SUMMARY
Neuronal function requires axon-dendrite membrane polarity, which depends on sorting of membrane traffic during entry into axons. Due to a microtubule network of mixed polarity, dendrites receive vesicles from the cell body without apparent capacity for directional sorting. We found that, during entry into dendrites, axonally destined cargos move with a retrograde bias toward the cell body, while dendritically destined cargos are biased in the anterograde direction. A microtubule-associated septin (SEPT9), which localizes specifically in dendrites, impedes axonal cargo of kinesin-1/KIF5 and boosts kinesin-3/KIF1 motor cargo further into dendrites. In neurons and in vitro single-molecule motility assays, SEPT9 suppresses kinesin-1/KIF5 and enhances kinesin-3/KIF1 in a manner that depends on a lysine-rich loop of the kinesin motor domain. This differential regulation impacts partitioning of neuronal membrane proteins into axons-dendrites. Thus, polarized membrane traffic requires sorting during entry into dendrites by a septin-mediated mechanism that bestows directional bias on microtubules of mixed orientation.
In Brief
Polarized neuronal membrane traffic relies primarily on sorting at axonal entry, while dendrites appear to lack directional sorting due to mixed microtubule orientation. Karasmanis et al. show that a dendritically enriched microtubule-associated septin, SEPT9, impedes axonal kinesin-1/KIF5 but promotes dendritic kinesin-3/KIF1A motor cargo, thereby bestowing directional bias during dendritic entry.
Graphical Abstract
INTRODUCTION
Neural signaling and transmission requires the development and maintenance of axons and dendrites (Tahirovic and Bradke, 2009; Takano et al., 2015). A long-standing question is how neuronal membrane proteins are differentially transported to axons and dendrites (Craig et al., 1992; Hirokawa et al., 2010; Kapitein and Hoogenraad, 2011; Maday et al., 2014). In neuronal cell bodies, membrane proteins are sorted into distinct vesicle populations by clathrin-mediated mechanisms during exit from the Golgi complex (Bonifacino, 2014; Li et al., 2016). Subsequently, vesicles with axonal and somato-dendritic cargo are sorted during entry into axons, which contain unipolar microtubules (MTs) of plus-end-out orientation (Baas and Lin, 2011; Song et al., 2009; Yau et al., 2016). Axonal cargos of the MT motor kinesin-1/KIF5 move selectively through the pre-axonal exclusion zone and the axon initial segment (AIS), while somato-dendritic cargos are halted and reverse directionality (Al-Bassam et al., 2012; Farias et al., 2015; Kuijpers et al., 2016; Nakata and Hirokawa, 2003).
Owing to a MT network of mixed orientation, membrane traffic in dendrites is inherently bidirectional and presumed to lack unidirectional sorting (Baas and Lin, 2011; Bannai et al., 2004; Kapitein et al., 2010a; Yau et al., 2016). Dendrites are thought to rely on the sorting mechanisms of the cell body and axon as well as on retaining select cargo (Kapitein and Hoogenraad, 2011). In the cell body, vesicles are directed to dendrites by MT motors of the kinesin-3 and kinesin-4 subfamilies as well as dynein (Ghiretti et al., 2016; Kapitein et al., 2010a; Lipka et al., 2016). Interestingly, kinesin-3-driven transport of dense core vesicles was recently shown to depend on a microtubule-associated protein (MAP), pointing to MT-associated cues that may modulate membrane traffic in dendrites (Lipka et al., 2016). However, such cues remain elusive, and it is unclear whether they play a role in the sorting of membrane traffic in dendrites.
Septins are a large family of cytoskeletal GTP-binding proteins with evolutionarily conserved functions in the spatial organization and compartmentalization of membrane proteins (Caudron and Barral, 2009; Mostowy and Cossart, 2012). Acting as diffusion barriers and protein scaffolds, septins promote asymmetry across membrane bilayers (Caudron and Barral, 2009; Spiliotis and Gladfelter, 2012). In epithelial cells, septins associate with subsets of MTs and are required for vesicle transport from the Golgi complex to the plasma membrane (Bowen et al., 2011; Spiliotis et al., 2008). In neurons, septins are involved in the morpho-genesis of axons and dendrites, but it is unknown if they function in polarized membrane traffic into axons and dendrites (Ageta-Ishihara et al., 2013; Boubakar et al., 2017; Hu et al., 2012; Tada et al., 2007; Xie et al., 2007).
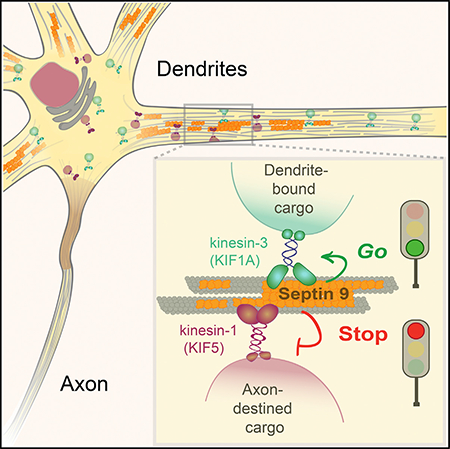
Here, we report that an MT-associated septin (SEPT9) that is specifically enriched in dendrites regulates differentially the motility of axonal and dendritic vesicles. We found that dendritic cargos of kinesin-3/KIF1A move with an anterograde bias in the initial 20-μm segments of dendrites, while axonal cargos of kinesin-1/KIF5 are biased toward the cell body. This directional sorting is mediated by SEPT9, which inhibits the motility of kinesin-1/KIF5 and its axonal cargo while enhancing the transport of kinesin-3/KIF1 motor cargo. SEPT9 distinguishes between kinesin-1/KIF5 and kinesin3/KIF1 based on the loop 12 (L12) of their motor domains and thereby regulates membrane traffic into axons and dendrites. Our results reveal that, upon entry into dendrites, axonal and dendritic cargos are directionally sorted by a SEPT9-mediated mechanism.
RESULTS
Neuronal Kinesin Motor Cargos Are Directionally Sorted during Entry into Dendrites
Axonally destined vesicles of kinesin-1/KIF5 enter axons selectively, but it is unknown if they move into dendrites and are subsequently sorted out (Farias et al., 2015; Huang and Banker, 2012; Kamal et al., 2000; Nakata and Hirokawa, 2003; Song et al., 2009). To examine this possibility, we expressed and imaged axonal cargo markers of kinesin-1/KIF5 in live primary rat hippocampal neurons. Using single particle-tracking analysis in the initial 20-mm segments of dendrites, we analyzed the trajectories of axonal cargos of kinesin-1/KIF5, such as the pre-synaptic matrix protein Bassoon, syntaxin-1, and the amyloid precursor protein (APP) (Cai et al., 2007; Kamal et al., 2000; Su et al., 2004). We observed that these axonal cargos enter dendrites, but the majority (>60%) are at a stationary state (Figure 1A). A small fraction (<15%) exhibited processive (i.e., uninterrupted) anterograde excursions. However, these events were outnumbered by processive retrograde events and anterograde movements that pause or reverse backwards to the cell body (Figures 1A and 1C, Video S1). Similar results were obtained by a frame-by-frame analysis of each vesicle trajectory, categorizing movements between individual frames into anterograde, retrograde, or stationary events (Figure 1B). By deriving the percentage of total time that each particle spent in these states, we found that axonal cargos spent 70%–80% of the total time in pausing, and displacements in the retrograde direction were 1.5- to 2-fold more than in the anterograde direction (Figure 1B). Therefore, the vast majority of membrane vesicles with axonal cargos stall or reverse directionality upon entry into dendrites.
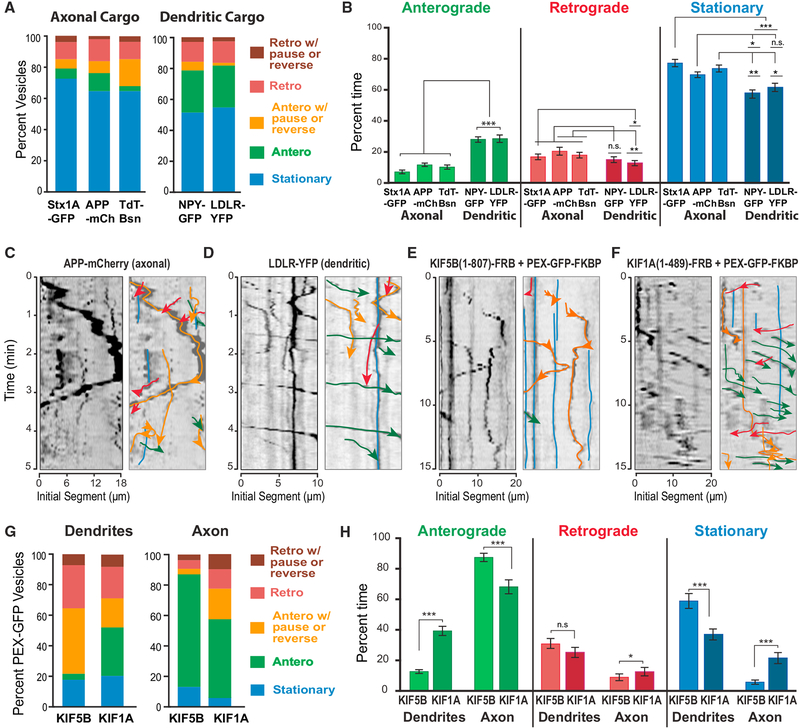
Figure 1. Axonal and Dendritic Motor Cargo Exhibit Differential Motility during Entry into Dendrites.
(A and B) The trajectories of vesicles that move from the cell body into the initial 20-μm segments of dendrites of hippocampal neurons were categorized into stationary, moving processively in the anterograde/retrograde direction, or pausing and reversing while moving anterogradely/retrogradely and graphed as percentage of total vesicles (A). A frame-by-frame analysis was also performed for individual vesicles, deriving the percentage of time spent in pausing, anterograde, or retrograde movement between consecutive frames of each trajectory (B). Quantifications were done for Syntaxin1a-GFP (Stx1A-GFP; n = 220), APP-mCherry (APP-mCh; n = 286) and TdT-Bassoon (Tdt-Bsn; n = 358), NPY-GFP (n = 264), and LDLR-YFP (n = 284) from 15 to 18 dendrites (seven neurons). (C–F) Kymographs show trajectories of vesicles with APP-mCherry (C) and LDLR-YFP (D), and peroxisomes (PEX-GFP) driven by kinesin-1/KIF5B(1–807) (E) and kinesin-3/KIF1A(1–489) (F) in the initial 20-μm segments of dendrites. Labels, line colors, and arrows are as denoted in (A).
(G and H) The trajectories of peroxisomes (G) that move from the cell body into the initial 20-μm segments of dendrites or axons of hippocampal neurons after treatment with rapalog were categorized and graphed as in (A). A frame-by-frame analysis was also performed for individual vesicles, deriving the percentage of time spent in pausing, anterograde, or retrograde movement between consecutive frames of each trajectory (H). Quantifications were done for peroxisomes driven by KIF5B(1–807) into dendrites (n = 86) and axons (n = 85), and by KIF1A(1–489) into axons (n = 66) and dendrites (n = 142) from four to five neurons. *p < 0.05; **p < 0.001; ***p < 0.0001; n.s., not significant (p > 0.05).
We next analyzed dendritic cargos of kinesin-3/KIF1A such as the low-density lipoprotein receptor (LDL-R) and neuropeptide Y (Jenkins et al., 2012; Lo et al., 2011). A significant fraction (~25%) of these cargos exhibited processive anterograde trajectories, which were 1.5- to 2-fold more frequent than processive retrograde trajectories and anterograde runs with pausing or reversing (Figures 1A and 1D, Video S2). Frame-by-frame analysis also showed that anterograde steps were 1.5- to 2-fold more than retrograde displacements and occupied more than twice the total time of movement compared with the anterograde events of axonal cargo (Figure 1B). Hence, dendrite-bound vesicles of kinesin-3/KIF1A move with an anterograde bias during entry into dendrites.
Because vesicles associate with multiple motors, we asked whether the motor domains of kinesin-1/KIF5 and kinesin-3/KIF1A move into dendrites with similar directional bias to their cargo. We performed an inducible trafficking assay that mobilizes peroxisomes from the cell body by rapalog-induced coupling of kinesin motor domains to a fluorescent peroxisome-resident protein (PEX-GFP) (Kapitein et al., 2010b). As previously reported, the constitutively active kinensin-1/KIF5B(1–807) directs peroxisomes to the axon (Lipka et al., 2016). In early stages of induction, however, peroxisomes were observed to traffic to dendrites. The majority of these peroxisomes exhibited processive retrograde or anterograde trajectories with pauses and reversals (Figures 1E and 1G, Video S3). Processive anterograde movement was minimal (~5% of peroxisomes), while this was the predominant (>80%) type of movement in the initial 20-μm segments of axons (Figure 1G). In striking contrast to kinesin-1, ~30% of peroxisomes that move into dendrites by kinesin-3/KIF1A(1–489) exhibited processive anterograde runs (Figures 1F and 1G, Video S4). This was also evident in the frame-by-frame analysis, which showed that kinesin-3-driven peroxisomes were less stationary and anterograde steps were 3-fold more than those of kinesin-1 (Figure 1H). Consistent with the anterograde bias of the dendritic cargo of kinesin-3/KIF1A, the motor domain of kinesin-3/KIF1A has an anterograde advantage over kinesin-1/KIF5. Thus, during entry into dendrites, cargos driven by kinesin-3/KIF1A are likely to move further along the dendrite, while cargos driven by kinesin-1/KIF5 move with retrograde bias toward to the cell body.
