Abstract
Cardiovascular disease is the leading cause of death worldwide. Although investment in drug discovery and development has been sky-rocketing, the number of approved drugs has been declining. Cardiovascular toxicity due to therapeutic drug use claims the highest incidence and severity of adverse drug reactions in late-stage clinical development. Therefore, to address this issue, new, additional, replacement and combinatorial approaches are needed to fill the gap in effective drug discovery and screening. The motivation for developing accurate, predictive models is twofold: first, to study and discover new treatments for cardiac pathologies which are leading in worldwide morbidity and mortality rates; and second, to screen for adverse drug reactions on the heart, a primary risk in drug development. In addition to in vivo animal models, in vitro and in silico models have been recently proposed to mimic the physiological conditions of heart and vasculature. Here, we describe current in vitro, in vivo, and in silico platforms for modelling healthy and pathological cardiac tissues and their advantages and disadvantages for drug screening and discovery applications. We review the pathophysiology and the underlying pathways of different cardiac diseases, as well as the new tools being developed to facilitate their study. We finally suggest a roadmap for employing these non-animal platforms in assessing drug cardiotoxicity and safety.
Keywords: Cardiovascular diseases, In vitro disease models, In silico disease models, In vivo disease models, Drug discovery, Human induced pluripotent stem cells, Organ-on-a-chip, Cardiomyocyte
1. Introduction
The cardiovascular system, with its broad interdependence with circulation, blood vessels and blood constituents, as well as renal and nervous systems, is especially complex to model [1]. The myriad of possible signaling pathways involved in both normal function and in pathogenesis makes it a challenge to track drug interactions, limiting our ability to make predictions about a drug’s effectiveness, safety, and global effects on the body. The motivation for developing accurate, predictive cardiac models is twofold: first, to study and discover new treatments for cardiac pathologies which are leading in worldwide morbidity and mortality rates; and second, to screen for adverse drug reactions on the heart, which is a primary risk in drug development.
Cardiovascular toxicity claims the highest incidence and severity of adverse drug reactions in late-stage clinical development [1]. For example, Vioxx (Rofecoxib), originally designed to treat pain related to osteoarthritis and approved by the Food and Drug Administration (FDA) in 1999, was linked to over 27,000 cardiovascular-related deaths and myocardial infarctions (MI). It was withdrawn from the market in 2004, although later relicensed for more specific indications, with implementation of regulatory and transparency safeguards [2]. In preliminary clinical investigations, the drug showed effectiveness in its target treatment and adverse events were not significant. It was not until four years of long-term clinical studies that it became evident that the risk of heart attack and stroke was actually two-fold higher with Vioxx compared to the control group [3]. Some other compounds, such as Micturin (Terodiline, for urinary incontince), Fen-phen (Fenfluramine/phentermine, anti-obesity treatment), Seldane (Terfenadine, allergy medication), Zelnorm (Tegaserod, for irritable bowel syndrome), Meridia (Sibutramine, appetite suppressant), and Darvon/Darvocet (Propoxyphene, analgesic drug), have all had a similar record in terms of adverse cardiovascular effects [4–6].
While up to 70% of human toxicity may be approximated in the preclinical stage, there are other subtler and higher-risk cardiovascular events that only emerge when drugs are administered to humans for longer periods of time and in larger populations [7]. Even though a step closer toward representing the human population than preclinical studies, clinical trials are also challenging. Clinical trials have relatively small sample sizes, narrow subgroups of patient demographics, and require non-invasive endpoints for study monitoring [8]. As a result, rare events may not be detected until after clinical trials, when the drug is already in the clinic. Similarly, the small patient sample sizes may not recapitulate the genetic and environmental variations that a target population may experience. There is also the issue of determining toxicity endpoints in a clinical trial. Although non-invasive, biomarkers assessed via biochemical assays, genomic markers, antibody-based methods and imaging techniques [9], often yield information that reflects already serious and acute toxicities, with limited insight available for long-term damage [8]. Identifying biomarkers that can signal both acute and long-term toxicities before irreversible organ damage is incurred would be ideal, but how to do this with current technologies remains a challenge, suggesting a need for new technologies.
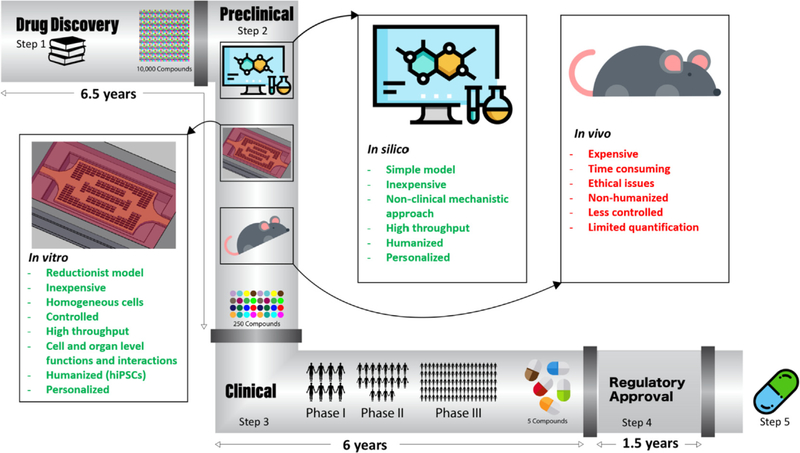
Using the current drug screening and safety testing paradigm, it takes approximately 12 years and over $2 billion before a single compound may reach patients [10]. Even so, late-stage attritions are still frequent [11]. Novel approaches are needed to make drug discovery affordable and effective. In this review, we describe current platforms for in vitro, in vivo, and in silico modelling of healthy and pathological cardiac tissues as well as their pros and cons for drug screening and discovery applications. To this end, we first briefly review the pathophysiology of different cardiovascular diseases (CVDs) and current knowledge about their underlying pathways, as well as the new tools being developed to facilitate their study. We finally suggest a roadmap for disease understanding and employing these platforms in assessing drug cardiotoxicity and safety.
2. Pathophysiology of cardiovascular diseases (CVDs)
Cardiovascular diseases (CVDs) including heart attack, stroke and heart failure (HF) are a leading cause of morbidity and mortality, contributing to an estimated 17 million deaths annually, around the world [12]. There are many underlying pathologies that lead to CVDs. Here, we will elucidate the pathophysiological pathways that play key roles in the development of different CVDs.
2.1. Atherosclerosis and myocardial infarction (MI)
2.1.1. Main players in atherosclerosis
Atherosclerosis is defined by cholesterol deposition in large- and medium-sized arteries. This accumulation leads to the proliferation of certain types of cells, such as macrophages and smooth muscle cells (SMCs), within the arterial wall. Enlargement of atherosclerotic plaques can gradually impinge vessels and block blood flow. Atherosclerosis is the most likely cause of MI and stroke [12–15].
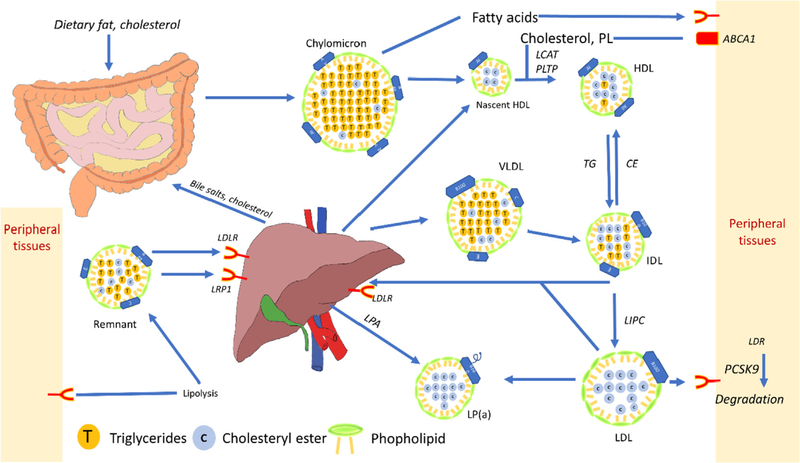
High levels of cholesterol and hypertriglyceridemia are risk factors for atherosclerosis. Both cholesterol and triglycerides (TG) can have endogenous or dietary sources and their metabolism is interrelated. Cholesterols are mainly carried in the blood by various lipoproteins. Chylomicron which carries dietary lipids from the intestines, is made up of a large portion of TG and a small portion of cholesterol and apolipoprotein(apo)B48 molecules that is secreted by the intestines and liver (Fig. 1). On its way to the liver, chylomicron undergoes lipolysis in peripheral tissues losing some TGs and gaining some other lipoproteins and becoming chylomicron remnant (Ch remnant) (Fig. 1). In the liver, Ch remnant can be converted to the very-low-density-lipoprotein (VLDL), a lipoprotein with higher cholesterol content and density than Ch, and with apoB100 instead of apoB48 (Fig. 1). Lipoprotein lipase (LPL), found in the circulation, converts lipoproteins to higher density particles by reducing free fatty acids from the lipoprotein, thereby converting it into intermediate density lipoprotein (IDL). IDL is taken up by the liver. In the liver, IDL is converted to LDL by LPL and TG lipase (Fig. 1) [16,17]. The LDL cholesterol level in circulation is regulated by the liver, via the LDL receptor. A protein, named proprotein convertase subtilisin/kexin type 9 (PCSK9), which is mainly expressed in the liver, controls LDL receptor degradation in the liver. The LDL receptor number on the surface of liver cells regulates the LDL cholesterol in circulation [5].
Fig. 1.
Schematic of lipoprotein metabolism and conversion in the body. Dietary lipid and cholesterol, digested from food with the help of cholesterol and bile acids in the intestines, pass through enterocytes and enter into circulation as chylomicrons. The molecules go through a series of lipolysis steps in the peripheral tissue and interact with nascent HDL on their way to the liver. They loose TG and get more cholesterol as they become denser and form remnants. Remnants enter the liver by interacting with LDL receptor-related protein 1 and LDL receptor, being modified to VLDL. VLDL communicates with HDL in circulation by Cholesterylester transfer protein (CETP) enzymatic activity and through lipolysis becomes IDL. Hepatic acid lipase (LIPC) converts IDL to LDL. IDL and LDL can go to the liver or peripheral tissues for further metabolism. In the liver and in circulation, LDL can be modified to form Lp(a). HDL acts as an acceptor of TG in circulation and from peripheral tissues, carrying TG to the liver for further metabolism and excretion. The concept of the figure is adapted from Ref. [39].
Atherosclerosis normally starts in arteries at points with high turbulent shear stress. Turbulent flow decreases nitric oxide (NO) generation, a mechanism for vascular relaxation, by enhancing endothelial nitric oxide synthase (eNOS) uncoupling and promoting reactive oxygen species generation. Moreover, uptake of lipoprotein and its modified forms in endothelial cells (ECs) perturbs NO bioavailability [18]. In contrast to high turbulent shear stress, low shear stress initiates endoplasmic reticulum (ER)-stress induced apoptosis of ECs [19]. Regeneration of endothelium by recruitment of blood-borne endothelial progenitor cells and by proliferation of adjacent ECs is not always perfect.
In healthy endothelium, lipid and inflammatory cells may enter into sub-endothelial spaces, through both intra- and trans-cellular mechanisms which are not completely understood, as reviewed in detail in other publications [20,21]. Damaged endothelium provides gaps in cell junctions which facilitate lipid and inflammatory cell migration to the sub-endothelial space, thereby promoting atherosclerosis. Reduced NO levels and perforation of blood vessels further promotes deposition of apoB containing lipids (LDL, VLDL, lipoprotein(a) and remnant lipoproteins), through receptor mediated endocytosis and transcytosis in sub-endothelial spaces [18,22,23]. ApoB containing lipoprotein in subendothelial space is considered to be atherogenic. The concentration of atherogenic lipoproteins in the circulation, their charge, particle size and cholesterol content, all govern their entry and retention in the sub-endothelial spaces [24]. In the sub-endothelium, positively charged apoB interacts with negatively charged extracellular matrix (ECM) proteoglycans [25], enhancing lipoprotein modification [26]. In vivo, LDL can be oxidized by enzymatic (e.g. 12/15 lipoxygenase) or non-enzymatic (e.g. free or heme-associated iron) pathways [27]. The occupation of sub-endothelial space with a high concentration of modified lipoprotein leads to lipoprotein spillover into resident ECs, SMCs and macrophages, and as a result a series of inflammatory reactions are initiated. Lipid uptake by resident cells is not a LDL receptor mediated process, instead sets of receptors with redundancy in function, that are called scavenger receptors, such as SRA, CD36 and LOX-1 or lipoprotein pinocytosis, are responsible for lipid uptake [27–29]. Macrophages are the most crucial cells in atherosclerosis [15,30]. Their phenotype is elastic in the plaque microenvironment, partly due to cholesterol deposition in the cells, creating a phenotype called foam cells. Foam cell formation is the result of an imbalance between lipoprotein uptake and cholesterol efflux. Irrespective of the uptake pathway, inside the cells in the late endosome and lysosome compartments, acidic cholesterol esterase generates free cholesterol from lipoproteins [31]. In the absence of adequate cholesterol efflux to prevent cholesterol-induced cytotoxicity, cholesterol is re-esterified by Acetyl-CoA acetyltransferase (ACAT-1) in the endoplasmic reticulum. To efflux cholesterol esters from intracellular lipid droplets, cholesterol esters need to be hydrolyzed to free cholesterol by a cholesterol ester hydrolase, associated with lipid droplets [31]. In another mechanism, when lipid droplets and autophagosomes are fused, the acid lipase involved in autophagy generates free cholesterol [32]. Several transporters in macrophages, including ATP-binding cassette (ABC) transporters, mediate efflux against cholesterol gradients. ABCA1, the most important transporter in cholesterol efflux, transfers cellular cholesterol to lipid-poor apoA-I and apoE to form pre-βHDL [33,34]. Other ABC transporters, such as ABCG1 and ABCG4 load more cholesterol on pre-HDL to form mature HDL [35].
HDL is formed in peripheral organs to help excrete cholesterol from the body in the form of bile acids. ApoA-I synthesized by the liver and intestines is a main component of most HDLs, but they may also contain apoE and apoC, which have distinct functions. ApoE, similar to ApoA1 in HDL is important in cholesterol efflux. As it is shown in Fig. 1, lipoproteins also carry triglyceride and apoC is regulating triglyceride homeostasis in HDL [36]. HDL has a number of anti-atherosclerotic effects other than cholesterol excretion, such as anti-inflammatory effects [37].
Besides macrophages, ECs and SMCs also take up modified lipoproteins and undergo phenotypic and metabolic changes leading to atherosclerotic plaque accumulation. These cells attract additional inflammatory cells with chemokines, cytokine secretion, and expression of adhesion molecules, thereby further amplifying atherosclerosis progression [38].
Complications arising from atherosclerosis mainly include narrowing of blood vessels and thrombosis. Thrombosis happens due to unstable plaque rupture, erosion, or calcified nodules in lesions, and may lead to infarction. Unresolved inflammation in atherosclerosis, together with lipid deposition and non-regulated lipid uptake in cells, leads to ER-induced stress and other mechanisms that trigger apoptosis in all stages of atherosclerosis. Impaired efferocytosis (i.e. clearance of apoptotic bodies) at later stages of atherosclerosis leads to secondary necrosis and the development of a necrotic core. Plaques with a large necrotic core and thin cap are considered unstable. The increased release of pro-inflammatory proteases in the necrotic milieu thins the fibrotic cap and degrades ECM, further enhancing plaque instability. Release of tissue factors into the circulation following plaque rupture leads to activation of coagulation pathways and promotes thrombosis [23,24]. Fig. 1 shows the schematic image of lipoprotein metabolism and conversions in body
2.1.2. Myocardial infarction
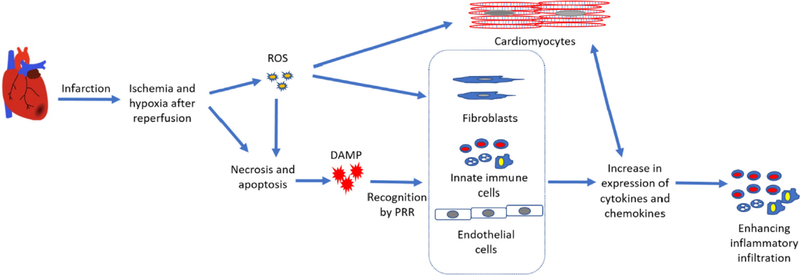
Unstable atherosclerotic plaque erosion leads to acute myocardial infarction (MI) (Fig. 2), where oxygen supply to myocytes becomes restricted. Myocyte contraction requires oxygen for energy supply, thus the limited oxygen supply following infarction results in cardiac damage and cell death. Reperfusion after ischemia restores blood flow; but over-oxygenation after ischemia, with the induction of reactive oxygen species (ROS), induces further damage to cardiac tissue by necrosis, apoptosis, autophagy, and secondary necrosis [40]. The extent of damage is related to the time of ischemia and reperfusion. Damaged tissue releases damage-associated molecular patterns (DAMPs), a set of danger molecules which signal for the infiltration of inflammatory cells, and for the clean-up of the damaged tissues. DAMPs bind to pattern recognition receptors (PRRs) of the innate immune system, on surrounding parenchymal cells and on infiltrating leukocytes, to recruit more inflammatory cells [41]. Border zone cardiomyocytes (CMs) also sense the ROS-inflicted damage through toll-like receptor (TLR) ligands and interleukins (IL), and respond by increasing their expression of cytokines, chemokines and adhesion molecules, to promote recruitment of more inflammatory cells [42,43]. ECs, which are abundant in cardiac tissue, also detect DAMPs and increase adhesion molecule expression, promoting neutrophil infiltration. Fibroblasts, similar to other resident cells in the heart, will also secrete inflammatory cytokines and chemokines in response to DAMPs, ROS and IL-1 signals [44].
Fig. 2.
Schematic of the consequences of myocardial infarction in the heart at a cellular level. Infarction restricts blood supply to the surrounding cells, leading to apoptosis and necrosis. After clog removal or reperfusion, surrounding live cells, which are at hypoxic states, start to generate ROS. ROS can trigger another phase of apoptosis and necrosis, as well as inflammation. Apoptotic and necrotic bodies release DAMPS. DAMPS, similar to ROS, are recognized by cells inside the heart (ECs, CMs, fibroblasts and immune cells) through a series of receptors (PRRs) and TLRs, triggering the expression of inflammatory cytokines and adhesion molecules. Enhanced inflammatory state is sensed by the surrounding cells, including CMs, thus leading to further expression of inflammatory molecules.
In the process of fibrotic repair, fibroblasts convert to myofibroblasts (myoFB). There is conflicting evidence about the source of fibroblasts participating in fibrotic repair. They may come from resident fibroblasts, bone marrow progenitor cells, ECs undergoing mesenchymal trans-differentiation, pericytes or epicardial epithelial cells [44]. MyoFBs develop stress fibers and express contractile proteins, such as the embryonic isoform of smooth muscle myosin and α-smooth muscle actin, which allows myoFBs to migrate and contract. MyoFBs also have large ER compartments which allow them to secrete elevated levels of ECM. MyoFBs secrete interstitial collagens (initially type III, then during infarction healing, type I). Collagen deposition increases tensile strength and prevents ventricular wall rupture. MyoFBs also generate more fibronectin and various matricellular proteins, such as thrombospondin which help myoFB migration and the healing response [45]. However, extensive cardiac fibrosis causes electro-mechanical disturbances in the heart and decreases the cardiac contractile reserve, which results in an attenuated ability of the heart to maintain cellular perfusion under normal cardiac filling pressure, eventually leading to HF [46]. Microvascular networks in the infarct area are formed by angiogenic signaling that causes EC infiltration and proliferation. Microvessels are crucial in supplying oxygen and nutrients during the repair process, thus motivating extensive tissue engineering efforts focused on revascularization [47].
2.2. Arrhythmia
Through excitation-contraction coupling, cardiac action potentials (AP) cause a coordinated contraction of CMs that pump the blood forward to peripheral tissues. AP is the result of a series of highly ordered opening and closing of channels in the CM cell membrane, thereby conducting the AP signal from one myocyte to another. Differential expression of channels in CMs of various regions of the heart results in a unidirectional electrical waves. However, disruption of the unidirectional ordered electrical wave, caused by physical obstacles like dead or ischemic tissues, fibrosis and inflammation, can cause arrhythmia [48]. Other causes of arrhythmia beyond physical obstacles include mutations in connecting molecules such as connexin 40, 45 and 43, as well as disturbances in parasympathetic innervation, hypovolemia and electrolyte disturbance [49]. An imbalance between outward and inward channel activity, as observed with mutated channels, can also trigger arrhythmia. Physical and phenotypic changes can alter the expression of ion channels. These changes affect AP duration and restitution, leading to arrhythmia and resulting in a heart that beats improperly [48].
2.3. Cardiomyopathy
Cardiomyopathy is a general term encompassing a variety of symptoms including heart muscle enlargement, thickening and rigidity. Cardiomyopathy of various phenotypes, resulting from known or unknown causes, leads to weakening of the heart and modification of its ejection fraction (EF) [50]. Common categories of cardiomyopathy include hypertrophic, dilated, arrhythmogenic, and left ventricular hypertrabeculation cardiomyopathies.
Hypertrophic cardiomyopathy (HCM) is characterized by cardiac hypertrophy, a non-dilated left ventricle (LV), and a normal or increased EF. The hypertrophy is usually asymmetrical, deriving most commonly from the basal interventricular septum adjacent to the aortic valve. At the cellular level, CMs are hypertrophied, disorganized, and separated by areas of interstitial fibrosis. This type of hypertrophy occurs due to many genetic or non-inheritable causes, leading to a wide range of phenotypes from asymptomatic to fibrillation, HF, and sudden cardiac death [51,52].
Dilated cardiomyopathy (DCM) is defined by an enlarged and poorly contractile LV. Genetic or non-genetic causes, including hypertension and valve diseases, may lead to dilated cardiomyopathy. This type of heart disease is the major cause of HF [51,53].
Arrhythmogenic cardiomyopathy (AC) is an inherited heart muscle disorder which causes sudden cardiac death, mostly in young patients and athletes. Loss of myocytes and replacement of right ventricular myocytes with fat deposition in fibroblasts are pathological features of this disease. Genetic abnormalities of cardiac desmosomes, leading to disconnection of myocytes and alteration of intracellular signal transduction are associated with arrhythmogenic cardiomyopathy [51,54].
Left ventricular hypertrabeculation (LVHT) or non-compaction, is a myocardial abnormality mostly associated with monogenic disorders or chromosomal defects, especially in neuromuscular genes. Presence of a thin, compacted, epicardial layer and a thick, noncompacted, spongy bi-layered myocardium in the endocardial layer is a common diagnosis for LVHT [55].
2.4. Cardiac fibrosis
Cardiac fibrosis is the scarring process that is characterized by cardiac fibroblast (CF) over-proliferation, myoFB activation, and increased deposition of fibrous ECM proteins [56]. It often naturally occurs following MI, dilated cardiomyopathy, and hypertension, and is a leading pathogenic factor in HF [57].
In healthy heart tissue, CFs are responsible for maintaining the ECM structure and forming an anisotropic syncytium [58]. In response to cardiac injury, fibroblasts often differentiate into myoFBs, which have a higher synthetic ability to produce ECM proteins, especially collagen [59]. Increased collagen deposition subsequently reduces compliance and increases stiffness of the affected tissue [59]. While the onset of fibrotic tissue formation may protect the heart from rupture by strengthening the injured site, continuous expansion of cardiac fibrosis will lead to progressive deterioration of cardiac contractile force and eventually result in the impairment of both systolic and diastolic functions of the heart [57].
Moreover, cardiac fibrosis is often associated with arrhythmogenicity by affecting electric signal propagation and causing rhythm disturbance [60]. Fibrotic myocardium displays distinctly altered electrophysiological properties that contribute to cardiac dysfunction. Previous studies showed that CFs could cause increased action potential duration (APD) and calcium transient duration (CTD) [61]. Despite the fact that they have multiple ion channels, fibroblasts and myoFBs are electrically non-excitable and cannot maintain AP propagation on their own. Thus, they often create conduction barriers by physically isolating myocytes, expanding the distance between neighboring myocyte membranes, and disrupting myocyte-to-myocyte coupling through gap junctions [60].
