Abstract
We previously developed red lettuce (Lactuca sativa L.) cultivars with high flavonoid and phenolic acid content and demonstrated their anti-diabetic effect. Here we report on developing three fertile and true-breeding lettuce lines enriched with flavonoids with reported beneficial health effects. These lines were identified in a segregating population of EMS-mutagenized red lettuce and characterized biochemically and genetically. Change in red coloration was used as a visual indicator of a mutation in a flavonoid pathway gene, leading to accumulation of flavonoid precursors of red anthocyanins. Pink-green kaempferol overproducing kfoA and kfoB mutants accumulated kaempferol to 0.6–1% of their dry weight, higher than in any vegetable reported. The yellow-green naringenin chalcone overproducing mutant (nco) accumulated naringenin chalcone, not previously reported in lettuce, to 1% dry weight, a level only observed in tomato peel. Kfo plants carried a mutation in the FLAVONOID-3′ HYDROXYLASE (F3′H) gene, nco in CHALCONE ISOMERASE (CHI). This work demonstrates how non-GMO approaches can transform a common crop plant into a functional food with possible health benefits.
Introduction
Fruits and vegetables are good dietary sources of phenolics, ubiquitous phytochemicals that include flavonoids and phenolic acids1. Epidemiological studies suggested that diets high in fruits and vegetables confer beneficial effects on chronic metabolic and cardiovascular diseases2–4. With some exceptions5, these benefits were confirmed by meta-analyses of cohort studies6–8. However, the average dietary flavonoid consumption in Europe and the US may be too low to confer health effects9,10.
Lettuce (Lactuca sativa, family Asteraceae) is a crop domesticated more than ten thousand years ago11. It is the third most commonly consumed vegetable in the US after potato and tomato, with US per capita consumption estimated at 11.7 kg/year12, and is considered a good source of fiber, iron, folic acid and vitamin C13. Common lettuce phenolics are caffeic acid derivatives, predominantly chicoric, chlorogenic, caffeoyltartaric and caffeoylmalic acids; and flavonol glycosides, predominantly quercetin 3-O-malonylglucoside, quercetin 3-O-glucoside and quercetin 3-O-glucuronide14,15. In addition, red varieties contain the anthocyanin cyanidin 3-O-malonylglucoside14,15. Flavonoid and total phenolics levels vary widely between lettuce types: crisphead varieties, commonly consumed in the US, have low levels of phenolics, while red leaf and red oak lettuces have the highest levels13,15. Thus, there is potential to develop cultivars with enhanced nutritional or functional value (see e.g.16). Earlier we have developed three Rutgers Scarlet Lettuce (RSL) lines from existing red cultivars using tissue culture selection for deep purple color, an indicator of high anthocyanin content17. In addition to the anthocyanin cyanidin 3-O-malonylglucoside, RSL lines accumulated high levels of common phenolics reported in lettuce14,15,18, such as quercetin glycosides and chlorogenic acids, resulting in a total phenolic content of >9% dry leaf weight, the highest reported in the literature17. RSL leaf and extract showed in vivo anti-diabetic effect in a mouse model of type 2 diabetes17,18. Specifically, daily oral administration of RSL extract to obese C57BL/6 mice kept on High Fat Diet (HFD) for 28 days resulted in improved oral glucose tolerance and decreased liver lipid levels compared to control17,18. Thirteen-week diet supplementation with RSL powder resulted in improved glucose tolerance in obese C57BL/6 mice on HFD, even though other measured physiological parameters did not change19. In another study, 4-week supplementation with red leaf lettuce powder resulted in decreased levels of total blood cholesterol and triglycerides in HFD-fed mice20.
The aim of this study was to develop fertile and true-breeding lettuce varieties enriched in specific flavonoids beneficial for human health, such as kaempferol and naringenin chalcone, present only in small or undetectable amounts in wild type lettuce, as even structurally similar flavonoids can produce markedly different health effects21,22.
As flavonoid biosynthesis genes have been characterized in Arabidopsis thaliana (Fig. 1) for review, see23–25, we designed primers based on lettuce homologs of Arabidopsis genes, and identified mutations in lettuce CHALCONE ISOMERASE (CHI) and FLAVONOID-3′ HYDROXYLASE (F3′H) genes responsible for the novel phenotypes. These mutations increased the levels of targeted flavonoids to levels higher than in any other vegetable. Recent sequencing of the lettuce genome26 and identification of loci associated with the flavonoid biosynthesis pathway in lettuce11 allowed us to compare the CHI and F3′H sequences we determined to those putatively identified by Zhang et al.11.
Figure 1.
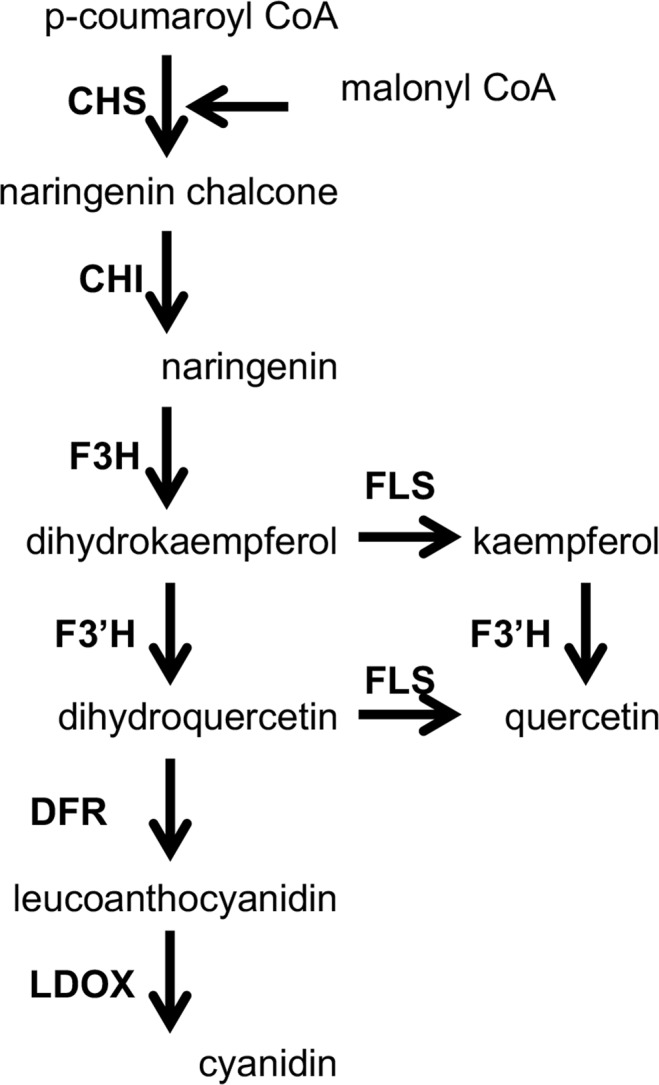
Enzymes of the flavonoid biosynthesis pathway in A. thaliana adapted from25. All enzymes but FLS are coded by a single gene in A. thaliana. Genes coding for the enzymes responsible for glycosylation, acylation, and intracellular transport of flavonoids are not shown. Abbreviations: CHS, CHALCONE SYNTHASE; CHI, CHALCONE ISOMERASE; F3H, FLAVANONE 3-HYDROXYLASE; F3′H, FLAVONOID 3′-HYDROXYLASE; FLS, FLAVANOL SYNTHASE; DFR, DIHYDROFLAVONOL REDUCTASE; LDOX, LEUCOANTHOCYANIN DEHYDROGENASE.
Results
Isolation of flavonoid biosynthesis mutants
An ethyl methanesulfonate (EMS)-mutagenized cv. Firecracker red leaf lettuce segregating population derived from seeds of self-pollinated mutagenized plants was screened for anthocyanin (cyanidin 3-O-malonylglucoside) loss manifested by changes in color. 1522 mutagenized (M1) plants were grown from seed mutagenized by 0.10 or 0.15% EMS, selfed, and the mature dry inflorescences collected to obtain the M2 segregating population. 136 M1 lines were sterile. Seed from the remaining 1386 M1 lines were planted (12 seeds per plant, if available) in growth chambers equipped with cool fluorescent lights emitting high levels of both photosynthetically active radiation and ultraviolet (UV); lighting conditions known to induce strong anthocyanin accumulation, and, thus, red color. Forty-three lines harboring color variants were identified visually. Methanolic extracts of the twenty most prominent color mutants were biochemically profiled using an Ultra Performance Liquid Chromatography - Tandem Mass Spectrometer (UPLC-MS/MS) system. Three mutants were selected for further studies.
