Abstract
Lung cancer is the deadliest cancer worldwide. The mutational frequency of EGFR and KRAS genes in lung adenocarcinoma varies worldwide per ethnicity and smoking. The impact of EGFR and KRAS mutations in Brazilian lung cancer remains poorly explored. Thus, we investigated the frequency of EGFR and KRAS mutations in a large Brazilian series of lung adenocarcinoma together with patients’ genetic ancestry, clinicopathological and sociodemographic characteristics. The mutational frequency of EGFR was 22.7% and KRAS was 20.4%. The average ancestry proportions were 73.1% for EUR, 13.1% for AFR, 6.5% for AME and 7.3% for ASN. EGFR mutations were independently associated with never-smokers, high-Asian ancestry, and better performance status. KRAS mutations were independently associated with tobacco exposure and non-Asian ancestry. EGFR-exon 20 mutations were associated with worse outcome. The Cox regression model indicated a worse outcome for patients whose were older at diagnosis (>61 y), solid histological subtype, loss of weight (>10%), worse performance status (≥2), and presence of KRAS mutations and EGFR mutational status in TKi non-treated patients. In conclusion, we assessed the clinicopathological and ethnic impact of EGFR and KRAS mutations in the largest series reported of Brazilian lung adenocarcinomas. These findings can support future clinical strategies for Brazilian lung cancer patients.
Introduction
Lung cancer is the deadliest cancer worldwide and in Brazil1–3. The 5-year survival rate for lung cancer patients is lower than 20%, possibly due to the lack of successful early detection and limited treatment options4. Over the past decade, the treatment of advanced/metastatic non–small cell lung cancer (NSCLC) has experienced significant modification mainly due to development of molecular testing to determine a druggable mutation and due to the addition of immune-based therapies5,6. Histologically, lung cancer is divided into non-small cell lung cancer (NSCLC), which corresponds about 85% of all lung cancer cases and, less commonly, small cell lung cancer (SCLC). The most common histologic subtype of NSCLC is adenocarcinoma7.
Epidermal growth factor receptor (EGFR) gene has a key role in the pathogenesis of several tumors8–10. Activating EGFR mutations are involved in the pathogenesis of a significant subset of lung adenocarcinomas11. The frequency of EGFR mutations in lung adenocarcinomas worldwide varies (from 8–13% in European populations to 27–60% Asian populations) according to ethnicity, gender, and tobacco exposure12,13. EGFR gene has emerged as a critical therapeutic target and EGFR mutations status has successfully guided clinical management14. The presence of activating EGFR mutations, mainly in exons 18, 19, 20 and 21, which correspond to the tyrosine kinase domain, sensitizes lung adenocarcinomas to treatment with anti-EGFR tyrosine kinase inhibitors (TKi), such as erlotinib, gefitinib, afatinib, dacomitinib and osimertinib15,16. However, basically, all TKi-treated lung adenocarcinoma patients will experience disease progression due to resistance mechanisms17,18. The most well-known resistance mechanism is the presence of EGFR p. T790M mutation and patients harboring this mutation are eligible for treatment with second and third generation of anti-EGFR TKi, such as osimeritinib19.
Another important oncogene in lung cancer is the KRAS (Kirsten rat sarcoma viral oncogene homolog), which codifies an EGFR downstream GTPase20. Hotspot KRAS mutations, at codons 12/13, are described in approximately 20% lung adenocarcinomas and are known to be associated with tobacco consumption21. Since KRAS and EGFR mutations are mutually exclusive events, molecular testing based on KRAS mutational status for treatment decisions was not currently recommended for anti-EGFR TKi22. However, combined strategies for targeting KRAS mutations can be promising23. Moreover, KRAS mutational status was described to be associated with the immune microenvironment implying that the molecular testing for this gene should be considered for stratifying patients aimed at immunotherapy24.
Taken together, the molecular testing of EGFR and KRAS mutations for lung cancer patients has offering advantages for guiding personalized therapy, providing a better patient selection and stratification for clinical management.
Reports of the prevalence of EGFR and KRAS mutations among Brazilian patients remain limited25,26. Moreover, it is also poorly explored the impact of EGFR and KRAS mutations in patients’ clinicopathological features. Therefore, the present study aims to investigate the frequency of EGFR and KRAS mutations in a large Brazilian series of lung adenocarcinoma, and to correlate the presence of these mutations with patients’ genetic ancestry and clinicopathological and sociodemographic characteristics.