The mechanisms associated with fibrosis have been extensively studied over the past decade. A variety of signals such as cytokines, growth factors (GFs), and hormones contribute to the pathogenesis of cardiac fibrosis [62]. Effector hormones of the renin-angiotensin-aldosterone system (RAAS) and cytokines such as TGF-β have relatively well-identified profibrotic roles in the heart [63]. TGF-β is the critical mediator of pathological fibrosis after myocardial injury [64]. TGF-β signaling facilitates persistence of CFs and activation of myoFBs through Smad-mediated pathways [65]. The RAAS is also closely linked to the onset and progression of cardiac fibrosis. It is thought that angiotensin II (Ang II) is the predominant mediator of RAAS-associated cardiac remodelling. Ang II is often elevated post injury, causing CF proliferation and collagen overexpression [66]. However, numerous upstream and downstream factors of the pro-fibrotic cascades are still not fully understood [67]. Further studies of the mechanism of cardiac fibrosis will lead to a deeper understanding of fibrosis-associated arrhythmia and HF, and open new doors for potential therapy.
Table 1 summarizes the above-mentioned CVDs and their causes, symptoms, associated complications, prevalence and mortality rates.
Table 1.
Summary of CVDs and their causes, symptoms and associated complications.
| Disease | Cause(s) | Consequence | Prevalence (%) | Annual mortality rate(%) |
|---|---|---|---|---|
| Atherosclerosis | Cholesterol and lipid deposition in subendothelial space and sustained inflammatory reactions | Arterial blockage, arterial stiffness, calcification, thrombosis and myocardial infarction | 30[68] | 5–6% (per 100 000 population)[68] |
| Myocardial infarction | Arterial blockage by atherosclerotic plaques or by thrombotic plaques | Dead myocardial tissue, fibrosis, decrease in ejection fraction and systemic circulation, arrhythmia | 5% (age: 40–59)15% (age: 60–79)30% (age > 80)[69] | 34%–42% [69] |
| Arrhythmia | Improper function of cardiac ion channels or cell junctions, physical obstacles (e.g. infarcted tissue) disturbing electrical wave propagation | Atrial and ventricular fibrillation leading to improper blood pumping in heart | 15–25% (relative to age)[70] | 15[70] |
| Cardiomyopathy | Genetic predisposition and as yet unknown causes | Various types with ranges of symptoms. CMs can become enlarged, thick or rigid decreasing pumping function | 1:200–500(0.2–0.5%)[71] | 6–8[71] |
3. Quest for CVD models in the drug development pipeline
CVD models including laboratory animal models, in vitro disease platforms, and in silico/computational models have been used to assess the efficacy and safety of new drugs earlier in the drug development pipeline. Here, we summarize the advantages and limitations of each platform in the context of CVD modelling for the development of safer and more efficient drugs.
3.1. In vivo models
Various animal models have historically been used to further our knowledge on the etiology, pathogenesis, pathophysiology, progression, and underlying mechanisms of CVDs. They offer valuable tools in disease modelling, drug discovery and therapeutic interventions [76,77]. However, differences between the human condition and experimentally-induced pathology in animals in aspects such as cardiovascular physiology and pathophysiology, along with genetic and environmental factors, have made it rather difficult to recapitulate the complexities of CVD conditions with just a single experimental model [78]. The choice of an appropriate animal model to efficiently and reliably study any disease (e.g. CVD) is a challenging process, especially since the outcome would ultimately need to be translated to humans [11,79–81]. To determine the most appropriate animal model several factors, including research questions, number of animals, quality of the anticipated results, and the relevance of the outcomes to complications, need to be meticulously considered [82]. More importantly, it is also vital for the scientific community to reduce animal use from animal welfare and research ethics standpoints, and to comply with the 3Rs principles (replacement, reduction, and refinement) [83]. In this section, we review commonly used animal models (i.e. small and large animal models) in cardiac research and discuss their advantages and limitations in terms of translational applicability to humans.
3.1.1. Small animal models
Rodent models are widely used in cardiovascular research due to their easy handling and housing, short gestation time, low maintenance costs, and more importantly ability for genetic manipulation. These advantages make them a more appropriate choice for high-throughput studies than large animal models [84]. However, the main disadvantage of these small animal models (i.e. mice, rats, rabbits, and cats) is the relevance of the obtained results to humans due to the distinct physiological differences between these species and humans [84–86]. Ligation-induced MI in mice and rats is one of the most commonly used and well-established models to investigate cardiac regeneration capacity which illuminate the cellular and molecular mechanisms of cardiac regeneration. In addition to this commonly used technique, electrocautery and infarction caused by cryoprobes has also been utilized to induce lesions on the epicardial surface and throughout the ventricular wall, respectively. Despite its widespread use in cardiac research, difficulties in standardizing the injury size of the left anterior descending artery (LAD) ligation model is one of the limitations of this model [88].
Cryogenic injury through open thoracotomy (e.g. LV lesion) or abdominal incision through a transverse laparotomy (e.g. RV lesion) are models which induce confluent necrosis in the heart in order to investigate its regeneration and remodelling [89]. It has been observed that macrophages residing in the lesion site could contribute to regeneration of the extensive lesion after cryoinjury of the LV myocardium in adult mice [89]. Doxorubicin (DOX)-induced HF is a suitable model to perform mechanistic evaluation of non-ischemic CVD. It mimics severe non-ischemic human cardiomyopathy and LV dysfunction. It is achieved by giving a single dose of DOX (20 mg/kg) to the animal, causing clinical symptoms reminiscent of non-ischemic HF [90].
Moreover, transgenic lines can be produced by manipulating genes in animals. Due to the short gestation time and lower cost of the mouse models compared to larger species, these models have gained great attention in cardiovascular research [91,92]. For example, genetically modified mice have been extensively used for feasibility and proof-of-concept studies. However, translational aspects need to be more carefully addressed due to the enormous differences in the heart function and size between mice and humans [93]. Among others, two strains of mice have been engineered by deletion of two of the main proteins such as actin-associated cytoskeletal protein (e.g. MLP), and calcium-sequestration (e.g. CQS) which are involved in formation of actin filaments [94] and myocardial relaxation [95], respectively. These changes simulate the development, progression, and relapse of DCM in humans. The former shows the disease phenotype (i.e. contractile dysfunction) as early as 4–6 months due to hypertrophy leading to HF with a progressive increase in the amount of the connective tissue and decreased myocardial mechanical compliance, whereas the latter contributes to a rapid-onset phenotype leading to animal death by 9–14 weeks [96,97]. Two other transgenic strains have also been developed to study HCM: one by ablating cMyBP-C (e.g. cMyBP-C−/− mice) and another by overexpressing human myotrophin gene (i.e. Tg mice). They have both shown the development of hypertrophy that progressively led to HF, with highly compromised function [98,99]. Although these genetically modified models could capture some of the features of the CVD phenotype in humans, they typically do not resemble all aspects of CVDs in humans [86,87].
Models of autoimmune cardiomyopathy (AICM) have been engineered by crossing two different transgenic mice: the DQ8 transgenic non-obese diabetic (NOD) mouse with an NOD Major Histocompatibility Complex (MHC) class II β-chain knockout (KO) line, which leads to premature death through development of progressive DCM and HF [100,101]. Duchenne muscular dystrophy (DMD) is a neuromuscular disorder initiated by a mutation in the dystrophin gene [102]. The double knock-out dystrophin/utrophin mouse model caused severe cardiac dysfunction at 8 weeks of age, which resulted in mortality due to respiratory issues or HF [103]. The atrial fibrillation (AF) mouse model has been generated with CM-specific liver kinase B1 (LKB1) KO mice. Among others, this model has shown bi-atrial enlargement along with AF, LV hypertrophy, and cardiac dysfunction between 4 and 12 weeks of age [104].
In addition to these models in mice, several other models have also been generated in rats which show numerous similarities to cardiac abnormalities in humans, and represent the most widely used models for investigating the role and mechanisms of therapeutic strategies, such as stem cell (SC) therapy. Examples of these rat models include ligation-induced MI [105], overload-induced cardiac hypertrophy (e.g. ascending aortic banding) [106], diabetic cardiomyopathy (DbCM) [107], transgenic strains (e.g. hypertensive rats, type II diabetic rats (Goto-Kakizaki (GK) rats)) [108], and DMD rats. The spontaneously hypertensive rat (SHR), a transgenic line of hypertensive rats, have been the animal of choice for screening antihypertensive compounds [109]. In addition, Dahl salt-sensitive rats have been generated by administering animals with a high salt diet [110]. Other hypertension rat models can be categorized as: environmental, pharmacological, renal and stress-induced hypertension, activated sympathetic nervous system, NO-deficient models, and one/two-kidney(s) one/two-clip(s) hypertension models, among others [111]. It is noteworthy that the surgical procedure and invasive hemodynamic assessments are more straightforward to perform on rats than on mice. In addition, due to the larger size of rat myocardium, the number of post-mortem histological and molecular biological analyses that may be obtained are much higher for rats than for mice.
Experimental models have also been generated in animals to study atherosclerosis and thrombosis. Although the use of the high-fat diet in animals (e.g. atherosclerosis models) has been a well-established method to generate these models, not all experimental models (i.e. mice and rats) are acceptable for atherosclerosis due to intrinsic genetic differences, their higher resistance to atherogenesis and the high cholesterol diet [112,113]. This occurs due to the differences in human versus murine lipid processing (i.e. LDL-based versus HDL-based, respectively) [113]. To overcome these deficiencies, as well as to reduce the use of non-human primates and large animal models, genetically modified murine models have been produced. For example, genetically modified ApoE- and Ldlr-deficient mice have higher plasma cholesterol levels (5 times and 2–3 times higher than controls, respectively) [114]. Aside from these, several other transgenic models have also been introduced as suitable experimental animals for studies of plaque deposition and formation, arterial calcification, ulceration, hemorrhage, plaque rapture, thrombosis, and stenosis [115]. For example, Fatty Zucker rats [116], cholesteryl ester transfer protein (CETP) transgenic rats, LDL receptor-knockout (KO) mice [117], and db/db mice [118] are a few of the genetically modified models developed for this purpose. However, the transgenic lines are relatively costly, limiting researchers from using optimal numbers of subjects in their studies [119]. Therefore, ovariectomized adult female Sprague-Dawley rats with suppressed resistance to atherosclerosis were developed by putting the animal on a 15% w/w of heated vegetable oil diet. The same diet over a longer period (16–24 weeks) promoted atherosclerosis in male subjects [120,121]. In addition, it has been shown that vitamin D3 administration may accelerate atherogenic processes.
Rabbits have been used as intermediate-size models, due to the fact that rabbit myocardium shares more similarities with humans than what small rodents do [122]. The spontaneous Watanabe heritable hyperlipidemic MI (WHHL-MI) model has been developed in rabbits for cardiac research purposes [122]. Although the cost of acquiring and housing rabbits is high, it is still much lower than for other larger species.
3.1.2. Large animal models
Small animal models provide significant insight about CVD phenotypes and pathways. However, there are key differences in physiology and pathophysiology of the heart and cardiovascular system of small animals (e.g. mice) and humans. For example, human heart architecture, heart rates, oxygen consumption, contractility, protein expression, and stem cell populations are different from those of rodents [123]. Therefore, large animal models (e.g. dogs, pigs, and sheep), which more faithfully resemble human physiology, function, and structural features, have been employed as experimental models to translate the cellular and biological findings from murine models into the clinic [123]. For example, the canine model has been used to study post-MI remodelling mechanisms of LV and infarct expansion, e.g. MI, ischemic cardiomyopathy, left-sided coronary artery microembolization, and pressure overload models, as well as to study the impact of therapeutic agents [124]. However, one of the main drawbacks of canine models is the inconsistency and heterogeneity in myocardial lesions and responses, due to the collateral coronary circulation which makes the comparison of the outcomes difficult post-injury [125]. In addition, there are growing efforts to minimize the use of canine models due to ethical concerns. Due to these complexities, alternative animal species such as porcine and ovine have been used for translational purposes, due to their greater anatomical similarity to humans, and the ability to make lesions of the same size [126]. For example, the arterial anatomy and collateral coronary circulation in pigs can better mimic those of humans. In addition, infarct size in porcine models can be precisely predicted [84]. Among porcine models, balloon occlusion of LAD coronary artery is the most commonly used model. In this model, infarction is induced by balloon inflation by means of a catheter through the femoral artery and placing an angioplasty balloon in the artery. For example, to quantify progressive infarct expansion over a 2-month period in adult porcine models, MI induction was tracked by radio-opaque markers. This model has shown promise for studying pharmacological effects on infarct expansion [127]. The described models have also been used to investigate the impact of stem cell transplantation on LV contractile function [128] as well as delivery of angiogenic and arteriogenic GFs [129]. Transplanted stem cells and delivered GF were able to preserve contractile function [128], as well as to restore stable collateral networks and enhance myocardial perfusion and function in the infarcted myocardium [129], respectively. More recently, our team also reported a minimally invasive delivery of human cell-derived polymeric cardiac patches to the epicardium in a porcine model [126]. Despite the keyhole access, this model still requires surgical facilities, specialized equipment, and expert personnel, limiting the feasibility of the procedure [130]. Table 2 summarize the pros and cons of each model.
Table 2.
Pros and cons of animal models of CVD.
| Small Animal Models (Rodents) | Large Animal Models (Swine) | |
|---|---|---|
| Pros | ||
| ✓ Easy breading and handling | ✓ Closer to human anatomy, better tissue availability and more accurate minimally invasive measurements | |
| ✓ Short reproductive cycle | ✓ Closer lipoprotein profile to human except for human HDL subclasses | |
| ✓ Relatively cheap | ✓ Moderately atherosclerosis sensitive on normal diet | |
| ✓ Well-defined genome | ✓ Similar vascular lesion structure and lesion distribution to humans | |
| ✓ Ease of genetic manipulation | ✓ Rare thrombosis due to plaque rupture | |
| ✓ Large litter number | ✓Suitable for translational research | |
| Cons | ||
| × Resistance to atherosclerosis development in Wild type (need for transgenic model) | × Costly and difficult maintenance and handling | |
| × Different gross anatomy compared to humans | × No genetic modifications | |
| × Different lipoprotein profile to humans/high level of lipid | × Limited genetic models available | |
| × Compromised lesion formation | × Rare thrombosis due to plaque rupture | |
| × Absence of plaque rupture and thrombosis | × Ethical concerns | |
Although large animal models are more physiologically relevant for studying disease phenotypes, they are costly and impose several ethical restrictions. Therefore, the bioengineering and tissue engineering communities have been implementing extensive efforts to develop in vitro and in silico CVD models for more physiologically and clinically relevant readouts of CVDs.
3.2. Cardiac tissue engineering approaches
3.2.1. Cell sources
Finding an appropriate cardiac cell source is the starting point of any attempt to engineer in vitro platforms. Generating high quality cells, that can retain their phenotype and resemble native heart tissue without losing their biological functions is critical [131]. The sources of suitable cardiac cells include primary cells, cell lines, and undifferentiated human pluripotent SCs [131].
3.2.1.1. Primary cells.
Primary cells are cells that are directly harvested from the tissue; these cells have a limited lifespan as they are not genetically or virally transformed, in contrast to transformed, immortalized cell lines. Primary cells retain relatively similar qualities to their in vivo phenotype [132]. For example, CMs isolated from embryonic chicken and neonatal rats/mice, among other animal species, have been mainly used as in vitro cardiac models [133]. The usage of human primary cells is restricted by some important limitations [134]. For example, primary cells derived from different donors can behave differently depending on age, sex, and genetics. Moreover, in addition to their low proliferation rate, limited expansion capacity, and finite lifespan, it is difficult to use primary cells more broadly in drug discovery, but the advent of commercially available, fully authenticated sources of primary cells is starting to address this issue [135]. There are also no abundant sources of viable primary human CMs, as they can only be isolated from small cardiac biopsies, which are only performed rarely in cardiac disease. At the fundamental level, primary human CMs have an extremely limited ability to proliferate, thus they cannot be expanded to appreciable numbers from cardiac biopsies.
3.2.1.2. Cell lines.
Most in vitro models use cell lines due to their availability and unlimited expansion capacity [136]. Common cell lines include fibroblast, SMC and EC lines. Their widespread, standardised use is one great advantage of the inclusion of authenticated cell lines in engineering of vasculature in vitro or in a model of cardiac tissue regeneration [132]. However, there are some issues associated with the use of cell lines in vitro. For instance, cell responses may be altered at different passage numbers. Similarly, cellular behavior can depend on culture medium and serum as well as the genetic manipulation required for immortalisation, impacting cellular phenotype, function and behavior. In addition, cell lines are not similar to primary cells (i.e. in vivo cells) in several important aspects [136]. For example, many cell lines are derived from tumors affecting their functional properties [136].
Primary CMs naturally cannot proliferate; however, there are a few cell lines such as AC16 [137] and HL-1 [138] which can. AC16 is an immortalized human ventricular cell line, developed by fusing primary ventricular CMs with a simian virus 40 transformed fibroblast cell line. It was reported that these cells can proliferate, express cardiac-specific markers and differentiate under specific culture conditions [137]. The HL-1 cardiac cell line, derived from atrial sarcoma, exhibits a well-organized sarcomere structure and the ability to maintain contractile function after passaging, similar gene expression to the adult CMs, spontaneous depolarization and good ion channel expression [138,139].
Progenitor cells are relatively immature and partially specialized cells that are precursors to entirely differentiated cells of the same tissue type. Some researchers believe that there is no significant difference between progenitors and SCs. They are typically formed by SCs and are able to differentiate to one or more types of cells, but are not capable of generating all cell types [140,141]. Various types of cardiac progenitor cells (such as c-Kit+, SSEA-1+, Scar-1+ and cardiosphere cells) have been used for in vitro and in vivo experiments and revealed some potential for therapeutic purposes [142–145]. Although these cells are relatively easy to identify in rodent models, it is considerably more difficult to demonstrate their relevance in primates and humans.
3.2.1.3. Stem cells.
Limitations of primary cells and cell lines have stimulated scientists to focus on SCs for modelling of tissues and organs. SCs are defined as undifferentiated cells with the ability of self-renewal and differentiation into various types of specialized cells [135]. This facilitates their applications for replacing lost tissues and curing devastating diseases. Controlling SC fate in order to generate differentiated cells of a desired lineage is still a challenge. Their proliferation and differentiation depends on a variety of factors, such as GFs, amino acids, proteins, active ions, and co-culturing with relevant cells or tissue types [146].
CMs can reliably be derived from human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) using protocols that involve timed application of GFs or small molecules, that are designed to recapitulate developmental pathways. The timed application of biomolecules is designed to drive the cells from the undifferentiated state, to mesoderm, to cardiovascular progenitors, and finally to CMs [147–151]. In general, these protocols can be applied to embryoid bodies or monolayer culture.
Since directed differentiation yields a heterogeneous cell population with purity of CMs between 50 and 90% on average, it is necessary to enrich CMs for some applications. This can be achieved by centrifugation [152], lactate switch [153] or cell sorting with an antibody against SIRPA [154]. iPSCs have also been demonstrated to differentiate into various cell types, overcoming the ethical issues associated with the use of human ESCs [155]. As they can be patient derived, iPSCs hold a great promise in providing patients with a significant number of cells, without immune rejection as well as providing improved models for disease modelling. This makes them suitable substrates for drug discovery [156]. Recent advances in iPSC technology [157] have tackled limitations related to low reprogramming efficiency, lengthy differentiation processes, high cost, and variability among iPSC lines [158], enabling the use of these cells for in vitro disease platforms. For example, iPSCs derived from patients with Barth syndrome (BTHS) were successfully differentiated to CMs to develop an in vitro microfabricated disease platform to model cardiomyopathy in patients with BTHS [159].
3.2.2. Maturation of stem cell derived CMs
In vitro engineered cardiac tissues were introduced with the ultimate goal of resembling human heart morphology and function for various purposes including disease modelling, compound testing, and patient specific drug screening. During embryonic development of the heart, from early stages to adult phenotype, not only do CMs go through many structural changes, but they also get exposed to various factors such as mechanical, biochemical, electrical, topographical, cell-cell and cell-ECM signals [160].
iPSC-CMs have become the most appropriate cell source for fabricating functional cardiac tissues [161]. However, due to the lack of a global standard to define and evaluate CM and cardiac tissue maturity, scientists have been comparing engineered cardiac tissue characteristics with adult CMs in regard to different parameters such as morphology [162], structural properties, gene expression [163] and electrophysiology [162].
From the structural point of view, whereas rod-like adult CMs have an aspect ratio between 7:1 to 9.5:1 [164], heterogeneous hiPSC-CMs are much smaller with lower aspect ratios (2–3:1) [165,166]. SCs maintain their proliferation capacity, although this capacity decreases during differentiation [167], but mature CMs are known to have no proliferation capacity [168]. The average length for adult human- and hiPSCs-CMs’ sarcomeres is about 2.0–2.2 μm and 1.6–1.7 μm, respectively [169].
Moreover, hiPSC-CMs generally exhibit no T-tubules, they largely have a single nucleus and microscopic analysis can detect mainly their Z-discs and I-bands. On the other hand, adult CMs have prominent T-tubules, they are multi-nuclear cells and transmission electron microscopy clearly shows their H-, A-bands, and M-lines in addition to Z-discs and I-bands [166]. In addition to their structural differences in comparison to the adult human CMs, iPSC-CMs have major gene expression and cell function diversity. Researchers utilized various methods to induce maturation in hiPSC-CMs to achieve morphology and function similar to the adult CMs.
One of the common approaches is adding different biochemical factors to the culture medium, such as triiodothyronine (T3) [170], ascorbic acid [171] and neuregulin-1β [172]. Among these, T3 is known to have an essential role in cardiac development. For example, hiPSC-CMs were incubated with T3 for a week and exhibited an increase in CM sarcomere length, size and anisotropy ratio, besides increase in consumption rate of oxygen and contractile force [170].
Electrical stimulation is among the most important biophysical cues that induce CM growth and maturation [173–175]. In an excitation-contraction coupling mechanism, an electrical stimulus is converted to contraction of CMs. In a recent study, early initiation of electrical stimulation (day 12 after differentiation) was compared to a late-stage initiation (day 28 after differentiation) in hiPSC-CMs based cardiac tissues [175]. The tissues were subjected to a constant stimulation frequency, or the intensity training by electrical stimulation of ramping frequency. In this study, early initiation (day 12 after differentiation) of intensity training exhibited a remarkable maturation efficiency of the tissue, resulting in sarcomeric structures indistinguishable from those of the adult human heart. hiPSC-CMs were reported to have a sarcomere length of 2.2 μm, 30% mitochondrial density, the presence of T-tubules and highly organized ultrastructure. While functional properties (such as resting membrane potential (−70.0 mV) and conduction velocity (25.0 cm/s)) showed considerable improvement, a less mature phenotype was observed compare to adult CMs [175].
Since CMs experience different mechanical stress such as cyclic stretch (from hemodynamic load), static stretch (cell-ECM interaction), and shear stress (laminar blood flow), scientists mimicked these mechanical cues in vitro to induce CM maturation [176]. Mihic et al. seeded hESC-CMs into gelatin sponge and utilized mechanical stretch to enhance cellular maturation [177]. The structure experienced uniaxial cyclic stretch for 3 days. This sample exhibited an increase in the size, number and elongation of cells compared to the unstretched sample. Moreover, the stretched sample showed a lower calcium cycle duration, higher rate of contraction and increase in the expression of ion channels and gap junctional proteins [177].
