Abstract
Bioremediation is the degradation potential of microorganisms to dissimilate the complex chemical compounds from the surrounding environment. The genetics and biochemistry of biodegradation processes in datasets opened the way of systems biology. Systemic biology aid the study of interacting parts involved in the system. The significant keys of system biology are biodegradation network, computational biology, and omics approaches. Biodegradation network consists of all the databases and datasets which aid in assisting the degradation and deterioration potential of microorganisms for bioremediation processes. This review deciphers the bio-degradation network, i.e., the databases and datasets (UM-BBD, PAN, PTID, etc.) aiding in assisting the degradation and deterioration potential of microorganisms for bioremediation processes, computational biology and multi omics approaches like metagenomics, genomics, transcriptomics, proteomics, and metabolomics for the efficient functional gene mining and their validation for bioremediation experiments. Besides, the present review also describes the gene editing tools like CRISPR Cas, TALEN, and ZFNs which can possibly make design microbe with functional gene of interest for degradation of particular recalcitrant for improved bioremediation.
Keywords: systems biology, xenobiotics, bioremediation, metabolomics, pollutant, metabolic network, gene editing
Introduction
Due to ever-increasing world population and their corresponding food commodities also need to be enhanced (Cazalis et al., 2018; Drangert et al., 2018). This is achieved by lowering down the damage to agricultural crops by pests (Schmidt-Jeffris and Nault, 2018). Pests can be weed, herb, insect, rodent, nematode, and microorganisms (bacteria, fungi, and algae) (Bottrell and Schoenly, 2018; Duke, 2018). Pesticides are classified according to their target and show an excellent role in the production of crop yield and lowering down the rate of agricultural losses due to pest (Allmaras et al., 2018). The intensive use of pesticides at unmanageable rate has led to decontaminate the soil, and agricultural runoffs are being biomagnifying water bodies and increase the toxicity level at each trophic level in food web, i.e., DDT (dichlorodiphenyltrichloroethane) (Plattner et al., 2018; Silva-Barni et al., 2018; Thomas et al., 2008). Besides this, pesticidal compounds also have ill effects on health affecting the function of organs and damage the DNA at molecular level leading to neurological diseases and cancer, i.e., azoxystrobin and atrazine (Fatima et al., 2018; Singh N.S. et al., 2018; Vidart d’Egurbide Bagazgoïtia et al., 2018). Pesticides are also reached in foodstuff beginning from the agricultural field to serving tables (Bakirhan et al., 2018; Kumar D. et al., 2018; Sarlio, 2018). Reports are noted encountering the presence of pesticides in fruit juices, milk, seaweeds (food supplement), and other food items (Bedi et al., 2018; Hamedi et al., 2018; Kumar D. et al., 2018; Leprêtre and Merten-Lentz, 2018). Thus, it is mandatory that the use of synthetic pesticides must be lowered down and organic farming should be done (Tal, 2018). The removal and degradation of pesticidal residues are being done with conventional methods of bioremediation (John et al., 2018; Moorman, 2018). Bioremediation points out environmental decontamination of pesticides by microbiological processes whether in situ or ex situ (Ortiz-Hernández et al., 2018). In situ bioremediation (bioventing, biosparging, and bioaugmentation) decontaminates without removal of soil from the site and ex situ (landfarming, biopiling, composting, bioreactors, and electrodialysis) treat the unearth soil at the site (Parween et al., 2018). It is an excellent environmental friendly option for degrading the pollutants (Muangchinda et al., 2018). With the advancement of scientific research methodologies, the gene editing and systemic biology tools are being applied in bioremediation of heavy metal, POPs (persistent organic pollutants), petroleum, acid drainage, xenobiotics (Basu et al., 2018; Dai et al., 2018; Gaur et al., 2018; Malla et al., 2018; Boudh and Singh, 2019). Bioremediation is the involvement of chemical components tends to degrade and tangled microbial metabolic web at the contaminated scenario (Nikolopoulou and Kalogerakis, 2018). Systems biology approach provides information about the microbial system (Hall et al., 2018). These microbial systems under different subsets of conditions respond differently (Montiel-Rozas et al., 2018). Microbial interactions within the communities are also observed with systems biology approaches (Kong et al., 2018). Also, this approach is very helpful in understanding the existence of microbes under different environmental conditions of extreme temperature and pressure (Borja, 2018). Omics such as genomics, transcriptomics, metabolomics, and proteomics aid the systems biology studies of microbes for analyzing the genetic level regulation for bioremediation (De Sousa et al., 2018). Advancement in sequencing with high throughput sequencing (HTS) and next-generation sequencing resolve the novel genes involved in biodegradation pathways of various persistent pollutants (Suenaga, 2012; von Netzer et al., 2018).
Gene Editing Tools
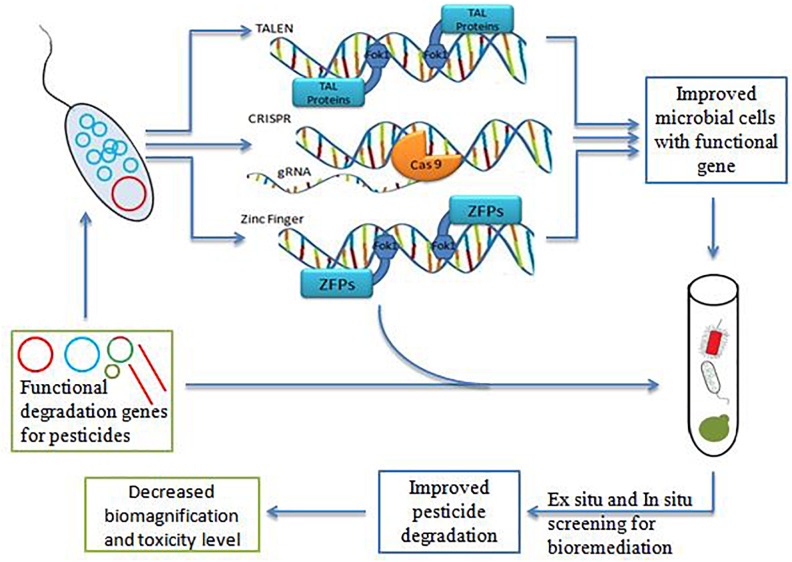
Gene editing is remarkable approach having the ability to manipulate DNA by using engineered nucleases named as molecular scissors. Molecular scissors have an immense application in wide range of research areas related to plant, animals, and microorganism (Butt et al., 2018). The process of editing involves targeting by self-designed guide sequence complementary to sequence of gene of interest assisting break at a site, repaired by homologous recombination, making manipulation (insertion or deletion) of desired sequence fragment (Bier et al., 2018). The genome engineering by gene editing tools led to next level application of microorganisms in various areas like feed, food, agriculture and medical, etc. (Yadav et al., 2018). The gene editing tools have potent capacity to improve the bioremediation processes (Figure 1) such as the elimination of xenobiotics, conversion of toxic compounds to less toxic compounds, and degradation of pesticide to simple components (Basu et al., 2018; Hussain et al., 2018).
FIGURE 1.
Gene editing tools for bioremediation.
The main gene editing tools are CRISPR-Cas, ZFN and TALEN can possibly make the above expectations fulfill (Singh V. et al., 2018; Waryah et al., 2018; Wong, 2018). The collective action of these gene editing tools is to establish double stranded break (DSB) in the target gene sequence, repaired via homology – directed repair (HRD) and error-prone non- homologous end joining (NHEJ) pathway (Arazoe et al., 2018; Yadav et al., 2018). Artificial restriction enzymes are utilized by ZFNs and TALEN which cleave the specific target DNA sequence by Zinc finger DNA binding domain and TAL effector DNA binding domain respectively (Banerjee et al., 2018; Shah et al., 2018). These gene editing tools aim to create better microbe having more complex genes and designing microorganism with maximum quality (Basu et al., 2018; Dangi et al., 2018). It is the root point of altered genetic makeup different from wild types for obtaining the desirable new microbes with functional gene of interest (Dai et al., 2018; Stein et al., 2018). The repercussion thus obtained are off target mutations, lethal mutations and risk of accidental or intentional release of modified organism in environment, etc. make the limitation boundary of using the above gene editing tools (Canver et al., 2018).
CRISPR-Cas
CRISPR-Cas describe the most effective and productive gene editing (McMahon et al., 2018; Yadav et al., 2018). There are three types, i.e., Types I, II, and III (Zhu Y. et al., 2018) and also numerous subtypes of the CRISPR-Cas systems present (Behler et al., 2018). Each system has specific Cas according to system acting, i.e., model organisms (Cooper et al., 2018). Cas9, a DNA endonuclease is guided by RNA to target foreign DNA for obstruction (Mahas and Mahfouz, 2018). CRISPR is 30–40 bp direct repeat sequence separated by spacer sequence complementing the foreign sequence then after processing and transcription crRNA is formed (Zhang C. et al., 2018). The gRNA (guide RNA) are then obtained by CRISPRs (Listgarten et al., 2018). crRNA and Cas protein together form crRNP (Ribonucleoprotein) which introduce a break in DNA/RNA of the intruder (Majumdar et al., 2017). gRNA makes specific binding to the target DNA site constitute the CRISPR particularity and specificity in function (Shah et al., 2018). The gene of interest can be manipulated (deleted or inserted) from the system with the help of CRISPR/Cas9 by introducing double strand break (DSB) at the target site (Shapiro et al., 2018). The suitable expression system is for the attainment of CRISPR-Cas sgRNA sequence, the codon optimizes variant of Cas9 and ideal promoters for transcription of sgRNA and Cas9 (Rico et al., 2018). The CRISPR technique is obtaining attention by molecular biologist due to the reason, it is highly suited with archeal and bacterial systems (Greene, 2018).
Transcription Activator-Like Effector Nucleases
TALENs stand for Transcription activator-like effector nucleases. It is an innovative tool for gene modification and editing. TALENs have TAL proteins. These proteins are originally secreted from Xanthomonas, a pathogenic bacterial genus. TAL proteins are so effective that they can bind to even very short sequence, i.e., 1–2 nucleotides. Furthermore, the nucleases involved are very efficient in binding due to the presence of 34 amino acids tandem repeats. Gene knock out (non-homologous end joining), and gene knock in (Homology directed repair) of the target gene or gene of interest are now preferred with TALENs. Two protein domains, one for sequence cleavage and second for recognizing and binding the very particular and specific site make the TALENs robust gene editing tool. It is applied to many eukaryotic targets like mammalian cells, frogs, zebrafish, rats, and chickens.
