Abstract
Recent advances in high-throughput genome sequencing technologies are now making the genetic dissection of the complex genome of cultivated strawberry easier. We sequenced Maehyang (short-day cultivar) × Albion (day-neutral cultivar) crossing populations using double digest restriction-associated DNA (ddRAD) sequencing technique that yielded 978,968 reads, 80.2% of which were aligned to strawberry genome allowing the identification of 13,181 high quality single nucleotide polymorphisms (SNPs). Total 3051 SNPs showed Mendelian segregation in F1, of which 1268 were successfully mapped to 46 linkage groups (LG) spanning a total of 2581.57 cM with an average interval genetic distance of 2.22 cM. The LGs were assigned to the 28 chromosomes of Fragaria × ananassa as determined by positioning the sequence tags on F. vesca genome. In addition, seven QTLs namely, qRU-5D, qRU-3D1, qRU-1D2, qRU-4D, qRU-4C, qRU-5C and qRU-2D2 were identified for runner production with LOD value ranging from 3.5–7.24 that explained 22–38% of phenotypic variation. The key candidate genes having putative roles in meristem differentiation for runnering and flowering within these QTL regions were identified. These will enhance our understanding of the vegetative vs sexual reproductive behavior in strawberry and will aid in setting breeding targets for developing perpetual flowering and profuse runnering cultivar.
Introduction
Cultivated strawberry is favored across the globe for its characteristic appearance, flavor, nutritional qualities and health benefitting anti-oxidative properties1 which is evident by the continual increase in the cultivation and production of the crop in the last two decades (FAOSTAT 2018). Moreover, strawberry is an interesting model crop due to its unique reproductive dynamics, having intricate relationship between sexual reproduction via flowering and asexual reproduction via runnering, which is greatly modulated by environmental cues2–5. Strawberry cultivars were characterized as seasonal flowering (also known as short-day, June bearing or non-remontant) or perpetual flowering (also known as day-neutral, long-day, everbearing, rebloomer or remontant) based on the flowering response that heavily depends on photoperiod and temperature6,7. In seasonal flowering (SF) genotypes, flowering is initiated by short-day lengths (<14 h) and low temperatures (<15 °C) of autumn which remains dormant during winter and emerges in spring, leading to a single harvest season in summer. The perpetual flowering (PF) genotypes, on the other hand, can initiate flowers at any photoperiod with a moderate range (15–21 °C) of temperature, leading to an extended harvest season from spring until late autumn7–9. In SF genotypes, the period of flowering is followed by runnering when primary stolons emerge from the basal axillary buds that elongate and give rise to new clonal plants at varyingly spaced nodes. Whereas, in PF genotypes, these sexual and asexual reproduction period overlaps in summer4,5,9. Evidences from both wild and cultivated strawberry show that flowering and runnering are genetically distinct but mutually exclusive processes as the predominance of one over the other is intricately related with genetic and environmental factors3–5,7. Longer days and higher temperatures promote runnering in SF genotypes, which in short and cooler days produce branch crowns that are differentiated to inflorescence and hence, increases flowering and crop productivity5,10,11. Higher temperatures also increase runnering in PF genotypes, however, the effect of photoperiod were found to be variable by different experiments3,12–14. PF genotypes are, in general, preferred for an extended commercial harvest season of fresh berries. However, the poor runnering habit of the PF genotypes compared to SF genotypes hinders their propagation, making it one of the prioritized breeding targets which require wider genetic understanding of the trait3,15–17.
The studies on the diploid strawberry identified that the PF and RU trait are controlled by different alleles18, while in cultivated strawberry, common genetic control with opposing effect has been reported3. Fewer studies reported single dominant gene19–21, while many studies favored the polygenic control of the PF trait in cultivated strawberry17,22. Developing the genetic maps and fine mapping of the loci in the genome will be helpful to genetically disentangle the trait. However, the complex allo-octoploid (2n = 8x = 56) genome of cultivated strawberry, which consists of four relatively similar sub-genomic chromosome sets from diploid donors posed as a constraint in this regard23–26. Several studies have constructed linkage maps using different suite of molecular markers such as RAPD, AFLP and SSR, etc.27–32 and few of those studies reported QTLs with various additive effects for PF and RU traits in cultivated strawberry3,4,15,17,33–35. However, these markers largely suffer from either non-transferability across investigations, insufficient genome-wide coverage, poor density or higher developmental and screening costs3,15,36.
Recent release of strawberry virtual reference genome and advances in reduced sequencing technologies such as ddRAD-seq (restriction site associated DNA sequencing), DArT-Seq (Diversity Arrays Technology) and GBS (genotyping by sequencing) offer the potentiality of rapid and cost effective detection of genome wide single nucleotide polymorphisms (SNPs) that can be used in construction of high density linkage maps and fine mapping of the traits of interest23,26,37. Such studies have recently been reported in few polyploid species such as wheat, Brassica napus, sugarcane, sweet potato and peanut etc.38,39. Very recently, the efficacy of DArT24, ddRAD-seq40 and GBS41 technologies have been reported for developing SNP based linkage maps in cultivated strawberry which showed the way for development of linkage maps with increased marker density and increased genomic span. None of these studies, however, attempted to identify QTLs using these SNP based linkage maps. We thus primarily aimed at developing high density and greater genome covering linkage map using ddRAD-seq derived SNPs and in addition, mapping QTLs for runner production in a population raised by crossing parents with contrasting runner producing capability which will be helpful in understanding the genetics of runnering in strawberry.
Results
ddRAD-seq based SNP discovery and genotyping
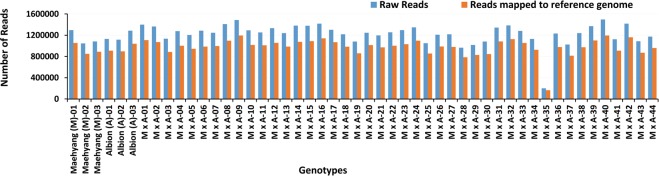
The ddRAD (PstI and MspI) representation libraries, prepared from the genomic DNA of Maehyang × Albion (M × A) populations were successfully sequenced using the Illumina HiSeq platform. The paired-end sequencing of individuals generated a total of 61,065,374 reads (10 GB sequence data). On average, 978,968 high quality reads per sample (80.2% of total reads) were aligned against the Fragaria × ananassa genome version FAN_r1.1 (Fig. 1). The statistics of raw reads, cleaned reads, reads mapped to the reference genome and alignment ratio for individual accessions were summarized in Supplementary Table S1. The mapped reads were further investigated to select a total of 13,181 high quality SNPs based on strict SNP filtration criteria (SNP quality score ≥ 999, minimum depth = 5, minimum allele frequency = 0.05 and minimum proportion of missing data = 0.5) through VCFTools program. The SNPs were distributed as follows, 678 (5%) C/A, 1914 (14%) G/A, 508 (4%) G/C, 889 (7%) T/A, 1765 (13%) T/C and 683 (5%) T/G (Fig. 2A). Cumulatively, the SNP transitions (A/G or C/T) were higher (3728) compared to transversions (G/T, A/C, A/T or C/G) (3016) and the transition/tranversion ratio was 1.23 (Fig. 2B). Overall, the highest SNP frequencies were observed for transition C/T (1941) followed by A/G (1787). In addition, transition/tranversion (TS/TV) ratio of >0.5 was used for calculating divergence and restructuring the phylogenetic tree42. The high quality SNPs were annotated using SnpEff tool (version 4.4) and the generated database was used to annotate possible SNPs based on their genomic, exonic and functional classes. A total of 6,094 SNPs (34.7%) were exonic while 1,844 (10.5%) and 3,245 (18.7%) were intronic and intergenic SNPs, respectively (Supplementary Fig. S1). The lowest number of SNPs (1.9%) were located in the 5′UTR region followed by 4.3% (759) located in 3′UTR region.
