Abstract
The human hippocampus can be subdivided into subfields with unique functional properties and differential vulnerability to disease or neuropsychiatric conditions. Identifying genes that confer susceptibility to such processes is an important goal in developing treatments. Recent advances in automatic subfield segmentation from magnetic resonance images make it possible to use these measures as phenotypes in large‐scale genome‐wide association studies. Such analyses are likely to rely largely on standard resolution (~1 mm isotropic) T1‐weighted images acquired on 3.0T scanners. Determining whether the genetic architecture of subfields can be detected from such images is therefore an important step. We used Freesurfer v6.0 to segment hippocampal subfields in two large twin studies, the Vietnam Era Twin Study of Aging and the Human Connectome Project. We estimated heritability of subfields and the genetic overlap with total hippocampal volume. Heritability was similar across samples, but little genetic variance remained after accounting for genetic influences on total hippocampal volume. Importantly, we examined genetic relationships between subfields to determine whether subfields can be grouped based on a smaller number of underlying, genetically independent factors. We identified three genetic factors in both samples, but the high degree of cross loadings precluded formation of genetically distinct groupings of subfields. These results confirm the reliability of Freesurfer v6.0 generated subfields across samples for phenotypic analyses. However, the current results suggest that it will be difficult for large‐scale genetic analyses to identify subfield‐specific genes that are distinct from both total hippocampal volume and other subfields using segmentations generated from standard resolution T1‐weighted images.
Keywords: automated segmentation, genetic correlation, genetic variance, heritability, hippocampal subregions, twins
1. INTRODUCTION
The hippocampus is a medial temporal lobe structure that plays a critical role in relational encoding (Shimamura, 2010), episodic memory retrieval (Tulving & Markowitsch, 1998), and spatial navigation (Burgess, Maguire, & O'Keefe, 2002). Alterations in hippocampal structure or function have been implicated in a variety of diseases and psychiatric conditions such as Alzheimer's disease (AD) (Sabuncu et al., 2011), schizophrenia (Harrison, 2004), temporal lobe epilepsy (Blumcke et al., 2013), posttraumatic stress disorder (Logue et al., 2018), and depression (Schmaal et al., 2016). Changes in hippocampal volume are also observed during normal aging (Fjell et al., 2014).
Although often treated as a unitary structure in magnetic resonance imaging (MRI) studies, the hippocampus is composed of multiple subfields with distinct structural and functional properties (Amaral & Witter, 1989; Bakker, Kirwan, Miller, & Stark, 2008; de Flores et al., 2017; Lacy, Yassa, Stark, Muftuler, & Stark, 2011; Leal & Yassa, 2015; Mankin, Diehl, Sparks, Leutgeb, & Leutgeb, 2015; Small, Schobel, Buxton, Witter, & Barnes, 2011; Witter, Wouterlood, Naber, & Van Haeften, 2000). These subfields include the cornu ammonis fields (CA1, CA2, CA3, and CA4), the dentate gyrus (DG), and the subiculum (Duvernoy, 2005). Some prior research has found that subfields are differentially associated with AD and mild cognitive impairment (La Joie et al., 2013; Mueller et al., 2010; Wolk et al., 2017; Yassa et al., 2010), schizophrenia and bipolar disorder (Baglivo et al., 2018; Cao et al., 2017; Haukvik et al., 2015; Ho et al., 2017; Mathew et al., 2014; Tannous et al., 2018), PTSD (Wang et al., 2010), and normal aging (Krogsrud et al., 2014; Yassa, Mattfeld, Stark, & Stark, 2011). Twin studies have shown that hippocampal volume is under significant genetic influence, with estimates of heritability ranging from 45 to 64% (Blokland, de Zubicaray, McMahon, & Wright, 2012; Kremen et al., 2010). Large‐scale consortia have used hippocampal volume as a quantitative trait in genome‐wide association analyses (GWAS), identifying novel genes associated with this structure (Bis et al., 2012; Hibar et al., 2015; Hibar et al., 2017; Stein et al., 2012). Identifying the specific genes that confer susceptibility to disease processes may facilitate development of new treatments. Given the structural and functional heterogeneity demonstrated by hippocampal subfields, it is reasonable to postulate that each subfield may have distinct genetic influences and that some subfields may contribute more or less to the overall genetic variation in total hippocampal volume.
Recent studies have examined the heritability of subfields generated by automatic segmentation techniques and have found that, for the most part, these regions are under significant genetic influence (Greenspan, Arakelian, & van Erp, 2016; Patel et al., 2017; Whelan et al., 2016). However, a few important questions remain. First, although the genetic correlation between subfields and total hippocampal volume have been examined, it is critical to directly test whether this correlation is significantly different from 1.0. In other words, are there independent genetic influences on these structures that are not captured by simply assessing the hippocampal formation as a whole? Second, are subfields genetically distinct from each other? Identifying subfield‐specific genes would require that these genes are not in common with the total hippocampal volume or other subfields. If subfields are not all genetically distinct, can we identify groupings of subfields that are genetically independent? The genetic relationship between different subfields has not previously been examined. Thus, the degree to which this segmentation scheme is delineating regions belonging to a small set of genetically distinct factors is unknown. If individual subfields or groupings of subfields provide additional genetic information beyond that of the total hippocampus, they would represent useful targets for further genetic analyses. If not, then total hippocampal volume may be the best phenotype for genetic analysis as it can be reliably and efficiently captured by various image processing software packages. Determining the genetic architecture of hippocampal subfields may therefore indicate that: (a) each subfield represents a genetically distinct feature; (b) subfields can be grouped based on a smaller number of underlying, genetically independent factors; or (c) all subfields are associated with a single underlying genetic factor. Answering these questions will help to inform large‐scales GWAS by determining the feasibility of identifying specific genes associated with individual subfields that may confer differential vulnerability to disease, cognitive deficits, or neuropsychiatric disorders.
Reliable and accurate segmentation of these small structures poses a significant technical challenge. Although several manual techniques to delineate these subfields have been proposed (as compared in Yushkevich et al., 2015), these techniques are labor intensive, susceptible to rater bias, and not scalable to large cohort studies. Recent advances in automatic segmentation methods provide a potential opportunity for more consistent and reproducible measurements of hippocampal subfields (Chakravarty et al., 2013; Iglesias et al., 2015; Pipitone et al., 2014; Van Leemput et al., 2009). The Freesurfer software package (Fischl, 2012) is widely used and includes an automated method to delineate hippocampal subfields. Updates to Freesurfer v6.0 have improved the reliability of the subfield segmentations (Iglesias et al., 2015), and may represent a compelling method to generate quantitative phenotypes for imaging genetics consortia such as ENIGMA (Thompson et al., 2014; Whelan et al., 2016) or CHARGE (Psaty et al., 2009). It is important to note that studies utilizing manual marking techniques often collect high‐resolution T2‐weighted images acquired perpendicular to the body of the hippocampus (Yushkevich et al., 2015). Given the relative rarity with which such scans are collected, large‐scale multisite projects may be limited to 3.0T high field resolution (~1 mm isotropic) T1‐weighted images. Although automated methods such as the one included with Freesurfer will accept such images, the authors have warned that the resulting segmentations may be largely influenced by the ex vivo 7.0 T ultra‐high field resolution atlas (0.13 mm isotropic) that guides the automatic segmentation. Thus, it is important to determine whether subfields automatically segmented from commonly acquired images (T1‐weighted, ~1 mm isotropic) represent useful phenotypes for a genetic analysis.
Here, we defined hippocampal subfields using Freesurfer v6.0 in two large imaging studies: the Vietnam Era Twin Study of Aging (VETSA) and the Human Connectome Project (HCP). Both studies included twin pairs in very different age ranges, making it possible to decompose variance into genetic and environmental influences using biometrical twin models. The HCP data are somewhat higher resolution than the VETSA data (0.7 mm2 vs. 1 mm2 in‐plane resolution), which also allows use to assess the impact of these differences on estimates of heritability. We examined whether the genetic architecture of subfields is consistent across studies, and the extent to which unique genetic influences can be detected in 3.0T standard resolution T1‐weighted images.
2. METHODS
2.1. Vietnam Era Twin Study of Aging
2.1.1. Participants
Participants were from the VETSA project (Kremen et al., 2006; Kremen, Franz, & Lyons, 2013). VETSA participants comprise a national, community‐dwelling sample of male–male twins who are similar to American men in their age range with respect to health and lifestyle characteristics based on Center for Disease Control and Prevention data (Schoenborn & Heyman, 2009). All participants served in the military service sometime between 1965 and 1975, but nearly 80% reported no combat exposure. The study was approved by the Institutional Review Boards at the University of California, San Diego (UCSD), Boston University, and the Massachusetts General Hospital (MGH).
Of the 447 individuals that underwent MRI scanning in wave 2 of the VETSA, data from 41 individuals were excluded due to incomplete data acquisition or poor quality (e.g., movement, artifacts, etc.), resulting in a final sample of 406 for this study. The sample had a mean age of 61.8 years (range 56–66; SD = 2.6), was primarily Caucasian (88.4%) and had a mean education of 13.8 years (SD = 2.1). The twin models were based on 89 monozygotic (MZ) pairs, 63 dizygotic (DZ) pairs, and 102 unpaired individuals (i.e., participants whose co‐twin either was not scanned or whose data were not useable).