Tailoring surface topography is another strategy to induce CM alignment [178–192]. Myofiber orientation in the native heart, especially in the LV, shows highly aligned myocardial strands. It has been shown that CM alignment has a significant impact on electromechanical coupling and production of contractile force [193,194]. It was demonstrated that sarcomere alignment and organization affect APD, contractile stress, and kinetics of Ca2+ transients (e.g CTD) [195,196]. Various surface topographies such as microgrooves [187,197], nanopatterns [180,198] and microposts/pillars [179,185] have been investigated to guide CM alignment. For instance, our team reported the role of microgrooves/micropatterns and hydrogel stiffness on the alignment and elongation of cardiac cells [188].
Furthermore, some other effective methods have been reported to induce CM maturity, including long term culture of CMs in vitro [169], genetic manipulation [199], and modulation of microRNAs [200]. Recently, the combinations of mentioned strategies have been used to improve CM maturity [173,201].
3.3. In vitro models in CVD research
3.3.1. 2. D in vitro models
Pharmaceutical companies have been utilizing 2D cardiac in vitro models to assess functional properties and test cardiotoxicity in preclinical stages for decades. For example, electrophysiology and rhythm disorders are among the main parameters that have been measured in 2D models [202]. Fabrication of 2D tissues with aligned CMs have been extensively used in order to engineer native-like cardiac monolayers and to model various diseases such as conduction disorders [203,204]. Alignment of CMs is critical for maintenance of high conduction velocity throughout the native myocardium [205]. Various techniques have been proposed to meet this requirment in the 2D environment, including ECM protein micro-contact printing [206], coverslip microabrasion [207], and soft substrate micro-molding [202].
Ion channels play an important role in rhythmic and effective cardiac contraction [208]. Many cardiac diseases, such as long QT syndrome (LQTS) [209], are due to ion channel dysfunction. Various high throughput assays including ligand binding assays, voltage-sensitive dye assays, flux-based assays, fluorescence-based assays, and automated electrophysiological assays (e.g. patch-clamping), have been conducted to investigate ion channel activity [210]. hiPSC-CMs from patients with specific inherited cardiac arrhythmias are used as a promising cell source to model these disorders [211–213]. LEOPARD syndrome [214], familial dilated cardiomyopathy [215], Timothy syndrome [216], familial hypertrophic cardiomyopathy [217], aldehyde dehydrogenase 2 genetic polymorphism [218] and long QT [211,212] are some examples of patient-specific iPSC-derived models. For instance, an in vitro model of congenital long QT syndrome was developed [212] by reprogramming dermal fibroblasts from a 28-year-old patient with type 2-long QT syndrome. Recorded data from patch-clamp and extracellular electrodes showed prolonged APD compared with healthy hiPSC-CMs, and more than a 60% decline in the cardiac potassium current (IKr) of long QT cells indicating appropriate phenotypic characteristic [212].
Another important tool used in investigating electrophysiological properties is the microelectrode array (MEA), which enables researchers to measure impulse propagation of CMs seeded on top of a substrate with micro-electrodes. For example, MEA recordings of cardiac ectopic activity detected cardiac arrhythmogenic activities in 38% of patient-specific samples compared to 6% of healthy ones [212].
Although using 2D models enables researchers to investigate cardiotoxicity at the cellular level, these models suffer from lack of suitable environmental factors including 3D ECM-cell and cell-cell interactions. To overcome these challenges and provide a more realistic microenvironment similar to the native tissue, 3D models have been developed and investigated in depth [219].
3.3.2. 3. D in vitro models
To mimic the physiological and anatomical structure of the native heart, researchers have used various techniques to build up more complex 3D microenvironments [220]. To this end, four main approaches were used to fabricate 3D scaffolds in vitro including encapsulating cells inside hydrogels [221,222], seeding cells into prefabricated structures [223], utilizing decellularized ECM of the native heart tissue [224], and overlaying 2D cell sheets on top of each other [225].
Among others, cell encapsulation inside a hydrogel is currently a main approach for creating engineered heart tissue (EHT) (Fig. 3) [226]. Development of the first EHTs goes back to 1997 when embryonic chick CMs were cultured in a collagen matrix, and resultant tissue exhibited a direct relationship between increase of applied electrical pacing frequency (from 0.8 to 2.0 Hz) and force generation [226]. Each EHT platform requires three components: 1) isolated heart cells (these might be from chicken, mouse, rat, hiPSC or hESC) [221,227]; 2) a hydrogel that is able to form suitable ECM for cells after gelation [228]; and 3) a chamber to provide an aseptic environment for further culture of the cell-containing hydrogel (i.e. bioreactor) [229]. These EHTs offer a platform with a high-level of monitoring capacity and a suitable approach toward enhancing the maturity of hiPSC-CMs [229]. A modified version of the EHT platform has been placed in a 24-well plate, to facilitate the need for repeated measurements of different functions of the tissues. Using elastic silicon posts, this modified EHT has the capacity to apply mechanical load without requiring any extra stretching devices. Moreover, the cell density per EHT was 15% lower than the original EHTs [222,230–232]. Myriamed and ETH-Technologies have commercialized the EHT platforms.
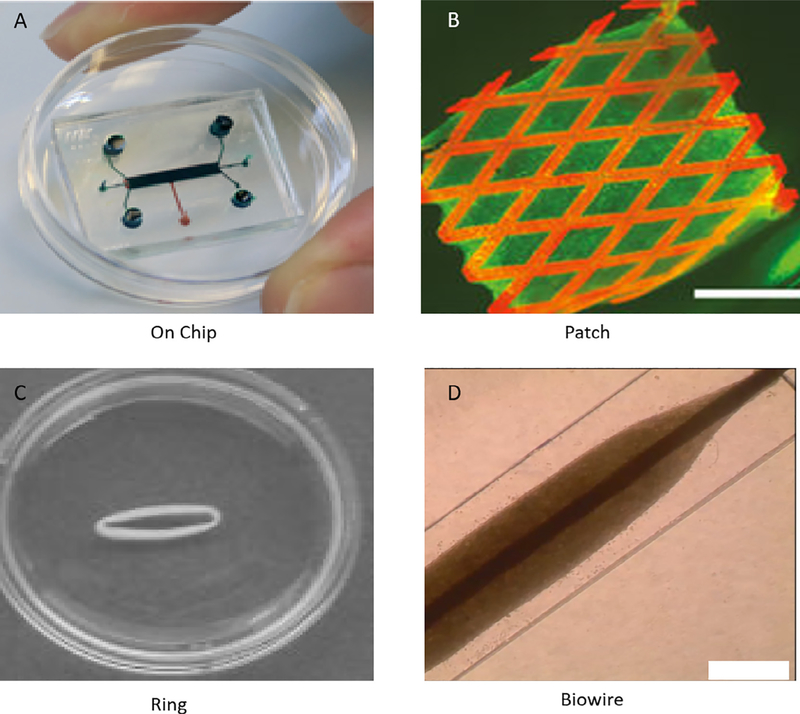
Fig. 3.
Different approaches to fabricate engineered heart/cardiac tissue: A) Organ on chip [250], B) Cardiac patch; scale bar: 2.5 mm [126], C) Circular EHT to apply mechanical stimulation [251], D) Rod-shaped Biowire to apply electrical stimulation; scale bar: 0.5 mm [252]. (Reprinted with permission from Ref. [126,250–252]).
3D models have several advantages compared to 2D models such as cell-cell, cell-ECM interactions, physiological cues, more complex microenvironments and higher similarity to native tissue. In addition to these general advantages, EHTs provide the opportunity to measure contractile function, Frank-Starling mechanism, and investigate the effects of mechanical and electrical stimulation [233,234]. Moreover, patient-specific and various disease models [235] can be designed using EHT platforms which allow one to assess their functions for a long period of time. For example, Eschenhagen and Zimmermann have developed EHTs in a two-post system that allows for easy tissue formation and contractile force evaluation [236,237].
These models have been adapted for different studies in the fields of cardiac and skeletal muscle tissue engineering [238–241]. Although EHTs show a great promise in cardiac research, one of the limitations includes the need for a large number of cells per EHT (0.4–2.5 million CMs) [227,232]. In a recent study, Shadrin et al. [242] combined hydrogel molding [243,244] and dynamic culture [245] techniques to develop a platform for culturing and maturing hiPSC-CMs to a clinically relevant size (from 7 mm × 7 mm–40 mm × 40 mm). Their 50 μm-thick platform was grown without any exogenous stimulation and exhibited mature functional properties including conduction velocity of 25.1 cm/s [242].
Motivated by a native cardiac niche, we developed Biowire [246], by seeding cardiac cells encapsulated in a hydrogel into a PDMS microwell, where the cardiac cell suspension self-organized around a suture template situated in the middle of the microwell. Since CMs were aligned toward the suture, the overall intercellular organization significantly improved compared to the age-matched embryoid bodies. Although the platform did not allow for contractile force measurement, the incorporation of electrical stimulation of increasing ramping frequency improved the maturation level of Biowire significantly in terms of both electrophysiological and calcium evaluations. Because most drug candidates target adult human tissues, Biowire, with a more matured phenotype and a closer recapitulation of the adult native cardiac tissue, is considered to have more predictive power for cardiotoxicity screening [247]. In addition, since this model lacks vasculature the tissue diameter was minimized on purpose to ~600 μm to allow for appropriate oxygen and nutrient diffusion [248]. Biowire is being commercialized by TARA Biosystems which enables the maturation of cardiac tissue in vitro by electrical stimulation [173,249]. Matured cardiac tissue is an essential factor for translation and commercialization. Table 3 summarizes the pros and cons of each 3D in vitro model.
3.3.3. Vasculature-on-a-chip
As already discussed, large 3D organs cannot be fully represented by a cell monolayer which lacks the sophisticated, highly organized structures that are integral to organ function. One of the main missing components is the vasculature, which is responsible for adequate nutrient and oxygen delivery to maintain cellular activity [254]. For example, after oral or intravenous injection, drugs circulate through the body in the bloodstream and enter tissue after penetrating the capillary endothelium. The interaction between drugs and endothelium may release paracrine signals, such as NO, which indirectly influences cellular behavior [255]. To tackle these challenges, the vasculature-on-a-chip platform may be an ideal tool to recapitulate in vivo drug-tissue interactions but in the in vitro setup. For example, drug candidates that can induce vascular injury, which typically manifest in preclinical animal studies through inflammation and changes in vascular tone [256], may be identified prior to animal testing with vasculature-on-a-chip platforms, reducing unnecessary expenses and casualties.
One of the challenges for vasculature-on-a-chip is to fabricate functional open lumen structures that can withstand continuous perfusion after cell seeding. ECs were exposed to molecules, such as VEGF [257] or thymosin β−4 [258] to create self-assembled or guided vasculature. However, these structures were not stable enough, making the direct perfusion challenging and necessitating the use of microfabrication to create a more stable vascular structure. Microfabrication enables the use of biomaterials to 1) support long-lasting open EC lumens for perfusion; 2) define sophisticated capillary networks to mimic the hierarchy of native vasculature; 3) guide and support EC growth and maintain their cellular functions [259–263]. Many fabrication methods have been recently explored. Zheng et al. [259] used the soft lithography technique to create a patterned collagen I hydrogel and obtain a fully endothelialized lumen network with physiologically relevant geometry. The network was simple but was capable of sprouting angiogenesis with GFs and adequate platelet responses after interaction with whole blood. Kolesky et al. [260,261,264] developed a 3D printing procedure that realized a more complex lumen network with multiple materials/cell mixtures. The printing method was able to give rise to a thick human tissue (> 1 cm) that integrates parenchymal, stromal cells and ECs. It was possible to cultivate the final construct up to 6 weeks. Miller et al. developed 3D printed rigid fiber networks of carbohydrate glass, which were then used as a sacrificial layer to create vascular networks in the hydrogels. These networks could be perfused with blood under high-pressure pulsatile flow [262]. In another study, Zhu et al. reported a rapid, mask-free digital light processing based bioprinting method to create pre-vascularized tissue structures directly with unique resolution and speed. This emerging technique could overcome challenges such as blocking and overflowing that occur during nozzle-based 3D printing and offer greater resolution, speed, scalability and flexibility [265]. More recently, our team has developed a multidimensional scaffold with a built-in vascular network using a 3D stamping technique (AngioChip). With long-term perfusion under gravity driven flow, the system was able to improve the viability of cells embedded in the parenchymal space. The system also enabled vascular sprouting through the micro-holes of the scaffold into the parenchymal spaces (Fig. 4 A-I). The cells in the parenchymal space demonstrated physiological responses to drugs applied through the vasculature, specifically engineered cardiac tissue perfused with epinephrine increased the spontaneous beating rate, and engineered metabolised terfenadine to fexofenadine [266]. The successes of these studies demonstrated the physiological relevance of vasculature-on-a-chip and its potential in drug testing.
Fig. 4.
Vasculature-on-a-chip (A–C), SEM of lumen networks for A) a 1D tube (scale bar = 1.5 mm and 500 μm), B) a 2D AngioChip scaffold (scale bar = 1 mm and 300 μm) and C) a multi-layer 3D AngioChip scaffold with 20 μm micro-holes (scale bar = 1 mm and 400 μm) created with the 3D stamping technique. SEM of parenchymal spaces of D) an AngioChip scaffold with 10 μm micro-holes on the channel walls (Scale bar = 200 μm); E) the 3D lattice matrix in between the microchannels (scale bar = 100 μm); F) the cross-section of a 10 μm micro-hole on the channel wall (scale bar: 50 μm). Red arrows point to the micro-holes. (G–H) SEM of the parenchymal space of an AngioChip scaffolds with 20 μm micro-holes on the top and side walls of the micro-channels. Red arrows point to the micro-holes on the top and side walls. Scale bar: G) 400μm, and H) 100 μm. I), Schematic of the assembly of the bioreactor showing inlet, main and outlet wells. (Reprinted with permission from Ref. [266]). J) schematic diagram that summarizes the key aspects of the InVADE platform (Reprinted with permission from Ref. [274]).
In addition, recent studies described the development of high throughput vasculature-on-a-chip systems, to facilitate drug testing on various organ types [266–271]. For example, Loskill et al. presented a Lego®-like plug & play system, μOrgano, which enabled both individual culture of single organ-on-a-chip platforms, as well as customization with integration of multi-organ systems [272]. Another vascularized and perfusable system was developed in a conventional 96-well plate format [273]. The platform facilitates perfusion using hydrostatic pressure and was theoretically capable of connecting multiple mini-organs (cardiac, liver, etc.) through the vasculature. Similarly, our team developed a multi-organ system, inVADE platform, with an integrated vasculature that enabled functional readouts, such as the contractile behavior of cardiac tissue (Fig. 4 J(1–6)) [274]. In the inVADE platform, cardiac tissues were formed from hiPSCs-derived CMs encapsulated in fibrin gel. Two cantilevers were also used to determine the frequency and force of tissue contraction in a non-invasive way. We also reported the impact of biochemical (a β-adrenergic agonist epinephrine) and electrical stimulation on cardiac tissue contraction. Immediate increase in tissue contraction frequency was observed after drug perfusion through the microvasculature. Electrical stimulation in this 96-well plate platform showed the gradual increase in tissue contraction under pacing which is known to improve cardiac tissue maturation [275].
These platforms are versatile tools to investigate specific drug effects on individual tissue models or to link multiple different tissue types together via integrated vasculature, and therefore potentially enable modelling of systemic exposure to either a drug and/or its metabolic by-products. Thus, micro-tissues corresponding to various organs would provide valuable information to aid pre-clinical and clinical drug screening. However, it remains a challenge to develop functional evaluations that allow systematic assessment of the risks and efficacy of drug candidates in such platforms.
Several vessel-on-a-chip platforms are currently being commercialized. For examples, Nortis developed a vessel-on-a-chip platform for drug testing [276]. The chip contains a tubular microchannel embedded inside a hydrogel. These channels are endothelialized and surrounded by pericytes. Robust endothelial sprouting and pericyte interaction have been reported [276]. Another example is an artery-on-a-chip device [277,278] by Quorum Technology which enables ex vivo probing of structural and functional properties of small diameter blood vessels. Organos Inc. has also commercialized μOrgano platform, which may be used to build a vascularized cardiac tissue [279].
3.3.4. Thrombosis on-a-chip
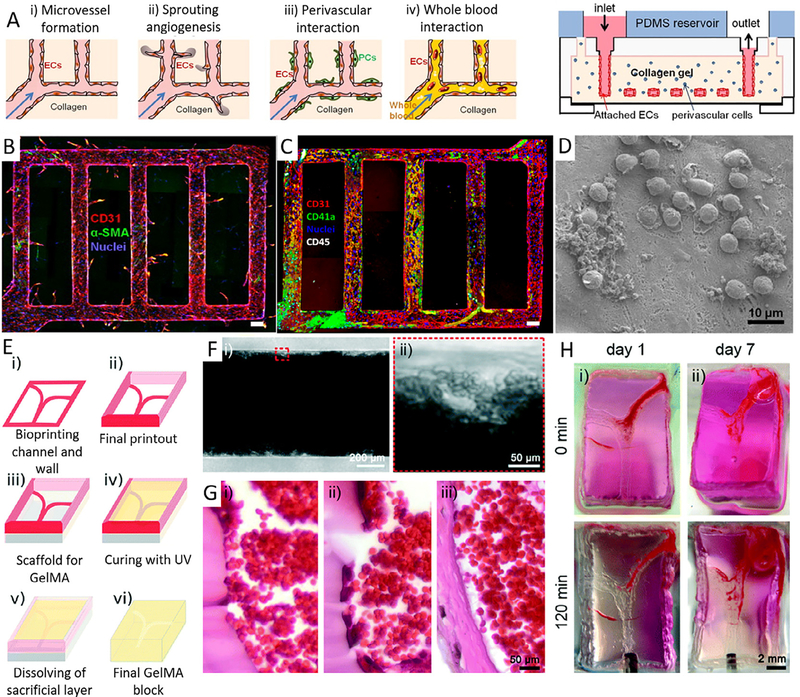
Fluid shear stresses and recirculation zones at stenosis or vessel bifurcations are critical contributors to atherogenesis [280–282]. The most frequently-used in vitro thrombosis models are cone-and-plate, parallel plate flow chamber viscometers and orbital shaker-based flow systems [283,284] (reviewed elsewhere in detail, for example [285,286]). These models can promote our understanding about the pathobiology and pathophysiology of thrombosis by inducing physiological shear stresses on cultured ECs or by recirculating blood flow to study the mechanism of shear stress on coagulation. However, they are incapable of faithfully mimicking 3D micro-physiological structure and hemodynamics of the vascular, arterial and microvascular networks. Therefore, microfluidic devices (i.e. thrombosis-on-chip devices) are of interest to mimic a range of flows and shear stresses, starting from low (i.e. in veins) to high values (i.e. in arteries) [287–290]. Microfluidic perfusion models that are used to model early stage atherosclerosis and later thrombosis have a significant potential in drug development [291–294]. These devices possess endothelialized lumen that can reliably capture several features of blood vessels such as atherosclerosis (i.e. fatty plaque deposition and formation) and bifurcations (i.e. branches and bends) by changing the channel geometries and flow patterns, ranging from laminar steady flow to pulsatile flow [287–290,295–304]. For example, platelet adhesion and thrombosis have been studied in a microfluidic device fabricated by soft lithography which resembles vascular networks [305]. In this work, endothelialized perfusable microchannels were fabricated with collagen hydrogels to study whole blood interactions with ECs (Fig. 5 A). Significant reduction in platelet adhesion on the lumen of the collagen hydrogel microchannels was observed as compared to a device fabricated from phorbol12-myristate-13-acetate (Fig. 5 B-D). Platelet adhesion to the lumen wall is known to trigger the secretion of von Willebrand factor (vWF) and the subsequent significant increase in platelet recruitment.
Fig. 5.
Thrombosis on-a-chip. A) Endothelialized perfusable microchannels fabricated with collagen hydrogel to study whole blood interactions with ECs, B) Z-stack confocal image of endothelialized microfluidic vessels showing endothelial sprouting from the walls, Red, CD31; blue, nuclei. (Scale bar: 100 μm), C) Leukocytes and platelet adhesion on stimulated microfluidic vessels after perfusion with of whole blood for 1 h. Red, CD31; green, CD41a; white, CD45; and blue, nuclei. (Scale bar: 100 μm), D) SEM of leukocyte adhesion on and migration through stimulated microfluidic vessels after 1 h of whole blood perfusion (Scale bar: 10 μm) (Reprinted with permission from Ref. [305]), E) Schematic of the bioprinting process: i, ii) bioprinting of a Pluronic mold; iii) assembly of the dried mold on PDMS; iv) filling the mold with GelMA followed by UV crosslinking; v) washing off the sacrificial channels to create vi) the final construct with hollow channels, F) Optical image representing the formed thrombus in a microchannel, where aggregated RBCs were clearly observed (Scale bar: 50 μm), (G) Optical image showing H&E-stained transverse sections of (i) a thrombus in control channel and (ii) a thrombus in endothelialized channel with HUVECs, both after 7 days, and (iii) a thrombus formed in vivo at 7 days (Scale bar: 50 μm), (H) Time-lapse photographs showing the thrombolysis of (i) a 1 day clot and (ii) a 7 day clot (Scale bar: 2 mm) (Reprinted with permission from Ref. [296]).
In a recent study [296], a 3D-printed thrombosis-on-a-chip model has been developed by using GelMA hydrogel (Fig. 5 E-H). ECs were seeded on the channel walls and perfused with human whole blood to understand the cellular interactions and clinical relevance of this coagulation model. Thus, 3D printing could provide an alternative technology to soft lithography, which is often burdened by high costs, lengthy multistep procedures and difficulty in curvature fabrication. Although microfluidic and 3D-printed platforms can complement the use of in vivo models, they may not fully predict biological processes especially in a longer period of time due to the coagulation issue. Long-term studies will inevitably be needed to systematically evaluate thrombotic processes using whole blood. To address this issue, Qiu et al. developed an endothelialized, agarose-gelatin IPN hydrogel-based microvasculature-on-a-chip platform. This platform can be used for longterm real-time visualization, with high spatiotemporal resolution, of microvascular obstruction and endothelial permeability under physiological flow conditions [303].
Some of these chips are now being commercialized and implemented by pharmaceutical companies. For example, Johnson & Johnson is validating the thrombosis-on-chip device by Emulate for the studies of pulmonary thrombosis.
3.3.5. Cardiac fibrosis-on-a-chip
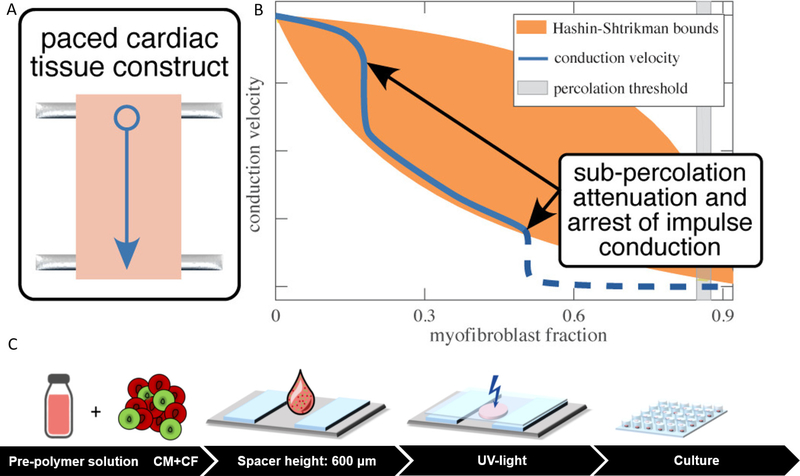
A majority of disease models of cardiac fibrosis are based on 2D coculture and thus lack the intricate microenvironmental properties of the fibrotic ECMs [306,307]. A 3D in vitro microenvironment provides a promising non-animal platform to elucidate pathways governing ECM remodelling, electrophysiological properties and contractile functions. A few tissue engineered fibrosis models have also been reported. van Spreeuwel et al. developed an in vitro cardiac fibrosis model by systematically manipulating the number of fibroblasts and collagen concentration in the engineered cardiac tissues, to mimic fibrotic myocardial composition [308]. Increased fibroblast number was shown to significantly reduce contractile force and alter beating frequency [308]. In another study, Spencer et al. generated a reconstituted tissue model of fibrotic cardiomyopathy by modulating CM and myofibroblast volume fractions (Fig. 6 A) [309]. The results demonstrated a significant role of myofibroblast population in reducing impulse conduction velocity in the fibrotic cardiac tissue (Fig. 6 B) [309]. Sadeghi et al. reported a 3D in vitro cardiac fibrosis model in a GelMA hydrogel-based platform (Fig. 6 C) [310]. By tuning mechanical properties of GelMA hydrogel and applying subsequent biochemical stimulation by transforming growth factor-β1 (TGF-β1), fibrotic-like tissues with activated myofibroblasts were created. The diseased tissues presented asynchronous beating behavior and increased mechanical stiffness. Figtree et al. also developed vascularized cardiac spheroids as a 3D in vitro model to investigate the mechanisms involved in cardiac fibrosis, by coculturing CMs, ECs, and CFs isolated from dissociated neonatal rat hearts in hanging drop cultures. A significant rise in collagen deposition following TGFβ1 treatment in spheroids was observed. In addition, cell death and disturbed vascular networks in spheroids were observed after addition of a cardiotoxic and profibrotic agent (i.e. Doxorubicin). These findings demonstrate that cardiac spheroids can be used to elucidate the underlying pathways as well as therapeutics for preventing and treating cardiac fibrosis in vitro [311].
