Abstract
Objective(s):
Previous studies have reached different conclusions regarding an association between apolipoprotein E (APOE) polymorphisms and depression. This meta-analysis was designed to clarify these controversies.
Materials and Methods:
Literatures were identified reviewing the national and international databases. The eligible articles for meta-analysis were determined by quality assessment and implementation of inclusion/exclusion criteria. This meta-analysis was performed using Review Manager 5.2 software. The odds ratios (ORs) with corresponding 95% confidence interval (CIs) were calculated using a fixed effects model. Funnel plots and Egger’s regression tests were used to assess the publication bias.
Results:
A total of nine studies that met the inclusion criteria were identified by performing a comprehensive search on the association between APOE polymorphisms and depression. APOE ε4 allele was significantly associated with depression (allele: OR=1.36, 95%CI=1.11-1.66, P=0.003; dominant: OR=1.34, 95%CI=1.06-1.68, P=0.001; recessive: OR= 1.11, 95%CI =0.45-2.76, P=0.82). HAMD scores were higher in depression patients with-APOE ε4 genotype than who without-APOE ε4 genotype (OR=0.96, 95%CI=0.16-1.76, P=0.02).
Conclusion:
APOE ε4 allele increased the depression risk; depressive patients carrying APOE ε4 allele had more severe depressive symptoms.
Key Words: Apolipoprotein E, Depression, Meta-analysis, Polymorphism, ε4 allele
Introduction
Depression is a serious mental disorder affecting human health that is characterized by mood or emotional dysfunction (1). The World Health Organization speculated that by 2020, depression will become one of the main reasons that individuals are unable to work (2). The depression incidence rate has risen in recent years. The current prevalence of depression is 3-5%, it accounts for the second-highest economic burden of disease (3, 4).
Figure 2.
Forest plots for estimation of the association between APOE ε4 allele and depression in different genetic model
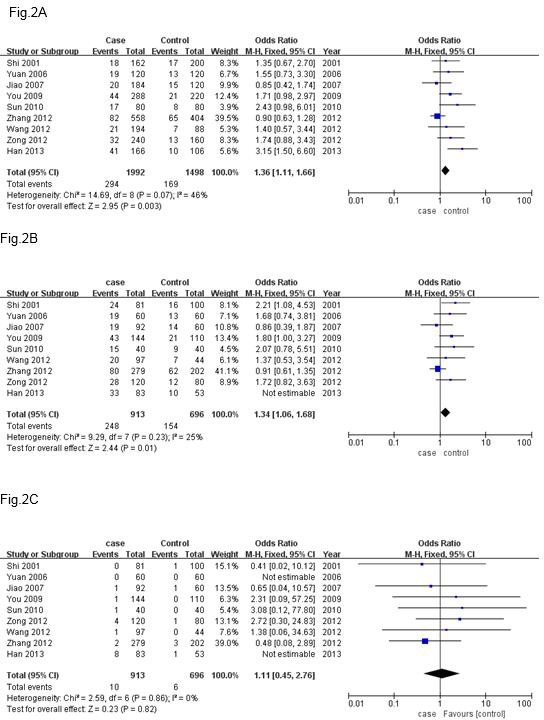
A: Allele model; B: Dominant model; C: Recessive model
The relationship between genetic makeup and susceptibility to depression had long been researched on the basis of family and twin studies. A meta-analysis of seven twin studies on depression shown the heritability impact of depression accounted for 37% while environmental accounting for 63% (5).The tremendous advances in the areas of molecular genetics, biotechnology and sophisticated statistical methods for analyzing complex mode of inheritance created new opportunities to detect so-called susceptibility alleles that increase the risk for depression. Apolipoprotein E (APOE) included ε2, ε3, and ε4 three alleles might be a susceptibility gene of depression. The molecular basis of the APOE gene polymorphisms are differences at two sites in the amino acid sequence (residues 112 and 158). Some have reported that APOE gene polymorphisms were a risk factor for triggering depression (6-9). Interestingly, APOE ε4 allele impact has been found to differ across ethnic groups. The relative frequencies of the ε2, ε3, and ε4 alleles were 5.3%, 88.4%, and 6.3%, respectively in human (10), the relative frequency of ε4 was the lowest among all ethnic groups (11).