2.1.2. VETSA image acquisition
Images were acquired at two sites, UCSD (n = 256) and MGH (n = 164). At UCSD, images were acquired with a GE 3T Discovery 750× scanner (GE Healthcare, Waukesha, WI) with an eight‐channel phased array head coil. The imaging protocol included a sagittal 3D fast spoiled gradient‐echo T1‐weighted volume (TE = 3.164 ms, TR = 8.084 ms, TI = 600 ms, flip angle = 8°, pixel bandwidth = 244.141, field of view [FOV] = 256 × 192, in‐plane resolution = 1 × 1 mm2, slice thickness = 1.2 mm, slices = 172). At MGH, images were acquired with a Siemens Tim Trio, (Siemens, Washington, DC) with a 32‐channel head coil. The imaging protocol included a 3D magnetization‐prepared rapid gradient‐echo (MPRAGE) T1‐weighted volume (TE = 4.33 ms, TR = 2,170 ms, TI = 1,100 ms, flip angle = 7°, pixel bandwidth = 140, FOV = 256 × 256, in‐plane resolution = 1 × 1 mm2, slice thickness = 1.2 mm, slices = 160). Members within twin pairs were scanned on the same MRI scanner and pairs of each zygosity type were equally represented at both sites. Thus, heritabilities and genetic correlations will be unaffected by the scanner differences.
2.1.3. VETSA image processing
Cortical reconstruction and volumetric segmentation of VETSA data were performed with the Freesurfer v6.0 (http://surfer.nmr.mgh.harvard.edu/) (Dale, Fischl, & Sereno, 1999; Fischl et al., 2002; Fischl, van der Kouwe, et al., 2004; Fischl, Sereno, & Dale, 1999). Briefly, this involves correction of distortion due to gradient nonlinearity (Jovicich et al., 2006), removal of nonbrain tissue using a hybrid watershed/surface deformation procedure (Segonne et al., 2004), intensity normalization (Sled, Zijdenbos, & Evans, 1998), rigid registration into standard orientation with 1 mm isotropic voxel size, segmentation of the subcortical white matter and deep gray matter volumetric structures (Fischl et al., 2002; Fischl, Salat, et al., 2004), tessellation of the gray matter white matter boundary, automated topology correction (Fischl, Liu, & Dale, 2001; Segonne, Pacheco, & Fischl, 2007), and surface deformation following intensity gradients to optimally place the gray/white and gray/cerebrospinal fluid borders at the location where the greatest shift in intensity defines the transition to the other tissue class (Dale et al., 1999; Fischl & Dale, 2000).
2.2. Human Connectome Project
2.2.1. HCP participants
Preprocessed structural data from the HCP 1200 Subjects Data Release were downloaded from ConnectomeDB (https://db.humanconnectome.org; Marcus et al., 2011). The HCP is a large‐scale consortium with the aim of investigating structural and functional properties of the human brain. The study has recruited 1,200 healthy adults, which includes both twin and nontwin siblings (for details, see Van Essen et al., 2013). Only same‐sex MZ and DZ twin pairs were included in the current study and processed images were reviewed for quality, resulting in a final sample size of 556 individuals. The sample had a mean age of 29.3 years (range 22–36; SD = 3.4), was majority female (60.8%), was primarily Caucasian (82.4%) and had a mean education of 15.0 years (SD = 1.8). The twin models were based on 146 MZ pairs, 81 DZ pairs, and 104 unpaired individuals (i.e., participants whose co‐twin either was not scanned or whose data were not useable).
2.2.2. HCP image acquisition
Images were acquired at Washington University with a Siemens 3T Skyra scanner with a 32‐channel head coil. The imaging protocol included a 3D MPRAGE T1‐weighted volume (TE = 2.14 ms, TR = 2,400 ms, TI = 1,000 ms, flip angle = 8°, GRAPPA = 2, pixel bandwidth = 210, FOV = 224 × 224, in‐plane resolution = 0.7 × 0.7 mm, slice thickness = 0.7 mm, slices = 256). For more details, see https://www.humanconnectome.org/storage/app/media/documentation/s1200/HCP_S1200_Release_Appendix_I.pdf on the HCP website.
2.2.3. HCP image processing
T1‐weighted structural images were processed with the HCP minimal preprocessing pipeline which uses a combination of FSL (Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012) and Freesurfer v5.3 (Fischl, 2012) tools. These methods are described in detail in Glasser et al. (2013) and processing scripts are available from https://github.com/Washington-University/Pipelines/releases. Briefly, obtaining subcortical segmentations involves gradient distortion correction, skull‐stripping, bias field correction, registration to MNI space, and automated segmentation of subcortical structures (Fischl et al., 2002).
2.3. Hippocampal subfield segmentation
Automated hippocampal subfield segmentation was performed using the revised procedure available in Freesurfer v6.0 (Iglesias et al., 2015). A probabilistic atlas was built from a combination of 7.0T ultra‐high field resolution (0.13 mm isotropic) ex vivo MRI images used to delineate hippocampal subfields as well as a separate data set of in vivo T1‐weighted MRI scans of the whole brain (1 mm isotropic) used to delineate surrounding structures. Twelve subfields are included in the atlas: CA1, CA2/3, and CA4, presubiculum, parasubiculum, subiculum, fimbria, hippocampal fissure, molecular layer (ML), granule cells of the DG (GC‐DG), hippocampal amygdala transition area (HATA), and the hippocampal tail. Segmentations were visually checked for quality and statistical outliers were further examined to verify accuracy.
2.4. Statistical analysis
Previous work from our group found no significant differences of genetic influences on hippocampal volume across hemispheres (Eyler et al., 2011; Eyler et al., 2014; Kremen et al., 2010). To verify that this was also the case for individual subfields, we directly examined the genetic correlation between left and right subfields. We found complete genetic overlap (r g = 1.0) between subfields from each hemisphere (see Supporting Information for details on analysis and Table S1 for results). Therefore, bilateral averages were generated for all volumes. All analyses, both univariate and multivariate, were performed using the raw data application of the maximum‐likelihood‐based structural equation modeling software OpenMx v2.7.12 (Boker et al., 2011; Neale et al., 2016). Prior to twin analysis, measures from VETSA data were adjusted by regressing out the effects of age and MRI scanner (one for each site); measures from HCP data were adjusted by regressing out the effects of age and gender. All measures were then standardized with a z‐transform (separately for each study).
2.4.1. Analysis of genetic and environmental influences on hippocampal subfields
To determine the relative influence of genetic and environmental factors on hippocampal subfields, we fit univariate biometrical models (also referred to as ACE models) for each subfield (Figure 1a) (Neale & Cardon, 1992). In the classical twin design, the variance of a phenotype is decomposed into additive genetic (A) influences, common or shared environmental (C) influences (i.e., environmental factors that make members of a twin pair similar to one another), and unique environmental (E) influences (i.e., environmental factors that make members of a twin pair different from one another, including measurement error). Additive genetic influences are assumed to correlate perfectly (1.0) between MZ twins because they are generally genetically identical. DZ twins, on the other hand, share on average 50% of their segregating DNA, and are therefore assumed to correlate .50 for additive genetic influences. The shared environment, by definition, correlates 1.0 between both members of a twin pair, regardless of their zygosity, and because nonshared environments (E) are, by definition, uncorrelated, E includes measurement error. The proportion of the overall variance in a phenotype that is attributable to additive genetic influences is the narrow‐sense heritability (a2/[a2 + c2 + e2]). This modeling assumes mean and variance homogeneity both within twin pairs and across zygosity. Prior to analysis, mean and variance homogeneity was satisfied at each region. The full ACE model was used for both univariate and multivariate analyses.
Figure 1.
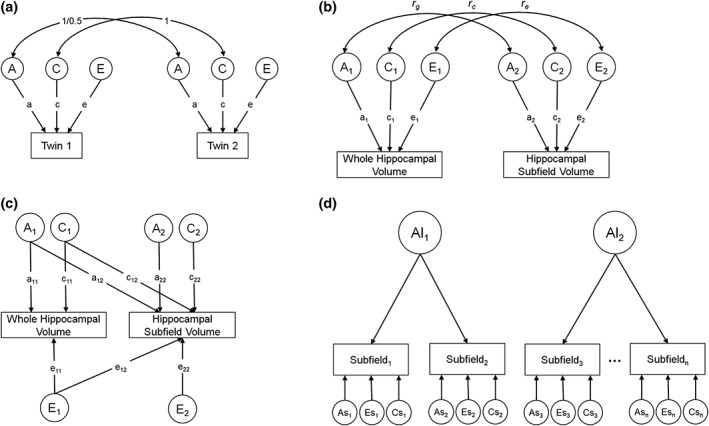
Path diagrams of twin models. (a) Univariate ACE model used to test heritability (a2) of each subfield. (b) Bivariate ACE model used to test genetic (r g) and environmental (r c and r e) correlations between whole hippocampal volume and each hippocampal subfield. (c) Bivariate Cholesky decomposition, which is a reparameterization of the bivariate ACE. Parameter a22 represents the genetic variance of each subfield after accounting for genetic covariance with the whole hippocampus. (d) Independent pathway model of (a). Al parameters represent latent genetic factors and As parameters represent residual A variance. Residual genetic variance was constrained to zero when nonsignificant
To determine whether there were any differences in heritability estimates between samples, we ran a multiple group model which included data from both HCP and VETSA. This model simultaneously estimates A, C, and E components for each group as described above. Significant differences in heritability between samples are tested by constraining estimates of additive genetic influences (A) to be equal across groups. A significant change in fit indicates that heritability is different between groups, and conversely, a nonsignificant change in fit indicates heritability estimates are not significantly different between groups.
2.4.2. Analysis of genetic overlap between total hippocampus and hippocampal subfields
To examine the degree of genetic overlap between total hippocampal volume and the volume of each hippocampal subfield, we conducted a set of bivariate ACE models. When extended beyond the univariate case, the twin design can further decompose the covariance between phenotypes into genetic and environmental components. Doing so allows for the estimation of genetic and environmental correlations between variables representing the degree of shared genetic and shared environmental variance, respectively (Figure 1b). Correlations were determined to be significantly different from 0 or 1 by examining 95% confidence intervals (CIs). A correlation with an upper bound of 1 indicates that there are no independent genetic influences between two phenotypes. This model also estimates genetic influences that are specific to each subfield after accounting for the genetic influences on total hippocampal volume (Figure 1c). We chose the bivariate modeling method over examining heritability of an adjusted phenotype that is calculated by residualizing subfield volumes for total hippocampal volume (e.g., Greenspan et al., 2016). The latter approach assumes proportional contributions to the phenotypic covariance from the A, C, and E components of total hippocampal volume. However, these contributions are not necessarily proportional, and the bivariate twin model allows for that possibility.