Results
EGFR and KRAS mutational status
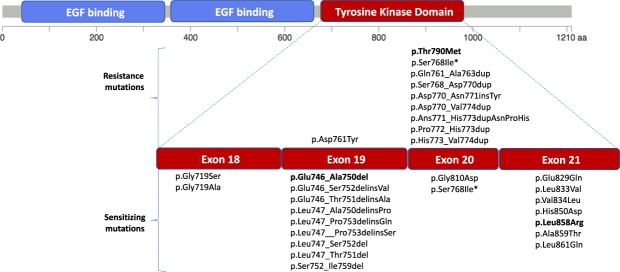
EGFR mutations were detected in 101 patients (22.7%) and mutations were distributed at tyrosine kinase domain predominantly located at exons 19 and 21 (Fig. 1 and Supplementary Fig. 1 and Supplementary Table 1). The most common EGFR mutation was the p. Leu858Arg (n = 32) followed by a substitution the p. Glu746_Ala750del (n = 31; Supplementary Fig. 1 and Supplementary Table 1). Additional EGFR sensibility mutations include mutations located at exons 18 (p. Gly719Ser and p. Gly719Ala), 19 (p.Glu746_Ser752delinsVal, p.Glu746_Thr751delinsAla, p.Leu747_Ala750delinsPro, p.Leu747_Pro753delinsGln, p.Leu747__Pro753delinsSer, p.Leu747_Ser752del, p.Leu747_Thr751del, p.Ser752_Ile759del), 20 (p. Gly810Asp) and 21 (p.Glu829Gln, p.Leu833Val, p.Val834Leu, p.His850Asp, p.Leu858Arg, p.Ala859Thr, p.Leu861Gln) (Fig. 1 and Supplementary Table 1).
Figure 1.
EGFR mutations and exon distribution. EGFR protein structure highlighting the tyrosine kinase domain (red), which is the hotspot region (red) for the EGFR mutations with predictive value. All resistance and sensitizing mutations detected in the present series of Brazilian lung adenocarcinoma are presented. *Controversial results in the literature.
Among EGFR positive patients, 12 presented mutations associated with drug resistance, namely p.Tyr790Met the most common one (n = 3; exon 20), followed by p.Gln761_Ala763dup (n = 2; exon 20), p.Asp761Tyr (n = 1; exon 19), p.Ser768_Asp770dup, p.Asp770_Asn771insTyr, p.Asp770_Val774dup, p.Ans771_His773dupAsnProHis, p.Pro772_His773dup and p.His773_Val774dup (n = 1/each; exon 20) (Supplementary Table 1 and Fig. 1). The p.Ser768Ile mutation has been described as both sensitivity and resistance mutation depends on the used drug (Fig. 1)27,28. Among the p. Tyr790Met-mutated cases, 2 of them were concomitant with p.Leu858Arg and p.Leu861Gln mutations.
Furthermore, we also tested a real time-PCR based commercial assay Cobas® EGFR Mutation Test (Roche), in the cases that exhibited uncommon mutations, namely, the p.Leu861Gln, p.Ans771_His773dupAsnProHis, p.Asp770_Val774dup, p.Pro772_His773dup, and p.His850Asp. The COBAS assay showed an absence of a mutation in those samples. Of note, the p.Leu861Gln was later included in the expanded version Cobas® EGFR Mutation v2 Test.
KRAS mutations (codons 12/13) were detected in 90 patients (20.4%) (Supplementary Fig. 1 and Supplementary Table 2). The most common KRAS mutation was p.Gly12Cys (n = 32), followed by p.Gly12Val (n = 21), p.Gly12Asp (n = 19), p.Gly13Cys (n = 6), p.Gly12Ala (n = 5), p.Gly12Ser (n = 2) and p.Gly12Phe, p.Gly13Asp, p.Gly13Glu, p.Gly13Ser and p.Gly13Val (n = 1/each) (Supplementary Table 2).
All EGFR and KRAS mutations were mutually exclusive.
Genetic ancestry component and its association with EGFR and KRAS mutations
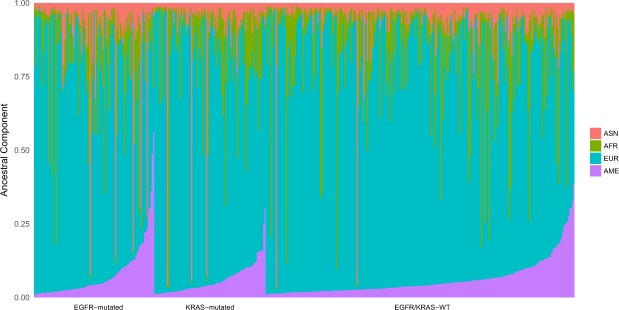
We further assessed the ancestry background by an AIM-INDEL panel that allowed to estimates the AFR, EUR, ASN and AME ancestral proportions in 427 out of 444 patients (Fig. 2). The average ancestry proportions for all individuals were 73.1% for EUR, 13.1% for AFR, 6.5% for AME and 7.3% for ASN (Fig. 2). Ancestry proportions were further categorically defined as low, intermediate and high based on terciles (Supplementary Table 3). Most of our patients were self-declared as white (Table 1) and likewise most of our cases presented high EUR background (Fig. 2).
Figure 2.
Individual ancestry proportion of Brazilian lung adenocarcinoma series (n = 427 out of 444). The pinkish-orange, green, blue and purple colors represent the Asian, African, European and Native American ancestry proportions, respectively.
Table 1.
