Abstract
Human platelets express two protease-activated receptors, PAR1 (F2R) and PAR4 (F2RL3), which are activated by a number of serine proteases that are generated during pathological events and cause platelet activation. Recent interest has focused on PAR4 as a therapeutic target, given PAR4 seems to promote experimental thrombosis and procoagulant microparticle formation, without a broadly apparent role in hemostasis. However, it is not yet known whether PAR4 activity plays a role in platelet-leukocyte interactions, which are thought to contribute to both thrombosis and acute or chronic thrombo-inflammatory processes. We sought to determine whether PAR4 activity contributes to granule secretion from activated platelets and platelet-leukocyte interactions. We performed in vitro and ex vivo studies of platelet granule release and platelet-leukocyte interactions in the presence of PAR4 agonists including PAR4 activating peptide, thrombin, cathepsin G, and plasmin in combination with small-molecule PAR4 antagonists. Activation of human platelets with thrombin, cathepsin G, or plasmin potentiated platelet dense granule secretion that was specifically impaired by PAR4 inhibitors. Platelet-leukocyte interactions and platelet P-selectin exposure following stimulation with PAR4 agonists were also impaired by activated PAR4 inhibition in either a purified system or in whole blood. These results indicate PAR4-specific promotion of platelet granule release and platelet-leukocyte aggregate formation and suggest that pharmacological control of PAR4 activity could potentially attenuate platelet granule release or platelet-leukocyte interaction-mediated pathological processes.
Introduction
Platelets become activated upon vessel injury or inflammation by serine proteases such as thrombin that cleave platelet protease-activated receptors (PARs) and initiate intracellular signaling pathways. Human platelets express PAR1 and PAR4, G-protein coupled receptors (GPCRs) that are activated by proteolytic cleavage of an N-terminal site to reveal a tethered ligand that binds the receptor itself and initiates intracellular G-protein signaling.1 PAR1 and PAR4 activation of G proteins leads to signaling cascades causing release of calcium stores, secretion of dense granule contents, and platelet shape change, culminating in platelet activation, adhesion, and aggregation.
Structural differences between PAR1 and PAR4 result in differing outputs in platelet function. PAR1 contains a negatively-charged N-terminal sequence that binds the anion-binding exosite I of thrombin,2 which allosterically enhances thrombin’s activity and enables it to activate both PAR1 and PAR4 while tethered to PAR1. PAR4 lacks this thrombin binding sequence, and higher concentrations of thrombin are required to activate PAR4 compared to PAR1.3 Thrombin binds PAR1 transiently, causing robust platelet activation that is carefully constrained by rapid phosphorylation, internalization and degradation of the receptor.4 PAR4 is also internalized to terminate its activity, but this internalization occurs via a different route than that of PAR1, in a manner hypothesized to enable prolonged signaling.5 These differences in PAR4 result in a response to thrombin that is slower but more sustained over time, with varying functional effects, including described roles in enhancing clot stability and procoagulant microparticle release that suggest a more pro-thrombotic effect of platelet PAR4 activity.6,7 Moreover, platelet PAR4 plays a described role in the activation of PKC substrates, which are required for platelet dense granule release.7 Release of platelet dense granule contents, which include a variety of biologically active molecules, is a physiologically important phenomenon,8 but it has also been implicated in the pathomechanism of certain diseases.9,10
PAR1 and PAR4 are cleaved by overlapping but distinct sets of proteases, leading to diverse functional outputs. PAR1 is known to be cleaved at its canonical N-terminal site (R41/S42) by thrombin, factor Xa, plasmin, and MMP1/13, and it also can be cleaved at different noncanonical sites by elastase, APC, and proteinase-3.11 Meanwhile, PAR4 is known to be cleaved only at its canonical site (R47/G48) by thrombin, trypsin, tissue kallikrein, plasmin, and cathepsin G.12–15 Given the unique role of neutrophil cathepsin G in cleavage of PAR4, this suggests involvement of PAR4 in facilitating interactions between platelets and leukocytes, including neutrophils, a subset of granulocytes, and monocytes, a subset of peripheral blood mononuclear cells (PBMCs). Platelet-leukocyte interactions increase during pathological conditions such as atherosclerosis and may detrimentally affect disease outcomes.16,17
In addition to observations that the neutrophil releasate cathepsin G activates platelets via PAR4 cleavage, studies have shown that PAR4 activity promotes leukocyte recruitment in animal models of inflammation and pain.18 Current interest is focused on PAR4 as a potential target against thrombosis, with the PAR4 inhibitor BMS-986120 showing promise in animal models of thrombosis as well as a completed phase 2 clinical trial in combination with aspirin for the prevention of recurrent stroke.19,20 However, investigations into the role of PAR4 activity on human platelet-leukocyte interactions are lacking. Therefore, in this study, we investigated the effect of PAR4 activity on platelet dense granule release and platelet-leukocyte interactions to interrogate the potential role of platelet PAR4 in inflammation and innate immunity.
Methods
Reagents
Activated PAR4 antagonists were synthesized, characterized, dissolved in dimethyl sulfoxide (DMSO), and stored refrigerated as described previously.21 Structure and characterization of PAR4 antagonists are described in Supplemental Figures S1-S2. All other reagents were from Sigma-Aldrich (St. Louis, MO, USA), unless stated otherwise. Hanks’ Balanced Salt Solution (HBSS) was from Corning cellgro (Manassas, VA, USA). Polymorphprep was from Axis-Shield (Oslo, Norway). PGI2 and the PAR1 inhibitor SCH 79797 were from Cayman Chemical (Ann Arbor, MI, USA). Collagen-related peptide (CRP) was from R. Farndale (Cambridge University, UK). TRAP-6 (SFLLRN-NH2) was obtained from Tocris (Bristol, UK). PPACK (D-Phe-Pro-Arg-chloromethylketone) and RBC Lysis Buffer were from Santa Cruz (Dallas, TX, USA). PAR4 activating peptide (AYPGKF-NH2) was from Abgent (San Diego, CA, USA). Human α-thrombin and human plasmin were from Haematologic Technologies (Essex Junction, VT, USA). Human cathepsin G was from Innovative Research (Novi, MI, USA). For human flow cytometry studies, anti-CD66b-PE and anti-CD14-PE/Cy7 were from BD Biosciences (Franklin Lakes, NJ, USA), and anti-CD41-FITC was from Invitrogen (Carlsbad, CA, USA). For nonhuman primate flow cytometry studies, anti-CD41-FITC was from Invitrogen, and anti-CD62P-PE and anti-CD45-APC were from BD Biosciences. Chronolume detection agent was from Chrono-Log Corporation (Havertown, PA, USA).