2.4.3. Factor analysis of genetic influences on hippocampal subfields
We fit a set of multivariate twin models to elucidate the genetic and environmental relationships among each of the hippocampal subfields. This model comparison analysis proceeded in a stepwise fashion. First, we fit the full multivariate ACE Cholesky, which included all subfields, to serve as the comparison model for all subsequent models. The multivariate ACE Cholesky provides a saturated representation of the genetic and environmental relationships among the variables, and is therefore ideal for comparing competing representations of the genetic factor structure. Second, we conducted separate tests to determine if the A, C, or E covariance between subfields could be set to zero. A significant reduction in model fit would indicate that they cannot be set to zero and that there is significant covariance between subfields for that variance component.
Third, we fit common pathway models (Kendler et al., 1987; McArdle & Goldsmith, 1990). The common pathways model assumes a single underlying latent phenotype accounts for the genetic and environmental covariation among measures in which the phenotypic covariance is equally apportioned into genetic and environmental components for all combinations of variables. None of the common pathway models provided a good fit to the data, and they are not discussed further.
Fourth, we fit independent pathway models separately for the A and E variance components (Figure 1d presents an example of an independent pathway model of genetic variance). The independent pathways model assumes that the genes and environments influencing covariation among measures operate directly on each variable through independent genetic and environmental pathways. The covariation between different pairs of variables can be due, in differing parts, to genetic and environmental influences. The genetic and environmental factor structure can thus be tested separately from one another so that they are not forced to adhere to the same factor pattern. Independent pathway models were fit with respect to one variance component at a time (i.e., the genetic factor structure was tested first, followed by the unique environment) with saturated models that impose no structure for the remaining components. For example, we fit a series of models with a factor structure for the genetic component (A), but saturated models of common (C) and unique environment (E).
Fifth, to determine the optimal factor model, we started with six factors (the maximum number at which the model was identified, represented as parameters Al at the top of Figure 1d), and proceeded to drop factors until we found the model with the smallest number of factors that: (a) did not result in a significant change in fit compared to the model previous (e.g., the comparison between a model with n factors and one with n + 1 factors), and (b) did not result in a significant decrement in fit relative to the full ACE Cholesky. Once the optimal number of factors was identified, we tested the significance of the residual/variable‐specific A parameters (genetic covariance specific to each individual subfield, represented as parameters Asn at the bottom of Figure 1d). Nonsignificant residual A parameters were constrained to zero, which allows the latent independent genetic pathway factors to capture more of the total genetic variance. Finally, to simplify the factor structure, we performed Varimax rotation on the factor loading matrix and fixed all loadings <0.1–0. We then refit the resulting model and fixed all nonsignificant loadings (as determined by 95% CIs) to zero to obtain a final model.
Evaluation of model fit was performed using the likelihood‐ratio chi‐square test (LRT), which is calculated as the difference in the −2 log‐likelihood (−2LL) of a model relative to that of a comparison model. Significance of individual model parameters was determined by examining 95% CIs.
3. RESULTS
3.1. Heritability of hippocampal subfields
MZ and DZ cross‐twin correlations, as well as the genetic and environmental variance components are presented in Table 1. The degree to which MZ twins (who share 100% of their segregating DNA) are more highly correlated than DZ twins (who share, on average, 50% of their segregating DNA) can be used to determine heritability via the ACE univariate biometrical modeling approach describe above. In VETSA, univariate estimates of twin correlations of hippocampal subfields in MZ twins ranged from 0.55 in the hippocampal fissure to 0.85 in CA1. Correlations between DZ twins ranged from −0.01 in the fimbria to 0.44 in the subiculum. Estimates of MZ twin correlations in HCP data ranged from 0.60 in the hippocampal fissure to 0.89 in the ML. Corresponding DZ correlations ranged from 0.12 in the hippocampal fissure to 0.43 in the presubiculum, fimbria, and HATA. Volume was significantly heritable in all subfields for both data sets, except for the hippocampal fissure in VETSA. Among these, VETSA maximum likelihood estimates of narrow sense heritability of subfields ranged from 0.57 in the fimbria to 0.84 in CA1, and HCP heritability estimates ranged from 0.37 in the HATA to 0.89 in the ML. Multiple group models did not show a significant change in fit when heritability estimates were constrained to be equal across groups. That is, estimated heritability was comparable across samples for all subfields. Estimates of common environmental variance (c2) were nonsignificant in all subfields of both studies. These estimates were zero for most regions in both studies, with a small number ranging from 0.10 to 0.34. However, this variance component was retained in the model to avoid inflating estimates of heritability.
Table 1.
Univariate ACE model for hippocampal subfields. Maximum likelihood estimates and 95% CIs for within pair correlations, additive genetic (a2), shared/common environmental (c2), and unique environmental (e2) influences. Estimates are presented separately for the VETSA, and HCP
| Subfield | rMZ | rDZ | a2 | Standardized variance components | ||||
|---|---|---|---|---|---|---|---|---|
| 95% CI | c2 | 95% CI | e2 | 95% CI | ||||
| VETSA | ||||||||
| Hippocampal tail | 0.70 | 0.30 | 0.73 | (0.41, 0.80) | 0.00 | (0.00, 0.29) | 0.27 | (0.20, 0.38) |
| Subiculum | 0.77 | 0.44 | 0.78 | (0.43, 0.84) | 0.00 | (0.00, 0.32) | 0.22 | (0.16, 0.31) |
| CA1 | 0.85 | 0.34 | 0.84 | (0.62, 0.88) | 0.00 | (0.00, 0.21) | 0.16 | (0.12, 0.23) |
| Hippocampal fissure | 0.55 | 0.32 | 0.40 | (0.00, 0.63) | 0.10 | (0.00, 0.48) | 0.50 | (0.37, 0.65) |
| Presubiculum | 0.78 | 0.33 | 0.78 | (0.52, 0.84) | 0.00 | (0.00, 0.24) | 0.22 | (0.16, 0.30) |
| Parasubiculum | 0.73 | 0.14 | 0.71 | (0.53, 0.79) | 0.00 | (0.00, 0.14) | 0.29 | (0.21, 0.41) |
| ML | 0.82 | 0.37 | 0.83 | (0.62, 0.87) | 0.00 | (0.00, 0.19) | 0.17 | (0.13, 0.24) |
| GC‐DG | 0.81 | 0.39 | 0.81 | (0.52, 0.86) | 0.00 | (0.00, 0.27) | 0.19 | (0.14, 0.27) |
| CA3 | 0.83 | 0.33 | 0.81 | (0.62, 0.86) | 0.00 | (0.00, 0.18) | 0.19 | (0.14, 0.26) |
| CA4 | 0.80 | 0.40 | 0.80 | (0.51, 0.85) | 0.00 | (0.00, 0.27) | 0.20 | (0.15, 0.28) |
| Fimbria | 0.60 | −0.01 | 0.57 | (0.36, 0.69) | 0.00 | (0.00, 0.14) | 0.43 | (0.31, 0.60) |
| HATA | 0.64 | 0.26 | 0.58 | (0.14, 0.68) | 0.00 | (0.00, 0.39) | 0.42 | (0.32, 0.54) |
| HCP | ||||||||
| Hippocampal tail | 0.83 | 0.42 | 0.84 | (0.58, 0.88) | 0.00 | (0.00, 0.26) | 0.16 | (0.12, 0.21) |
| Subiculum | 0.88 | 0.29 | 0.86 | (0.69, 0.89) | 0.00 | (0.00, 0.18) | 0.14 | (0.11, 0.18) |
| CA1 | 0.85 | 0.28 | 0.85 | (0.67, 0.89) | 0.00 | (0.00, 0.18) | 0.15 | (0.11, 0.19) |
| Hippocampal fissure | 0.60 | 0.12 | 0.57 | (0.35, 0.67) | 0.00 | (0.00, 0.19) | 0.43 | (0.33, 0.54) |
| Presubiculum | 0.80 | 0.43 | 0.70 | (0.40, 0.84) | 0.10 | (0.00, 0.39) | 0.20 | (0.16, 0.27) |
| Parasubiculum | 0.72 | 0.35 | 0.59 | (0.25, 0.77) | 0.11 | (0.00, 0.43) | 0.30 | (0.23, 0.38) |
| ML | 0.89 | 0.32 | 0.89 | (0.70, 0.91) | 0.00 | (0.00, 0.19) | 0.11 | (0.09, 0.15) |
| GC‐DG | 0.86 | 0.28 | 0.86 | (0.71, 0.89) | 0.00 | (0.00, 0.15) | 0.14 | (0.11, 0.18) |
| CA3 | 0.74 | 0.29 | 0.75 | (0.49, 0.81) | 0.00 | (0.00, 0.24) | 0.25 | (0.19, 0.33) |
| CA4 | 0.85 | 0.25 | 0.85 | (0.70, 0.88) | 0.00 | (0.00, 0.14) | 0.15 | (0.12, 0.20) |
| Fimbria | 0.61 | 0.43 | 0.51 | (0.18, 0.73) | 0.14 | (0.00, 0.43) | 0.35 | (0.27, 0.45) |
| HATA | 0.73 | 0.43 | 0.37 | (0.07, 0.75) | 0.34 | (0.00, 0.61) | 0.29 | (0.22, 0.37) |
95% CI = corrected 95% confidence intervals; CA = cornu ammonis; GC‐DG = granule cells of the dentate gyrus; HATA = hippocampal amygdala transition area; HCP = Human Connectome Project; ML = molecular layer; rDZ = correlation between dizygotic twins; rMZ = correlation between monozygotic twins; VETSA = Vietnam Era Twin Study of Aging.