Fig. 6.
Engineered disease models of cardiac fibrosis. A) An engineered cardiac tissue model of fibrotic myocardium based on modulation of CM and myofibroblast volume fractions. The fibrotic EHTs were constructed by encapsulating chicken embryonic cardiomyocytes and myofibroblasts in Type I rat tail collagen. The fibrotic tissue models were generated by replacing cardiomyocytes with myofibroblasts. B) The effects of cellular composition on impulse conduction in this fibrotic model. The CM and myofibroblast volume fractions determined the impulse propagation velocity [309]. C) A simplified 3D hydrogel platform to study cardiac fibrosis. Primary neonatal rat CMs and CFs were encapsulated within a GelMA-based pre-polymer solution to generate in vitro EHTs. Cell-laden hydrogel was placed into a customized UV-chamber and subsequently crosslinked by UV light. The fibrotic EHTs were treated by TGF-β1 [310]. (Reprinted with permission from Ref. [309,310]).
These studies highlighted the opportunity to use engineered cardiac disease models in the studies of fibrosis pathogenesis and molecular pathways underlying cardiac remodelling. However, the aforementioned models were engineered with either rat or mouse CMs. A highfidelity human cardiac tissue model using CMs derived from hiPSCs would be an asset in our progress toward generating human-relevant cardiac disease models enabling personalized medicine and preclinical drug screening.
Moreover, advances in RNA-Sequencing (RNA-Seq) analysis and mass spectrometry-based proteomics have facilitated the examination of gene expression and global protein profile to better understand the transcriptional and translational regulation underlying human cardiac disease. There are several human signaling pathway databases distributing comprehensive datasets from systematic collections of compiled experimental results [67]. They constitute a helpful resource that can guide targeted investigations of future pathway studies. Therefore, combining these technologies with 3D in vitro platforms will provide valuable insights into the pathological processes responsible for cardiac fibrosis.
3.4. In silico approaches
Recent developments in statistical/empirical and mechanistic cardiac modelling have opened new avenues improving drug-induced cardiotoxicity screening. In silico approaches have been implemented by both regulatory agencies as well as pharmaceutical companies. The paradigm change in cardiotoxicity assessment started with the mechanistic modelling of electrophysiology of cardiac cells (i.e. a modification to Hodgkin–Huxley formulation) [312]. These studies could delineate the contribution of specific ion channels and transporters to normal and disrupted heart rhythm [313–316]. In silico modelling also provides a robust tool for demonstrating drug-ion channel interactions and their effects on heart function. For example, one of the potential adverse effects of a drug is the proarrhythmic effect due to the interaction between drug compounds and cardiac membrane ion channels [317]. Several theoretical models have been developed to address these phenomena. Receptor theory is one of the well-established and important frameworks for computational modelling of pharmacological actions on cardiac ion channels [318]. A receptor is modelled using the known laws of physical chemistry to delineate binding of molecules to cellular receptors [319]. In addition, other modelling frameworks, such as the Markov chain model, were developed for simulating interactions between drugs and Na+ and Ca+2 channels [320,321]. These mathematical models have been used to explain the effect of β-adrenoceptor on ventricular arrhythmogenesis in association with long QT syndrome [322,323].
Current preclinical screening assays typically involve in vitro and in vivo models to detect structural, contractile and electrophysiological toxicity, which could lead to myocyte impairment and loss, heart failure and proarrhythmia, respectively. These simple and non-specific assays for the early screening of cardiac toxicity can lead to unjustified attrition of new drug entities due to false-positive findings. This risk can also be augmented by the differences among the effects of compounds in animals and humans. Therefore, more-comprehensive multi-parametric approaches, which combine in vitro and in silico technologies have been implemented to detect such cardiotoxicities. More recently, this concept has evolved through the Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative [324,325]. CiPA provides a mechanistic evaluation of drug effects on ventricular myocyte electrophysiology based on in silico reconstructions of human ventricular electrical activity and multiple cardiac ion currents (i.e. delayed repolarization, repolarization instability and early after-depolarizations) to predict the proarrhythmic risk. In the subsequent steps of testing, the results are then validated with hiPSC-CMs platforms [326]. The combination of the human ion channels, cardiac currents, in silico reconstructions of human cellular proarrhythmia and hiPSC-CM models to assess contractile and structural cardiotoxicity is expected to lead to significantly diminished drug attrition rates and to the reduced number of animals in pre-clinical drug discovery.
Furthermore, a new quantitative systems pharmacology (QSP) discipline has recently emerged, that uses mathematical computational models to describe biological systems, disease progression and drug pharmacology in a single modelling framework [327–329]. QSP combines elements of translational drug pharmacokinetics (PK), pharmacodynamics (PD) and systems biology to address clinical needs, such as representation of patient variability for dose selection, demonstration of long-term disease progression to find optimal treatment, and facilitation of modelling accessibility for clinicians and pharmaceutical companies in order to understand the diseases [330]. The main goal of the QSP modelling approach is the identification of drug targets in a biological system. QSP is based on dynamic computational modelling that integrates experimental evidences and data from multiple studies. This approach has been used in several cases related to CVDs [330,331]. QSP modelling can also be used to investigate cardiotoxic events. In addition, QSP computational modelling attempts to define the mechanisms underlying the drug-induced arrhythmic events, in order to find the drug’s therapeutic window where the occurrence of such events may be avoided [332]. For example, an in silico platform was developed by combining a PK model with a cardiac model to simulate AP at data points along the PK profile [321,333]. In silico reconstruction of known cardiac biology can be integrated with the observed pre-clinical and clinical data to predict CVD progression in individuals or populations. In this regard, the physiologically based pharmacokinetic model (PBPK) modelling approaches can also be implemented to predict the concentration time courses in the tissue of interest (e.g. myocardium) and PK of drug-drug interactions. For example, PBPK was integrated with the response model of CM ion channels [334,335] in order to predict the drug-drug interaction of domperidone and ketoconazole on QT prolongation.
Another area that QSP modelling can have significant impact is the development of tyrosine kinase inhibitors (TKIs). TKIs are cancer therapeutics that often exhibit cardiotoxic effects, including LV malfunction and HF. It would be ideal to establish a mechanistic approach first, to assess and evaluate new TKIs efficacy, especially on signaling networks in CMs, as well as to identify new mechanisms of action to minimize accompanying cardiotoxicity. These can be addressed by the QSP approach, where large-scale measurements are combined with mechanism-based mathematical modelling [336].
In addition, data driven quantitative modelling has also been used in the drug screening process. For example, Conant et al. [337] have developed an artificial neural network model to analyze the experimental results in an efficient and unbiased manner to select kinase inhibitors with maximum efficacy and minimum side effects on cardiac cell viability and function. This model helps identify cardiotoxic effects for some available kinase inhibitors that were not detected during clinical trials.
Overall, these examples demonstrate the potential use of mechanistic and data driven modelling in dose selection, studies of drug-drug interaction, and identification of the most efficient therapies [338].
4. Roadmaps for CVD drug screening
CVDs are among the most prevalent diseases worldwide accounting for approximately 30% of deaths annually [12]. Common strategies for prevention and treatment of such complications have been the administration of drugs in early stages, and interventional procedures in later stages of the diseases [339]. Therefore, there is a growing need for new therapeutic agents with improved efficacy and safety. Pharmaceutical companies invest on average 2 billion dollars for each new drug candidate, taking over 12 years to develop an approved drug and place it on the market [10,340,341]. Despite high demand and ongoing R&D efforts, the number of new regulatory-approved therapeutic compounds has been declining over the past decade [10,341]. New drugs must pass strict regulatory standards before entering the market, which are upheld by national and international agencies (e.g. FDA, Health Canada, EMA, etc) [342,343]. However, despite the exhaustive, timely and costly efforts in the early stages of drug development, many of those therapeutic drug candidates cannot enter the market due to safety (e.g. cardiotoxicity or hepatoxicity) or efficacy concerns in the late stages of the drug development pipeline. The attrition rates in Phase II and Phase III development stages have been reported to be as high as 80% [75,344]. In addition, some drugs have also been withdrawn after approval due to their cardiotoxicity [345]. Over 60% of expenditures in the drug development process occur during early stages of development [346]. This fact emphasizes the importance of investing in reliable, economical, safe preclinical assays to screen for effective drug candidates in early stages, in order to reduce the tremendous development costs as well as replace and optimize the less efficient steps of the drug development process [347].
Lack of the appropriate human disease models has been reported as a key limitation in early stage drug discovery [348]. Even though animal models have conventionally been used to elucidate the disease response, there are vast limitations in terms of predictability and reliability of these models [115]. Although significant efforts have been made in terms of developing alternative animal models to shed light on the mechanisms of human diseases and to advance novel and more effective therapies and diagnostics [349], there are still unmet needs in order to overcome the challenges of moving drugs from the bench to bedside. Current animal models, including genetically modified ones for CVDs, are incapable of fully recapitulating human physiology and pathophysiology, and thus imprecisely predict the biology and mechanisms involved in human cardiac dysfunctions [84]. Therefore, pharmaceutical companies use alternative systems such as the relatively simple 2D monolayer cell cultures of cell lines, and, more recently, CMs derived from iPSCs [350] which are unable to accurately mimic aspects of the 3D microenvironment of tissues and organs of interest [351], where the main functions and physiological features are more complex. Therefore, questions have been raised about their clinical predictability and accuracy [350].
3D tissue models, such as in vitro tissue-engineered disease models have shown promise for addressing the current obstacles faced with 2D in vitro cultured cells and animal models [352–356]. Engineered 3D in vitro disease models that use cells from patients differentiated to the specific cell fate under standard culture conditions, provide a deeper understanding of the breadth and complexity of human disease mechanisms, and can be used for testing the efficacy and safety of emerging therapeutic drugs during preclinical testing [156,357].
During the past decade, some important advances have been reported for mimicking features of native tissue physicochemical microenvironments in vitro. These more relevant physiological structures can promote cell phenotype, metabolic activity, maturation, and functionality, leading to more reliable and more predictive models [202].
Recent advances in tissue engineering and microfabrication technologies have contributed to the emergence of organ-on-a-chip models for emulating human systems biology [358]. These novel techniques could be a valid support for the development of 3D biomimetic systems displaying the intricate structural and functional characteristics of human organs [359]. These platforms contain more complex and sophisticated topographical and biomechanical cues for human tissue modelling than a thin layer of cells [360]. However, engineering of disease models still remains a challenge [361].
In parallel, in silico modelling and computational medicine are emerging fields focused on developing quantitative approaches to help make decisions, reduce drug discovery costs and enhance the chances of success in therapy [365,366]. In silico modelling is a low-cost, ethical and practical way to quickly investigate multiple hypotheses [367]. One of the well-established approaches in computational medicine is mechanistic modelling or knowledge-based method, which translated biological events to mathematical expressions [368–370]. For example, QSP is a powerful tool for developing a dynamic mechanistic model capturing the interactions between drugs and disease networks [371]. These in silico drug-disease models have received significant attention in the attempts to find optimal treatment strategies for CVD patients with multiple risk factors [332].
The in vitro and in silico disease models have so far been used to complement the use of laboratory animal models, however it is foreseen that the combination of these two fast-growing and emerging platforms with less costly, more ethical and accurate outcomes, would be a promising substitute, which can reduce animal model consumption and acquire high-quality results in the future. Fig. 7 represents different steps in the drug development pipeline that are required to receive a regulatory approval.
Fig. 7.
Drug development pipeline. Step 1: Discovery: Search for a drug candidate starts in the R&D setting; Step 2: Preclinical Research: Drugs undergo in vitro and in vivo testing to address basic concerns about safety; Step 3: Clinical Research: Drugs are evaluated on people in terms of their safety and efficacy; Step 4: FDA Review: Regulatory teams meticulously evaluate all master files related to a new drug or device and decide on their approval; Step 5: FDA Post-Market Safety Monitoring: FDA monitors all device and drug safety once products are marketed.
5. Future perspective
Engineered in vitro disease models have already made incredible progress in academic and pharmaceutical R&D settings, thanks to the high-level engineering approaches which enable one to replicate organ-like complexity and functionality in vitro [202,372–374]. The development of a multi-functional platform that combines knowledge about pathophysiology and etiology of CVDs, ever-growing engineering technologies (i.e. micro/nanofabrication) and stem cell biology, brings hope to the mandate of reducing the use of experimental animals in preclinical research and improving translation and drug discovery. For example, the ultimate goal would be to develop a multi-organ-on-chip platform which integrates different functional organs to assess efficacy, toxicity, and PK at the same time in a controlled manner [375,376]. Despite these advantages, these platforms currently face some limitations; for example, real-time high-resolution imaging (due to the low transparency and high thickness of the chips) and biomarker detection in the chips are difficult. Current studies are focused on solving these issues by integrating on-chip analytical techniques (e.g. HPLC and mass spectrometer, electrophoresis, etc.) [377–379] and in situ imaging tools to improve the suitability of organ-on-a-chip platforms in high-throughput screening and to generate high quality, reliable, results [376,380,381]. However, further improvements and optimization are needed.
Another challenge in these platforms is the incorporation of different cell types (i.e. co-cultured cells) in a single tissue (i.e. heterocellular tissues) to better resemble native tissue environments. This could be a costly and difficult process due to the complexity of the environment and the requirement for tissue stability in a longer period of time [382,383]. In addition, emerging additive manufacturing technology (e.g. 3D printing, electrospinning, biotextiles, etc.) could tackle the issues regarding the timely, costly, multi-step and labor-intensive microfabrication process [384–393]. However, these technologies are still in their infancy and need further improvement and optimization.
As we reported in this review, the majority of cardiac disease research has so far been conducted in animal models, with primary cell cultures, or cell lines. However, animal models are not entirely capable of representing human disease states, and the phenotypes of isolated primary cells often change drastically during culture [115]. On the other hand, human-derived cells are often difficult to obtain [394]. Accessing limited cell populations such as CMs from living patients is particularly impractical. The advent of iPSC technology represents a landmark breakthrough in regenerative medicine, as it enabled acquisition of hiPSCs without ethical concerns and their targeted differentiation into any cell type [395,396]. For example, various cell types that make up the heart can be produced via directed differentiation. The derivation of CMs can be achieved by controlling the expression of Wnt and BMP at specific times from the mesoderm stage [397,398]. Although few protocols reported differentiation of different subtypes of CMs including the ventricular [399,400], atrial [401,402], and pacemaker cells [172,398,403], they still need standardization and harmonization as they result in heterogeneous cell populations. This will enable more precise modelling of specific diseases in the ventricles, or the atria and will open new insights for differentiating patient-specific bioengineered pacemakers. Moreover, hiPSCs could provide an unlimited source of cells for the construction of diseased tissue. Differentiating iPSCs into disease-relevant mature cells and constructing diseased tissues using these cells combined with pathological cues have important implications for understanding disease mechanisms [404]. On the other hand, the maturation of hiPSCs-derived cells is vital to being able to fully mimic the adult phenotype in humans. This process can be expedited by applying chemical, physical and electro-mechanical cues to the differentiating cells. More studies are needed to better understand paracrine signaling, the possible influence of non-CMs (e.g. fibroblasts), and scaffold stiffness [405,406] on CM differentiation. Although hiPSCs could provide an infinite source of cells, the differentiated cell population (i.e. ventricular-, atrial-, and pacemaker-like cells) is not homogeneous [152]. This issue could be solved by cell sorting techniques such as microfluidic cell sorting [407–409].
It is known that patient-specific genotype can significantly affect disease onset and progression, as well as influence drug responses [211,214,410]. Further, hiPSCs can be generated from patients harboring genetic diseases, thereby providing target cells that will possess relevant pathological genetic backgrounds [411].
Patient-on-chip platforms including engineered in vitro tissue models associated with patient derived cells (i.e. individual- and disease-specific hiPSCs) have the potential to revolutionize the drug discovery paradigm, and bring us closer towards the reality of personalized medicine [160,359,412]. Towards this aim, there are efforts to systematically generate iPSCs from hundreds of phenotypically healthy donors, plus several cohorts of donors with inherited genetic diseases, using a standardised experimental pipeline. For example, the International Stem Cell Banking Initiative (ISCBI) and other counterparts in Europe (human induced pluripotent stem cell initiative (HIPSCI)) and around the world have coordinated efforts to regulate the derivation, culture, and characterization methods of hiPSCs [413–418].
Moreover, genome editing technology (e.g. the clustered regularly interspaced short palindromic repeats (CRISPR) technology) promises to be the most important translational perspective in the future [419–421]. Accurate genome editing of mammalian cells using CRISPR technology could change the landscape of personalized medicine. Combining tissue engineering and organ-on-a-chip technology with genome editing/cellular reprogramming to generate new human cell-based models for CVDs can promote accuracy and predictability of these platforms.
Furthermore, there is an unmet need for high throughput toxicity screening for drug testing in which many compounds can be tested with minimized costs and time. There are many drugs that have received U.S. FDA approval, such as TKI-related drugs for cancer, that were developed based on high-throughput screening technologies. However, development of a suitable platform for high-throughput drug screening with reliable, repeatable results and sufficient physiological function to the native cardiac system is challenging, due to the technical limitations as well as the resultant tissue maturity. To assess in vitro cardiotoxicity of a new drug, the FDA has requested companies to investigate inhibition of the cardiac hERG gene that encodes a potassium ion channel of cardiac cells. Inhibition of the hERG channel can be an indicator of the APD prolongation and cardiotoxicity in early stages of drug development [11]. In addition, using patient-specific hiPSC-CMs, researchers are able to model various CVDs such as long QT syndrome, LV non-compaction, hypertrophic cardiomyopathy and dilated cardiomyopathy [217,423,424].
In addition to the recent breakthroughs with engineered in vitro models, in silico models (i.e. theoretical and computational modelling such as QSP modelling and in silico modelling of electrophysiology) have enabled us to capture dynamic systems at variable scales, simulating the impact of drugs and therapeutic strategies used in clinical settings, and testing the validity of current biological understanding and clinical outcomes [325,425]. These computational models are far less costly solutions for making predictions about drug PK, PD and patient population responses [426]. They also provide novel insights into underlying biology which augment our knowledge of diseases [329]. For example, the CiPA initiative has revolutionized the regulatory decision-making paradigm by using a mechanistic assessment to eliminate the risk of a rare and life-threatening arrhythmia which is induced by a newly discovered drug. Thus, the implementation of these new technologies in early stages of drug development (e.g. preclinical and clinical-phase I stages) could help prevent later-stage drug attritions [330]. Similarly, bringing together academic and industrial scientists with regulatory agencies, to make decisions about standardization, regulation and validation of the current platforms to ensure accuracy, specificity, and reproducibility could prevent late-stage drug failures. Furthermore, the synergy between in vitro and in silico CVD models based on individual genomics, environmental factors and lifestyle choices would lead to more reliable in vivo predictions, which would benefit CVD patients who would have access to safer and more effective drugs. The new drug discovery paradigm can also reform the current approach of utilizing animal models in the preclinical stage.
6. Conclusion
In this review, we have reported several types of CVDs that lead to cardiac complications and reviewed current understanding of their underlying pathophysiological pathways. We have also summarized conventional disease models such as experimental animal models and evaluated their relevance towards modelling human disease, as well as translational success and limitations. We also discussed current human-specific tools including in vitro and in silico approaches to model human cardiac tissue and vasculature in both healthy and pathological states, along with the their advantages and limitations.
Emergence of human-based engineered cardiovascular platforms has revolutionized preclinical cardiac regulatory assessments. This paradigm change has emerged from our ever-growing knowledge of the basic molecular and cellular pathways underlying CVDs, the advent of hiPSC-CMs, the availability of high-throughput screening platforms and in silico models which take into account complex physiological functions. These combined human-based in vitro and in silico approaches will complement preclinical in vivo assessments and can fine-tune decision making steps in drug discovery and development pipelines.
Table 3.
Summary of current in vitro cardiac models.
| Shape | Cell source | Scaffold | Culture time | Cell number | Stimulation method | Pros | Cons | Ref. |
|---|---|---|---|---|---|---|---|---|
| tab | Embryonic chick ventricular CMs | Collagen | 6–11d | 1M | None | -Some structural and functional maturation -Genetic modification in cardiac tissue |
-Instability in culture | [253] |
| Ring | Neonatal rat-CMs | Collagen | 14d | 1–10M | Cyclic mechanical stretch | - Some structural and functional maturation | -High number of cells -Specialized set-up is needed for mechanical stimulation |
[222] |
| Cylinder | hESC-CMs, hiPSC-CMs | Collagen | 1wk | 0.5M | Biphasic electrical stimulation | -Lower cell number -Structural and functional maturation |
- Inability to measure contraction force | [173] |
| 2D culture | hESC-CMs,hiPSC-CMs | Matrigel coated plate | 80–120d | 0.175M | None | -Universal culture technique -High level of maturation | - Long culture time -2D shape -Inability to measure contraction force |
[169] |
| Patch | hiPSC-CMs | Matrigel/Fibrinogen | 3wks | 0.5–1.75 and 8.5 M depending on patch size | None | -Therapeutic approach in animal model | -Relatively thin structure | [242] |
| Ring | hESC-CMs | Gelatin sponge | 5d | 6M | Cyclical stretch | -Therapeutic approach in animal model -Biodegradable scaffold |
- Relatively limited maturation | [177] |
| Cylinder | hiPSC-CMs | Fibrin | 5–6wks | 2M | Biphasic electrical stimulation | -Highly structured mature tissue | -Shape not ideal for animal transplantation | [201] |
Acknowledgments
Our work is supported by the Canadian Institutes of Health Research (CIHR) Operating Grants (MOP-126027 and MOP-137107), National Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (RGPIN-2015–05952), NSERC Steacie Fellowship (SMFSU 4620), Heart and Stroke Foundation Grant-in-Aid (G-16–00012), NSERC-CIHR Collaborative Health Research Grant (CHRPJ 4937) to M.R. and National Institutes of Health Grant 2R01 HL076485. H.S. gratefully acknowledges the Fonds de Recherche du Québec -Nature et Technologies (FRQNT) and the Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowships. M.H.M, M.KH., E.Y.W., Y.ZH., A.K. acknowledge the Natural Science and Engineering Council of Canada (NSERC) Doctoral Scholarship. We also acknowledge financial support of the Humane Society International (HSI) and Humane Society of the United States (HSUS). We also thank Dr. Lindsey Marshall (HSI) and Dr. Hisham Ibrahim (Laboratory of Cellular Physiology and Immunology, U of T, Canada) for critical reading of this manuscript.