Major clinicopathological features of NSCLC adenocarcinomas.
| Variables | Parameters | n |
|---|---|---|
| Agea, years | 61 (22–87) | 444 |
| Gender | Male | 232 |
| Female | 212 | |
| Self-reported raceb | White | 342 |
| Brown | 64 | |
| Black | 20 | |
| Yellow | 7 | |
| Smoking history | Never smoker | 135 |
| Current Smoker | 171 | |
| Former smoker | 131 | |
| Alcohol consuming | Never | 272 |
| Current | 114 | |
| Former | 35 | |
| Metastasis at diagnosis | No | 114 |
| One site | 129 | |
| Multiple sites | 200 | |
| Disease staging | I | 28 |
| II | 15 | |
| III | 71 | |
| IV | 329 | |
| PS ECOG | 0 | 46 |
| 1 | 233 | |
| 2 | 73 | |
| 3 or 4 | 88 | |
| Loss of weightc | No | 209 |
| <10% | 145 | |
| >10% | 69 | |
| Histologyd | Adenocarcinoma NOS | 120 |
| Acinar | 163 | |
| Solid | 112 | |
| Papillary | 45 | |
| Lepidic | 12 | |
| Mucinous | 7 |
n, number of patients; PS ECOG, performance status ECOG (Eastern Cooperative Oncology Group).aAverage age (range); bSelf-reported race according to Brazilian Institute of Geography and Statistics (IBGE). cLoss of weight <10% and >10% of total body weight. dAdenocarcinoma predominant subtypes; When histology subtype was not determined, the case was considered as only adenocarcinoma.
We then correlated the genetic ancestry with the molecular features. EGFR mutations were associated with high ASN (p = 0.03; Supplementary Table 4). In the multivariate analysis, the high ASN background was independently associated with the presence of EGFR mutations [OD = 2.01 (1.09–3.71); p = 0.03; Table 2]. On the other hand, in the multivariate analysis, the low ASN background was independently associated with the presence of KRAS mutations [OD = 1.93 (1.06–3.52); p = 0.03; Table 2].
Table 2.
Multivariate analysis of the association between clinicopathological characteristics and ancestry background and EGFR and KRAS mutations.
| Variables | Parameters | Total (n) | OR | 95% CI | p-value | |
|---|---|---|---|---|---|---|
| EGFR | Gender | Male | 224 | 1 | Ref. | Ref. |
| Female | 196 | 1.67 | 0.98–2.85 | 0.058 | ||
| Tobacco | Never | 126 | 5.11 | 2.71–9.62 | <0.0001 | |
| Current smoker | 166 | 0.59 | 0.28–1.24 | 0.16 | ||
| Former smoker | 128 | 1 | Ref. | Ref. | ||
| ASN Ancestry | Low | 140 | 1 | Ref. | Ref. | |
| Intermediate | 138 | 1.05 | 0.54–2.04 | 0.88 | ||
| High | 142 | 2.01 | 1.09–3.71 | 0.03 | ||
| PS ECOG | 0 | 44 | 3.94 | 1.47–10.57 | 0.006 | |
| 1 | 222 | 1.67 | 0.79–3.53 | 0.18 | ||
| 2 | 67 | 1.2 | 0.46–3.13 | 0.72 | ||
| 3 or 4 | 84 | 1 | Ref. | Ref | ||
| KRAS | Tobacco | Never | 126 | 1 | Ref. | Ref. |
| Current smoker | 166 | 3.42 | 1.67–7.00 | 0.001 | ||
| Former smoker | 128 | 3.74 | 1.79–7.81 | <0.0001 | ||
| ASN Ancestry | Low | 140 | 1.93 | 1.06–3.52 | 0.03 | |
| Intermediate | 138 | 1.31 | 0.69–2.46 | 0.41 | ||
| High | 142 | 1 | Ref. | Ref. |
n, number of patients; OR, odds ratio; 95% CI, 95% confidence interval; p-value: significance of Wald test.; ASN, Asian race; Ref., reference group; PS ECOG, performance status ECOG (Eastern Cooperative Oncology Group). Significant associations are indicated in bold.
The additional ethnic groups, AFR, EUR, and AME, were not associated with the presence of EGFR and KRAS mutations (Supplementary Table 4).
Association of patients’ clinicopathological features and molecular features
The presence of EGFR mutations was associated with female gender (p < 0.0001), absence of smoking habit (p < 0.0001), absence of alcohol consuming (p = 0.002); and better ECOG PS (p = 0.03) (Supplementary Table 4). Age, self-reported race, disease staging, metastasis at diagnosis, weight loss at diagnosis, and differentiation grade were not associated with the presence of EGFR mutations (Supplementary Table 4). EGFR-mutated patients harboring exon 20 mutations presented the lowest OS (exon 19 = 3.4 months; exon 20 = 0.5 months; exon 21 = 3.0 months; Fig. 3). In addition, EGFR-mutated patients harboring exon 20 mutations mostly presented with disease progression (exon 19 = 22.2%; exon 20 = 55.6%; exon 21 = 22.2%), only one patient harboring exon 20 mutation presented partial response to TKi (exon 19 = 85.3%; exon 20 = 2.9%; exon 21 = 11.8%) and none of the patients harboring exon 20 mutations presented with stable disease (exon 19 = 55.6%; exon 20 = 0%; exon 21 = 44.4%; p < 0.0001; Supplementary Table 5). Exon 18 was not included in the analysis because only one patient with disease stage IV carries EGFR exon 18 mutation.