Zinc Finger Nucleases
ZFNs stand for Zinc Finger Nucleases. It is most commonly used endonuclease. These are artificial restriction enzyme. Zinc Finger Nucleases have ZFPs (Zinc Finger Proteins). ZFPs are basically eukaryotic transcription factors having the ability to act as DNA binding domain. ZFNs also have Folk1 (nucleotide cleavage domain) originated from Flavobacterium okeanokoites. Numerous ZFPs (usually four to six) surrounds the cleavage domain depending upon the target site. These ZFPs have 18 bp specificity possibly making the accurate target specific gene editing. ZFPs are 30 amino acids long with alpha-helix in opposition to two antiparallel β-sheets. This gene editing tool is mentioned with gene knock out (non-homologous end joining) and knock in (Homology directed repair) for successful prokaryotic and eukaryotic gene editing.
Advantages and Disadvantages of Gene Editing Tools
Among the above gene editing tools, CRISPR-Cas is cheap, simple, and easy for researchers to apply in comparison with TALENs and ZFNs (Ju et al., 2018). It has the advantage to evaluate the gene interaction and their genetic and phenotypic relationship along with the gene knock out system replaced with another gene of interest (VanderSluis et al., 2018). The limitation of CRISPR-Cas system is off target mutation leading to lethality, genomic disintegration and hindrance in applicability (Sun et al., 2018). Moreover, unlike CRISPR-Cas is more reliable in terms of specificity in target binding, the TALENs and ZFNs lead to strategies for mutagenesis due to random binding to DNA sequence (Stein et al., 2018).
Keys of Systems Biology
Biodegradation Network
The execution of computational tools and bioinformatics resources is an advanced approach toward the pesticide bioremediation (Malla et al., 2018; Vanacek et al., 2018). It takes the online platform of biodegradative databases publicly accessed for retrieving information on biodegradation of xenobiotic (pesticide) by microorganisms and biodegradation pathways of persistent chemicals (Nolte et al., 2018). These databases comprise the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD), Biodegradation Network-Molecular Biology database (Bionemo), Pesticide Target interaction database (PTID), Microbial Genome Database (MBGD), Biodegradative Oxygenases Database (OxDBase), BioCyc and MetaCyc compatible with both windows as well as Linux operating systems (Arora and Bae, 2014). UMBBD-Pathway Prediction database1 displays the data concerning microbial biocatalytic reactions and biodegradation pathways (Erythropel et al., 2018) explored for various types of pesticidal compounds as mentioned in Table 1. The ambition of the UM-BBD is to provide data on microbial enzyme-catalyzed reactions that are important for bioremediation (Ellis et al., 2006). Besides, it also gives the information of intermediate compounds obtained during degradation by microorganisms (Dvořák et al., 2017). With the advancement of synthetic pesticides due to pest resistance, PTID was developed by Gong et al. (2012). This database contains annotation of 1347 pesticides and 13738 pesticide target interactions. By text mining PTID aid to design novel agrochemical products and identification of pesticide targets (Gong et al., 2012). Another database is a Microbial Genome Database (MBGD) an open door for comparative investigation at the genomic level used for evaluating the gene arrangement, ortholog recognition, and collection of paralog data (Bhatt, 2018; Selzer et al., 2018). A broad information resource associated to bioremediation and biodegradation is MetaRouter which allows data mining. It is an established database providing the foundation for bioremediation laboratories, consulting biodegradative routes of different chemical compounds persistent in nature (Korjus, 2014). Synthetic pesticides are called as xenobiotics. OxDBase is biodegradative oxygenase database (Arora et al., 2009). Oxygenase, a class of enzyme, transfers the O2 for oxidation of chemical compound (Guengerich and Yoshimoto, 2018). Oxidation is responsible for aromatic ring cleavage breaking down the persistent organic compounds of xeno pesticides (Westcott and Cessna, 2018). OxDBase database also aids the biodegradation network by providing information of oxygenase catalyzed reactions (Shah et al., 2012). Biodegradation and bioconversion of recalcitrant compounds by oxygenases make the bioremediation possible (Sharma et al., 2018). Another database named Bionemo (Biodegradation Network Molecular Biology) have entries of sequences encoding for biodegradation genes (BDGs) and their transcription and regulation (Arora and Bae, 2014). The retrieved data is worthy important for robust biodegradation network (Oladipo et al., 2018). Other components/ databases for robust biodegradation network are mentioned in Table 2. Biodegradation pathways of persistent pesticides, i.e., DDT (dichlorodiphenyltrichloroethane), HCH (hexachlorocyclohexane), and ATZ (atrazine) present under different conditions have been studied (Das et al., 2016). Fang et al. (2014) studied the pesticide biodegradation pathways of isolates from marine and freshwater sediments.
Table 1.
Classification of the pesticides.
| S. no. | Classification of pesticide (examples) | Target (examples) | Reference |
|---|---|---|---|
| 1 | Herbicide (Benazolin, Bentazone, Imazapyr, Atrazine, Triclopyr, Glyphosate) | Herbs (Cenchrus macrourus, Eragrostis curvula, Kalanchoe delagoensis) | Guerra-García et al., 2018 |
| 2 | Weedicide (Borax, Nitrofan) | Weeds (Galium spurium, Selaginella kraussiana, Alternanthera philoxeroides, Evolvulus nummularius, Verbesina encelioides, Euphorbia thymifolia) | Paynter et al., 2015; Kaur et al., 2018 |
| 3 | Insecticide (DDT, BHC, Chloropyrifos, HCH) | Insects (Grasshopper, Aphid, Beetle, Thrips, Mealybug) | Kumar D. et al., 2018; Qian et al., 2018; van Lenteren et al., 2018 |
| 4 | Rodenticide (Warfarin, Zinc phosphide) | Rodents (Anas, Platyrhynchos, Aves, Sciurus, Tamias, Rattus, Mus) | Hogue et al., 2018; Mushtaq, 2018 |
| 5 | Nematicide (Phorate, Fenamiphos, Ethoprop, Dibromochloropropane, Carbamate) | Nematodes (Meloidogyne incognita, M. javanica) | Mohamed, 2018; Visagie et al., 2018; Warmerdam et al., 2018 |
| 6 | Bactericide (Difenoconazole, Mefenoxam, Benzovindiflupyr, Mancozeb, Azoxystrobin, Tebuconazole, Copper sulfate, Pehtahydrate) | Bacteria (Agrobacterium tumefaciens, Clavibacter, Erwinia amylovora, Xanthomonas campestris, Ralstonia solanacearum, Pseudomonas) | Milijašević-Marčić, 2018; Scala et al., 2018 |
| 7 | Fungicide (Monozeb, Methasulfocarb, Prothiocarb, Quinacetol, Sulfuryl fluoride, Trichlamide, Zineb) | Fungi (Fusarium, Rhizoctonia, Pythium, Phytophthora, Trichoderma, Aspergillus, Penicillium) | Aladdin et al., 2018; Delaney et al., 2018; Halo et al., 2018 |
| 8 | Algaecide (Diuron, Copper sulfate, Benzalkonium chloride, Cybutryne, Bethoxazin, Dichlone, Endothal, Fentin) | Algae (Microcystis, Cyanobacteria, Cephaleuros virescens) | Bishop et al., 2017; Crafton et al., 2018; Vasconcelos et al., 2018; Wonglom et al., 2018 |
Table 2.
Biodegradation databases and their significance.
| S. no. | Biodegradation databases | Link | Significance in pesticide bioremediation | Reference |
|---|---|---|---|---|
| 1 | University of Minnesota Biocatalysis/Biodegradation Database (UMBBD) | https://www.msi.umn.edu/content/university-minnesota-biocatalysis-and-biodegradation-database | Give information about molecular mechanisms involved in biodegradation pathways and tells about biotransformation rules, enzymes, genes, and reactions involved in microbial degradation of xeno pesticidal compounds | Arora and Bae, 2014; Malla et al., 2018 |
| 2 | Biodegradation Network- Molecular Biology Database (Bionemo) | http://bionemo.bioinfo.cnio.es | Tells about dynamic regulation of metabolic pathways and transcription factors in degradation pathways | Arora and Bae, 2014; Koch et al., 2018 |
| 3 | Oxygenase Database (OxDBase) | http://crdd.osdd.net/raghava/oxdbase/ | Give information regarding oxygenases, i.e., aromatic ring-hydroxylating dioxygenases (ARHD) and aromatic ring cleavage dioxygenases (ARCD) involved in breaking down pesticidal compounds | Arora and Bae, 2014; Wołejko et al., 2016 |
| 4 | Pathway/Genome Databases (BioCyc) | https://biocyc.org/ | Enable access to information related to biochemistry and genetics of microbial degradation | Arora and Bae, 2014 |
| 5 | Metabolic Pathway Database (MetaCyc) | https://metacyc.org/ | Predict metabolic pathways and reconstruction of catabolic pathways | Millacura et al., 2017; Ali et al., 2018 |
| 6 | Pesticide Target Interaction Database (PTID) | http://lilab.ecust.edu.cn/ptid | Interaction of pesticides with their target | Ning et al., 2018 |
| 7 | Microbial Genome Database (MBGD) | http://mbgd.genome.ad.jp | Comparative analysis of microbial genome | Fory et al., 2014; Uchiyama, 2017 |
| 8 | Metarouter | http://pdg.cnb.uam.es/MetaRouter | Maintain diverse information related to biodegradation | Paliwal et al., 2012; Kumavath and Deverapalli, 2013 |
| 9 | Pesticide Action Network (PAN) | http://pesticideinfo.org/Index.html | Give informative data on the toxicity of pesticides | Echeverría-Sáenz et al., 2018; Zhu J. et al., 2018 |
| 10 | The Environmental Contaminant Biotransformation Pathway (EAWAGBBD/PPS) | https://envipath.org/ | Give informative from bulk data of multi-omics approaches | Malla et al., 2018 |
Computational Tools
Upward elevation of scientific technologies and system biology approaches (Figure 2) represent tools to investigate the interaction of microbe with chemical compounds and their application for bioremediation (Basu et al., 2018). The integrative approach of various computational methods can be applied for the betterment of the bioremediation process to improve soil health (De Sousa et al., 2018). These in silico approaches are helpful in the construction of contemporary enzyme based mechanisms for bioremediation (Malla et al., 2018). Computational biology is in silico approach for genes and proteins study and dealing with cell system (Purohit et al., 2018). It is feasible to perceive complex metabolic pathways of biodegradation and bioremediation by computational techniques (Liu et al., 2018). In silico metabolic engineering of microbes has been done in the various field of microbiology related to agriculture, medical as well as industrial (Babar et al., 2018). There are many in silico tools accessible which are used by users for data mining and understanding the metabolic pathways of a cellular metabolic network applied to foster cellular processes, i.e., biodegradation and bioremediation (Ostrem Loss and Yu, 2018; Ravikrishnan et al., 2018; Zhang S. et al., 2018). Flux balance analysis (FBA), metabolic flux analysis (MFA), and metabolic pathway analysis (MPA) are most widely used tools for stoichiometric analysis of metabolic networks (Zhang and Xiu, 2009; Gonzalez-Garcia et al., 2017). Flux can be described as the flow of material with the edges carrying a value (Gomez and Barton, 2018). Knowing flux and organizing it led to alter the biological process dynamics by metabolic engineering (Cuperlovic-Culf, 2018). The consumption of pesticide compounds can also be enhanced, and properties of degrading bacteria can also be manipulated (Sulpice and McKeown, 2015). The reconstruction of quantitative structure-activity relationship (QSAR) and 3DQSAR (Dreher et al., 2018) of chemical atoms created models to assume and predict the interactions of pesticidal compounds with bioremediating and degrading microbes at the molecular level. For instance, QSAR and 3DQSAR is applied to study the toxicity level of xeno-pesticidal compounds at different environmental conditions, i.e., marine ecosystem, terrestrial ecosystem and accumulation or biomagnification of pesticides (DDT) in the food web. 3DQSAR also aids the molecular level study of atomic interactions with different atoms, ligands, and compounds. OptKnock is another interesting computational tool for not only to get gene knock outs but also to get incorporate the genes encoding the novel enzymes for bioremediation as well. A snapshot of integrated approaches of systems biology tools in biodegradation network is depicted in Figure 3. This gene mining from a diversity of microbial strains would be done for integration in a particular and specific microbial GEM (Genome Scale Model) is possible by OptStrain. Moreover, OptReg allows in silico regulation and manipulation (positive or negative) of metabolic pathways and enzymes involved in the pathway for functional pesticidal bioremediation efficiency. The above mentioned computational tools help to account for the understanding of the vast amount of genome scale models, their interacting genes and genomic data.