Figure 1.
Frequency of mapped and unmapped raw reads obtained from the ddRAD-seq of the strawberry cultivars, Maehyang and Albion, and their F1 offspring on the reference genome.
Figure 2.
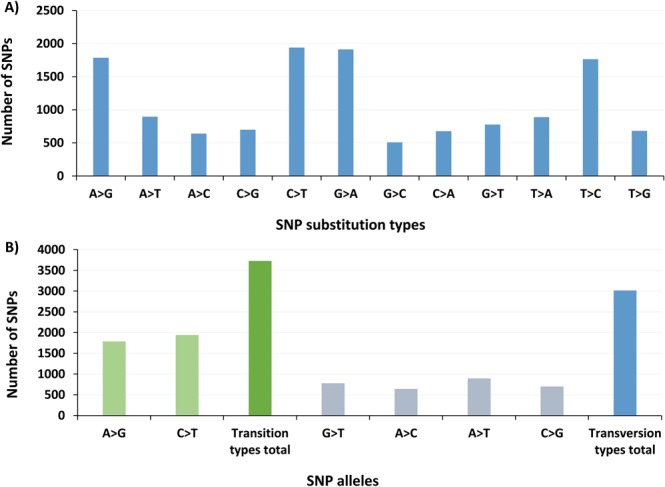
Distribution types (A) and transition/transversion ratios (B) of SNPs identified from ddRAD-sequencing of strawberry parental and F1 population.
Construction of high density linkage map
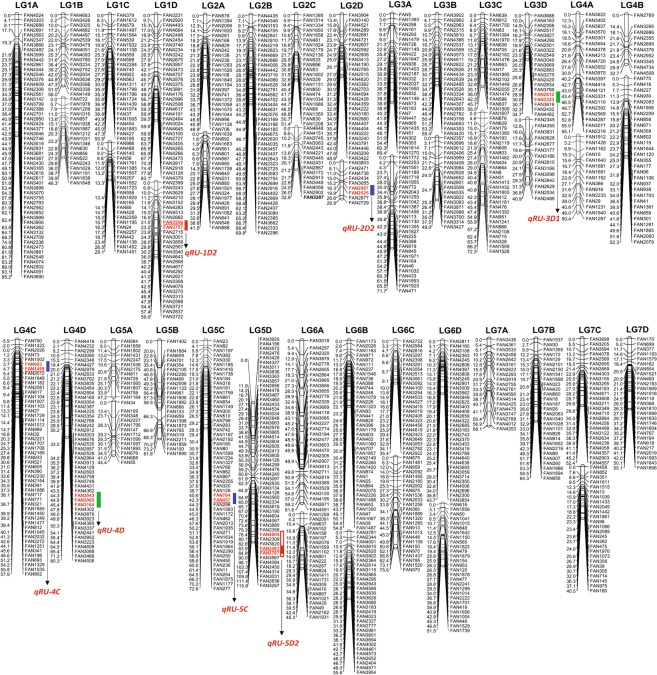
Among the 13,181 high quality SNPs identified, a total of 4738 were selected based on the presence of heterozygous loci either in maternal or paternal or both parents for linkage analysis. A total of 1687 markers showed distortion from Mendelian segregation. Among the 3051 markers showing significant Mendelian segregation, 1064 pairs (consisting of 932 unique markers) showed exactly identical (Supplementary Table S2) and 1010 pairs (consisting of 961 unique markers) showed highly similar (Supplementary Table S3) patterns of segregation. Finally, 1268 SNP markers were mapped to the 46 linkage groups spanning a genetic distance of 2581.57 cM along the length of the 7 diploid F. vesca chromosomes representing all 28 chromosomes of octoploid Fragaria × ananassa (Table 1; Fig. 3). The remaining markers that were not included were either not linked to any of the recognized LGs, were mapped in small groups or showed conflicting segregation pattern with other markers of the same linkage group at the selected LOD threshold. The lengths of linkage groups ranged from 28.93 cM (LG2D2) to 115.84.1 cM (LG5D) with an average length of 56.12 cM. The map resolution corresponded to an average marker density of one marker every 0.48 cM with the longest marker interval of 20.94 cM observed in LG5B. The physical mapping of the SNP markers on the F. vesca genome (v4.0.a1) sequences revealed a total coverage of 76.8% (184.33 Mb) of the diploid genome (240 Mb) which represents one of the four sub-genomes of the cultivated octoploid Fragaria × ananassa. The details of linkage group-wise markers, marker resolutions, genetic distance on Fragaria × ananassa and corresponding physical position on diploid F. vesca are shown in Table 1 and Supplementary Table S4 and the corresponding gene IDs, the SNP positions and genotypes are shown in Supplementary Table S5. High degree of collinearity between the genetic distances of the mapped SNP markers of each linkage group and their corresponding physical position on the F. vesca chromosomes were observed for most of the LGs except for few shorter and less dense LGs such as LG1B2, LG2D2, LG4B1, LG5A1 and LG5A2 (Supplementary Fig. S2).
Table 1.
Summary statistics of the linkage groups constructed using the population arising from the cross ‘Maehyang × Albion’ along with their physical distances on the F. vesca_v4.0.a1 genome sequences.