References
- [1].Laverty H, Benson C, Cartwright E, Cross M, Garland C, Hammond T, Holloway C, McMahon N, Milligan J, Park B, How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol. 163 (4) (2011) 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ross JS, Krumholz HM, Bringing Vioxx Back to Market, British Medical Journal Publishing Group, 2018. [DOI] [PubMed] [Google Scholar]
- [3].Waxman HA, The lessons of Vioxx—drug safety and sales, N. Engl. J. Med 352 (25) (2005) 2576–2578. [DOI] [PubMed] [Google Scholar]
- [4].Onakpoya IJ, Heneghan CJ, Aronson JK, Worldwide withdrawal of medicinal products because of adverse drug reactions: a systematic review and analysis, Crit. Rev. Toxicol 46 (6) (2016) 477–489. [DOI] [PubMed] [Google Scholar]
- [5].Shah RR, Can Pharmacogenetics Help Rescue Drugs Withdrawn from the Market? (2006). [DOI] [PubMed]
- [6].Alex A, Harris CJ, Smith DA, Attrition in the Pharmaceutical Industry: Reasons, Implications, and Pathways Forward, John Wiley & Sons, 2015. [Google Scholar]
- [7].Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Concordance of the toxicity of pharmaceuticals in humans and in animals, Regul. Toxicol. Pharmacol. 32 (1) (2000) 56–67. [DOI] [PubMed] [Google Scholar]
- [8].Li AP, Accurate prediction of human drug toxicity: a major challenge in drug development, Chem. Biol. Interact 150 (1) (2004) 3–7. [DOI] [PubMed] [Google Scholar]
- [9].Stevens JL, Baker TK, The future of drug safety testing: expanding the view and narrowing the focus, Drug Discov. Today 14 (3–4) (2009) 162–167. [DOI] [PubMed] [Google Scholar]
- [10].DiMasi JA, Grabowski HG, Hansen RW, Innovation in the pharmaceutical industry: new estimates of R&D costs, J. Health Econ 47 (2016) 20–33. [DOI] [PubMed] [Google Scholar]
- [11].Kola I, Landis J, Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov 3 (8) (2004) 711. [DOI] [PubMed] [Google Scholar]
- [12].Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, Floyd J, Fornage M, Gillespie C, Isasi C, Heart disease and stroke statistics-2017 update: a report from the American Heart Association, Circulation 135 (10) (2017) e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Förstermann U, Xia N, Li H, Roles of vascular oxidative stress and nitric oxide in the pathogenesis of atherosclerosis, Circ. Res. 120 (4) (2017) 713–735. [DOI] [PubMed] [Google Scholar]
- [14].Rosenfeld ME, Ross R, Macrophage and smooth muscle cell proliferation in atherosclerotic lesions of WHHL and comparably hypercholesterolemic fat-fed rabbits, Arteriosclerosis: Off. J. Am. Heart Assoc. Inc 10 (5) (1990) 680–687. [DOI] [PubMed] [Google Scholar]
- [15].Robbins CS, Hilgendorf I, Weber GF, Theurl I, Iwamoto Y, Figueiredo J-L, Gorbatov R, Sukhova GK, Gerhardt LM, Smyth D, Local proliferation dominates lesional macrophage accumulation in atherosclerosis, Nat. Med. 19 (9) (2013) 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Morita S.-y., Metabolism and modification of apolipoprotein B-containing lipoproteins involved in dyslipidemia and atherosclerosis, Biol. Pharm. Bull 39 (1) (2016) 1–24. [DOI] [PubMed] [Google Scholar]
- [17].Toth PP, Triglyceride-rich lipoproteins as a causal factor for cardiovascular disease, Vasc. Health Risk Manag 12 (2016) 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pircher A, Treps L, Bodrug N, Carmeliet P, Endothelial cell metabolism: a novel player in atherosclerosis? Basic principles and therapeutic opportunities, Atherosclerosis 253 (2016) 247–257. [DOI] [PubMed] [Google Scholar]
- [19].Heo K-S, Fujiwara K, J.-i. Abe, Shear stress and atherosclerosis, Mol. Cell 37 (6) (2014) 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Muller WA, Transendothelial migration: unifying principles from the endothelial perspective, Immunol. Rev 273 (1) (2016) 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tavares JC, Muscará MN, Chapter 14 - adhesion molecules and endothelium, in: Da Luz PL, Libby P, Chagas ACP, Laurindo FRM (Eds.), Endothelium and Cardiovascular Diseases, Academic Press, 2018, pp. 189–201. [Google Scholar]
- [22].Xu Q, Disturbed flow-enhanced endothelial turnover in atherosclerosis, Trends Cardiovasc. Med 19 (6) (2009) 191–195. [DOI] [PubMed] [Google Scholar]
- [23].Twickler T, Dallinga-Thie G, Chapman M, Cohn J, Remnant lipoproteins and atherosclerosis, Curr. Atherosclerosis Rep 7 (2) (2005) 140–147. [DOI] [PubMed] [Google Scholar]
- [24].Berneis KK, Krauss RM, Metabolic origins and clinical significance of LDL heterogeneity, J. Lipid Res 43 (9) (2002) 1363–1379. [DOI] [PubMed] [Google Scholar]
- [25].Skålén K, Gustafsson M, Rydberg EK, Hultén LM, Wiklund O, Innerarity TL, Borén J, Subendothelial retention of atherogenic lipoproteins in early atherosclerosis, Nature 417 (6890) (2002) 750–754. [DOI] [PubMed] [Google Scholar]
- [26].Fogelstrand P, Boren J, Retention of atherogenic lipoproteins in the artery wall and its role in atherogenesis, Nutr. Metab. Cardiovascular Dis 22 (1) (2012) 1–7. [DOI] [PubMed] [Google Scholar]
- [27].Miller YI, Choi S-H, Fang L, Tsimikas S, Lipoprotein modification and macrophage uptake: role of pathologic cholesterol transport in atherogenesis, Cholesterol Binding Cholesterol Transport Prot. (2010) 229–251 Springer. [DOI] [PubMed] [Google Scholar]
- [28].Ylä-Herttuala S, Rosenfeld ME, Parthasarathy S, Sigal E, Särkioja T, Witztum JL, Steinberg D, Gene expression in macrophage-rich human atherosclerotic lesions. 15-lipoxygenase and acetyl low density lipoprotein receptor messenger RNA colocalize with oxidation specific lipid-protein adducts, J. Clin. Investig 87 (4) (1991) 1146–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kruth HS, Receptor-independent fluid-phase pinocytosis mechanisms for induction of foam cell formation with native LDL particles, Curr. Opin. Lipidol 22 (5) (2011) 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cybulsky MI, Cheong C, Robbins CS, Macrophages and dendritic cells: partners in atherogenesis, Circ. Res 118 (4) (2016) 637–652. [DOI] [PubMed] [Google Scholar]
- [31].Ghosh S, Macrophage cholesterol homeostasis and metabolic diseases: critical role of cholesteryl ester mobilization, Expet Rev. Cardiovasc. Ther 9 (3) (2011) 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ouimet M, Franklin V, Mak E, Liao X, Tabas I, Marcel YL, Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase, Cell Metabol. 13 (6) (2011) 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Voloshyna I, Reiss AB, The ABC transporters in lipid flux and atherosclerosis, Prog. Lipid Res 50 (3) (2011) 213–224. [DOI] [PubMed] [Google Scholar]
- [34].Greenow K, Pearce NJ, Ramji DP, The key role of apolipoprotein E in atherosclerosis, J. Mol. Med 83 (5) (2005) 329–342. [DOI] [PubMed] [Google Scholar]
- [35].Tarling EJ, Edwards PA, ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter, Proc. Natl. Acad. Sci. U. S. A 108 (49) (2011) 19719–19724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Getz GS, Reardon CA, Apoprotein E as a lipid transport and signaling protein in the blood, liver, and artery wall, J. Lipid Res 50 (Supplement) (2009) S156–S161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Papachristou NI, Blair HC, Kypreos KE, Papachristou DJ, High-density lipoprotein (HDL) metabolism and bone mass, J. Endocrinol 233 (2) (2017) R95–R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Linton MF, Yancey PG, Davies SS, Jerome WGJ, Linton EF, Vickers KC, The Role of Lipids and Lipoproteins in Atherosclerosis, (2015).
- [39].Lusis AJ, Fogelman AM, Fonarow GC, Genetic basis of atherosclerosis: part I: new genes and pathways, Circulation 110 (13) (2004) 1868–1873. [DOI] [PubMed] [Google Scholar]
- [40].Eltzschig HK, Carmeliet P, Hypoxia and inflammation, N. Engl. J. Med 364 (7) (2011) 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Arslan F, De Kleijn DP, Pasterkamp G, Innate immune signaling in cardiac ischemia, Nat. Rev. Cardiol 8 (5) (2011) 292–300. [DOI] [PubMed] [Google Scholar]
- [42].Gwechenberger M, Mendoza LH, Youker KA, Frangogiannis NG, Smith CW, Michael LH, Entman ML, Cardiac myocytes produce interleukin-6 in culture and in viable border zone of reperfused infarctions, Circulation 99 (4) (1999) 546–551. [DOI] [PubMed] [Google Scholar]
- [43].Tarzami ST, Chemokines and inflammation in heart disease: adaptive or maladaptive? Int. J. Clin. Exp. Med 4 (1) (2011) 74. [PMC free article] [PubMed] [Google Scholar]
- [44].Talman V, Ruskoaho H, Cardiac fibrosis in myocardial infarction—from repair and remodeling to regeneration, Cell Tissue Res. 365 (3) (2016) 563–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Frangogiannis NG, Fibroblasts and the extracellular matrix in right ventricular disease, Cardiovasc. Res 113 (12) (2017) 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Suthahar N, Meijers WC, Silljé HH, de Boer RA, From inflammation to fibrosis—molecular and cellular mechanisms of myocardial tissue remodelling and perspectives on differential treatment opportunities, Curr. Heart Fail. Rep 14 (4) (2017) 235–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Frangogiannis NG, The inflammatory response in myocardial injury, repair, and remodelling, Nat. Rev. Cardiol 11 (5) (2014) 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tse G, Mechanisms of cardiac arrhythmias, J. Arrhythmia 32 (2) (2016) 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].John RM, Kumar S, Sinus node and atrial arrhythmias, Circulation 133 (19) (2016) 1892–1900. [DOI] [PubMed] [Google Scholar]
- [50].Sisakian H, Cardiomyopathies: evolution of pathogenesis concepts and potential for new therapies, World J. Cardiol 6 (6) (2014) 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Braunwald E, Cardiomyopathies: an overview, Circ. Res 121 (7) (2017) 711–721. [DOI] [PubMed] [Google Scholar]
- [52].Marian AJ, Braunwald E, Hypertrophic cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy, Circ. Res 121 (7) (2017) 749–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McNally EM, Mestroni L, Dilated cardiomyopathy: genetic determinants and mechanisms, Circ. Res 121 (7) (2017) 731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Corrado D, Basso C, Judge DP, Arrhythmogenic cardiomyopathy, Circ. Res 121 (7) (2017) 784–802. [DOI] [PubMed] [Google Scholar]
- [55].Captur G, Nihoyannopoulos P, Left ventricular non-compaction: genetic heterogeneity, diagnosis and clinical course, Int. J. Cardiol 140 (2) (2010) 145–153. [DOI] [PubMed] [Google Scholar]
- [56].Krenning G, Zeisberg EM, Kalluri R, The origin of fibroblasts and mechanism of cardiac fibrosis, J. Cell. Physiol 225 (3) (2010) 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Leask A, Getting to the heart of the matter: new insights into cardiac fibrosis, Circ. Res 116 (7) (2015) 1269–1276. [DOI] [PubMed] [Google Scholar]
- [58].Camelliti P, Borg TK, Kohl P, Structural and functional characterisation of cardiac fibroblasts, Cardiovasc. Res 65 (1) (2005) 40–51. [DOI] [PubMed] [Google Scholar]
- [59].Davis J, Molkentin JD, Myofibroblasts: trust your heart and let fate decide, J. Mol. Cell. Cardiol 70 (2014) 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vasquez C, Morley GE, The origin and arrhythmogenic potential of fibroblasts in cardiac disease, J. Cardiovascular Transl. Res 5 (6) (2012) 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kohl P, Gourdie RG, Fibroblast–myocyte electrotonic coupling: does it occur in native cardiac tissue? J. Mol. Cell. Cardiol 70 (2014) 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Moore-Morris T, Cattaneo P, Puceat M, Evans SM, Origins of cardiac fibroblasts, J. Mol. Cell. Cardiol 91 (2016) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Weber KT, Fibrosis in hypertensive heart disease: focus on cardiac fibroblasts, J. Hypertens 22 (1) (2004) 47–50. [DOI] [PubMed] [Google Scholar]
- [64].Leask A, Abraham DJ, TGF-β signaling and the fibrotic response, FASEB J 18 (7) (2004) 816–827. [DOI] [PubMed] [Google Scholar]
- [65].Vivar R, Humeres C, Ayala P, Olmedo I, Catalán M, García L, Lavandero S, Díaz-Araya G, TGF-β1 prevents simulated ischemia/reperfusion-induced cardiac fibroblast apoptosis by activation of both canonical and non-canonical signaling pathways, Biochim. Biophys. Acta (BBA) - Mol. Basis Dis 1832 (6) (2013) 754–762. [DOI] [PubMed] [Google Scholar]
- [66].Schorb W, Booz GW, Dostal DE, Conrad KM, Chang KC, Baker KM, Angiotensin II is mitogenic in neonatal rat cardiac fibroblasts, Circ. Res 72 (6) (1993) 1245–1254. [DOI] [PubMed] [Google Scholar]
- [67].Accornero F, van Berlo JH, Correll RN, Elrod JW, Sargent MA, York A, Rabinowitz JE, Leask A, Molkentin JD, Genetic analysis of connective tissue growth factor as an effector of transforming growth factor β signaling and cardiac remodeling, Mol. Cell Biol 35 (12) (2015) 2154–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S, Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease, Circ. Res 118 (4) (2016) 535–546. [DOI] [PubMed] [Google Scholar]
- [69].Aso S.-i., Imamura H, Sekiguchi Y, Iwashita T, Hirano R, Ikeda U, Okamoto K, Incidence and mortality of acute myocardial infarction a population-based study including patients with out-of-hospital cardiac arrest, Int. Heart J 52 (4) (2011) 197–202. [DOI] [PubMed] [Google Scholar]
- [70].Mehra R, Global public health problem of sudden cardiac death, J. Electrocardiol 40 (6, Supplement 1) (2007) S118–S122. [DOI] [PubMed] [Google Scholar]
- [71].Semsarian C, Ingles J, Maron MS, Maron BJ, New perspectives on the prevalence of hypertrophic cardiomyopathy, J. Am. Coll. Cardiol 65 (12) (2015) 1249–1254. [DOI] [PubMed] [Google Scholar]
- [75].Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, Pairaudeau G, Pennie WD, Pickett SD, Wang J, An analysis of the attrition of drug candidates from four major pharmaceutical companies, Nat. Rev. Drug Discov 14 (7) (2015) 475. [DOI] [PubMed] [Google Scholar]
- [76].Patten RD, Hall-Porter MR, Small animal models of heart failure: development of novel therapies, past and present, Circulation: Heart Fail. 2 (2) (2009) 138–144. [DOI] [PubMed] [Google Scholar]
- [77].Dixon JA, Spinale FG, Large animal models of heart failure: a critical link in the translation of basic science to clinical practice, Circulation: Heart Fail. 2 (3) (2009) 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ribas J, Sadeghi H, Manbachi A, Leijten J, Brinegar K, Zhang YS, Ferreira L, Khademhosseini A, Cardiovascular organ-on-a-chip platforms for drug discovery and development, Appl. in vitro Toxicol 2 (2) (2016) 82–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Tricklebank MD, Garner JP, The possibilities and limitations of animal models for psychiatric disorders, Drug Discov. Psychiatric Disorders (2012) 534–557. [Google Scholar]
- [80].Cummings JL, Morstorf T, Zhong K, Alzheimer’s disease drug-development pipeline: few candidates, frequent failures, Alzheimer’s Res. Ther 6 (4) (2014) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hearse DJ, Sutherland FJ, Experimental models for the study of cardiovascular function and disease, Pharmacol. Res 41 (6) (2000) 597–603. [DOI] [PubMed] [Google Scholar]
- [82].Wilson AJ, Reale D, Clements MN, Morrissey MM, Postma E, Walling CA, Kruuk LE, Nussey DH, An ecologist’s guide to the animal model, J. Anim. Ecol 79 (1) (2010) 13–26. [DOI] [PubMed] [Google Scholar]
- [83].Waxenecker G, Binder R, Regulatory animal testing for the development of medicines, Comp. Med (2017) 209–218 Springer. [Google Scholar]
- [84].Milani-Nejad N, Janssen PM, Small and large animal models in cardiac contraction research: advantages and disadvantages, Pharmacol. Therapeut 141 (3) (2014) 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tarnavski O, Mouse surgical models in cardiovascular research, Cardiovascular Genom. (2009) 115–137 Springer. [DOI] [PubMed] [Google Scholar]
- [86].Houser SR, Margulies KB, Murphy AM, Spinale FG, Francis GS, Prabhu SD, Rockman HA, Kass DA, Molkentin JD, Sussman MA, Animal models of heart failure: a scientific statement from the American Heart Association, Circ. Res 111 (1) (2012) 131–150. [DOI] [PubMed] [Google Scholar]
- [87].Leong X-F, Ng C-Y, Jaarin K, Animal models in cardiovascular research: hypertension and atherosclerosis, BioMed Res. Int 2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Camacho P, Fan H, Liu Z, He J-Q, Small mammalian animal models of heart disease, Am. J. Cardiovascular Dis 6 (3) (2016) 70. [PMC free article] [PubMed] [Google Scholar]
- [89].van Amerongen MJ, Harmsen MC, Petersen AH, Popa ER, van Luyn MJ, Cryoinjury: a model of myocardial regeneration, Cardiovasc. Pathol 17 (1) (2008) 23–31. [DOI] [PubMed] [Google Scholar]
- [90].Agbulut O, Menot M-L, Li Z, Marotte F, Paulin D, Hagege AA, Chomienne C, Samuel J-L, Menasche P, Temporal patterns of bone marrow cell differentiation following transplantation in doxorubicin-induced cardiomyopathy, Cardiovasc. Res 58 (2) (2003) 451–459. [DOI] [PubMed] [Google Scholar]
- [91].Carmeliet P, Collen D, Transgenic mouse models in angiogenesis and cardiovascular disease, J. Pathol 190 (3) (2000) 387–405. [DOI] [PubMed] [Google Scholar]
- [92].Nerbonne JM, Mouse models of arrhythmogenic cardiovascular disease: challenges and opportunities, Curr. Opin. Pharmacol 15 (2014) 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Rosenthal N, Brown S, The mouse ascending: perspectives for human-disease models, Nat. Cell Biol 9 (9) (2007) 993. [DOI] [PubMed] [Google Scholar]
- [94].Dos Remedios C, Chhabra D, Kekic M, Dedova I, Tsubakihara M, Berry D, Nosworthy N, Actin binding proteins: regulation of cytoskeletal microfilaments, Physiol. Rev 83 (2) (2003) 433–473. [DOI] [PubMed] [Google Scholar]
- [95].Zima AV, Bovo E, Mazurek SR, Rochira JA, Li W, Terentyev D, Ca handling during excitation–contraction coupling in heart failure, Pflueg. Arch. Eur. J. Physiol 466 (6) (2014) 1129–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Unsöld B, Schotola H, Jacobshagen C, Seidler T, Sossalla S, Emons J, Klede S, Knöll R, Guan K, El-Armouche A, Age-dependent changes in contractile function and passive elastic properties of myocardium from mice lacking muscle LIM protein (MLP), Eur. J. Heart Fail 14 (4) (2012) 430–437. [DOI] [PubMed] [Google Scholar]
- [97].Bullard TA, Protack TL, Aguilar F, Bagwe S, Massey HT, Blaxall BC, Identification of Nogo as a novel indicator of heart failure, Physiol. Genom 32 (2) (2008) 182–189. [DOI] [PubMed] [Google Scholar]
- [98].de Lange WJ, Grimes AC, Hegge LF, Ralphe JC, Ablation of cardiac myosin–binding protein-C accelerates contractile kinetics in engineered cardiac tissue, J. Gen. Physiol 141 (1) (2013) 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Sarkar S, Chawla-Sarkar M, Young D, Nishiyama K, Rayborn ME, Hollyfield JG, Sen S, Myocardial cell death and regeneration during progression of cardiac hypertrophy to heart failure, J. Biol. Chem 279 (50) (2004) 52630–52642. [DOI] [PubMed] [Google Scholar]
- [100].Elliott JF, Liu J, Yuan Z-N, Bautista-Lopez N, Wallbank SL, Suzuki K, Rayner D, Nation P, Robertson MA, Liu G, Autoimmune cardiomyopathy and heart block develop spontaneously in HLA-DQ8 transgenic IAβ knockout NOD mice, Proc. Natl. Acad. Sci. U. S. A 100 (23) (2003) 13447–13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Aoki CA, Borchers AT, Ridgway WM, Keen CL, Ansari AA, Gershwin ME, NOD mice and autoimmunity, Autoimmun. Rev 4 (6) (2005) 373–379. [DOI] [PubMed] [Google Scholar]
- [102].McNally EM, Kaltman JR, Benson DW, Canter CE, Cripe LH, Duan D, Finder JD, Groh WJ, Hoffman EP, Judge DP, Contemporary cardiac issues in Duchenne muscular dystrophy, Circulation 131 (18) (2015) 1590–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Janssen PM, Hiranandani N, Mays TA, Rafael-Fortney JA, Utrophin deficiency worsens cardiac contractile dysfunction present in dystrophin-deficient mdx mice, Am. J. Physiol. Heart Circ. Physiol 289 (6) (2005) H2373–H2378. [DOI] [PubMed] [Google Scholar]
- [104].Ozcan C, Battaglia E, Young R, Suzuki G, LKB1 knockout mouse develops spontaneous atrial fibrillation and provides mechanistic insights into human disease process, J. Am. Heart Assoc 4 (3) (2015) e001733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].van den Bos EJ, Mees BM, de Waard MC, de Crom R, Duncker DJ, A novel model of cryoinjury-induced myocardial infarction in the mouse: a comparison with coronary artery ligation, Am. J. Physiol. Heart Circ. Physiol 289 (3) (2005) H1291–H1300. [DOI] [PubMed] [Google Scholar]
- [106].Barretti D, Melo S, Oliveira E, Barauna V, Resistance training attenuates salt overload-induced cardiac remodeling and diastolic dysfunction in normotensive rats, Braz. J. Med. Biol. Res 50 (9) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Nunes S, Rolo AP, Palmeira CM, Reis F, Diabetic cardiomyopathy: focus on oxidative stress, mitochondrial dysfunction and inflammation, in: Kirali Kaan (Ed.), Cardiomyopathies, IntechOpen, 2017, pp. 235–257,, 10.5772/65915. [DOI] [Google Scholar]
- [108].Gralinski M, Neves LA, Tiniakova O, Methods to induce experimental hypertension, Drug Discov. Eval.: Pharmacol. Assays (2016) 135–164. [Google Scholar]
- [109].Kodavanti UP, Schladweiler MC, Ledbetter AD, Watkinson WP, Campen MJ, Winsett DW, Richards JR, Crissman KM, Hatch GE, Costa DL, The spontaneously hypertensive rat as a model of human cardiovascular disease: evidence of exacerbated cardiopulmonary injury and oxidative stress from inhaled emission particulate matter, Toxicol. Appl. Pharmacol 164 (3) (2000) 250–263. [DOI] [PubMed] [Google Scholar]
- [110].Fernandes R, Bonilla PAP, Garver H, Galligan JJ, Fink GD, Xu H, High fat diet increases salt sensitivity and promotes metabolic disorder-independent hypertension in Dahl salt sensitive rats, FASEB J. 31 (Supplement 1) (2017) 1025.9–1025.9. [Google Scholar]
- [111].Török J, Kristek F, Functional and morphological pattern of vascular responses in two models of experimental hypertension, Exp. Clin. Cardiol 6 (3) (2001) 142. [PMC free article] [PubMed] [Google Scholar]
- [112].Badimon L, Atherosclerosis and thrombosis: lessons from animal models, Thromb. Haemostasis 86 (01) (2001) 356–365. [PubMed] [Google Scholar]
- [113].Vilahur G, Padro T, Badimon L, Atherosclerosis and thrombosis: insights from large animal models, BioMed Res. Int 2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Getz GS, Reardon CA, Do the Apoe−/− and Ldlr−/–mice yield the same insight on atherogenesis? Highlights, Arterioscler. Thromb. Vasc. Biol 36 (9) (2016) 1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].McGonigle P, Ruggeri B, Animal models of human disease: challenges in enabling translation, Biochem. Pharmacol 87 (1) (2014) 162–171. [DOI] [PubMed] [Google Scholar]