Figure 3.
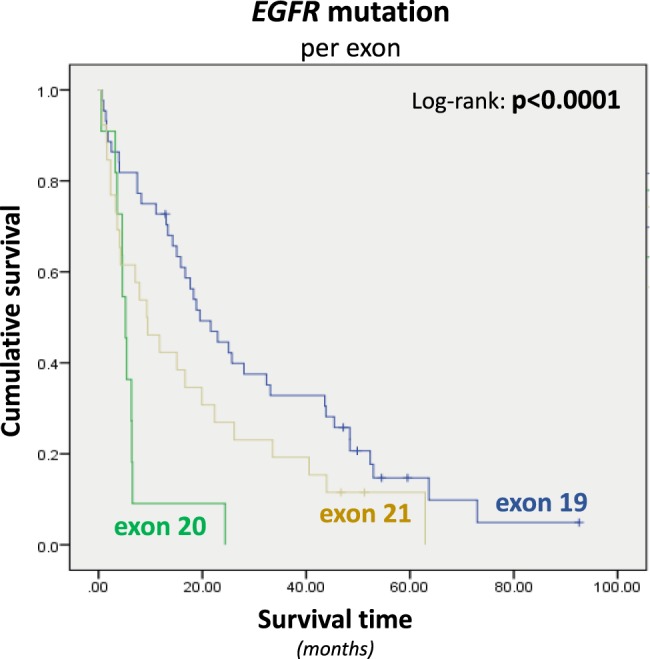
Kaplan-Meier curves for overall survival of lung adenocarcinoma patients (stage IV) according to EGFR mutations categorized by exon distribution (Median OS exon 19 = 19.5 months; Median OS exon 20 = 5.2 months; Median OS exon 21 = 9.3 months). Survival time is presented in months; p values are related to Log-rank test results. Exon 18 was not included in the analysis because only one patient with disease stage IV carries EGFR exon 18 mutation.
A multivariate analysis indicated the following independent variables were associated with the presence of EGFR mutations: absence of smoking habit (OR = 5.11; p < 0.0001; Table 2), high ASN (OR = 2.01; p = 0.03; Table 2) and better ECOG PS (OR = 3.94; p = 0.006; Table 2). Female gender was marginally associated with the presence of EGFR mutations (OR = 1.67; p = 0.058; Table 2).
Concerning KRAS status, the presence of mutations was associated with the presence of smoking habit (p < 0.0001). Age, self-reported race, disease staging, metastasis at diagnosis, weight loss at diagnosis, and differentiation grade were not associated with the presence of KRAS mutations (Supplementary Table 4). A multivariate analysis indicated the following independent variables as associated with the presence of KRAS mutations: tobacco exposure (current: OR = 3.42; p = 0.001/former: OR = 3.74; p < 0.0001; Table 2) and low Asian ancestry (OR = 1.93; p = 0.03; Table 2).
Since most patients were diagnosed at disease stage IV, and they exhibit a very distinct outcome from stage I, II and III (Supplementary Fig. 2), we only analyzed this group of patients in the multivariate analysis of disease outcome (Table 3). Unfavorable outcome was independently associated with age at diagnosis higher than 61 years old (OR = 1.45; p = 0.01), solid histological subtype (OR = 1.91; p < 0.0001), increased weight loss (OR = 1.72; p = 0.006), ECOG PS 2 and 3 or 4 (OR = 2.43 and OR = 6.28, respectively; p = 0.03 and p < 0.0001, respectively) (Table 3). Metastasis at diagnosis in the central nervous system was considered a risk factor for an unfavorable outcome (Table 3). Clinical outcome was not independently associated with self-reported race and alcohol consuming (Table 3).
Table 3.
Multivariate analysis of the association between clinicopathological characteristics and overall survival.