| SL | Linkage group | Length (cM) | First and last mapped marker | Total no. of mapped markers | Average interval (cM/locus) | Longest gap (cM) | F. vesca chromo-some | LG physical start (bp)a | LG physical end (bp)b | Physical Span (bp)c |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LG1A | 95.25 | FAN4524-FAN3690 | 54 | 1.76 | 9.64 | Fvb1 | 112529 | 16227481 | 16114952 |
| 2 | LG1B1 | 55.99 | FAN4683-FAN4149 | 17 | 3.29 | 7.78 | Fvb1 | 144009 | 4479442 | 4335433 |
| 3 | LG1B2 | 46.33 | FAN722-FAN1648 | 18 | 2.57 | 6.47 | Fvb1 | 1383360 | 5403172 | 4019812 |
| 4 | LG1C1 | 50.53 | FAN379-FAN195 | 26 | 1.94 | 8.31 | Fvb1 | 6183315 | 10129223 | 3945908 |
| 5 | LG1C2 | 29.13 | FAN986-FAN1451 | 23 | 1.27 | 7.36 | Fvb1 | 7969400 | 20650547 | 12681147 |
| 6 | LG1D1 | 83.11 | FAN3231-FAN4379 | 34 | 2.44 | 9.88 | Fvb1 | 3921057 | 18970557 | 15049500 |
| 7 | LG1D2 | 58.98 | FAN2399-FAN3722 | 28 | 2.11 | 6.76 | Fvb1 | 7438667 | 19123178 | 11684511 |
| 8 | LG2A1 | 48.88 | FAN578-FAN708 | 24 | 2.04 | 7.14 | Fvb4 | 804175 | 11904624 | 11100449 |
| 9 | LG2A2 | 41.55 | FAN1639-FAN888 | 20 | 2.08 | 6.38 | Fvb4 | 425782 | 13276588 | 12850806 |
| 10 | LG2B | 69.78 | FAN4435-FAN2296 | 45 | 1.55 | 7.09 | Fvb2 | 267888 | 28873870 | 28605982 |
| 11 | LG2C1 | 51.38 | FAN1395-FAN1208 | 23 | 2.23 | 7.86 | Fvb2 | 5674815 | 29167427 | 23492612 |
| 12 | LG2C2 | 32.94 | FAN4648-FAN3287 | 15 | 2.20 | 8.59 | Fvb2 | 15750516 | 19803671 | 4053155 |
| 13 | LG2D1 | 48.82 | FAN3904-FAN3422 | 30 | 1.63 | 13.42 | Fvb2 | 24250389 | 29150635 | 4900246 |
| 14 | LG2D2 | 28.93 | FAN3803-FAN2729 | 9 | 3.21 | 8.09 | Fvb2 | 21804777 | 26832106 | 5027329 |
| 15 | LG3A1 | 69.38 | FAN5-FAN1270 | 29 | 2.39 | 11.75 | Fvb3 | 26607 | 12531139 | 12504532 |
| 16 | LG3A2 | 71.72 | FAN1772-FAN471 | 28 | 2.56 | 19.38 | Fvb3 | 2288930 | 37706799 | 35417869 |
| 17 | LG3B1 | 55.36 | FAN3902-FAN3420 | 25 | 2.21 | 4.12 | Fvb3 | 3610045 | 14574195 | 10964150 |
| 18 | LG3B2 | 48.96 | FAN2357-FAN3427 | 19 | 2.58 | 11.49 | Fvb3 | 8122733 | 13333825 | 5211092 |
| 19 | LG3C1 | 56.48 | FAN639-FAN1500 | 20 | 2.82 | 10.76 | Fvb3 | 12328117 | 36511410 | 24183293 |
| 20 | LG3C2 | 72.33 | FAN1607-FAN1528 | 29 | 2.49 | 6.38 | Fvb3 | 17089842 | 38167538 | 21077696 |
| 21 | LG3D1 | 34.68 | FAN3688-FAN4482 | 21 | 1.65 | 10.02 | Fvb3 | 33191108 | 37766726 | 4575618 |
| 22 | LG3D2 | 40.92 | FAN2345-FAN2466 | 14 | 2.92 | 12.77 | Fvb3 | 30017669 | 38165007 | 8147338 |
| 23 | LG4A1 | 55.10 | FAN3622-FAN2319 | 21 | 2.62 | 12.32 | Fvb4 | 1372325 | 19654576 | 18282251 |
| 24 | LG4A2 | 45.95 | FAN1912-FAN1940 | 15 | 3.28 | 6.75 | Fvb4 | 572887 | 22338700 | 21765813 |
| 25 | LG4B1 | 32.73 | FAN2769-FAN4509 | 10 | 3.27 | 14.41 | Fvb4 | 2824575 | 4495688 | 1671113 |
| 26 | LG4B2 | 52.26 | FAN770-FAN2079 | 29 | 1.80 | 5.34 | Fvb4 | 6686516 | 27802611 | 21116095 |
| 27 | LG4C | 57.91 | FAN2026-FAN802 | 51 | 1.14 | 3.89 | Fvb4 | 344401 | 33602364 | 33257963 |
| 28 | LG4D | 80.28 | FAN4418-FAN4506 | 66 | 1.22 | 8.99 | Fvb4 | 1119936 | 33498276 | 32378340 |
| 29 | LG5A1 | 47.20 | FAN564-FAN1164 | 13 | 3.63 | 13.32 | Fvb5 | 319932 | 2289061 | 1969129 |
| 30 | LG5A2 | 44.43 | FAN34-FAN55 | 13 | 3.42 | 13.37 | Fvb5 | 7581891 | 9861778 | 2279887 |
| 31 | LG5B | 81.79 | FAN1402-FAN1895 | 24 | 3.41 | 20.94 | Fvb5 | 3768759 | 18410022 | 14641263 |
| 32 | LG5C | 72.80 | FAN23-FAN277 | 54 | 1.35 | 4.47 | Fvb5 | 130730 | 15080365 | 14949635 |
| 33 | LG5D | 115.84 | FAN3920-FAN3267 | 73 | 1.59 | 11.00 | Fvb5 | 21228806 | 27416727 | 6187921 |
| 34 | LG6A1 | 61.62 | FAN3018-FAN4591 | 38 | 1.62 | 6.24 | Fvb6 | 2083928 | 7251058 | 5167130 |
| 35 | LG6A2 | 45.34 | FAN1259-FAN1931 | 23 | 1.97 | 10.04 | Fvb6 | 70354 | 11645796 | 11575442 |
| 36 | LG6B1 | 54.78 | FAN1173-FAN1487 | 41 | 1.34 | 5.05 | Fvb6 | 1660733 | 10248262 | 8587529 |
| 37 | LG6B2 | 55.64 | FAN3781-FAN3954 | 31 | 1.79 | 7.67 | Fvb6 | 5864047 | 15255602 | 9391555 |
| 38 | LG6C1 | 35.35 | FAN2722-FAN2922 | 25 | 1.41 | 3.31 | Fvb6 | 10630696 | 20387841 | 9757145 |
| 39 | LG6C2 | 75.55 | FAN675-FAN570 | 25 | 3.02 | 10.68 | Fvb6 | 3657849 | 27953227 | 24295378 |
| 40 | LG6D1 | 43.69 | FAN3811-FAN3854 | 35 | 1.25 | 4.88 | Fvb6 | 30923227 | 39197465 | 8274238 |
| 41 | LG6D2 | 51.45 | FAN616-FAN1739 | 28 | 1.84 | 6.17 | Fvb6 | 15955826 | 37293159 | 21337333 |
| 42 | LG7A | 68.14 | FAN3436-FAN4253 | 19 | 3.59 | 10.79 | Fvb7 | 1700487 | 8698128 | 6997641 |
| 43 | LG7B | 64.28 | FAN1537-FAN656 | 29 | 2.22 | 7.15 | Fvb7 | 7860566 | 22021634 | 14161068 |
| 44 | LG7C1 | 52.37 | FAN3699-FAN4557 | 26 | 2.01 | 7.16 | Fvb7 | 5168144 | 20476274 | 15308130 |
| 45 | LG7C2 | 40.51 | FAN458-FAN180 | 26 | 1.56 | 7.56 | Fvb7 | 17500684 | 24064058 | 6563374 |
| 46 | LG7D | 55.15 | FAN170-FAN1606 | 27 | 2.04 | 8.11 | Fvb7 | 16557672 | 24078363 | 7520691 |
| Total | n/a | 2581.57 | n/a | 1268 | 2.22 | n/a | n/a | n/a | n/a | 184.33 Mb d |
a,bThe overall physical start and end positions of all markers of one linkage group (that are usually mapped to a particular F. vesca chromosome) in the F. vesca_v4.0.a1 genome.
cThe distance between the LG physical start and LG physical end that indicates the overall physical span of all the markers of one linkage group in a particular chromosome of the F. vesca_v4.0.a1 genome.
dThe sum of chromosome-wise overall physical distances (span between the first and last mapped markers in a particular F. vesca chromosome) covered by the markers in all seven F. vesca (v4.0.a1 genome) chromosomes.
Figure 3.
A SNP-based linkage map of a Fragaria × ananassa mapping population derived from the progeny of the cross ‘Maehynag × Albion’ comprising of 1292 SNP markers. The genetic distances are given in centi-Morgans (cM) in left. Green, blue and red bars indicate QTLs detected by CIM method, multi-locus GWAS methods (mrMLM, FASTmrMLM, pLARmEB and ISIS EM-BLASSO) and all of these methods, respectively. Underlined marker indicate the central marker within the QTLs that are identified by multi-locus GWAS methods.
