Abstract
The island-like distribution of subalpine habitats across mountain ranges can trigger the parallel evolution of locally adapted ecotypes. Such naturally replicated scenarios allow testing hypotheses on how elevational differentiation structures genetic diversity within species. Nevertheless, the parallel colonization of subalpine habitats across different mountain ranges has only rarely been documented with molecular data. We chose Primula elatior (Primulaceae), naturally spanning entire elevation range in multiple mountain regions of central Europe, to test for the origin of its scattered subalpine populations. Nuclear microsatellite variation revealed three genetic groups corresponding with the distinct study regions. We found that genetic differentiation between foothill and subalpine populations within each region was relatively low, suggesting that the colonization of subalpine habitats occurred independently within each mountain range. Furthermore, the strongest differentiation was usually found between the subalpine populations suggesting that mountain ridges may act as migration barriers that can reduce gene flow more strongly than elevational differences between foothill and subalpine populations. Finally, we found that subalpine colonization did not result in a loss of genetic diversity relative to foothill populations in agreement with the high migration rates that we document here between the subalpine and the foothill populations. In summary, our study shows subalpine Primula elatior populations are genetically diverse and distinct results of parallel colonization events from multiple foothill gene pools.
Introduction
Alpine and subalpine habitats represent challenging and often unpredictable environments for plants. Plants have to face a complex set of stresses in such environments including freezing, fluctuating temperatures, increased UV radiation, and terrain disturbances implying various selection pressures1. Local adaptation to these environmental stresses often results in the evolution of specific morphological and/or physiological traits that confer increased fitness in these challenging environments2,3. Populations characterized by these adaptations are often called “ecotypes” and their level of overall genetic differentiation from ancestral populations may be still very low2,4. In cases where reproductive barriers arise between ecotypes and their ancestral populations, either as a consequence of natural selection or simply due to spatial isolation, the ecotype may represent the first step towards the founding of a new species5,6. In such cases, we can observe speciation as a continuum of divergence along an elevational gradient leading to textbook examples of ecotypic differentiation7,8. The restriction of gene flow between lower and higher elevations as well as the reduction or absence of gene flow among mountain ridges can lead to the accumulation of reproductive barriers and/or hybrid incompatibilities9. This process is but one of several potential explanations for relatively high species diversity in alpine/subalpine habitats throughout the world10.
The island-like distribution of alpine/subalpine habitats can trigger allopatric differentiation and parallel colonization from lower elevations independently in each mountain range11. It can also lead to phenotypic differentiation potentially resulting in the convergent evolution of traits that confer local adaptation (i.e. ecotypes)2. Although the mountains provide an ideal model system for studying the evolution of recurrently adapted ecotypes, the parallel colonization of alpine/subalpine habitats has only been rarely documented with molecular data. Rare exceptions to this are represented by studies in Arabidopsis halleri12, Zea mays13, and Populus trichocarpa14. The alternative scenario of a single origin of alpine/subalpine ecotype followed by dispersal has also been documented, for example in Senecio halleri whose alpine populations represent a single lineage that colonized the Alps in a stepwise manner15.
The influence of habitat differentiation on the genetic diversity of populations has been previously studied mainly at lower elevations16–18. It is poorly known how the colonization of alpine/subalpine h abitats has shaped the genetic diversity of populations, but it is hypothesized that alpine/subalpine populations would be genetically depauperate relative to lowland populations due to the effects of genetic bottleneck, genetic drift, reduced gene flow, and habitat fragmentation19, all of which can promote the genetic isolation of alpine/subalpine populations from their foothill relatives20. This has indeed been observed in alpine populations of Arabidopsis thaliana from the Italian Alps, which exhibit reduced genetic diversity relative to foothill populations21, but in contrast, alpine A. thaliana populations from the Swiss Alps show no evidence of reduced genetic diversity22. In Primula merrilliana from eastern China, alpine populations exhibit even higher genetic diversity, moreover, they are larger and more inter-connected with gene flow than foothill populations23. This pattern implies that the foothill populations may have been colonized by alpine populations. In summary, studies of recurrently originated ecotypes of a single species may provide valuable replicates to test the generality of how alpine conditions shape population genetic diversity.
