Correction to: Nature Communications; 10.1038/ncomms16065; published online 19 July 2017
This Article contains errors in Fig. 1, Table 1 and the Methods section. In panel c, the labels for PmScsC and EcDsbC in the upper two curves are interchanged. In Table 1 and the Methods section entitled ‘Extended structure’, the space group of the extended PmScsC structure is incorrectly referred to as H32 and should read H32. Correct versions of Fig. 1 and Table 1 are presented below; the errors have not been corrected in the Article.
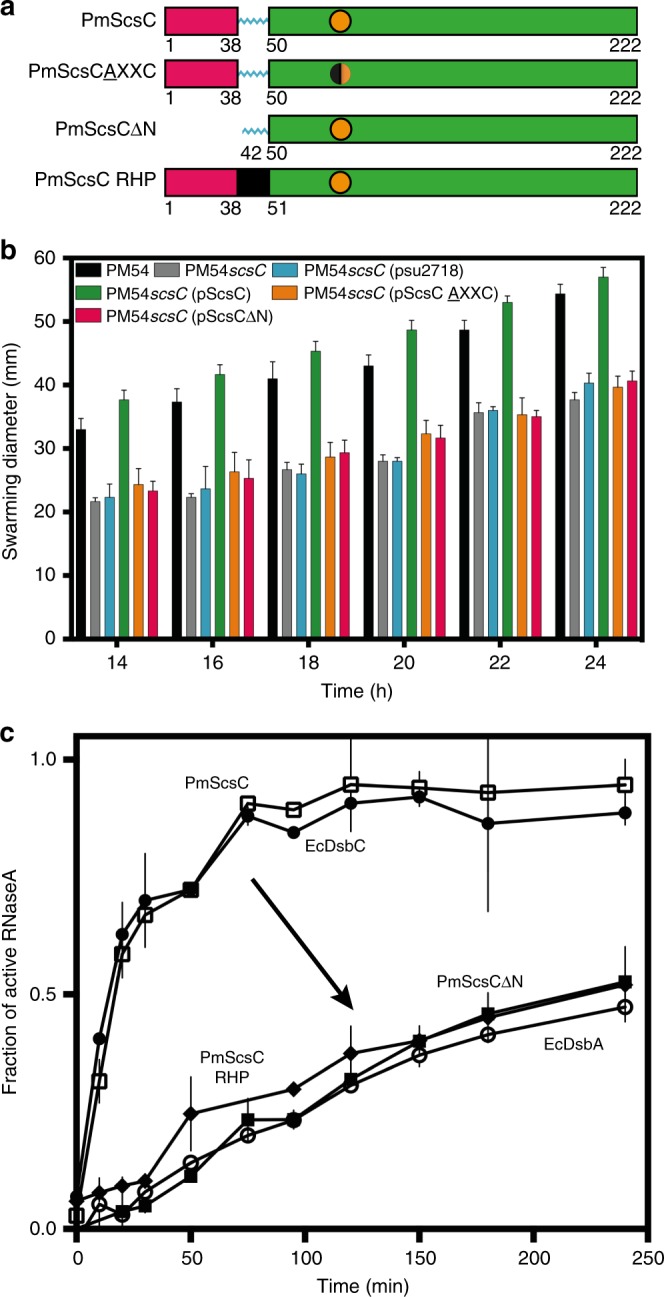
Fig. 1
Table 1.
PmScsC crystal structure statistics
| Compact (4XVW) | Transitional (5IDR) | Extended (5ID4) | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | I4 | H32 |
| Cell dimensions | |||
| a, b, c (Å) | 137.5, 163.9, 181.9 | 193.1, 193.1, 105.8 | 86.7, 86.7, 330.9 |
| α, β, γ (º) | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 91.15–2.60 (2.74–2.60) | 136.51–2.56 (2.57–2.56) | 110.29–2.92 (2.93–2.92) |
| Rmerge | 0.072 (0.617) | 0.083 (0.741) | 0.059 (0.625) |
| I /σI | 11.0 (2.0) | 14.9 (2.2) | 14.2 (2.8) |
| Completeness (%) | 98.6 (95.4) | 99.4 (100.0) | 99.2 (100.0) |
| Redundancy | 3.8 (3.7) | 4.1 (4.1) | 4.1 (4.2) |
| Refinement | |||
| Resolution (Å) | 91.15–2.60 | 42.82–2.56 | 40.36–2.92 |
| No. of reflections | 243,409 | 62,069 | 10,652 |
| Rwork/Rfree (%) | 24.8/28.2 | 17.1/22.2 | 25.1/26.3 |
| No. of atoms | |||
| Protein | 40,850 | 10,262 | 1720 |
| Ligand/ion | NA | NA | NA |
| Water | 281 | 82 | 0 |
| B factors (Å2) | |||
| Protein | 59.7 | 50.6 | 122.2 |
| Ligand/ion | NA | NA | NA |
| Water | 41.5 | 43.0 | NA |
| RMS deviations | |||
| Bond length (Å) | 0.006 | 0.008 | 0.010 |
| Bond angles (º) | 1.21 | 1.05 | 1.17 |
Single crystals were used to collect each dataset. Values for highest resolution shell are shown in parentheses
Contributor Information
Andrew E. Whitten, Email: awh@ansto.gov.au
Mark A. Schembri, Email: m.schembri@uq.edu.au
Jennifer L. Martin, Email: jlm@griffith.edu.au