Pink-green kaempferol overproducer kfoA had high levels of kaempferol glycosides (mostly kaempferol 3-O-malonylglucoside, low amounts of kaempferol 3-O-glucoside and kaempferol 3-O-glucuronide) but lacked quercetin or cyanidin derivatives. Another kaempferol overproducer, kfoB, accumulated the same kaempferol derivatives as kfoA, and had low but detectable cyanidin and quercetin glycoside content. The yellow-green naringenin chalcone overproducer nco line had high levels of glycosylated compounds (hexosides and malonylhexoside) (Supplementary Fig. S1g,h,i), with a shared aglycone ion of m/z 273 [M + H] (Supplementary Fig. S1m,n), which corresponds to the isomers naringenin chalcone (Supplementary Fig. S1l) and naringenin (Supplementary Fig. S1j). However, naringenin and naringenin chalcone have characteristically different UV spectra, naringenin chalcone having its absorption maximum at 365 nm (Supplementary Fig. S1q), and naringenin at 289 nm (Supplementary Fig. S1o). Additionally, the UV absorbance spectra of naringenin glycosides and naringenin chalcone glycosides are similar to the spectra of their aglycones27 and Supplementary Fig. S1p. Both glycosides in nco lettuce had the characteristic UV absorbance spectra of naringenin chalcone (Supplementary Fig. S1r,s). Based on these data and on genetic data below, we concluded that nco lettuce accumulated naringenin chalcone glycosides. Nco lacked detectable kaempferol or cyanidin derivatives and had greatly reduced quercetin level compared to cv. Firecracker. Supplementary Fig. S2 shows major peaks of cv. Firecracker, kfo and nco extract chromatograms. Accumulation of high levels of kaempferol or naringenin chalcone is a novel trait in lettuce28, therefore, kfoA, kfoB and nco were further characterized. Figure 2 shows representative photos of 15-week old cv. Firecracker, kfoA, kfoB and nco plants grown under UV-emitting, cool fluorescent lights. Under these conditions, Firecracker plants were deep red (Fig. 2a,b), kfoA (Fig. 2c,d) and kfoB (Fig. 2e,f) were pink-green, and nco (Fig. 2g,h) were yellow-green color. All mutants grew slower than wild type cv. Firecracker plants under fluorescent lights (UV light intensity 0.4 ± 0.1 mol/m2d), a trait previously described in A. thaliana flavonoid biosynthesis mutants29,30.
Figure 2.
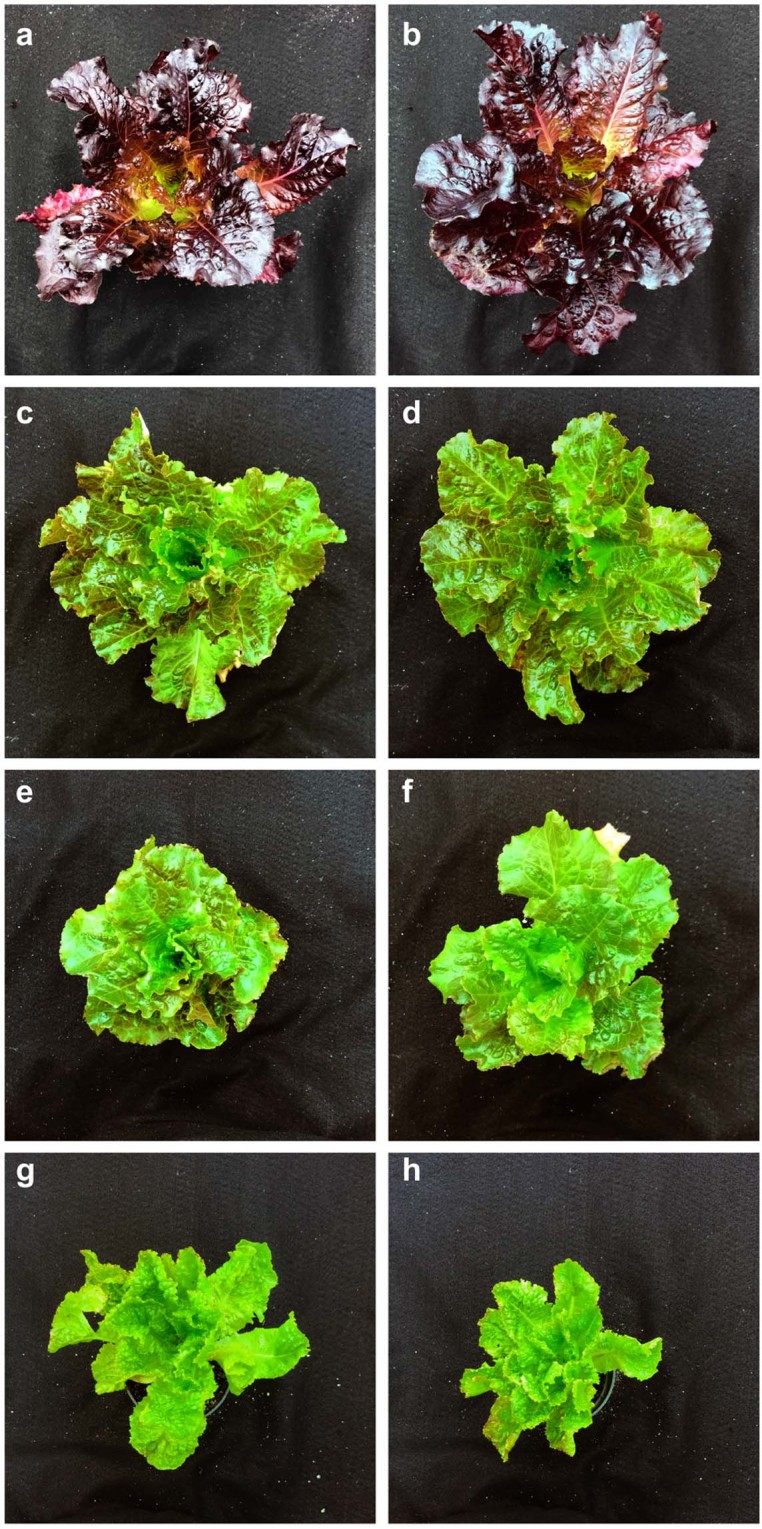
Representative 15-week old cv. Firecracker (wild type) and flavonoid biosynthesis mutants, grown under cool fluorescent lights. Two representative plants per line are shown photographed to scale. (a,b) cv. Firecracker; (c,d), kfoA (e,f); kfoB; (g,h), nco.
KfoA and nco accumulate high amounts of flavonoid compounds missing from parental line cv. Firecracker
KfoA, kfoB and nco mutants and wild type cv. Firecracker were grown under identical conditions illuminated by cool fluorescent lights and subjected to further UPLC-MS/MS analysis. Leaves were harvested from 18-week old plants, lyophilized, mixed with HCl-acidified methanol, and subjected to acid hydrolysis, based on the method of Hertog et al.31. This treatment results in the removal of glycosylation from all flavonoids and chalcones, allowing for the quantification of aglycones, or, in case of nco, their derivatives using UPLC-MS/MS (Table 1). Supplementary Fig. S3 shows representative chromatograms of cv. Firecracker, kfoA, kfoB and nco acid hydrolyzed extracts.
Table 1.
Flavonoid aglycones in 18-week old red cv. Firecracker, kfoA, kfoB and nco lettuce grown under cool fluorescent lights. Acid hydrolysis was used to convert compounds to aglycones.
| Lines | Cyanidin | Quercetin | Kaempferol | Naringenin | Pelargonidin | Total polyphenol |
|---|---|---|---|---|---|---|
| cv. Firecracker | 5.3 ± 2.5 (57.49 ± 26.41) | 26.0 ± 2.5 (281.53 ± 47.83) | <0.02 BQ | <0.04 BQ | <0.2 BQ | 45.00 ± 8.51 |
| kfoA | <0.02 BQ (<0.18) | <0.02 BQ (<0.18) | 10.9 ± 2.9 (102.62 ± 32.47) | <0.04 BQ | 0.30 ± 0.1 (3.38 ± 2.18) | 23.33 ± 7.45 |
| kfoB | 0.14 ± 0.01 (1.80 ± 0.29) | 0.3 ± 0.2 (3.79 ± 2.15) | 6.4 ± 1.5 (83.41 ± 25.31) | <0.04 BQ | 0.58 ± 0.2 (7.42 ± 2.41) | 44.87 ± 3.75 |
| nco | <0.02 BQ (<0.18) | 0.6 ± 0.2 (5.59 ± 1.34) | <0.02 BQ (<0.18) | 10.4 ± 2.5 (92.73 ± 22.81) | <0.02 BQ | 36.29 ± 8.57 |
Mean mg compound/ g dry leaf weight, and, in parenthesis, as mg compound/ 100 g fresh leaf weight ± standard deviation is shown for cv. Firecracker (n = 6), kfoA, kfoB and nco (n = 10). Pelargonidin was quantified in cyanidin equivalents. Total polyphenol content is calculated as gallic acid (GA) equivalent in mg GA/ g dry leaf weight. BQ, below quantification limits. Naringenin chalcone glycosides were converted to naringenin during acid hydrolysis.