Phenotypic evaluation
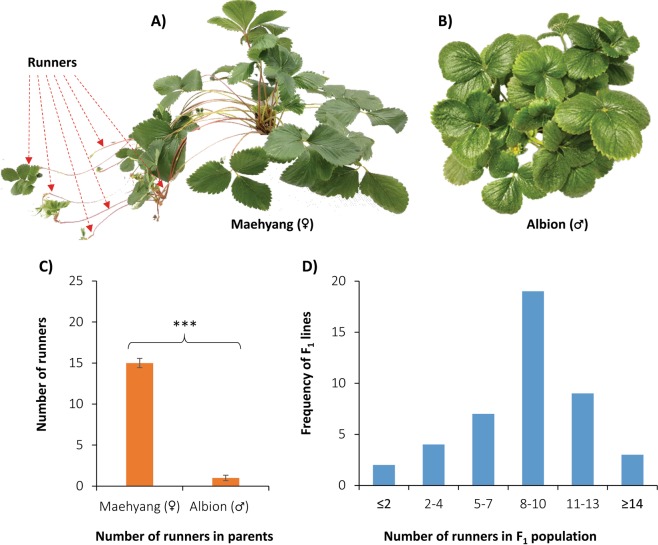
The two cultivars exhibited significant variation (p < 0.001) in terms of number of runner production in one growing season (Fig. 4A–C). The seasonal flowering cultivar ‘Maehyang’ produced 15 runners while the perpetual flowering cultivar produced only one runner during the entire growing season (Fig. 4C). In the segregating F1 population derived from crossing these two contrasting cultivars, continuous variation was observed for total number of runners (Fig. 4D). The minimum (0) and the maximum (15) number of runners were produced by one and two F1 lines, respectively while a moderate 8–10 runners were produced by 19 F1 lines (Fig. 4D).
Figure 4.
Runnering behavior of the profuse runner producing and seasonal flowering parent, Maehyang (A) and poor runner producing and perpetual flowering parent, Albion (B); and the frequency distribution of the total number of runners in parents (C) and in their F1 population (D). For figure C, data presented as mean ± SE (n = 5) and asterisks (***) represent significant differences between the two parental lines for total number of runners at the end of the growing season at P < 0.001 by Student’s T-test.
Identification of QTLs
Using the composite interval mapping and several multi-locus GWAS methods, we identified five negative effect QTLs namely, qRU-5D, qRU-3D1, qRU-1D2, qRU-4D and qRU-2D2 located on linkage groups LG5D, LG3D1, LG1D2, LG4D and LG2D2, respectively and two positive effect QTLs namely, qRU-4C and qRU-5C located on linkage groups LG5C and LG5C, respectively related to the number of runners in the F1 population raised from crossing Maehyang × Albion (Table 2; Fig. 3). The QTL qRU-5D identified on LG5D had the highest LOD score of 7.24 (p < 0.05) and explained the highest proportion (38%) of phenotypic variation (R2) among the identified QTLs. This QTL was associated with the flanking markers, FAN4605 and FAN3787 (83.88–84.81 cM) on the linkage group LG5D. The next highest LOD (7.15) was observed for the QTL qRU-3D1 (additive effect = −8.14; R2 = 0.36; flanking markers = FAN2812-FAN3323).
Table 2.
Map positions and genetic effects of QTLs detected for the number of runner (RU) in F1 population derived from crossing octoploid strawberry cultivars Maehyang and Albion.
| QTL name | Linkage Group | Marker | Genetic distance (cM)a | Fragaria × ananassa genome ID | F. vesca Physical position (bp) | Threshold LODb | LODc | Additive Effect | R2d |
|---|---|---|---|---|---|---|---|---|---|
| qRU-5D** | LG5D | FAN4605-FAN3787 | 83.88–84.81 | FAN_iscf00235992.1-FAN_iscf00019356.1 | Fvb5:23272711-23422418 | 2.7 | 7.24 | −9.79 | 0.38 |
| qRU-3D1 | LG3D1 | FAN2812-FAN3323 | 28.9–30.04 | FAN_iscf00310724.1-FAN_iscf00161563.1 | Fvb3:37021445-37232303 | 2.9 | 7.15 | −8.14 | 0.36 |
| qRU-1D2** | LG1D2 | FAN3556-FAN3757 | 20.99–23.23 | FAN_iscf00315119.1-FAN_iscf00003334.1 | Fvb1:9879715-9988189 | 2.9 | 5.54 | −8.54 | 0.37 |
| qRU-4D | LG4D | FAN3943-FAN3164 | 43.51–45.28 | FAN_iscf00171554.1-FAN_iscf00369931.1 | Fvb4:23126333-24533627 | 2.9 | 3.5 | −5.12 | 0.22 |
| qRU-4C* | LG4C | FAN603-FAN1459 | 3.74–4.73 | FAN_iscf00089616.1-FAN_iscf00083346.1 | Fvb2:16350821-17032825 | 2.8 | 4.11 | 3.663 | 0.32 |
| qRU-5C* | LG4C | FAN764-FAN457 | 40.58–42.31 | FAN_iscf00280849.1-FAN_iscf00206802.1 | Fvb5:8718088- 10445687 | 2.7 | 3.97 | 3.182 | 0.24 |
| qRU-2D2* | LG2D2 | FAN2967-FAN2525 | 16.31–19.90 | FAN_iscf00134314.1-FAN_iscf00342145.1 | Fvb2:23649400-23980136 | 2.8 | 3.96 | −4.37 | 0.28 |
No asterisk, * and ** indicate QTLs detected by composite interval mapping (CIM), multi-locus GWAS methods (mrMLM, FASTmrMLM, pLARmEB and ISIS EM-BLASSO method) and all of these techniques, respectively.
aThe distance of the QTL in cM (expressed in Kosambi) from the top of the linkage group.
bThe significant threshold logarithm of the odds (LOD) score calculated by the composite interval mapping using Kosambi’s map function with 1000 permutation (p ≤ 0.05).
cThe peak LOD score.
dThe percentage of phenotypic variance explained by the QTLs as determined by QTL Cartographer version 2·5 (NC, USA).
Candidate genes within the QTL regions
Altogether 587 genes were found within the seven QTL regions, with a maximum of 196 genes within QTL qRU-5C and minimum of 19 genes within QTL qRU-5D and qRU-1D2 each (Supplementary Table S6). Among these, the key candidate genes having putative roles in the vegetative vs reproductive differentiation of shoot apical meristems and regulation of flowering are listed in supplementary Table S7. For example, WUSCHEL related homeobox 1 (FvH4_5g17270) and AGAMOUS-like 71 (FvH4_4g20680) within QTL qRU-4D, CLAVATA3/ESR (CLE)-related protein coding gene FvH4_2g19210 within qRU-4C and Subtilisin-like protease (FvH4_1g17150, FvH4_1g17160 and FvH4_1g17170) within qRU-1D2 may have roles in the CLAVATA-WUSCHEL signaling pathway that specifies meristematic stem cell fate during differential of shoot and floral buds. Several genes encoding cytokinin dehydrogenase 7 (FvH4_2g30990, FvH4_2g31000 and FvH4_2g31010), auxin response factor 17-like (FvH4_2g19860), auxin efflux carrier family protein (FvH4_5g173100), NAC domain-containing protein 86 (FvH4_5g18130) and homeobox protein knotted-1-like gene KNOXI (FvH4_5g17670) could be involved in cytokinin-auxin mediated meristem differentiation. In addition, a total of 12 tetratricopeptide repeat (TPR)-like superfamily protein genes such as FvH4_5g32370; FvH4_5g32380 and FvH4_2g30790 etc. are found within all but qRU-3D1 QTLs which may have roles in the maintenance of meristem cell organization. Two phytochrome related genes, FvH4_4g19750 and FvH4_2g19790 identified within QTLs qRU-4D and qRU-4C, respectively, could be involved in the photoperiodic control of flowering. In addition, six genes namely, FvH4_4g20210.1 encoding ‘flowering time control protein FCA-like isoform X1’ within the QTL qRU-4D, FvH4_2g19520.1 encoding ‘zinc finger protein KNUCKLES-like protein’ within the QTL qRU-4C, FvH4_2g30770.1 encoding ‘flowering-promoting factor 1-like protein (FPF1)’ within the QTL qRU-2D2, FvH4_5g17130 encoding ‘phosphatidylethanolamine-binding protein (PEBP)- mother of FT (FLOWERING LOCUS T) and TF1 (TERMINAL FLOWER 1)’ within qRU-5C and FvH4_5g18150 and FvH4_5g18160 encoding ‘protein argonaute 1 (AGO1) within qRU-5C may have potential roles in control of flowering time. Three translation elongation factor genes EF1B (FvH4_4g20660), EIF3A (FvH4_4g19950) and EF1A (FvH4_5g17000) could be involved in meristem stability and organogenesis.