FIGURE 2.
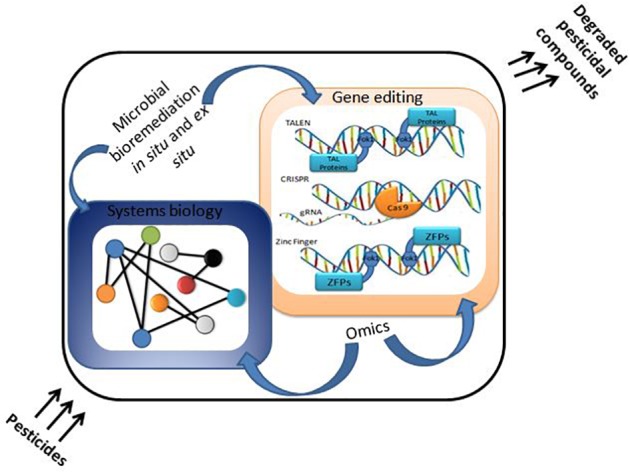
Advancement for pesticide bioremediation through gene editing tools and systems biology.
FIGURE 3.
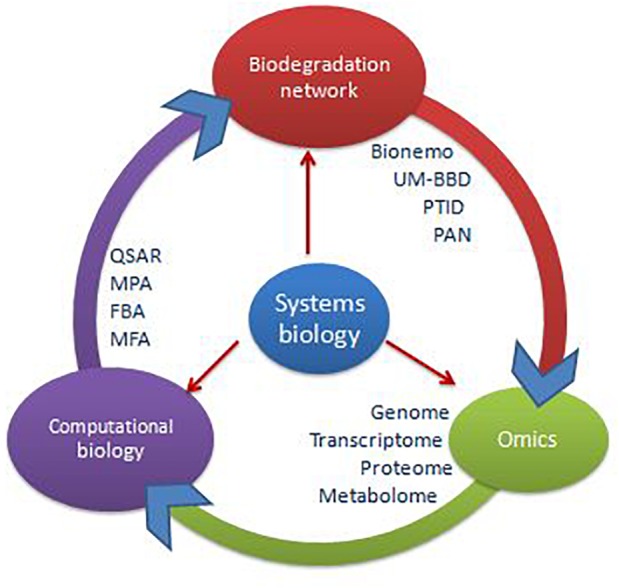
Integrated approaches of systems biology tools in biodegradation network.
The Multi Omics Approach
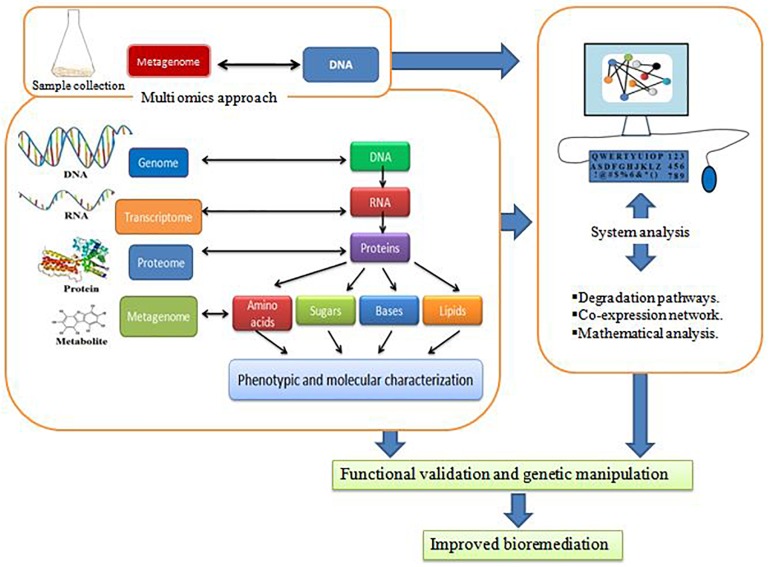
With the revolution of computer applications to every possible biological study, it is possible to study the interactions of genes encoding proteins within a cellular model or model organisms via multi omics approach. Thus, it makes possible to study metabolic pathways of biodegrading microorganisms. Genomics deals with the study of DNA for various molecular genetics approaches. These traditional approaches are also applied in the field of bioremediation. Table 3 shows the genomics tools for studying bioremediation of different contaminants in the environment. Environmental scientists consider the metagenomics as the ladder for stepping up in the field of bioremediation (Guerra et al., 2018; Jeffries et al., 2018; Plewniak et al., 2018; Roy et al., 2018). They mentioned the metagenomic appeal for finding the microbial potential degrading heavy metals, oil, petroleum, and other hydrocarbons (Bacosa et al., 2018; Guerra et al., 2018; Logeshwaran et al., 2018; Napp et al., 2018; Ramadass et al., 2018). Metagenomics is the direct analysis of genome (Zolfo et al., 2018). The microbial DNA is extracted directly from the soil sample (Gupta et al., 2018; Jeffries et al., 2018). This repute of metagenomics is the major advantage enabling the DNA analysis of non-culturable microbes present in the sample, i.e., culture independent approach (Perito and Cavalieri, 2018). The DNA thus obtained is sequenced and analyzed for expression (De Sousa et al., 2018). This appeal is now becoming an extension of all life sciences research (Tekle et al., 2018). Metaproteomics is the protein study derived from environmental samples (Biswas and Sarkar, 2018; Bhatt, 2018). Recent reports highlighted metaproteomics approach to observe the bacterial adaptation tactics in various contaminated sites, i.e., heavy metals, oil, xenobiotics, POPs, and other pollutants (Covino et al., 2016; Gianfreda and Rao, 2017; Das and Osborne, 2018). Besides this, the effect of these contaminants on bacterial communities can also be revealed (Mason et al., 2014). Gillan et al. (2015) observed that heavy metal contaminated sites harbouring bacterial community have diversity at the genetic level in terms of release of exopolymers and enzymes. The fast evolution of gene sequencing technology, i.e., HTS has mentioned a vast number of microbes with biodegradation potential. Also, it is adopted with metagenomics to screen particular metagenome of interest, i.e., detection of interacting bacterial species in a community (Gao et al., 2018; Mesuere et al., 2018). Fang et al., 2014 studied biodegradation pathways of persistent pesticides present in ecosystems of marine and freshwater sediments. They did analysis of metagenomic DNA for determining the BDGs involved in degradation of three pesticides namely DDT (dichlorodiphenyltrichloroethane), HCH (hexachlorocyclohexane), and ATZ (atrazine) and created data of 3 giga base pairs. They generated clean data after reducing the garbage data after HTS. Then clean data was analyzed. They get the functional gene annotates of DDT degradation (rrat, cpo, dhc, sds, dcl, ods, dhg, hdl, doa, rdh, hdt, dhc, and ort), HCH degradation (ccd, dog, dcn, rdg, hdg, cbd, rdt, mog, dhg, and dhc) and atrazine degradation (atza, atzB, atzC, atzD, thc, apobec, triA, trzA, and trzB). The genome annotation now allowed the identification of functional genes involved in bioremediation and biodegradation. Furthermore, it also enhances the description of existing metabolic pathways for consumption of pesticidal compounds as substrate or metabolites. There are several bacterial strains known for pesticide bioremediation whose whole genome has been sequenced, i.e., Pseudomonas putida and Rhodococcus sp. Transcriptomics, proteomics and metabolomics study (Table 4) data make us to understand the genotype and phenotype of particular biodegrading microbes. This prediction aids in determining genome scale model (GEM). This model would give the best microorganisms for bioremediation of pesticides and other xenobiotics. These GEMs have created possibilities to utilize the bacterial species with bioremediation potential, i.e., Pseudomonas putida KT2440 with functional genes (mpd, opd, vgb, gfp, pnpA, linC, pnpB, linB, linD, and linA, etc.) (Gong et al., 2016) incorporated for achieving the greater rate of bioremediation at different conditions of pH, temperature, and even at different ecosystems. The above mentioned omics constitutes the multi omics approach (Figure 4) whose action is possible via system analysis. The system analysis would give output to researchers as functional validation and genetic manipulation for improved and efficient bioremediation of contaminants.
Table 3.