Platelet preparation
Venous blood was obtained from healthy volunteers in accordance with an Oregon Health & Science University (OHSU) IRB-approved protocol. No demographic data was collected on volunteers. For washed platelet preparation, blood was drawn into 3.8% trisodium citrate 9:1 (v:v), and acid-citrate dextrose (ACD) was added at 1:10 (v:v). Platelet-rich plasma (PRP) was isolated by centrifugation at 200 g for 20 minutes, and platelets were separated from PRP at 1000 g for 10 minutes in the presence of prostacyclin (0.1 μg/ml). Platelets were resuspended in modified HEPES/Tyrode buffer (129 mM NaCl, 0.34 mM Na2HPO4, 2.9 mM KCl, 12 mM NaHCO3, 20 mM HEPES, 1 mM MgCl2, pH 7.3, supplemented with 5 mM glucose), washed by centrifugation at 1000 g for 10 minutes, and resuspended again in modified HEPES/Tyrode buffer to the specified platelet count. Of note, exogenous CaCl2 was not added to any of the assays.
Platelet dense granule secretion assay
Platelet dense granule secretion was measured as luminescence in an ATP-luciferin-luciferase reaction, as previously described.22 Briefly, washed platelets (20 × 107/ml; 70 μl) were incubated in a white, flat bottom Corning 96-well plate with 10 μl inhibitor or vehicle (1% DMSO) for 15 minutes at 37 °C with orbital shaking. Agonists (10 μl) were added and incubation continued for an additional 10 minutes. Finally, 10 μl Chronolume detection agent was added and luminescence measured on an Infinite M200 spectrophotometer (TECAN, Switzerland).
Granulocyte preparation
Human granulocytes were isolated as previously described,23 with minor modifications. Venous blood was drawn in accordance with an OHSU IRB-approved protocol at 7:1 (v:v) into citrate phosphate dextrose (CPD). Blood was layered onto an equal volume of Polymorphprep and centrifuged at 500 g for 45 min, and the middle layer containing granulocytes was removed and washed in Hanks’ Balanced Salt Solution (HBSS) at 400 g for 10 minutes at 19 °C. The pellet was resuspended in cold sterile H2O for 30 seconds to lyse remaining red blood cells, followed by dilution in 10× PIPES buffer (250 mM PIPES, 1.1 mM CaCl2, 50 mM KCl, pH 7.4) and HBSS buffer and centrifugation at 400 g for 10 minutes at 19 °C. The granulocyte pellet was resuspended in HBSS buffer to the specified cell count.
Flow cytometry – platelets and granulocytes
Isolated platelets and granulocytes from the same donor were prepared as described above and combined in equal volume to a final concentration of 2 × 106/ml granulocytes and 20 × 107/ml platelets (1:100) and incubated with inhibitor or vehicle (0.2% DMSO) for 15 minutes at 37 °C. Treated cells (60 μl) were added to FACS tubes containing 20 μl antibody and agonist mixtures and incubated at room temperature 20 minutes. Antibody dilutions were 1:50 for anti-CD41, 1:80 for anti-CD62P, and 1:400 for anti-CD66b. Samples were fixed in BD Cytofix and modified HEPES/Tyrode buffer (1% PFA final) for 10 minutes before dilution to 300 μl in PBS containing 0.5% fatty acid-free bovine serum albumin (BSA). Samples were collected for 30 seconds at medium flow rate in a BD FACSCantoII and analyzed on FlowJo software v. 10.2 (Ashland, OR).
Flow cytometry – whole blood
We developed a whole blood flow cytometry assay based on several approaches.24,25 Venous blood from healthy volunteers under an OHSU IRB-approved protocol was drawn 9:1 (v:v) into 3.8% trisodium citrate containing PPACK (50 μM final) and subsequently diluted 1:1 (v:v) in PBS. Blood was incubated with inhibitor or vehicle (0.2% DMSO) for 15 minutes at room temperature. Treated blood (60 μl) was added to FACS tubes containing 20 μl of antibody and agonist mixture and incubated 20 minutes at room temperature. Antibody dilutions were 1:80 for anti-CD41 and anti-CD62P, 1:200 for anti-CD66b, and 1:400 for anti-CD14. Samples were fixed and red blood cells lysed in a mixture of BD Cytofix (1% PFA final) and RBC Lysis Buffer (0.67× final) in PBS for 10 minutes before dilution to 300 μl in RBC lysis buffer in PBS (1× final). Samples were collected for 60 seconds at low flow rate in a BD FACSCantoII and analyzed on FlowJo software v. 10.2 (Ashland, OR).
Electron microscopy
Isolated platelets and granulocytes from the same donor were prepared as described above and combined in equal volume to a final concentration of 2 × 106/ml granulocytes and 20 × 107/ml platelets (1:100) and incubated with agonist for 10 minutes at 37 °C. The sample was then centrifuged at 2500 g for 15 minutes, the pellet was fixed in 2.5% glutaraldehyde, and sections were prepared using standard methods in the OHSU Department of Pathology Electron Microscopy Research Laboratory, similar to previous studies demonstrating neutrophil-platelet adhesion.26
Nonhuman primate studies
Male baboons (Papio anubis) were housed and cared for at the OHSU Oregon National Primate Research Center (ONPRC), a Category I facility. All experiments described herein were approved by the OHSU West Campus Animal Care and Use Committee according to the Guide for the Care and Use of Laboratory Animals by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council (ISBN 0–309-05377–3, 1996). The Laboratory Animal Care and Use Program at the ONPRC is fully accredited by the American Association for Accreditation of Laboratory Animal Care and has an approved assurance (no. A3304–01) for the care and use of animals from the Office for
Protection from Research Risks at the National Institutes of Health.
For whole blood flow cytometry experiments in nonhuman primates, methods were similar to the human experiments, with a few modifications. Venous baboon blood was drawn 9:1 (v:v) into 3.8% trisodium citrate containing PPACK (50 μM final) and subsequently diluted 1:1 (v:v) in phosphate-buffered saline (PBS). Blood was incubated with inhibitor or vehicle (0.2% DMSO) for 15 minutes at room temperature. Treated blood (60 μl) was added to FACS tubes containing 20 μl of antibody mixture and incubated 20 minutes at room temperature. Antibody dilutions were 1:80 for anti-CD41, 1:50 for anti-CD62P and 1:300 for anti-CD45. Samples were fixed and red blood cells lysed in a mixture of BD Cytofix (1% PFA final) and RBC Lysis Buffer (0.67× final) in PBS for 10 minutes before dilution to 300 μl in RBC lysis buffer in PBS (1× final). Samples were collected for 30 seconds at low flow rate in a BD LSR II and analyzed on FlowJo software v. 10.2 (Ashland, OR).
Data analysis
Data in bar graphs have been normalized to vehicle (with agonist) to facilitate comparison of inhibitor treatments across agonist conditions. For statistical analysis, a one-way ANOVA was employed with post-hoc analysis using the Dunnett test to compare treatments to vehicle. P < 0.05 was considered statistically significant. Analysis was performed in GraphPad Prism software v. 6 (La Jolla, CA).