Discussion
The intricate interplay between vegetative and sexual reproduction with the regards to the environmental cues have both biological and economic significance. This makes strawberry an important model system to study flowering behavior. Runner and flower arise from different meristem crowns (basal and terminal, respectively) based on competitive investment of resources triggered mainly by environmental factors4,7,13,43. Genetics of these two mutually exclusive and antagonistic processes of wild and cultivated strawberry has long been investigated and several hypotheses, mostly favoring polygenic over monogenic control has been put forwarded by previous studies2,5,15,18,19,44. Inheritance studies and mapping of the traits of interest at molecular level necessitated the construction of linkage maps. Here, we report the construction of a linkage map and identification of QTLs for number of runners based on the SNPs detected in the F1 progeny developed from the cross of profuse and poor runnering cultivars, Maehyang and Albion, respectively by aligning the ddRAD-seq reads against strawberry genome (FAN_r1.1).
History of construction of LGs in strawberry followed distinct phases, starting from the initial low density and non-transferrable marker based LGs17,27 followed by the use of a comprehensive suite of sequence characterized and transferrable markers21,28,32 which were further updated upon the release of the diploid F. vesca genome and octoploid Fragaria × ananassa draft genome29,36,45 and finally the development of high-throughput SNP based LGs24,40,41,46. The first genetic linkage map in octoploid strawberry were constructed on the full-sib progeny of ‘Capitola’ × ‘CF1116’, where 235 and 280 single dose restriction fragment (SDRF) markers were mapped on the 43 co-segregating groups spanning a total of 1604 and 1494 cM in female and male parents, respectively27. Rousseau-Gueutin et al.32 included an additional 306 SSR and 5 STS and SCAR markers in the original AFLP marker set of Lerceteau-Köhler et al.27 and constructed 28 and 26 LGs spanning 2582 and 2165 cM in female and male parents, respectively in the extended ‘Capitola × ‘CF1116’ population. Weebadde et al.17 further identified 43 linkage groups using 429 AFLP markers on a population of the cross ‘Tribute’ (day-neutral) × ‘Honeoye’ (short-day). Using 71 RAPD markers in a population of 199 F1 plants of the cross ‘Ever Berry’ (Japanese everbearing cultivar) and ‘Toyonoka’ (Japanese seasonal flowering cultivar), Sugimoto et al.21 identified a linkage group of 39.7 cM that harbor the everbearing gene in strawberry. Using a combination of 315 AFLP, RAPD, SSR and gene specific markers on the 174 F1 progeny of the cross ‘Redgauntlet’ × ‘Hapil’, Sargent et al.28 constructed 37 and 32 LGs spanning 1440.7 and 1675.3 cM in the male and female parents, respectively. The observation of high collinearity between the diploid and octoploid strawberry linkage maps28,32 prompted others29,36,45 to further populate the linkage maps of octoploid strawberry. More markers were designed from the freshly released diploid strawberry genome in 201126 which enabled to saturate the gaps and increase the genomic coverage of previous linkage maps in different populations.
The release of octoploid reference genome23 enabled the development of linkage groups based on SNPs detected by various techniques24,25,40,41,46. Davik et al.40 first demonstrated the effectiveness of ddRAD-seq derived SNPs by constructing a 1518.5 cM robust, high resolution linkage map consisting of 31 linkage groups from a population of Sonata × Babette. We have identified 46 linkage group fragments that spanned a higher genetic distance (2581.57 cM) with an average marker space of only 0.48 cM in M × A crossing population. Mapping of the marker associated sequence tags of each of the LGs to the updated F. vesca genome assembly (v4.0.a1) revealed a reasonable genome-wide coverage (76.8% of the total 240 Mb) of the diploid wild relative’s genome. The markers of a particular LG were usually mapped to a single F. vesca chromosome. However, for some LGs, few markers (a total of 185 out of 1268 mapped markers with an average of 4.02 marker per LG) were mapped to different chromosomes (Supplementary Table S4). The highest 20 markers (out of 73) of LG5D were mapped to a different chromosome followed by LG4C (18 out of 51). This is in agreement with the report of highly conserved macro-synteny between diploid and octoploid strawberry with few inter-chromosome rearrangements25,29,32. These high degree of collinearity further indicates the reliability of the constructed linkage map. However, it is noteworthy that 41% (1268) of the total markers (3051) which showed significant Mendelian segregation ratio were assigned to LGs and there are five LGs with less than 15 markers and two LGs with a longest gap of ~20 cM. This could be attributed to the stringent analysis parameters such as a high LOD (>5) aiming at constructing a representative linkage map by avoiding spurious linkage that can facilitate accurate detection of QTL.
Using the SNP marker based linkage group, we are the first to report QTL in Fragaria × ananassa. Besides the composite interval mapping, we used multi-locus GWAS methods such as mrMLM, FASTmrMLM, pLARmEB and ISIS EM-BLASSO which is shown to detect minor effect QTLs47–50. Altogether, seven QTLs, two with relatively high LOD scores (>7.0) were identified which further strengthens the polygenic control of runnering hypothesis8,17. Gaston et al.3 demonstrated common genetic control of flowering and runnering by a major dominant locus (FaPFRU) which positively influences flowering and exerts a negative effect on runnering. Of the seven QTLs identified in this study, five were negative effect QTLs. Using SSR marker derived LGs, a single major QTL for runnering and perpetual flowering reported by Castro et al.15 was further confirmed by Sooriyapathirana et al.33 using an extended population. Using three separate Japanese population, Honjo et al.35 mapped the PF trait to similar region of previously detected QTL regions.