3.2. Genetic relationship between total hippocampal volume and subfields
Phenotypic, genetic, and unique environmental correlations from the bivariate analysis of total hippocampal volume and individual subfields are presented in Table 2. Phenotypic correlations between total hippocampal volume and subfields were all significant in both studies. These correlations ranged from 0.30 in the subiculum and 0.93 in the ML in VETSA; correlations in the HCP ranged from 0.50 in the fimbria to 0.97 in the ML.
Table 2.
Relationships between whole hippocampal volume and hippocampal subfields from bivariate ACE model. Maximum likelihood estimates and 95% CIs for phenotypic (r p), additive genetic (r g), and unique environmental (r e) correlations between total hippocampal volume and specific hippocampal subfields. Residual genetic variance (A residual) after accounting for whole hippocampal volume is presented and corresponds to path a22 in Figure 1c. Estimates are presented separately for the VETSA, and HCP. Common environment correlations were nonsignificant and are not shown
| Subfield | r p | 95% CI | r g | 95% CI | r e | 95% CI | A residual | 95% CI |
|---|---|---|---|---|---|---|---|---|
| VETSA | ||||||||
| Hippocampal tail | 0.67 | (0.60, 0.72) | 0.68 | (0.47, 0.83) | 0.63 | (0.49, 0.74) | 0.39 | (0.21, 0.50) |
| Subiculum | 0.78 | (0.73, 0.81) | 0.81 | (0.67, 0.89) | 0.64 | (0.50, 0.74) | 0.26 | (0.15, 0.33) |
| CA1 | 0.90 | (0.88, 0.92) | 0.95 | (0.90, 1.00) | 0.78 | (0.69, 0.85) | 0.08 | (0.00, 0.13) |
| Hippocampal fissure | 0.47 | (0.38, 0.54) | 0.51 | (−1.00, 1.00) | 0.32 | (0.13, 0.48) | 0.28 | (0.00, 0.47) |
| Presubiculum | 0.57 | (0.49, 0.64) | 0.62 | (0.42, 0.80) | 0.43 | (0.25, 0.58) | 0.49 | (0.26, 0.62) |
| Parasubiculum | 0.30 | (0.20, 0.40) | 0.33 | (0.10, 0.56) | 0.22 | (0.01, 0.41) | 0.64 | (0.44, 0.82) |
| ML | 0.93 | (0.92, 0.95) | 0.98 | (0.95, 1.00) | 0.81 | (0.73, 0.87) | 0.03 | (0.00, 0.08) |
| GC‐DG | 0.87 | (0.84, 0.89) | 0.99 | (0.91, 1.00) | 0.75 | (0.65, 0.83) | 0.01 | (0.00, 0.13) |
| CA3 | 0.70 | (0.64, 0.75) | 0.76 | (0.63, 0.92) | 0.55 | (0.39, 0.68) | 0.34 | (0.12, 0.45) |
| CA4 | 0.86 | (0.83, 0.89) | 0.98 | (0.89, 1.00) | 0.74 | (0.64, 0.82) | 0.03 | (0.00, 0.16) |
| Fimbria | 0.31 | (0.22, 0.41) | 0.46 | (0.21, 0.76) | 0.05 | (−0.16, 0.25) | 0.45 | (0.20, 0.64) |
| HATA | 0.56 | (0.48, 0.63) | 0.71 | (0.36, 1.00) | 0.50 | (0.34, 0.64) | 0.26 | (0.00, 0.47) |
| HCP | ||||||||
| Hippocampal tail | 0.61 | (0.54, 0.66) | 0.63 | (0.49, 0.71) | 0.42 | (0.28, 0.55) | 0.51 | (0.37, 0.61) |
| Subiculum | 0.87 | (0.85, 0.90) | 0.92 | (0.88, 0.96) | 0.56 | (0.43, 0.66) | 0.13 | (0.06, 0.17) |
| CA1 | 0.92 | (0.90, 0.93) | 0.94 | (0.91, 0.96) | 0.80 | (0.73, 0.85) | 0.10 | (0.07, 0.13) |
| Hippocampal fissure | 0.47 | (0.40, 0.54) | 0.86 | (0.59, 1.00) | 0.06 | (−0.10, 0.22) | 0.14 | (0.00, 0.41) |
| Presubiculum | 0.77 | (0.73, 0.81) | 0.84 | (0.74, 0.94) | 0.48 | (0.35, 0.60) | 0.21 | (0.06, 0.30) |
| Parasubiculum | 0.55 | (0.48, 0.61) | 0.55 | (0.31, 0.71) | 0.24 | (0.08, 0.39) | 0.32 | (0.11, 0.48) |
| ML HP | 0.97 | (0.97, 0.98) | 0.98 | (0.97, 0.99) | 0.93 | (0.90, 0.95) | 0.04 | (0.02, 0.04) |
| GC‐DG | 0.92 | (0.90, 0.93) | 0.94 | (0.91, 0.96) | 0.77 | (0.70, 0.83) | 0.10 | (0.07, 0.13) |
| CA3 | 0.71 | (0.66, 0.76) | 0.76 | (0.66, 0.85) | 0.56 | (0.43, 0.66) | 0.31 | (0.19, 0.39) |
| CA4 | 0.89 | (0.87, 0.91) | 0.92 | (0.89, 0.96) | 0.71 | (0.62, 0.78) | 0.12 | (0.07, 0.16) |
| Fimbria | 0.50 | (0.43, 0.57) | 0.60 | (0.39, 0.93) | 0.14 | (−0.03, 0.30) | 0.30 | (0.03, 0.51) |
| HATA | 0.71 | (0.66, 0.76) | 0.85 | (0.67, 1.00) | 0.38 | (0.23, 0.51) | 0.10 | (0.00, 0.29) |
95% CI = corrected 95% confidence intervals; CA = cornu ammonis; GC‐DG = granule cells of the dentate gyrus; HATA = hippocampal amygdala transition area; HCP = Human Connectome Project; ML = molecular layer; VETSA = Vietnam Era Twin Study of Aging.
Genetic correlations provide a measure of the shared genetic influence between two traits. In general, genetic correlations with total hippocampal volume were high and, in many cases, did not differ from 1.0. This suggests that there is a high degree of overlap between the genetic influences on the total hippocampus and each individual subfield. In VETSA, genetic correlations between total hippocampal volume and the subfields ranged from 0.33 in the parasubiculum to 0.99 in the GC‐DG. The 95% CIs included 1.0 (meaning that the genetic correlation was not distinguishable from unity) in six of these regions: CA1, CA4, hippocampal fissure, ML, GC‐DG, and HATA. Equivalently, examining the A residual term demonstrates that only half of the subfields have any significant unique genetic variance after accounting for the genetic covariance with total hippocampal volume. Genetic correlations in the HCP between total hippocampal volume and subfields range from 0.55 in the parasubiculum to 0.98 in the ML. The 95% CIs in two of these regions did not differ from 1.0, with the upper bounds of another seven being greater than 0.90.
Unique environmental correlations with total hippocampal volume were significant in all regions except the fimbria in both studies, and the hippocampal fissure in the HCP. In the VETSA, these values ranged from 0.22 in the parasubiculum to 0.81 in the ML. Significant environmental correlations in the HCP ranged from 0.24 in the parasubiculum to 0.93 in the ML.
No regions of interest showed any significant common environment covariance between measures in either study. However, as noted before, we retained the common environment variance component in all further models to avoid inflating estimates of genetic variance.
3.3. Factor analysis of hippocampal subfields
The omnibus test fixing A covariances (but not variances) to zero resulted in a significant change of fit in both samples (VETSA: LRT = 134.86, Δdf = 66, p < .001; HCP: LRT = 188.71, Δdf = 66, p < .01). These results indicate that there is significant genetic covariance between subfields. We also found evidence for significant unique environmental covariance, but not common environmental influences (see Table 3 for results).
Table 3.
Model fit summaries of multivariate ACE and independent pathway models: VETSA data. Models tested to identify genetic relationship between subfields in the VETSA sample. For omnibus tests of a given variance component (A, C, or E), the covariance (but not variance) was constrained to zero while the covariance of the remaining variance components was estimated freely. The independent pathways section presents results of the model identification procedure. All models except the one‐factor model provided a good fit relative to the ACE Cholesky. While there was no significant reduction in fit between the four‐ and three‐factor models, the comparison of the three‐ versus two‐factor models resulted in a significant reduction in fit, indicating that the three‐factor solution provided the best model for further testing
| Model | Estimated parameters | −2LL | df | ΔLL | Δdf | p |
|---|---|---|---|---|---|---|
| Omnibus tests of covariance | ||||||
| ACE Cholesky | 246 | 7,008.06 | 4,626 | – | – | – |
| ACE no A covariance | 180 | 7,142.92 | 4,692 | 134.86 | 66 | 0 |
| ACE no C covariance | 180 | 7,026.17 | 4,692 | 18.11 | 66 | 1 |
| ACE no E covariance | 180 | 8,882.51 | 4,692 | 1874.45 | 66 | 0 |
| Independent pathway models of A | ||||||
| ACE Cholesky | 246 | 7,008.06 | 4,626 | – | – | – |
| Ind. path. A six factors | 243 | 7,009.62 | 4,635 | 1.56 | 9 | 1 |
| Ind. path. A five factors | 235 | 7,011.14 | 4,642 | 3.08 | 16 | 1 |
| Ind. path. A four factors | 226 | 7,016.43 | 4,650 | 8.37 | 24 | 1 |
| Ind. path. A three factors | 216 | 7,033.59 | 4,659 | 25.54 | 33 | 0.82 |
| Ind. path. A two factors | 205 | 7,053.31 | 4,669 | 45.26 | 43 | 0.38 |
| Ind. path. A one factor | 193 | 7,089.09 | 4,680 | 81.03 | 54 | 0.01 |
| Independent pathway models of A (three vs. two factors) | ||||||
| Ind. path. A three factors | 216 | 7,033.59 | 4,659 | – | – | – |
| Ind. path. A two factors | 205 | 7,053.31 | 4,669 | 19.72 | 10 | 0.03 |
−2LL = −2 log‐likelihood; df = degrees of freedom; LRT = likelihood‐ratio chi‐square test; p = significance of the LRT; VETSA = Vietnam Era Twin Study of Aging.