| Variables | Parameters | Total (n) | OR | 95% CI | p-value |
|---|---|---|---|---|---|
| Age* | ≤61 years | 126 | 1 | Ref. | Ref. |
| >61 years | 107 | 1.45 | 1.09–1.93 | 0.01 | |
| Histology | Acinar | 119 | 1 | Ref. | Ref. |
| Mucinous | 5 | 0.77 | 0.27–2.20 | 0.63 | |
| Lepidic | 8 | 1.01 | 0.43–2.39 | 0.98 | |
| Papillary | 27 | 1.22 | 0.76–1.97 | 0.41 | |
| Solid | 74 | 1.91 | 1.36–2.68 | <0.0001 | |
| Loss of weight** | No | 105 | 1 | Ref. | Ref. |
| <10% | 81 | 1.11 | 0.80–1.53 | 0.55 | |
| >10% | 47 | 1.72 | 1.17–2.54 | 0.006 | |
| PS ECOG | 0 | 13 | 1 | Ref. | Ref |
| 1 | 122 | 1.64 | 0.78–3.46 | 0.16 | |
| 2 | 42 | 2.43 | 1.07–5.51 | 0.03 | |
| 3 or 4 | 56 | 6.28 | 2.80–14.08 | <0.0001 | |
| Metastasis at diagnosis | CNS | 74 | 1 | Ref. | Ref. |
| Other sites | 159 | 0.62 | 0.45–0.85 | 0.004 | |
| TKi_EGFR | Yes_WT | 13 | 1 | Ref. | Ref. |
| Yes_Mutated | 48 | 0.91 | 0.45–1.84 | 0.926 | |
| No_WT | 152 | 1.75 | 0.92–3.31 | 0.052 | |
| No_Mutated | 20 | 3.79 | 1.73–8.33 | 0.001 | |
| KRAS mutations | WT | 194 | 1 | Ref. | Ref. |
| Mutated | 39 | 2.93 | 1.94–4.42 | <0.0001 |
*Only patients diagnosed at stage IV were included in this analysis. n, number of patients; OR, odds ratio; 95% CI, 95% confidence interval; p-value: significance of Cox Regression; Ref., reference group; *age was categorized into two groups considering the average age of the entire series as the cutoff; **Loss of weight <10% and >10% of total body weight; PS ECOG, performance status ECOG (Eastern Cooperative Oncology Group); CNS, central nervous system; TKi_EGFR, combination of two variables (TKi treatment and EGFR mutation). Significant associations are indicated in bold.
The Cox regression analysis indicated the presence of EGFR mutations in TKi non-treated patients were independently associated with unfavorable outcome (OR = 3.79; p = 0.001; Median OS TKi non-treated = 6.7 months; Median OS TKi-treated = 19.9 months) (Table 3 and Supplementary Fig. 2). Of note, among the EGFR-mutated patients that were not treated with TKi (n = 30), 15 received Best Support of Care (BSC), 9 received only chemotherapy, and 6 of them had localized disease and they were submitted to local treatment.
In addition, the Cox regression analysis also showed that the presence of KRAS mutations was independently associated with unfavorable outcome (OR = 2.93; p < 0.0001; Table 3).
Discussion
Genetic testing is crucial for molecular-targeted therapies in NSCLC. In Brazil, less than half of the Brazilian cancer patients are tested for therapeutic targets and the public health system does not cover the greatest majority of molecular testing for NSCLC29. Conversely, at the Barretos Cancer Hospital, a non-profit cancer center where 100% of patients are from the public health system30, all non-squamous NSCLC patients are benefited with the molecular testing for tailored therapies. Herein, we reported the association of EGFR and KRAS mutational status with clinicopathological features from approximately 500 Brazilian lung adenocarcinoma patients attended at the Barretos Cancer Hospital.
Overall, regarding clinical and histopathological characteristics, adenocarcinoma solid subtype was strongly associated with worse disease outcome in a multivariate analysis irrespective the presence of EGFR mutations. Since only a subset of tumors was suitable for histological subclassification, few histological subgroups have a small number of cases. Nevertheless, solid histological subtype had been previously associated with a worse prognosis25,31.
Although the role of EGFR has been well established in the last few years, data on Brazilian populations remains limited. In the present study, the frequency of EGFR mutations was 22.7%. Previous findings from smaller Brazilian cohorts described frequencies of EGFR mutations between 22% (27/125) to 30% (63/207)25,26. The most recurrently EGFR mutation in the present adenocarcinoma series was a deletion in exon 19 followed by a substitution in exon 21 (p.L858R), similarly to previously reported in Brazilian patients25. These mutations are known to be sensitive to TKi. Interestingly, in the present study, the EGFR-mutated patients that were not treated with TKi, due to poor PS at diagnosis or death before receive the result of molecular test, presented a worse outcome compared with those EGFR-mutated patients TKi-treated, supporting the clinical benefit of TKi in EGFR mutated patients. As expected, the most recurrently EGFR resistance mutation was the p. Tyr790Met and further EGFR resistance mutations were mostly located at exon 2032. In accordance, EGFR-mutated patients harboring mutations located at exon 20 presented lower overall survival and compromised response to TKi.
Several commercial assays are available for EGFR testing, such as COBAS (Roche), and Therascreen (Qiagen) among others, which are realtime-PCR based and are built to harbor the major mutations reported in the literature. In this context, we can hypothesize that approximately 13% of the mutations identified in our series, mainly located in exon 20 and 21, would not be detected by these commercial assays that are widely used. Thus, these results emphasize the importance of the knowledge of the mutational profile of each population to better guide the methodology used for routine practice.
We next interrogate the impact of EGFR status in patients’ clinicopathological features. In a multivariate analysis, we observed that EGFR mutated cases was associated with never smokers, better PS, and higher Asian ancestry, and a tangentially with the female gender. These results are in accordance with the literature16,26,33–35. Interestingly, the association of EGFR mutation with higher Asian ancestry observed in our Brazilian cases is related with the admixture of Asian background in our Brazilian cases, probably due to Japanese/Korean/Chinese immigration wave in the 1940’s.