APOE gene polymorphism status is hypothesized to be a risk factor for depression (7). However, studies have reached inconsistent conclusions regarding APOE polymorphism frequencies in subjects with depression. We performed a meta-analysis of recent studies to assess the relationship between APOE ε4 allele and depression.
Materials and Methods
Literature search
The databases PubMed, Web of Science, Wiley Online Library, EMBASE, CBM (Chinese Biomedical Database, http://www.sinomed.ac.cn/), CNKI (Chinese National Knowledge Infrastructure, http://oversea.cnki.net/), VIP (http://www.cqvip.com/), and Wanfang (http://www.wanfangdata.com.cn/) were used to search the literature with the following keywords: (˝Apoprotein E˝ OR ˝apolipoprotein E˝ OR ˝APOE˝) AND (˝polymorphism˝ OR ˝mutation˝ OR ˝variant˝) AND depression. We did not impose a language restriction, and we searched the literature published up to February 2018. The most recent results were included when there were multiple publications from the same research group.
Inclusion and exclusion criteria
Eligible studies fulfilled the following inclusion criteria: (a) a case-control study design; (b) a focus on the association between APOE polymorphisms and depression; (c) the original data, genotype, and allele data are provided; (d) ages of onset < 60, the objects are diagnosed no cognitive impairment (such as Alzheimer’s disease et al.) through long-term follow-up studies; and (e) a control group with no history of mental disorders and genotypes in Hardy–Weinberg equilibrium (HWE).
The exclusion criteria were: (a) insufficient information for data extraction, (b) missing information of APOE genotype/allele frequency, (c) reviews or case reports.
The data included in this meta-analysis were obtained from the published literature, so written consent by the subjects and ethics committee approval were not required.
Data extraction
According to the inclusion and exclusion criteria, two authors (WW Wang and TH Bao) independently extracted data, including the first author, year of publication, ethnicity, frequencies of APOE polymorphisms, and information of research objects in cases and controls. Disagreements on study eligibility were resolved by consulting another author (Ye Ruan).
Statistical analysis
The statistical analysis was conducted using Review Manager 5.2 software. A chi-square test was used to test whether the control group conformed to HWE. Differences were considered statistically significant at P<0.05.
Heterogeneity between the studies was detected using Cochran’s Q test and I2 statistics (12), with P>0.10 or I2<50% indicating homogeneity. A fixed effects model was used to calculate the odds ratio (OR) and 95% confidence interval (95% CI). These values were used to determine the association strength between APOE polymorphisms and depression. A random effects model was used if I2>50% (13). In this meta-analysis, we studied on an association between APOE ε4 allele and depression, so we used genetic models as that the allele model (ε4 vs. ε2+ε3), the dominant model (ε4/ε4+ε4/ε3+ε4/ε2 vs. ε2/ε3+ε2/ε2+ε3/ε3), and the recessive model (ε4/ε4 vs. ε2/ε2+ε2/ε3ε+ε3/ε3+ε2/ε4+ε3/ε4).
We deleted one study from our meta-analysis at a time to evaluate the potential influences of any single study included in our research. Funnel plots and Egger’s tests were used to assess for possible publication bias. Egger’s test (14) was performed by STATA12.0 software. Potential publication bias was noted when a funnel plot was asymmetric or Egger’s test yielded P<0.05.
Results
Included studies
A total of 398 papers were retrieved in a preliminary literature search. After applying our inclusion and exclusion criteria, 9 case–control studies including 903 cases with depression and 737 controls were eligible (15-23). The study selection flow chart is depicted in Figure 1. None of the control groups in this meta-analysis departed from HWE (P>0.05). The characteristics of the included papers are presented in Table 1-2.
Figure 1.