Genomics tools for studying bioremediation of different contaminants.
| S. no. | Tool | Purpose | Microorganisms involved | Contaminant | Reference |
|---|---|---|---|---|---|
| 1 | Cloning and sequencing of ribosomal DNA | Identification of #BGD genes in community members of contaminated sites | Stenotrophomonas maltophilia | Pesticides, Heavy metals, Acid mine drainage | Raman et al., 2018; Simfukwe and Tindwa, 2018; Shukla et al., 2018 |
| 2 | Second generation sequencing | Identification of community members having #BGD genes | Cycloclasticus, Pseudomonas, Halomonas, Pseudoalteromonas, Marinomonas, Bacillus, Dietzia, Colwellia, Acinetobacter, Alcanivorax, Salinisphaera, and Shewanella | Polycyclic aromatic hydrocarbons (PAHs) | Dong et al., 2015; Lozada and Dionisi, 2016 |
| 3 | Quantitative PCR (polymerase chain reaction), RT-qPCR (real time quantitative PCR) | Quantification of #BGD genes and their expression | Pseudomonas and Rhodococcus | Diesel | Yergeau et al., 2012; Denaro et al., 2014 |
| 4 | RFLP (restriction fragment length polymorphism), fingerprinting methods | Bacterial communities involved in biodegradation of persistent compounds | Thermoanaerobacteraceae, Desulfobulbaceae | Naphthalene | Marozava et al., 2018 |
| 5 | FISH (fluorescent in situ hybridization) | In situ identification of metabolites involved in bioremediation | Dehalococcoides | Chlorinated solvents | Matturro et al., 2012 |
| 6 | SIP (stable isotope probing) | Uptake of labeled compounds as substrate under defined conditions | Rhodoplanes, Kaistobacter, Pseudomonas, Flavobacterium, Mycobacterium | Naphthenic acids, phenanthrene, and atrazine | Fenner et al., 2013; Ahad et al., 2018; Lin et al., 2018; Subashchandrabose et al., 2018 |
#BGD, biodegradation.
Table 4.
The multi omics applied for bioremediation study.
| S.no. | Omics approach | Center of study | Method | Marker | Application | Microorganism involved | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Genomics | Genomic study | DNA sequencing | Gene promoters | Polycyclic aromatic hydrocarbons (PAHs), organophosphate, para-nitrophenol, and phenanthrene compounds of pesticides | Mycobacterium, Rhodococcus wratislaviensis strain 9 | Birolli et al., 2018; Li J. et al., 2018; Subashchandrabose et al., 2018 |
| 2 | Metagenomics | Genetic study of sample | Sequencing and pyrosequencing | 16S rDNA | Oil, xenobiotics, and heavy metals | Marinobacterium, Marinobacter, Cycloclasticus, Sphingomonas, Candidatus Solibacter, Flexibacter, Arthrobacter sp., Pseudomonas putida, Alcaligenes eutrophus, Dehalospirilum multivorans | Desai et al., 2010; Dos Santos et al., 2011; Gołêbiewski et al., 2014 |
| 3 | Metabolomics | Metabolites study of cellular reactions | HPLC, GC–MS | Metabolites | Insecticides, i.e., diazinon, malathion, chlorpyrifos, permethrin, cyfluthrin, cypermethrin, deltamethrin, and pyrethroids | Streptomyces aureus strain HP-S-01, Bacillus megaterium JCm2, Sphingobium sp. JQL4-5, Aspergillus niger, Aspergillus terricola, and Candida pelliculosa strain ZS-02 | Cycoń and Piotrowska-Seget, 2016; Radford et al., 2018 |
| 4 | Proteomics | Study of proteins and their application | X-ray crystallography | Protein, peptides, and oligopeptides | Organophosphorus insecticides | Aspergillus, Pseudomonas, Chlorella, and Arthrobacter | Kumar S. et al., 2018; Li X. et al., 2018 |
| 5 | Transcriptomics | Study of transcripts and their function | RNA sequencing, Q- and RT-PCR | miRNA, siRNA, and RNAi transcripts | Organophosphates, pyrethroids, and carbamates | Pseudomonas putida KT2440, Sphingobium sp. strain 1017-1 | Gong et al., 2018; Yan et al., 2018 |
FIGURE 4.
Multi omics approach for improved bioremediation.
Conclusion and Future Perspectives
Dissimal and removal of persistent pesticides by gene editing tools and systems biology have come forth as the outstanding option. Many bioremediation approaches are present to solve the difficulties in the field of bioremediating recalcitrant pollutants from the environment. The environment is constantly being harmed by the continuous use of synthetic pesticides (xeno-pesticidal compounds). These synthetic pesticides are organic and inorganic compounds whose remediation processes vary from one another thus it is obvious that individual or single bioremediation pathway is not enough. Therefore, empathizing on metabolic pathways for gene editing and application of systems biology is very important. This will undertake the existing metabolic pathways towards the increased and efficient microbial remediation of pesticides. Acceptable improvements have been witnessed for bioremediation of pesticides by applying gene tools. Furthermore, genomics, metagenomics, metabolomics, transcriptomics, proteomics, and biodegradation network pave the path of pesticide bioremediation. TALEN, ZFNs, and CRISPR Cas9 are auspicious gene editing tools to get the function specific microorganisms with particular genes and enzymes responsible for pesticide bioremediation. The multi omics approach also contributed to the logical identification of microbial host having degradation strength.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Maharshi Dayanand University, Rohtak, India for providing infrastructure facility. PS acknowledges the infrastructural support from Department of Science and Technology, New Delhi, Government of India, FIST grant (Grant No. 1196 SR/FST/LS-I/2017/4) and Department of Biotechnology, Government of India (Grant No. BT/PR27437/BCE/8/1433/2018). PS acknowledges Department of Microbiology, Barkatullah University, Bhopal, India for their infrastructural support for D.Sc. work.
Footnotes
References
- Ahad J. M., Pakdel H., Gammon P. R., Siddique T., Kuznetsova A., Savard M. M. (2018). Evaluating in situ biodegradation of 13 C-labelled naphthenic acids in groundwater near oil sands tailings ponds. Sci. Total Environ. 643 392–399. 10.1016/j.scitotenv.2018.06.159 [DOI] [PubMed] [Google Scholar]
- Aladdin A., Dib J. R., Malek R. A., El Enshasy H. A. (2018). “Killer yeast, a novel biological control of soilborne diseases for good agriculture practice,” in Sustainable Technologies for the Management of Agricultural Wastes, (Singapore: Springer; ), 71–86. 10.1007/978-981-10-5062-6_6 [DOI] [Google Scholar]
- Ali M., Ali S., Ishrat R. (2018). “In silico biochemical pathways for bacterial metabolite synthesis,” in In Silico Approach for Sustainable Agriculture, (Singapore: Springer; ), 239–250. 10.1007/978-981-13-0347-0_14 [DOI] [Google Scholar]
- Allmaras R. R., Wilkins D. E., Burnside O. C., Mulla D. J. (2018). “Agricultural technology and adoption of conservation practices,” in Advances in soil and water conservation, (Abingdon: Routledge; ), 99–158. 10.1201/9781315136912-6 [DOI] [Google Scholar]
- Arazoe T., Kondo A., Nishida K. (2018). Targeted nucleotide editing technologies for microbial metabolic engineering. Biotechnol. J. 13:e1700596. 10.1002/biot.201700596 [DOI] [PubMed] [Google Scholar]
- Arora P. K., Bae H. (2014). Integration of bioinformatics to biodegradation. Biol. Proc. Online 16:8. 10.1186/1480-9222-16-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P. K., Kumar M., Chauhan A., Raghava G. P., Jain R. K. (2009). OxDBase: a database of oxygenases involved in biodegradation. BMC Res. Notes 2:67. 10.1186/1756-0500-2-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar M. M., Afzaal H., Pothineni V. R., Najam-us-Sahar S. Z., Ali Z., Zahid M. A., et al. (2018). “Omics approaches in industrial biotechnology and bioprocess engineering,” in Omics Technologies and Bio-Engineering, eds Barh D., Azevedo V. (Cambridge, MA: Academic Press; ), 251–269. 10.1016/B978-0-12-815870-8.00014-0 [DOI] [Google Scholar]
- Bacosa H. P., Erdner D. L., Rosenheim B. E., Shetty P., Seitz K. W., Baker B. J., et al. (2018). Hydrocarbon degradation and response of seafloor sediment bacterial community in the northern Gulf of Mexico to light Louisiana sweet crude oil. ISME J. 12 2532–2543. 10.1038/s41396-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakirhan N. K., Uslu B., Ozkan S. A. (2018). “The detection of pesticide in foods using electrochemical sensors,” in Food Safety and Preservation, eds Grumezescu A., Holban A.-M. (Cambridge, MA: Academic Press; ), 91–141. [Google Scholar]
- Banerjee A., Banerjee C., Negi S., Chang J.-S., Shukla P. (2018). Improvements in algal lipid production: a systems biology and gene editing approach. Crit. Rev. Biotechnol. 38 369–385. 10.1080/07388551.2017.1356803 [DOI] [PubMed] [Google Scholar]
- Basu S., Rabara R. C., Negi S., Shukla P. (2018). Engineering PGPMOs through gene editing and systems biology: a solution for phytoremediation? Trends Biotechnol. 36 499–510. 10.1016/j.tibtech.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Bedi J. S., Gill J. P. S., Kaur P., Aulakh R. S. (2018). Pesticide residues in milk and their relationship with pesticide contamination of feedstuffs supplied to dairy cattle in Punjab (India). J. Anim. Feed Sci. 27 18–25. 10.22358/jafs/82623/2018 [DOI] [Google Scholar]
- Behler J., Sharma K., Reimann V., Wilde A., Urlaub H., Hess W. R. (2018). The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR–Cas subtype III-Bv system. Nat. Microbiol. 3:367. 10.1038/s41564-017-0103-5 [DOI] [PubMed] [Google Scholar]
- Bhatt P. (2018). “Insilico tools to study the bioremediation in microorganisms,” in Handbook of Research on Microbial Tools for Environmental Waste Management, (Harrisburg, PA: IGI Global; ), 389–395. 10.4018/978-1-5225-3540-9.ch018 [DOI] [Google Scholar]
- Bier E., Harrison M. M., O’Connor-Giles K. M., Wildonger J. (2018). Advances in engineering the fly genome with the CRISPR-Cas system. Genetics 208 1–18. 10.1534/genetics.117.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birolli W. G., Santos D. D. A., Alvarenga N., Garcia A. C., Romão L. P., Porto A. L. (2018). Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Mar. Pollut. Bull. 129 525–533. 10.1016/j.marpolbul.2017.10.023 [DOI] [PubMed] [Google Scholar]