The anthocyanin cyanidin and the flavonol quercetin were detected in cv. Firecracker extracts, as expected in red leaf lettuce14,15,17. Additionally, low levels of pelargonidin were observed (Table 1). In kfoA plants cyanidin and quercetin were not detectable. Instead, accumulation of the flavonol kaempferol and the anthocyanidin pelargonidin was observed. While kaempferol has been reported in lettuce28,32–34, kfoA plants accumulated >10 mg kaempferol/ g dry weight, or ~103 mg kaempferol/100 g fresh weight, two orders of magnitude higher than previously reported. Additionally, kfoA and kfoB plants contained more pelargonidin (0.33 and 0.60 mg pelargonidin/g dry weight), the predominant anthocyanin in strawberries35, than cv. Firecracker (<0.2 mg pelargonidin/g dry weight). KfoB plants accumulated >6 mg kaempferol/g dry weight, lower than kfoA. However, they accumulated more pelargonidin than kfoA, and contained quantifiable cyanidin and quercetin. To our best knowledge, this is the first report on the accumulation of pelargonidin in lettuce leaves.
Nco acid hydrolyzed extracts lacked cyanidin, kaempferol or pelargonidin, but contained >10 mg naringenin/g dry weight, and, on average, 0.6 mg quercetin/g dry weight. As naringenin chalcone glycosides, but not naringenin glycosides were observed in non-hydrolyzed nco extracts (see previous section), we tested the effect of acid hydrolysis on pure naringenin chalcone and observed full conversion to naringenin. Therefore, the levels of naringenin in hydrolyzed extracts of nco correspond to the levels of naringenin chalcone glycosides in the plant. Small amounts of quercetin observed in nco were also likely derived from naringenin formed spontaneously in planta from naringenin chalcone, as naringenin chalcone can spontaneously isomerize by C ring closure to naringenin36. To our best knowledge, naringenin chalcone has not been described in lettuce before.
Total polyphenol levels were measured in ten plants per line, using a modified Folin-Ciocalteu assay17. Wild type cv. Firecracker and kfoB both had 45 mg gallic acid equivalent/g dry weight. KfoA and nco plants had somewhat lower total polyphenol levels: 23 and 36 mg gallic acid equivalent/g dry weight, respectively (Table 1).
Nco is a chalcone isomerase mutant
The nco flavonoid profile (Table 1) resembled A. thaliana tt5 null mutants, which have nonfunctional CHALCONE ISOMERASE (CHI), an enzyme that converts naringenin chalcone to naringenin29,37. Therefore, primers designed based on lettuce Expressed Sequence Tags (ESTs) homologous to the A. thaliana CHI gene (TAIR AT3G55120) were used to amplify the full coding sequence (CDS) of the putative lettuce CHI from cDNA in cv. Firecracker and nco. The wild type cv. Firecracker CHI (CHI+, NCBI MG981123) was predicted to code for a 235-amino acid protein, and the CDS was identical to XM_023891334, a predicted CHALCONE ISOMERASE from green crisphead lettuce cv. Salinas. Additionally, it was identical to LG9_805610, identified as the only CHI expressed (of two putative CHI genes) in the lettuce genome11. Nco plants were homozygous for an allele (chi1, NCBI MG981124) that harbors a premature stop codon caused by a G to A mutation in codon 120, truncating the CHI enzyme. The CHI1 truncated protein lacks two conserved residues of the naringenin binding cleft, as well as a residue of the active site hydrogen bond network38; therefore, it is expected to be nonfunctional.
Of the M2 population, one nco mutant and 4 wild type siblings were genotyped. The mutant was homozygous for the chi1 allele, whereas wild type plants were heterozygous or homozygous for CHI+ allele. The M2 mutant and its wild type red siblings were selfed, and segregation ratios in M3 individuals were observed. In addition, selfed seed from two M3 mutants were planted. (Table 2; Supplementary Table S1 for segregation ratios of individual parents). Homozygous chi1 mutants always produced yellow-green offspring, heterozygotes produced yellow-green and red offspring, and homozygous CHI+ plants always produced red offspring, indicating that the mutant allele is recessive and responsible for the observed phenotype.
Table 2.
Phenotype segregation ratios in kfoA, kfoB and nco lines. Summary of all lines is shown; segregation data for individual lines is shown in Supplementary Table S1.
| Line | Parent genotype | Number of mutant offspring | Number of wild type offspring | Total number of offspring | Percentage of mutants |
|---|---|---|---|---|---|
| kfoA | +/M | 32 | 53 | 85 | 37.6% |
| +/+ | n/a | n/a | n/a | n/a | |
| M/M | 385 | 0 | 385 | 100.0% | |
| kfoB | +/M | 14 | 46 | 60 | 23.3% |
| +/+ | 0 | 38 | 38 | 0.0% | |
| M/M | 67 | 0 | 67 | 100.0% | |
| nco | +/M | 18 | 46 | 64 | 28.1% |
| +/+ | 0 | 27 | 27 | 0.0% | |
| M/M | 112 | 0 | 112 | 100.0% |
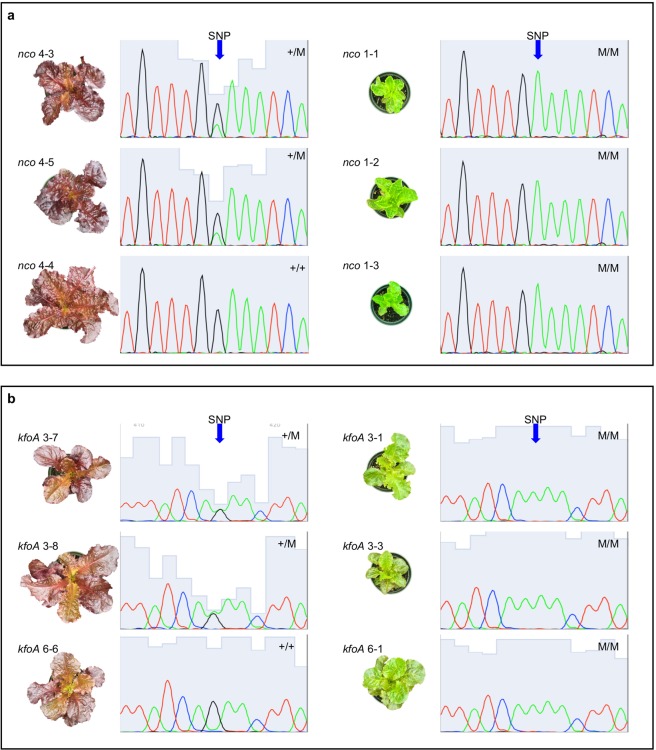
We then genotyped 5 mutants and 14 wild-type siblings from self-pollinated offspring of nco M2 plants (M3 generation) and found that only yellow-green mutants were homozygous for chi1 (Fig. 3a). Additionally, we amplified the full CDS of the putative lettuce F3H gene in cv. Firecracker and nco from cDNA using primers designed based on lettuce ESTs homologous to the A. thaliana FLAVANONE-3-HYDROXYLASE (F3H) gene (TAIR AT3G51240). F3H converts naringenin to dihydrokaempferol (Fig. 1), and, in A. thaliana, f3h mutants accumulate a mix of chalcones, flavonols and anthocyanins39. As in Arabidopsis, F3H in lettuce is a single-copy gene11. The F3H coding sequence of nco was found to be identical to that of cv. Firecracker. Our data suggest that though two putative CHI copies exist in the lettuce genome11, losing both functional copies of the LG9_805610 gene leads to low anthocyanin (yellow-green) phenotype, and that nco is a chi mutant.
Figure 3.