MT-Associated SEPT9 Is Enriched in Dendrites
We reasoned that an MT-associated cue that is specific to dendrites underlies the directional sorting of the kinesin-1 and −3 motors and their cargo. To date, MAP2 is the only dendritic MAP known to inhibit kinesin-1 binding to MTs, but this function was discovered in the axons of pseudounipolar neurons (Gumy et al., 2017). In polarized hippocampal neurons, MAP2 is also present along the initial segment of the axon that precedes the AIS, with which kinesin-1 associates preferentially (Farias et al., 2015; van Beuningen et al., 2015). Hence, MAP2 alone cannot account for the dendrite-specific halting and reversal of kinesin-1/KIF5 and its axonal cargo.
Septins localize to subsets of MTs in epithelia (Bai et al., 2013; Bowen et al., 2011), but their localization and functions in neuronal cells are little understood. In the septin family, SEPT9 binds and bundles MTs directly through N-terminal repeat motifs, resembling the MT-binding domains (MTBDs) of bona fide MAPs (e.g., tau, MAP2) (Bai et al., 2013). SEPT9 is a ubiquitously expressed septin that is essential for embryonic development as SEPT9 knockout mice die by embryonic day 10 (Fuchtbauer et al., 2011). In adult mice, SEPT9 is highly expressed in the CA and dentate gyrus regions of the hippocampus (Allen, 2004). In cultured hippocampal neurons, SEPT9 protein levels increase during the developmental stage in which post- and pre-synaptic membrane proteins polarize into dendrites and axons (Frese et al., 2017). In fully differentiated hippocampal neurons, SEPT9 localizes preferentially to dendrites and is more enriched in dendritic segments that are proximal to the cell body (Figures 2A–2C). Interestingly, SEPT9 enrichment in dendrites occurs after polarization of MAP2 in dendrites and takes place concomitantly with the segregation of the post-synaptic density protein 95 (PSD95) and synapsin into dendrites and axons, respectively (Figure S1A).
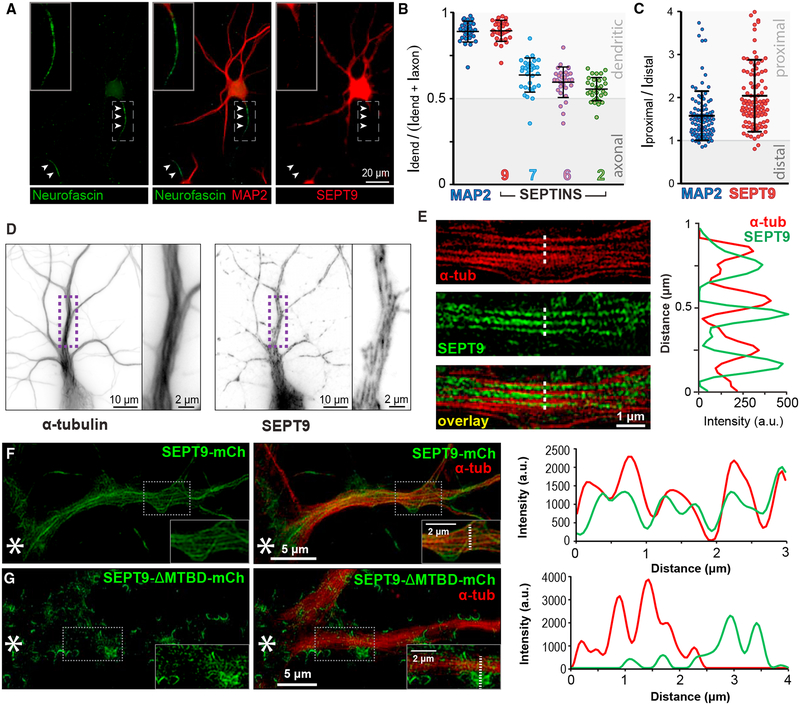
Figure 2. MT-Associated SEPT9 Is Enriched in Dendrites.
(A) Immunofluorescence microscopy of rat hippocampal neurons (DIV14) stained for neurofascin (axon; arrowheads), MAP2 (dendrites), and SEPT9.
(B) Scatter dot plots show the polarity index (dendritic intensity over sum of axonal and dendritic intensities) of MAP2, SEPT9, SEPT7, SEPT6, and SEPT2 (n = 30 cells). Horizontal lines show mean (±SD).
(C) Plot shows proximal-to-distal ratio of the dendritic fluorescence intensities of MAP2 and SEPT9 (n = 101 dendrites; 20 neurons).
(D and E) Shallow-angle TIRF (D) and super-resolution SIM (E) images of hippocampal neurons stained for α-tubulin and SEPT9. Plot shows MT and SEPT9 fluorescence intensity across the dotted line.
(F and G) SIM images of hippocampal neurons that were stained for α-tubulin after transfection with a plasmid that expresses simultaneously SEPT9 shRNA1 and rat shRNA-resistant SEPT9-mCherry (F) or SEPT9-ΔMTBD-mCherry (G) that lacks the MTBD (amino acids [aa] 1–147) and localizes to areas of membrane curvature. Plots show MT and SEPT9 fluorescence intensities across the dotted lines. Asterisks denote cell body location and insets show a larger magnification of outlined regions.
See also Figure S1.
Imaging by total internal reflection fluorescence (TIRF) microscopy at shallow incident angles revealed that SEPT9 localizes as filaments that are closely apposed to dendritic MTs (Figure 2D). To determine if SEPT9 decorates the MTs of proximal dendrites, we performed super-resolution structured illumination microscopy (SIM), which achieves lateral resolution of ~100 nm; four times the width of a single MT. SEPT9 was observed to overlap laterally with MTs (Figure 2E). We observed similar localization for SEPT9-mCherry in neurons that were depleted of endogenous SEPT9 and imaged by SIM, while SEPT9-mCherry that lacked the MTBD localized largely to sites of membrane curvature and not MTs (Figures 2F and 2G). Note that SEPT9-mCherry co-aligns more extensively with the width of MTs than endogenous SEPT9, which might be due to poor antibody accessibility to sites of tight MT-SEPT9 binding.
Because, in non-neuronal cells, SEPT9 hetero-oligomerizes with SEPT7, SEPT6, and SEPT2 (Kim et al., 2011; Sellin et al., 2011), we also imaged and quantified the localization of these septins. SEPT2, SEPT6, and SEPT7 were less enriched in dendrites (Figure 2B) and did not colocalize significantly with SEPT9 (Figures S1B–S1D). Additionally, knockdown of SEPT9 did not affect the expression levels of its hetero-dimeric partner SEPT7 and vice versa (Figures S1E–S1H). Hence, SEPT9 appears to localize uniquely to dendritic MTs and might be critical for the polarity of membrane traffic.
SEPT9 Is Required for the Axonal Selectivity of Kinesin-1 Motor Cargo
To test if SEPT9 affects the polarized trafficking of kinesin-1/KIF5, hippocampal neurons were treated with two different small hairpin RNAs (shRNAs) that target rat SEPT9 (Figures S1E–S1G). SEPT9 depletion abolished the axonal polarity of the kinesin-1 motor domain KIF5C(1–560)-mCit, which translocated to the tips of both axons and dendrites (Figures 3A and 3B). This phenotype was specific to SEPT9 and its N-terminal MTBD because it was rescued with an shRNA-resistant SEPT9-mCherry but not with SEPT9-Δ(1–147)-mCherry (Figure 3B). The axonal polarity of KIF5C(1–560)-mCit was also partially rescued by the MTBD of SEPT9 (Figure 3B). Note that SEPT7 depletion did not affect the axonal polarity of KIF5C(1–560)-mCit (Figures S2A–S2C).
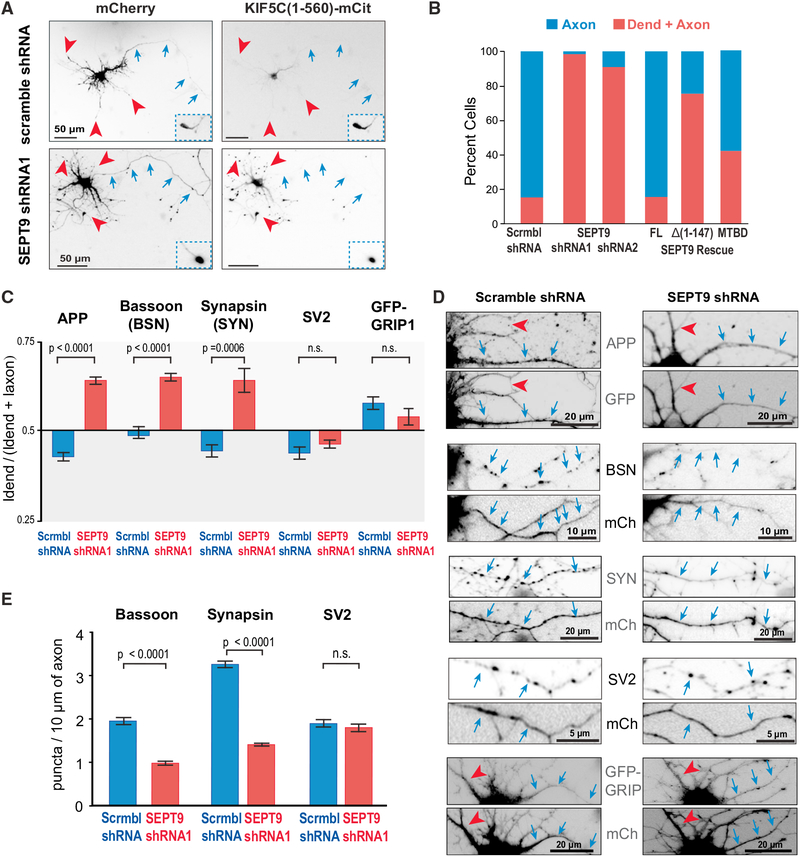
Figure 3. SEPT9 Is Required for the Axonal Preference of Kinesin-1 Motor Cargo.
(A and B) Images (A) show hippocampal neurons that were co-transfected with KIF5C(1–560)-mCit and plasmids encoding for mCherry and scramble or SEPT9 shRNAs. Blue arrows and red arrowheads point to axons and dendritic tips, respectively. Insets show axonal tips outside the field of view. Bar graph (B) shows percentage of cells (n = 43–77) with KIF5C(1–560)-mCit in axons or dendrites. Rescue experiments were performed by co-expressing SEPT9 shRNA with shRNA-resistant SEPT9-mCherry, SEPT9-Δ(1–147)-mCherry or SEPT9(1–125)-mCherry (MTBD).
(C) Mean polarity index (±SEM) of APP, Bassoon, Synapsin, SV2, and GFP-GRIP1 in hippocampal neurons (n = 21–29) transfected with scramble or SEPT9 shRNAs.
(D) Images show dendritic (red arrowheads) and axonal segments (blue arrows) of neurons expressing scramble or SEPT9 shRNAs (mCherry/GFP) and stained for endogenous APP, Bassoon, Synapsin, SV2, or co-transfected with GFP-GRIP1.
(E) Bar graphs show the mean number (±SEM) of Bassoon, synapsin, or SV2 puncta per 10 mm of axonal length in cells expressing scramble or SEPT9 shRNAs. See also Figures S2–S4.
Using the inducible peroxisome-trafficking assay, we tested if SEPT9 depletion also affects the axonal polarity of peroxisomes driven by the kinesin-1 motor domain. The majority (~70%) of SEPT9-depleted neurons contained peroxisomes in both axons and dendrites and a significant fraction (~20%) exhibited a complete reversal of polarity as peroxisomes were present in dendrites only (Figures S2D and S2E). Consistent with these data, the localization of endogenous kinesin-1 became more dendritic upon SEPT9 depletion (Figure S2F). We concluded that the MT-associated SEPT9 is required for the axonal selectivity of the kinesin-1 motor domain.
Next, we sought to determine whether SEPT9 affects the axonal localization of endogenous APP, Bassoon, and synapsin, which are kinesin-1 cargos that stall and reverse directionality upon entry into dendrites (Figure 1). By quantifying the mean fluorescence intensity of these membrane proteins in axons and dendrites after background subtraction using a pixel-wide line scan, we derived a polarity index: the dendritic fluorescence value as a fraction of the sum of dendritic and axonal values (Li et al., 2016). SEPT9 knockdown increased the polarity index of APP, Bassoon, and synapsin, indicating a shift from axons to dendrites (Figures 3C and 3D). This loss in axonal localization was accompanied by a significant reduction (>50%) in the number of Bassoon and synapsin puncta per 10-μm axon length (Figures 3D and 3E). Notably, SEPT9 depletion did not alter the axonal localization of the synaptic vesicle glycoprotein 2 (SV2), a pre-synaptic marker whose transport into axons might be compensated by kinesin-3 (Zhao et al., 2001) (Figures 3C–3E). As an additional control, we also tested if SEPT9 depletion affects the dendritic localization of the glutamate receptor interacting protein 1 (GRIP1), which is an adaptor protein that shifts the directionality of kinesin-1 into dendrites by possibly altering the interactions of kinesin-1 with MTs (Heisler et al., 2014; Setou et al., 2002). We observed no change in the polarity index of GRIP1 (Figures 3C and 3D), concluding that SEPT9 impedes specifically axonal cargos of kinesin-1 from entering into dendrites.
SEPT9 Impedes the Motility of Kinesin-1 and Its Axonal Cargo
We hypothesized that SEPT9 directly impedes the motility of kinesin-1 and its axonal cargo. Before exploring this idea, we considered the possibility that SEPT9 affected kinesin-1 motility indirectly by altering MT orientation and/or post-translational modifications (PTMs), which can influence the trafficking of MT motor cargo (Janke and Kneussel, 2010). Time-lapse microscopy of the neuronal MT plus-end protein EB3-GFP in the initial segments of axons and proximal dendrites did not show any change in the orientation of MT growth upon SEPT9 knockdown (Figure S3A). We also did not observe any alterations in the axon-dendritic distribution of acetylated and glutamylated MTs (Figures S3B–S3E). Given that SEPT9 depletion does not alter the translocation of the kinesin-1 motor domain to axonal tips (Figures 3A and 3B), abnormal entry of kinesin-1 motor cargo into dendrites does not appear to be due to alterations in MT orientation and PTMs. Moreover, the axonal localization of dephosphorylated tau was unaffected and no changes were observed in the fluorescence intensity or length of the AIS marker proteins ankyrin-G and neurofascin (Figure S4).