- Bishop W. M., Lynch C. L., Willis B. E., Cope W. G. (2017). Copper-based aquatic algaecide adsorption and accumulation kinetics: influence of exposure concentration and duration for controlling the cyanobacterium Lyngbya wollei. Bull. Environ. Contamin. Toxicol. 99 365–371. 10.1007/s00128-017-2134-2 [DOI] [PubMed] [Google Scholar]
- Biswas R., Sarkar A. (2018). “‘Omics’ tools in soil microbiology: the state of the art,” in Advances in Soil Microbiology: Recent Trends and Future Prospects, (Singapore: Springer; ), 35–64. 10.1007/978-981-10-6178-3_3 [DOI] [Google Scholar]
- Borja A. (2018). Testing the efficiency of a bacterial community-based index (microgAMBI) to assess distinct impact sources in six locations around the world. Ecol. Indicat. 85 594–602. 10.1016/j.ecolind.2017.11.018 [DOI] [Google Scholar]
- Bottrell D. G., Schoenly K. G. (2018). Integrated pest management for resource-limited farmers: challenges for achieving ecological, social and economic sustainability. J. Agric. Sci. 156 1–19. 10.1017/S0021859618000473 [DOI] [Google Scholar]
- Boudh S., Singh J. S. (2019). “Pesticide contamination: environmental problems and remediation strategies,” in Emerging and Eco-Friendly Approaches for Waste Management, eds Bharagava R. M., Chowdhary P. (Singapore: Springer; ), 245–269. [Google Scholar]
- Butt H., Jamil M., Wang J. Y., Al-Babili S., Mahfouz M. M. (2018). Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 18:174. 10.1186/s12870-018-1387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver M. C., Joung J. K., Pinello L. (2018). Impact of genetic variation on CRISPR-Cas targeting. CRISPR J. 1 159–170. 10.1089/crispr.2017.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalis V., Loreau M., Henderson K. (2018). Do we have to choose between feeding the human population and conserving nature? Modelling the global dependence of people on ecosystem services. Sci. Total Environ. 634 1463–1474. 10.1016/j.scitotenv.2018.03.360 [DOI] [PubMed] [Google Scholar]
- Cooper L. A., Stringer A. M., Wade J. T. (2018). Determining the specificity of cascade binding, interference, and primed adaptation In Vivo in the Escherichia coli type IE CRISPR-cas system. mBio 9:e 02100-17. 10.1128/mBio.02100-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covino S., Stella T., Cajthaml T. (2016). “Mycoremediation of organic pollutants: principles, opportunities, and pitfalls,” in Fungal Applications in Sustainable Environmental Biotechnology, ed. Diane P. (Cham: Springer; ), 185–231. 10.1007/978-3-319-42852-9_8 [DOI] [Google Scholar]
- Crafton E. A., Glowczewski J., Ott D. W., Cutright T. J. (2018). In situ field trial to evaluate the efficacy of Cutrine Ultra to manage a cyanobacteria population in a drinking water source. Environ. Sci. 4 863–871. 10.1039/C8EW00124C [DOI] [Google Scholar]
- Cuperlovic-Culf M. (2018). Machine learning methods for analysis of metabolic data and metabolic pathway modeling. Metabolites 8:4. 10.3390/metabo8010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cycoń M., Piotrowska-Seget Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front. Microbiol. 7:1463. 10.3389/fmicb.2016.01463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Zhang S., Yang Q., Zhang W., Qian X., Dong W., et al. (2018). Genetic tool development and systemic regulation in biosynthetic technology. Biotechnol. Biofuels 11:152. 10.1186/s13068-018-1153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi A. K., Sharma B., Hill R. T., Shukla P. (2018). Bioremediation through microbes: systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 39 79–98. 10.1080/07388551.2018.1500997 [DOI] [PubMed] [Google Scholar]
- Das A., Osborne J. W. (2018). “Bioremediation of Heavy Metals,” in Nanotechnology, Food Security and Water Treatment, eds Gothandam K. M., Ranjan S., Dasgupta N., Ramalingam C., Lichtfouse E. (Cham: Springer; ), 277–311. 10.1007/978-3-319-70166-0_9 [DOI] [Google Scholar]
- Das S., Dash H. R., Chakraborty J. (2016). Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 100 2967–2984. 10.1007/s00253-016-7364-4 [DOI] [PubMed] [Google Scholar]
- De Sousa C. S., Hassan S. S., Pinto A. C., Silva W. M., De Almeida S. S., Soares S. D. C., et al. (2018). “Microbial omics: applications in biotechnology,” in Omics Technologies and Bio-Engineering, eds Barh D., Azevedo V. (Cambridge, MA: Academic Press; ), 3–20. 10.1016/B978-0-12-815870-8.00001-2 [DOI] [Google Scholar]
- Delaney M., ArchMiller A. A., Delaney D. P., Wilson A. E., Sikora E. J. (2018). Effectiveness of fungicide on soybean rust in the southeastern united states: a meta-analysis. Sustainability 10 1–15. 10.3390/su1006178430607262 [DOI] [Google Scholar]
- Denaro R., Crisafi F., Russo D., Genovese M., Messina E., Genovese L., et al. (2014). Alcanivorax borkumensis produces an extracellular siderophore in iron-limitation condition maintaining the hydrocarbon-degradation efficiency. Mar. Genomics 17 43–52. 10.1016/j.margen.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Desai C., Pathak H., Madamwar D. (2010). Advances in molecular and “-omics” technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresource Technol. 101 1558–1569. 10.1016/j.biortech.2009.10.080 [DOI] [PubMed] [Google Scholar]
- Dong C., Bai X., Sheng H., Jiao L., Zhou H., Shao Z. (2015). Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences 12 2163–2177. 10.5194/bg-12-2163-2015 [DOI] [Google Scholar]
- Dos Santos H. F., Cury J. C., Do Carmo F. L., Dos Santos A. L., Tiedje J., Van Elsas J. D., et al. (2011). Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS One 6:e16943. 10.1371/journal.pone.0016943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drangert J. O., Tonderski K., McConville J. (2018). Extending the European Union Waste Hierarchy to Guide Nutrient-Effective Urban Sanitation toward Global Food Security—Opportunities for Phosphorus Recovery. Front. Sustain. Food Syst. 2:3 10.3389/fsufs.2018.00003 [DOI] [Google Scholar]
- Dreher J., Scheiber J., Stiefl N., Baumann K. (2018). xMaP? an interpretable alignment-free four-dimensional quantitative structure–activity relationship technique based on molecular surface properties and conformer Ensembles. J. Chem. Inform. Model. 58 165–181. 10.1021/acs.jcim.7b00419 [DOI] [PubMed] [Google Scholar]
- Duke S. O. (2018). Interaction of chemical pesticides and their formulation ingredients with microbes associated with plants and plant pests. J. Agric. Food Chem. 66 7553–7561. 10.1021/acs.jafc.8b02316 [DOI] [PubMed] [Google Scholar]
- Dvořák P., Nikel P. I., Damborský J., de Lorenzo V. (2017). Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35 845–866. 10.1016/j.biotechadv.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Echeverría-Sáenz S., Mena F., Arias-Andrés M., Vargas S., Ruepert C., Van den Brink P. J., et al. (2018). In situ toxicity and ecological risk assessment of agro-pesticide runoff in the Madre de Dios River in Costa Rica. Environ. Sci. Pollut. Res. 25 13270–13282. 10.1007/s11356-016-7817-4 [DOI] [PubMed] [Google Scholar]
- Ellis L. B., Roe D., Wackett L. P. (2006). The University of Minnesota biocatalysis/biodegradation database: the first decade. Nucleic Acids Res. 34(Suppl. 1), D517–D521. 10.1093/nar/gkj076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erythropel H. C., Zimmerman J. B., de Winter T. M., Petitjean L., Melnikov F., Lam C. H., et al. (2018). The green chemisTREE: 20 years after taking root with the 12 principles. Green Chem. 20 1929–1961. 10.1039/C8GC00482J [DOI] [Google Scholar]
- Fang H., Cai L., Yang Y., Ju F., Li X., Yu Y., et al. (2014). Metagenomic analysis reveals potential biodegradation pathways of persistent pesticides in freshwater and marine sediments. Sci. Total Environ. 470 983–992. 10.1016/j.scitotenv.2013.10.076 [DOI] [PubMed] [Google Scholar]
- Fatima S. A., Hamid A., Yaqub G., Javed A., Akram H. (2018). Detection of volatile organic compounds in blood of farmers and their general health and safety profile. Nat. Environ. Pollut. Technol. 17 657–660. [Google Scholar]
- Fenner K., Canonica S., Wackett L. P., Elsner M. (2013). Evaluating pesticide degradation in the environment: blind spots and emerging opportunities. Science 341 752–758. 10.1126/science.1236281 [DOI] [PubMed] [Google Scholar]
- Fory P. A., Triplett L., Ballen C., Abello J. F., Duitama J., Aricapa M. G., et al. (2014). Comparative analysis of two emerging rice seed bacterial pathogens. Phytopathology 104 436–444. 10.1094/PHYTO-07-13-0186-R [DOI] [PubMed] [Google Scholar]
- Gao X., Huynh B.-T., Guillemot D., Glaser P., Opatowski L. (2018). Inference of significant microbial interactions from longitudinal metagenomics data. Front. Microbiol. 9:2319. 10.3389/fmicb.2018.02319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N., Narasimhulu K., Pydisetty Y. (2018). Recent advances in the bio-remediation of persistent organic pollutants and its effect on environment. J. Clean. Prod. 198 1602–1631. 10.1016/j.jclepro.2018.07.076 [DOI] [Google Scholar]
- Gianfreda L., Rao M. A. (2017). “Soil microbial and enzymatic diversity as affected by the presence of xenobiotics,” in Xenobiotics in the Soil Environment, eds Zaffar H. M., Vivek K., Ajit V. (Cham: Springer; ),153–169. [Google Scholar]
- Gillan D. C., Roosa S., Kunath B., Billon G., Wattiez R. (2015). The long-term adaptation of bacterial communities in metal-contaminated sediments: a metaproteogenomic study. Environ. Microbiol. 17 1991–2005. 10.1111/1462-2920.12627 [DOI] [PubMed] [Google Scholar]
- Gołêbiewski M., Deja-Sikora E., Cichosz M., Tretyn A., Wróbel B. (2014). 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microbial Ecol. 67 635–647. 10.1007/s00248-013-0344-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. A., Barton P. I. (2018). Dynamic flux balance analysis using DFBAlab. Methods Mol. Biol. 1716 353–370. 10.1007/978-1-4939-7528-0_16 [DOI] [PubMed] [Google Scholar]