Phenotype of kfo and nco plants is determined by F3′H and CHI genotypes. (a) Representative photos, to scale, and genotyping chromatograms of wild type and yellow-green nco mutants. CHI homozygous recessive mutant genotype results in mutant yellow-green phenotype. Heterozygous and homozygous wild type genotypes result in wild type red phenotype. (b) Representative photos, to scale, and genotyping chromatograms of wild type and pink-green mutant kfoA line plants. F3′H splice site homozygous mutant genotype results in mutant pink-green phenotype. Heterozygous and homozygous wild type genotypes result in wild type red phenotype. Genotypes: +/+, homozygous wild type; +/M, heterozygous mutant; M/M, homozygous mutant. Note single peak in homozygotes, and double peak in heterozygotes at SNP site. Fifteen-week-old plants are shown, grown under cool fluorescent lights in identical conditions.
KfoA and kfoB are flavonoid 3′-hydroxylase mutants
The kfoA flavonoid profile (Table 1) resembled Arabidopsis thaliana tt7 null mutants, which have a nonfunctional FLAVONOID-3′ HYDROXYLASE (F3′H) enzyme40,41. Therefore, primers designed based on lettuce ESTs homologous to the A. thaliana F3′H gene (TAIR AT5G07990) were used to amplify the full CDS of the putative lettuce F3′H in cv. Firecracker, kfoA and kfoB. The wild type cv. Firecracker F3′H (F3′H+, NCBI MG981125) was a gene containing three exons, predicted to code for a 512-amino acid protein, and was identical to XM_023887166, a predicted FLAVONOID 3′-MONOOXYGENASE-LIKE gene from green crisphead lettuce cv. Salinas. Additionally, it was identical to LG5_471950, one of five putative F3′H genes in the lettuce genome identified by Zhang et al.11.
KfoA plants were homozygous for an allele (f3′h1, NCBI MG981126) harboring a G to A mutation in the splice acceptor site of intron 2, while kfoB plants were homozygous for an allele (f3′h2, NCBI MG981127) harboring a premature stop codon caused by a C to T mutation in codon 233. Translated kfo F3′H proteins harbor the CR1 active site (amino acids 171–186) responsible for the hydroxylating activity, but lack three substrate recognition sites as well as the EXXR motif necessary for core stabilization42, thus, it is expected that both kfoA and kfoB mutant F3′H proteins are nonfunctional. Of the M2 population, one kfoA mutant and seven wild type siblings were genotyped, as well as one kfoB and five wild type siblings. Mutants were homozygous for f3′h1 or f3′h2, while wild type plants were heterozygous or homozygous for the wild type F3′H+. Mutant plants and wild type red siblings were selfed, and segregation ratios in M3 and M4 individuals were observed (Table 2; Supplementary Table S1 for segregation ratios of individual parents). Homozygous f3′h mutants always produced pink-green offspring, heterozygotes produced pink-green and red offspring, and homozygous wild type plants always produced red offspring, indicating that the mutant alleles are recessive and responsible for the observed phenotype. Genotyping 15 mutants and 5 wild-type siblings from self-pollinated offspring of kfoA M2 plants (M3 generation), we found that only pink-green mutants were homozygous for f3′h1 (Fig. 3b). Our data suggest that kfoA and kfoB are mutants for one of the five F3′H gene copies in lettuce (LG5_471950 in11), and that this gene is predominantly responsible for the synthesis of dihydroquercetin, a precursor in anthocyanin biosynthesis in wild type cv. Firecracker.
Discussion
Nco lettuce plants have a characteristic yellow-green leaf color due to the accumulation of yellow-colored naringenin chalcone glycosides and the lack of red anthocyanins. Naringenin chalcone has not been reported in lettuce before. Tomato (Solanum lycopersicum) skin is the best-known food source of this compound, where it accumulates up to 1% dry weight43, a level similar to that of nco lettuce. Naringenin chalcone is anti-inflammatory, anti-allergic (e.g.44,45) and anti-obesity46 in vitro and in vivo, and was found to improve symptoms of perennial allergic rhinitis in a clinical trial47. Therefore, nco lettuce could be a useful dietary source for naringenin chalcone, with one US leaf lettuce serving of 85 g containing ~79 mg of the compound.
The color phenotype in nco lettuce is caused by a nonsense mutation in the CHI gene. To our knowledge, nco is the first chi mutant in lettuce. While CHI is ubiquitous in higher plants, chi mutants have been characterized from just a handful of species, with individual flavonoids from these mutants not quantified. Chi mutants have been described in ornamental flowers such as Petunia hybrida48, Callistephus chinensis49 and Dianthus caryophillus50, in crops such as barley (Hordeum vulgare)51,52, rice (Oryza sativa)53 and onion (Allium cepa)54 and inA. thaliana29,37,55. In all species, the chi mutant phenotype results in yellowish tissues: hull and internodes in rice53, bulb color in onion54, petals in C. chinensis49 and D. caryophillus50, seed coat in A. thaliana56 and pollen in P. hybrida48. P. hybrida chi mutants accumulate naringenin chalcone as aglycone48, while C. chinensis49, D. caryophillus50 and barley (Hordeum vulgare)51 chi mutants accumulate naringenin chalcone 2′-glucoside (isosalipurposide). Detailed metabolome analysis of A. thaliana chi mutants revealed the presence of multiple naringenin chalcone glycosides37, while in chi onion54 and rice53 the compound responsible for the yellowish or golden color was not identified. We found that nco lettuces accumulate naringenin chalcone hexoside and malonylhexoside, but not the aglycone, similarly to most chi mutants.
Like nco lettuce, chi mutants of A. thaliana30,37, P. hybrida48, C. chinensis49 and D. caryophyllus50 had low but detectable levels of flavonols. In Arabidopsis, Peer et al.39 hypothesized that spontaneous isomerisation of naringenin chalcone in planta to naringenin, the substrate of the next enzyme in the anthocyanidin biosynthesis pathway, F3H, was responsible for the presence of flavonols.
KfoA and kfoB lettuce plants are pink-green and accumulate high levels of kaempferol glycosides. Kaempferol has well-documented anti–diabetic, pancreatic β-cell protecting and anti-inflammatory effects57–59. It has been reported in lettuce; however, quercetin is the dominant flavonol in most cultivars28,32–34. Reported kaempferol levels in lettuce range from 0.0–2.36 mg/100 g fresh weight28,32–34, while kfoA and kfoB lettuce has 103 and 83 mg /100 g fresh weight, respectively, two orders of magnitude higher. Unlike naringenin chalcone, kaempferol is a ubiquitous flavonoid, described from over 400 species (for review, see57). However, kaempferol accumulation in kfo lettuces is higher than the amounts reported in vegetables considered high in kaempferol, e.g. endive (Cichorium endiva), 1.5–9.560; leek (Allium porrum), 1.1–5.660 and 11.834; shallot (Allium fistulosum), 11.734; potherb mustard (Brassica juncea), 48.234; kale (Brassica oleracea var. acephala), 5.134, 21.160 and 47.061; broccoli (Brassica oleracea var. italica), 2.134 and 6.061; choi sum cabbage (Brassica rapa var. parachinensis), 2.0–3.732; turnip tops (Brassica campestris), 3.1–6.460; spinach (Spinacia oleracea), 4.9–9.032; radish root (Raphanus sativus), 0–4.134; toona leaf (Toona sinensis), 41.734 and 60.433; Chinese boxthorn shoot (Lycium barbarum), 44.633, rocket (Eruca sativa) 36.533 and water cress (Nasturtium officinale) 35.133 mg kaempferol/100 g fresh weight. The only natural source higher in kaempferol than kfo lettuce is caper flower buds (Capparis ssp.), which have 85–295 mg kaempferol /100 g fresh weight, providing 8.5–29.5 mg kaempferol per 10 g serving62. Therefore, kfo lettuces could be valuable dietary sources of kaempferol with one US leaf lettuce serving (85 g) providing ~71–87 mg kaempferol.
Kfo phenotypes were caused by mutations in the F3′H gene: a mutation of the intron 2 splice acceptor site in kfoA, and a nonsense mutation in kfoB. In lettuce, no f3′h mutant has been described, but in a study of 240 lettuce accessions five genes were identified as F3′H, three of which were expressed and two of which (including the F3′H gene mutant in kfoA and kfoB) carried expressed Qualitative Trait Loci (eQTL) for flavonoid composition11. The lettuce f3′h phenotype is very similar to A. thaliana f3′h mutants (called tt7), which accumulate kaempferol, and the anthocyanin pelargonidin40,41 that differs from cyanidin by the lack of 3′-hydroxylation. Kfo f3′h mutant lettuces also accumulate pelargonidin, although pelargonidin levels in kfo are much lower than cyanidin levels in red parent line cv. Firecracker (Table 1). This difference suggests reduced substrate specificity of the DFR enzyme for its substrate in f3′h mutants, dihydrokaempferol, compared to its substrate in wild type lettuce, dihydroquercetin (Fig. 1). Interestingly, f3′h mutants in morning glory (Ipomoea ssp.) accumulate pelargonidin derivatives producing magenta, pink or fuschia flowers63. In carnation (D. caryophillus), f3′h mutants have pink petals, accumulating a pelargonidin glycoside, while plants with functional F3'H have purple petals accumulating a cyanidin glycoside64.