Results
PAR4 activity potentiates platelet dense granule secretion
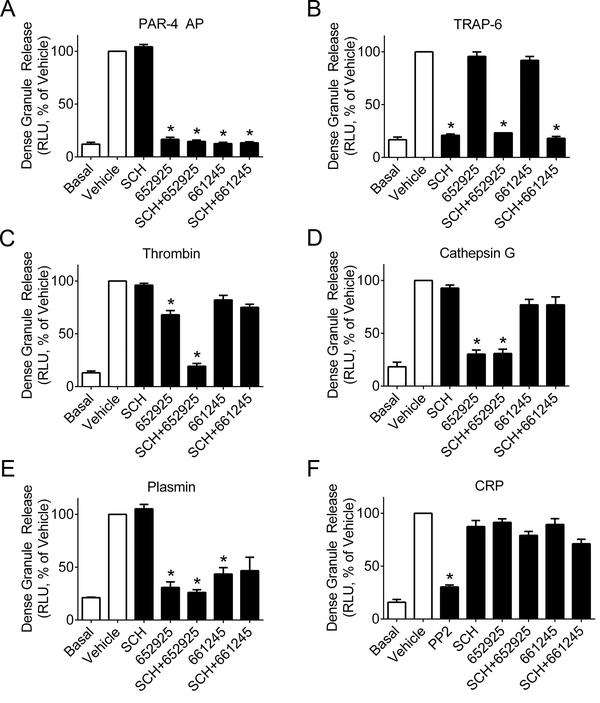
Platelet dense granules store a number of immunomodulatory molecules such as ADP, serotonin, glutamate, and polyphosphates, and dense granule secretion has been implicated in the proinflammatory recruitment of leukocytes and vascular remodeling in the progression of atherosclerosis.9,27,28 Dense granule secretion follows PAR1- and PAR4-mediated activation of protein kinase C (PKC),29 but PAR4 has been shown to activate PKC substrates more robustly than PAR1, suggesting a greater role for PAR4 in dense granule secretion.7 To test whether PAR4 plays a distinct role in platelet dense granule release, we performed a luminescent ATP-luciferin-luciferase assay on washed human platelets stimulated with PAR4 activating peptide (AP, AYPGKF-NH2), α-thrombin, or two other activators of PAR4, neutrophil cathepsin G and plasmin, which are proteases involved in inflammation and fibrinolysis, respectively. For comparison, platelets were stimulated with the PAR1 activating peptide TRAP-6 or the platelet GPVI receptor agonist CRP. In select experiments, platelets were pre-incubated with either the PAR1 inhibitor SCH 79797 (CAS # 1216720–69-2) or one of two small-molecule PAR4 antagonists, VU0652925, an analogue of BMS-986120, and VU0661245, an intermediate generated in an effort to define the minimum pharmacophore of VU0652925 (structures shown in Supplemental Figure S1).21 Both PAR4 antagonists have been characterized to inhibit PAR4 activity by either PAR-4 AP or the PAR4 tethered ligand generated by thrombin, but at higher thrombin concentrations (316 nM γ-thrombin), VU0652925 inhibits platelet activation more potently than VU0661245, as shown in Supplemental Figure S2.
In the dense granule release assay, either VU0652925 or VU0661245 blocked dense granule release by PAR4 AP, while the PAR1 inhibitor SCH 79797 had no effect (Figure 1A). Conversely, dense granule release by the PAR1 agonist TRAP-6 was blocked by SCH 79797 but not by either PAR4 inhibitor (Figure 1B). Dense granule release in response to a moderately high concentration of α-thrombin (5 nM, 0.7 U/ml) was partially blocked by the PAR4 inhibitor VU062925 but not by the PAR1 inhibitor SCH 79797, while VU0652925 and SCH 79797 combined together caused full blockage of dense granule release (Figure 1C). When platelets were stimulated by cathepsin G, VU0652925 completely blocked dense granule secretion, while the PAR1 inhibitor SCH 79797 did not provide any inhibition either alone or in combination with either PAR4 inhibitor (Figure 1D). Upon stimulation with plasmin, either PAR4 inhibitor blocked dense granule release, while SCH 79797 had no effect, either alone or in combination with a PAR4 inhibitor (Figure 1E). This suggests these potentially inflammatory and profibrinolytic proteases, cathepsin G and plasmin, facilitate platelet dense granule release through cleavage of PAR4 but not PAR1. Finally, none of the PAR4 or PAR1 inhibitors blocked dense granule release by collagen-related peptide (CRP), suggesting no involvement in signaling downstream of the collagen receptor GPVI (Figure 1F).
Figure 1.
PAR4 inhibitors impair platelet dense granule release. Washed human platelets (2 × 108/ml) were pretreated with the PAR1 inhibitor SCH 79797 (3 μM), PAR4 inhibitors VU0652925 (10 μM) and VU0661245 (10 μM), Src kinase inhibitor PP2 (10 μM, only shown in CRP condition), or vehicle (0.2% DMSO), then stimulated with the agonists (A) PAR4 AP (activating peptide, 200 μM), (B) TRAP-6 (20 μM), (C) human α-thrombin (5 nM = 0.7 U/ml), (D) human cathepsin G (340 nM = 0.1 U/ml), (E) human plasmin (260 nM = 0.3 U/ml), or (F) CRP (10 μg/ml) and assessed for dense granule release by luminescent ATP assay. Basal = no agonist; n ≥ 3–6 independent experiments; * designates p < 0.05 versus vehicle.