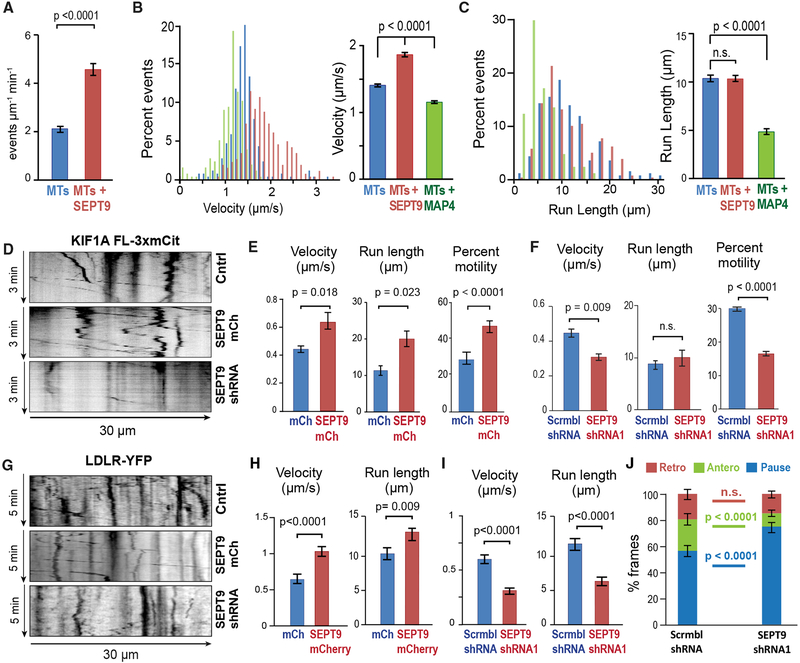
To test if MT-associated SEPT9 impedes directly the motility of kinesin-1, we performed an in vitro (cell-free) single-molecule motility assay using stable pre-polymerized MTs that were coated with recombinant His-mCherry-SEPT9 (10 nM), which remains stably bound to MTs for the duration of the assay (Figures S5A and S5B). Strikingly, MT-associated SEPT9 reduced the landing rates of kinesin-1 by 80% and increased pausing by more than 2-fold (Figures 4A and 4B). The velocity was also decreased by ~50%, while run lengths remained unchanged (Figures 4C and 4D). This effect was specific to kinesin-1 as SEPT9 did not affect the motility of kinesin-2/KIF17 in vitro or its localization and motility in neurons (Figures S5C–S5F) (Bai et al., 2016).
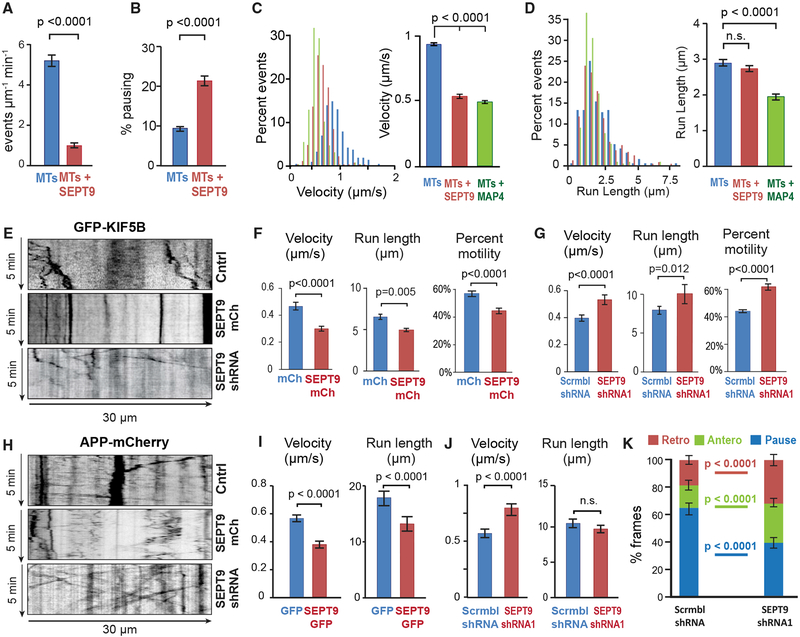
Figure 4. SEPT9 Impedes the Motility of Kinesin-1 and Its Axonal Cargo.
(A and B) Mean (±SEM) landing rate (A) and percentage pausing (B) of KIF5C(1–560)-mCit on MTs alone (n = 4603 events; 17 MTs) or MTs coated with HismCherry-SEPT9 (10 nM; n = 856 events; 21 MTs).
(C and D) Histograms show the distribution of velocities (C) and run lengths (D) of KIF5C(1–560)-mCit on MTs alone (n = 257 events), MTs coated with HismCherry-SEPT9 (10 nM; n = 271), or His-mCherry-MAP4-MTBD (10 nM; n = 254). Bar graphs show mean velocity and run length (±SEM).
(E) Representative kymographs of GFP-KIF5B in the dendrites of hippocampal neurons (DIV12) transfected with scramble (Cntrl) or SEPT9 shRNAs with mCherry, or SEPT9-mCherry.
(F and G) Mean (±SEM) percentage motility (n > 300; 7–10 cells), velocity, and run length (n = 50–102; 7–10 cells) of GFP-KIF5B in dendrites of neurons transfected with mCherry or SEPT9-mCherry (F) and scramble or SEPT9 shRNAs (G).
(H) Kymographs of APP-mCherry in the dendrites of hippocampal neurons (DIV12) transfected with scramble or SEPT9 shRNAs with mCherry, or SEPT9-mCherry.
(I and J) Mean (±SEM) velocities and run lengths of APP-mCherry vesicles (n > 80) in dendrites (n = 15–20; seven or eight neurons) transfected with GFP or SEPT9-GFP (I) or scramble or SEPT9 shRNAs (J).
(K) The movement of each APP-mCherry vesicle (n = 220–290; 16 dendrites, seven or eight neurons) was tracked frame by frame in the initial 20-μm segments of dendrites and categorized into pausing, retrograde or anterograde displacements between consecutive frames, and then quantified as percentage of total time. Graph shows mean percentage (±SEM).
See also Figure S5.
Next, we tested whether the motility of kinesin-1 and its axonal cargo (e.g., APP) are affected by SEPT9 over-expression and depletion in the dendrites of living neurons. SEPT9 over-expression reduced the velocity, run length, and percentage motility of GFP-KIF5B, while SEPT9 knockdown had the opposite effect (Figures 4E–4G). Analysis of APP vesicle motility showed a marked decrease in velocity and run lengths in neurons that over-expressed SEPT9 (Figures 4H and 4I). In contrast, SEPT9 depletion accelerated APP-carrying vesicles (Figures 4H and 4J). Notably, in the initial 20-μm segments of dendrites, frame-by-frame analysis of APP vesicles showed a ~40% decrease in pausing and >1.5-fold increase in anterograde movement (Figure 4K). Taken together, these data indicate that MT-associated SEPT9 promotes the axonal targeting of kinesin-1 cargos by inhibiting their motility upon entry into dendrites.
SEPT9 Is Required for Kinesin-3/KIF1A-Cargo Transport from the Cell Body to Distal Dendrites
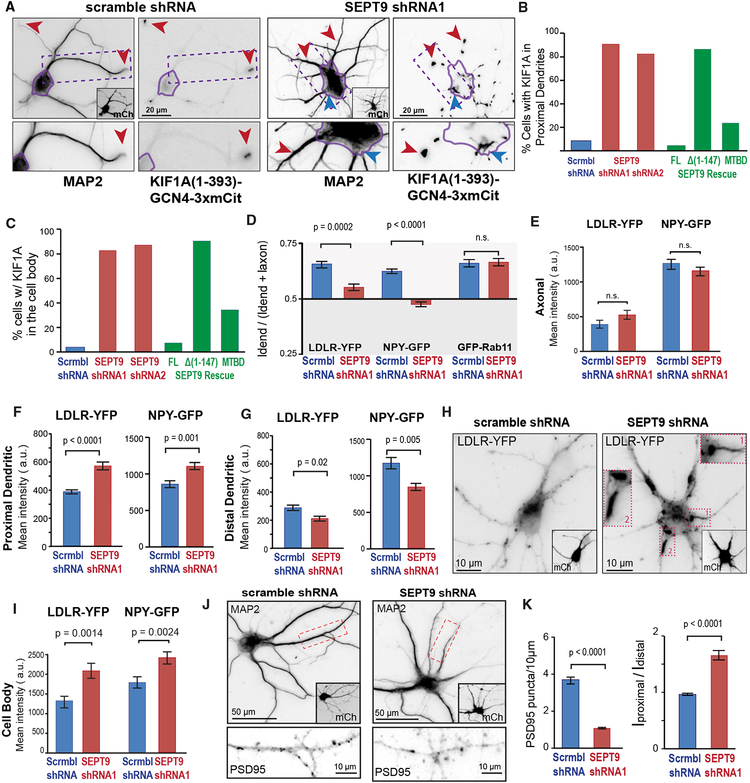
During entry into dendrites, dendritic cargos of kinesin-3/KIF1A are more motile and move with an anterograde bias (Figure 1). We therefore sought to examine whether SEPT9 distinguishes between kinesin-3/KIF1A and kinesin-1/KIF5 motility, favoring the former while impeding the latter. First, we tested if SEPT9 affects the localization of the dimeric kinesin-3 motor domain KIF1A(1–393)-GCN4–3xmCit, which translocates to both axonal and dendritic tips (Huang and Banker, 2012; Lipka et al., 2016). SEPT9 knockdown did not affect axonal localization, but in dendrites KIF1A(1–393)-GCN4–3xmCit accumulated at segments that were proximal to the cell body (Figures 5A and 5B). Additionally, there was a marked increase in the percentage of cells with KIF1A(1–393)-GCN4–3xmCit localizing in the cell body (Figure 5C) and particularly in regions that precede dendrites, as if there were a traffic bottleneck at sites of dendritic entry (blue arrowhead in Figure 5A). This phenotype was observed with two different shRNAs against SEPT9 and rescued with a shRNA-resistant mCherry-SEPT9 (Figures 5B and 5C); SEPT7 depletion had no effect on KIF1A(1–393)-GCN4–3xmCit localization (Figures S6A–S6C). Notably, the MTDB of SEPT9 was necessary and partially sufficient in restoring the localization of the kinesin-3/KIF1A motor domain (Figures 5B and 5C). In peroxisome-trafficking assays, SEPT9 knockdown also shifted KIF1A(1–489)-driven peroxisomes to proximal dendrites and the same was observed for endogenous kinesin-3/KIF1A (Figures S6D–S6F).
Figure 5. SEPT9 Is Required for Kinesin-3/KIF1A-Cargo Transport from Initial-Proximal to Distal Segments of Dendrites.
(A–C) Images (A) show hippocampal neurons stained for MAP2 after co-transfection with KIF1A(1–393)-GCN4–3xmCit and scramble or SEPT9 shRNAs with mCherry (insets). Cell bodies are outlined in purple. Red arrowheads point to KIF1A accumulation at dendritic tips (scramble shRNA) and dendritic segments proximal to the cell body (SEPT9 shRNAs). Blue arrowhead points to KIF1A accumulation at cell body exit site (shRNA). Bar graphs show the percentage of cells (n = 53–75) with KIF1A in proximal dendrites (B) and in the cell body (C). Rescue experiments were done by co-expressing SEPT9 shRNA with shRNA-resistant SEPT9-mCherry, SEPT9-Δ(1–147)-mCherry, or SEPT9(1–125)-mCherry (MTBD).
(D) Mean polarity index (±SEM) of LDLR-YFP, NPY-GFP, and GFP-Rab11 in hippocampal neurons (n = 20–30, 20–30 neurons) transfected with scramble or SEPT9 shRNAs.
(E–I) Mean intensity (±SEM) of LDLR-YFP and NPY-GFP in axons (E), proximal dendrites (F), distal dendrites (G), and cell bodies (I) (n > 100 dendrites, 20–30 neurons). Images (H) show the localization of LDLR-YFP in neurons transfected with scramble or SEPT9 shRNAs (mCherry; insets). Outlined dendritic segments are shown in higher magnification.
(J and K) Images (J) show the localization of PSD95 in outlined dendritic segments (MAP2) from neurons that express scramble or SEPT9 shRNAs (mCherry, insets). Bar graphs (K) show the mean number (±SEM) of PSD95 puncta per 10 μm of dendritic length and mean proximal-to-distal ratio of dendritic fluorescence intensity (n = 27–29; 93–221 dendrites).
See also Figures S6 and S7.