- Gong J., Liu X., Cao X., Diao Y., Gao D., Li H., et al. (2012). PTID: an integrated web resource and computational tool for agrochemical discovery. Bioinformatics 29 292–294. 10.1093/bioinformatics/bts651 [DOI] [PubMed] [Google Scholar]
- Gong T., Liu R., Zuo Z., Che Y., Yu H., Song C., et al. (2016). Metabolic engineering of Pseudomonas putida KT2440 for complete mineralization of methyl parathion and γ-hexachlorocyclohexane. ACS Synth. Biol. 5 434–442. 10.1021/acssynbio.6b00025 [DOI] [PubMed] [Google Scholar]
- Gong T., Xu X., Dang Y., Kong A., Wu Y., Liang P., et al. (2018). An engineered Pseudomonas putida can simultaneously degrade organophosphates, pyrethroids and carbamates. Sci. Total Environ. 628 1258–1265. 10.1016/j.scitotenv.2018.02.143 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia R. A., Aispuro-Castro R., Salgado-Manjarrez E., Aranda-Barradas J., Garcia-Pena E. I. (2017). Metabolic pathway and flux analysis of H2 production by an anaerobic mixed culture. Int. J. Hydrogen Energy 42 4069–4082. 10.1186/1475-2859-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene A. C. (2018). CRISPR-Based Antibacterials: transforming Bacterial Defense into Offense. Trends Biotechnol. 36 127–130. 10.1016/j.tibtech.2017.10.021 [DOI] [PubMed] [Google Scholar]
- Guengerich F. P., Yoshimoto F. K. (2018). Formation and cleavage of C–C bonds by enzymatic oxidation–reduction reactions. Chem. Rev. 118 6573–6655. 10.1021/acs.chemrev.8b00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra A. B., Oliveira J. S., Silva-Portela R. C., Araujo W., Carlos A. C., Vasconcelos A. T. R., et al. (2018). Metagenome enrichment approach used for selection of oil-degrading bacteria consortia for drill cutting residue bioremediation. Environ. Pollut. 235 869–880. 10.1016/j.envpol.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Guerra-García A., Barrales-Alcalá D., Argueta-Guzmán M., Cruz A., Mandujano M. C., Arévalo-Ramírez J. A., et al. (2018). Biomass allocation, plantlet survival, and chemical control of the invasive chandelier plant (Kalanchoe delagoensis)(Crassulaceae). Invasive Plant Sci. Manag. 11 33–39. 10.1017/inp.2018.6 [DOI] [Google Scholar]
- Gupta N., Vats S., Bhargava P. (2018). “Sustainable agriculture: role of metagenomics and metabolomics in exploring the soil microbiota,” in In Silico Approach for Sustainable Agriculture, eds Choudhary D. K., Kumar M., Prasad R., Kumar V. (Singapore: Springer; ), 183–199. [Google Scholar]
- Hall E., Bernhardt E. S., Bier R. L., Bradford M. A., Boot C. M., Cotner J. B., et al. (2018). Understanding how microbiomes influence the systems they inhabit. Understanding how microbiomes influence the systems they inhabit. Nat. Microbiol. 3 977–982. 10.1038/s41564-018-0201-z [DOI] [PubMed] [Google Scholar]
- Halo B. A., Al-Yahyai R. A., Al-Sadi A. M. (2018). Aspergillus terreus inhibits growth and induces morphological abnormalities in Pythium aphanidermatum and suppresses pythium-induced damping-off of cucumber. Front. Microbiol. 9:95. 10.3389/fmicb.2018.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamedi R., Bg Aghaie A., Hadjmohammadi M. R. (2018). Magnetic core micelles as a nanosorbent for the efficient removal and recovery of three organophosphorus pesticides from fruit juice and environmental water samples. J. Separat. Sci. 41 2037–2045. 10.1002/jssc.201701090 [DOI] [PubMed] [Google Scholar]
- Hogue M. M., Sanchez F. F., Benigno E. A. (2018). “Rodent problems in selected countries in Southeast Asia and islands in the Pacific,” in Rodent Pest Management, ed. Prakash I. (Boca Raton, FL: CRC Press; ), 95–110. [Google Scholar]
- Hussain I., Aleti G., Naidu R., Puschenreiter M., Mahmood Q., Rahman M. M., et al. (2018). Microbe and plant assisted-remediation of organic xenobiotics and its enhancement by genetically modified organisms and recombinant technology: a review. Sci. Total Environ. 628 1582–1599. 10.1016/j.scitotenv.2018.02.037 [DOI] [PubMed] [Google Scholar]
- Jeffries T. C., Rayu S., Nielsen U. N., Lai K., Ijaz A., Nazaries L., et al. (2018). Metagenomic functional potential predicts degradation rates of a model organophosphorus xenobiotic in pesticide contaminated soils. Front . Microbiol. 9:147. 10.3389/fmicb.2018.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John E. M., Varghese E. M., Krishnasree N., Jisha M. S. (2018). In situ bioremediation of Chlorpyrifos by Klebsiella sp. Isolated from pesticide contaminated agricultural soil. Int. J. Curr. Microbiol. App. Sci. 7 1418–1429. 10.20546/ijcmas.2018.703.170 [DOI] [Google Scholar]
- Ju X. D., Xu J., Sun Z. S. (2018). CRISPR editing in biological and biomedical investigation. J. Cell. Biochem. 119 52–61. 10.1002/jcb.26154 [DOI] [PubMed] [Google Scholar]
- Kaur S., Barua I. C., Kaur T., Kaur N., Kaul A., Bhullar M. S. (2018). Appearance of new weeds in Punjab. Indian J. Weed Sci. 50 59–63. 10.5958/0974-8164.2018.00013.8 [DOI] [Google Scholar]
- Koch M., Pandi A., Delépine B., Faulon J. L. (2018). A dataset of small molecules triggering transcriptional and translational cellular responses. Data Brief. 17 1374–1378. 10.1016/j.dib.2018.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Meldgin D. R., Collins J. J., Lu T. (2018). Designing microbial consortia with defined social interactions. Nat. Chem. Biol. 14 821–829. 10.1038/s41589-018-0091-7 [DOI] [PubMed] [Google Scholar]
- Korjus H. (2014). “Polluted soils restoration,” in Climate Change and Restoration of Degraded Land, eds Numben S. T., Almaraz R. A., Eswaran H. (Madrid: Colegio de Ingenieros de Montes; ), 411–480. [Google Scholar]
- Kumar D., Rai D., Porwal P., Kumar S. (2018). Compositional quality of milk and its contaminants on physical and chemical concern: a review. Int. J. Curr. Microbiol. App. Sci. 7 1125–1132. 10.20546/ijcmas.2018.705.137 [DOI] [Google Scholar]
- Kumar S., Kaushik G., Dar M. A., Nimesh S., Lopez-Chuken U. J., Villarreal-Chiu J. F. (2018). Microbial degradation of organophosphate pesticides: a review. Pedosphere 28 190–208. 10.1016/S1002-0160(18)60017-7 [DOI] [Google Scholar]
- Kumavath R. N., Deverapalli P. (2013). “Scientific swift in bioremediation: an overview,” in Applied Bioremediation-Active and Passive Approaches, ed. Patil Y. (London: IntechOpen Limited; ), 375–388. 10.5772/56409 [DOI] [Google Scholar]
- Leprêtre C., Merten-Lentz K. (2018). The forthcoming 12th meeting of the Codex Alimentarius Committee on Contaminants (and Toxins) in Foods (CCCF12) has a loaded agenda, including the adoption of new and revised maximum tolerable levels for contaminants and toxins, such as lead, cadmium, methyl mercury, total aflatoxins, and ochratoxin A in several foods. CCCF12 will also consider the adoption of two codes of practices concerning the reduction of (i) dioxins, PCB-like dioxins, and (ii) 3-MCPD and glycidyl esters in refined vegetable oils. World Food Regulat. Rev. 27 24–30. [Google Scholar]
- Li J., Luo C., Zhang D., Song M., Cai X., Jiang L., et al. (2018). Autochthonous bioaugmentation-modified bacterial diversity of phenanthrene degraders in PAH-contaminated wastewater as revealed by DNA-stable isotope probing. Environ. Sci. & Technol. 52 2934–2944. 10.1021/acs.est.7b05646 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang D., Liu Z., Xu Y., Wang D. (2018). Synthesis, characterization of a ternary Cu (II) Schiff base complex with degradation activity of organophosphorus pesticides. Inorgan. Chim. Acta 471 280–289. 10.1016/j.ica.2017.11.024 [DOI] [Google Scholar]
- Lin Z., Zhen Z., Ren L., Yang J., Luo C., Zhong L., et al. (2018). Effects of two ecological earthworm species on atrazine degradation performance and bacterial community structure in red soil. Chemosphere 196 467–475. 10.1016/j.chemosphere.2017.12.177 [DOI] [PubMed] [Google Scholar]
- Listgarten J., Weinstein M., Kleinstiver B. P., Sousa A. A., Joung J. K., Crawford J., et al. (2018). Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng. 2:38. 10.1038/s41551-017-0178-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liu Y., Zeng G., Shao B., Chen M., Li Z., et al. (2018). Application of molecular docking for the degradation of organic pollutants in the environmental remediation: a review. Chemosphere 203 139–150. 10.1016/j.chemosphere.2018.03.179 [DOI] [PubMed] [Google Scholar]
- Logeshwaran P., Megharaj M., Chadalavada S., Bowman M., Naidu R. (2018). Petroleum hydrocarbons (PH) in groundwater aquifers: an overview of environmental fate, toxicity, microbial degradation and risk-based remediation approaches. Environ. Technol. Innovat. 10 175–193. 10.1016/j.eti.2018.02.001 [DOI] [Google Scholar]
- Lozada M., Dionisi H. M. (2016). “Molecular biological tools for the assessment of hydrocarbon-degrading potential in coastal environments,” in Biology and Biotechnology of Patagonian Microorganisms, eds Nelda Lila O., Diego L., Edgardo D. (Cham: Springer; ), 15–29. [Google Scholar]
- Mahas A., Mahfouz M. (2018). Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virol. 32 1–8. 10.1016/j.coviro.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Majumdar S., Ligon M., Skinner W. C., Terns R. M., Terns M. P. (2017). Target DNA recognition and cleavage by a reconstituted Type IG CRISPR-Cas immune effector complex. Extremophiles 21 95–107. 10.1007/s00792-016-0871-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malla M. A., Dubey A., Yadav S., Kumar A., Hashem A., Abd_Allah E. F. (2018). Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 9:1132. 10.3389/fmicb.2018.01132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozava S., Mouttaki H., Müller H., Laban N. A., Probst A. J., Meckenstock R. U. (2018). Anaerobic degradation of 1-methylnaphthalene by a member of the Thermoanaerobacteraceae contained in an iron-reducing enrichment culture. Biodegradation 29 23–39. 10.1007/s10532-017-9811-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason O. U., Scott N. M., Gonzalez A., Robbins-Pianka A., Bælum J., Kimbrel J., et al. (2014). Metagenomics reveals sediment microbial community response to Deepwater Horizon oil spill. ISME J. 8:1464. 10.1038/ismej.2013.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matturro B., Aulenta F., Majone M., Papini M. P., Tandoi V., Rossetti S. (2012). Field distribution and activity of chlorinated solvents degrading bacteria by combining CARD-FISH and real time PCR. New Biotechnol. 30 23–32. 10.1016/j.nbt.2012.07.006 [DOI] [PubMed] [Google Scholar]