In plants, many environmental stresses trigger the accumulation of antioxidants including flavonoids and other phenolics65. Flavonoids are hypothesized to act as UV absorbers and reduce the levels of damaging reactive oxygen species66. In lettuce, exposure to UV or blue light increases flavonoid levels, but reduces yield (e.g.67–73). This effect was observed during different months in the field growth season71, and in field67,69,70 and greenhouse68 experiments, where levels of UV exposure were controlled using UV-blocking cover foils, as well as in controlled growth chambers supplemented by UV or blue light emitting LED diodes72,73. UV-induced increase in flavonoid and total phenolic content was observed across different green and red cultivars74, indicating that it is a universal phenomenon in lettuce. Armas Gutierrez75 reported that continuous exposure to cool fluorescent lights resulted in high accumulation of total phenolics, total antioxidants and total anthocyanins. Therefore, in our experiment, we replicated the growth conditions optimal for high phenolic content determined by Armas Gutierrez75.
A. thaliana flavonoid biosynthesis mutants are more sensitive to high UV29,30,76 and visible light stress77 than wild type plants. As in lettuce, wild type A. thaliana plants have a decreased rate of biomass accumulation under high UV stress compared to low UV conditions, but the effects are more severe in flavonoid biosynthesis mutants29,30. This sensitivity has been attributed to enhanced photoinhibition77 and increased lipid and protein peroxidation76,77 in mutants lacking flavonoids that absorb UV and scavenge reactive oxygen species. However, the UV sensitivity of the different A. thaliana mutants is not equal. The kaempferol-accumulating f3′h mutant is less UV sensitive than chs, chi and f3h mutants, which accumulate low levels of flavonols30. We found that nco and kfo lettuces grew somewhat slower than wild type cv. Firecracker plants under UV-emitting cool fluorescent lights, though we did not observe visible growth retardation under greenhouse conditions (natural light plus supplemental white light). Under cool fluorescent lights, kfo (f3′h) lettuce grew faster than nco (chi) but not as fast as wild type cv. Firecracker, similarly to A. thaliana f3′h and chi mutants30. Potentially, desirable high biomass and flavonoid levels could be obtained by growing nco and kfo under low UV conditions, and subjecting them to higher levels UV or blue light before harvest. 3-day supplemental UV treatment for 16 h/day has significantly increased total anthocyanin and antioxidant levels in red leaf lettuce, with no effect on the total leaf biomass73. Similarly, 6-day pre-harvest exposure to UV resulted in a 4.6x increase in total anthocyanin content and 2.3x increase in total phenolic content in red leaf lettuce field grown under UV-blocking foil70, while total biomass of these plants was not significantly different from those not exposed to UV.
In conclusion, we created and characterized flavonoid biosynthetic mutants in lettuce with potential health benefits. The modified flavonoid profile characterized by record high accumulation of kaempferol and naringenin chalcone may transform lettuce into a food with health benefits. However, animal studies and human clinical trials will be needed to confirm the health benefits of the high flavonoid lettuce varieties described here. Innovative mutagenizing and selection strategies producing higher levels of beneficial phytochemicals could be an important strategy for adding value added output traits to common crops.
Methods
EMS mutagenesis of cv. Firecracker lettuce seeds
Lettuce cv. Firecracker (Johnny’s Selected Seeds) seeds were mutagenized with 0.10 or 0.15% EMS, using the protocol in75. In short, seeds (M0) were soaked in distilled water containing 0.1% or 0.15% (v/v) EMS and incubated for 12 h at room temperature in a rotary shaker. Thereafter, the EMS solution was decanted, the seeds were washed five times with 50 ml of distilled water and dried. Mutagenized M1 seeds were planted and grown under standard greenhouse conditions at the Rutgers New Jersey Agricultural Experiment Station (NJAES) glass research greenhouse under the following settings: 25 °C/19 °C day/night temperature, 16 h light/8 h dark photoperiod with natural light supplemented with 400 W high pressure sodium lamps. Inflorescences were individually collected from 1,522 mature M1 plants, and the M2 seed was threshed, dried out in the greenhouse and placed in paper coin envelopes. The envelopes were placed in re-sealable plastic storage bags with desiccant and stored at 4 °C.
Visual screen of the segregating M2 lettuce population
Selfed and segregated M2 seeds were planted in Sun Gro Propagation Mix (Sun Gro Horticulture, Agawam, MA, USA) in plastic trays. At least 12 M2 seeds were planted if there were more than 12 seeds per M2 line. The trays were placed in growth chambers equipped with cool fluorescent lights (Sylvania F96T12/CW/VHO, Osram Sylvania, Danvers, MA, USA), providing a PAR light intensity of 26.3 mol/m2d and UV light intensity of 0.7 mol/m2d75. Growth chambers were programmed for 16 h light/8 h dark photoperiod, 18 °C/15 °C day/night temperature and 60% relative humidity. Under these conditions, wild type cv. Firecracker develops a deep red color. At 4–6 weeks, segregating putative mutants exhibiting color variations, and their siblings with wild-type phenotype were identified and transplanted to 10 cm diameter pots filled with Sun Gro Professional Growing Mix (Sun Gro Horticulture, Agawam, MA, USA). At 12–18 weeks, tissue samples were collected from putative mutants for genetic and phytochemical analysis, and the plants were transplanted to 23 cm diameter, 6 l pots (Nursery Supplies, USA) filled with Sun Gro Professional Growing Mix (Sun Gro Horticulture, Agawam, MA, USA), and placed in the Rutgers New Jersey Agricultural Experiment Station (NJAES) research greenhouse under the following conditions: 25 °C/19 °C day/night temperature, 16 h light/8 h dark photoperiod, with natural light supplemented provided by 400 W high pressure sodium lamps. Individual inflorescences were collected from mature M2 plants; the seeds (M3 generation) were dried, threshed, and stored at 4 °C under the same conditions as M2 seeds.
Growing Firecracker, kfo and nco plants for phytochemical analysis
Seeds from selfed cv. Firecracker, and from homozygous mutant kfoA, kfoB and nco plants were planted in Sun Gro Propagation Mix (Sun Gro Horticulture, Agawam, MA, USA) in plastic trays. The trays were placed in growth chambers under conditions described above. 26 days after planting, seedlings were transplanted to 10 cm-diameter pots filled with Sun Gro Professional Growing Mix (Sun Gro Horticulture, Agawam, MA, USA), and kept in the growth chamber. 130 days (18 weeks) after seeding, a tissue sample was taken from 6 cv. Firecracker, 10 kfoA, kfoB and nco plants, weighed, frozen at −80 °C, then lyophilized.
Extraction and UPLC-MS/MS analysis of flavonoids from putative flavonoid mutants
To determine the flavonoid composition of putative color mutants, extracts were prepared from lyophilized and ground leaf tissue, using the method described in18. In short, leaves were kept at −80 °C prior to lyophilization. Freeze-dried leaves were ground to a fine powder with a mortar and pestle. 50 or 100 mg lyophilized leaf powder was placed in a 15 ml plastic tube, protected from light, then, respectively, 1.5 ml or 3 ml solvent (methanol/water/acetic acid; 85:14.5:0.5 v/v), was added. The leaf powder was vortexed with the solvent for 30 sec, sonicated for 5 min, vortexed for another 30 sec, kept for 10 min at room temperature, and centrifuged at 1700 rcf for 5 min. The supernatant was decanted, then the extraction was repeated twice, and the decanted extracts pooled. The decanted solution was centrifuged at 1700 rcf for 8 min and filtered through 0.45 µm polytetrafluoroethylene (PTFE) filters (Fisher Scientific) for UPLC-MS/MS analysis.
Extracts were separated and analyzed by a UPLC-MS/MS system using the protocol described in78. Since this protocol results in the co-elution of chlorogenic acid and cyanidin 3-O-malonylglucoside, we used a modified gradient elution to separate these two compounds. For this protocol, the mobile phase consisted of two components: Solvent A (0.5% ACS grade acetic acid in double distilled de-ionized water, pH 3–3.5), and Solvent B (100% acetonitrile). The initial conditions of the gradient were 95% A and 5% B; for 20 minutes the proportion reached linearly 80% A and 20% B. Within the next 3 minutes the proportion was 5% A and 95% B, which was maintained for 4 minutes. Within the following 3 minutes the gradient was adjusted to initial conditions, and 5 additional minutes were included for equilibration before subsequent injections.