Next, we tested how SEPT9 affects the trafficking of LDLR and NPY, membrane cargos of kinesin-3/KIF1A that move with anterograde bias into dendrites. We found that SEPT9 depletion reduced the dendritic localization of LDLR-YFP and NPY-GFP, but without a shift to axons; mean fluorescence intensities remained unchanged in axons (Figures 5D and 5E). However, LDLR-YFP and NPY-GFP intensities were elevated in the cell bodies and the proximal segments of dendrites, and decreased in distal dendrites (Figures 5F–5I and S6G). Phenocopying the mislocalization of the kinesin-3/KIF1A motor domain, LDLR-YFP was observed to accumulate at dendritic entry sites of the cell body and proximal dendrites (Figure 5H; insets 1 and 2). As kinesin-3/KIF1B is known to associate with the post-synaptic density protein 95 (PSD95), we also examined the localization of PSD95 in SEPT9-depleted neurons (Mok et al., 2002). Quantification of PSD95 puncta in dendrites showed a ~75% reduction, and, like the NPY and LDLR cargos, the dendritic distribution of PSD95 shifted to segments proximal to the cell body (Figures 5J and 5K). To rule out the possibility that SEPT9 affects the localization of all dendritic cargos and not just those of kinesin-3/KIF1, we asked if SEPT9 knockdown alters the localization of GFPRab11, a membrane cargo of KIF13 (Delevoye et al., 2014). We observed no change in the distribution of GFP-Rab11 (Figures S6H and S6I). Taken together, these data indicate that SEPT9 is required for the directional trafficking of kinesin-3/KIF1 motor cargo from the cell body to distal dendrites.
SEPT9 Enhances the Motility of Kinesin-3/KIF1A and Its Dendritic Cargo
Given that SEPT9 depletion results in traffic jam-like accumulations of kinesin-3/KIF1A and its cargo in the initial-proximal segments of dendrites, we tested whether SEPT9 directly affects the motility of kinesin-3/KIF1A. In vitro single-molecule motility as-says showed that the motor domain of kinesin-3/KIF1A lands on SEPT9-coated MTs with a 1.5-fold higher frequency and moves with increased velocity compared with SEPT9-free MTs (Figures 6A and 6B). This effect was specific to SEPT9 as kinesin-3/KIF1A velocity and run lengths were reduced on MTs decorated with the MTBD of MAP4 (Figures 6B and 6C). Notably, MAP4-MTBD reduced the velocity and run length of both kinesin-3/KIF1A and kinesin-1/KIF5 (Figures 4B and 4C), demonstrating that SEPT9 is unique in its ability to enhance kinesin-3/KIF1A, while impeding kinesin-1/KIF5.
Figure 6. SEPT9 Enhances the Motility of Kinesin-3/KIF1A and LDLR Cargo.
(A) Bar graph shows the mean (±SEM) landing rate of KIF1A(1–393)-GCN4–3xmCit on uncoated MTs (n = 18) or MTs coated with His-mCherry-SEPT9 (10 nM; n = 16).
(B and C) Histograms show the distribution of velocities (B) and run lengths (C) of KIF1A(1–393)GCN4–3xmCit on MTs alone (n = 250) and MTs coated with HismCherry-SEPT9 (n = 257) or His-mCherry-MAP4-MTBD (n = 250). Bar graphs shows mean (±SEM) velocity and run length.
(D) Kymographs of KIF1A-3xmCit in dendrites of neurons (DIV12) transfected with scramble or SEPT9 shRNAs with mCherry, or SEPT9-mCherry alone. (E and F) Mean (±SEM) percentage motility (n > 240; six or seven cells), velocity, and run length (n = 50–73; six or seven cells) of KIF1A-3xmCit in dendrites of neurons transfected with mCherry or SEPT9-mCherry (E) and scramble or SEPT9 shRNAs (F).
(G) Kymographs of LDLR-YFP vesicles in dendrites of neurons transfected with scramble (Cntrl) or SEPT9 shRNAs with mCherry, or SEPT9-mCherry alone. (H and I) Mean velocity and run length (±SEM) of LDLR-YFP vesicles (n = 86–155) in neurons (n = seven or eight; 16–18 dendrites) transfected with mCherry or SEPT9-mCherry (H) and scramble or SEPT9 shRNAs (I).
(J) The movement of each LDLR-YFP vesicle (n = 220–290; 16–18 dendrites, seven or eight neurons) was tracked frame by frame in the initial 20-μm segments of dendrites and categorized into pausing, retrograde, or anterograde displacements between consecutive frames, and then quantified as percentage of total time. Graph shows mean percentage (±SEM).
See also Figure S7.
To further test if kinesin-3/KIF1A motility is enhanced by SEPT9, we analyzed the motility of full-length KIF1A-3xmCit in live neurons under conditions of SEPT9 over-expression and depletion. Over-expression of SEPT9-mCherry increased the percentage motility, velocity, and run lengths of KIF1A-3xmCit (Figures 6D and 6E). Conversely, SEPT9 knockdown resulted in significant reduction of percentage motility and velocity (Figures 6D and 6F). Taken together with the in vitro motility data, these results show that SEPT9 promotes the transport of kinesin-3/KIF1A.
Next, we examined how SEPT9 affects the motility and directionality of LDLR-carrying vesicles, which move with an anterograde bias in the initial segments of dendrites. We found that the velocity and run length of LDLR-carrying vesicles increased in the dendrites of neurons that over-expressed SEPT9-mCherry, while SEPT9 knockdown had the opposite effect (Figures 6G–6I). Notably, SEPT9 depletion reduced the motility of LDLR in the initial 20-mm segment of dendrites, increasing pausing by ~30% and decreasing the percentage time spent in anterograde displacements by more than 50% (Figure 6J). Therefore, SEPT9 promotes the forward movement of dendritic cargo of kinesin-3/KIF1A from initial-proximal to distal dendrites. This function appears to be critical for preventing traffic jam-like accumulations of dendritic cargo in the initial segments of dendrites and sorting them away from axonally destined vesicles.
Because in parallel to kinesin-3/KIF1A, dynein also directs cargo from the soma into dendrites, we tested if SEPT9 affects dynein-driven motility. In vitro motility assays showed that the velocity and run lengths of dynein-dynactin-BicD2 were reduced on MTs coated with SEPT9 (Figures S7A and S7B). To test whether SEPT9 affects dynein-driven movement in neurons, we used the peroxisome-trafficking assay; the N-terminal domain of the dynein-dynactin adaptor Bicaudal D (BICDN) was targeted to peroxisomes (Kapitein et al., 2010a). SEPT9 knockdown did not alter the polarity index of peroxisomes (Figure S7C), but we observed a subtle change (~10%) in the distribution of peroxisomes in dendrites, which became more distal than proximal upon SEPT9 knockdown; SEPT9 over-expression had the opposite effect (Figure S7D). These data suggest that SEPT9 does not have a strong effect on the dynein-driven motility in neurons, but further studies are required to explore the role of SEPT9 in dynein-driven motility in neurons as the BicD2 adaptor has also been reported to interact with kinesin-1/KIF5 (Grigoriev et al., 2007). Overall, SEPT9 appears to favor movement by kinesin-3/KIF1 over kinesin-1/KIF5, kinesin-2/KIF17, and dynein.
SEPT9 Distinguishes between the L12 Loops of Kinesin-1/KIF5 and Kinesin-3/KIF1A
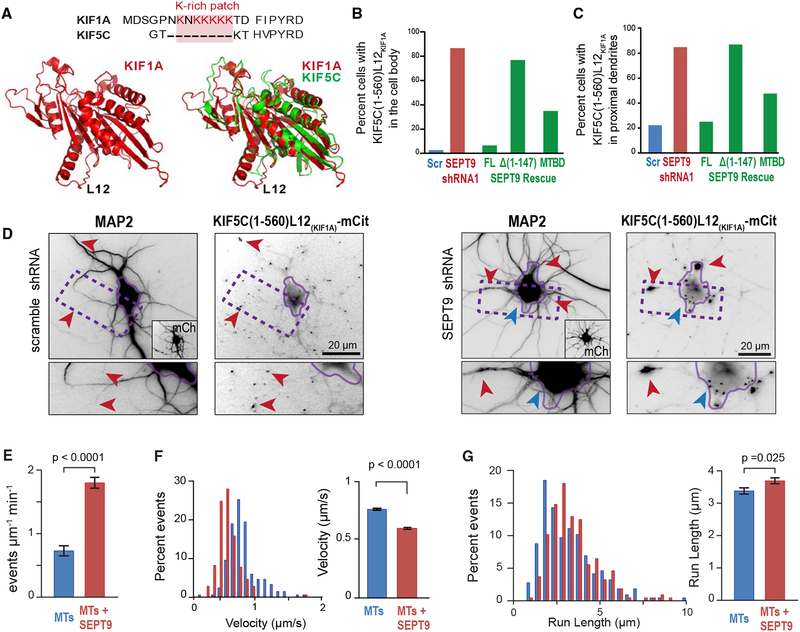
To gain an insight into the mechanism by which SEPT9 impedes axonal cargos of kinesin-1 and boosts dendritic cargos of kinesin-3/KIF1 into dendrites, we sought to identify structural features of kinesin-1/KIF5 and/or kinesin-3/KIF1A that enable SEPT9 to distinguish between them. A salient difference between kinesin-1/KIF5A and kinesin-3/KIF1A is the presence of a lysine-rich patch in the L12 loop of kinesin-3/KIF1A (Figure 7A). Remarkably, the L12 of kinesin-3/KIF1A enables entry into dendrites. A chimeric kinesin-1/KIF5C containing the lysine-rich L12 of kinesin-3/KIF1A (KIF5C-L12KIF1A) enters both axons and den drites rather than axons only, and deletion of the lysine patch of L12 from kinesin-3/KIF1A impairs its translocation to dendritic tips (Huang and Banker, 2012).
Figure 7. SEPT9 Distinguishes between the L12 Loops of Kinesin-1/KIF5 and Kinesin-3/KIF1A.
(A) Sequence alignment (top) of the L12 of KIF1A and KIF5C, highlighting the lysine-rich patch. Cartoon models (bottom) of the structure of the motor domain of KIF1A alone (left) or superimposed onto KIF5C (right).
(B–D) Bar graphs show percentage neurons (n = 61–110) with KIF5C(1–560)-L12KIF1A-mCit in the cell body (B) and proximal dendrites (C). Images (D) show KIF5C(1–560)-L12KIF1A-mCit localization in hippocampal neurons transfected with scramble or SEPT9 shRNAs. Red arrowheads point to KIF1A accumulation at dendritic tips (scramble shRNA), and blue and red arrowheads point to the base and proximal segments of dendrites, respectively (SEPT9 shRNA). Rescue experiments were performed by co-expressing SEPT9 shRNA with shRNA-resistant SEPT9-mCherry, SEPT9-Δ(1–147)-mCherry or SEPT9(1–125)-mCherry (MTBD).
(E) Bar graph shows the mean (±SEM) landing rate of KIF5C(1–560)-L12KIF1A-mCit on MTs alone (n = 17) or His-mCherry-SEPT9 (10 nM)-coated MTs (n = 19). (F and G) Histograms shows the distribution of KIF5C(1–560)-L12KIF1A-mCit velocities (F) and run lengths (G) on MTs alone (n = 217) or MTs coated with 10 nM HismCherry-SEPT9 (n = 210 events). Bar graph shows mean (±SEM) velocity and run length.
To test whether SEPT9 regulates entry into dendrites in a manner that is dependent on the L12 of the kinesin motor domain, we asked if SEPT9 knockdown affects the localization of the chimeric KIF5C(1–560)-L12KIF1A-mCit. In control cells, KIF5C(1–560)-L12KIF1A-mCit localized to the tips of axons and dendrites, resembling the motor domain of kinesin-3/KIF1A (Figures 7B–7D). Strikingly, SEPT9 depletion caused abnormal accumulation of KIF5C(1–560)-L12KIF1A-mCit in initial-proximal dendrites and regions of the cell body at the base of dendritic processes (Figures 7B–7D). This phenotype was rescued with an shRNA-resistant SEPT9 and required the N-terminal MTBD of SEPT9, which alone was capable of partial rescue (Figures 7B and 7C). Therefore, MT-associated SEPT9 appears to distinguish between kinesin-1/KIF5 and kinesin-3/KIF1A based on the presence of a lysine-rich L12, which serves as a token for moving forward into dendrites.
To better understand how SEPT9 promotes the movement of kinesin motors with a K-rich L12, we used in vitro motility assays to analyze the motility of KIF5C(1–560)-L12KIF1A on SEPT9-coated versus SEPT9-free MTs. We found that MT-associated SEPT9 increased by 2-fold the landing rates of KIF5C(1–560)-L12KIF1A (Figure 7E), similar to native KIF1A (Figure 6A). A subtle increase in run length was observed, but velocity decreased as observed for the native kinesin-1/KIF5C (Figures 7F and 7G); the reduction, however, was dampened (Figure 7F). Collectively, these results suggest that SEPT9 selectively promotes the movement of kinesin-3/KIF1A in dendrites owing in part to the presence of a lysine-rich L12, which is critical for productive attachment to MTs.
DISCUSSION
Polarized traffic of membrane proteins into axons and dendrites is thought to depend largely on sorting of vesicles at axonal entry. Here, we found that, upon entry into dendrites, axonally destined vesicles of kinesin-1/KIF5 move with a retrograde bias toward the cell body, while dendritically destined vesicles of kinesin-3/KIF1A move with an anterograde bias deeper into the dendrite. Our data show that selective entry of kinesin-1 motor cargo into axons does not suffice for polarized trafficking, but additional sorting is required during entry into dendrites.