The breeding industry requires both perpetual flowering and profuse runnering traits but the PF genotypes are poor runner producers. Several putative candidates for PF have already been reported in wild and cultivated strawberry such as TFL1, KSN, FT1, FT2, FT3, FvSOC1 and TCP734,51–55. However, their exact roles in the intricate interplay between meristem differentiation leading to inflorescence or runners are yet to be fully established. In this study, we have mined the genes within the QTL regions and identified several key candidate genes having putative roles in CLAVATA-WUSCHEL signaling, cytokinin–auxin mediated meristem differentiation, directional cell division, meristem stability and organogenesis, gravitropism, vernalization and photoperiodic regulation of flowering etc. that ultimately determines the stem cell fate towards shoot or floral meristem differentiation56–59 (Table S7). Functional characterization of these genes will enhance our understanding of the complex interplay between vegetative vs sexual propagation in strawberry. In addition, the sequence polymorphisms such as the peak QTL SNP markers identified in this study could be used for marker assisted breeding (Table 2). Very recently, the gene FveRGA1 (gene06210 in F. vesca_v1.0) encoding a putative DELLA protein GAIP-B has been shown to play a role in stolon production in diploid strawberry60. It’s corresponding homologue located in FAN_iscf00392249 (Fvb4:32063138..32064973) of Fragaria × ananassa did not inhabit any SNP in our study population. However, one SNP was found in their closest genetic locus FAN_iscf00207522.1 (Fvb4: 32318115..32316672) in the linkage group LG4D at 19.19 cM apart from our qTL qRU-4D.
In conclusion, we report a linkage map with higher density and greater genome-wide span using the ddRAD-seq derived SNPs from the progeny of two strawberry cultivars with contrasting flowering and runnering attributes. In addition, we have also identified QTLs for total number of runners. This high-throughput genotyping based linkage map will serve as a reference for precise sequence scaffold anchoring and orientations in this species and the approach can be replicated in other genetically complicated species. The candidate genes identified within the QTLs, upon functional validation, can be targeted for biotechnological manipulations to develop genotypes with desirable flowering and runnering habit. Besides, the large number of SNPs will supply abundant choices of transferrable markers for future genetic studies and will assist in identifying QTLs, causal genes and linked markers for agronomically important traits which in turn will accelerate strawberry genetic improvement programs via molecular breeding.
Materials and Methods
Plant materials and phenotyping
The experimental population included 44 F1 hybrid seedlings raised by crossing the seasonal flowering (SF) short-day cultivar Maehyang (♀) and the perpetual flowering day-neutral cultivar Albion (♂) (M × A). The F1 seeds were germinated in growth chamber before being transplanted to large rectangular pots in the farm facility of Damyang-gun Agricultural Technology Center, South Korea specialized for strawberry cultivation. The young leaves from the F1 progenies and three sets of both the parents were chosen for further procedure. The cultivars Maehyang and Albion are economically important and harbor several other contrasting growth and fruit qualitative traits in them. Total number of runners were recorded by counting and removing the newly emerged primary stolons twice a week during one growing season under natural temperature and photoperiodic conditions.
DNA extraction
Newly emerged fresh leaves from the parents and F1 plants were disrupted in TissueLyser II (Qiagen, CA, USA). The DNA extracted using DNeasy Plant Mini Kit (Qiagen) following manufacturer’s instructions. The quality of DNA was assessed in agarose gel (1.2%) electrophoresis. The concentration and purity of the extracted DNA were evaluated using Nanodrop-2000 (Nanodrop Technologies, Wilmington, DE, USA).
Double digest restriction site associated DNA (ddRAD) sequencing
Genomic DNA of each strawberry accession was double-digested with the restriction enzymes, PstI and MspI to prepare the ddRAD-seq libraries. The unique barcode of 8 nucleotide base pairs with Illumina adaptor was assigned to each strawberry accession for tracing the samples (Supplementary Table S1). The adaptor-ligated DNA amplicons were pooled and DNA fragments of 300–900 base pair length were separated with BluePippin (Sage Science, Beverly, MA, USA). The constructed ddRAD-seq libraries were sequenced on HiSeq platform (Illumina, USA) using 93 base paired-end (PE) mode as described by Shirasawa et al.38.
Sequence data analysis and SNP detection
Primary data processing of ddRAD-seq was performed as per the procedures described in Shirasawa et al.38 with minor modifications. The sequencing data of strawberry accessions were examined for their quality using FastQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The low-quality sequences and adaptor sequences were trimmed using PRINSEQ (http://prinseq.sourceforge.net)61 and fastx_clipper in FASTX-Toolkit (version 0.10.1; http://hannonlab.cshl.edu/fastx_toolkit), respectively. The obtained high quality reads from each accession were then mapped to the genome of Fragaria × ananassa (FAN_r1.1, http://strawberry-garden.kazusa.or.jp/) as reference using Bowtie 2 tool (version 2.1.0; parameters: -minins 100 -no-mixed)62. The resulting sequence alignment/map format (SAM) files were converted into Binary Alignment Map (BAM) files and thereafter, SAMtools (version 0.1.19; parameters: -Duf) was used for sorting, indexing and removal of duplicates63. Thereafter, genomic variants (SNPs) were called out for each strawberry accessions against reference genome using mpileup module from SAM tools (parameters: -Duf) and BCF tools (parameters: -vcg). In addition, the produced variant call format (VCF) files including SNP details were further filtered as per procedures described by Shirasawa et al., (2017) using VCFtools (version 0.1.11)64. Furthermore, missing data were imputed using Beagle4 software package (version 5.0)65. The effect of SNP annotations on gene functions were predicted using SnpEff tool (version 4.11)66.
Construction of linkage map
The linkage analysis was performed using the SNP-based markers that were homozygous in one parent and heterozygous in the other parent (segregation type code lm × ll and nn × np in JoinMap version 4.1) and heterozygous in both parents (segregation type code hk × hk). Markers with more than 10% missing data and markers that did not fit significantly with the segregation ratio of 1:1 or 1:2:1 in the progeny as per the chi-square test of goodness of fit (p < 0.05) were excluded from map construction. The regression mapping algorithm and Kosambi’s mapping function with a maximum recombination fraction of 0.45, goodness-of-fit jump threshold of 5 and a ripple value of 1 were used for calculating marker order and the map distances were expressed in centi-Morgans (cM) using JoinMap v4.1 (Kyazma, Wageningen, The Netherlands). Linkage groups with a minimum logarithm of odds (LOD) score limit of 5.0 were visualized in MapChart (version 2.32)67. All the linkage groups except the smaller ones (<10 markers) were mapped on the F. vesca (genome v4.0.a1) genome. The LGs were named LG1 to LG7 based on the number of F. vesca chromosome to which the marker associated sequences of each of the LGs were mapped. The suffixes ‘A’-‘D’ were assigned to the LGs mapped to a particular chromosome based on their sequential physical position throughout the length of that chromosome. If any two small linkage groups were found to be mapped to a particular segment of a F. vesca chromosome sequentially, they were considered as one linkage group.
QTL identification
QTLs were mapped based on the marker and phenotypic information of the parents and each individual of the mapping population by the composite interval mapping (CIM) analysis with the following parameters (linkage map method = 10, segregation test size = 0.01, linkage test size = 0.35, mapping function = Kosambi, objective function = SAL and step size = 1 cM) using Windows QTL Cartographer (version 2.5_011) program68. In addition, QTLs were also identified using several ‘multi-locus mixed linear model’ such as mrMLM, FASTmrMLM, pLARmEB and ISIS EM-BLASSO methods in R program47–50. Putative QTLs were declared based on the significant logarithm of odds (LOD) threshold determined by 1000 permutations69. The square of the partial correlation coefficient (R2) indicates the proportion of phenotypic variation explained by a QTL. The genes along with their putative functions that lie within the QTL regions were identified against Fragaria vesca annotated genome version v4.0.a1 using the Fragaria × ananassa genome IDs corresponding to the flanking markers of the QTLs as blast queries.