Acid hydrolysis, UPLC-MS/MS analysis and quantification of flavonoid aglycones and total polyphenol content
Lyophilized lettuce leaves from 18-week old cv. Firecracker, kfoA, kfoB and nco plants were ground using a mortar and pestle. Fifty mg leaf powder was placed in a plastic tube and subjected to acid hydrolysis, based on the method of Hertog et al.31. In short, 4 ml solvent (methanol/water; 62.5:37.5 v/v, 2 g/l tert-butylhydroquinone, Sigma Aldrich) was added, then the mix was acidified with 1.0 ml 6 M HCl, vortexed for a few seconds, and placed in a 90 °C water bath for 2 h. Afterwards, 100% methanol was used to make up the volume of the extract to 10 ml. The extract was then sonicated for 5 min, centrifuged at 2500 rpm for 8 min, and filtered through 0.45 μm PTFE filters (Fisher Scientific) for UPLC-MS/MS analysis. Total polyphenol content of the extracts was measured by a modified Folin-Ciocalteu assay17 based on79 and80.
Extracts were separated and analyzed by a UPLC-MS/MS consisting of the Dionex® UltiMate 3000 RSLC ultra-high-pressure liquid chromatography system, consisting of a workstation with ThermoFisher Scientific’s Xcalibur v. 4.0 software package combined with Dionex®’s SII LC control software, solvent rack/degasser SRD-3400, pulseless chromatography pump HPG-3400RS, autosampler WPS-3000RS, column compartment TCC-3000RS, and photodiode array detector DAD-3000RS. After passing through the photodiode array detector, the eluent flow was guided to a Q Exactive Plus Orbitrap high-resolution high-mass-accuracy mass spectrometer (MS). Mass detection was full MS scan with low energy collision induced dissociation (CID) from 100 to 1000 m/z in either positive, or negative ionization mode with electrospray (ESI) interface. Sheath gas flow rate was 30 arbitrary units, auxiliary gas flow rate was 7, and sweep gas flow rate was 1. The spray voltage was 3500 volts (−3500 for negative ESI) with a capillary temperature of 275 °C. The mass resolution was 140,000. Column and run conditions were identical to78 apart from that the average pump pressure was 3900 psi for the initial conditions.
Putative formulas of flavonoids and other compounds were determined by isotope abundance analysis on the high-resolution mass spectral data with Xcalibur v.4.0 software and reporting the best fitting empirical formula. Database searches were performed using reaxys.com (Elsevier RELX Intellectual Properties SA) and SciFinder (American Chemical Society).
Quantification was based on external standards of commercially available compounds. Naringenin and naringenin chalcone were purchased from Cerilliant, quercetin from Tocris, kaempferol and cyanidin chloride from Sigma Aldrich. Standards were dissolved in anhydrous methanol (naringenin, naringenin chalcone) or ethanol (cyanidin chloride, quercetin, kaempferol). Additionally, pelargonidin was quantified in cyanidin equivalents.
Nucleic acid isolation, and genotyping kfo and nco lettuces
Total cellular DNA was isolated from leaves of lettuces grown in growth chambers, using a modified cetyltrimethylammonium bromide (CTAB) method81. Total RNA was isolated using the QIAGEN RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s instructions. Nucleic acids were quantified using a NanoDrop UV-Vis spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed from total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Primers (Supplementary Table S2) were designed based on lettuce ESTs or genomic DNA homologs of A. thaliana CHI (TAIR AT3G55120), F3′H (TAIR AT5G07990) and F3H (TAIR AT3G51240). PCR-amplification of the full CDS of F3H and CHI was performed on nco cDNA, and of F3′H was performed on kfoA and kfoB genomic DNA, with the following PCR program: 5 min at 94 °C; 34 cycles of 30 sec at 94 °C, 30 sec at 60 °C, 90 sec at 72 °C; 10 min at 72 °C. PCR products were treated with ExoSAP-IT (Affymetrix), and Sanger sequenced. Raw sequence reads were assembled using SeqMan Pro (DNASTAR). Of the M2 generation, one kfoA mutant and seven wild type siblings, as well as one kfoB and five wild type siblings were genotyped for their F3′H alleles. Fifteen kfoA mutants and five wild-type siblings were genotyped for their F3′H alleles in the M3 generation. One M2 generation nco mutant and four wild type siblings were genotyped for their CHI alleles. Five nco mutants and fourteen wild-type siblings were genotyped for their CHI alleles in the M3 generation.
Supplementary information
Acknowledgements
We thank Dr. Natalia Pogrebnyak for helping to create the EMS-mutagenized lettuce population, and NJAES Research Greenhouse staff for plant maintenance. C.G. was supported by the National Institutes of Health under a T32 Fellowship from the National Center for Complementary and Integrative Health (AT004094). I.A. was partially supported by a fellowship from the USA–Spain Fulbright Commission and a research assistantship from Rutgers University. S.S. was supported by the Fogarty International Center of the National Institutes of Health (D43TW009672) and I.R. was partially funded by grant 2-P50 AT002776-06 from the NIH-NCCIH and NIH Office of Dietary Supplements (NIH-ODS).
Author Contributions
C.G., I.A. and I.R. conceived the research plans; C.G. designed all experiments and performed most of them, analyzed data and wrote the manuscript with contributions from all authors; I.A. created the EMS-mutagenized M2 seed collection; S.S. and M.T. provided technical assistance to C.G.; A.P. performed all LC-MS experiments and analysis; I.R. supervised the project and complemented writing the manuscript.
Data Availability
The following sequences have been deposited in the NCBI database: cv. Firecracker CHI (MG981123), nco chi1 (MG981124), cv. Firecracker F3′H (MG981125), kfoA f3′h1 (MG981126), kfoB f3′h2 (MG981127).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39287-y.
References
- 1.Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009;26:1001–1043. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- 2.Hertog MG, Feskens EJ, Hollman PC, Katan MB, Kromhout D. Dietary antioxidant flavonoids and risk of coronary heart disease: the Zutphen Elderly Study. Lancet. 1993;342:1007–1011. doi: 10.1016/0140-6736(93)92876-U. [DOI] [PubMed] [Google Scholar]
- 3.Hertog MG, Feskens EJ, Kromhout D. Antioxidant flavonols and coronary heart disease risk. Lancet. 1997;349:699. doi: 10.1016/S0140-6736(05)60135-3. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y, Je Y. Flavonoid intake and mortality from cardiovascular disease and all causes: A meta-analysis of prospective cohort studies. Clin. Nutr. ESPEN. 2017;20:68–77. doi: 10.1016/j.clnesp.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZM, et al. Flavonols intake and the risk of coronary heart disease: a meta-analysis of cohort studies. Atherosclerosis. 2012;222:270–273. doi: 10.1016/j.atherosclerosis.2012.02.026. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Ouyang YY, Liu J, Zhao G. Flavonoid intake and risk of CVD: a systematic review and meta-analysis of prospective cohort studies. Br. J. Nutr. 2014;111:1–11. doi: 10.1017/S000711451300278X. [DOI] [PubMed] [Google Scholar]
- 7.Muraki I, et al. Fruit consumption and risk of type 2diabetes: results from three prospective longitudinal cohort studies. Br. Med. J. 2013;347:f5001. doi: 10.1136/bmj.f5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, X.M, et al. Dietary total flavonoids intake and risk of mortality from all causes and cardiovascular disease in the general population: A systematic review and meta-analysis of cohort studies. Mol. Nutr. Food Res. 61 (2017). [DOI] [PubMed]
- 9.Bai W, Wang C, Ren C. Intakes of total and individual flavonoids by US adults. Int. J. Food Sci. Nutr. 2014;65:9–20. doi: 10.3109/09637486.2013.832170. [DOI] [PubMed] [Google Scholar]
- 10.Vogiatzoglou A, et al. Flavonoid intake in European adults (18 to 64 years) Plos One. 2015;10:e0128132. doi: 10.1371/journal.pone.0128132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L, et al. RNA sequencing provides insights into the evolution of lettuce and the regulation of flavonoid biosynthesis. Nat. Commun. 2017;8:2264. doi: 10.1038/s41467-017-02445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parr, B., Minor, K. & Bond, J. K. Vegetable and Pulses Yearbook Data. (USDA Economic Research Service, 2018).