Our results indicate that SEPT9 slows down and halts kinesin-1/KIF5, while it mobilizes and speeds up kinesin-3/KIF1A during entry into dendrites. SEPT9 differentiates between kinesin-1/KIF5- and kinesin-3/KIF1A-driven motility based on the presence of a lysine-rich patch in the L12 of the kinesin motor domain. The lysine-rich patch is present in kinesin-3/KIF1A but absent from kinesin-1/KIF5 (Atherton et al., 2014). Transplantation of the lysine-rich L12 to kinesin-1/KIF5 enables entry into dendrites (Huang and Banker, 2012), which in turn requires SEPT9 for moving further into dendrites. In cell-free motility as-says, MT landing rates of the native kinesin-1/KIF5 are reduced by SEPT9, but addition of the lysine-rich L12 enables SEPT9 to enhance productive attachment to MTs, which appears to be critical for moving motor-cargo complexes farther into dendrites. The lysine-rich L12 interacts with the C-terminal tails (CTT) of β-tubulin, which are also bound by SEPT9, and increases the affinity of kinesin-3/KIF1A to MTs (Atherton et al., 2014; Bai et al., 2013; Okada and Hirokawa, 1999, 2000; Soppina and Verhey, 2014). Given that the L12, tubulin CTTs and the MTBD of SEPT9 are structurally disordered, the precise mechanism of SEPT9-mediated entry into dendrites may involve conformational dynamics, which are inherently challenging to determine. Nevertheless, it appears that SEPT9 affects motor-cargo motility with a high degree of specificity as it does not affect the motility of Rab11, which is transported by KIF13A (Delevoye et al., 2014), a kinesin-3 motor that shares only three out of the 15 residues of the L12 lysine-rich patch of KIF1A. Additionally, SEPT9 does not affect dendritic cargo (e.g., GRIP1), which alters the intrinsic selectivity of kinesin-1/KIF5 for axonal MTs, redirecting it to dendrites (Setou et al., 2002). Under these cargos, kinesin-1/KIF5 may adapt conformations, which evade hindrance by SEPT9.
Unexpectedly, kinesin-3/KIF1A motor cargo was found to move with a directional anterograde bias on the MTs of initial-proximal dendrites, which contain plus- and minus-out MTs at roughly equal percentages (Baas and Lin, 2011). It is puzzling how the processive anterograde events of the kinesin-3/KIF1A motor domain are a lot more frequent than kinesin-1/KIF5 and how the anterograde bias of kinesin-3/KIF1A motor cargo depends on SEPT9. A parsimonious explanation is that SEPT9 marks and/or bridges MTs of plus-end-out orientation. While this article was under review, it was reported that kinesin-1/KIF5 associates preferentially with acetylated MTs of minus-end-out orientation and kinesin-3/KIF1A interacts with dynamic tyrosinated MTs, which are oriented with their plus ends out (Tas et al., 2017). Hence, SEPT9 may have a role in the regulation of membrane traffic on the plus-end-out subsets of MTs. Alternatively, by enhancing the MT binding and velocity of kinesin-3/KIF1A motor cargo, SEPT9 may promote a directional bias that arises stochastically. Differential cargo velocities drive spatial segregation in axons and differential cellular motilities can in part account for the cell sorting events underlying tissue development and patterning (Gumy et al., 2017; Mori et al., 2009).
Previous work has shown that kinesin-3/KIF1A-driven motility is aided in dendrites by doublecortin-like kinase 1 (DCLK1), a dendritic MAP that is hypothesized to promote the processivity of kinesin-3/KIF1A (Lipka et al., 2016; Liu et al., 2012). Hence, kinesin-3/KIF1-driven motility is favored on the MTs of the geometrically complex dendrites. By favoring movement by the faster and super-processive kinesin-3/KIF1, dendritic MAPs may not only prevent membrane traffic jams but also enhance the probability of membrane cargo moving from one dendritic arbor to the next with higher efficiency.
While septins are known to associate with subsets of MTs in non-neuronal cells, their roles in intracellular traffic are little understood. In epithelial cells, where SEPT9 forms heteromeric complexes with SEPT2/6/7, septins are required for transport from the Golgi to the plasma membrane (Spiliotis et al., 2008). In the dendrites of hippocampal neurons, SEPT9 differentially regulates the motility of kinesin motors. Lacking a typical colocalization with SEPT2/6/7, SEPT9 may function in complex with other members of the septin family, such as SEPT4 and SEPT11, which belong to the septin groups of SEPT2 and SEPT6, respectively (Sudo et al., 2007). Alternatively, owing to potentially higher levels of expression in the hippocampus than other septins (Allen, 2004), SEPT9 may function as a homomer; SEPT2 homomerizes into filaments in vitro and we have observed similar homoassembly for SEPT9 (Mendoza et al., 2002). Previous studies have shown that MT acetylation and polyglutamylation, which can modulate MT interactions with motors and MAPs, are functionally dependent on SEPT7 and SEPT2 (Ageta-Ishihara et al., 2013; Froidevaux-Klipfel et al., 2015; Reed et al., 2006; Sirajuddin et al., 2014; Spiliotis et al., 2008). Here, we found no significant changes in the axon-dendrite distribution of MT acetylation and polyglutamylation after SEPT9 knockdown, which suggests that SEPT9 may function independently of SEPT7 and SEPT2. We envisage that various members of the septin family, alone or in combination, could play different roles in the spatial control of membrane traffic, being part of the tubulin and MAP codes of intra-cellular traffic (Atherton et al., 2013; Janke and Kneussel, 2010; Verhey and Gaertig, 2007).
In addition to modulating motor movement on MT tracks, sep-tins are implicated in MT motor-cargo interactions. We recently reported that SEPT9 inhibits the interaction of the NMDA receptor subunit 2 (NR2B) with kinesin-2/KIF17, a motor whose motility is not affected by SEPT9 (Bai et al., 2016). Hence, similar to other MT-binding proteins (e.g., MAP6, MAP1A light chain 3, Hook1), SEPT9 might serve as a membrane-bound cargo or cargo adaptor (Maday et al., 2014). Of note, SEPT9 contains a polybasic membrane-binding domain, which is conserved among several septins. Hence, SEPT9 may shift between MTs and membranes through regulatory mechanisms that elude our present understanding. This shift might be critical for targeting SEPT9 to a subset of MTs as it was recently shown for MAP6, whose palmitoylation is critical for its association with axonal MTs (Tortosa et al., 2017). Future studies will further explore how the septin family regulates microtubule-dependent transport and the underlying mechanisms involved.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Elias T. Spiliotis (ets33@drexel.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary Cultures and Transfection
Primary rat embryonic hippocampal neurons were purchased from the Neuron Culture Service Center (University of Pennsylvania). Neurons were obtained by trypsinization of hippocampi that were isolated from the brains of mixed sex embryos, which were taken from timed pregnant Sprague-Dawley rats at 18 days of gestation. Cells were plated in 24 well plates containing a 12 mm round glass coverslip, coated with 0.5 mg/mL poly-L-Lysine (Sigma) at a cell density of 60,000 cells/well and cultured in Neurobasal medium supplemented with 2% B27 (ThermoFischer), 2 mM GlutaMax, 100 units/mL penicillin/100 mg/mL streptomycin and 2.4 mM cytosine d-D-arabinofuranoside (Sigma). Cells were maintained at 37°C in a 5% CO2 incubator. Neurons were cultured 8–10 days in vitro (DIV) and transfected with Lipofectamine 3000 (Invitrogen) for 48–72 h (DIV10–12) prior to imaging.
METHOD DETAILS
Plasmids and Cloning
Plasmids for PEX-GFP-FKBP, HA-KIF5B(1–807)FRB, KIF1A(1–489)-HA-FRB, KIF17(1–547)-HA-FRB, BicDN(1–594)FRB were previously made and described (Kapitein et al., 2010a). In brief, KIF5B(1–807)-GFP-FRB, KIF1A(1–489)-GFP-FRB and KIF17(1–547)-HA-FRB were amplified from human Kif5B (NM_004521), human KIF1A (NM_004321), and human KIF17 (NM_020816.3) cDNAs and inserted into the pβactin-GFP-FRB vector. HA-BICD2(1–594)-FRB was created by inserting mouse BICD2 (NM_001039179) and the FRB fragment into the pCI-neo vector.
Plasmids encoding for the fluorescently tagged truncated motor constructs KIF5C(1–560)-mCit, KIF1A(1–393)GCN4–3xmCit and the full length KIF1A-3xmCit motor construct were a gift from Dr. Kristen Verhey, University of Michigan and were created by amplifying the rat KIF5C (NM_001107730.1) and rat KIF1A (XM_017596912.1) sequences and inserting them into the mCitrine-N1 or 3xmCitrine-N1 vectors, respectively (Soppina and Verhey, 2014). GFP-KIF17(1–738) which encodes for the human KIF17 (NM_020816.3) was made by truncating the full length pEGFP-C1-KIF17 (Bai et al., 2016) with the QuickChange II site directed muta-genesis kit (Agilent Technologies) using the primer sequences 5’-CTGCCCGTTGTGGACCAGTG AGGTACCGCGGGCCCG-3’ and 5’-CGGGCCCGCGGTACCTCACTGGTCCACAACGGGCAG-3’. GFP-Rab11 (NM_004218.3) cloned under the chicken β-actin vector was a gift from Dr. Gary Banker (Petersen et al., 2014). Fluorescently labeled syntaxin1A-GFP was purchased from addgene (pEGFP(N1)-DCMV-rSyntaxin1a-meGFP; #34631, deposited by the Wolfhard Almers lab). pEGFP-N1-EB3 was a gift from Dr. Peter Baas, Drexel University College of Medicine (Stepanova et al., 2003). The plasmid encoding for NPY-EGFP (JPA5-NPY-EGFP) (Lo et al., 2011) was a gift form Dr. Michael Silverman, Simon Fraser University. pEGFP-C1-GRIP1 (NM_032069.1) (Heisler et al., 2014) was a gift from Dr Matthias Kneussel, University of Hamburg Medical School, Germany. TdT-Bassoon (NM_019146.2) was a gift from Dr Michael Akins, Drexel University and was created by replacing EGFP with tdTomato in the EGFP-Bassoon95–3938 construct (Matz et al., 2010). The plasmid pmCherry-N1-APP695 (NM_000484.3) was a gift from Dr. Aleister Saunders, Drexel University. The Full length human GFP-KIF5B (NM_004521.2) (Jaulin et al., 2007) and YFP-tagged human LDLR (NM_000527) construct (Hunziker et al., 1991), were a gift from Dr. Geri Kreitzer, The City College of New York.
The construction of pmCherry-N1-SEPT9_i1, where SEPT9 (GenBank accession number NP_001106963; isoform a) was inserted into the pmCherry-N1 vector has been described previously (Bai et al., 2016). His-GFP-SEPT9_i1 (GenBank accession number NP_001106963; isoform a) was created by PCR amplifying GFP-SEPT9 and inserting it into the pET-28a(+) vector and has been described previously (Bai et al., 2013). His-mCherry-SEPT9_i1 (NP_001106963; isoform a) was made by two-step cloning. First, SEPT9_i1 was PCR amplified using the primer sequences 5’-CGTAAGCTTATGAAGAAGTCTTACTC-3’ and 5’-GTAGTCGACCTA CATCTCTGGGGC-3’ and inserted into the HindIII and SalI sites of the pmCherry-C1 vector. Next, the mCherry-SEPT9 fragment was PCR amplified from the mammalian construct described above using the primers 5’-ACTCATATGATGGTGAGCAAGGGC GAGG-3’ and 5’-GTAGTCGACCTACATCTCTGGGGC-3’and inserted into the NdeI and SalI sites of the pET28a(+) vector. The recombinant mCherry-tagged MT binding domains of MAP4, His-mCherry-MAP4 (654–1090), was made by two step cloning. First, the eGFP fragment of pEGFP-C1-MAP4(654–1090aa) (Spiliotis et al., 2008) was cut out using the Ase I and BglII enzymes and swapped for mCherry which was cut out from pmCherry-C1, using the same enzymes. Then mCherry-MAP4 was cut out from the latter construct using the NdeI and EcoRI enzymes and pasted in the pET28a vector. Rat SEPT9 transcript variant 1 (GenBank accession number NM_031837) was targeted with scramble shRNA (5’-AGAACGACCGTATGTAATC-3’), SEPT9 shRNA1 (5’-GACCATGAG TATCAAGTCA-3’), and shRNA2 (5’-GACCGACTGGTGAACGAGAAGTT-3’). ShRNA sequences were inserted in the GFP- and mCherry-expressing pG-SUPER and pSuper vectors, respectively. The plasmid for rescue of rat SEPT9 expression was generated by a three-step molecular cloning process. First, the shRNA1 oligos 5’-GATCCCCGACCATGAGTATCAAGTCA TTCAAGAGATG ACTTGATACTCATGGTCTTTTTA-3’ and 5’-AGCTTAAAAAGACCATG AGTATCAAGTCATCTCTTGAATGACTTGATACTCATGGT CGGG-3’ were annealed into the BamHI and HindIII sites of pGFP-V-RS vector. Next, the rat-SEPT9 (GenBank accession number NM_031837)-mCherry sequence was cloned into the BglII and MluI sites of the same vector and finally, the rat SEPT9 sequence was mutated at six different sites to create silent mutations that would not be recognized by the shRNA1. Mutations were done using the QuickChange II Site-directed mutagenesis kit (Agilent Technologies) using the primers 5’-CATTTGCCGTGGTGGGCAGCGATCACGAATACCAGGTAAATGGCAAGAGGATTCTGGGAA-3’ and 5’-TTCCCAGAATCCTCT TGCCATTTACCTGGTATTCGTGATCGCTGCCCACCACGGCAAATG-3’. The plasmid for rescue of rat SEPT9 lacking the MT-binding domain, SEPT9-Δ(1–147), was generated by PCR amplifying the rat SEPT9 (148–576aa)-mCherry sequence from the rescue construct for the full length rat SEPT9 using the primer sequences 5’-CATAGATCTATGGTAGAGACCCCTGCC’3 and 5’-CTAACGCGTCTACTTG TACAGCTCGTC-3’ and inserting SEPT9-Δ(1–147)-mCherry, into the BglII and MluI sites of the pGFP V-RS vector. The construction of the rescue construct containing only the SEPT9 MT-binding N-terminus fragment (1–125 aa) was generated using two step cloning. First rat SEPT9(1–125) was PCR amplified from the full length rat SEPT9 using the primers 5’-CAGCTCGAGATGGAGAGAGATCGC-3’ and 5’-CTAAAGCTTAGTCTTATGGCCCAG-3’ and ligated into the XhoI and HindIII sites of pmCherry-N1. Then rat SEPT9(1–125)-mCherry was cut and pasted into the BglII and NotI sites of pGFP-V-Rs vector that contains SEPT9 shRNA1. The loop L12 chimera KIF5C(1–560)L12KIF1A-mCit was made by inserting the lysine rich patch of the loop L12 of KIF1A (KNKKKKK) into the Loop12 of KIF5C(1–560)-mCit using the QuickChange II Site-directed mutagenesis kit (Agilent Technologies) and the primers 5’-GCCTTGGCA GAAGGGACAAAAAATAAAAAAAAAAAAAAAAAAACACATGTGCCATAC-3’ and 5’-GTATGGCACATGT GTTTTTTTTTTTTTTTTTTTATTTTTTGTCCCTTCTGCC AAGGC-3’.