We fit a set of independent pathway models to determine whether there were a smaller number of latent factors that can be used to generate genetically distinct groupings of subfields. In VETSA, a model with three latent genetic factors satisfied both of our selection criteria by providing a good fit relative to the (a) Cholesky (LRT = 25.54, ΔDF = 33, p = .82) and (b) to the four‐factor model (LRT = 17.17, Δdf = 9, p = .05). Table 3 provides a summary of model fit indices.
We next examined the residual genetic variance (A s parameters in Figure 1d) on each subfield to determine which of these parameters could be trimmed from the model. We were able to fix the residual genetic variance to 0 on all subfields except for the parasubiculum without resulting in a significant change in fit compared to the ACE Cholesky (LRT = 41.36, Δdf = 44, p = .59) or the full three‐factor model (LRT = 15.82, Δdf = 11, p = .15). Next, to identify a simpler factor structure, we performed Varimax rotation on the genetic factor matrix from this trimmed three‐factor model and refit the model with any loadings of 0.1 or less constrained to zero. This resulted in a nonsignificant change in fit relative to the full three‐factor model (LRT = 16.48, Δdf = 13, p = .22). To further simplify the structure, we then constrained any nonsignificant loadings to 0. However, we were not able to identify a truly simple structure, as four subfields maintained cross‐loadings with multiple factors (although loadings on factor 3 were nonsignificant for all subfields except the hippocampal tail). In addition, the parasubiculum did not significantly load onto any of the factors. See Figure 2a for the final model and Table S2 (Supporting Information) for factor loadings.
Figure 2.
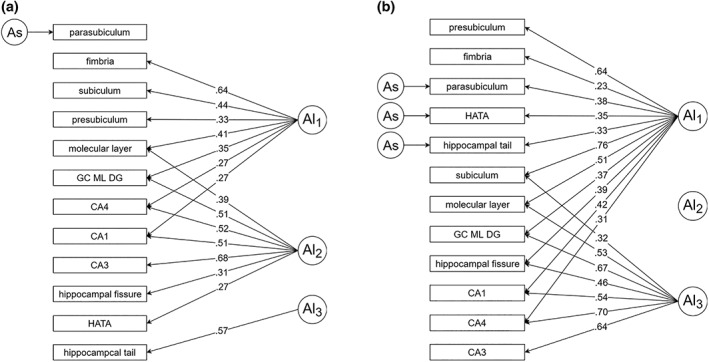
Independent pathway model of genetic variance. In the three‐factor model, we fit a factor structure on the genetic component (A), but saturated models of common (C) and unique environmental (E). Al parameters represent latent genetic factors and As parameters represent residual A variance. Significant loadings only are shown along the paths from latent genetic factors to hippocampal subfields. (a) Final model for the VETSA sample. (b) Final model for the HCP sample
In the HCP, a model with three latent genetic factors also provided a good fit relative to: (a) the Cholesky (LRT = 30.57, Δdf = 33, p = .59); and (b) the four‐factor model (LRT = 14.42, Δdf = 9, p = .05). Table 4 provides a summary of model fit indices.
Table 4.
Model fit summaries of multivariate ACE and independent pathway models: HCP data. Models tested to identify genetic relationship between subfields in the VETSA sample. For omnibus tests of a given variance component (A, C, or E), the covariance (but not variance) was constrained to zero while the covariance of the remaining variance components was estimated freely. The independent pathways section presents results of the model identification procedure. All models except the one‐factor model provided a good fit relative to the ACE Cholesky. While there was no significant reduction in fit between the four‐ and three‐factor models, the comparison of the three‐ versus two‐factor models resulted in a significant reduction in fit, indicating that the three‐factor solution provided the best model for further testing
| Model | Estimated parameters | −2LL | df | ΔLL | Δdf | p |
|---|---|---|---|---|---|---|
| Omnibus tests of covariance | ||||||
| ACE Cholesky | 246 | 8,635.54 | 6,426 | – | – | – |
| ACE no A covariance | 180 | 8,824.25 | 6,492 | 188.71 | 66 | 0 |
| ACE no C covariance | 180 | 8,669.35 | 6,492 | 33.81 | 66 | 1 |
| ACE no E covariance | 180 | 11,299.3 | 6,492 | 2,663.75 | 66 | 0 |
| Independent pathway models of A | ||||||
| ACE Cholesky | 246 | 8,635.54 | 6,426 | – | – | |
| Ind. path. A six factors | 243 | 8,638.17 | 6,435 | 2.63 | 9 | 0.98 |
| Ind. path. A five factors | 235 | 8,641.62 | 6,442 | 6.07 | 16 | 0.99 |
| Ind. path. A four factors | 226 | 8,651.69 | 6,450 | 16.15 | 24 | 0.88 |
| Ind. path. A three factors | 213 | 8,666.11 | 6,459 | 30.57 | 33 | 0.59 |
| Ind. path. A two factors | 205 | 8,687.65 | 6,469 | 52.11 | 43 | 0.16 |
| Ind. path. A one factor | 193 | 8,717.5 | 6,480 | 81.96 | 54 | 0.01 |
| Independent pathway models of A (three vs. two factors) | ||||||
| Ind. path. A three factors | 213 | 8,666.11 | 6,459 | – | – | – |
| Ind. path. A two factors | 205 | 8,687.65 | 6,469 | 21.55 | 10 | 0.02 |
−2LL = −2 log‐likelihood; df = degrees of freedom; HCP = Human Connectome Project; LRT = likelihood‐ratio chi‐square test; p = significance of the LRT; VETSA = Vietnam Era Twin Study of Aging.
We next examined the residual genetic variance (As in Figure 1d) on each subfield for the HCP data. These intervals were nonsignificant (i.e., included 0) for all subfields except the hippocampal tail, parasubiculum, and HATA. Fixing these parameters to 0 resulted in a nonsignificant change in fit relative to the ACE Cholesky (LRT = 45.17, Δdf = 42, p = .34) and the full three‐factor model (LRT = 14.6, Δdf = 9, p = .10). As with the VETSA data, to identify a simpler factor structure, we performed Varimax rotation on the genetic factor matrix and refit the model with any loadings of 0.1 or less constrained to zero. This resulted in a nonsignificant change in fit relative to the full three‐factor model (LRT = 15.75, Δdf = 11, p = .15). However, we were unable to simplify the factor structure any further. None of the loadings on factor 2 was individually significant, whereas all loadings on factors 1 and 3 were significant, and six subfields maintained cross‐loadings with multiple factors. Note that, while no loadings on factor 2 were significant, this factor could not be dropped without a significant change in model fit. See Figure 2b for the final model and Table S2 (Supporting Information) for factor loadings.
4. DISCUSSION
Heritability estimates for most subfields obtained from univariate twin models ranged from 0.51 to 0.89, consistent with previous studies (Greenspan et al., 2016; Patel et al., 2017; Whelan et al., 2016). The hippocampal fissure measured in the VETSA data was the sole region to demonstrate nonsignificant heritability. A previous study found this subfield to have much lower test–retest reliability (Whelan et al., 2016), which could explain the low heritability estimate. The samples included in the present study differed in age, sex, scanner manufacturer, scan sequences, and image resolution. Although hippocampal subfields were segmented with Freesurfer v6.0 in both data sets, initial structural preprocessing was carried out with different versions (VETSA: Freesurfer v6.0; HCP: Freesurfer v5.3). However, heritability estimates did not significantly differ across samples, and the genetic factor structures identified in each sample were broadly similar. Thus, the subfield measures appear to represent reliable phenotypes across studies that are robust to differences in image acquisition parameters of T1‐weighted images of the magnitude tested here (0.7 mm2 vs. 1 mm2 in‐plane resolution), as well as demographic and preprocessing differences between these data sets. Taken together, these results suggest that hippocampal subfields as delineated by Freesurfer v6.0 represent reliable measures that may be useful in large‐scale phenotypic analyses by consortia such as ENIGMA or CHARGE.
Although previous studies have examined the genetic correlations between subfields and total hippocampal volume (Greenspan et al., 2016; Patel et al., 2017), here we focus on determining whether significant genetic variance remains after accounting for the total hippocampal volume. Genetic correlations between total hippocampus and subfields were very high (>0.90), and the upper 95% CIs included 1 in some cases (Table 2). In addition, the genetic correlations were very similar between samples and the CIs were highly overlapping, thus we do not consider the differences in point estimates to be meaningful. Put another way, the residual genetic variance was quite low after accounting for genetic variance attributable to the whole hippocampal volume. This genetic overlap should not come as a surprise, as the total hippocampal volume is composed of the subfields. Overlapping genetic influences likely reflect genes related to structural properties of cells common the entire hippocampal formation. However, our findings are important in that they indicate that in standard 3.0T standard resolution T1‐weighted images; hippocampal subfield volumes do not provide substantial additional genetic information beyond what is can be detected from measures of total hippocampal volume.