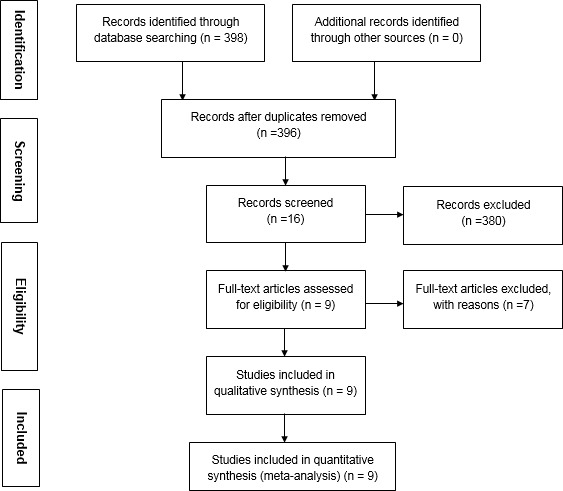
Flow diagram of the study selection process
Table 1.
Characteristics of the studies included in this meta-analysis
| Author | Year | Ethnicity | Cases |
Controls |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ε2/ε2 | ε3/ε3 | ε4/ε4 | ε2/ε3 | ε2/ε4 | ε3/ε4 | ε2/ε2 | ε3/ε3 | ε4/ε4 | ε2/ε3 | ε2/ε4 | ε3/ε4 | |||
| Shi et al (18) | 2001 | Han | 2 | 55 | 0 | 6 | 1 | 8 | 11 | 69 | 1 | 4 | 0 | 15 |
| Yuan et al (19) | 2006 | Han | 1 | 35 | 0 | 5 | 4 | 15 | 2 | 41 | 0 | 4 | 2 | 11 |
| Jiao et al (20) | 2007 | Han | 1 | 62 | 1 | 10 | 2 | 16 | 0 | 39 | 1 | 7 | 2 | 11 |
| You et al (16) | 2009 | Han | 3 | 76 | 1 | 22 | 11 | 31 | 2 | 55 | 0 | 32 | 6 | 15 |
| Sun et al (17) | 2010 | Han | 0 | 21 | 1 | 4 | 3 | 11 | 1 | 26 | 0 | 4 | 1 | 8 |
| Zhang et al (22) | 2012 | Han | 4 | 147 | 2 | 48 | 24 | 54 | 2 | 108 | 3 | 30 | 15 | 44 |
| Zong et al (15) | 2012 | Han | 2 | 74 | 4 | 16 | 4 | 20 | 2 | 54 | 1 | 12 | 2 | 9 |
| Wang et al (21) | 2012 | Han | 2 | 61 | 1 | 14 | 5 | 14 | 1 | 26 | 0 | 10 | 0 | 7 |
| Han et al (23) | 2015 | Han | 0 | 41 | 8 | 9 | 1 | 24 | 1 | 36 | 1 | 7 | 1 | 7 |
Cases: Patients diagnosed with depression; Controls: Healthy people.
Table 2.
Information of the Research object included in this meta-analysis
| Cases |
Controls |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Diagnostic criteria | Male | Female | Age | Comorbidity | MMSE OR HAMD(SCORE) | Male | Female | Age | HAMD (SCORD) |
| Shi et al (18) | 2001 | CCMD-2R | 30 | 51 | 21-78 | No | - | 38 | 62 | 23-83 | - |
| Yuan et al (19) | 2006 | DSM-IV&CCMD-3 | 15 | 45 | 60-82 | Yes | - | 18 | 42 | 60-78 | <7 |
| Jiao et al (20) | 2007 | CCMD-3 | 52 | 40 | 18-65 | No | HAMD>17 | 29 | 31 | 27-50 | - |
| You et al (16) | 2009 | DSM-IV | 66 | 78 | 62-85 | No | MMSE>24;HAMD>30 | 50 | 60 | 63-78 | <7 |
| Sun et al (17) | 2010 | DSM-IV&CCMD-3 | 18 | 22 | 60-86 | No | MMSE:10-24;HAMD>20 | 16 | 24 | 60-81 | - |
| Zhang et al (22) | 2012 | DSM-IV&CCMD-3 | - | - | 15-80 | - | HAMD>17 | 51 | 151 | 16-75 | <7 |
| Zong et al (15) | 2012 | DSM-IV | 36 | 84 | 60-75 | no | HAMD>20 | 25 | 55 | 60-74 | <8 |
| Wang et al (21) | 2012 | DSM-IV | 33 | 64 | 55-78 | no | MMSE>24;HAMD>17 | 21 | 23 | 55-78 | <7 |
| Han et al (23) | 2015 | DSM-IV | 32 | 51 | 43-80 | no | HAMD>20 | 26 | 27 | 42-78 | <8 |
Cases: Patients diagnosed with depression; Controls: Healthy people.-: this information was not provided in the primary papers.MMSE OR HAMD (SCORE): Some primary papers provided HAMD (SCORE) only when others provided MMSE (SCORE).Comorbidity: Refers to cardiovascular, cerebrovascular disease and other mental disorders.CCMD: Chinese classification of mental disorders; DSM: Diagnostic and statistical manual of mental disorders.