Protein Expression and Purification of Recombinant SEPT9 and MAP4-MTBD (654–1090)
Plasmids encoding for recombinant SEPT9 (pET28a-GFP-SEPT9 and pET28a-mCherry-SEPT9) or His-mCherry-MAP4(654–1090) (microtubule binding domain; MTBD) were transformed into bacterial cells of the strain Escherichia coli BL21 (DE3) (Invitrogen). After bacterial cultures reached an OD600 of 0.8, protein expression was induced with 0.5 mM IPTG for 3 h at 25°C. Bacteria were centrifuged at 3500 rpm for 10 min at 4°C. Pellets were resuspended in lysis buffer (1% Triton X-100, 50 mM Tris pH 8.0, 2 mM MgCl2, 150 mM NaCl, 10% glycerol, and 20 mM imidazole) and lysed by sonication (8 sets of 15 pulses). Cell lysates were clarified by centrifuging at 21,000 g for 30 min at 4°C. Protein was bound to columns packed with 250 μL of Ni-NTA beads (QIAGEN). Columns were washed 10 ml lysis buffer and protein was eluted with elution buffer (50 mM Tris pH 8.0, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, and 250 mM imidazole) and dialyzed overnight in BRB80 (80 mM K-PIPES, 2 mM MgCl2, 1 mM EGTA, 10% Glycerol pH 6.9).
Single Molecule Motility Assays
Motility chambers were constructed using acid washed glass coverslips (0.15 mm thick) which were mounted on a slide with double sided-tape creating a ~15–20 mL volume chamber. Motility chambers were coated (5 min in room temperature) sequentially with 5 μg/mL biotinylated BSA (Sigma) and 0.5 mg/mL Neutravidin (ThermoFiscer) diluted in BRB80. Microtubules were polymerized in BRB80 (80 mM Pipes, pH 6.9, 1 mM EGTA, 1 mM MgCl2, 10% glycerol) supplemented with 1 mM GTP, for 30 min at 37°C. Then 10 μM of paclitaxel was added and the microtubule mix was incubated in 37°C for an additional 5min. Pre-polymerized taxol-stabilized MTs were made with a mix of unlabeled (80%), biotin-conjugated (10%) and rhodamine- or HiLyte647- labeled (10%) bovine brain tubulin (Cytoskeleton Inc) or a mixture of unlabeled (85%) and biotin (15%) tubulin in BRB80 supplemented with 1mM GTP and 10μM paclitaxel, which were then flooded into the chamber for 15 minutes at a concentration of ~300 nM. Excess tubulin was washed out and the chamber was blocked for 10 min using blocking buffer (1% w/V Pluronic F-127 (Sigma), 1 mg/mL BSA in BRB80). Unlabeled-biotinylated or 647Hilyte-biotinylated MTs were coated for 5 min with 10 nM of His-mCherry SEPT9 (Figures 3,5, and 7) or His-GFP-SEPT9 (Figure S2F) diluted in BRB12 (12 mM K-PIPES, 2 mM MgCl2, 1 mM EGTA, 10% Glycerol pH 6.9).
COS-7 cells were transfected with the motor domain constructs KIF5C(1–560)-3xmCit, GFP-KIF17(1–738), KIF1A(1–393) GCN4–3xmCit or KIF5C(1–560)-L12KIF1A-mCit and were lysed in ice cold lysis buffer (BRB12, 0.5% Triton X-100, 1 mM PMSF, EDTA-free protease inhibitor III (Calbiochem) and 1 mM ATP, pH 6.9). Lysates were clarified by centrifugation (16000 g), and diluted in motility buffer (BRB12, 1mg/mL BSA, 10 μM taxol, 2 mM ATP, 0.5% Pluronic F-127) with an oxygen scavenging system (3.5 μg/mL Catalase, 20 μg/mL Glucose Oxidase, 4.5 mg/mL D-Glucose, 30 mM β-mercaptoethanol). Time-lapse TIRF microscopy (5–10 frames/sec) was done at room temperature using the Deltavision OMX V4 inverted microscope equipped with the Olympus Plan-Apo 60X/1.49 objective. Dynein-Dynactin-BicD2 (DDB) purification and motility assays were performed as described previously (McKenney et al., 2014).
Immunofluorescence
Rat hippocampal neurons were fixed with fixation buffer (PBS, 4% PFA, 4% sucrose) for 10 min, permeabilized with GDB (30 mM sodium phosphate pH 7.4, 0.2% gelatin, 450 mM NaCI) containing 0.05% Triton X-100 for 10 min and blocked with GDB for an additional 20 min. Primary antibodies were diluted in GDB and spun at 100,000 g for 10 min at 4°C before adding on cells. Primary antibodies were incubated for 3 h at room temperature or 4°C overnight. Secondary antibodies were similarly diluted in GDB, spun before adding on cells and incubated for 1 h in room temperature. Samples were mounted with FluorSave hard mounting medium (EMD Millipore).
To stain for MT-associated SEPT9 (Figures 2A, 2D, and 2E), neurons were incubated in extraction buffer (80 mM K-PIPES pH 6.9, 1 mM EGTA, 150 mM NaCl, 7 mM MgCl2, 5 mM D-Glucose, 0.25% glutaraldehyde and 0.1% Triton X-100) for 20 s and then immediately fixed with fixation buffer (PBS, 4% PFA, 4% sucrose). Samples were quenched with 75 mM NH4Cl, permeabilized with GDB containing 0.05% triton X-100 and blocked with GDB for 20 minutes prior to staining.
Antibodies and Reagents
See also the Key Resources Table. DIV12-DIV14 hippocampal neurons were immunostained with the following antibodies: chicken anti-MAP2 (1:2000; EMD Millipore), rabbit anti-SEPT9 (1:200; Proteintech Group), rat anti-SEPT9 (1:200; SIGMA), mouse anti-α-tubulin (DM1α, 1:500; SIGMA), rabbit anti-SEPT2 (a5N5 1:300; gift from Makoto Kinoshita, Nagoya University, Japan), rabbit anti- SEPT7 (1:300; IBL America), rabbit anti-SEPT6 (1:300; gift from Makoto Kinoshita, Nagoya University, Japan), mouse anti-KHC (1:200; EMD Millipore), mouse anti-KIF1A (1:200; BD Biosciences), mouse anti-KIF17 (1:200; M20- Santa Cruz), mouse anti-tau-1 (1:500; EMD Millipore), mouse anti-acetylated tubulin (1:500; T7451 SIGMA), mouse anti-glutamylated tubulin (GT335 1:500; a gift from Dr. C. Janke, institute Curie, France), rabbit anti-APP (1:500; A8717 SIGMA), rabbit anti-Bassoon (1:1000; D63B6 Cell Signaling), rabbit anti-synapsin (1:1000; D12G5 Cell Signaling), mouse anti-SV2 (1:1000; SV2 Iowa DSHB), mouse anti-PSD95 (1:1000; NeuroMAb, Antibodies Incorporated), mouse anti-Ankyrin G (1:50; SC-12719 Santa Cruz) and mouse anti-extracellular pan-neurofascin (1mg/mL; NeuroMab, Antibodies Incorporated). F(ab’)2 fragment affinity-purified secondary antibodies (1:200) were purchased from Jackson ImmunoResearch Laboratories and included donkey anti-mouse, -rabbit, -chicken and -rat antibodies conjugated with AMCA, Alexa488, Alexa594 or Alexa647.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-MAP2 | EMD Millipore | Cat#AB5543 ; RRID:AB_571049 |
| Rabbit anti-septin 9 | PTG labs | Cat#10769-IAP; RRID:AB_2185012 |
| Rat anti-septin 9 | Sigma | Cat#SAB4200191; RRID:AB_10646426 |
| Mouse anti-α-tubulin (DM1 α) | Sigma | Cat#T6199; RRID:AB_477583 |
| Rabbit anti-Septin 2 (clone: a5N5) | Kind gift from M. Kinoshita (Nagoya University, Japan) | N/A |
| Rabbit anti-Septin 6 | Kind gift from M. Kinoshita (Nagoya University, Japan) | N/A |
| Rabbit anti-Septin 7 | IBL America | Cat#18991; RRID:AB_1630825 |
| Mouse anti-KHC (kinesin heavy chain) | EMD Millipore | Cat#MAB1614; RRID:AB_94284 |
| Mouse anti-KIF1A | BD Biosciences | Cat#612094; RRID:AB_399465 |
| Mouse anti-KIF17 (clone: M-20) | Santa Cruz | Cat#sc-26850; RRID:AB_2131285 |
| Mouse anti-Tau-1 | EMD Millipore | Cat#MAB3420; RRID:AB_94855 |
| Mouse anti-glutamylated tubulin (clone: GT335) | Kind gift from C. Janke (Institute Curie, France) | N/A |
| Mouse anti-acetylated tubulin | Sigma | Cat#T7451; RRID:AB_609894 |
| Rabbit anti-APP (C-terminus) | Sigma | Cat#A8717; RRID:AB_258409 |
| Rabbit anti-Bassoon | Cell Signaling | Cat# D63B6; RRID:AB_10828496 |
| Rabbit anti-synapsin | Cell Signaling | Cat# D12G5; RRID:AB_2616578 |
| Mouse anti-SV2 | Iowa DSHB | Cat#SV2; RRID:AB_2315387 |
| Mouse anti-PSD95 | NeuroMAb; Antibodies Incorporated | Cat# 75–028; RRID:AB_2307331 |
| Mouse anti-Ankyrin G | Santa Cruz | Cat#SC-12719 |
| Mouse anti-extracellular Neurofascin | NeuroMAb; Antibodies Incorporated | Cat# 75–172; RRID:AB_626674 |
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) | Invitrogen | Cat#C600003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| A/C Heterodimerizer (Rapamycin analog) | Clontech | Cat#635057 |
| Paclitaxel (Taxol) | ThermoFisher | Cat#P3456 |
| Porcine Tubulin (unlabeled) | Cytoskeleton Inc. | Cat#T240-A |
| Porcine Tubulin (Rhodamine) | Cytoskeleton Inc. | Cat# TL590M |
| Porcine Tubulin (HiLyte647) | Cytoskeleton Inc. | Cat# TL670M |
| Porcine Tubulin Biotin | Cytoskeleton Inc. | Cat# T333P |
| Purified 6XHis-mCherry-hSEPT9 (GenBank accession number NP_001106963; isoform a) protein | This paper | N/A |
| Purified 6XHis-GFP-hSEPT9 (GenBank accession number NP_001106963; isoform a) | Baietal., 2013 | N/A |
| Purified 6XHis-mCherry-MAP4-MTBD(654–1090) protein (GenBank accession number NP_001192259.1) | This paper | N/A |
| Critical Commercial Assays | ||
| Quick Change II Site-directed mutagenesis kit | Agilent Technologies | Cat# 200523 |
| Experimental Models: Cell Lines | ||
| Monkey: COS7 cells | ATCC | CRL-1651 |
| Cultured primary rat hippocampal neurons isolated from embryonic day 18 mixed sex embryos | “Neurons R Us”: the neuron culture service center at the University of Pennsylvania | N/A |
| Oligonucleotides | ||
| Rat SEPT9 scramble shRNA targeting sequence: 5’-AGAACGACCGTATGTAATC-3’ | This paper | N/A |
| SEPT9 shRNA1 targeting sequence: 5’-GACCATGAGTATCAAGTCA-3’ | This paper | N/A |
| SEPT9 shRNA2 targeting sequence: 5’-GACCGACTGGTGAACGAGAAGTT-3’ | This paper | N/A |
| Primer for shRNA resistant SEPT9-FL rescue construct (Forward and Reverse): 5’-CATTTGCCGTGGTGGGCAGCGATCACGAATAC CAGGTAAATGGCAAGAGGATTCTGGGAA-3’ and 5’-TTCCCAGAATCCTCTTGCCATTTACCTG GTATTCGTGATCGCTGCCCACCACGGCAAATG-3’ | This paper | N/A |
| Primer for Rescue SEPT9 ΔMTBD(1–147) construct (Forward and Reverse): 5’-CATAGATCTATGGTA GAGACCCCTGCC’3 and 5’-CTAACGCGTCTACTTG TACAGCTCGTC-3’ | This paper | N/A |
| Primer for Rescue SEPT9 (1–125) construct (Forward and reverse): 5’-CAGCTCGAGATGGAGAGAGAT CGC-3’ and 5’-CTAAAGCTTAGTCTTATGGCCCAG-3’ | This paper | N/A |
| Primer for KIF5C(1–560)-L12KIF1A domain swap: 5’-GCCTTGG CAGAAGGGACAAAAAATAAAAAAAAAAAAAA AAAA ACACATGTGCCATAC-3’ | This paper | N/A |
| Primers for pmCherry-C-SEPT9 (Forward and Reverse): 5’-CGTAAGCTTATGAAGAAGTCTTACTC-3’ and 5’-GT AGTCGACCTACATCTCTGGG-3; | This paper | N/A |
| Primers for His-mCherry-SEPT9 (Forward and Reverse): 5’-ACTCAT ATGATGGTGAGCAAGGGCGAGG-3’ and 5’-GTAGTCGACCTACATCTCTGGGGC-3’ | This paper | N/A |
| Primers for GFP-KIF17(1 −738) Forward and Reverse: 5’-CTGCCCGTTGTGGACCAGTGAGGTACCGCGG GCCCG-3’ and 5’-CGGGCCCGCGGTACCTCACT GGTCCAC AACGGGCAG-3’ | This paper | N/A |
| Recombinant DNA | ||
| Pex-GFP-FKBP | Kapitein et al., 2010b | N/A |
| HA-KIF5B(1–807)FRB | Kapitein et al., 2010b | N/A |
| KIF1A(1–489)-HA-FRB | Kapitein et al., 2010b | N/A |
| KIF17(1–547)-HA-FRB | Kapitein et al., 2010b | N/A |
| HA-BicDN(1–594)FRB | Kapitein et al., 2010b | N/A |
| KIF5C(1–560)-mCit | Kind gift from K.Verhey, University of Michigan | N/A |
| KIF1A(1–393)-GCN4–3xmCit | Kind gift from K.Verhey, University of Michigan | N/A |
| KIF1A-FL-3xmCit | Kind gift from K.Verhey, University of Michigan | N/A |
| pEGFP-C-KIF17(1–178) (GFP-KIF17(1–178) | This paper | N/A |
| GFP-KIF5B | Kind gift by G.Kreitzer, The City College of New York | N/A |
| KIF5C(1–560)L12KIFA −3xmCit | This paper | N/A |
| GFP-RAB11 | Petersen et al., 2014; Gift from G Banker, Ohio State University | N/A |
| pESGP(N1)-ΔCMV-rSyntaxin1a-meGFP(Syntaxin 1A-GFP) | Addgene | Cat#34631, deposited by the Wolfhard Aimers lab |
| pEGFP-N1-EB3 (EB3-GFP) | Kind gift from P.Baas, Drexel University | N/A |
| JPA5-NPY-EGFP | Kind gift from M.Silverman, Simon Frasier University | N/A |
| pEGFP-C1-GRIP1 | Kind gift by M. Kneussel, University of Hamburg Medical School, Germany | N/A |
| TdT-Bassoon | Kind gift by M.Akins, Drexel University | N/A |
| pmCherry-N1-APP | Kind gift by A.Saunders, Drexel University | N/A |
| YFP-hLDLR | Kind gift by G.Kreitzer, The City College of New York | N/A |
| pmCherry-C-hSEPT9 | This paper | N/A |
| pET28a-mCherry-hSEPT9 (6xHIS-mCherry-SEPT9) | This paper | N/A |