Our estimates of residual genetic influence on hippocampal subfields after accounting for shared genetic influences on total hippocampal volume are somewhat lower than what has been reported previously (Greenspan et al., 2016). This is likely due to differences in the approaches one can take to estimate residual genetic variance. A common approach is to residualize the phenotype of interest by regressing out another phenotype. In this case, hippocampal subfield volumes would be residualized for total hippocampal volume. Regressing out the phenotypic variance of the total hippocampal volume from each subfield assumes proportional contributions to the phenotypic covariance based on the A, C, and E estimates of total hippocampal volume. Our results indicate that this assumption does not hold, as additive genetic influences constitute a much higher proportion of the phenotypic covariance than do common or environmental influences (see Tables 1 and 2). Thus, regressing out phenotypic variance may not sufficiently remove the proper amount of genetic variance, and can result in higher estimates of residual variance. The bivariate Cholesky approach does not make this assumption. Instead, it directly estimates the proportions of genetic and environmental variance and covariance such that shared genetic influences can be more accurately controlled for. When we follow the residual approach, heritability estimates are higher and similar to previously reported heritability estimates (Greenspan et al., 2016). The same logic applies to approaches that create a ratio of each subfield to the total hippocampal volume. Heritability estimates of subfield ratios are almost identical to those resulting from the residual approach. However, we believe that the results obtained from the bivariate Cholesky provide more precise estimates of residual genetic variance.
The lack of distinct genetic influences on each subfield coupled with the low image resolution relative to subfields' volumes leads to a logical follow‐up question: are there combinations of subfields that reflect a smaller number of genetically distinct regions that could be used in future genetic analysis? This requires determining the genetic relationship between subfields themselves, something that has not previously been explored. We fit an independent pathways model that attempts to find a smaller number of genetically independent factors comprising subfields with high degrees of genetic overlap. If genetically independent factors can be identified, these could represent new targets for genetic association studies. The results indicate that an underlying genetic architecture can be detected to a certain extent. For the most part, these genetic factors seem to reflect the CA fields plus DG in one factor, and medial subfields including the fimbria and subiculum‐related structures in the second factor. Internal layers such as the ML and GC‐DG exhibited loadings on both of these factors, likely due to their adjacent position to structures in each of these factors and small size. The third factor was much less clear, although it should be noted that the three‐factor solution did provide a significantly better fit relative to a two‐factor solution. Although this general pattern of segregation was apparent when we examined models with different numbers of genetic factors, we were not able to find a simple factor structure. That is, no set of genetically distinct factors emerged. There were many cross loadings, reflecting a large amount of shared genetic variance between subfields. Unfortunately, the overlap between genetic factors precludes our ability to identify combinations of subfields that would represent independent genetic factors.
A GWAS of hippocampal subfields was conducted using standard 3.0T resolution T1‐weighted sequences segmented with Freesurfer 6.0 (van der Meer et al., 2018). This analysis found 15 significant loci, eight of which had not previously been associated with the hippocampus. However, all of the loci that were significantly associated with subfields were also associated with either the total hippocampal volume or other subfields. Of the loci that were not associated with the total hippocampal volume, all but one was associated with the hippocampal tail. The hippocampal tail is not a subfield per se, but rather an umbrella label assigned to the posterior portion of the hippocampus where it is difficult to define reliable subfields (Iglesias et al., 2015). Thus, we do not believe the results of this GWAS are inconsistent with our conclusions. The detection of additional loci associated with the hippocampus is certainly an important finding, and these results indicate that a rough division of the hippocampal structure into anterior and posterior regions may be useful for further genetic analysis. However, the high degree of genetic covariance among subfields in our sample suggests that it will be difficult for future studies that use standard T1‐weighted sequences to detect genes that are distinct from both the total hippocampal volume and neighboring subfields.
Taken together, these findings suggest that there is little independent genetic variation specific to hippocampal subfields when using the automated Freesurfer segmentation method on standard 3.0T resolution T1‐weighted sequences. This is consistent with a warning by the authors of the algorithm that, in these cases, segmentation is largely driven by atlas priors. Thus, caution is warranted when using such data in genetic association analyses. The HCP data are acquired with an in‐plane resolution of 0.7 mm2, which does fall within the upper end of the range typically acquired for manual and automated segmentation protocols (Wisse, Biessels, & Geerlings, 2014). However, important anatomical features such as the “dark band” separating CA fields from the DG are not visible in T1‐weighted scans as they are in T2‐weighted scans (Wisse et al., 2017). Thus, inclusion of higher resolution T2‐weighted images is likely necessary to extract more precise genetic parcellations, and such data should be sought for the purposes of large‐scale GWAS of subfields. While it is possible that there truly is little genetic influence specific to individual subfields, it is also possible that another segmentation scheme or approach may delineate more genetically distinct regions on standard resolution T1‐weighted images. To this point, there is an international effort by the Hippocampal Subfields Group to develop harmonized segmentation protocols (Wisse et al., 2017), and the recommendations of this group should be considered carefully when choosing data sets appropriate for further genetic analysis.
Given the variability in development, function, and differential susceptibility to disease, there is ample reason to further investigate the hippocampus at the level of subfields (Amaral & Witter, 1989; Leal & Yassa, 2015; Small et al., 2011; Witter et al., 2000). Thus far, manual tracings of the subfields on high‐resolution images acquired for this specific purpose remain the gold standard (de Flores et al., 2015; Yushkevich et al., 2015). Yet it does appear that automated subfield segmentation on standard resolution images can be useful in phenotypic analyses, as multiple studies find subfields defined in this way are differentially affected by aging and disease (Haukvik et al., 2015; Iglesias et al., 2016; Krogsrud et al., 2014; Kurth, Cherbuin, & Luders, 2017; Pereira et al., 2013). Thus, automated segmentation methods are important tools that can lower the barriers to studying disease and age‐related phenomena at the subfield level in large‐scale studies and for researchers who may not otherwise have the resources to generate manual segmentations. In the present study, the high heritabilities of hippocampal subfields in two different samples suggest that automated segmentation methods provide reliable estimates of subfield volumes across age groups and acquisition parameters. On the other hand, our results suggest that little additional information about the genetic underpinnings of hippocampal volume is likely to be gained by examining individual subfields automatically segmented on 3.0T standard resolution T1‐weighted images rather than the hippocampus as a whole.
CONFLICT OF INTERESTS
L.K.M. has stock options in CorTechs Laboratories, Inc. A.M.D. is a Founder of and holds equity in CorTechs Laboratories, Inc., and serves on its Scientific Advisory Board. He is a member of the Scientific Advisory Board of Human Longevity, Inc. and receives funding through research agreements with General Electric Healthcare and Medtronic, Inc. The terms of these arrangements have been reviewed and approved by University of California, San Diego in accordance with its conflict of interest policies. The other authors report no conflict of interests.
Supporting information
Table S1 Relationships between left and right hemisphere volumes from bivariate ACE model. Maximum likelihood estimates and 95% confidence intervals for additive genetic (rg) correlations between left and right hemisphere structures of the total hippocampal volume and hippocampal specific subfields. All structures show complete overlap in genetic influences, i.e., 95% confidence intervals overlap with 1. Estimates are presented separately for the Vietnam Era Twin Study of Aging (VETSA), and Human Connectome Project (HCP). 95% CI = corrected 95% confidence intervals. CA = cornu ammonis; GC‐DG = granule cells in the dentate gyrus; HATA = hippocampal amygdala transition area
ACKNOWLEDGMENTS
The content of this manuscript is the responsibility of the authors and does not represent official views of NIA/NIH, or the Veterans' Administration. Numerous organizations provided invaluable assistance in the conduct of the VET Registry, including: U.S. Department of Veterans Affairs, Department of Defense; National Personnel Records Center, National Archives and Records Administration; Internal Revenue Service; National Opinion Research Center; National Research Council, National Academy of Sciences; and the Institute for Survey Research, Temple University. The authors gratefully acknowledge the continued cooperation of the twins and the efforts of many staff members. The study was supported by awards from the National Institutes of Health/National Institute on Aging (R01s AG018386, AG022381, AG050595 to W.S.K.; R01 AG018384 to M.J.L.; and K08 AG047903 to M.S.P). Data were provided (in part) by the Human Connectome Project, WU‐Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Elman JA, Panizzon MS, Gillespie NA, et al. Genetic architecture of hippocampal subfields on standard resolution MRI: How the parts relate to the whole. Hum Brain Mapp. 2019;40:1528–1540. 10.1002/hbm.24464