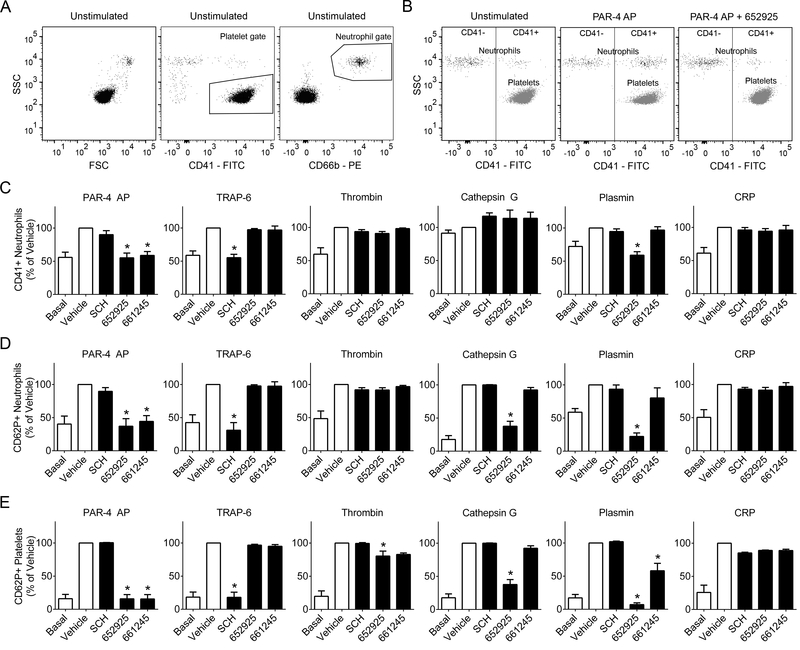
PAR4 activity promotes platelet-granulocyte interactions in a purified system
Platelet dense granule secretion has been linked to platelet-leukocyte interactions, which are a marker of inflammation and hypothesized to prime leukocyte-endothelial interactions underlying the progression of disease states such as atherosclerosis.9,16 Given our results demonstrating the role of PAR4 in facilitating dense granule release via plasmin and cathepsin G, we next investigated whether PAR4 also promotes platelet-leukocyte interactions. Isolated platelets and granulocytes from the same donor were combined, pre-incubated with inhibitors, and stimulated with agonists before analysis. Ultrastructural interactions between activated platelets and neutrophils were observed by electron microscopy (Supplemental Figure S3). Platelet-neutrophil interactions were analyzed via flow cytometry (Figure 2) as expression of the platelet marker CD41 in the granulocyte gate (Figure 2C). Platelet-granulocyte interactions were diminished by PAR4 inhibitors upon stimulation with PAR4 AP (AYPGKF-NH2), while SCH 79797 had no effect. Platelet-granulocyte interactions following platelet activation with TRAP-6 were blocked by the PAR1 inhibitor SCH 79797 but not either PAR4 inhibitor. Following stimulation by 5 nM α-thrombin, platelet-granulocyte interactions were not significantly impaired by PAR4 or PAR1 inhibitors. When stimulated by cathepsin G, platelet-granulocyte interactions did not increase above the baseline level; the presence of either PAR4 or PAR1 inhibitors had no effect on platelet-granulocyte interactions in the presence of cathepsin G. Meanwhile, platelet-granulocyte interactions stimulated by plasmin were blocked by VU0652925 but not VU0661245 or SCH 79797. Finally, PAR4 or PAR1 inhibitors did not impair platelet-granulocyte interactions induced by the GPVI agonist CRP. These results were recapitulated for the activation state of platelets bound to granulocytes, as quantified by P-selectin (CD62P) surface exposure in the granulocyte gate (Figure 2D), except for in the cathepsin G stimulated condition, in which VU0652925 impaired activated platelet-granulocyte interactions. Finally, platelet activation via α-granule release was measured as P-selectin (CD62P) exposure in the platelet gate (Figure 2E). P-selectin exposure following stimulation by PAR4 AP was fully blocked by PAR4 inhibitors, while SCH 79797 had no effect. Meanwhile, upon stimulation with TRAP-6, P-selectin surface exposure was blocked by SCH 79797 but not PAR4 inhibitors. P-selectin exposure induced by human α-thrombin was slightly impaired by VU0652925 but not by SCH 79797. Stimulation of platelet P-selectin exposure by cathepsin G was blocked by VU0652925 but not VU0661245 or the PAR1 inhibitor. Meanwhile, stimulation of platelet P-selectin exposure by plasmin was fully blocked by VU0652925 and partially blocked by VU0661245, while SCH 79797 had no effect. Neither PAR4 inhibitor nor SCH 79797 impaired P-selectin exposure stimulated by the GPVI agonist CRP.
Figure 2.
PAR4 inhibitors diminish platelet-granulocyte interactions and platelet alpha granule release. Washed human platelets (2 × 108/ml) and purified granulocytes (2 × 106/ml) from the same donor were combined and pretreated with the PAR1 inhibitor SCH 79797 (3 μM), PAR4 inhibitors VU0652925 (10 μM) and VU0661245 (10 μM), or vehicle (0.2% DMSO), then stimulated with the agonists PAR4 AP (activating peptide, 200 μM), TRAP-6 (20 μM), human α-thrombin (5 nM = 0.7 U/ml), human cathepsin G (340 nM = 0.1 U/ml), human plasmin (260 nM = 0.3 U/ml), or CRP (10 μg/ml) and stained with markers for granulocytes (CD66b), platelets (CD41), and activated platelets (CD62P). (A) All samples were gated for platelets by CD41+ and granulocytes by CD66b+. (B) Granulocytes (CD66b+) were plotted against CD41, and a line was drawn to designate the CD41+ region. Platelets (CD41+) are shown for comparison. Bar graphs designate (C) percent of CD41+ granulocytes and (D) percent of activated (CD62P+) platelets for each treatment. Basal = no agonist; n ≥ 3 independent experiments; * designates p < 0.05 versus vehicle.
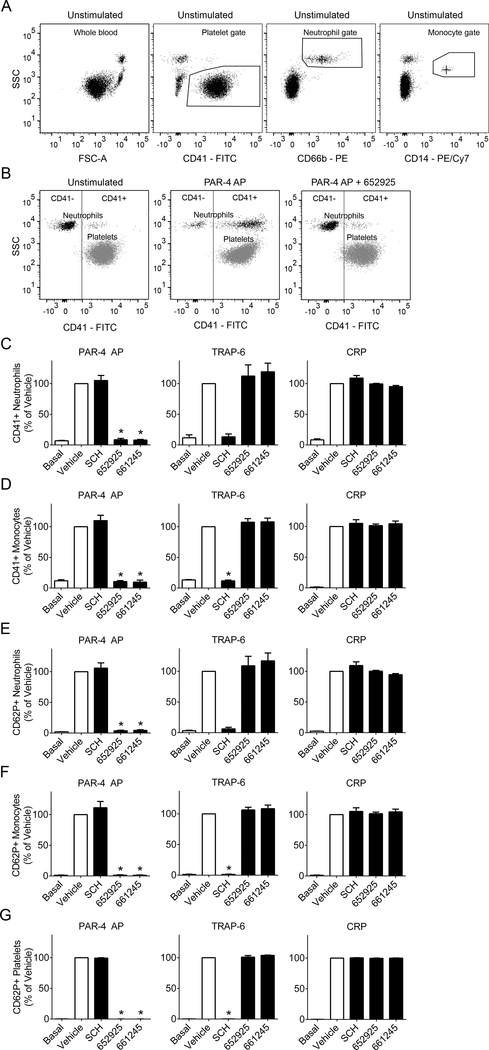
PAR4 activity promotes platelet-leukocyte interactions in whole human blood
To investigate platelet-leukocyte interactions in the context of whole blood, we developed and utilized a whole blood flow cytometry assay (Figure 3) using blood drawn directly into the protease inhibitor PPACK to prevent fibrin formation while allowing platelet activation and platelet-leukocyte interactions to occur. The addition of PPACK precluded the use of thrombin, cathepsin G, and plasmin as agonists; instead, the agonists PAR4 AP, TRAP-6, and CRP were utilized to validate the selectivity of the inhibitors in whole blood. Platelet-granulocyte or platelet-monocyte interactions were measured as expression of the platelet marker CD41 in the granulocyte or monocyte gate (Figure 3C and D, respectively). Upon stimulation with PAR4 AP (AYPGKF-NH2), PAR4 inhibitors blocked interactions between platelets and granulocytes and platelets and monocytes, while no inhibition was seen with SCH 79797. Conversely, stimulation of platelet-granulocyte or platelet-monocyte interactions with TRAP-6 was blocked with SCH but not PAR4 inhibitors. Neither the PAR4 inhibitors nor SCH 79797 blocked platelet-granulocyte or platelet-monocyte interactions induced by the platelet GPVI agonist CRP. These results were recapitulated for the activation state of platelets bound to granulocytes or monocytes, measured as P-selectin (CD62P) surface exposure in the granulocyte or monocyte gate (Figure 3E and F, respectively). Lastly, platelet activation was measured as expression of P-selectin (CD62P) in the platelet gate (Figure 3G). For platelet P-selectin exposure, stimulation with PAR4 AP was also blocked by PAR4 inhibitors but not SCH 79797. TRAP-6-induced P-selectin exposure on platelets was blocked by SCH 79797 but not PAR4 inhibitors. CRP-stimulated P-selectin exposure on platelets was not impaired by either PAR4 inhibitor or the PAR1 inhibitor SCH 79797. These results demonstrate that both platelet-granulocyte interactions and platelet-monocyte interactions are stimulated by PAR4 activity and can be specifically blocked by PAR4 inhibition in a whole blood setting.