| pET28a-mCherry-MAP4 MTBD (654–1090) (6xHIS-mCherry-MAP4(654–1090)) | This paper | N/A |
| pET28a-GFP-hSEPT9 (6xHIS-GFP-SEPT9) | Bai et al., 2013 | N/A |
| pmCherry-N-RatSEPT9_i1 (SEPT9 mch; GenBank accession number NM_031837.1) | This paper | N/A |
| pGFP-VR-S-shRNA1-rescueSEPT9-FL-mCherry | This paper | N/A |
| pGFP-VR-S-shRNA1-rescueSEPT9Δ(1–147)-mCherry | This paper | N/A |
| pGFP-VR-S-shRNA1-rescueSEPT9 (1–125)-mCherry | This paper | N/A |
| Software and Algorithms | ||
| Fiji (ImageJ) | NIH | https://imagej.nih.gov/ij/; RRID:SCR_002285 |
| Slidebook | 3i | https://www.intelligent-imaging.com/; RRID:SCR_014300 |
| ICY | Institut Pasteur | http://icy.bioimageanalysis.org/; RRID:SCR_010587 |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID:SCR_002798 |
| PyMOL | PyMOL | https://pymol.org ; RRID:SCR_000305 |
For induced coupling of immotile peroxisomes to truncated motor domains via chemically dimerizing the mTOR fragments FKBP and FRB that were attached to the PEX tags and motor domains respectively, rapalog A/C (AP21967, Clontech) was used at a concentration of 2 μM for 75–105 minutes.
Time-Lapse Imaging
Live-cell imaging of hippocampal neurons was performed on DIV10–12, 48 h after transfection. Coverslips were mounted on a bottomless 35 mm dish with a 7 mm microwell using vacuum grease silicone (Beckman Coulter). Cell media was replaced with phenol red-free Neurobasal media supplemented with 2% B27 (Invitrogen) and 30 mM HEPES, and dishes were sealed using ParaFilm. Prior to imaging, axons were labeled with a pan-neurofascin antibody (1 μg/mL), which was first diluted in neurobasal media supplemented with 2% B27 and mixed with an anti-mouse secondary (1:200) conjugated to Alexa-647. The mix was incubated at 37°C, 5% CO2 for 15 min and then added to cells for 10 min. After washing, cells were mounted for live-imaging.
In live cell imaging experiments of cells expressing motor and cargo constructs (Figures 1A–1D, 4E–4K, and 6D–6J, Videos S1, S2, S3, and S4), cells were imaged at 10–60 frames/minute for 5–10 minutes.
For live-cell imaging of motor-coupled-to-peroxisome motility (Figures 1E–1H), cells were transfected with GFP-PEX-FKBP and KIF1A(1–489)-FRB or KIF5B(1–807)-FRB for 48h, live-stained with neurofascin and mounted for live-imaging. Transfected cell positions were marked and 2 mM of rapalog was added to the cell chamber which was subsequently sealed with parafilm. Cells were imaged at a rate of 4–6 frames/min for 90 minutes.
Microscopy
Fixed samples were imaged on a Zeiss AxioObserver Z1 inverted microscope equipped with a Zeiss 20X/0.8 dry objective, a 40X/1.2 NA water objective, a 63x/1.4 NA oil objective, a Hamamatsu Orca-R2 CCD camera and the Slidebook 6.0 software. In Figure S4, AnkG and SEPT9 were imaged on an Olympus FV1000 laser scanning confocal inverted microscope equipped with an Olympus 60X/1.42 NA oil objective. Super resolution 3D structured illumination microscopy (SIM) imaging was performed with the OMX V4 microscope (GE Healthcare) using a 60X/1.42 NA objective, a z-step size of 0.125 μm and immersion oil with refractive index 1.514 (GE Healthcare). Total internal reflection fluorescence (TIRF) imaging (20–60 frames per min) of live neurons was performed at 37°C using the TIRF module on the DeltaVision OMX V4 inverted microscope equipped with an Olympus 60x/1.49 objective and a temperature controlled stage-top incubator. Images were acquired with sCMOS pco.edge cameras (PCO) and reconstructed with softWoRx software (Applied Precision).
QUANTIFICATION AND STATISTICAL ANALYSIS
In Vitro Motor Motility
Single motor particles were tracked manually frame-by-frame using Fiji. All counted events were at least 0.6 seconds (or 3–6 frames) in duration and had at least 7mm of microtubule available to run on. Each event start and end frame were recorded to extract event duration. The trajectory of each particle was saved as a region of interest (ROI) and used to measure run lengths. Each event start, end frame, and run length were logged in Microsoft Excel. Event duration was calculated by subtracting the start frame from the end frame and multiplying the difference by the time interval of the motility time-lapse. Velocities were calculated by dividing the run length with duration values. Histograms (Figures 4C, 4D, 6B, 6C, S5C, S5D, S7A, and S7B) were generated by calculating percent frequencies of events. Velocity and run length values were binned and their frequencies were measured by using the frequency function of Excel. Percent frequencies were calculated by dividing the frequencies with the total n value of events and multiplying by 100. Landing rate of motors were measured for each microtubule. Each microtubule was hand-traced and its length was recorded and saved as an ROI. The number of motors that landed and walked processively at least 0.4 mm were counted. The number of events was then divided by the length of the MT (mm) and the duration of the video (s) to extract the landing rates (events* μm−1* min−1). Percent pausing of events (Figure 4B) was calculated by measuring the number of events where the motor paused for more than two frames (400 ms) during a processive run. Those events were then divided by the total number of events per microtubule and multiplied by 100.
Single Particle Tracking Analysis
Single particle tracking (Figures 1A, 1B, 1G, 1H, 4K and 6J) was done manually using Fiji free hand selection tool. Cells were transfected with the plasmids denoted in each figure for 48 hours and then were mounted and imaged live for 5 or 10 minutes (Figures 1A, 1B, 4K, and 6J) or 90 minutes for induced peroxisome motility assays (Figures 1G and 1H). Neurons were stained live with anti-neurofascin in order to distinguish between axons and dendrites. Each particle within the 20 mm proximal to the cell body dendritic region was tracked frame by frame and the directionality and duration of pausing, reversing or processive behavior was recorded. By performing a frame-by–frame tracking of each individual vesicle, the percent frames (time) spent in pausing, anterograde or retrograde movement between consecutive frames was derived for each trajectory (Figures 1B and 1H). The overall trajectory of each vesicle was characterized as moving processively in the anterograde or retrograde direction, being stationary, moving anterogradely with pausing and/or reversing towards the cell body, and moving retrogradely with pausing and/or reversing away from the cell body (Figures 1A and 1G). Note that the kymographs in Figures 1C–1F provide a visual depiction of representative movements, which can also be seen in Videos S1, S2, S3, and S4, and were not used for analysis, which was done by tracking single particles frame-by-frame.
Motility of Motors and Their Cargo in Neurons
The motility of MT motors and their membrane cargo was analyzed using Fiji software. Motor and cargo particles with processive motility were tracked manually using Fiji’s free hand selection tool as described in “in vitro motility” section above. The total run length, duration and average velocity of each motile particle was derived from the start and end points of processive runs. To measure percent motility, the multiple kymograph plugin (with a thickness of 1) was used to generate kymographs and distinguish between motile and immotile particles. EB3 comet directionality was calculated using manual tracking in ICY software or Fiji for 8 mm of dendritic length and 20 mm of axonal length proximal to the cell body.
Image Analysis and Fluorescence Quantifications
Polarity Index Analysis
The polarity index (Idend/Idend+Iaxon; Figures 2B, 3C, 5D, S1A, S2F, S3C, S3D, S4D, S5E, S5F, and S7C) was derived by drawing 1 pixel width ROIs throughout the length of each dendrite, axon and extracellular (background fluorescence) areas using Fiji’s free hand selection tool. The mean fluorescence intensity for dendrites and axon was derived and mean background intensity was subtracted from their values. Using the formula (D-B)/[(D-B)+(A-B)], where D is the mean fluorescence of the motor or cargo in dendrites, A the mean fluorescence in the axon and B is the mean background fluorescence, the polarity index value ranges between 0 (exclusively axonal) and 1 (exclusively dendritic).
Proximal to Distal Dendritic Ratios
In Figure 2C, cells were transfected with pEGFP empty vector and 48 h later were fixed and stained for MAP2 and endogenous SEPT9. GFP was used as a volume marker to generate a distinct mask for each dendrite. The dendritic length of each dendrite was also measured and then divided into two halves (proximal and distal to the cell body). Using these proximal and distal dendrite masks, the sum fluorescence intensity per area were measured for SEPT9 and MAP2 for proximal and distal dendrites, and their ratio was derived for each dendrite.
In Figures 2C, S5E, S5F, S6E, S6F, and S7D, the mCherry protein co expressed with the scramble or SEPT9 shRNAs was used as the cell volume marker and MAP2 was used to distinguish the dendrites from the axon. A single pixel line was drawn across the middle of each dendrite using the free hand tracing tool in Fiji. The dendritic length was divided by two and a single line across the proximal and distal halves were recorded and used to calculate the mean intensity across them. Then, the ratio of the proximal mean intensity was divided by the distal mean intensity and the ratio for each dendrite was recorded. In Figure S7D where the proximal to distal ratio of BicD2-driven peroxisome accumulations were measured, only cells which had peroxisomes in their dendrites were considered; cells with axonal or cell body accumulation were not included for this analysis.
Other Fluorescence Intensity Quantifications
To measure the mean axonal, dendritic and cell body intensities (Figures 5E–5G, 5I, and S6I), the respective cell areas were outlined using the threshold function and wand and free hand tracing tools in Fiji and the mCherry cell volume marker as a guide. The selections were stored as ROIs and their mean intensities were recorded. Intensities of SEPT9 and SEPT7 in Figure S1 were measured in Slidebook. Neurons expressing mCherry-scramble or SEPT9 or SEPT7 shRNAs were outlined using the segmented mask tool in Slidebook. Mask statistics were used to measure the sum intensity of the septin channel and area of cells. The sum intensity was divided by the area of each cell to generate mean intensities. Localization of truncated motors and peroxisomes to axonal and dendritic processes were assessed visually, as was done previously (Huang and Banker, 2012). Each cell was scored based on the loci of motor accumulations as: in axon only, in axons and dendrites, in cell body, in proximal or distal dendrites and in dendritic tips. Pre-synaptic Bassoon, SV2 and synapsin or post synaptic PSD95 punta (Figures 3D and 5J, respectively) were counted manually in cells expressing scramble or SEPT9 shRNAs, and the number of puncta was divided by the length of the neuronal process. Axonal initial segments (neurofascin live-staining and AnkG staining) were outlined using Fiji’s polygon selection tool and their perimeter, area and mean intensity was measured. Line scan graphs (Figures 2E–2G) were generated using the line intensity function in Slidebook 6.0 software.
Kinesin 1/KIF5C and Kineisn3/KIF1A Motor Structures
Ribbon cartoon structures in Figure 7A were generated by inserting the structures for the mouse KIF1A (PDB ID: 1I5S) and KIF5C (PDB ID: 3X2T) motors into the 3D structure visualization software PyMOL. The structures were rotated identically to highlight the Loop 12 and superimposed for comparison.