- [116].Lum-Naihe K, Toedebusch R, Mahmood A, Bajwa J, Carmack T, Kumar SA, Ardhanari S, DeMarco VG, Emter CA, Pulakat L, Cardiovascular disease progression in female Zucker Diabetic Fatty rats occurs via unique mechanisms compared to males, Sci. Rep 7 (1) (2017) 17823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Liao J, Huang W, Liu G, Animal models of coronary heart disease, J. Biomed. Res 31 (1) (2017) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Papinska A, Mordwinkin N, Meeks C, Jadhav S, Rodgers K, Angiotensin–(1–7) administration benefits cardiac, renal and progenitor cell function in db/db mice, Br. J. Pharmacol 172 (18) (2015) 4443–4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Pinkert CA, Introduction to Transgenic Animal Technology, Transgenic Animal Technology, third ed., Elsevier, 2014, pp. 3–13. [Google Scholar]
- [120].Leong XF, Aishah A, Aini UN, Das S, Jaarin K, Heated palm oil causes rise in blood pressure and cardiac changes in heart muscle in experimental rats, Arch. Med. Res 39 (6) (2008) 567–572. [DOI] [PubMed] [Google Scholar]
- [121].Jaarin K, Mustafa MR, Leong X-F, The effects of heated vegetable oils on blood pressure in rats, Clinics 66 (12) (2011) 2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Fan J, Kitajima S, Watanabe T, Xu J, Zhang J, Liu E, Chen YE, Rabbit models for the study of human atherosclerosis: from pathophysiological mechanisms to translational medicine, Pharmacol. Therapeut 146 (2015) 104–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tsang H, Rashdan N, Whitelaw C, Corcoran B, Summers K, MacRae V, Large animal models of cardiovascular disease, Cell Biochem. Funct 34 (3) (2016) 113–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yue L, Melnyk P, Gaspo R, Wang Z, Nattel S, Molecular mechanisms underlying ionic remodeling in a dog model of atrial fibrillation, Circ. Res 84 (7) (1999) 776–784. [DOI] [PubMed] [Google Scholar]
- [125].Lindsey ML, Bolli R, Canty JM, Du X-J, Frangogiannis NG, Frantz S, Gourdie RG, Holmes JW, Jones SP, Kloner R, Guidelines for experimental models of myocardial ischemia and infarction, Am. J. Physiol. Heart Circ. Physiol 314 (4) (2018) H812–H838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Montgomery M, Ahadian S, Huyer LD, Rito ML, Civitarese RA, Vanderlaan RD, Wu J, Reis LA, Momen A, Akbari S, Flexible shape-memory scaffold for minimally invasive delivery of functional tissues, Nat. Mater 16 (10) (2017) 1038. [DOI] [PubMed] [Google Scholar]
- [127].Mukherjee R, Brinsa TA, Dowdy KB, Scott AA, Baskin JM, Deschamps AM, Lowry AS, Escobar GP, Lucas DG, Yarbrough WM, Myocardial infarct expansion and matrix metalloproteinase inhibition, Circulation 107 (4) (2003) 618–625. [DOI] [PubMed] [Google Scholar]
- [128].Zeng L, Hu Q, Wang X, Mansoor A, Lee J, Feygin J, Zhang G, Suntharalingam P, Boozer S, Mhashilkar A, Bioenergetic and functional consequences of bone marrow–derived multipotent progenitor cell transplantation in hearts with postinfarction left ventricular remodeling, Circulation 115 (14) (2007) 1866–1875. [DOI] [PubMed] [Google Scholar]
- [129].Lu H, Xu X, Zhang M, Cao R, Bråkenhielm E, Li C, Lin H, Yao G, Sun H, Qi L, Combinatorial protein therapy of angiogenic and arteriogenic factors remarkably improves collaterogenesis and cardiac function in pigs, Proc. Natl. Acad. Sci. U. S. A 104 (29) (2007) 12140–12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Galindo CL, Kasasbeh E, Murphy A, Ryzhov S, Lenihan S, Ahmad FA, Williams P, Nunnally A, Adcock J, Song Y, Anti-remodeling and anti-fibrotic effects of the Neuregulin-1β glial growth factor 2 in a large animal model of heart failure, J. Am. Heart Assoc 3 (5) (2014) e000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Emmert MY, Hitchcock RW, Hoerstrup SP, Cell therapy, 3D culture systems and tissue engineering for cardiac regeneration, Adv. Drug Deliv. Rev 69 (2014) 254–269. [DOI] [PubMed] [Google Scholar]
- [132].Stacey G, Primary Cell Cultures and Immortal Cell Lines, eLS, 2006. [Google Scholar]
- [133].Natarajan A, Stancescu M, Dhir V, Armstrong C, Sommerhage F, Hickman JJ, Molnar P, Patterned cardiomyocytes on microelectrode arrays as a functional, high information content drug screening platform, Biomaterials 32 (18) (2011) 4267–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Eglen R, Reisine T, Primary cells and stem cells in drug discovery: emerging tools for high-throughput screening, Assay Drug Dev. Technol 9 (2) (2011) 108–124. [DOI] [PubMed] [Google Scholar]
- [135].Polak JM, Bishop AE, Stem cells and tissue engineering: past, present, and future, Ann. N. Y. Acad. Sci 1068 (1) (2006) 352–366. [DOI] [PubMed] [Google Scholar]
- [136].Pan C, Kumar C, Bohl S, Klingmueller U, Mann M, Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions, Mol. Cell. Proteomics 8 (3) (2009) 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Davidson MM, Nesti C, Palenzuela L, Walker WF, Hernandez E, Protas L, Hirano M, Isaac ND, Novel cell lines derived from adult human ventricular cardiomyocytes, J. Mol. Cell. Cardiol 39 (1) (2005) 133–147. [DOI] [PubMed] [Google Scholar]
- [138].White SM, Constantin PE, Claycomb WC, Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function, Am. J. Physiol. Heart Circ. Physiol 286 (3) (2004) H823–H829. [DOI] [PubMed] [Google Scholar]
- [139].Claycomb WC, Lanson NA, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ, HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte, Proc. Natl. Acad. Sci. U. S. A 95 (6) (1998) 2979–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Naderi H, Matin MM, Bahrami AR, Review Article: critical issues in tissue engineering: biomaterials, cell sources, angiogenesis, and drug delivery systems, J. Biomater. Appl 26 (4) (2011) 383–417. [DOI] [PubMed] [Google Scholar]
- [141].Cells S, Scientific Progress and Future Research Directions, Department of health and human services, 2008. June 2001. [Google Scholar]
- [142].Tillmanns J, Rota M, Hosoda T, Misao Y, Esposito G, Gonzalez A, Vitale S, Parolin C, Yasuzawa-Amano S, Muraski J, Formation of large coronary arteries by cardiac progenitor cells, Proc. Natl. Acad. Sci. U. S. A 105 (5) (2008) 1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Tian S, Liu Q, Gnatovskiy L, Ma PX, Wang Z, Heart regeneration with embryonic cardiac progenitor cells and cardiac tissue engineering, J. Stem Cell Transplant. Biol 1 (1) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Oh H, Bradfute SB, Gallardo TD, Nakamura T, Gaussin V, Mishina Y, Pocius J, Michael LH, Behringer RR, Garry DJ, Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction, Proc. Natl. Acad. Sci. U. S. A 100 (21) (2003) 12313–12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Tang X-L, Rokosh G, Sanganalmath SK, Yuan F, Sato H, Mu J, Dai S, Li C, Chen N, Peng Y, Intracoronary administration of cardiac progenitor cells alleviates left ventricular dysfunction in rats with a 30-day-old infarction, Circulation 121 (2) (2010) 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Howard D, Buttery LD, Shakesheff KM, Roberts SJ, Tissue engineering: strategies, stem cells and scaffolds, J. Anat 213 (1) (2008) 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Burridge PW, Holmström A, Wu JC, Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells, Curr. Protocols Hum. Genet (2015) 21.3. 1–21.3. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population, Nature 453 (7194) (2008) 524. [DOI] [PubMed] [Google Scholar]
- [149].Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP, Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling, Proc. Natl. Acad. Sci. U. S. A 109 (27) (2012) E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP, Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions, Nat. Protoc 8 (1) (2013) 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Zhao M-T, Chen H, Liu Q, Shao N-Y, Sayed N, Wo H-T, Zhang JZ, Ong S-G, Liu C, Kim Y, Molecular and functional resemblance of differentiated cells derived from isogenic human iPSCs and SCNT-derived ESCs, Proc. Natl. Acad. Sci. U. S. A (2017) 201708991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Xu C, Police S, Rao N, Carpenter MK, Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells, Circ. Res 91 (6) (2002) 501–508. [DOI] [PubMed] [Google Scholar]
- [153].Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes, Cell Stem Cell 12 (1) (2013) 127–137. [DOI] [PubMed] [Google Scholar]
- [154].Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G, SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells, Nat. Biotechnol 29 (11) (2011) 1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Lu X, Zhao T, Clinical therapy using iPSCs: hopes and challenges, Dev. Reprod. Biol 11 (5) (2013) 294–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Avior Y, Sagi I, Benvenisty N, Pluripotent stem cells in disease modelling and drug discovery, Nat. Rev. Mol. Cell Biol 17 (3) (2016) 170. [DOI] [PubMed] [Google Scholar]
- [157].Geraili A, Jafari P, Hassani MS, Araghi BH, Mohammadi MH, Ghafari AM, Tamrin SH, Modarres HP, Kolahchi AR, Ahadian S, Controlling differentiation of stem cells for developing personalized organ-on-chip platforms, Adv. Healthcare Mater 7 (2) (2018) 1700426. [DOI] [PubMed] [Google Scholar]
- [158].Singh VK, Kalsan M, Kumar N, Saini A, Chandra R, Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery, Front. Cell Dev. Biol 3 (2015) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies, Nat. Med 20 (6) (2014) 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Mathur A, Ma Z, Loskill P, Jeeawoody S, Healy KE, In vitro cardiac tissue models: current status and future prospects, Adv. Drug Deliv. Rev 96 (2016) 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Burridge PW, Keller G, Gold JD, Wu JC, Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming, Cell Stem Cell 10 (1) (2012) 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Robertson C, Tran DD, George SC, Concise review: maturation phases of human pluripotent stem cell-derived cardiomyocytes, Stem Cell. 31 (5) (2013) 829–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Synnergren J, Améen C, Jansson A, Sartipy P, Global transcriptional profiling reveals similarities and differences between human stem cell-derived cardiomyocyte clusters and heart tissue, Physiol. Genom 44 (4) (2011) 245–258. [DOI] [PubMed] [Google Scholar]
- [164].Gerdes AM, Kellerman SE, Moore JA, Muffly KE, Clark LC, Reaves PY, Malec KB, McKeown PP, Schocken DD, Structural remodeling of cardiac myocytes in patients with ischemic cardiomyopathy, Circulation 86 (2) (1992) 426–430. [DOI] [PubMed] [Google Scholar]
- [165].Maillet M, Van Berlo JH, Molkentin JD, Molecular basis of physiological heart growth: fundamental concepts and new players, Nat. Rev. Mol. Cell Biol 14 (1) (2013) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Denning C, Borgdorff V, Crutchley J, Firth KS, George V, Kalra S, Kondrashov A, Hoang MD, Mosqueira D, Patel A, Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform, Biochim. Biophys. Acta Mol. Cell Res 1863 (7) (2016) 1728–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Soonpaa MH, Field LJ, Survey of studies examining mammalian cardiomyocyte DNA synthesis, Circ. Res 83 (1) (1998) 15–26. [DOI] [PubMed] [Google Scholar]
- [168].Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Evidence for cardiomyocyte renewal in humans, Science 324 (5923) (2009) 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Lundy SD, Zhu W-Z, Regnier M, Laflamme MA, Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells, Stem Cell. Dev 22 (14) (2013) 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yang X, Rodriguez M, Pabon L, Fischer KA, Reinecke H, Regnier M, Sniadecki NJ, Ruohola-Baker H, Murry CE, Tri-iodo-l-thyronine promotes the maturation of human cardiomyocytes-derived from induced pluripotent stem cells, J. Mol. Cell. Cardiol 72 (2014) 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Cao N, Liu Z, Chen Z, Wang J, Chen T, Zhao X, Ma Y, Qin L, Kang J, Wei B, Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells, Cell Res. 22 (1) (2012) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Zhu W-Z, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA, Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells novelty and significance, Circ. Res 107 (6) (2010) 776–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Massé S, Gagliardi M, Hsieh A, Biowire: a platform for maturation of human pluripotent stem cell–derived cardiomyocytes, Nat. Methods 10 (8) (2013) 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Ruan J-L, Tulloch NL, Razumova MV, Saiget M, Muskheli V, Pabon L, Reinecke H, Regnier M, Murry CE, Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue, Circulation 114 (2016) 014998CIRCULATIONAHA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature (2018) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Shyu K-G, Cellular and molecular effects of mechanical stretch on vascular cells and cardiac myocytes, Clin. Sci 116 (5) (2009) 377–389. [DOI] [PubMed] [Google Scholar]
- [177].Mihic A, Li J, Miyagi Y, Gagliardi M, Li S-H, Zu J, Weisel RD, Keller G, Li R-K, The effect of cyclic stretch on maturation and 3D tissue formation of human embryonic stem cell-derived cardiomyocytes, Biomaterials 35 (9) (2014) 2798–2808. [DOI] [PubMed] [Google Scholar]
- [178].Pasqualini FS, Sheehy SP, Agarwal A, Aratyn-Schaus Y, Parker KK, Structural phenotyping of stem cell-derived cardiomyocytes, Stem Cell Rep. 4 (3) (2015) 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Palankar R, Glaubitz M, Martens U, Medvedev N, von der Ehe M, Felix SB, Münzenberg M, Delcea M, 3D micropillars guide the mechanobiology of human induced pluripotent stem cell-derived cardiomyocytes, Adv. Healthcare Mater 5 (3) (2016) 335–341. [DOI] [PubMed] [Google Scholar]
- [180].Kim D-H, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh K-Y, Tung L, Levchenko A, Nanoscale cues regulate the structure and function of macroscopic cardiac tissue constructs, Proc. Natl. Acad. Sci. U. S. A 107 (2) (2010) 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Mengsteab PY, Uto K, Smith AS, Frankel S, Fisher E, Nawas Z, Macadangdang J, Ebara M, Kim D-H, Spatiotemporal control of cardiac anisotropy using dynamic nanotopographic cues, Biomaterials 86 (2016) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Bettinger CJ, Langer R, Borenstein JT, Engineering substrate topography at the micro-and nanoscale to control cell function, Angew. Chem. Int. Ed 48 (30) (2009) 5406–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Annabi N, Tsang K, Mithieux SM, Nikkhah M, Ameri A, Khademhosseini A, Weiss AS, Highly elastic micropatterned hydrogel for engineering functional cardiac tissue, Adv. Funct. Mater 23 (39) (2013) 4950–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].McCain ML, Sheehy SP, Grosberg A, Goss JA, Parker KK, Recapitulating maladaptive, multiscale remodeling of failing myocardium on a chip, Proc. Natl. Acad. Sci. U. S. A 110 (24) (2013) 9770–9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Bian W, Jackman CP, Bursac N, Controlling the structural and functional anisotropy of engineered cardiac tissues, Biofabrication 6 (2) (2014) 024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].van Spreeuwel A, Bax N, Bastiaens A, Foolen J, Loerakker S, Borochin M, van der Schaft D, Chen C, Baaijens F, Bouten C, The influence of matrix (an) isotropy on cardiomyocyte contraction in engineered cardiac microtissues, Integr. Biol 6 (4) (2014) 422–429. [DOI] [PubMed] [Google Scholar]
- [187].Rao C, Prodromakis T, Kolker L, Chaudhry UA, Trantidou T, Sridhar A, Weekes C, Camelliti P, Harding SE, Darzi A, The effect of microgrooved culture substrates on calcium cycling of cardiac myocytes derived from human induced pluripotent stem cells, Biomaterials 34 (10) (2013) 2399–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Al-Haque S, Miklas JW, Feric N, Chiu LL, Chen WLK, Simmons CA, Radisic M, Hydrogel substrate stiffness and topography interact to induce contact guidance in cardiac fibroblasts, Macromol. Biosci 12 (10) (2012) 1342–1353. [DOI] [PubMed] [Google Scholar]
- [189].Lücker PB, Javaherian S, Soleas JP, Halverson D, Zandstra PW, McGuigan AP, A microgroove patterned multiwell cell culture plate for highthroughput studies of cell alignment, Biotechnol. Bioeng 111 (12) (2014) 2537–2548. [DOI] [PubMed] [Google Scholar]
- [190].Abadi PP, Garbern JC, Behzadi S, Hill MJ, Tresback JS, Heydari T, Ejtehadi MR, Ahmed N, Copley E, Aghaverdi H, Engineering of mature human induced pluripotent stem cell‐derived cardiomyocytes using substrates with multiscale topography, Adv. Funct. Mater 28 (19) (2018) 1707378. [Google Scholar]
- [191].Navaei A, Moore N, Sullivan RT, Truong D, Migrino RQ, Nikkhah M, Electrically conductive hydrogel-based micro-topographies for the development of organized cardiac tissues, RSC Adv. 7 (6) (2017) 3302–3312. [Google Scholar]
- [192].Navaei A, Saini H, Christenson W, Sullivan RT, Ros R, Nikkhah M, Gold nanorod-incorporated gelatin-based conductive hydrogels for engineering cardiac tissue constructs, Acta Biomater. 41 (2016) 133–146. [DOI] [PubMed] [Google Scholar]
- [193].Black III LD, Meyers JD, Weinbaum JS, Shvelidze YA, Tranquillo RT, Cell-induced alignment augments twitch force in fibrin gel–based engineered myocardium via gap junction modification, Tissue Eng. Part A 15 (10) (2009) 3099–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].CHUNG CY, Bien H, Entcheva E, The role of cardiac tissue alignment in modulating electrical function, J. Cardiovasc. Electrophysiol 18 (12) (2007) 1323–1329. [DOI] [PubMed] [Google Scholar]
- [195].Ribeiro MC, Tertoolen LG, Guadix JA, Bellin M, Kosmidis G, D’Aniello C, Monshouwer-Kloots J, Goumans M-J, Wang Y.-l., Feinberg AW, Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro–correlation between contraction force and electrophysiology, Biomaterials 51 (2015) 138–150. [DOI] [PubMed] [Google Scholar]
- [196].Feinberg AW, Alford PW, Jin H, Ripplinger CM, Werdich AA, Sheehy SP, Grosberg A, Parker KK, Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture, Biomaterials 33 (23) (2012) 5732–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Aubin H, Nichol JW, Hutson CB, Bae H, Sieminski AL, Cropek DM, Akhyari P, Khademhosseini A, Directed 3D cell alignment and elongation in microengineered hydrogels, Biomaterials 31 (27) (2010) 6941–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Macadangdang J, Lee HJ, Carson D, Jiao A, Fugate J, Pabon L, Regnier M, Murry C, Kim D-H, Capillary force lithography for cardiac tissue engineering, Jo. Visual. Exp.: JoVE 88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Lieu DK, Fu J-D, Chiamvimonvat N, Tung KWC, McNerney GP, Huser T, Keller G, Kong C-W, Li RA, Mechanism-based facilitated maturation of human pluripotent stem cell-derived cardiomyocytes, Circulation: Arrhythmia Electrophysiol. 112 (2013) 973420CIRCEP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Fu J-D, Rushing SN, Lieu DK, Chan CW, Kong C-W, Geng L, Wilson KD, Chiamvimonvat N, Boheler KR, Wu JC, Distinct roles of microRNA-1 and-499 in ventricular specification and functional maturation of human embryonic stem cell-derived cardiomyocytes, PloS One 6 (11) (2011) e27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G, Advanced maturation of human cardiac tissue grown from pluripotent stem cells, Nature 556 (7700) (2018) 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang K-J, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, Engineered in vitro disease models, Annu. Rev. Pathol. Mech Dis 10 (2015) 195–262. [DOI] [PubMed] [Google Scholar]
- [203].Thomas SP, Kucera JP, Bircher-Lehmann L, Rudy Y, Saffitz JE, Kléber AG, Impulse propagation in synthetic strands of neonatal cardiac myocytes with genetically reduced levels of connexin43, Circ. Res 92 (11) (2003) 1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Beauchamp P, Choby C, Desplantez T, de Peyer K, Green K, Yamada KA, Weingart R, Saffitz JE, Kléber AG, Electrical propagation in synthetic ventricular myocyte strands from germline connexin43 knockout mice, Circ. Res 95 (2) (2004) 170–178. [DOI] [PubMed] [Google Scholar]
- [205].Chang MG, Zhang Y, Chang CY, Xu L, Emokpae R, Tung L, Marbán E, Abraham MR, Spiral waves and reentry dynamics in an in vitro model of the healed infarct border zone, Circ. Res 105 (11) (2009) 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Thompson SA, Copeland CR, Reich DH, Tung L, Mechanical coupling between myofibroblasts and cardiomyocytes slows electric conduction in fibrotic cell monolayers clinical perspective, Circulation 123 (19) (2011) 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Bursac N, Parker K, Iravanian S, Tung L, Cardiomyocyte cultures with controlled macroscopic anisotropy: a model for functional electrophysiological studies of cardiac muscle, Circ. Res 91 (12) (2002) e45–e54. [DOI] [PubMed] [Google Scholar]
- [208].Nattel S, Maguy A, Le Bouter S, Yeh Y-H, Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation, Physiol. Rev 87 (2) (2007) 425–456. [DOI] [PubMed] [Google Scholar]
- [209].Ackerman MJ, The long QT syndrome: ion channel diseases of the heart, Mayo Clinic Proceedings, Elsevier, 1998, pp. 250–269. [DOI] [PubMed] [Google Scholar]
- [210].Yu H.-b., Li M, Wang W.-p., Wang X.-l., High throughput screening technologies for ion channels, Acta Pharmacol. Sin 37 (1) (2016) 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Patient-specific induced pluripotent stem-cell models for long-QT syndrome, N. Engl. J. Med 363 (15) (2010) 1397–1409. [DOI] [PubMed] [Google Scholar]
- [212].Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Modelling the long QT syndrome with induced pluripotent stem cells, Nature 471 (7337) (2011) 225–229. [DOI] [PubMed] [Google Scholar]
- [213].Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE, Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome, Nature 471 (7337) (2011) 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang Y-S, Schaniel C, Lee D-F, Yang L, Kaplan AD, Adler ED, Rozov R, Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome, Nature 465 (7299) (2010) 808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Tse H-F, Ho JC, Choi S-W, Lee Y-K, Butler AW, Ng K-M, Siu C-W, Simpson MA, Lai W-H, Chan Y-C, Patient-specific induced-pluripotent stem cells-derived cardiomyocytes recapitulate the pathogenic phenotypes of dilated cardiomyopathy due to a novel DES mutation identified by whole exome sequencing, Hum. Mol. Genet 22 (7) (2013) 1395–1403. [DOI] [PubMed] [Google Scholar]
- [216].Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, Dolmetsch RE, Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome, Nature 471 (7337) (2011) 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells, Cell Stem Cell 12 (1) (2013) 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Ebert AD, Kodo K, Liang P, Wu H, Huber BC, Riegler J, Churko J, Lee J, De Almeida P, Lan F, Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system, Sci. Transl. Med 6 (255) (2014) 255ra130–255ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Ulmer BM, Stoehr A, Schulze ML, Patel S, Gucek M, Mannhardt I, Funcke S, Murphy E, Eschenhagen T, Hansen A, Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes, Stem Cell Rep. 10 (3) (2018) 834–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Rezaei Kolahchi A, Khadem Mohtaram N, Pezeshgi Modarres H, Mohammadi MH, Geraili A, Jafari P, Akbari M, Sanati-Nezhad A, Microfluidic-based multi-organ platforms for drug discovery, Micromachines 7 (9) (2016) 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Zimmermann W-H, Melnychenko I, Wasmeier G, Didié M, Naito H, Nixdorff U, Hess A, Budinsky L, Brune K, Michaelis B, Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts, Nat. Med 12 (4) (2006) 452–458. [DOI] [PubMed] [Google Scholar]