APOE ε4 allele was associated with depression
In this meta-analysis, an association was detected between APOE ε4 allele and depression in three genetic models (allele model, dominant model, and recessive model). The allele and dominant models showed statistically significant differences. There were no statistically significant differences in the recessive model (Table 3, allele: OR=1.36, 95%CI=1.11-1.66, P=0.003; dominant: OR=1.34, 95%CI=1.06-1.68, P=0.001; recessive: OR= 1.11, 95%CI =0.45-2.76, P=0.82). These results showed significant associations between APOE ε4 genotype and depression. The Forest plots were presented in 2.
Table 3.
Pooled odds ratio for APOE ε4 allele in meta-analyses
| Gene model | Gene formula | Pooled OR(95%CI) | Z score | P-value a | P-value b | P-value c | I 2 |
|---|---|---|---|---|---|---|---|
| Allele | ε4 vs. ε2+ε3 | 1.36(1.11-1.66) | 2.95 | 0.003 | 0.047 | 0.07 | 46% |
| Dominant | ε4/ε4+ε4/ε3+ε4/ε2 vs. ε2/ε3+ε2/ε2+ε3/ε3 | 1.34(1.06-1.68) | 2.44 | 0.001 | 0.146 | 0.23 | 25% |
| Recessive | ε4/ε4 vs. ε2/ε2+ε2/ε3ε+ε3/ε3+ε2/ε4+ε3/ε4 | 1.11(0.45-2.76) | 0.23 | 0.82 | 0.874 | 0.86 | 0% |
OR: odd ratio; CI: confidence interval
: Fix effect model was used in the meta-analysis to calculate Z score, P<0.05 was considered statistically significant.
: Egger’s test to assess publication bias, P<0.05 was considered potential publication bias.
: Heterogeneity test was assess using Cochran’s (Q) x2 test, P<0.01 was considered potential heterogeneity.
The fixed effects model was used for the allele, dominant, and recessive models because the heterogeneities were not significant (I2=46%, 25%, and 0%, respectively). The sensitivity analysis indicated what that deleting one study at a time did not affect the outcomes of the three genetic models.
We did not find any evidence of publication bias in the dominant and recessive models on funnel plots (Figure 3) or by Egger’s tests (P=0.047, 0.146, and 0.874, respectively).
Figure 3.
Funnel plots for estimation of the association between APOE ε4 allele and depression in different genetic model
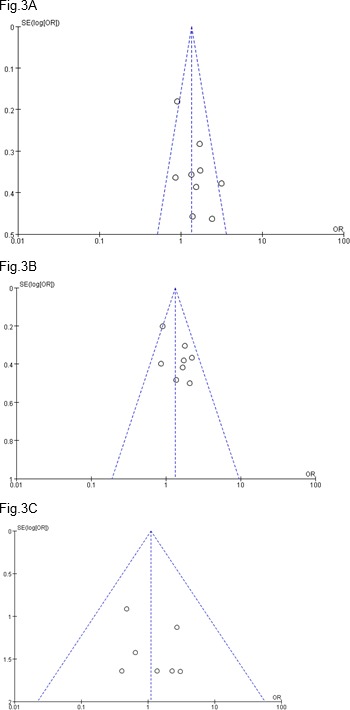
A: Allele model; B: Dominant model; C: Recessive model
Depression patients carrying APOE ε 4 genotype got a higher HAMD score
Depression patients were divided into with-APOE ε4 genotype group and without-APOEε4 genotype group. The differences of HAMD score between the two groups were observed. For I2=91%, the random effects model was used. Only four studies provided HAMD scores of depression patients with and without APOE ε4 genotype. It shown depression patients carrying APOE ε4 genotype got higher HAMD score, the difference was statistically significant (OR=0.96, 95%CI=0.16-1.76, P=0.02). This indicated that APOE ε4 genotype associated with severity of depression. The Forest plot was presented in Figure 4A. The publication bias was not seen in Figure 4B.