- 13.Kim MJ, Moon Y, Tou JC, Mou BQ, Waterland NL. Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.) J. Food Compos. Anal. 2016;49:19–34. doi: 10.1016/j.jfca.2016.03.004. [DOI] [Google Scholar]
- 14.Ferreres F, Gil MI, Castaner M, Tomas-Barberan FA. Phenolic metabolites in red pigmented lettuce (Lactuca sativa). Changes with minimal processing and cold storage. J. Agric. Food Chem. 1997;45:4249–4254. doi: 10.1021/jf970399j. [DOI] [Google Scholar]
- 15.Llorach R, Martinez-Sanchez A, Tomas-Barberan FA, Gil MI, Ferreres F. Characterisation of polyphenols and antioxidant properties of five lettuce varieties and escarole. Food Chem. 2008;108:1028–1038. doi: 10.1016/j.foodchem.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 16.Damerum A, et al. Elucidating the genetic basis of antioxidant status in lettuce (Lactuca sativa) Hortic. Res. 2015;2:15055. doi: 10.1038/hortres.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng DM, et al. Development and Phytochemical Characterization of High Polyphenol Red Lettuce with Anti-Diabetic Properties. Plos One. 2014;9:e91571. doi: 10.1371/journal.pone.0091571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng DM, et al. Polyphenol-rich Rutgers Scarlet Lettuce improves glucose metabolism and liver lipid accumulation in diet-induced obese C57BL/6 mice. Nutrition. 2014;30:S52–S58. doi: 10.1016/j.nut.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng DM, et al. High phenolics Rutgers Scarlet Lettuce improves glucose metabolism in high fat diet-induced obese mice. Mol. Nutr. Food Res. 2016;60:2367–2378. doi: 10.1002/mnfr.201600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JH, et al. Effects of dietary supplementation with red-pigmented leafy lettuce (Lactuca sativa) on lipid profiles and antioxidant status in C57BL/6J mice fed a high-fat high-cholesterol diet. Br. J. Nutr. 2009;101:1246–1254. doi: 10.1017/S0007114508073650. [DOI] [PubMed] [Google Scholar]
- 21.Da-Silva WS, et al. The small polyphenolic molecule kaempferol increases cellular energy expenditure and thyroid hormone activation. Diabetes. 2007;56:767–776. doi: 10.2337/db06-1488. [DOI] [PubMed] [Google Scholar]
- 22.Overall, J. et al. Metabolic Effects of Berries with Structurally Diverse Anthocyanins. Int. J. Mol. Sci. 18 (2017). [DOI] [PMC free article] [PubMed]
- 23.Shirley BW, et al. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 1995;8:659–671. doi: 10.1046/j.1365-313X.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- 24.Bowerman PA, Ramirez MV, Price MB, Helm RF, Winkel BS. Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia. BMC Res. Notes. 2012;5:485. doi: 10.1186/1756-0500-5-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito K, et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Reyes-Chin-Wo S, et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017;8:14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimura M, Sano A, Kamei J, Obata A. Identification and quantification of metabolites of orally administered naringenin chalcone in rats. J. Agric. Food Chem. 2009;57:6432–6437. doi: 10.1021/jf901137x. [DOI] [PubMed] [Google Scholar]
- 28.Bilyk A, Sapers GM. Distribution of Quercetin and Kaempferol in Lettuce, Kale, Chive, Garlic Chive, Leek, Horseradish, Red Radish, and Red Cabbage Tissues. J. Agric. Food Chem. 1985;33:226–228. doi: 10.1021/jf00062a017. [DOI] [Google Scholar]
- 29.Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. Arabidopsis Flavonoid Mutants Are Hypersensitive to UV-B Irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan KG, Swinny EE, Winefield C, Markham KR. Flavonoids and UV photoprotection in Arabidopsis mutants. Z. Naturforsch. C. 2001;56:745–754. doi: 10.1515/znc-2001-9-1013. [DOI] [PubMed] [Google Scholar]
- 31.Hertog MGL, Hollman PCH, Venema DP. Optimization of a Quantitative HPLC Determination of Potentially Anticarcinogenic Flavonoids in Vegetables and Fruits. J. Agric. Food Chem. 1992;40:1591–1598. doi: 10.1021/jf00021a023. [DOI] [Google Scholar]
- 32.Franke AA, Custer LJ, Arakaki C, Murphy SP. Vitamin C and flavonoid levels of fruits and vegetables consumed in Hawaii. J. Food Compos. Anal. 2004;17:1–35. doi: 10.1016/S0889-1575(03)00066-8. [DOI] [Google Scholar]
- 33.Yang RY, Lin S, Kuo G. Content and distribution of flavonoids among 91 edible plant species. Asia Pac. J. Clin. Nutr. 2008;17(Suppl 1):275–279. [PubMed] [Google Scholar]
- 34.Cao J, Chen W, Zhang Y, Zhang YQ, Zhao XJ. Content of Selected Flavonoids in 100 Edible Vegetables and Fruits. Food Sci. Technol. Res. 2010;16:395–402. doi: 10.3136/fstr.16.395. [DOI] [Google Scholar]
- 35.Nyman NA, Kumpulainen JT. Determination of anthocyanidins in berries and red wine by high-performance liquid chromatography. J. Agric. Food Chem. 2001;49:4183–4187. doi: 10.1021/jf010572i. [DOI] [PubMed] [Google Scholar]
- 36.Mol JNM, Robbins MP, Dixon RA, Veltkamp E. Spontaneous and Enzymic Rearrangement of Naringenin Chalcone to Flavanone. Phytochemistry. 1985;24:2267–2269. doi: 10.1016/S0031-9422(00)83023-X. [DOI] [Google Scholar]
- 37.Bottcher C, et al. Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 2008;147:2107–2120. doi: 10.1104/pp.108.117754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jez JM, Bowman ME, Dixon RA, Noel JP. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol. 2000;7:786–791. doi: 10.1038/79025. [DOI] [PubMed] [Google Scholar]
- 39.Peer WA, et al. Flavonoid Accumulation Patterns of Transparent Testa Mutants of Arabidopsis. Plant Physiol. 2001;126:536–548. doi: 10.1104/pp.126.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koornneef M, Luiten W, de Vlaming P, Schram A. A Gene Controlling Flavonoid 3′-Hydroxylation in Arabidopsis. Arabid. Inf. Serv. 1982;19:113–115. [Google Scholar]
- 41.Schoenbohm C, Martens S, Eder C, Forkmann G, Weisshaar B. Identification of the Arabidopsis thaliana flavonoid 3′-hydroxylase gene and functional expression of the encoded P450 enzyme. Biol. Chem. 2000;381:749–753. doi: 10.1515/BC.2000.095. [DOI] [PubMed] [Google Scholar]
- 42.Falginella L, et al. Expansion and subfunctionalisation of flavonoid 3′,5′-hydroxylases in the grapevine lineage. BMC Genomics. 2010;11:562. doi: 10.1186/1471-2164-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muir SR, et al. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nat. Biotechnol. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto T, et al. Anti-allergic activity of naringenin chalcone from a tomato skin extract. Biosci. Biotechnol. Biochem. 2004;68:1706–1711. doi: 10.1271/bbb.68.1706. [DOI] [PubMed] [Google Scholar]
- 45.Iwamura C, et al. Naringenin chalcone suppresses allergic asthma by inhibiting the type-2 function of CD4 T cells. Allergol. Int. 2010;59:67–73. doi: 10.2332/allergolint.09-OA-0118. [DOI] [PubMed] [Google Scholar]
- 46.Horiba T, Nishimura I, Nakai Y, Abe K, Sato R. Naringenin chalcone improves adipocyte functions by enhancing adiponectin production. Mol. Cell. Endocrinol. 2010;323:208–214. doi: 10.1016/j.mce.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Yoshimura M, et al. An evaluation of the clinical efficacy of tomato extract for perennial allergic rhinitis. Allergol. Int. 2007;56:225–230. doi: 10.2332/allergolint.O-06-443. [DOI] [PubMed] [Google Scholar]
- 48.Forkmann G, Kuhn B. Genetic control of chalcone isomerase activity in anthers of Petunia hybrida. Planta. 1979;144:189–192. doi: 10.1007/BF00387269. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn B, Forkmann G, Seyffert W. Genetic control of chalcone-flavanone isomerase activity in Callistephus chinensis. Planta. 1978;138:199–203. doi: 10.1007/BF00386811. [DOI] [PubMed] [Google Scholar]