- [222].Zimmermann W-H, Schneiderbanger K, Schubert P, Didie M, Munzel F, Heubach J, Kostin S, Neuhuber W, Eschenhagen T, Tissue engineering of a differentiated cardiac muscle construct, Circ. Res 90 (2) (2002) 223–230. [DOI] [PubMed] [Google Scholar]
- [223].MacQueen LA, Sheehy SP, Chantre CO, Zimmerman JF, Pasqualini FS, Liu X, Goss JA, Campbell PH, Gonzalez GM, Park S-J, A tissue-engineered scale model of the heart ventricle, Nat. Biomed. Eng (2018) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Ott HC, Matthiesen TS, Goh S-K, Black LD, Kren SM, Netoff TI, Taylor DA, Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart, Nat. Med 14 (2) (2008) 213–221. [DOI] [PubMed] [Google Scholar]
- [225].Shimizu T, Yamato M, Kikuchi A, Okano T, Cell sheet engineering for myocardial tissue reconstruction, Biomaterials 24 (13) (2003) 2309–2316. [DOI] [PubMed] [Google Scholar]
- [226].Mohammadi MH, Obregón R, Ahadian S, Ramón-Azcón J, Radisic M, Engineered muscle tissues for disease modeling and drug screening applications, Curr. Pharmaceut. Des 23 (20) (2017) 2991–3004. [DOI] [PubMed] [Google Scholar]
- [227].Stoehr A, Neuber C, Baldauf C, Vollert I, Friedrich FW, Flenner F, Carrier L, Eder A, Schaaf S, Hirt MN, Automated analysis of contractile force and Ca2+ transients in engineered heart tissue, Am. J. Physiol. Heart Circ. Physiol 306 (9) (2014) H1353–H1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Vedadghavami A, Minooei F, Mohammadi MH, Khetani S, Kolahchi AR, Mashayekhan S, Sanati-Nezhad A, Manufacturing of hydrogel biomaterials with controlled mechanical properties for tissue engineering applications, Acta Biomater. 62 (2017) 42–63. [DOI] [PubMed] [Google Scholar]
- [229].Eder A, Vollert I, Hansen A, Eschenhagen T, Human engineered heart tissue as a model system for drug testing, Adv. Drug Deliv. Rev 96 (2016) 214–224. [DOI] [PubMed] [Google Scholar]
- [230].Schaaf S, Shibamiya A, Mewe M, Eder A, Stöhr A, Hirt MN, Rau T, Zimmermann W-H, Conradi L, Eschenhagen T, Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology, PloS One 6 (10) (2011) e26397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Stoehr A, Neuber C, Baldauf C, Vollert I, Friedrich FW, Flenner F, Carrier L, Eder A, Schaaf S, Hirt MN, Automated analysis of contractile force and Ca 2+ transients in engineered heart tissue, Am. J. Physiol. Heart Circ. Physiol 306 (9) (2014) H1353–H1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Hansen A, Eder A, Bönstrup M, Flato M, Mewe M, Schaaf S, Aksehirlioglu B, Schwörer A, Uebeler J, Eschenhagen T, Development of a drug screening platform based on engineered heart tissue, Circ. Res 107 (1) (2010) 35–44. [DOI] [PubMed] [Google Scholar]
- [233].Hirt MN, Sörensen NA, Bartholdt LM, Boeddinghaus J, Schaaf S, Eder A, Vollert I, Stöhr A, Schulze T, Witten A, Increased afterload induces pathological cardiac hypertrophy: a new in vitro model, Basic Res. Cardiol 107 (6) (2012) 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Ralphe JC, de Lange WJ, 3D engineered cardiac tissue models of human heart disease: learning more from our mice, Trends Cardiovasc. Med 23 (2) (2013) 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Stöhr A, Friedrich FW, Flenner F, Geertz B, Eder A, Schaaf S, Hirt MN, Uebeler J, Schlossarek S, Carrier L, Contractile abnormalities and altered drug response in engineered heart tissue from Mybpc3-targeted knock-in mice, J. Mol. Cell. Cardiol 63 (2013) 189–198. [DOI] [PubMed] [Google Scholar]
- [236].Zimmermann WH, Schneiderbanger K, Schubert P, Didie M, Munzel F, Heubach JF, Kostin S, Neuhuber WL, Eschenhagen T, Tissue engineering of a differentiated cardiac muscle construct, Circ. Res 90 (2) (2002) 223–230. [DOI] [PubMed] [Google Scholar]
- [237].Schaaf S, Shibamiya A, Mewe M, Eder A, Stohr A, Hirt MN, Rau T, Zimmermann WH, Conradi L, Eschenhagen T, Hansen A, Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology, PloS One 6 (10) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Sakar MS, Neal D, Boudou T, Borochin MA, Li YQ, Weiss R, Kamm RD, Chen CS, Asada HH, Formation and optogenetic control of engineered 3D skeletal muscle bioactuators, Lab a Chip 12 (23) (2012) 4976–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Boudou T, Legant WR, Mu A, Borochin MA, Thavandiran N, Radisic M, Zandstra PW, Epstein JA, Margulies KB, Chen CS, A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues, Tissue Eng. Part A 18 (9–10) (2012) 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Miklas JW, Nunes SS, Sofla A, Reis LA, Pahnke A, Xiao Y, Laschinger C, Radisic M, Bioreactor for modulation of cardiac microtissue phenotype by combined static stretch and electrical stimulation, Biofabrication 6 (2) (2014) 024113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Thavandiran N, Dubois N, Mikryukov A, Masse S, Beca B, Simmons CA, Deshpande VS, McGarry JP, Chen CS, Nanthakumar K, Keller GM, Radisic M, Zandstra PW, Design and formulation of functional pluripotent stem cell-derived cardiac microtissues, Proc. Natl. Acad. Sci. U. S. A 110 (49) (2013) E4698–E4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Shadrin IY, Allen BW, Qian Y, Jackman CP, Carlson AL, Juhas ME, Bursac N, Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues, Nat. Commun 8 (1) (2017) 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Zhang D, Shadrin IY, Lam J, Xian H-Q, Snodgrass HR, Bursac N, Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes, Biomaterials 34 (23) (2013) 5813–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Bian W, Liau B, Badie N, Bursac N, Mesoscopic hydrogel molding to control the 3D geometry of bioartificial muscle tissues, Nat. Protoc 4 (10) (2009) 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Jackman CP, Carlson AL, Bursac N, Dynamic culture yields engineered myocardium with near-adult functional output, Biomaterials 111 (2016) 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B, Jiang J, Masse S, Gagliardi M, Hsieh A, Thavandiran N, Laflamme MA, Nanthakumar K, Gross GJ, Backx PH, Keller G, Radisic M, Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes, Nat. Methods 10 (8) (2013) 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Yang X, Pabon L, Murry CE, Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes, Circ. Res 114 (3) (2014) 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Radisic M, Malda J, Epping E, Geng W, Langer R, Vunjak-Novakovic G, Oxygen gradients correlate with cell density and cell viability in engineered cardiac tissue, Biotechnol. Bioeng 93 (2) (2006) 332–343. [DOI] [PubMed] [Google Scholar]
- [249].Ronaldson K, Feric N, Zhao Y, Zhang B, Conant G, Panhke A, Aschar-Sobbi R, Vunjak-Novakovic G, Backx P, Radisic M, TARA Biosystems’ biowire TM II: engineering mature human cardiac tissues enables more predictive drug screening, J. Pharmacol. Toxicol. Methods 88 (2017) 242. [Google Scholar]
- [250].Marsano A, Conficconi C, Lemme M, Occhetta P, Gaudiello E, Votta E, Cerino G, Redaelli A, Rasponi M, Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues, Lab a Chip 16 (3) (2016) 599–610. [DOI] [PubMed] [Google Scholar]
- [251].Zimmermann W-H, Didié M, Wasmeier GH, Nixdorff U, Hess A, Melnychenko I, Boy O, Neuhuber WL, Weyand M, Eschenhagen T, Cardiac grafting of engineered heart tissue in syngenic rats, Circulation 106 (12 suppl 1) (2002) I-151–I-157. [PubMed] [Google Scholar]
- [252].Sun X, Nunes SS, Biowire platform for maturation of human pluripotent stem cell-derived cardiomyocytes, Methods 101 (2016) 21–26. [DOI] [PubMed] [Google Scholar]
- [253].Eschenhagen T, Fink C, Remmers U, Scholz H, Wattchow J, Weil J, Zimmermann W, Dohmen HH, Schäfer H, Bishopric N, Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: a new heart muscle model system, FASEB J. 11 (8) (1997) 683–694. [DOI] [PubMed] [Google Scholar]
- [254].Zhao Y, Feric NT, Thavandiran N, Nunes SS, Radisic M, The role of tissue engineering and biomaterials in cardiac regenerative medicine, Can. J. Cardiol 30 (11) (2014) 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Xiao Y, Zhang B, Liu H, Miklas JW, Gagliardi M, Pahnke A, Thavandiran N, Sun Y, Simmons C, Keller G, Microfabricated perfusable cardiac biowire: a platform that mimics native cardiac bundle, Lab a Chip 14 (5) (2014) 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Fernandez C, Yen R, Perez S, Bedell H, Povsic T, Reichert W, Truskey G, Human vascular microphysiological system for in vitro drug screening, Sci. Rep 6 (2016) 21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Coultas L, Chawengsaksophak K, Rossant J, Endothelial cells and VEGF in vascular development, Nature 438 (7070) (2005) 937–945. [DOI] [PubMed] [Google Scholar]
- [258].Chiu LL, Montgomery M, Liang Y, Liu H, Radisic M, Perfusable branching microvessel bed for vascularization of engineered tissues, Proc. Natl. Acad. Sci. U. S. A 109 (50) (2012) E3414–E3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, Lopez JA, Stroock AD, In vitro microvessels for the study of angiogenesis and thrombosis, Proc. Natl. Acad. Sci. U. S. A 109 (24) (2012) 9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs, Adv. Mater 26 (19) (2014) 3124–3130. [DOI] [PubMed] [Google Scholar]
- [261].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Three-dimensional bioprinting of thick vascularized tissues, Proc. Natl. Acad. Sci. U. S. A 113 (12) (2016) 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DH, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues, Nat. Mater 11 (9) (2012) 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Bertassoni LE, Cecconi M, Manoharan V, Nikkhah M, Hjortnaes J, Cristino AL, Barabaschi G, Demarchi D, Dokmeci MR, Yang Y, Hydrogel bioprinted microchannel networks for vascularization of tissue engineering constructs, Lab a Chip 14 (13) (2014) 2202–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs, Adv. Mater 26 (19) (2014) 3124–3130. [DOI] [PubMed] [Google Scholar]
- [265].Zhu W, Qu X, Zhu J, Ma X, Patel S, Liu J, Wang P, Lai CSE, Gou M, Xu Y, Direct 3D bioprinting of prevascularized tissue constructs with complex microarchitecture, Biomaterials 124 (2017) 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Zhang B, Montgomery M, Chamberlain MD, Ogawa S, Korolj A, Pahnke A, Wells LA, Massé S, Kim J, Reis L, Biodegradable scaffold with built-in vasculature for organ-on-a-chip engineering and direct surgical anastomosis, Nat. Mater 15 (6) (2016) 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Kim S, Kim W, Lim S, Jeon JS, Vasculature-on-a-chip for in vitro disease models, Bioengineering 4 (1) (2017) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Franco C, Gerhardt H, Tissue engineering: blood vessels on a chip, Nature 488 (7412) (2012) 465. [DOI] [PubMed] [Google Scholar]
- [269].Zhang W, Zhang YS, Bakht SM, Aleman J, Shin SR, Yue K, Sica M, Ribas J, Duchamp M, Ju J, Elastomeric free-form blood vessels for interconnecting organs on chip systems, Lab a Chip 16 (9) (2016) 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Zheng W, Jiang B, Wang D, Zhang W, Wang Z, Jiang X, A microfluidic flow-stretch chip for investigating blood vessel biomechanics, Lab a Chip 12 (18) (2012) 3441–3450. [DOI] [PubMed] [Google Scholar]
- [271].Islam MM, Beverung S, Steward R Jr., Bio-inspired microdevices that mimic the human vasculature, Micromachines 8 (10) (2017) 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Loskill P, Marcus SG, Mathur A, Reese WM, Healy KE, muOrgano: a Lego(R)-like plug & play system for modular multi-organ-chips, PloS One 10 (10) (2015) e0139587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Phan DT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LY, George SC, Lee AP, Hughes CC, A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications, Lab a Chip 17 (3) (2017) 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Lai BFL, Huyer LD, Lu RXZ, Drecun S, Radisic M, Zhang B, InVADE: integrated vasculature for assessing dynamic events, Adv. Funct. Mater 27 (46) (2017). [Google Scholar]
- [275].Lai BFL, Huyer LD, Lu RXZ, Drecun S, Radisic M, Zhang B, InVADE: integrated vasculature for assessing dynamic events, Adv. Funct. Mater 27 (46) (2017) 1703524. [Google Scholar]
- [276].Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T, Tissue-engineered microenvironment systems for modeling human vasculature, Exp. Biol. Med 239 (9) (2014) 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Günther A, Yasotharan S, Vagaon A, Lochovsky C, Pinto S, Yang J, Lau C, Voigtlaender-Bolz J, Bolz S-S, A microfluidic platform for probing small artery structure and function, Lab a Chip 10 (18) (2010) 2341–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Yasotharan S, Pinto S, Sled JG, Bolz S-S, Günther A, Artery-on-a-chip platform for automated, multimodal assessment of cerebral blood vessel structure and function, Lab a Chip 15 (12) (2015) 2660–2669. [DOI] [PubMed] [Google Scholar]
- [279].Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Human iPSC-based cardiac microphysiological system for drug screening applications, Sci. Rep 5 (2015) 8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Chatzizisis YS, Coskun AU, Jonas M, Edelman ER, Feldman CL, Stone PH, Role of endothelial shear stress in the natural history of coronary atherosclerosis and vascular remodeling: molecular, cellular, and vascular behavior, J. Am. Coll. Cardiol 49 (25) (2007) 2379–2393. [DOI] [PubMed] [Google Scholar]
- [281].Li Y-SJ, Haga JH, Chien S, Molecular basis of the effects of shear stress on vascular endothelial cells, J. Biomech 38 (10) (2005) 1949–1971. [DOI] [PubMed] [Google Scholar]
- [282].Malek AM, Alper SL, Izumo S, Hemodynamic shear stress and its role in atherosclerosis, Jama 282 (21) (1999) 2035–2042. [DOI] [PubMed] [Google Scholar]
- [283].Rennier K, Ji JY, Effect of shear stress and substrate on endothelial DAPK expression, caspase activity, and apoptosis, BMC Res. Notes 6 (1) (2013) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Van Kruchten R, Cosemans JM, Heemskerk JW, Measurement of whole blood thrombus formation using parallel-plate flow chambers–a practical guide, Platelets 23 (3) (2012) 229–242. [DOI] [PubMed] [Google Scholar]
- [285].Sukavaneshvar S, Device thrombosis and pre-clinical blood flow models for assessing antithrombogenic efficacy of drug-device combinations, Adv. Drug Deliv. Rev 112 (2017) 24–34. [DOI] [PubMed] [Google Scholar]
- [286].Coenen DM, Mastenbroek TG, Cosemans JM, Platelet interaction with activated endothelium: mechanistic insights from microfluidics, Blood 130 (2017) 2819–2828. [DOI] [PubMed] [Google Scholar]
- [287].Jain A, van der Meer AD, Papa A-L, Barrile R, Lai A, Schlechter BL, Otieno MA, Louden CS, Hamilton GA, Michelson AD, Assessment of whole blood thrombosis in a microfluidic device lined by fixed human endothelium, Biomed. Microdevices 18 (4) (2016) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Costa PF, Albers HJ, Linssen JE, Middelkamp HH, Van Der Hout L, Passier R, Van Den Berg A, Malda J, Van Der Meer AD, Mimicking arterial thrombosis in a 3D-printed microfluidic in vitro vascular model based on computed tomography angiography data, Lab a Chip 17 (16) (2017) 2785–2792. [DOI] [PubMed] [Google Scholar]
- [289].Tsai M, Kita A, Leach J, Rounsevell R, Huang JN, Moake J, Ware RE, Fletcher DA, Lam WA, In vitro modeling of the microvascular occlusion and thrombosis that occur in hematologic diseases using microfluidic technology, J. Clin. Investig 122 (1) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Mannino RG, Myers DR, Ahn B, Wang Y, Rollins M, Gole H, Lin AS, Guldberg RE, Giddens DP, Timmins LH, Do-it-yourself in vitro vasculature that recapitulates in vivo geometries for investigating endothelial-blood cell interactions, Sci. Rep 5 (2015) 12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Zheng W, Huang R, Jiang B, Zhao Y, Zhang W, Jiang X, An early-stage atherosclerosis research model based on microfluidics, Small 12 (15) (2016) 2022–2034. [DOI] [PubMed] [Google Scholar]
- [292].Khodabandehlou K, Masehi-Lano JJ, Poon C, Wang J, Chung EJ, Targeting cell adhesion molecules with nanoparticles using in vivo and flow-based in vitro models of atherosclerosis, Exp. Biol. Med 242 (8) (2017) 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Jain A, Graveline A, Waterhouse A, Vernet A, Flaumenhaft R, Ingber DE, A shear gradient-activated microfluidic device for automated monitoring of whole blood haemostasis and platelet function, Nat. Commun 7 (2016) 10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Stangeby DK, Ethier CR, Computational analysis of coupled blood-wall arterial LDL transport, J. Biomech. Eng 124 (1) (2002) 1–8. [DOI] [PubMed] [Google Scholar]
- [295].Barrile R, van der Meer AD, Park H, Fraser JP, Simic D, Teng F, Conegliano D, Nguyen J, Jain A, Zhou M, Organ‐on‐Chip recapitulates thrombosis induced by an anti‐CD154 monoclonal antibody: translational potential of advanced microengineered systems, Clin. Pharmacol. Therapeut (2018), 10.1002/bit.25160. [DOI] [PubMed] [Google Scholar]
- [296].Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell’Erba V, Miri AK, Albadawi H, Arneri A, Li X, Bioprinted thrombosis-on-a-chip, Lab a Chip 16 (21) (2016) 4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Lam WA, Thrombosis-on-a-Chip: a new way to model a complex process, Am. Soc. Hematol 130 (2017) SCI-10. [Google Scholar]
- [298].Pandian NK, Mannino RG, Lam WA, Jain A, Thrombosis-on-a-chip: prospective impact of microphysiological models of vascular thrombosis, Curr. Opin. Biomed. Eng 5 (2018) 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Diamond SL, New microfluidic paths to test for bleeding or clotting, Cell. Mol. Bioeng 10 (1) (2017) 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Zhang YS, Oklu R, Albadawi H, Bioengineered in vitro models of thrombosis: methods and techniques, Cardiovasc. Diagn. Ther 7 (3) (2017) S329–S335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Zhang C, Neelamegham S, Application of microfluidic devices in studies of thrombosis and hemostasis, Platelets 28 (5) (2017) 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Hastings SM, Griffin MT, Ku DN, Hemodynamic studies of platelet thrombosis using microfluidics, Platelets 28 (5) (2017) 427–433. [DOI] [PubMed] [Google Scholar]
- [303].Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Microvasculature-on-a-chip for the long-term study of endothelial barrier dysfunction and microvascular obstruction in disease, Nat. Biomed. Eng (2018) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Li Y, Pan C, Li Y, Kumacheva E, Ramachandran A, An exploration of the reflow technique for the fabrication of an in vitro microvascular system to study occlusive clots, Biomed. Microdevices 19 (4) (2017) 82. [DOI] [PubMed] [Google Scholar]
- [305].Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, In vitro microvessels for the study of angiogenesis and thrombosis, Proc. Natl. Acad. Sci. U. S. A 109 (24) (2012) 9342–9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Zhao H, Li X, Zhao S, Zeng Y, Zhao L, Ding H, Sun W, Du Y, Microengineered in vitro model of cardiac fibrosis through modulating myofibroblast mechanotransduction, Biofabrication 6 (4) (2014) 045009. [DOI] [PubMed] [Google Scholar]
- [307].Soares CP, Midlej V, de Oliveira MEW, Benchimol M, Costa ML, Mermelstein C, 2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression, PloS One 7 (5) (2012) e38147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].van Spreeuwel AC, Bax NA, van Nierop BJ, Aartsma-Rus A, Goumans M-JT, Bouten CV, Mimicking Cardiac Fibrosis in a Dish: fibroblast density rather than collagen density weakens cardiomyocyte function, J. Cardiovascular Transl. Res 10 (2) (2017) 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Spencer TM, Blumenstein RF, Pryse KM, Lee S-L, Glaubke DA, Carlson BE, Elson EL, Genin GM, Fibroblasts slow conduction velocity in a reconstituted tissue model of fibrotic cardiomyopathy, ACS Biomater. Sci. Eng 3 (11) (2016) 3022–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Sadeghi AH, Shin SR, Deddens JC, Fratta G, Mandla S, Yazdi IK, Prakash G, Antona S, Demarchi D, Buijsrogge MP, Engineered 3D cardiac fibrotic tissue to study fibrotic remodeling, Adv. Healthcare Mater 6 (11) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Figtree GA, Bubb KJ, Tang O, Kizana E, Gentile C, Vascularized cardiac spheroids as novel 3D in vitro models to study cardiac fibrosis, Cells Tissues Organs 204 (3) (2017) 191–198. [DOI] [PubMed] [Google Scholar]
- [312].Noble D, A modification of the Hodgkin—Huxley equations applicable to Purkinje fibre action and pacemaker potentials, J. Physiol 160 (2) (1962) 317–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Rodríguez B, Li L, Eason JC, Efimov IR, Trayanova NA, Differences between left and right ventricular chamber geometry affect cardiac vulnerability to electric shocks, Circ. Res 97 (2) (2005) 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Plank G, Burton RA, Hales P, Bishop M, Mansoori T, Bernabeu MO, Garny A, Prassl AJ, Bollensdorff C, Mason F, Generation of histo-anatomically representative models of the individual heart: tools and application, Phil. Trans. Roy. Soc. Lond.: Math. Phys. Eng. Sci 367 (1896) (2009) 2257–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Vigmond E, Vadakkumpadan F, Gurev V, Arevalo H, Deo M, Plank G, Trayanova N, Towards predictive modelling of the electrophysiology of the heart, Exp. Physiol 94 (5) (2009) 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Bishop MJ, Plank G, Burton RA, Schneider JE, Gavaghan DJ, Grau V, Kohl P, Development of an anatomically detailed MRI-derived rabbit ventricular model and assessment of its impact on simulations of electrophysiological function, Am. J. Physiol. Heart Circ. Physiol 298 (2) (2009) H699–H718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Li Z, Dutta S, Sheng J, Tran PN, Wu W, Chang K, Mdluli T, Strauss DG, Colatsky T, Improving the in silico assessment of proarrhythmia risk by combining hERG (human ether-à-go-go-related gene) channel–drug binding kinetics and multichannel pharmacology, Circulation: Arrhythmia Electrophysiol. 10 (2) (2017) e004628. [DOI] [PubMed] [Google Scholar]
- [318].Zhang Y, Barocas VH, Berceli SA, Clancy CE, Eckmann DM, Garbey M, Kassab GS, Lochner DR, McCulloch AD, Tran-Son-Tay R, Multi-scale modeling of the cardiovascular system: disease development, progression, and clinical intervention, Ann. Biomed. Eng 44 (9) (2016) 2642–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Kenakin T, Principles: receptor theory in pharmacology, Trends Pharmacol. Sci 25 (4) (2004) 186–192. [DOI] [PubMed] [Google Scholar]