Figure 3.
PAR4 inhibitors diminish platelet-leukocyte interactions and platelet alpha granule release in whole human blood. Whole human blood was pretreated with the PAR1 inhibitor SCH 79797 (3 μM), PAR4 inhibitors VU0652925 (10 μM) and VU0661245 (10 μM), or vehicle (0.2% DMSO), then stimulated with the agonists PAR4 AP (activating peptide, 200 μM), TRAP-6 (20 μM), or CRP (10 μg/ml) and stained with markers for granulocytes (CD66b), platelets (CD41), activated platelets (CD62P), and monocytes (CD14). (A) All samples were gated for platelets by CD41+, granulocytes by CD66b+, and monocytes by CD14+. (B) Granulocytes (CD66b+) or monocytes (CD14+, not shown) were plotted against CD41, and a line was drawn to designate the CD41+ region. Platelets (CD41+) are shown for comparison. Bar graphs designate (C) percent of CD41+ granulocytes, (D) percent of CD41+ monocytes, and (E) percent of activated (CD62P+) platelets for each treatment. Basal = no agonist; n ≥ 3 independent experiments; * designates p < 0.05 versus vehicle.
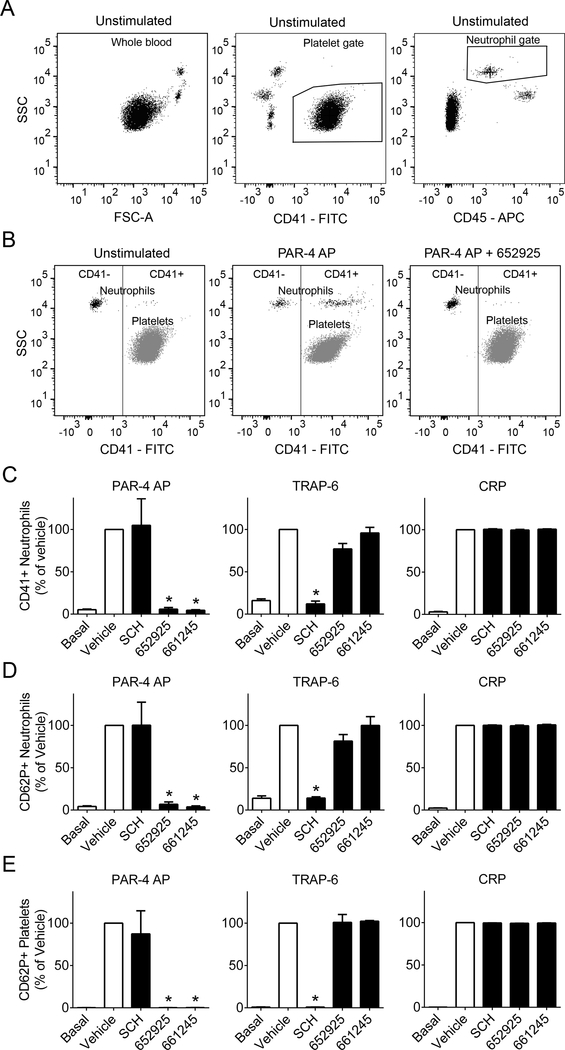
PAR4 activity promotes platelet-leukocyte interactions in whole baboon blood
Human and nonhuman primate platelets express PAR1 and PAR4, while murine platelets express PAR-3 and PAR4 and demonstrate other differences in PAR function, making non-human primates a preferred model for in vivo studies of PAR4 function for translational studies evaluating safety versus efficacy.30,31 In order to validate our whole blood flow cytometry approach in nonhuman primates before further studies, we performed flow cytometry analysis of whole baboon blood ex vivo (Figure 4). Platelet-granulocyte interactions were measured as expression of the platelet marker CD41 in the granulocyte gate (Figure 4C). In alignment with our results in human blood, PAR4 inhibitors blocked interactions between baboon platelets and granulocytes stimulated by PAR4 AP (AYPGKF-NH2), while SCH 79797 had no effect. Platelet-granulocyte interactions stimulated by TRAP-6 were blocked by SCH 79797 but not PAR4 inhibitors. Neither inhibitor blocked platelet-granulocyte interactions stimulated by CRP. These results were recapitulated for the activation state of platelets bound to granulocytes, measured as platelet P-selectin (CD62P) exposure in the granulocyte gate (Figure 4D). Finally, platelet activation was measured as P-selectin (CD62P) exposure in the platelet gate (Figure 4E). For platelet P-selectin exposure, PAR4 AP stimulation was blocked by PAR4 inhibitors but not SCH 79797. TRAP-6-stimulated P-selectin exposure was blocked by SCH 79797 but not PAR4 inhibitors. Neither PAR4 inhibitor nor the PAR1 inhibitor blocked P-selectin exposure stimulated by CRP. These results demonstrate efficacy of these inhibitors and feasibility of future studies in a nonhuman primate model.
Figure 4.
PAR4 inhibitors diminish platelet-leukocyte interactions and platelet alpha granule release in whole baboon (Papio anubis) blood. Whole baboon blood was pretreated with the PAR1 inhibitor SCH 79797 (3 μM), PAR4 inhibitors VU0652925 (10 μM) and VU0661245 (10 μM), or vehicle (0.2% DMSO), then stimulated with the agonists PAR4 AP (activating peptide, 200 μM), TRAP-6 (1 mM), or CRP (10 μg/ml) and stained with markers for leukocytes (CD45), platelets (CD41), and activated platelets (CD62P). (A) All samples were gated for platelets by CD41+ and granulocytes by CD45+ and high side scatter (SSC, granularity). (B) Granulocytes (CD45+, high SSC) were plotted against CD41, and a line was drawn to designate the CD41+ region. Platelets (CD41+) are shown for comparison. Bar graphs designate (C) percent of CD41+ granulocytes and (D) percent of activated (CD62P+) platelets for each treatment. Basal = no agonist; n ≥ 3–4 independent experiments; * designates p < 0.05 versus vehicle.