Funding information: McDonnell Center for Systems Neuroscience; NIH Blueprint for Neuroscience Research, Grant/Award Number: 1U54MH091657; National Institute on Aging, Grant/Award Number: AG018384AG018386AG022 381AG047903AG050595; National Institutes of Health; Temple University; National Academy of Sciences; National Research Council; National Archives and Records Administration; Department of Defense; U.S. Department of Veterans Affairs
REFERENCES
- Amaral, D. G. , & Witter, M. P. (1989). The three‐dimensional organization of the hippocampal formation: A review of anatomical data. Neuroscience, 31, 571–591. [DOI] [PubMed] [Google Scholar]
- Baglivo, V. , Cao, B. , Mwangi, B. , Bellani, M. , Perlini, C. , Lasalvia, A. , … GET UP Group . (2018). Hippocampal subfield volumes in patients with first‐episode psychosis. Schizophrenia Bulletin, 44, 552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker, A. , Kirwan, C. B. , Miller, M. , & Stark, C. E. L. (2008). Pattern separation in the human hippocampal CA3 and dentate gyrus. Science, 319, 1640–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis, J. C. , DeCarli, C. , Smith, A. V. , van der Lijn, F. , Crivello, F. , Fornage, M. , … Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium . (2012). Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nature Genetics, 44, 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland, G. A. M. , de Zubicaray, G. I. , McMahon, K. L. , & Wright, M. J. (2012). Genetic and environmental influences on neuroimaging phenotypes: A meta‐analytical perspective on twin imaging studies. Twin Research and Human Genetics, 15, 351–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumcke, I. , Thom, M. , Aronica, E. , Armstrong, D. D. , Bartolomei, F. , Bernasconi, A. , … Spreafico, R. (2013). International consensus classification of hippocampal sclerosis in temporal lobe epilepsy: A task force report from the ILAE commission on diagnostic methods. Epilepsia, 54, 1315–1329. [DOI] [PubMed] [Google Scholar]
- Boker, S. , Neale, M. , Maes, H. , Wilde, M. , Spiegel, M. , Brick, T. , … Fox, J. (2011). OpenMx: An open source extended structural equation modeling framework. Psychometrika, 76, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, N. , Maguire, E. A. , & O'Keefe, J. (2002). The human hippocampus and spatial and episodic memory. Neuron, 35, 625–641. [DOI] [PubMed] [Google Scholar]
- Cao, B. , Passos, I. C. , Mwangi, B. , Amaral‐Silva, H. , Tannous, J. , Wu, M. J. , … Soares, J. C. (2017). Hippocampal subfield volumes in mood disorders. Molecular Psychiatry, 22, 1352–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, M. M. , Steadman, P. , van Eede, M. C. , Calcott, R. D. , Gu, V. , Shaw, P. , … Lerch, J. P. (2013). Performing label‐fusion‐based segmentation using multiple automatically generated templates. Human Brain Mapping, 34, 2635–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, A. M. , Fischl, B. , & Sereno, M. I. (1999). Cortical surface‐based analysis. I. Segmentation and surface reconstruction. NeuroImage, 9, 179–194. [DOI] [PubMed] [Google Scholar]
- de Flores, R. , La Joie, R. , Landeau, B. , Perrotin, A. , Mezenge, F. , de La Sayette, V. , … Chetelat, G. (2015). Effects of age and Alzheimer's disease on hippocampal subfields: Comparison between manual and FreeSurfer volumetry. Human Brain Mapping, 36, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores, R. , Mutlu, J. , Bejanin, A. , Gonneaud, J. , Landeau, B. , Tomadesso, C. , … Chetelat, G. (2017). Intrinsic connectivity of hippocampal subfields in normal elderly and mild cognitive impairment patients. Human Brain Mapping, 38, 4922–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy, H. M. (2005). The human hippocampus: Functional anatomy, vascularization and serial sections with MRI. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- Eyler, L. T. , Prom‐Wormley, E. , Fennema‐Notestine, C. , Panizzon, M. S. , Neale, M. C. , Jernigan, T. L. , … Kremen, W. S. (2011). Genetic patterns of correlation among subcortical volumes in humans: Results from a magnetic resonance imaging twin study. Human Brain Mapping, 32, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler, L. T. , Vuoksimaa, E. , Panizzon, M. S. , Fennema‐Notestine, C. , Neale, M. C. , Chen, C.‐H. , … Kremen, W. S. (2014). Conceptual and data‐based investigation of genetic influences and brain asymmetry: A twin study of multiple structural phenotypes. Journal of Cognitive Neuroscience, 26, 1100–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. (2012). FreeSurfer. NeuroImage, 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. , & Dale, A. M. (2000). Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences of the United States of America, 97, 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl, B. , Liu, A. , & Dale, A. M. (2001). Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Transactions on Medical Imaging, 20, 70–80. [DOI] [PubMed] [Google Scholar]
- Fischl, B. , Salat, D. H. , Busa, E. , Albert, M. , Dieterich, M. , Haselgrove, C. , … Dale, A. M. (2002). Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron, 33, 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl, B. , Salat, D. H. , van der Kouwe, A. J. , Makris, N. , Segonne, F. , Quinn, B. T. , & Dale, A. M. (2004). Sequence‐independent segmentation of magnetic resonance images. NeuroImage, 23(Suppl. 1), S69–S84. [DOI] [PubMed] [Google Scholar]
- Fischl, B. , Sereno, M. I. , & Dale, A. M. (1999). Cortical surface‐based analysis. II: Inflation, flattening, and a surface‐based coordinate system. NeuroImage, 9, 195–207. [DOI] [PubMed] [Google Scholar]
- Fischl, B. , van der Kouwe, A. , Destrieux, C. , Halgren, E. , Segonne, F. , Salat, D. H. , … Dale, A. M. (2004). Automatically parcellating the human cerebral cortex. Cerebral Cortex, 14, 11–22. [DOI] [PubMed] [Google Scholar]
- Fjell, A. M. , McEvoy, L. , Holland, D. , Dale, A. M. , Walhovd, K. B. , & Alzheimer's Disease Neuroimaging Initiative . (2014). What is normal in normal aging? Effects of aging, amyloid and Alzheimer's disease on the cerebral cortex and the hippocampus. Progress in Neurobiology, 117, 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Sotiropoulos, S. N. , Wilson, J. A. , Coalson, T. S. , Fischl, B. , Andersson, J. L. , … Jenkinson, M. (2013). The minimal preprocessing pipelines for the human connectome project. NeuroImage, 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan, K. S. , Arakelian, C. R. , & van Erp, T. G. M. (2016). Heritability of hippocampal formation sub‐region volumes. Journal of Neurology and Neuroscience, 7, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P. J. (2004). The hippocampus in schizophrenia: A review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology, 174, 151–162. [DOI] [PubMed] [Google Scholar]
- Haukvik, U. K. , Westlye, L. T. , Morch‐Johnsen, L. , Jorgensen, K. N. , Lange, E. H. , Dale, A. M. , … Agartz, I. (2015). In vivo hippocampal subfield volumes in schizophrenia and bipolar disorder. Biological Psychiatry, 77, 581–588. [DOI] [PubMed] [Google Scholar]
- Hibar, D. P. , Adams, H. H. , Jahanshad, N. , Chauhan, G. , Stein, J. L. , Hofer, E. , … Ikram, M. A. (2017). Novel genetic loci associated with hippocampal volume. Nature Communications, 8, 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar, D. P. , Stein, J. L. , Renteria, M. E. , Arias‐Vasquez, A. , Desrivieres, S. , Jahanshad, N. , … Medland, S. E. (2015). Common genetic variants influence human subcortical brain structures. Nature, 520, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, N. F. , Iglesias, J. E. , Sum, M. Y. , Kuswanto, C. N. , Sitoh, Y. Y. , De Souza, J. , … Holt, D. J. (2017). Progression from selective to general involvement of hippocampal subfields in schizophrenia. Molecular Psychiatry, 22, 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, J. E. , Augustinack, J. C. , Nguyen, K. , Player, C. M. , Player, A. , Wright, M. , … Alzheimer's Disease Neuroimaging Initiative . (2015). A computational atlas of the hippocampal formation using ex vivo, ultra‐high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage, 115, 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias, J. E. , Van Leemput, K. , Augustinack, J. , Insausti, R. , Fischl, B. , Reuter, M. , & Alzheimer's Disease Neuroimaging Initiative . (2016). Bayesian longitudinal segmentation of hippocampal substructures in brain MRI using subject‐specific atlases. NeuroImage, 141, 542–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). FSL. NeuroImage, 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Jovicich, J. , Czanner, S. , Greve, D. , Haley, E. , van der Kouwe, A. , Gollub, R. , … Dale, A. (2006). Reliability in multi‐site structural MRI studies: Effects of gradient non‐linearity correction on phantom and human data. NeuroImage, 30, 436–443. [DOI] [PubMed] [Google Scholar]
- Kendler, K. S. , Heath, A. C. , Martin, N. G. , & Eaves, L. J. (1987). Symptoms of anxiety and symptoms of depression. Same genes, different environments? Archives Of General Psychiatry, 44, 451–457. [DOI] [PubMed] [Google Scholar]
- Kremen, W. S. , Franz, C. E. , & Lyons, M. J. (2013). VETSA: The Vietnam era twin study of aging. Twin Research and Human Genetics, 16, 399–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen, W. S. , Prom‐Wormley, E. , Panizzon, M. S. , Eyler, L. T. , Fischl, B. , Neale, M. C. , … Fennema‐Notestine, C. (2010). Genetic and environmental influences on the size of specific brain regions in midlife: The VETSA MRI study. NeuroImage, 49, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen, W. S. , Thompson‐Brenner, H. , Leung, Y. M. , Grant, M. D. , Franz, C. E. , Eisen, S. A. , … Lyons, M. J. (2006). Genes, environment, and time: The Vietnam era twin study of aging (VETSA). Twin Research and Human Genetics, 9, 1009–1022. [DOI] [PubMed] [Google Scholar]
- Krogsrud, S. K. , Tamnes, C. K. , Fjell, A. M. , Amlien, I. , Grydeland, H. , Sulutvedt, U. , … Walhovd, K. B. (2014). Development of hippocampal subfield volumes from 4 to 22 years. Human Brain Mapping, 35, 5646–5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth, F. , Cherbuin, N. , & Luders, E. (2017). The impact of aging on subregions of the hippocampal complex in healthy adults. NeuroImage, 163, 296–300. [DOI] [PubMed] [Google Scholar]