Supplementary information
Acknowledgements
This study was supported by the Strawberry Project (Grant No. 315047-3) and the Golden Seed Project of Center for Horticultural Seed Development (Grant No. 213007-05-3-CG100) of the Ministry of Agriculture, Food and Rural affairs in the Republic of Korea (MAFRA). We acknowledge Kenta Shirasawa and Sachiko Isobe, Kazusa DNA Research Institute, Japan for their technical support in ddRAD-seq and sequence data analysis. We thank Professor Jundae Lee, Chonbuk National University, South Korea for technical support with JoinMap analysis.
Author Contributions
I.-S.N., J.-I.P. and H.-T.K. conceived the study. M.R.H. and S.N. designed the experiments. C.-G.L. managed plants in the farm and recorded phenotypic data. M.R.H. prepared the samples, extracted DNA, conducted linkage map and QTL analysis and written the manuscript. S.N. did SNP analysis and written the related part of the manuscript. D.M.I.J. performed batch blast and Marey map analysis. All authors have read and approved the final manuscript.
Data Availability
The ddRAD-seq raw reads generated in this study were deposited into Sequence Read Archive (SRA) under NCBI accession PRJNA478299 accessible from (https://www.ncbi.nlm.nih.gov/sra/PRJNA478299).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mohammad Rashed Hossain and Sathishkumar Natarajan contributed equally.
Contributor Information
Hoy-Taek Kim, Email: htkim@sunchon.ac.kr.
Ill-Sup Nou, Email: nis@sunchon.ac.kr.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39808-9.
References
- 1.Giampieri F, Alvarez-Suarez JM, Battino M. Strawberry and Human Health: Effects beyond Antioxidant Activity. J. Agric. Food Chem. 2014;62:3867–3876. doi: 10.1021/jf405455n. [DOI] [PubMed] [Google Scholar]
- 2.Dong X, et al. Genetic control of flowering time in woody plants: Roses as an emerging model. Plant Divers. 2017;39:104–110. doi: 10.1016/j.pld.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaston A, et al. PFRU, a single dominant locus regulates the balance between sexual and asexual plant reproduction in cultivated strawberry. J. Exp. Bot. 2013;64:1837–1848. doi: 10.1093/jxb/ert047. [DOI] [PubMed] [Google Scholar]
- 4.Perrotte J, Guédon Y, Gaston A, Denoyes B. Identification of successive flowering phases highlights a new genetic control of the flowering pattern in strawberry. J. Exp. Bot. 2016;67:5643–5655. doi: 10.1093/jxb/erw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hytönen T, Elomaa P. Genetic and environmental regulation of flowering and runnering in strawberry. Genes, Genomes and Genomics. 2011;5:56–64. [Google Scholar]
- 6.Stewart PJ, Folta KM. A Review of Photoperiodic Flowering Research in Strawberry (Fragaria spp.) CRC. Crit. Rev. Plant Sci. 2010;29:1–13. doi: 10.1080/07352680903436259. [DOI] [Google Scholar]
- 7.Heide OM, Stavang JA, Sønsteby A. Physiology and genetics of flowering in cultivated and wild strawberries – a review. J. Hortic. Sci. Biotechnol. 2013;88:1–18. doi: 10.1080/14620316.2013.11512930. [DOI] [Google Scholar]
- 8.Hancock JF, et al. Utilizing wild Fragaria virginiana in strawberry cultivar development: Inheritance of photoperiod sensitivity, fruit size, gender, female fertility and disease resistance. Euphytica. 2002;126:177–184. doi: 10.1023/A:1016309724998. [DOI] [Google Scholar]
- 9.Savini G, Giorgi V, Scarano E, Neri D. Strawberry plant relationship through the stolon. Physiol. Plant. 2008;134:421–429. doi: 10.1111/j.1399-3054.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 10.Heide OM. Photoperiod and Temperature Interactions in Growth and Flowering of Strawberry. Physiol. Plant. 1977;40:21–26. doi: 10.1111/j.1399-3054.1977.tb01486.x. [DOI] [Google Scholar]
- 11.Hytönen T, Palonen P, Mouhu K, Junttila O. Crown branching and cropping potential in strawberry (Fragaria x ananassaxananassa Duch.) can be enhanced by daylength treatments. J. Hortic. Sci. Biotechnol. 2004;79:466–471. doi: 10.1080/14620316.2004.11511791. [DOI] [Google Scholar]
- 12.Sønsteby A, Heide OM. Long-day control of flowering in everbearing strawberries. J. Hortic. Sci. Biotechnol. 2007;82:875–884. doi: 10.1080/14620316.2007.11512321. [DOI] [Google Scholar]
- 13.Bradford E, Hancock JF, Warner RM. Interactions of temperature and photoperiod determine expression of repeat flowering in strawberry. J. Amer. Soc. Hort. Sci. 2010;135:102–107. doi: 10.21273/JASHS.135.2.102. [DOI] [Google Scholar]
- 14.Konsin M, Voipio I, Palonen P. Influence of photoperiod and duration of short-day treatment on vegetative growth and flowering of strawberry (Fragaria x ananassa Duch.) J. Hortic. Sci. Biotechnol. 2001;76:77–82. doi: 10.1080/14620316.2001.11511330. [DOI] [Google Scholar]
- 15.Castro, P. et al. Genetic mapping of day-neutrality in cultivated strawberry. Mol. Breed. 35 (2015).
- 16.Samad, S. et al. Additive QTLs on three chromosomes control flowering time in woodland strawberry (Fragaria vesca L.). Hortic. Res. 4 (2017). [DOI] [PMC free article] [PubMed]
- 17.Weebadde CK, et al. Using a linkage mapping approach to identify QTL for day-neutrality in the octoploid strawberry. Plant Breed. 2008;127:94–101. [Google Scholar]
- 18.Brown T, Wareing PF. The genetical control of the everbearing habit and three other characters in varieties of fragaria vesca. Euphytica. 1965;14:97–112. [Google Scholar]
- 19.Bringhurst RS, Ahmadi H, Voth V. Inheritance of the day-neutral trait in strawberries. Acta Hortic. 1989;265:35–42. doi: 10.17660/ActaHortic.1989.265.2. [DOI] [Google Scholar]
- 20.Shaw, D. V. & Famula, T. R. Complex segregation analysis of day-neutrality in domestic strawberry (Fragaria × ananassa Duch.). Euphytica145, 331–338 (2005).