Discussion
Release of dense granule contents from activated platelets facilitates recruitment of leukocytes to the site of injury; this process is often dysregulated in inflammatory disease.9,10 Given the described role of platelet PAR4 in supporting the phosphorylation and activation of a number of PKC substrates requisite for platelet dense granule release,7 we examined whether PAR4 plays a disproportionate role relative to PAR1 in dense granule release. Intriguingly, PAR4 inhibitors diminished dense granule release in response to α-thrombin, while the PAR1 inhibitor SCH 79797 on its own had no effect. Furthermore, upon stimulation with the inflammatory mediators cathepsin G or plasmin, PAR4 inhibitors fully blocked dense granule release, while SCH had no effect either alone or in combination with a PAR4 inhibitor. Given that cathepsin G and plasmin are proteases known to specifically cleave PAR413,14 and are released during inflammation mediated by leukocytes or during fibrinolysis,32,33 this suggests a unique and important role for PAR4 in facilitating platelet dense granule release under inflammatory conditions.
Our results also suggest that platelet PAR4 may perform a unique role in the steps of thrombus formation. Other studies have shown that inhibition of PAR1 with a vorapaxar analog was fully rescued with 10 nM but not 2 nM thrombin, suggesting PAR4 can act without PAR1 only at thrombin levels above 2 nM,21 which we also observed in our assay of dense granule release and our flow cytometry assay of platelet P-selectin using 5 nM thrombin. Higher concentrations of thrombin are likely present in the inner core of a thrombus,34 supporting the physiological relevance of PAR4 in promoting thrombus formation. Furthermore, PAR4 may play a more prominent role than PAR1 in the release of platelet microparticles and preactivated factor V from platelet α-granules, both events representing a procoagulant phenotype.7 Platelet stimulation with PAR4 AP also supports thrombin generation in plasma up to 5 minutes faster than stimulation with TRAP-6.7 These prior observations combined with our findings that PAR4 mediated dense granule release by 5 nM thrombin suggest that PAR4 plays a distinct role apart from PAR1 to promote thrombus formation.
Activated platelets express surface P-selectin, which binds to the PSGL-1 receptor on circulating leukocytes such as neutrophils and monocytes; importantly, these interactions are upregulated in cardiovascular disease, inflammation, and sepsis, suggesting platelet-leukocyte interactions may be a potential disease biomarker.35–40 Given our observation that platelet PAR4 plays a specific role in dense granule release via neutrophil cathepsin G, we hypothesized that PAR4 activity also plays a role in platelet-leukocyte interactions. We first investigated the role of PAR4 in facilitating platelet-granulocyte interactions in a flow cytometry assay of isolated platelets and granulocytes, which are largely composed of neutrophils. Our results demonstrate that the PAR4 inhibitor VU0652925 impaired platelet α-granule release (P-selectin exposure) following stimulation with α-thrombin, cathepsin G, or plasmin, while the PAR1 inhibitor SCH 79797 had no effect. Furthermore, platelet-granulocyte interactions stimulated by plasmin were completely blocked by the PAR4 inhibitor VU0652925, suggesting a PAR4 specific effect. Surprisingly, addition of cathepsin G did not stimulate platelet-granulocyte interactions, as the basal (no agonist) condition was similar to the vehicle (with agonist) condition. This could be due to a saturation effect in part from cathepsin G released by neutrophils in the platelet-granulocyte mixture. Meanwhile, unlike the strong inhibition seen with VU0652925, the second PAR4 inhibitor VU0661245 was unable to block platelet-granulocyte interactions by plasmin and platelet activation by thrombin or cathepsin G. Our data show that the IC50 values for activation by 100 nM γ-thrombin are very different for the two PAR4 inhibitors: 229 pM for VU0652925 and 8.42 nM for VU0661245 (Supplemental Figure S2). Thus, we believe that VU0661245 has a lower affinity for PAR4, possibly due to its smaller size (Supplemental Figure S1), and therefore it is more easily outcompeted by the tethered ligand than VU0652925, despite being effective against PAR4 AP.
Next, we expanded our investigation to a whole blood flow cytometry assay in order to validate the potency and specificity of the PAR4 inhibitors in whole blood. In this assay, we examined platelet P-selectin along with platelet-granulocyte and platelet-monocyte interactions, the two most prevalent platelet-leukocyte interactions and potential markers of inflammation. Both PAR4 inhibitors impaired platelet P-selectin, platelet-granulocyte interactions, and platelet-monocyte interactions upon stimulation with PAR4 AP but not TRAP-6 or CRP, demonstrating validation of these PAR4 inhibitors in whole blood. This inhibition of platelet P-selectin and platelet-granulocyte interactions was recapitulated in a flow cytometry assay with whole baboon blood ex vivo, demonstrating feasibility for future in vivo studies with PAR4 inhibitors. Given the differences between VU0652925 and VU0661245 seen in our assays with purified cells, future work will focus on exploring the differential effects of these PAR4 inhibitors in vivo. Overall, these results demonstrate that in platelet-granulocyte preparations or in whole blood, inhibition of PAR4 impairs platelet α-granule release and platelet-leukocyte interactions. This suggests that PAR4 may be a new druggable target against platelet-leukocyte interactions to reduce inflammation, including thrombo-inflammation.
This novel finding that PAR4 activity promotes platelet-leukocyte interactions extends the role of platelet PAR4 beyond thrombosis to interactions between platelets and immune cells that are activated in inflammatory disease states. The pathological role of platelet-leukocyte interactions was highlighted years ago when clinical trials of the platelet GPIIb/IIIa inhibitor eptifibatide was halted due to increased cardiac events; studies determined that despite inhibiting platelet aggregation, the drug also triggered platelet-neutrophil and platelet-monocyte interactions.41,42 More recently, race-specific PAR4 polymorphisms have been identified that confer a pro-atherothrombotic phenotype, pointing to potential use of PAR4 inhibitors for targeting inflammation in specific populations. While donor demographics were not taken into account as a variable in the current study, the effect of race-related PAR4 variants on the pharmacodynamics of PAR4 inhibitors is an important area of future investigation.43 Meanwhile, the literature supports roles for platelet PAR4 in platelet-leukocyte interactions and inflammation, though many studies have been limited by the use of murine models. Some studies have shown roles for PAR4 activity in leukocyte recruitment in animal models of inflammation and pain but do not define the role of platelet PAR4 in these conditions.18 Others have shown that PAR4 activity by cathepsin G plays a role in the inflammatory bowel disease ulcerative colitis, and either a PAR4 inhibitor or a cathepsin G inhibitor were beneficial in a mouse model of the disease.44,45 In addition to cathepsin G, high levels of plasmin can also activate platelets through PAR4,13 and elevated plasmin may be partly responsible for the cytokine storm in the deadly macrophage activation syndrome,46 raising questions about the role of PAR4 in inflammation that involves plasmin activity. Our results show for the first time that inhibition of PAR4 but not PAR1 with small molecule antagonists impairs cathepsin G- and plasmin-induced human platelet granule release and plasmin-induced platelet-granulocyte interactions. This study provides a new rationale for the use of PAR4 activity inhibitors as therapeutic agents in disease conditions that are driven by platelet-leukocyte interactions.