In this study, we address the genetic consequences of elevational differentiation in Primula elatior (Primulaceae), a species with broad ecological preferences including a large elevational range in several mountain ranges in Europe24. The wide ecological breadth is linked to high morphological variation primarily in leaf and calyx shape24,25. Individuals originally from subalpine populations tended to have urceolate calyxes (the narrowest in the top part of calyx), compared to individuals from foothill populations mainly with tubular calyxes (the same wide along the calyx)26. The morphological traits are plastic, except the shape of calyx, which remained stable after cultivation of subalpine population under uniform foothill conditions in a common garden experiment in one of the mountain ranges (the Krkonoše mountain range, V. Konečná unpubl.). In contrast to morphological investigations, the genetic structure of P. elatior remains unknown except studies at fine-scale in Belgium16,27–29. We focus on three mountain ranges in central Europe, where P. elatior grows along an elevational gradient from foothill meadows, river banks, and forest edges, up to subalpine meadows, snowbeds, and rocky outcrops in the glacial cirques. In the Krkonoše and the Jeseníky mountains, subalpine populations are restricted to glacial cirques. In the Tatry mountains, subalpine populations grow in valley meadows and snowbeds. Generally, the difference between foothill and subalpine populations across mountain ranges is defined by treeline, foothill populations occur below the treeline in contrast to subalpine populations, which occur above the treeline. We assume colonization of subalpine habitats from foothill habitats during warmer periods of the postglacial Holocene. Our assumption of upslope colonization is likely because temperate species are highly unlikely to have survived past glaciations that affected these subalpine habitats30.
Using P. elatior as a suitable system, we examined how elevation shapes genetic structure within a species, and by comparing these results across three distinct mountain ranges (the Jeseníky, the Krkonoše, and the Tatry), we evaluated how generally applicable these patterns of differentiation might be in P. elatior. First, we tested whether the subalpine populations in three mountain ranges represent parallel colonization events of subalpine habitats occurring independently in each mountain range or if the subalpine ecotype evolved once and later spread across the different mountain ranges. Second, we tested if the elevation acts as a barrier to gene flow between foothill and subalpine populations, and whether gene flow is asymmetric; e.g. are migration events from high to low elevations following the downslope transport of seeds and pollen more common than upslope migration events. Finally, we tested for a reduction of genetic diversity associated with the colonization of subalpine habitats relative to lowland habitats.
Results
Genetic structure and evolutionary relationships among populations
By genotyping 12 nuclear microsatellite loci in 202 individuals from 16 populations we detected a total of 120 alleles with maximum of 24 and minimum of three alleles per locus.
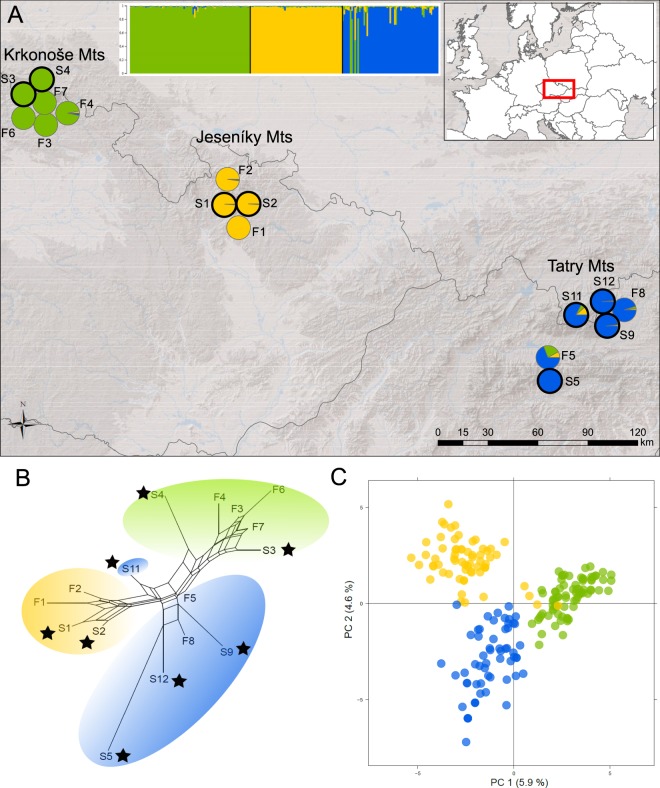
We explored the genetic structure of P. elatior populations across the three target mountain ranges, where it occupies both foothill and subalpine habitats, using Bayesian clustering (structure), distance networks (Neighbor-joining networks), and ordinations (principal component analysis, PCA). The results of the structure analyses showed that populations from each mountain range (the Jeseníky, the Krkonoše, and the Tatry) formed a separate cluster, regardless of their foothill-subalpine differentiation, under the corresponding partition of K = 3 (Fig. 1A). The model of K = 3 also exhibited the highest similarity among the replicated runs and at this partition the rise of likelihood values started to flatten, suggesting that the inclusion of additional parameters do not significantly improve the fit of the model beyond K = 3 (Supplementary Fig. S1). We observed three clusters according to the three mountain ranges in PCA (Fig. 1C) as well as in Neighbor-joining network based on FST distances (Fig. 1B). The separation of populations into three clusters that largely corresponded to geographical range and the absence of any genetic structure associated with elevation strongly suggested the parallel evolution of the subalpine populations in each mountain range.