Figure 4.
Estimation of the association between APOE ε4 allele and HAMD score in depression
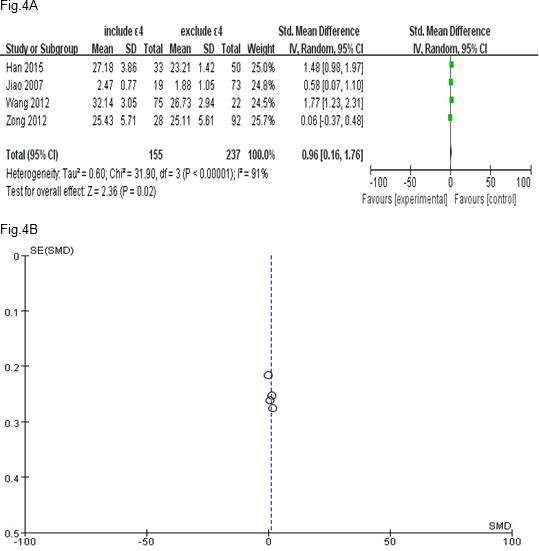
A: Forest plots for the effects of APOE ε4 allele on HAMD score in depression
B: Funnel plots for publication bias, there was no publication bias
Discussion
Depression was a kind of mental disorders with a high recurrence rate that poses serious hazards to human health. The pathogenesis of depression was complex and could be affected by factors such as personality, genetics, and social environments Hypotheses of depression pathogenesis included inflammation, monoamine neurotransmitters and their receptors, hypothalamic-pituitary-adrenal (HPA) axis dysfunction, and the neurotropic factor (24), however the exact pathogenesis’ mechanisms of depression remain unclear.
APOE was playing a role in nervous system growth and injury restoration. APOE ε2, ε3, and ε4 alleles can combine to yield six genotypes (homozygotes for ε2, ε3, and ε4; heterozygotes for ε2/3, ε2/4, and ε3/4). The frequencies of alleles and genotypes were difference among different ethnic populations and geographic areas (25). Plasma concentrations of total cholesterol and low-density lipoprotein in convalescing depressed patients could predict the risk of suicide during the next depressive episode (26). Low concentrations of cholesterol in serum were associated with depression risk (27). Many depressed patients exhibit neuronal loss and brain structural abnormalities, which might be a result of dyslipidemia (28). Depressed patients had a higher APOE ε4 allele frequency, the APOE ε4 polymorphism was a risk factor of depression (29-30). Compared with patients who did not carry a ε4 allele, patients with the ε4 allele exhibited more obvious depressive symptoms (31).
This meta-analysis reviewed the existing eligible studies and examined the association between APOE ε4 allele and depression. In this study, 9 papers were included to observant the affection of APOE ε4 status on depression. The results shown APOE ε4 allele was associated with depression significantly. Depression patients carrying ε4 allele had higher HAMD score. The higher ε4 frequency the more severe of depressive symptoms. This paper reported the association between depression and APOE ε4 allele, indicated that APOE ε4 allele increased the depression risk, and depressive patients carrying APOE ε4 allele had more severe depressive symptoms.
The objects of this study were Han ethnicity, the frequency of the APOE ε4 allele was 7.5% in Han, which were comparable to values found in other studies of Asian populations (32). The lowest concentration of serum APOE was found in the ε4 group (33); ε4 carriers showed increased levels of TNF-α, IL-6, and IL-1β when compared with the ε2 and ε3 carriers in Han population (34). Inflammation and dyslipidemia might be one of hypotheses of depression pathogenesis.