Conclusions
This study points to an expanded role for the platelet receptor PAR4 in facilitating platelet granule release and platelet-leukocyte interactions. We demonstrate that PAR4 activity via PAR4 AP, thrombin, cathepsin G and plasmin can be targeted with PAR4 inhibitors to block platelet granule release and platelet-leukocyte interactions. These results suggest broader roles for platelet PAR4 than previously presumed and open up new possibilities for PAR4 as a therapeutic target for inhibition in diseases where platelet-leukocyte interactions play a pathogenic role.
Supplementary Material
Acknowledgements
The authors thank Jennifer Johnson for technical assistance with the baboon studies. This work was supported by grants from the National Institutes of Health (R01HL101972 and R01GM116184 to O.J.T.M., R01HL133923 to H.E.H., R21HD16037 to T.K.M., and T32AI007472 to L.D.H.) and the American Heart Association (13EIA12630000 to O.J.T.M. and 17SDG33350075 to J.E.A.). R.A.R. is a Whitaker International Fellow. T.T. Chu was supported in part by the Johns Hopkins University Department of Biomedical Engineering. This project was supported in part by funds from Oregon Health & Science University and Oregon State University as part of the OHSU/OSU Cancer Prevention and Control Initiative.
Footnotes
Disclosures
A. Gruber and OHSU have financial interest in Aronora, Inc., a company that may have a commercial interest in the result of this research. This potential conflict of interest has been reviewed and managed by the OHSU Conflict of Interest in Research Committee.
References
- 1.Nieman MT. Protease-activated receptors in hemostasis. Blood 2016;128(2):169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vu TK, Wheaton VI, Hung DT, Charo I, Coughlin SR. Domains specifying thrombin-receptor interaction. Nature 1991;353(6345):674–7. [DOI] [PubMed] [Google Scholar]
- 3.Nieman MT, Schmaier AH. Interaction of thrombin with PAR1 and PAR4 at the thrombin cleavage site. Biochemistry 2007;46(29):8603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arora P, Ricks TK, Trejo J. Protease-activated receptor signalling, endocytic sorting and dysregulation in cancer. J Cell Sci 2007;120(Pt 6):921–8. [DOI] [PubMed] [Google Scholar]
- 5.Smith TH, Coronel LJ, Li JG, Dores MR, Nieman MT, Trejo J. Protease-activated Receptor-4 Signaling and Trafficking Is Regulated by the Clathrin Adaptor Protein Complex-2 Independent of beta-Arrestins. J Biol Chem 2016;291(35):18453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covic L, Singh C, Smith H, Kuliopulos A. Role of the PAR4 thrombin receptor in stabilizing platelet-platelet aggregates as revealed by a patient with Hermansky-Pudlak syndrome. Thromb Haemost 2002;87(4):722–7. [PubMed] [Google Scholar]
- 7.Duvernay M, Young S, Gailani D, Schoenecker J, Hamm HE. Protease-activated receptor (PAR) 1 and PAR4 differentially regulate factor V expression from human platelets. Mol Pharmacol 2013;83(4):781–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golebiewska EM, Poole AW. Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev 2015;29(3):153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King SM, McNamee RA, Houng AK, Patel R, Brands M, Reed GL. Platelet dense-granule secretion plays a critical role in thrombosis and subsequent vascular remodeling in atherosclerotic mice. Circulation 2009;120(9):785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manne BK, Xiang SC, Rondina MT. Platelet secretion in inflammatory and infectious diseases. Platelets 2017;28(2):155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao P, Metcalf M, Bunnett NW. Biased signaling of protease-activated receptors. Front Endocrinol (Lausanne) 2014;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu WF, Andersen H, Whitmore TE, Presnell SR, Yee DP, Ching A, Gilbert T, Davie EW, Foster DC. Cloning and characterization of human protease-activated receptor 4. Proc Natl Acad Sci U S A 1998;95(12):6642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinton TM, Kim S, Derian CK, Jin J, Kunapuli SP. Plasmin-mediated activation of platelets occurs by cleavage of protease-activated receptor 4. J Biol Chem 2004;279(18):18434–9. [DOI] [PubMed] [Google Scholar]
- 14.Sambrano GR, Huang W, Faruqi T, Mahrus S, Craik C, Coughlin SR. Cathepsin G activates protease-activated receptor-4 in human platelets. J Biol Chem 2000;275(10):6819–23. [DOI] [PubMed] [Google Scholar]
- 15.Oikonomopoulou K, Hansen KK, Saifeddine M, Tea I, Blaber M, Blaber SI, Scarisbrick I, Andrade-Gordon P, Cottrell GS, Bunnett NW and others. Proteinase-activated receptors, targets for kallikrein signaling. J Biol Chem 2006;281(43):32095–112. [DOI] [PubMed] [Google Scholar]
- 16.Totani L, Evangelista V. Platelet-leukocyte interactions in cardiovascular disease and beyond. Arterioscler Thromb Vasc Biol 2010;30(12):2357–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smyth SS, McEver RP, Weyrich AS, Morrell CN, Hoffman MR, Arepally GM, French PA, Dauerman HL, Becker RC. Platelet functions beyond hemostasis. J Thromb Haemost 2009;7(11):1759–66. [DOI] [PubMed] [Google Scholar]
- 18.Vergnolle N Protease-activated receptors as drug targets in inflammation and pain. Pharmacol Ther 2009;123(3):292–309. [DOI] [PubMed] [Google Scholar]
- 19.Wong PC, Seiffert D, Bird JE, Watson CA, Bostwick JS, Giancarli M, Allegretto N, Hua J, Harden D, Guay J and others. Blockade of protease-activated receptor-4 (PAR4) provides robust antithrombotic activity with low bleeding. Sci Transl Med 2017;9(371). [DOI] [PubMed] [Google Scholar]
- 20.Safety and Efficacy Study of a Protease Activated Receptor-4 Antagonist Being Tested to Reduce the Chances of Having Additional Strokes or “Mini Strokes”. 2017. Retrieved from http://clinicaltrials.gov (Identification No. NCT02671461).