- [320].Hund TJ, Rudy Y, Rate dependence and regulation of action potential and calcium transient in a canine cardiac ventricular cell model, Circulation 110 (20) (2004) 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].O’Hara T, Virág L, Varró A, Rudy Y, Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation, PLoS Comput. Biol 7 (5) (2011) e1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Faber GM, Silva J, Livshitz L, Rudy Y, Kinetic properties of the cardiac L-type Ca2+ channel and its role in myocyte electrophysiology: a theoretical investigation, Biophys. J 92 (5) (2007) 1522–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Faber GM, Rudy Y, Calsequestrin mutation and catecholaminergic polymorphic ventricular tachycardia: a simulation study of cellular mechanism, Cardiovasc. Res 75 (1) (2007) 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Chi KR, Revolution Dawning in Cardiotoxicity Testing, Nature Publishing Group, 2013. [DOI] [PubMed] [Google Scholar]
- [325].Gintant G, Sager PT, Stockbridge N, Evolution of strategies to improve preclinical cardiac safety testing, Nat. Rev. Drug Discov 15 (7) (2016) 457. [DOI] [PubMed] [Google Scholar]
- [326].Gintant G, Fermini B, Stockbridge N, Strauss D, The evolving roles of human iPSC-derived cardiomyocytes in drug safety and discovery, Cell Stem Cell 21 (1) (2017) 14–17. [DOI] [PubMed] [Google Scholar]
- [327].Kumar N, Hendriks BS, Janes KA, de Graaf D, Lauffenburger DA, Applying computational modeling to drug discovery and development, Drug Discov. Today 11 (17) (2006) 806–811. [DOI] [PubMed] [Google Scholar]
- [328].Leil TA, Ermakov S, The emerging discipline of quantitative systems pharmacology, Front. Pharmacol 6 (2015) 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Sorger PK, Allerheiligen SR, Abernethy DR, Altman RB, Brouwer KL, Califano A, D’Argenio DZ, Iyengar R, Jusko WJ, Lalonde R, Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms, An NIH White Paper by the QSP Workshop Group, NIH Bethesda,, 2011. [Google Scholar]
- [330].Vicini P, Graaf P, Systems pharmacology for drug discovery and development: paradigm shift or flash in the pan? Clin. Pharmacol. Therapeut 93 (5) (2013) 379–381. [DOI] [PubMed] [Google Scholar]
- [331].Yang JH, Saucerman JJ, Computational models reduce complexity and accelerate insight into cardiac signaling networks, Circ. Res 108 (1) (2011) 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Davies MR, Wang K, Mirams GR, Caruso A, Noble D, Walz A, Lave T, Schuler F, Singer T, Polonchuk L, Recent developments in using mechanistic cardiac modelling for drug safety evaluation, Drug Discov. Today 21 (6) (2016) 924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Redfern W, Carlsson L, Davis A, Lynch W, MacKenzie I, Palethorpe S, Siegl P, Strang I, Sullivan A, Wallis R, Relationships between preclinical cardiac electrophysiology, clinical QT interval prolongation and torsade de pointes for a broad range of drugs: evidence for a provisional safety margin in drug development, Cardiovasc. Res 58 (1) (2003) 32–45. [DOI] [PubMed] [Google Scholar]
- [334].Mishra H, Polak S, Jamei M, Rostami-Hodjegan A, Interaction between domperidone and ketoconazole: toward prediction of consequent QTc prolongation using purely in vitro information, CPT Pharmacometrics Syst. Pharmacol 3 (8) (2014) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Wagner C, Zhao P, Pan Y, Hsu V, Grillo J, Huang S, Sinha V, Application of physiologically based pharmacokinetic (PBPK) modeling to support dose selection: report of an FDA public workshop on PBPK, CPT Pharmacometrics Syst. Pharmacol 4 (4) (2015) 226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Shim JV, Chun B, van Hasselt JG, Birtwistle MR, Saucerman JJ, Sobie EA, Mechanistic systems modeling to improve understanding and prediction of cardiotoxicity caused by targeted cancer therapeutics, Front. Physiol 8 (2017) 651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Conant G, Ahadian S, Zhao Y, Radisic M, Kinase inhibitor screening using artificial neural networks and engineered cardiac biowires, Sci. Rep 7 (1) (2017) 11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK, Systems biology and combination therapy in the quest for clinical efficacy, Nat. Chem. Biol 2 (9) (2006) 458–466. [DOI] [PubMed] [Google Scholar]
- [339].A.D. Association, 8. Cardiovascular disease and risk management, Diabetes Care 39 (Supplement 1) (2016) S60–S71. [DOI] [PubMed] [Google Scholar]
- [340].Fordyce CB, Roe MT, Ahmad T, Libby P, Borer JS, Hiatt WR, Bristow MR, Packer M, Wasserman SM, Braunstein N, Pitt B, DeMets DL, Cooper-Arnold K, Armstrong PW, Berkowitz SD, Scott R, Prats J, Galis ZS, Stockbridge N, Peterson ED, Califf RM, Cardiovascular drug development, is it dead or just, Hibernating? 65 (15) (2015) 1567–1582. [DOI] [PubMed] [Google Scholar]
- [341].Beierlein JM, McNamee LM, Walsh MJ, Kaitin KI, DiMasi JA, Ledley FD, Landscape of innovation for cardiovascular pharmaceuticals: from basic science to new molecular entities, Clin. Therapeut 39 (7) (2017) 1409–1425 e20. [DOI] [PubMed] [Google Scholar]
- [342].Levaggi R, Moretto M, Pertile P, The dynamics of pharmaceutical regulation and R&D investments, J. Publ. Econ. Theor 19 (1) (2017) 121–141. [Google Scholar]
- [343].Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL, How to improve R&D productivity: the pharmaceutical industry’s grand challenge, Nat. Rev. Drug Discov 9 (3) (2010) 203. [DOI] [PubMed] [Google Scholar]
- [344].Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J, Clinical development success rates for investigational drugs, Nat. Biotechnol 32 (1) (2014) 40. [DOI] [PubMed] [Google Scholar]
- [345].Siramshetty VB, Nickel J, Omieczynski C, Gohlke B-O, Drwal MN, Preissner R, WITHDRAWN—a resource for withdrawn and discontinued drugs, Nucleic Acids Res. 44 (D1) (2015) D1080–D1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Nicolaou K, Advancing the drug discovery and development process, Angew. Chem 126 (35) (2014) 9280–9292. [DOI] [PubMed] [Google Scholar]
- [347].Colatsky TJ, Emerging Technologies and their Role in Regulatory Review, Drug Discovery Toxicology: From Target Assessment to Translational Biomarkers, (2016), p. 1. [Google Scholar]
- [348].Khanna I, Drug discovery in pharmaceutical industry: productivity challenges and trends, Drug Discov. Today 17 (19–20) (2012) 1088–1102. [DOI] [PubMed] [Google Scholar]
- [349].Denayer T, Stöhr T, Van Roy M, Animal models in translational medicine: validation and prediction, New Horizons Transl. Med 2 (1) (2014) 5–11. [Google Scholar]
- [350].Breslin S, O’Driscoll L, Three-dimensional cell culture: the missing link in drug discovery, Drug Discov. Today 18 (5–6) (2013) 240–249. [DOI] [PubMed] [Google Scholar]
- [351].Edmondson R, Broglie JJ, Adcock AF, Yang L, Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors, Assay Drug Dev. Technol 12 (4) (2014) 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Esch EW, Bahinski A, Huh D, Organs-on-chips at the frontiers of drug discovery, Nat. Rev. Drug Discov 14 (4) (2015) 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Ryan AJ, Brougham CM, Garciarena CD, Kerrigan SW, O’Brien FJ, Towards 3D in vitro models for the study of cardiovascular tissues and disease, Drug Discov. Today 21 (9) (2016) 1437–1445. [DOI] [PubMed] [Google Scholar]
- [354].Nam K-H, Smith AS, Lone S, Kwon S, Kim D-H, Biomimetic 3D tissue models for advanced high-throughput drug screening, J. Lab. Autom 20 (3) (2015) 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Skardal A, Shupe T, Atala A, Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling, Drug Discov. Today 21 (9) (2016) 1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [356].Peng W, Unutmaz D, Ozbolat IT, Bioprinting towards physiologically relevant tissue models for pharmaceutics, Trends Biotechnol. 34 (9) (2016) 722–732. [DOI] [PubMed] [Google Scholar]
- [357].Tzatzalos E, Abilez OJ, Shukla P, Wu JC, Engineered heart tissues and induced pluripotent stem cells: macro-and microstructures for disease modeling, drug screening, and translational studies, Adv. Drug Deliv. Rev 96 (2016) 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Shuler ML, Organ-, body-and disease-on-a-chip systems, Lab a Chip 17 (14) (2017) 2345–2346. [DOI] [PubMed] [Google Scholar]
- [359].Zheng F, Fu F, Cheng Y, Wang C, Zhao Y, Gu Z, Organ-on-a chip systems: microengineering to biomimic living systems, Small 12 (17) (2016) 2253–2282. [DOI] [PubMed] [Google Scholar]
- [360].Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E, Davenport-Huyer L, Lai B, Zhang B, Zhao Y, Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies, Adv. Healthcare Mater. 7 (2018) 1700506. [DOI] [PubMed] [Google Scholar]
- [361].Gomes ME, Rodrigues MT, Domingues RM, Reis RL, Tissue engineering and regenerative medicine: new trends and directions—a year in review, Tissue Eng. Part B Rev 23 (3) (2017) 211–224. [DOI] [PubMed] [Google Scholar]
- [365].Geris L, Guyot Y, Schrooten J, Papantoniou I, In silico regenerative medicine: how computational tools allow regulatory and financial challenges to be addressed in a volatile market, Interface Focus 6 (2) (2016) 20150105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Winslow RL, Trayanova N, Geman D, Miller MI, Computational medicine: translating models to clinical care, Sci. Transl. Med 4 (158) (2012) 158rv11158rv11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Kapetanovic I, Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach, Chem. Biol. Interact 171 (2) (2008) 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Jones H, Chen Y, Gibson C, Heimbach T, Parrott N, Peters S, Snoeys J, Upreti V, Zheng M, Hall S, Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective, Clin. Pharmacol. Therapeut 97 (3) (2015) 247–262. [DOI] [PubMed] [Google Scholar]
- [369].Jamei M, Recent advances in development and application of physiologically-based pharmacokinetic (PBPK) models: a transition from academic curiosity to regulatory acceptance, Curr. Pharmacol. Rep 2 (3) (2016) 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Kirchmair J, Göller AH, Lang D, Kunze J, Testa B, Wilson ID, Glen RC, Schneider G, Predicting drug metabolism: experiment and/or computation? Nat. Rev. Drug Discov 14 (6) (2015) 387. [DOI] [PubMed] [Google Scholar]
- [371].Knight-Schrijver V, Chelliah V, Cucurull-Sanchez L, Le Novère N, The promises of quantitative systems pharmacology modelling for drug development, Comput. Struct. Biotechnol. J 14 (2016) 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Arrigoni C, Gilardi M, Bersini S, Candrian C, Moretti M, Bioprinting and organ-on-chip applications towards personalized medicine for bone diseases, Stem Cell Rev. Rep 13 (3) (2017) 407–417. [DOI] [PubMed] [Google Scholar]
- [373].Kodzius R, Schulze F, Gao X, Schneider MR, Organ-on-Chip Technology: Current State and Future Developments, Multidisciplinary Digital Publishing Institute, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].Memic A, Navaei A, Mirani B, Cordova JAV, Aldhahri M, Dolatshahi-Pirouz A, Akbari M, Nikkhah M, Bioprinting technologies for disease modeling, Biotechnol. Lett 39 (9) (2017) 1279–1290. [DOI] [PubMed] [Google Scholar]
- [375].Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, Interconnected microphysiological systems for quantitative biology and pharmacology studies, Sci. Rep 8 (1) (2018) 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, Massa S, Riahi R, Chae S, Hu N, Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors, Proc. Natl. Acad. Sci. U. S. A (2017) 201612906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [377].Mirasoli M, Guardigli M, Michelini E, Roda A, Recent advancements in chemical luminescence-based lab-on-chip and microfluidic platforms for bioanalysis, J. Pharmaceut. Biomed. Anal 87 (2014) 36–52. [DOI] [PubMed] [Google Scholar]
- [378].Dietze C, Schulze S, Ohla S, Gilmore K, Seeberger P, Belder D, Integrated onchip mass spectrometry reaction monitoring in microfluidic devices containing porous polymer monolithic columns, Analyst 141 (18) (2016) 5412–5416. [DOI] [PubMed] [Google Scholar]
- [379].Beulig R, Warias R, Heiland J, Ohla S, Zeitler K, Belder D, A droplet-chip/mass spectrometry approach to study organic synthesis at nanoliter scale, Lab a Chip 17 (11) (2017) 1996–2002. [DOI] [PubMed] [Google Scholar]
- [380].Zhang YS, Ribas J, Nadhman A, Aleman J, Selimović Š, Lesher-Perez SC, Wang T, Manoharan V, Shin S-R, Damilano A, A cost-effective fluorescence mini-microscope for biomedical applications, Lab a Chip 15 (18) (2015) 3661–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [381].Cho S, Islas-Robles A, Nicolini AM, Monks TJ, Yoon J-Y, In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope, Biosens. Bioelectron 86 (2016) 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [382].Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, Lindner M, Drewell C, Bauer S, Thomas A, A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents, Lab a Chip 15 (12) (2015) 2688–2699. [DOI] [PubMed] [Google Scholar]
- [383].Materne E-M, Maschmeyer I, Lorenz AK, Horland R, Schimek KM, Busek M, Sonntag F, Lauster R, Marx U, The multi-organ chip-a microfluidic platform for long-term multi-tissue coculture, Jo. Visual. Exp.: JoVE 98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [384].Ronaldson-Bouchard K, Vunjak-Novakovic G, Organs-on-a-Chip: a fast track for engineered human tissues in drug development, Cell Stem Cell 22 (3) (2018) 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Peng W, Datta P, Ayan B, Ozbolat V, Sosnoski D, Ozbolat IT, 3D bioprinting for drug discovery and development in pharmaceutics, Acta Biomater. 57 (2017) 26–46. [DOI] [PubMed] [Google Scholar]
- [386].Savoji H, Maire M, Lequoy P, Liberelle B, De Crescenzo G, Ajji A, Wertheimer MR, Lerouge S, Combining electrospun fiber mats and bioactive coatings for vascular graft prostheses, Biomacromolecules 18 (1) (2016) 303–310. [DOI] [PubMed] [Google Scholar]
- [387].Park J, Wetzel I, Dréau D, Cho H, Human organ miniaturization: 3D miniaturization of human organs for drug discovery (Adv. Healthcare mater. 2/2018), Adv. Healthcare Mater 7 (2) (2018). [DOI] [PubMed] [Google Scholar]
- [388].Wu J, Xie L, Lin WZY, Chen Q, Biomimetic nanofibrous scaffolds for neural tissue engineering and drug development, Drug Discov. Today 22 (9) (2017) 1375–1384. [DOI] [PubMed] [Google Scholar]
- [389].Akbari M, Tamayol A, Bagherifard S, Serex L, Mostafalu P, Faramarzi N, Mohammadi MH, Khademhosseini A, Textile technologies and tissue engineering: a path toward organ weaving, Adv. Healthcare Mater 5 (7) (2016) 751–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Kharaziha M, Memic A, Akbari M, Brafman DA, Nikkhah M, Nano‐enabled approaches for stem cell‐based cardiac tissue engineering, Adv. Healthcare Mater 5 (13) (2016) 1533–1553. [DOI] [PubMed] [Google Scholar]
- [391].Savoji H, Hadjizadeh A, Maire M, Ajji A, Wertheimer MR, Lerouge S, Electrospun nanofiber scaffolds and plasma polymerization: a promising combination towards complete, stable endothelial lining for vascular grafts, Macromol. Biosci 14 (8) (2014) 1084–1095. [DOI] [PubMed] [Google Scholar]
- [392].Hadjizadeh A, Savoji H, Ajji A, A facile approach for the mass production of submicro/micro poly (lactic acid) fibrous mats and their cytotoxicity test towards neural stem cells, BioMed Res. Int (2016) 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [393].Savoji H, Lerouge S, Ajji A, Wertheimer MR, Plasma‐etching for controlled modification of structural and mechanical properties of electrospun PET scaffolds, Plasma Process. Polym 12 (4) (2015) 314–327. [Google Scholar]
- [394].Giebel LB, Stem cells—a hard sell to investors, Nat. Biotechnol 23 (7) (2005) 798. [DOI] [PubMed] [Google Scholar]
- [395].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell 126 (4) (2006) 663–676. [DOI] [PubMed] [Google Scholar]
- [396].Matsa E, Ahrens JH, Wu JC, Human induced pluripotent stem cells as a platform for personalized and precision cardiovascular medicine, Physiol. Rev 96 (3) (2016) 1093–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Witty AD, Mihic A, Tam RY, Fisher SA, Mikryukov A, Shoichet MS, Li R-K, Kattman SJ, Keller G, Generation of the epicardial lineage from human pluripotent stem cells, Nat. Biotechnol 32 (10) (2014) 1026–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Protze SI, Liu J, Nussinovitch U, Ohana L, Backx PH, Gepstein L, Keller GM, Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker, Nat. Biotechnol 35 (1) (2017) 56. [DOI] [PubMed] [Google Scholar]
- [399].Karakikes I, Senyei GD, Hansen J, Kong C-W, Azeloglu EU, Stillitano F, Lieu DK, Wang J, Ren L, Hulot J-S, Small molecule‐mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes, Stem Cells Transl. Med 3 (1) (2014) 18–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [400].Weng Z, Kong C-W, Ren L, Karakikes I, Geng L, He J, Chow MZY, Mok CF, Chan HY, Webb SE, A simple, cost-effective but highly efficient system for deriving ventricular cardiomyocytes from human pluripotent stem cells, Stem Cell. Dev 23 (14) (2014) 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Josowitz R, Lu J, Falce C, D’Souza SL, Wu M, Cohen N, Dubois NC, Zhao Y, Sobie EA, Fishman GI, Identification and purification of human induced pluripotent stem cell-derived atrial-like cardiomyocytes based on sarcolipin expression, PloS One 9 (7) (2014) e101316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Tanwar V, Bylund JB, Hu J, Yan J, Walthall JM, Mukherjee A, Heaton WH, Wang WD, Potet F, Rai M, Gremlin 2 promotes differentiation of embryonic stem cells to atrial fate by activation of the JNK signaling pathway, Stem Cell. 32 (7) (2014) 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [403].Kapoor N, Liang W, Marbán E, Cho HC, Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18, Nat. Biotechnol 31 (1) (2013) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Matsa E, Burridge PW, Wu JC, Human stem cells for modeling heart disease and for drug discovery, Sci. Transl. Med 6 (239) (2014) 239ps6–239ps6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [405].Karakikes I, Ameen M, Termglinchan V, Wu JC, Human induced pluripotent stem cell–derived cardiomyocytes: insights into molecular, cellular, and functional phenotypes, Circ. Res 117 (1) (2015) 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].Czirok A, Isai DG, Rajasingh S, Filla M, Johnson R, Cardiac ECM Environment Improves IPSC-Derived Cardiomyocyte Differentiation, Am Heart Assoc, 2017. [Google Scholar]
- [407].Murthy SK, Sethu P, Vunjak-Novakovic G, Toner M, Radisic M, Size-based microfluidic enrichment of neonatal rat cardiac cell populations, Biomed. Microdevices 8 (3) (2006) 231–237. [DOI] [PubMed] [Google Scholar]
- [408].Zhang B, Green JV, Murthy SK, Radisic M, Label-free enrichment of functional cardiomyocytes using microfluidic deterministic lateral flow displacement, PloS One 7 (5) (2012) e37619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [409].Adams JD, Kim U, Soh HT, Multitarget magnetic activated cell sorter, Proc. Natl. Acad. Sci. U. S. A 105 (47) (2008) 18165–18170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [410].Jung CB, Moretti A, y Schnitzler MM, Iop L, Storch U, Bellin M, Dorn T, Ruppenthal S, Pfeiffer S, Goedel A, Dantrolene rescues arrhythmogenic RYR2 defect in a patient‐specific stem cell model of catecholaminergic polymorphic ventricular tachycardia, EMBO Mol. Med 4 (3) (2012) 180–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [411].Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, Navarrete EG, Hu S, Wang L, Lee A, Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy, Sci. Transl. Med 4 (130) (2012) 130ra47–130ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [412].Zhang YS, Zhang Y-N, Zhang W, Cancer-on-a-chip systems at the frontier of nanomedicine, Drug Discov. Today 22 (9) (2017) 1392–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [413].Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, Common genetic variation drives molecular heterogeneity in human iPSCs, Nature 546 (7658) (2017) 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, Patel M, Goncalves A, Ferreira R, Benn CL, Molecular and functional variation in iPSC-derived sensory neurons, Nat. Genet 50 (1) (2018) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [415].Kim JH, Kurtz A, Yuan BZ, Zeng F, Lomax G, Loring JF, Crook J, Ju JH, Clarke L, Inamdar MS, Report of the international stem cell banking initiative workshop activity: current hurdles and progress in seed‐stock banking of human pluripotent stem cells, Stem Cells Transl. Med 6 (11) (2017) 1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [416].DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D’Antonio M, Large-scale profiling reveals the influence of genetic variation on gene expression in human induced pluripotent stem cells, Cell Stem Cell 20 (4) (2017) 533–546 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [417].Panopoulos AD, D’Antonio M, Benaglio P, Williams R, Hashem SI, Schuldt BM, DeBoever C, Arias AD, Garcia M, Nelson BC, iPSCORE: a resource of 222 iPSC lines enabling functional characterization of genetic variation across a variety of cell types, Stem Cell Rep. 8 (4) (2017) 1086–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Aoi T, Stacey G, Impact of national and international stem cell banking initiatives on progress in the field of cell therapy: IABS-JST joint workshop: summary for session 5, Biologicals 43 (5) (2015) 399–401. [DOI] [PubMed] [Google Scholar]
- [419].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity, Science (2012) 1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [420].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini L, Multiplex genome engineering using CRISPR/Cas systems, Science (2013) 1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat. Protoc 8 (11) (2013) 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [423].Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Epigenetic regulation of phosphodiesterases 2A and 3A underlies compromised β-adrenergic signaling in an iPSC model of dilated cardiomyopathy, Cell Stem Cell 17 (1) (2015) 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC, Human induced pluripotent stem cell–derived cardiomyocytes as an in vitro model for coxsackievirus B3–induced myocarditis and antiviral drug screening platform, Circ. Res 115 (6) (2014) 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Musante C, Ramanujan S, Schmidt B, Ghobrial O, Lu J, Heatherington A, Quantitative systems pharmacology: a case for disease models, Clin. Pharmacol. Therapeut 101 (1) (2017) 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [426].Helmlinger G, Al-Huniti N, Aksenov S, Peskov K, Hallow KM, Chu L, Boulton D, Eriksson U, Hamrén B, Lambert C, Drug-disease modeling in the pharmaceutical industry-where mechanistic systems pharmacology and statistical pharmacometrics meet, Eur. J. Pharmaceut. Sci 109 (2017) S39–S46. [DOI] [PubMed] [Google Scholar]