- McMahon M. A., Prakash T. P., Cleveland D. W., Bennett C. F., Rahdar M. (2018). Chemically modified Cpf1-CRISPR RNAs mediate efficient genome editing in mammalian cells. Mol. Ther. 26 1228–1240. 10.1016/j.ymthe.2018.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesuere B., Van der Jeugt F., Willems T., Naessens T., Devreese B., Martens L., et al. (2018). High-throughput metaproteomics data analysis with Unipept: a tutorial. J. Proteomics 171 11–22. 10.1016/j.jprot.2017.05.022 [DOI] [PubMed] [Google Scholar]
- Milijašević-Marčić S., Todorović V., Stanojević O., Berić T., Stanković S., Todorović B., et al. (2018). Antagonistic potential of Bacillus spp. isolates against bacterial pathogens of tomato and fungal pathogen of pepper. Pesticidi Fitomedicina 33 9–18. 10.2298/PIF1801009M [DOI] [Google Scholar]
- Millacura F. A., Cardenas F., Mendez V., Seeger M., Rojas L. A. (2017). Degradation of benzene by the heavy-metal resistant bacterium Cupriavidus metallidurans CH34 reveals its catabolic potential for aromatic compounds. bioRxiv [Preprint]. 10.1101/164517 [DOI] [Google Scholar]
- Mohamed M. (2018). Comparative study on the integrated application of environmental friendly compounds and a chemical-nematicide in controlling root knot nematode Meloidogyne incognita infecting sunflower plants; a field study. Agric. Eng. Int. 5 132–137. [Google Scholar]
- Montiel-Rozas M. M., Domínguez M. T., Madejón E., Madejón P., Pastorelli R., Renella G. (2018). Long-term effects of organic amendments on bacterial and fungal communities in a degraded Mediterranean soil. Geoderma 332 20–28. 10.1016/j.geoderma.2018.06.022 [DOI] [Google Scholar]
- Moorman T. B. (2018). “Pesticide degradation by soil microorganisms: environmental, ecological, and management effects,” in Soil Biology, ed. Hatfield J. L. (Boca Raton, FL: CRC Press; ), 127–172. [Google Scholar]
- Muangchinda C., Rungsihiranrut A., Prombutara P., Soonglerdsongpha S., Pinyakong O. (2018). 16S metagenomic analysis reveals adaptability of a mixed-PAH-degrading consortium isolated from crude oil-contaminated seawater to changing environmental conditions. J. Hazard. Mater. 357 119–127. 10.1016/j.jhazmat.2018.05.062 [DOI] [PubMed] [Google Scholar]
- Mushtaq S. (2018). Smart agriculture system based on iot and image processing. Int. J. Adv. Res. Comput. Sci. 9. 10.26483/ijarcs.v9i1.5278 [DOI] [Google Scholar]
- Napp A. P., Pereira J. E. S., Oliveira J. S., Silva-Portela R. C., Agnez-Silva L. F., Peralba M. C., et al. (2018). Comparative metagenomics reveals different hydrocarbon degradative abilities from enriched oil-drilling waste. Chemosphere 209 7–16. 10.1016/j.chemosphere.2018.06.068 [DOI] [PubMed] [Google Scholar]
- Nikolopoulou M., Kalogerakis N. (2018). “Biostimulation strategies for enhanced bioremediation of marine oil spills including chronic pollution,” in Consequences of Microbial Interactions with Hydrocarbons, Oils, and Lipids: Biodegradation and Bioremediation, ed. Steffan R. (Berlin: Springer; ), 1–10. [Google Scholar]
- Ning X., Ku T., Gao R., Ji X., Li G., Sang N. (2018). In vitro PPARγ agonistic potential of chitin synthesis inhibitors and their energy metabolism-related hepatotoxicity. Sci. Total Environ. 615 1126–1132. 10.1016/j.scitotenv.2017.10.016 [DOI] [PubMed] [Google Scholar]
- Nolte T. M., Pinto-Gil K., Hendriks A. J., Ragas A. M., Pastor M. (2018). Quantitative structure–activity relationships for primary aerobic biodegradation of organic chemicals in pristine surface waters: starting points for predicting biodegradation under acclimatization. Environ. Sci. 20 157–170. 10.1039/c7em00375g [DOI] [PubMed] [Google Scholar]
- Oladipo A. A., Vaziri R., Abureesh M. A. (2018). Highly robust AgIO3/MIL-53 (Fe) nanohybrid composites for degradation of organophosphorus pesticides in single and binary systems: application of artificial neural networks modelling. J. Taiwan Ins. Chem. Eng. 83 133–142. 10.1016/j.jtice.2017.12.013 [DOI] [Google Scholar]
- Ortiz-Hernández M. L., Castrejón-Godínez M. L., Popoca-Ursino E. C., Cervantes-Dacasac F. R., et al. (2018). “Strategies for biodegradation and bioremediation of pesticides in the environment,” in Strategies for Bioremediation of Organic and Inorganic Pollutants, eds Fuentes M. S., Colin V. L., Saez J. M. (Boca Raton, FL: CRC Press; ), 95–115. [Google Scholar]
- Ostrem Loss E. M., Yu J.-H. (2018). Bioremediation and microbial metabolism of benzo(a)pyrene. Mol. Microbiol. 109 433–444. 10.1111/mmi.14062 [DOI] [PubMed] [Google Scholar]
- Paliwal V., Puranik S., Purohit H. J. (2012). Integrated perspective for effective bioremediation. Appl. Biochem. Biotechnol. 166 903–924. 10.1007/s12010-011-9479-5 [DOI] [PubMed] [Google Scholar]
- Parween T., Bhandari P., Sharma R., Jan S., Siddiqui Z. H., et al. (2018). “Bioremediation: a sustainable tool to prevent pesticide pollution,” in Modern Age Environmental Problems and their Remediation, eds Mohammad O., Mohammad Z. K., Iqbal M. I. I. (Cham: Springer; ), 215–227. 10.1007/978-3-319-64501-8_12 [DOI] [Google Scholar]
- Paynter Q., Fowler S. V., Hayes L., Hill R. L. (2015). Factors affecting the cost of weed biocontrol programs in New Zealand. Biol. Control 80 119–127. 10.1016/j.biocontrol.2014.10.008 [DOI] [Google Scholar]
- Perito B., Cavalieri D. (2018). “. Innovative metagenomic approaches for detection of microbial communities involved in biodeteriorattion of cultural heritage,” in IOP Conference Series: Materials Science and Engineering Vol. 364 (Bristol: IOP Publishing; ), 012074. 10.1088/1757-899X/364/1/012074 [DOI] [Google Scholar]
- Plattner J., Kazner C., Naidu G., Wintgens T., Vigneswaran S. (2018). Pesticide and microbial contaminants of groundwater and their removal methods: a mini review. J. Jaffna Sci. Assoc. 1 1–9. [Google Scholar]
- Plewniak F., Crognale S., Rossetti S., Bertin P. N. (2018). A genomic outlook on bioremediation: the case of arsenic removal. Front. Microbiol. 9:820. 10.3389/fmicb.2018.00820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit H. J., Tikariha H., Kalia V. C. (2018). “Current scenario on application of computational tools in biological systems,” in Soft Computing for Biological Systems, eds Purohit H. J., Kalia V. C., Prabhakar M. R. (Singapore: Springer; ), 1–12. [Google Scholar]
- Qian S., Wang Y., Lin H. (2018). A colorimetric sensor array based on sulfuric acid assisted KMnO4 fading for the detection and identification of pesticides. Talanta 181 305–310. 10.1016/j.talanta.2018.01.029 [DOI] [PubMed] [Google Scholar]
- Radford S. A., Panuwet P., Hunter R. E., Jr., Barr D. B., Ryan P. B. (2018). Degradation of organophosphorus and pyrethroid insecticides in beverages: implications for risk assessment. Toxics 6:11. 10.3390/toxics6010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass K., Megharaj M., Venkateswarlu K., Naidu R. (2018). Bioavailability of weathered hydrocarbons in engine oil-contaminated soil: impact of bioaugmentation mediated by Pseudomonas spp. on bioremediation. Sci. Total Environ. 636 968–974. 10.1016/j.scitotenv.2018.04.379 [DOI] [PubMed] [Google Scholar]
- Raman N. M., Asokan S., Sundari N. S., Ramasamy S. (2018). Bioremediation of chromium (VI) by Stenotrophomonas maltophilia isolated from tannery effluent. Int. J. Environ. Sci. Technol. 15 207–216. 10.1007/s13762-017-1378-z [DOI] [Google Scholar]
- Ravikrishnan A., Nasre M., Raman K. (2018). Enumerating all possible biosynthetic pathways in metabolic networks. Sci. Rep. 8:9932. 10.1038/s41598-018-28007-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico E., Jeacock L., Kovářová J., Horn D. (2018). Inducible high-efficiency CRISPR-Cas9-targeted gene editing and precision base editing in African trypanosomes. Sci. Rep. 8:7960. 10.1038/s41598-018-26303-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Dutta A., Pal S., Gupta A., Sarkar J., Chatterjee A., et al. (2018). Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresource Technol. 253 22–32. 10.1016/j.biortech.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Sarlio S. (ed.) (2018). “Serving sustainable and healthy food to consumers and decision makers: from commitments to action,” in Towards Healthy and Sustainable Diets (Cham: Springer; ), 63–82. 10.1007/978-3-319-74204-5_4 [DOI] [Google Scholar]
- Scala V., Pucci N., Loreti S. (2018). The diagnosis of plant pathogenic bacteria: a state of art. Front. Biosci. 10:449–460. [DOI] [PubMed] [Google Scholar]
- Schmidt-Jeffris R. A., Nault B. A. (2018). Crop spatiotemporal dominance is a better predictor of pest and predator abundance than traditional partial approaches. Agric. Ecosyst. Environ. 265 331–339. 10.1016/j.agee.2018.06.017 [DOI] [Google Scholar]
- Selzer P. M., Marhöfer R. J., Koch O. (2018). “Comparative genome analyses,” in Applied Bioinformatics (Cham: Springer; ), 123–140. 10.1007/978-3-319-68301-0_7 [DOI] [Google Scholar]
- Shah T., Andleeb T., Lateef S., Noor M. A. (2018). Genome editing in plants: advancing crop transformation and overview of tools. Plant Physiol. Biochem. 131 12–21. 10.1016/j.plaphy.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Shah V., Jain K., Desai C., Madamwar D. (2012). “Molecular analyses of microbial activities involved in bioremediation,” in Microorganisms in Environmental Management (Dordrecht: Springer; ), 221–247. [Google Scholar]