- 21.Duvernay MT, Temple KJ, Maeng JG, Blobaum AL, Stauffer SR, Lindsley CW, Hamm HE. Contributions of Protease-Activated Receptors PAR1 and PAR4 to Thrombin-Induced GPIIbIIIa Activation in Human Platelets. Mol Pharmacol 2017;91(1):39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitrugno A, Williams D, Kerrigan SW, Moran N. A novel and essential role for FcgammaRIIa in cancer cell-induced platelet activation. Blood 2014;123(2):249–60. [DOI] [PubMed] [Google Scholar]
- 23.Healy LD, Puy C, Fernandez JA, Mitrugno A, Keshari RS, Taku NA, Chu TT, Xu X, Gruber A, Lupu F and others. Activated protein C inhibits neutrophil extracellular trap formation in vitro and activation in vivo. J Biol Chem 2017;292(21):8616–8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mauler M, Seyfert J, Haenel D, Seeba H, Guenther J, Stallmann D, Schoenichen C, Hilgendorf I, Bode C, Ahrens I and others. Platelet-neutrophil complex formation-a detailed in vitro analysis of murine and human blood samples. J Leukoc Biol 2016;99(5):781–9. [DOI] [PubMed] [Google Scholar]
- 25.Zilberman-Rudenko J, Itakura A, Wiesenekker CP, Vetter R, Maas C, Gailani D, Tucker EI, Gruber A, Gerdes C, McCarty OJ. Coagulation Factor XI Promotes Distal Platelet Activation and Single Platelet Consumption in the Bloodstream Under Shear Flow. Arterioscler Thromb Vasc Biol 2016;36(3):510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantopoulos K, Neelamegham S, Burns AR, Hentzen E, Kansas GS, Snapp KR, Berg EL, Hellums JD, Smith CW, McIntire LV and others. Venous levels of shear support neutrophil-platelet adhesion and neutrophil aggregation in blood via P-selectin and beta2-integrin. Circulation 1998;98(9):873–82. [DOI] [PubMed] [Google Scholar]
- 27.Rondina MT, Weyrich AS, Zimmerman GA. Platelets as cellular effectors of inflammation in vascular diseases. Circ Res 2013;112(11):1506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrell CN, Aggrey AA, Chapman LM, Modjeski KL. Emerging roles for platelets as immune and inflammatory cells. Blood 2014;123(18):2759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin J, Mao Y, Thomas D, Kim S, Daniel JL, Kunapuli SP. RhoA downstream of G(q) and G(12/13) pathways regulates protease-activated receptor-mediated dense granule release in platelets. Biochem Pharmacol 2009;77(5):835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahn ML, Zheng YW, Huang W, Bigornia V, Zeng D, Moff S, Farese RV Jr.,Tam C, Coughlin SR. A dual thrombin receptor system for platelet activation. Nature 1998;394(6694):690–4. [DOI] [PubMed] [Google Scholar]
- 31.Hamilton JR, Cornelissen I, Coughlin SR. Impaired hemostasis and protection against thrombosis in protease-activated receptor 4-deficient mice is due to lack of thrombin signaling in platelets. J Thromb Haemost 2004;2(8):1429–35. [DOI] [PubMed] [Google Scholar]
- 32.Foley JH. Plasmin(ogen) at the Nexus of Fibrinolysis, Inflammation, and Complement. Semin Thromb Hemost 2017;43(2):135–142. [DOI] [PubMed] [Google Scholar]
- 33.Kessenbrock K, Dau T, Jenne DE. Tailor-made inflammation: how neutrophil serine proteases modulate the inflammatory response. J Mol Med (Berl) 2011;89(1):23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diamond SL. Systems Analysis of Thrombus Formation. Circ Res 2016;118(9):1348–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haselmayer P, Grosse-Hovest L, von Landenberg P, Schild H, Radsak MP. TREM-1 ligand expression on platelets enhances neutrophil activation. Blood 2007;110(3):1029–35. [DOI] [PubMed] [Google Scholar]
- 36.Barnard MR, Linden MD, Frelinger AL 3rd, Li Y, Fox ML, Furman MI, Michelson AD Effects of platelet binding on whole blood flow cytometry assays of monocyte and neutrophil procoagulant activity. J Thromb Haemost 2005;3(11):2563–70. [DOI] [PubMed] [Google Scholar]
- 37.Sarma J, Laan CA, Alam S, Jha A, Fox KA, Dransfield I. Increased platelet binding to circulating monocytes in acute coronary syndromes. Circulation 2002;105(18):2166–71. [DOI] [PubMed] [Google Scholar]
- 38.Zarbock A, Singbartl K, Ley K. Complete reversal of acid-induced acute lung injury by blocking of platelet-neutrophil aggregation. J Clin Invest 2006;116(12):3211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornerup KN, Salmon GP, Pitchford SC, Liu WL, Page CP. Circulating platelet-neutrophil complexes are important for subsequent neutrophil activation and migration. J Appl Physiol (1985) 2010;109(3):758–67. [DOI] [PubMed] [Google Scholar]
- 40.Nijm J, Wikby A, Tompa A, Olsson AG, Jonasson L. Circulating levels of proinflammatory cytokines and neutrophil-platelet aggregates in patients with coronary artery disease. Am J Cardiol 2005;95(4):452–6. [DOI] [PubMed] [Google Scholar]
- 41.Scholz T, Zhao L, Temmler U, Bath P, Heptinstall S, Losche W. The GPIIb/IIIa antagonist eptifibatide markedly potentiates platelet-leukocyte interaction and tissue factor expression following platelet activation in whole blood in vitro. Platelets 2002;13(7):401–6. [DOI] [PubMed] [Google Scholar]
- 42.Newby LK, Califf RM, White HD, Harrington RA, Van de Werf F, Granger CB, Simes RJ, Hasselblad V, Armstrong PW. The failure of orally administered glycoprotein IIb/IIIa inhibitors to prevent recurrent cardiac events. Am J Med 2002;112(8):647–58. [DOI] [PubMed] [Google Scholar]
- 43.Edelstein LC, Simon LM, Montoya RT, Holinstat M, Chen ES, Bergeron A, Kong X, Nagalla S, Mohandas N, Cohen DE and others. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat Med 2013;19(12):1609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dabek M, Ferrier L, Roka R, Gecse K, Annahazi A, Moreau J, Escourrou J, Cartier C, Chaumaz G, Leveque M and others. Luminal cathepsin g and protease-activated receptor 4: a duet involved in alterations of the colonic epithelial barrier in ulcerative colitis. Am J Pathol 2009;175(1):207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dabek M, Ferrier L, Annahazi A, Bezirard V, Polizzi A, Cartier C, Leveque M, Roka R, Wittmann T, Theodorouand others. Intracolonic infusion of fecal supernatants from ulcerative colitis patients triggers altered permeability and inflammation in mice: role of cathepsin G and protease-activated receptor-4. Inflamm Bowel Dis 2011;17(6):1409–14. [DOI] [PubMed] [Google Scholar]
- 46.Shimazu H, Munakata S, Tashiro Y, Salama Y, Dhahri D, Eiamboonsert S, Ota Y, Onoda H, Tsuda Y, Okada Y and others. Pharmacological targeting of plasmin prevents lethality in a murine model of macrophage activation syndrome. Blood 2017;130(1):59–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.