- 50.Forkmann G, Dangelmayr B. Genetic control of chalcone isomerase activity in flowers of Dianthus caryophyllus. Biochem. Genet. 1980;18:519–527. doi: 10.1007/BF00484399. [DOI] [PubMed] [Google Scholar]
- 51.Reuber S, Jende-Strid B, Wray V, Weissenbock G. Accumulation of the chalcone isosalipurposide in primary leaves of barley flavonoid mutants indicates a defective chalcone isomerase. Physiol. Plant. 1997;101:827–832. doi: 10.1111/j.1399-3054.1997.tb01070.x. [DOI] [Google Scholar]
- 52.Druka A, et al. Chalcone isomerase gene from rice (Oryza sativa) and barley (Hordeum vulgare): physical, genetic and mutation mapping. Gene. 2003;302:171–178. doi: 10.1016/S0378-1119(02)01105-8. [DOI] [PubMed] [Google Scholar]
- 53.Hong L, et al. A mutation in the rice chalcone isomerase gene causes the golden hull and internode 1 phenotype. Planta. 2012;236:141–151. doi: 10.1007/s00425-012-1598-x. [DOI] [PubMed] [Google Scholar]
- 54.Kim S, Jones R, Yoo KS, Pike LM. Gold color in onions (Allium cepa): a natural mutation of the chalcone isomerase gene resulting in a premature stop codon. Mol. Genet. Genomics. 2004;272:411–419. doi: 10.1007/s00438-004-1076-7. [DOI] [PubMed] [Google Scholar]
- 55.Shirley BW, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell. 1992;4:333–347. doi: 10.1105/tpc.4.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koornneef M. Mutations Affecting the Testa Colour in Arabidopsis. Arabid. Inf. Serv. 1990;28:1–4. [Google Scholar]
- 57.Calderon-Montano JM, Burgos-Moron E, Perez-Guerrero C, Lopez-Lazaro M. A review on the dietary flavonoid kaempferol. Mini Rev. Med. Chem. 2011;11:298–344. doi: 10.2174/138955711795305335. [DOI] [PubMed] [Google Scholar]
- 58.Alkhalidy H, et al. Small Molecule Kaempferol Promotes Insulin Sensitivity and Preserved Pancreatic Beta -Cell Mass in Middle-Aged Obese Diabetic Mice. J. Diabetes Res. 2015;2015:532984. doi: 10.1155/2015/532984. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Luo C, et al. Kaempferol alleviates insulin resistance via hepatic IKK/NF-kappaB signal in type 2 diabetic rats. Int. Immunopharmacol. 2015;28:744–750. doi: 10.1016/j.intimp.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 60.Hertog MGL, Hollman PCH, Katan MB. Content of Potentially Anticarcinogenic Flavonoids of 28 Vegetables and 9 Fruits Commonly Consumed in the Netherlands. J. Agric. Food Chem. 1992;40:2379–2383. doi: 10.1021/jf00024a011. [DOI] [Google Scholar]
- 61.Justesen U, Knuthsen P, Leth T. Quantitative analysis of flavonols, flavones, and flavanones in fruits, vegetables and beverages by high-performance liquid chromatography with photo-diode array and mass spectrometric detection. J. Chromatogr. A. 1998;799:101–110. doi: 10.1016/S0021-9673(97)01061-3. [DOI] [PubMed] [Google Scholar]
- 62.Inocencio C, Rivera D, Alcaraz F, Tomas-Barberan FA. Flavonoid content of commercial capers (Capparis spinosa, C. sicula and C. orientalis) produced in mediterranean countries. Eur. Food Res. Technol. 2000;212:70–74. doi: 10.1007/s002170000220. [DOI] [Google Scholar]
- 63.Hoshino A, et al. Spontaneous mutations of the flavonoid 3 ‘-hydroxylase gene conferring reddish flowers in the three morning glory species. Plant Cell Physiol. 2003;44:990–1001. doi: 10.1093/pcp/pcg143. [DOI] [PubMed] [Google Scholar]
- 64.Momose M, et al. An active hAT transposable element causing bud mutation of carnation by insertion into the flavonoid 3′-hydroxylase gene. Mol. Genet. Genomics. 2013;288:175–184. doi: 10.1007/s00438-013-0742-z. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Frei M. StressedFood – The impact of abiotic environmental stresses on crop quality. Agric. Ecosyst. Environ. 2011;141:271–286. doi: 10.1016/j.agee.2011.03.017. [DOI] [Google Scholar]
- 66.Agati G, Azzarello E, Pollastri S, Tattini M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012;196:67–76. doi: 10.1016/j.plantsci.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 67.Garcia-Macias P, et al. Changes in the flavonoid and phenolic acid contents and antioxidant activity of red leaf lettuce (Lollo Rosso) due to cultivation under plastic films varying in ultraviolet transparency. J. Agric. Food Chem. 2007;55:10168–10172. doi: 10.1021/jf071570m. [DOI] [PubMed] [Google Scholar]
- 68.Shioshita R, Enoka J, Aiona DK, Young CS, Sakai WS. Coloration and Growth of Red Lettuce Grown under UV-Radiation Transmitting and Non-Transmitting Covers. Acta Hortic. 2007;761:221–225. doi: 10.17660/ActaHortic.2007.761.28. [DOI] [Google Scholar]
- 69.Tsormpatsidis E, et al. UV irradiance as a major influence on growth, development and secondary products of commercial importance in Lollo Rosso lettuce ‘Revolution’ grown under polyethylene films. Environ. Exp. Bot. 2008;63:232–239. doi: 10.1016/j.envexpbot.2007.12.002. [DOI] [Google Scholar]
- 70.Tsormpatsidis E, Henbest RGC, Battey NH, Hadley P. The influence of ultraviolet radiation on growth, photosynthesis and phenolic levels of green and red lettuce: potential for exploiting effects of ultraviolet radiation in a production system. Ann. Appl. Biol. 2010;156:357–366. doi: 10.1111/j.1744-7348.2010.00393.x. [DOI] [Google Scholar]
- 71.Marin A, Ferreres F, Barbera GG, Gil MI. Weather variability influences color and phenolic content of pigmented baby leaf lettuces throughout the season. J. Agric. Food Chem. 2015;63:1673–1681. doi: 10.1021/acs.jafc.5b00120. [DOI] [PubMed] [Google Scholar]
- 72.Li Q, Kubota C. Effects of supplemental light quality on growth and phytochemicals of baby leaf lettuce. Environ. Exp. Bot. 2009;67:59–64. doi: 10.1016/j.envexpbot.2009.06.011. [DOI] [Google Scholar]
- 73.Goto E, Hayashi K, Furuyama S, Hikosaka S, Ishigami Y. Effect of UV light on phytochemical accumulation and expression of anthocyanin biosynthesis genes in red leaf lettuce. Acta Hortic. 2016;1134:293–300. [Google Scholar]
- 74.Sytar O, et al. Shift in accumulation of flavonoids and phenolic acids in lettuce attributable to changes in ultraviolet radiation and temperature. Sci. Hortic. (Amsterdam) 2018;239:193–204. doi: 10.1016/j.scienta.2018.05.020. [DOI] [Google Scholar]
- 75.Armas Gutierrez, I. Nutritional Enhancement of Lettuce Using Mutational Breeding. (MS Thesis, Rutgers University, New Brunswick, NJ, USA, 2015).
- 76.Landry LG, Chapple CC, Last RL. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Havaux M, Kloppstech K. The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta. 2001;213:953–966. doi: 10.1007/s004250100572. [DOI] [PubMed] [Google Scholar]
- 78.Poulev A, Chen MH, Cherravuru S, Raskin I, Belanger FC. Variation in levels of the flavone tricin in bran from rice genotypes varying in pericarp color. J. Cereal Sci. 2018;79:226–232. doi: 10.1016/j.jcs.2017.11.001. [DOI] [Google Scholar]
- 79.Singleton VL, Orthofer R, Lamuela-Raventos RM. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Method. Enzymol. 1999;299:152–178. doi: 10.1016/S0076-6879(99)99017-1. [DOI] [Google Scholar]
- 80.Sharma M, et al. Effects of fruit ellagitannin extracts, ellagic acid, and their colonic metabolite, urolithin A, on Wnt signaling. J. Agric. Food Chem. 2010;58:3965–3969. doi: 10.1021/jf902857v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following sequences have been deposited in the NCBI database: cv. Firecracker CHI (MG981123), nco chi1 (MG981124), cv. Firecracker F3′H (MG981125), kfoA f3′h1 (MG981126), kfoB f3′h2 (MG981127).