, Breda L, Rivella S et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res 2014; 74: 3104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Favaro FP, Alvizi L, Zechi-Ceide RM, Bertola D, Felix TM, De Souza J et al. A Noncoding expansion in EIF4A3 causes richieri-costa-pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet 2014; 94: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ge Z, Quek BL, Beemon KL, Hogg JR. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife 2016; 5: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet 2012; 44: 435–9, S1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]