In addition, according to the inclusion and exclusion criteria, only nine eligible articles originated from the Han ethnicity were included in this meta-analysis, it might be the limitations of current study. The truth is always being perfected. When other ethnological data are updated in the future, we will further improve our research on the association of APOE ε4 allele and depression.
Conclusion
Collectively, the results of this meta-analysis suggested that APOE ε4 allele might be associated with depression, it determined the severity of depression.
Conflicts of Interest
The authors declare no conflicts of interest.
Conflicts of Interest
The funding was obtained from Education Department of Yunnan Province, China (2016ZZX093); Yunnan Provincial Applied Fundamental Research (grant number 2017FE467 (-152); Yunnan Provincial Applied Fundamental Research (grant number 2018FE001 (-148).
References
- 1.Yu S, Holsboer F, Almeida OF. Neuronal actions of glucocorticoids: focus on depression. J Steroid Biochem Mol Biol. 2008;108:300–309. doi: 10.1016/j.jsbmb.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PloS Med. 2006;3:e442–446. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayberg HS, Lozano AM, Voon V, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–660. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 4.He YC, Zhong B. Progress in clinical studies of depression. Int J Lab Med. 2013;34:832–834. [Google Scholar]
- 5.Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- 6.Lavretsky H, Lesser IM, Wohl M, Miller BL, Mehringer CM, Vinters HV. Apolipoprotein-E and white-matter hyperintensities in late-life depression. Am J Geriatr Psychiatry. 2000;8:257–261. [PubMed] [Google Scholar]
- 7.Lavretsky H, Ercoli L, Siddarth P, Bookheimer S, Miller K, Small G. Apolipoprotein epsilon 4 allele status, depressive symptoms, and cognitive decline in middle-aged and elderly persons without dementia. Am J Geriatr Psychiatry. 2003;11:667–673. doi: 10.1176/appi.ajgp.11.6.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YL, Zhang HM, Pan HM, et al. The relationship between apolipoprotein E gene ε2/ε3/ε4 polymorphism and breast cancer risk: a systematic review and meta-analysis. Onco Targets Ther. 2016;9:1241–1249. doi: 10.2147/OTT.S94228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigaud AS, Traykov L, Caputo L, et al. Association of the apolipoprotein E epsilon 4 allele with late-onset depression. Neuroepidemiology. 2001;20:268–272. doi: 10.1159/000054801. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Zhang M. Apolipoprotein E and cerebrovascular disease. Cerebrovascular Diseases Volume of International Journal. 2000;8:139–141. [Google Scholar]
- 11.Seet WT, Mary ATJ, Yen TS. Apolipoprotein E genotyping in the Malay, Chinese and Indian ethnic groups in Malaysia-a study on the distribution of the different APOE alleles and genotypes. Clin Chim Acta. 2004;340:201–205. doi: 10.1016/j.cccn.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical researched.) 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dersimonian R, Laird N. Meta-analysis in clinical trials. Controlled clinical trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 14.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical researched.) 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong XF. Association of Apolipoprotein E Genetic Polymorphisms with Geriatric Depression. Paper of Suzhou University. 2012;38:129–135. [Google Scholar]
- 16.You JY. Association study of brain-derived neurotrophic factor (BDNF) haplotype and apolipoprotein E (APOE) gene polymorphism in late-onset depression. Paper of Nanjing Medical College. 2009;4:115–126. [Google Scholar]
- 17.Sun B. Serum Levels of BDNF and Apolipoprotein E Genotype among patients with Senile Depressive and Alzheimer’s. Paper of Suzhou University. 2010;13:109–132. [Google Scholar]
- 18.Shi SX, Jiang SD, Wang DX, et al. An association of apolipoprotein E gene polymorphisms between depression and dementia. Shanghai Archives of Psychiatry. 2001;13:196–198. [Google Scholar]