We observed KRAS mutations in 20.4% of lung adenocarcinomas. This frequency is in accordance with reported in international literature, that vary from 15–33% of cases21,33,34,36. Likewise, our results are in line with the two previous Brazilian reports, which showed 15% (30/207) and 26% (33/115) of KRAS mutations in lung cancer25,26. As well reported26, we found that KRAS mutations were more frequently found in patients who reported tobacco exposure. We also observed an independent association of KRAS mutation with a lower Asian background. Importantly, in our series, KRAS mutation was an independent factor for unfavorable outcome supporting the prognostic value of KRAS mutations. The prognostic role of KRAS in lung cancer is not consensual, with diverge reports37,38. Recently, it was reported that KRAS mutation induced upregulation of PD-L1, through p-ERK, mediated immune escape in lung adenocarcinoma, and induces the apoptosis of CD3-positive T cells, which were reversed by anti-PD-L1 or ERK inhibition39. In addition, it was reported that specific KRAS mutations could affect the immune microenvironment of lung adenocarcinoma patients, which affect the efficacy of immune checkpoint inhibitors, implying stratification of patients for immunotherapy should be tailored based on the specific mutant KRAS variants and tumor microenvironment24. Thus, following the advent of immunotherapy, KRAS mutations have rewarded new purposes with the promising clinical utility.
The present study harbors some limitations, mainly due to the retrospective nature of the study, therefore patients were not treated uniformly, which hamper proper outcome analysis and only patients diagnosed at stage IV were included in the survival analysis, since patients diagnosed at stages I, II, and III presented distinctive outcomes compared with stage IV.
Concluding, this is the largest study assessing EGFR and KRAS mutation status in the Brazilian lung adenocarcinoma population. EGFR mutation was associated with Asian ancestry background, confirming the known geographic disparities. In our series, KRAS mutation was an independent prognostic factor. Overall, these data provide important information about the role of some of the most important driver genes and tailored-guided treatment for lung adenocarcinoma in the Brazilian population.
Materials e Methods
Study population and design
This is a retrospective study conducted at the Center for Molecular Diagnosis, from patients diagnosed at Barretos Cancer Hospital from 2011 to 2014. Overall, 496 NSCLC cases, who underwent surgical resection or core biopsy were histopathologically re-evaluated. Of these, 52 cases with non-adenocarcinoma histology were excluded for further analysis. A subset of this series was previously published and tested for ALK translocations40. The major clinicopathological features of the 444 lung adenocarcinomas are summarized in Table 1. Overall, 232 were male (52%) and 212 female (48%) with an average age at the diagnosis of 61 years old (22–87 years). Most of the patients were self-reported as white (79%), were current or former smokers (77%) and were no alcohol consumers (61%). Most patients were diagnosed at stage IV (74%) and among these patients, most of them presented metastasis at multiple sites (61%). ECOG PS 1 was the most prevalent at diagnosis, and weight loss was observed in half of the patients (50.6%). The most predominant histological subtype was acinar, followed by solid, papillary, lepidic and mucinous but about a quarter of the tumors (27%) was not possible to determine the histological subtype (Table 1).
Considering the present study enrolls a retrospective series, patients were treated ununiformly. Detailed information about treatment regimens are described in supplementary material (Supplementary Tables 6–9).
The present study was approved by the Barretos Cancer Hospital IRB (Project n°. 630/2012), which bestowed the exemption of informed consent due to the retrospective nature of the study since most of the patients are dead. All methods were performed in accordance with the relevant guidelines and regulations.
DNA isolation
Serial 10 μm unstained sections of FFPE blocks were cut for DNA isolation and one hematoxylin and eosin-stained (H&E) section was taken for pathological evaluation and selection of the tumor area as previously reported41. Briefly, sections were heated at 80 °C and serial washes with xylene and ethanol (100, 70 and 50%) were performed for paraffin removal. Then, sections were macrodissected using a sterile needle and carefully collected into a microtube. Next, DNA was isolated from FFPE tissues using the QIAmp DNA micro kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA concentration and quality were evaluated by Nanodrop 2000 (Thermo Scientific, Wilmington, USA). DNA samples were diluted to a final concentration of 50 ng/μL and stored at −20 °C for further molecular analysis.
Mutational analysis for EGFR and KRAS hotspot regions
The mutational analysis for hotspots regions of EGFR (exons 18, 19, 20 and 21) and KRAS (exon 2, codons 12 and 13) genes was analyzed by PCR, followed by direct sequencing, as previously described9,42. Briefly, the PCR reaction was performed with 50 ng of DNA in a final volume of 15 µL, using 10 µM of both forward and reverse primers and 7,5 µl of the HotStart master mix (Qiagen, Hilden, Germany), following the manufacturer’s instructions. PCR conditions are following described: 96 °C for 15 minutes, 40 cycles of 96 °C for 45 seconds, 56.5 °C for 45 seconds, 72 °C for 45 seconds and a final extension of 72 °C for 10 minutes.