- La Joie, R. , Perrotin, A. , de La Sayette, V. , Egret, S. , Doeuvre, L. , Belliard, S. , … Chetelat, G. (2013). Hippocampal subfield volumetry in mild cognitive impairment, Alzheimer's disease and semantic dementia. NeuroImage. Clinical, 3, 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy, J. W. , Yassa, M. A. , Stark, S. M. , Muftuler, L. T. , & Stark, C. E. L. (2011). Distinct pattern separation related transfer functions in human CA3/dentate and CA1 revealed using high‐resolution fMRI and variable mnemonic similarity. Learning & Memory, 18, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal, S. L. , & Yassa, M. A. (2015). Neurocognitive aging and the hippocampus across species. Trends in Neurosciences, 38, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue, M. W. , van Rooij, S. J. H. , Dennis, E. L. , Davis, S. L. , Hayes, J. P. , Stevens, J. S. , … Morey, R. A. (2018). Smaller hippocampal volume in posttraumatic stress disorder: A multisite ENIGMA‐PGC study: Subcortical volumetry results from posttraumatic stress disorder consortia. Biological Psychiatry, 83, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankin, E. A. , Diehl, G. W. , Sparks, F. T. , Leutgeb, S. , & Leutgeb, J. K. (2015). Hippocampal CA2 activity patterns change over time to a larger extent than between spatial contexts. Neuron, 85, 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus, D. S. , Harwell, J. , Olsen, T. , Hodge, M. , Glasser, M. F. , Prior, F. , … Van Essen, D. C. (2011). Informatics and data mining tools and strategies for the human connectome project. Frontiers in Neuroinformatics, 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew, I. , Gardin, T. M. , Tandon, N. , Eack, S. , Francis, A. N. , Seidman, L. J. , … Keshavan, M. S. (2014). Medial temporal lobe structures and hippocampal subfields in psychotic disorders: Findings from the bipolar‐schizophrenia network on intermediate phenotypes (B‐SNIP) study. JAMA Psychiatry, 71, 769–777. [DOI] [PubMed] [Google Scholar]
- McArdle, J. J. , & Goldsmith, H. H. (1990). Alternative common factor models for multivariate biometric analyses. Behavior Genetics, 20, 569–608. [DOI] [PubMed] [Google Scholar]
- Mueller, S. G. , Schuff, N. , Yaffe, K. , Madison, C. , Miller, B. , & Weiner, M. W. (2010). Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Human Brain Mapping, 31, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale, M. , & Cardon, L. (1992). Methodology for genetic studies of twins and families (p. 530). Dordrecht, The Netherlands: Springer Science & Business Media. [Google Scholar]
- Neale, M. C. , Hunter, M. D. , Pritikin, J. N. , Zahery, M. , Brick, T. R. , Kirkpatrick, R. M. , … Boker, S. M. (2016). OpenMx 2.0: Extended structural equation and statistical modeling. Psychometrika, 81, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, S. , Park, M. T. M. , Devenyi, G. A. , Patel, R. , Masellis, M. , Knight, J. , & Chakravarty, M. M. (2017). Heritability of hippocampal subfield volumes using a twin and non‐twin siblings design. Human Brain Mapping, 38, 4337–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, J. B. , Junque, C. , Bartres‐Faz, D. , Ramirez‐Ruiz, B. , Marti, M. J. , & Tolosa, E. (2013). Regional vulnerability of hippocampal subfields and memory deficits in Parkinson's disease. Hippocampus, 23, 720–728. [DOI] [PubMed] [Google Scholar]
- Pipitone, J. , Park, M. T. , Winterburn, J. , Lett, T. A. , Lerch, J. P. , Pruessner, J. C. , … Alzheimer's Disease Neuroimaging Initiative . (2014). Multi‐atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. NeuroImage, 101, 494–512. [DOI] [PubMed] [Google Scholar]
- Psaty, B. M. , O'Donnell, C. J. , Gudnason, V. , Lunetta, K. L. , Folsom, A. R. , Rotter, J. I. , … CHARGE Consortium . (2009). Cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium: Design of prospective meta‐analyses of genome‐wide association studies from 5 cohorts. Circulation. Cardiovascular Genetics, 2, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu, M. R. , Desikan, R. S. , Sepulcre, J. , Yeo, B. T. , Liu, H. , Schmansky, N. J. , … Alzheimer's Disease Neuroimaging Initiative . (2011). The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Archives of Neurology, 68, 1040–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaal, L. , Veltman, D. J. , van Erp, T. G. , Samann, P. G. , Frodl, T. , Jahanshad, N. , … Hibar, D. P. (2016). Subcortical brain alterations in major depressive disorder: Findings from the ENIGMA major depressive disorder working group. Molecular Psychiatry, 21, 806–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn, C. A. , & Heyman, K. M. (2009). Health characteristics of adults aged 55 years and over: United States, 2004‐2007. National Health Statistics Report, 16, 1–31. [PubMed] [Google Scholar]
- Segonne, F. , Dale, A. M. , Busa, E. , Glessner, M. , Salat, D. , Hahn, H. K. , & Fischl, B. (2004). A hybrid approach to the skull stripping problem in MRI. NeuroImage, 22, 1060–1075. [DOI] [PubMed] [Google Scholar]
- Segonne, F. , Pacheco, J. , & Fischl, B. (2007). Geometrically accurate topology‐correction of cortical surfaces using nonseparating loops. IEEE Transactions on Medical Imaging, 26, 518–529. [DOI] [PubMed] [Google Scholar]
- Shimamura, A. P. (2010). Hierarchical relational binding in the medial temporal lobe: The strong get stronger. Hippocampus, 20, 1206–1216. [DOI] [PubMed] [Google Scholar]
- Sled, J. G. , Zijdenbos, A. P. , & Evans, A. C. (1998). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging, 17, 87–97. [DOI] [PubMed] [Google Scholar]
- Small, S. A. , Schobel, S. A. , Buxton, R. B. , Witter, M. P. , & Barnes, C. A. (2011). A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nature Reviews. Neuroscience, 12, 585–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, J. L. , Medland, S. E. , Vasquez, A. A. , Hibar, D. P. , Senstad, R. E. , Winkler, A. M. , … Enhancing Neuro Imaging Genetics through Meta‐Analysis Consortium . (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nature Genetics, 44, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous, J. , Amaral‐Silva, H. , Cao, B. , Wu, M. J. , Zunta‐Soares, G. B. , Kazimi, I. , … Soares, J. C. (2018). Hippocampal subfield volumes in children and adolescents with mood disorders. Journal of Psychiatric Research, 101, 57–62. [DOI] [PubMed] [Google Scholar]
- Thompson, P. M. , Stein, J. L. , Medland, S. E. , Hibar, D. P. , Vasquez, A. A. , Renteria, M. E. , … Saguenay Youth Study (SYS) Group . (2014). The ENIGMA consortium: Large‐scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior, 8, 153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulving, E. , & Markowitsch, H. J. (1998). Episodic and declarative memory: Role of the hippocampus. Hippocampus, 8, 198–204. [DOI] [PubMed] [Google Scholar]
- van der Meer, D. , Rokicki, J. , Kaufmann, T. , Cordova‐Palomera, A. , Moberget, T. , Alnaes, D. , … Neurocognition and Genetics Study . (2018). Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Molecular Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. , Yacoub, E. , Ugurbil, K. , & WU‐Minn HCP Consortium . (2013). The WU‐Minn human connectome project: An overview. NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput, K. , Bakkour, A. , Benner, T. , Wiggins, G. , Wald, L. L. , Augustinack, J. , … Fischl, B. (2009). Automated segmentation of hippocampal subfields from ultra‐high resolution in vivo MRI. Hippocampus, 19, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Neylan, T. C. , Mueller, S. G. , Lenoci, M. , Truran, D. , Marmar, C. R. , … Schuff, N. (2010). Magnetic resonance imaging of hippocampal subfields in posttraumatic stress disorder. Archives of General Psychiatry, 67, 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan, C. D. , Hibar, D. P. , van Velzen, L. S. , Zannas, A. S. , Carrillo‐Roa, T. , McMahon, K. , … Alzheimer's Disease Neuroimaging Initiative . (2016). Heritability and reliability of automatically segmented human hippocampal formation subregions. NeuroImage, 128, 125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. , Biessels, G. J. , & Geerlings, M. I. (2014). A critical appraisal of the hippocampal subfield segmentation package in FreeSurfer. Frontiers in Aging Neuroscience, 6, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse, L. E. M. , Daugherty, A. M. , Olsen, R. K. , Berron, D. , Carr, V. A. , Stark, C. E. L. , … Hippocampal Subfields Group . (2017). A harmonized segmentation protocol for hippocampal and parahippocampal subregions: Why do we need one and what are the key goals? Hippocampus, 27, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter, M. P. , Wouterlood, F. G. , Naber, P. A. , & Van Haeften, T. (2000). Anatomical organization of the parahippocampal‐hippocampal network. Annals of the New York Academy of Sciences, 911, 1–24. [DOI] [PubMed] [Google Scholar]
- Wolk, D. A. , Das, S. R. , Mueller, S. G. , Weiner, M. W. , Yushkevich, P. A. , & Alzheimer's Disease Neuroimaging Initiative . (2017). Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer's disease. Neurobiology of Aging, 49, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa, M. A. , Mattfeld, A. T. , Stark, S. M. , & Stark, C. E. (2011). Age‐related memory deficits linked to circuit‐specific disruptions in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America, 108, 8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassa, M. A. , Stark, S. M. , Bakker, A. , Albert, M. S. , Gallagher, M. , & Stark, C. E. (2010). High‐resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic mild cognitive impairment. NeuroImage, 51, 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Amaral, R. S. , Augustinack, J. C. , Bender, A. R. , Bernstein, J. D. , Boccardi, M. , … Hippocampal Subfields Group . (2015). Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. NeuroImage, 111, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Relationships between left and right hemisphere volumes from bivariate ACE model. Maximum likelihood estimates and 95% confidence intervals for additive genetic (rg) correlations between left and right hemisphere structures of the total hippocampal volume and hippocampal specific subfields. All structures show complete overlap in genetic influences, i.e., 95% confidence intervals overlap with 1. Estimates are presented separately for the Vietnam Era Twin Study of Aging (VETSA), and Human Connectome Project (HCP). 95% CI = corrected 95% confidence intervals. CA = cornu ammonis; GC‐DG = granule cells in the dentate gyrus; HATA = hippocampal amygdala transition area