- 19.Yuan YG, Ye Q, Chen Y, et al. A Study of Serum Lipid Concentrations and Apolipoprotein E Genotype among Patients with Senile Depression and Alzhemier Disease. Chinese General Practice. 2006;9:106–108. [Google Scholar]
- 20.Jiao YM, Song LS, Liu Y, et al. Apolipoprotein E gene polymorphisms and serum lipid characteristics in depressive patients. Shanghai Archives of Psychiatry. 2007;19:326–330. [Google Scholar]
- 21.Wang XL, Wang XQ, Yuan YG. Association study between cognitive function and apolipoprotein E polymorphism in patients with late-onset depression. Journal of Clinical Psychiatry. 2012;22:387–389. [Google Scholar]
- 22.Zhang J, Shen XH, Qian MC, Sun JS, Zhong H, Yang JH, et al. Effects of Apolipoprotein E Genetic Polymorphism on Susceptibility of Depression and Efficacy of antidepressants. ACTA ACADEMIAE MEDICINAE SINICAE. 2012;34:595–600. doi: 10.3881/j.issn.1000-503X.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Han YK, Li J, Zhang CH, Song JG, et al. Correlation between apolipoprotein E gene polymorphism and serum lipid levels in depressive patients after stroke. Chinese Journal of Gerontology. 2013;33:5230–5232. [Google Scholar]
- 24.Chen B, Wang XY, Miao MS. The Molecular Mechanisms of depression and characteristics of Traditional Chinese Medicine Treatment of depression. China journal of Chinese medicine. 2014;29:212–214. [Google Scholar]
- 25.Namura I, Hishikawa Y. [Cerebral damages in alcoholic patients visualized by MRI; with special reference to frontal atrophy, basal ganglia and white matter lesion] Nihon rinsho. Japanese journal of clinical medicine. 1997;55:288–298. [PubMed] [Google Scholar]
- 26.Rabe-Jablonska J, Szymanska A. Diurnal profile of melatonin secretion in the acute phase of major depression and in remission. Med Sci Monit. 2001;7:946–952. [PubMed] [Google Scholar]
- 27.Maes M, Smith R, Christophe A, et al. Lower serum high-density lipoprotein cholesterol (HDL-C) in major depression and in depressed men with serious suicidal attempts: relationship with immune-inflammatory markers. Acta Psychiatr Scand. 1997;95:212–221. doi: 10.1111/j.1600-0447.1997.tb09622.x. [DOI] [PubMed] [Google Scholar]
- 28.Butters MA, Sweet RA, Mulsant BH, et al. APOE is associated with age-of-onset, but not cognitive functioning, in late-life depression. Int J Geriatr Psychiatry. 2003;18:1075–1081. doi: 10.1002/gps.1006. [DOI] [PubMed] [Google Scholar]
- 29.Bertram L, Tanzi RE. The current status of Alzheimer's disease genetics: what do we tell the patients. Pharmacol Res. 2004;50:385–396. doi: 10.1016/j.phrs.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 30.Stewart R, Russ C, Richards M, Brayne C, Lovestone S, Mann A. Depression, APOE genotype and subjective memory impairment: a cross-sectional study in an African-Caribbean population. Psychol Med. 2001;31:431–440. [PubMed] [Google Scholar]
- 31.Niti M, Yap KB, Kua EH, Ng TP. APOE-epsilon 4, depressive symptoms, and cognitive decline in Chinese older adults: Singapore Longitudinal Aging Studies. J Gerontol A Biol Sci Med Sci. 2009;64:306–311. doi: 10.1093/gerona/gln013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia Y, Sass C, Shen X, Siest G, Visvikis S. Associations of apolipoprotein E concentration and polymorphism with lipids and apolipoprotein levels in Chinese from Beijing and Shanghai. Clin Chem Lab Med. 2000;38:655–659. doi: 10.1515/CCLM.2000.094. [DOI] [PubMed] [Google Scholar]
- 33.Han S, Xu Y, Gao M, et al. Serum apolipoprotein E concentration and polymorphism influence serum lipid levels in Chinese Shandong Han population. Medicine (Baltimore) 2016;95:e5639–5641. doi: 10.1097/MD.0000000000005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan YY, Cai QL, Gao ZY, et al. APOE ε4 allele elevates the expressions of inflammatory factors and promotes Alzheimer's disease progression: A comparative study based on Han and She populations in the Wenzhou area. Brain Res Bull. 2017;132:39–43. doi: 10.1016/j.brainresbull.2017.04.017. [DOI] [PubMed] [Google Scholar]