The amplification of PCR products was checked by electrophoresis in agarose gel and purified by enzymatic reaction (ExoSAP-it, ThermoFisher Scientific). Next, direct sequencing was carried out using BigDye Terminator v3.1 Cycle Sequencing kit (ThermoFisher Scientific) with the following conditions: 97 °C for 3 minutes, 28 cycles of 96 °C for 10 seconds, 50 °C for 5 seconds, and 60 °C for 4 minutes. Sequencing products were purified using BigDye Xterminator (ThermoFisher Scientific) and analyzed on a 3500 Genetic Analyzer, ABI capillary electrophoresis system (Applied Biosystems). Sequences were captured by the SeqScape software (Applied Biosystems) and manually compared to reference sequences collected from GenBank (EGFR: NG_007726.3; KRAS: NG_007524.1). All mutations were confirmed twice.
Cobas®EGFR Mutation
A subset of cases was processed using the Cobas® DNA Sample Preparation Kit for manual sample preparation and the Cobas z 480 analyzer for automated amplification and detection following Cobas® EGFR Mutation Test kit manual instructions.
Ancestry analysis
The ancestry analysis was performed using a set of 46 ancestry-informative markers (AIMs) among the most informative INDELs for Native American (AME), European (EUR), African (AFR), and East Asian (ASN) population groups as previously published43. Primer sequences and PCR conditions were previously described43,44. Multiplex PCR was performed and the amplified products were submitted to fragment analysis on a 3500xL Genetic Analyzer, ABI capillary electrophoresis system, according to the manufacturer’s instructions. The electropherograms were analyzed and genotypes were automatically assigned using GeneMapper v4.1 (Applied Biosystems).
Ancestry proportions were then assessed using the Structure v2.3.3 software45,46 considering the four major population groups as possible contributors to the current genetic background of the Brazilian population. Data from the Human Genome Diversity Panel (HGDP-CEPH) previously demonstrating no significant departures from Hardy-Weinberg equilibrium and linkage equilibrium43 were employed as a reference for the ancestral populations and a supervised analysis was performed to estimate ancestry membership proportions of the individuals involved in the present study. Structure v2.3.3 software runs considering K = 4 consisted of 100.000 burnin steps followed by 100.000 Markov Chain Monte Carlo iterations. The option ‘Use population Information to test for migrants’ was used with the Admixture model, considering allele frequencies correlated, and updating allele frequencies using only individuals with POPFLAG = 1.
Statistical analysis
Clinicopathological factors were used in univariate and multivariate to determine whether the mutations have a significant effect on the parameter. Ancestry proportions were defined as categorical variables according to Lima-Costa et al.47. The significance of multivariate analysis for association with the presence of the mutations was assessed by the Wald test. Survival analysis was carried out using the Kaplan-Meier method and the Log-rank test. The multivariate analysis was performed by the Cox proportional hazard model to determine whether they have a significant effect on overall survival. All statistical analyses were conducted using SPSS 21.0 (IBM Corp, Armonk, NY, USA) and the level of significance was 5%.
Supplementary information
Acknowledgements
We thank Luciane Sussuchi da Silva for graphical support in genetic ancestry component analysis. This work was supported by the Barretos Cancer Hospital; FINEP (MCTI/FINEP/MS/SCTIE/DECIT-CT-INFRA grant number 02/2010); Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer); and National Council for Scientific and Technological Development (CNPq, Brazil).
Author Contributions
L.F.L. and R.M.R. wrote the main manuscript text. F.E.P., G.N.B. and C.D.C. carried out the experiments. P.D.M., L.S.V., G.D.J.P., J.E.M. and C.M.S. performed clinical data collection. E.C.A.S. and C.S.N. performed the histopathological analysis. L.F.L. and M.A.O. performed statistical analysis. L.F.L., R.M.R., P.D.M., R.P. and C.S.N. contributed to the interpretation of the results. R.M.R. conceived the study and was in charge of overall direction and planning. All authors reviewed the manuscript.
Data Availability
The data that support the findings of this study are available from Dr. Rui Manuel Reis but restrictions apply to the availability of these data, which were used under ethics committee approval for the current study, and so are not publicly available because of patients’ personal data. Data are however available from the authors upon reasonable request and with permission of the Dr. Rui Manuel Reis (Scientific and Executive Director of the Molecular Oncology Research Center, Barretos Cancer Hospital).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Letícia Ferro Leal, Flávia Escremim de Paula and Pedro De Marchi contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39965-x.
References
- 1.IARC. Population Fact Sheets: World, (https://gco.iarc.fr/today/fact-sheets-populations?population=900&sex=0). (2012).
- 2.INCA. Estimate/2016–Cancer Incidence in Brazil. https://www.inca.gov.br/estimativa/2016/estimativa-2016-v11.pdf (2015).
- 3.Ferlay J et al. GLOBOCAN 2012v1.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 11, http://globocan.iarc.fr/. (2012).