Statistical Analysis
Statistical analysis of data was done using GraphPad Prism 6.0 software. Each data set was tested for normal distribution of variance using the D’Agostino and Pearson normality test. p values were derived using a student’s t-test for normally distributed data and the Mann Whitney U test for non-normally distributed data. P values lower than 0.05 were considered statistically significant. Data graphs were plotted in GraphPad Prism using the scatter dot plot function showing the mean and standard deviation (SD) of the mean or the bar graph plot function showing the standard error of the mean (SEM). N values and p values for each experiment are denoted in the corresponding figure and/or figure legend.
Supplementary Material
Highlights.
Kinesin motor cargos enter dendrites with differential motility and directional bias
SEPT9 is a dendritic MAP that impedes movement of kinesin-1/KIF5 and axonal cargo
SEPT9 promotes anterograde movement of kinesin-3/KIF1A and its dendritic cargo
SEPT9 distinguishes between loops L12 of KIF5 and KIF1A for entry into dendrites
ACKNOWLEDGMENTS
We are grateful to Drs. M. Akins, P. Baas, G. Banker, M. Kinoshita, M. Kneussel, G. Kreitzer, A. Saunders, M. Silverman, and K. Verhey for plasmids and antibodies. We thank Drs. R. Petrie, S. Mostowy, and L. Dolat for comments on the manuscript and Ms Ayana Suber for help with analysis. All microscopy was performed at Drexel University’s Cell Imaging Center. The work was funded by NIGMS grant R01GM097664 to E.T.S. and NCI pre-doctoral fellowship F31CA213776 to E.P.K. R.J.M. is supported by grants R00NS089428 from NINDS and R35GM124889 from NIGMS.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and four videos and can be found with this article online at https://doi.org/10.1016/j.devcel.2018.06.013.
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
REFERENCES
- Ageta-Ishihara N, Miyata T, Ohshima C, Watanabe M, Sato Y, Hamamura Y, Higashiyama T, Mazitschek R, Bito H, and Kinoshita M (2013). Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun 4, 2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam S, Xu M, Wandless TJ, and Arnold DB (2012). Differential trafficking of transport vesicles contributes to the localization of dendritic proteins. Cell Rep. 2, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen P (2004). Allen atlas: mouse brain (http://mouse.brain-map.org/gene/show/33155).
- Atherton J, Farabella I, Yu IM, Rosenfeld SS, Houdusse A, Topf M, and Moores CA (2014). Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife 3, e03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J, Houdusse A, and Moores C (2013). MAPping out distribution routes for kinesin couriers. Biol. Cell 105, 465–487. [DOI] [PubMed] [Google Scholar]
- Baas PW, and Lin S (2011). Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev. Neurobiol 71, 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Bowen JR, Knox TK, Zhou K, Pendziwiat M, Kuhlenbaumer G, Sindelar CV, and Spiliotis ET (2013). Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. J. Cell Biol. 203, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Karasmanis EP, and Spiliotis ET (2016). Septin 9 interacts with kinesin KIF17 and interferes with the mechanism of NMDA receptor cargo binding and transport. Mol. Biol. Cell 27, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Inoue T, Nakayama T, Hattori M, and Mikoshiba K (2004). Kinesin dependent, rapid, bi-directional transport of ER sub-compartment in dendrites of hippocampal neurons. J. Cell Sci. 117, 163–175. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS (2014). Adaptor proteins involved in polarized sorting. J. Cell Biol. 204, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakar L, Falk J, Ducuing H, Thoinet K, Reynaud F, Derrington E, and Castellani V (2017). Molecular memory of morphologies by septins during neuron generation allows early polarity inheritance. Neuron 95, 834–851.e5. [DOI] [PubMed] [Google Scholar]
- Bowen JR, Hwang D, Bai X, Roy D, and Spiliotis ET (2011). Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J. Cell Biol. 194, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Pan PY, and Sheng ZH (2007). Syntabulin-kinesin-1 family member 5B-mediated axonal transport contributes to activity-dependent presynaptic assembly. J. Neurosci 27, 7284–7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, and Barral Y (2009). Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell 16, 493–506. [DOI] [PubMed] [Google Scholar]
- Craig AM, Jareb M, and Banker G (1992). Neuronal polarity. Curr. Opin. Neurobiol 2, 602–606. [DOI] [PubMed] [Google Scholar]
- Delevoye C, Miserey-Lenkei S, Montagnac G, Gilles-Marsens F, Paul-Gilloteaux P, Giordano F, Waharte F, Marks MS, Goud B, and Raposo G (2014). Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 6, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias GG, Guardia CM, Britt DJ, Guo X, and Bonifacino JS (2015). Sorting of dendritic and axonal vesicles at the pre-axonal exclusion zone. Cell Rep. 13, 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese CK, Mikhaylova M, Stucchi R, Gautier V, Liu Q, Mohammed S, Heck AJR, Altelaar AFM, and Hoogenraad CC (2017). Quantitative map of proteome dynamics during neuronal differentiation. Cell Rep. 18, 1527–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froidevaux-Klipfel L, Targa B, Cantaloube I, Ahmed-Zaid H, Pous C, and Baillet A (2015). Septin cooperation with tubulin polyglutamylation contributes to cancer cell adaptation to taxanes. Oncotarget 6, 36063–36080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchtbauer A, Lassen LB, Jensen AB, Howard J, Quiroga Ade S, Warming S, Sorensen AB, Pedersen FS, and Fuchtbauer EM (2011). Septin9 is involved in septin filament formation and cellular stability. Biol. Chem 392, 769–777. [DOI] [PubMed] [Google Scholar]
- Ghiretti AE, Thies E, Tokito MK, Lin T, Ostap EM, Kneussel M, and Holzbaur ELF (2016). Activity-dependent regulation of distinct transport and cytoskeletal remodeling functions of the dendritic kinesin KIF21B. Neuron 92, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I, Splinter D, Keijzer N, Wulf PS, Demmers J, Ohtsuka T, Modesti M, Maly IV, Grosveld F, Hoogenraad CC, et al. (2007). Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13, 305–314. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Katrukha EA, Grigoriev I, Jaarsma D, Kapitein LC, Akhmanova A, and Hoogenraad CC (2017). MAP2 defines a pre-axonal filtering zone to regulate KIF1-versus KIF5-dependent cargo transport in sensory neurons. Neuron 94, 347–362.e7. [DOI] [PubMed] [Google Scholar]
- Heisler FF, Lee HK, Gromova KV, Pechmann Y, Schurek B, Ruschkies L, Schroeder M, Schweizer M, and Kneussel M (2014). GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. Proc. Natl. Acad. Sci. USA 111, 5030–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Niwa S, and Tanaka Y (2010). Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638. [DOI] [PubMed] [Google Scholar]
- Hu J, Bai X, Bowen JR, Dolat L, Korobova F, Yu W, Baas PW, Svitkina T, Gallo G, and Spiliotis ET (2012). Septin-driven coordination of actin and microtubule remodeling regulates the collateral branching of axons. Curr. Biol 22, 1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, and Banker G (2012). The translocation selectivity of the kine-sins that mediate neuronal organelle transport. Traffic 13, 549–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker W, Harter C, Matter K, and Mellman I (1991). Basolateral sorting in MDCK cells requires a distinct cytoplasmic domain determinant. Cell 66, 907–920. [DOI] [PubMed] [Google Scholar]
- Janke C, and Kneussel M (2010). Tubulin post-translational modifications: encoding functions on the neuronal microtubule cytoskeleton. Trends Neurosci. 33, 362–372. [DOI] [PubMed] [Google Scholar]
- Jaulin F, Xue X, Rodriguez-Boulan E, and Kreitzer G (2007). Polarization-dependent selective transport to the apical membrane by KIF5B in MDCK cells. Dev. Cell 13, 511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B, Decker H, Bentley M, Luisi J, and Banker G (2012). A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J. Cell Biol. 198, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Stokin GB, Yang Z, Xia CH, and Goldstein LS (2000). Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron 28, 449–459. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, and Hoogenraad CC (2011). Which way to go? Cytoskeletal organization and polarized transport in neurons. Mol. Cell. Neurosci 46, 9–20. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, and Hoogenraad CC (2010a). Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr. Biol 20, 290–299. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, van der Zwan WA, Wulf PS, Keijzer N, and Hoogenraad CC (2010b). Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys. J 99, 2143–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Froese CD, Estey MP, and Trimble WS (2011). SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers M, van de Willige D, Freal A, Chazeau A, Franker MA, Hofenk J, Rodrigues RJ, Kapitein LC, Akhmanova A, Jaarsma D, et al. (2016). Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron 89, 461–471. [DOI] [PubMed] [Google Scholar]
- Li P, Merrill SA, Jorgensen EM, and Shen K (2016). Two clathrin adaptor protein complexes instruct axon-dendrite polarity. Neuron 90, 564–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka J, Kapitein LC, Jaworski J, and Hoogenraad CC (2016). Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 35, 302–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JS, Schubert CR, Fu X, Fourniol FJ, Jaiswal JK, Houdusse A, Stultz CM, Moores CA, and Walsh CA (2012). Molecular basis for specific regulation of neuronal kinesin-3 motors by doublecortin family proteins. Mol. Cell 47, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, and Silverman MA (2011). KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci. Lett 491, 168–173. [DOI] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, and Holzbaur EL (2014). Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84, 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz J, Gilyan A, Kolar A, McCarvill T, and Krueger SR (2010). Rapid structural alterations of the active zone lead to sustained changes in neuro-transmitter release. Proc. Natl. Acad. Sci. USA 107, 8836–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney R, Huynh W, Tanenbaum M, Bhabha G, and Vale R (2014). Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M, Hyman AA, and Glotzer M (2002). GTP binding induces filament assembly of a recombinant septin. Curr. Biol 12, 1858–1863. [DOI] [PubMed] [Google Scholar]
- Mok H, Shin H, Kim S, Lee JR, Yoon J, and Kim E (2002). Association of the kinesin superfamily motor protein KIF1Balpha with postsynaptic density-95 (PSD-95), synapse-associated protein-97, and synaptic scaffolding molecule PSD-95/discs large/zona occludens-1 proteins. J. Neurosci 22, 5253–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Gjorevski N, Inman JL, Bissell MJ, and Nelson CM (2009). Self-organization of engineered epithelial tubules by differential cellular motility. Proc. Natl. Acad. Sci. USA 106, 14890–14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, and Cossart P (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194. [DOI] [PubMed] [Google Scholar]
- Nakata T, and Hirokawa N (2003). Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J. Cell Biol. 162, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, and Hirokawa N (1999). A processive single-headed motor: kinesin superfamily protein KIF1A. Science 283, 1152–1157. [DOI] [PubMed] [Google Scholar]
- Okada Y, and Hirokawa N (2000). Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA 97, 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JD, Kaech S, and Banker G (2014). Selective microtubule-based transport of dendritic membrane proteins arises in concert with axon specification. J. Neurosci 34, 4135–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, and Verhey KJ (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol 16, 2166–2172. [DOI] [PubMed] [Google Scholar]
- Sellin ME, Sandblad L, Stenmark S, and Gullberg M (2011). Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, and Hirokawa N (2002). Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417, 83–87. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, and Poo MM (2009). A selective filter for cytoplasmic transport at the axon initial segment. Cell 136, 1148–1160. [DOI] [PubMed] [Google Scholar]
- Soppina V, and Verhey KJ (2014). The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Mol. Biol. Cell 25, 2161–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, and Gladfelter AS (2012). Spatial guidance of cell asymmetry: septin GTPases show the way. Traffic 13, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, Hunt SJ, Hu Q, Kinoshita M, and Nelson WJ (2008). Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J. Cell Biol. 180, 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, and Galjart N (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci 23, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Cai Q, Gerwin C, Smith CL, and Sheng ZH (2004). Syntabulin is a microtubule-associated protein implicated in syntaxin transport in neurons. Nat. Cell Biol. 6, 941–953. [DOI] [PubMed] [Google Scholar]
- Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, and Nagata K (2007). SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum. Mutat 28, 1005–1013. [DOI] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, and Sheng M (2007). Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol 17, 1752–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, and Bradke F (2009). Neuronal polarity. Cold Spring Harb. Perspect. Biol 1, a001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T, Xu C, Funahashi Y, Namba T, and Kaibuchi K (2015). Neuronal polarization. Development 142, 2088–2093. [DOI] [PubMed] [Google Scholar]
- Tas RP, Chazeau A, Cloin BMC, Lambers MLA, Hoogenraad CC, and Kapitein LC (2017). Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron 96, 1264–1271.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa E, Adolfs Y, Fukata M, Pasterkamp RJ, Kapitein LC, and Hoogenraad CC (2017). Dynamic palmitoylation targets MAP6 to the axon to promote microtubule stabilization during neuronal polarization. Neuron 94, 809–825.e7. [DOI] [PubMed] [Google Scholar]
- van Beuningen SF, Will L, Harterink M, Chazeau A, van Battum EY, Frias CP, Franker MA, Katrukha EA, Stucchi R, Vocking K, et al. (2015). TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 88, 1208–1226. [DOI] [PubMed] [Google Scholar]
- Verhey KJ, and Gaertig J (2007). The tubulin code. Cell Cycle 6, 2152–2160. [DOI] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, and Kiebler MA (2007). The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol 17, 1746–1751. [DOI] [PubMed] [Google Scholar]
- Yau KW, Schatzle P, Tortosa E, Pages S, Holtmaat A, Kapitein LC, and Hoogenraad CC (2016). Dendrites in vitro and in vivo contain microtubules of opposite polarity and axon formation correlates with uniform plus-end-out microtubule orientation. J. Neurosci 36, 1071–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, et al. (2001). Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell 105, 587–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.