- Shapiro R. S., Chavez A., Collins J. J. (2018). CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat. Rev. Microbiol. 16:1. 10.1038/s41579-018-0002-7 [DOI] [PubMed] [Google Scholar]
- Sharma B., Dangi A. K., Shukla P. (2018). Contemporary enzyme based technologies for bioremediation: a review. J. Environ. Manag. 210 10–22. 10.1016/j.jenvman.2017.12.075 [DOI] [PubMed] [Google Scholar]
- Shukla A., Srivastava S., D’Souza S. F. (2018). An integrative approach toward biosensing and bioremediation of metals and metalloids. Int. J. Environ. Sci. Technol. 15 2701–2712. 10.1007/s13762-018-1766-z [DOI] [Google Scholar]
- Silva-Barni M. F., Gonzalez M., Wania F., Lei Y. D., Miglioranza K. S. B. (2018). Spatial and temporal distribution of pesticides and PCBs in the atmosphere using XAD-resin based passive samplers: a case study in the Quequén Grande River watershed. Argentina Atmospheric Pollut. Res. 9 238–245. 10.1016/j.apr.2017.09.008 [DOI] [Google Scholar]
- Simfukwe E. J., Tindwa H. J. (2018). Rock phosphate-solubilising potential of fungal and bacterial isolates from soils surrounding panda Hill and Minjingu phosphate rock deposits in Tanzania. Tropical Ecol. 59 109–118. [Google Scholar]
- Singh V., Gohil N., Ramírez García R., Braddick D., Fofié C. K. (2018). Recent advances in CRISPR-Cas9 genome editing technology for biological and biomedical investigations. J. Cell. Biochem. 119 81–94. 10.1002/jcb.26165 [DOI] [PubMed] [Google Scholar]
- Singh N. S., Sharma R., Parween T., Patanjali P. K. (2018). “Pesticide contamination and human health risk factor,” in Modern Age Environmental Problems and their Remediation, eds Mohammad O., Mohammad Z. K., Iqbal M. I. I. (Cham: Springer; ), 49–68. 10.1007/978-3-319-64501-8_3 [DOI] [Google Scholar]
- Stein H. P., Navajas-Pérez R., Aranda E. (2018). “Potential for CRISPR genetic engineering to increase xenobiotic degradation capacities in model fungi,” in Approaches in Bioremediation, eds Ram P., Elisabet A. (Cham: Springer; ), 61–78. [Google Scholar]
- Subashchandrabose S. R., Venkateswarlu K., Krishnan K., Naidu R., Lockington R., Megharaj M. (2018). Rhodococcus wratislaviensis strain 9: an efficient p-nitrophenol degrader with a great potential for bioremediation. J. Hazard. Mater. 347 176–183. 10.1016/j.jhazmat.2017.12.063 [DOI] [PubMed] [Google Scholar]
- Suenaga H. (2012). Targeted metagenomics: a high-resolution metagenomics approach for specific gene clusters in complex microbial communities. Environ. Microbiol. 14 13–22. 10.1111/j.1462-2920.2011.02438.x [DOI] [PubMed] [Google Scholar]
- Sulpice R., McKeown P. C. (2015). Moving toward a comprehensive map of central plant metabolism. Annu. Rev. Plant Biol. 66 187–210. 10.1146/annurev-arplant-043014-114720 [DOI] [PubMed] [Google Scholar]
- Sun J., Wang Q., Jiang Y., Wen Z., Yang L., Wu J., et al. (2018). Genome editing and transcriptional repression in Pseudomonas putida KT2440 via the type II CRISPR system. Microbial Cell Factor. 17:41. 10.1186/s12934-018-0887-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal A. (2018). Making conventional agriculture environmentally friendly: moving beyond the glorification of organic agriculture and the demonization of conventional agriculture. Sustainability 10:1078 10.3390/su10041078 [DOI] [Google Scholar]
- Tekle K. M., Gundersen S., Klepper K., Bongo L. A., Raknes I. A., Li X., et al. (2018). Norwegian e-Infrastructure for Life Sciences (NeLS). F1000Res. 7:ELIXIR–968. 10.12688/f1000research.15119.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. E., Ou L. T., Al-Agely A. (2008). “DDE remediation and degradation,” in Reviews of environmental contamination and toxicology, ed. Pim D. V. (New York, NY: Springer; ), 55–69. 10.1007/978-0-387-74816-0_3 [DOI] [PubMed] [Google Scholar]
- Uchiyama I. (2017). Ortholog identification and comparative analysis of microbial genomes using MBGD and RECOG. Methods Mol. Biol. 1611 147–168. 10.1007/978-1-4939-7015-5_12 [DOI] [PubMed] [Google Scholar]
- van Lenteren J. C., Bolckmans K., Köhl J., Ravensberg W. J., Urbaneja A. (2018). Biological control using invertebrates and microorganisms: plenty of new opportunities. BioControl 63 39–59. 10.1007/s10526-017-9801-4 [DOI] [Google Scholar]
- Vanacek P., Sebestova E., Babkova P., Bidmanova S., Daniel L., Dvorak P., et al. (2018). Exploration of enzyme diversity by integrating bioinformatics with expression analysis and biochemical characterization. ACS Catalysis 8 2402–2412. 10.1021/acscatal.7b03523 [DOI] [Google Scholar]
- VanderSluis B., Costanzo M., Billmann M., Ward H. N., Myers C. L., Andrews B. J., et al. (2018). Integrating genetic and protein–protein interaction networks maps a functional wiring diagram of a cell. Curr. Opin. Microbiol. 45 170–179. 10.1016/j.mib.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos C. V., Pereira F. T., Duarte E. A. A., de Oliveira T. A. S., Peixoto N., Carvalho D. D. C. (2018). physiological and molecular characterization of cephaleuros virescens occurring in mango trees. Plant Pathol. J. 34:157. 10.5423/PPJ.OA.09.2017.0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidart d’Egurbide Bagazgoïtia N., Bailey H. D., Orsi L., Lacour B., Guerrini-Rousseau L., Bertozzi A. I., et al. (2018). Maternal residential pesticide use during pregnancy and risk of malignant childhood brain tumors: a pooled analysis of the ESCALE and ESTELLE studies (SFCE). Int. J. Cancer 142 489–497. 10.1002/ijc.31073 [DOI] [PubMed] [Google Scholar]
- Visagie M., Mienie C. M., Marais M., Daneel M., Karssen G., Fourie H. (2018). Identification of Meloidogyne spp. associated with agri-and horticultural crops in South Africa. Nematology 20 397–401. 10.1163/15685411-00003160 [DOI] [Google Scholar]
- von Netzer F., Granitsiotis M. S., Szalay A. R., Lueders T. (2018). “Next-generation sequencing of functional marker genes for anaerobic degraders of petroleum hydrocarbons in contaminated environments,” in Anaerobic Utilization of Hydrocarbons, Oils, and Lipids. Handbook of Hydrocarbon and Lipid Microbiology, ed. Boll M. (Cham: Springer; ), 1–20. 10.1007/978-3-319-33598-8_15-1 [DOI] [Google Scholar]
- Warmerdam S., Sterken M. G., van Schaik C., Oortwijn M. E., Sukarta O. C., Lozano-Torres J. L., et al. (2018). Genome-wide association mapping of the architecture of susceptibility to the root-knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 218 724–737. 10.1111/nph.15034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waryah C. B., Moses C., Arooj M., Blancafort P. (2018). “Zinc fingers, TALEs, and CRISPR systems: a comparison of tools for epigenome editing,” in Epigenome Editing, eds Jeltsch A., Rots M. G. (New York, NY: Humana Press; ), 19–63. 10.1007/978-1-4939-7774-1_2 [DOI] [PubMed] [Google Scholar]
- Westcott N. D., Cessna A. J. (2018). “Fate of pesticides applied to cereals under field conditions,” in Pesticide Interactions in Crop Production, ed. Altman J. (Boca Raton, FL: CRC Press; ), 59–85. [Google Scholar]
- Wołejko E., Wydro U., Łoboda T. (2016). The ways to increase efficiency of soil bioremediation. Ecol. Chem. Eng. S 23 155–174. 10.1515/eces-2016-0011 [DOI] [Google Scholar]
- Wong D. W. S. (ed.) (2018). “Gene targeting and genome editing,” in The ABCs of Gene Cloning (Cham: Springer; ), 187–197. 10.1007/978-3-319-77982-9_20 [DOI] [Google Scholar]
- Wonglom P., Thithuan N., Bunjongsiri P., Sunpapao A. (2018). Plant-Parasitic Algae (Cephaleuros spp.) in Thailand, Including Four New Records. Pacific Sci. 72 363–371. 10.2984/72.3.7 [DOI] [Google Scholar]
- Yadav R., Kumar V., Baweja M., Shukla P. (2018). Gene editing and genetic engineering approaches for advanced probiotics: a review. Crit. Rev. Food Sci. Nutr. 58 1735–1746. 10.1080/10408398.2016.1274877 [DOI] [PubMed] [Google Scholar]
- Yan X., Huang J. W., Xu X. H., Chen D., Xie X. T., Tao Q., et al. (2018). Enhanced and complete removal of phenylurea herbicides by combinational transgenic plant-microbial remediation. Appl. Environ. Microbiol. 10.1128/AEM.00273-18 [Epub ahead of print]., [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E., Sanschagrin S., Beaumier D., Greer C. W. (2012). Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS One 7:e30058. 10.1371/journal.pone.0030058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Konermann S., Brideau N. J., Lotfy P., Novick S. J., Strutzenberg T., et al. (2018). Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 175 212–223.e17. 10.1016/j.cell.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Xiu Z. (2009). Metabolic pathway analysis of glycerol metabolism in Klebsiella pneumoniae incorporating oxygen regulatory system. Biotechnol. Prog. 25 103–115. 10.1002/btpr.70 [DOI] [PubMed] [Google Scholar]
- Zhang S., Hu Z., Wang H. (2018). A retrospective review of microbiological methods applied in studies following the deepwater horizon oil spill. Front. Microbiol. 9:520. 10.3389/fmicb.2018.00520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Wang J., Ding Y., Liu B., Xiao W. (2018). A systems-level approach for investigating organophosphorus pesticide toxicity. Ecotoxicol. Environ. Saf. 149 26–35. 10.1016/j.ecoenv.2017.10.066 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Klompe S. E., Vlot M., van der Oost J., Staals R. H. (2018). Shooting the messenger: RNA-targetting CRISPR-Cas systems. Biosci. Rep. 38:BSR20170788. 10.1042/BSR20170788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfo M., Asnicar F., Manghi P., Pasolli E., Tett A., Segata N. (2018). Profiling microbial strains in urban environments using metagenomic sequencing data. Biol. Dir. 13:9. 10.1186/s13062-018-0211-z [DOI] [PMC free article] [PubMed] [Google Scholar]