- 4.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. The New England journal of medicine. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 5.Artal Cortes A, Calera Urquizu L, Hernando Cubero J. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Transl Lung Cancer Res. 2015;4:191–197. doi: 10.3978/j.issn.2218-6751.2014.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 7.Travis WD, et al. The World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 8.Normanno N, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Reis-Filho JS, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–453. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 10.Sorich MJ, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 11.Thomas A, Liu SV, Subramaniam DS, Giaccone G. Refining the treatment of NSCLC according to histological and molecular subtypes. Nat Rev Clin Oncol. 2015;12:511–526. doi: 10.1038/nrclinonc.2015.90. [DOI] [PubMed] [Google Scholar]
- 12.Cho J, et al. The Association of EGFR Mutations with Stage at Diagnosis in Lung Adenocarcinomas. PloS one. 2016;11:e0166821. doi: 10.1371/journal.pone.0166821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma BB, Hui EP, Mok TS. Population-based differences in treatment outcome following anticancer drug therapies. The Lancet. Oncology. 2010;11:75–84. doi: 10.1016/S1470-2045(09)70160-3. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch FR, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 15.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 16.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackman D, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piotrowska Z, Sequist LV. Epidermal Growth Factor Receptor-Mutant Lung Cancer: New Drugs, New Resistance Mechanisms, and Future Treatment Options. Cancer J. 2015;21:371–377. doi: 10.1097/PPO.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 19.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Molecular cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colicelli J. Human RAS superfamily proteins and related GTPases. Science’s STKE: signal transduction knowledge environment. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalemkerian GP, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 23.Manchado E, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–651. doi: 10.1038/nature18600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk AT, et al. Effect of mutant variants of the KRAS gene on PD-L1 expression and on the immune microenvironment and association with clinical outcome in lung adenocarcinoma patients. Lung Cancer. 2018;121:70–75. doi: 10.1016/j.lungcan.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 25.de Melo AC, et al. Mutational Profile and New IASLC/ATS/ERS Classification Provide Additional Prognostic Information about Lung Adenocarcinoma: A Study of 125 Patients from Brazil. Oncology. 2015;89:175–186. doi: 10.1159/000376552. [DOI] [PubMed] [Google Scholar]
- 26.Bacchi CE, et al. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics (Sao Paulo) 2012;67:419–424. doi: 10.6061/clinics/2012(05)03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahina H, et al. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer. 2006;54:419–422. doi: 10.1016/j.lungcan.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Yang JC, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. The Lancet. Oncology. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 29.Araujo LH, et al. Lung cancer in Brazil. J Bras Pneumol. 2018;44:55–64. doi: 10.1590/S1806-37562017000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmero EI, et al. Oncogenetics service and the Brazilian public health system: the experience of a reference Cancer Hospital. Genet Mol Biol. 2016;39:168–177. doi: 10.1590/1678-4685-GMB-2014-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung JJ, et al. Prognostic value of the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification on death and recurrence in completely resected stage I lung adenocarcinoma. Ann Surg. 2013;258:1079–1086. doi: 10.1097/SLA.0b013e31828920c0. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda H, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med. 2013;5:216ra177. doi: 10.1126/scitranslmed.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varghese AM, et al. Lungs don’t forget: Comparison of the KRAS and EGFR mutation profile and survival of collegiate smokers and never smokers with advanced lung cancers. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:123–125. doi: 10.1097/JTO.0b013e31827914ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogan S, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shigematsu H, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 36.Capella G, Cronauer-Mitra S, Pienado MA, Perucho M. Frequency and spectrum of mutations at codons 12 and 13 of the c-K-ras gene in human tumors. Environ Health Perspect. 1991;93:125–131. doi: 10.1289/ehp.9193125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson ML, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2013;119:356–362. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roman M, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Molecular cancer. 2018;17:33. doi: 10.1186/s12943-018-0789-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen N, et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol Immunother. 2017;66:1175–1187. doi: 10.1007/s00262-017-2005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evangelista AF, et al. Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology. BMC Pulm Med. 2017;17:86. doi: 10.1186/s12890-017-0428-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berardinelli GN, et al. Advantage of HSP110 (T17) marker inclusion for microsatellite instability (MSI) detection in colorectal cancer patients. Oncotarget. 2018;9:28691–28701. doi: 10.18632/oncotarget.25611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamane LS, et al. KRAS and BRAF mutations and MSI status in precursor lesions of colorectal cancer detected by colonoscopy. Oncol Rep. 2014;32:1419–1426. doi: 10.3892/or.2014.3338. [DOI] [PubMed] [Google Scholar]
- 43.Pereira R, et al. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PloS one. 2012;7:e29684. doi: 10.1371/journal.pone.0029684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Campanella NC, et al. Optimization of a pentaplex panel for MSI analysis without control DNA in a Brazilian population: correlation with ancestry markers. Eur J Hum Genet. 2014;22:875–880. doi: 10.1038/ejhg.2013.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lima-Costa MF, et al. Genomic Ancestry, Self-Rated Health and Its Association with Mortality in an Admixed Population: 10 Year Follow-Up of the Bambui-Epigen (Brazil) Cohort Study of Ageing. PloS one. 2015;10:e0144456. doi: 10.1371/journal.pone.0144456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from Dr. Rui Manuel Reis but restrictions apply to the availability of these data, which were used under ethics committee approval for the current study, and so are not publicly available because of patients’ personal data. Data are however available from the authors upon reasonable request and with permission of the Dr. Rui Manuel Reis (Scientific and Executive Director of the Molecular Oncology Research Center, Barretos Cancer Hospital).