- 21.Sugimoto T, et al. Detection of RAPD markers linked to the everbearing gene in Japanese cultivated strawberry. Plant Breed. 2005;124:498–501. doi: 10.1111/j.1439-0523.2005.01144.x. [DOI] [Google Scholar]
- 22.Shaw DV. Heterogeneity of Segregation Ratios from Selfed Progenies Demonstrate Polygenic Inheritance for Day Neutrality in Strawberry (Fragaria × ananassa Duch.) J. AMER. SOC. HORT. SCI. 2003;128:504–507. doi: 10.21273/JASHS.128.4.0504. [DOI] [Google Scholar]
- 23.Hirakawa HI, et al. Dissection of the Octoploid Strawberry Genome by Deep Sequencing of the Genomes of Fragaria Species. DNA Res. 2014;21:169–181. doi: 10.1093/dnares/dst049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Sevilla, J. F. et al. Diversity arrays technology (DArT) marker platforms for diversity analysis and linkage mapping in a complex crop, the octoploid cultivated strawberry (Fragaria × ananassa). PLoS One10 (2015). [DOI] [PMC free article] [PubMed]
- 25.Tennessen JA, Govindarajulu R, Ashman TL, Liston A. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 2014;6:3295–3313. doi: 10.1093/gbe/evu261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat. Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lerceteau-Köhler E, Guérin G, Laigret F, Denoyes-Rothan B. Characterization of mixed disomic and polysomic inheritance in the octoploid strawberry (Fragaria x ananassa) using AFLP mapping. Theor. Appl. Genet. 2003;107:619–628. doi: 10.1007/s00122-003-1300-6. [DOI] [PubMed] [Google Scholar]
- 28.Sargent DJ, et al. A genetic linkage map of the cultivated strawberry (fragaria × ananassa) and its comparison to the diploid fragaria reference map. Mol. Breed. 2009;24:293–303. doi: 10.1007/s11032-009-9292-9. [DOI] [Google Scholar]
- 29.Sargent DJ, et al. A microsatellite linkage map for the cultivated strawberry (Fragaria x ananassa) suggests extensive regions of homozygosity in the genome that may have resulted from breeding and selection. Theor. Appl. Genet. 2012;124:1229–1240. doi: 10.1007/s00122-011-1782-6. [DOI] [PubMed] [Google Scholar]
- 30.Spigler R, Lewers K, Johnson AL, Ashman TL. Comparative Mapping Reveals Autosomal Origin of Sex Chromosome in Octoploid Fragaria virginiana. J. Hered. 2010;101:S107–S117. doi: 10.1093/jhered/esq001. [DOI] [PubMed] [Google Scholar]
- 31.Zorrilla-Fontanesi Y, et al. Development and bin mapping of strawberry genic-SSRs in diploid Fragaria and their transferability across the Rosoideae subfamily. Mol. Breed. 2011;27:137–156. doi: 10.1007/s11032-010-9417-1. [DOI] [Google Scholar]
- 32.Rousseau-Gueutin M, et al. Comparative genetic mapping between octoploid and diploid fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics. 2008;179:2045–2060. doi: 10.1534/genetics.107.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sooriyapathirana SS, et al. Identification of QTL associated with flower and runner production in octoploid strawberry (Fragaria × ananassa) J. Berry Res. 2015;5:107–116. doi: 10.3233/JBR-150095. [DOI] [Google Scholar]
- 34.Perrotte J, et al. Narrowing down the single homoeologous FaPFRU locus controlling flowering in cultivated octoploid strawberry using a selective mapping strategy. Plant Biotechnol. J. 2016;14:2176–2189. doi: 10.1111/pbi.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honjo M, et al. Simple sequence repeat markers linked to the everbearing flowering gene in long-day and day-neutral cultivars of the octoploid cultivated strawberry Fragaria x ananassa. Euphytica. 2016;209:291–303. doi: 10.1007/s10681-015-1626-6. [DOI] [Google Scholar]
- 36.Isobe SN, et al. Construction of an Integrated High Density Simple Sequence Repeat Linkage Map in Cultivated Strawberry (Fragaria x ananassa) and its Applicability. DNA Res. 2013;20:79–92. doi: 10.1093/dnares/dss035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edger PP, et al. Single-molecule sequencing and optical mapping yields an improved genome of woodland strawberry (Fragaria vesca) with chromosome-scale contiguity. Gigascience. 2018;7:1–7. doi: 10.1093/gigascience/gix124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirasawa K, et al. A high-density SNP genetic map consisting of a complete set of homologous groups in autohexaploid sweetpotato (Ipomoea batatas) Sci. Rep. 2017;7:1–8. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia AAF, et al. SNP genotyping allows an in-depth characterisation of the genome of sugarcane and other complex autopolyploids. Sci. Rep. 2013;3:3399. doi: 10.1038/srep03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davik J, et al. A ddRAD based linkage map of the cultivated Strawberry, Fragaria xananassaxananassa. PLoS One. 2015;10:1–10. doi: 10.1371/journal.pone.0137746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vining KJ, et al. Genotyping-by-sequencing enables linkage mapping in three octoploid cultivated strawberry families. PeerJ. 2017;5:e3731. doi: 10.7717/peerj.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Raskin L, Samuels DC, Shyr Y, Guo Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics. 2015;31:318–323. doi: 10.1093/bioinformatics/btu668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costes, E. et al. Bud structure, position and fate generate various branching patterns along shoots of closely related Rosaceae species: a review. Front. Plant Sci. 5 (2014). [DOI] [PMC free article] [PubMed]
- 44.Mookerjee, S. Genetics of remontancy in octoploid strawberry (Fragaria × ananassa). (Michigan State University, 2012).
- 45.van Dijk T, et al. Genomic rearrangements and signatures of breeding in the allo-octoploid strawberry as revealed through an allele dose based SSR linkage map. BMC Plant Biol. 2014;14:1–16. doi: 10.1186/1471-2229-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YR, Lee J. A genetic linkage map of allo-octoploid strawberry (Fragaria × ananassa Duch.) using SNP markers. Korean J. Breed. Sci. 2017;49:119–128. doi: 10.9787/KJBS.2017.49.3.119. [DOI] [Google Scholar]
- 47.Tamba CL, Ni Y-L, Zhang Y-M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput. Biol. 2017;13:e1005357. doi: 10.1371/journal.pcbi.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S-B, et al. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016;6:19444. doi: 10.1038/srep19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wen Y-J, et al. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2018;19:700–712. doi: 10.1093/bib/bbw145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, et al. pLARmEB: integration of least angle regression with empirical Bayes for multilocus genome-wide association studies. Heredity (Edinb). 2017;118:517. doi: 10.1038/hdy.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwata H, et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012;69:116–125. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- 52.Koskela EA, et al. Mutation in TERMINAL FLOWER1 Reverses the Photoperiodic Requirement for Flowering in the Wild Strawberry Fragaria vesca. Plant Physiol. 2012;159:1043–1054. doi: 10.1104/pp.112.196659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koskela EA, et al. TERMINAL FLOWER1 is a breeding target for a novel everbearing trait and tailored flowering responses in cultivated strawberry (Fragaria × ananassa Duch.) Plant Biotechnol. J. 2016;14:1852–1861. doi: 10.1111/pbi.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mouhu K, et al. Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biol. 2009;9:1–16. doi: 10.1186/1471-2229-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakano Y, Higuchi Y, Yoshida Y, Hisamatsu T. Environmental responses of the FT/TFL1 gene family and their involvement in flower induction in Fragaria × ananassa. J. Plant Physiol. 2015;177:60–66. doi: 10.1016/j.jplph.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of ¯owering in rice (a short-day plant) Plant J. 2000;22:391–399. doi: 10.1046/j.1365-313X.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- 57.Su YH, Liu YB, Zhang XS. Auxin-cytokinin interaction regulates meristem development. Mol. Plant. 2011;4:616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somssich M, Je BIl, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 59.Sotta N, Shantikumar L, Sakamoto T, Matsunaga S, Fujiwara T. TPR5 is involved in directional cell division and is essential for the maintenance of meristem cell organization in Arabidopsis thaliana. J. Exp. Bot. 2016;67:2401–2411. doi: 10.1093/jxb/erw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Caruana JC, Sittmann JW, Wang W, Liu Z. Suppressor of Runnerless Encodes a DELLA Protein that Controls Runner Formation for Asexual Reproduction in Strawberry. Mol. Plant. 2018;11:230–233. doi: 10.1016/j.molp.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Browning SR, Browning BL. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin). 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voorrips RE. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 68.Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. J. Infect. Dis. 2011;204(Suppl):198–199. [Google Scholar]
- 69.Churchill G, Doerge R. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ddRAD-seq raw reads generated in this study were deposited into Sequence Read Archive (SRA) under NCBI accession PRJNA478299 accessible from (https://www.ncbi.nlm.nih.gov/sra/PRJNA478299).