Figure 1.
Parallel origin of subalpine populations of Primula elatior in the three mountain ranges in central Europe. (A) Geographical distribution of the complete dataset of 16 foothill and subalpine (marked by bold circles) populations of P. elatior, pie charts and barplots show the proportional assignment of individuals from each population to the three clusters inferred by structure, (B) neighbor-joining network of the same populations based on among-population FST distances (subalpine populations marked by asterisks), (C) principal component analysis (PCA) of all 202 individuals (coloured according to structure results). Map was processed in ArcMap version 10.0 (http://desktop.arcgis.com/en/arcmap/) by V. Konečná, map layer was modified (http://desktop.arcgis.com/en/arcmap/10.3/manage-data/raster-and-images/hillshade-function.htm) by V. Konečná.
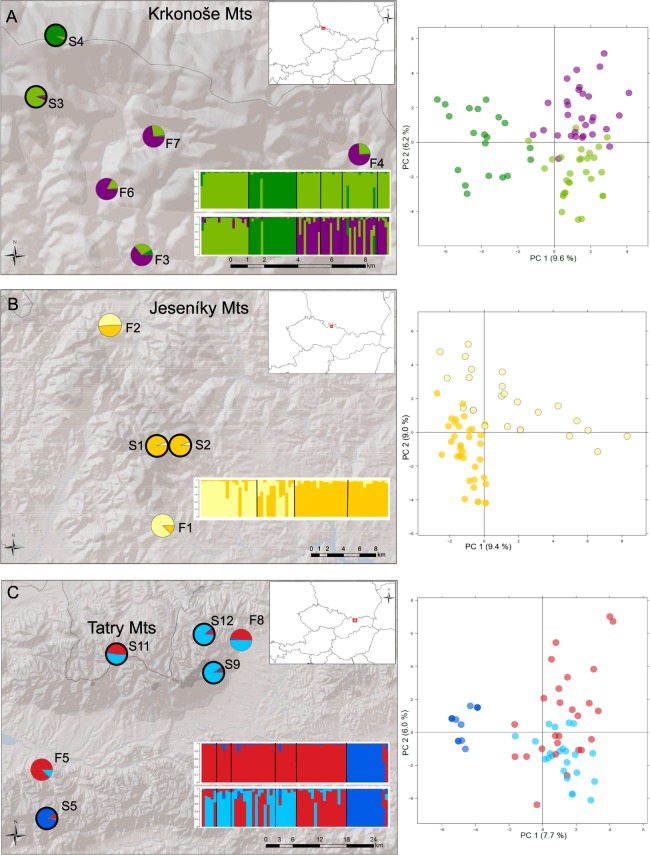
In subsequent analyses, we performed separate structure analyses for each region to investigate finer structure within each mountain range (Fig. 2). All the analyses tended to separate the subalpine and foothill populations within each region: this was apparent already under K = 2 in the Jeseníky and under K = 3 in the other two mountain ranges. In the Krkonoše and the Tatry, one subalpine population (S4 and S5, respectively) separated from the remaining populations under K = 2, suggesting that not only one group of subalpine populations exists in these mountain ranges.
Figure 2.
Genetic sub-structuring of P. elatior populations in each target mountain range. Pie charts and barplots show the proportional assignment of individuals from each population to clusters inferred by a separate structure analysis of (A) the Krkonoše (79 individuals), (B) the Jeseníky (60 individuals), and (C) the Tatry (63 individuals); accompanied by principal component analysis (PCA) of individuals. Subalpine populations are denoted by bold circles. Maps were processed in ArcMap version 10.0 (http://desktop.arcgis.com/en/arcmap/) by V. Konečná, map layers were modified (http://desktop.arcgis.com/en/arcmap/10.3/manage-data/raster-and-images/hillshade-function.htm) by V. Konečná.
Genetic diversity of populations and among population variation
The populations varied considerably in terms of within population variation. Allelic richness ranged from 2.29 to 3.72 with the highest values (3.33–3.72) being present in foothill populations in the Tatry (Table 1, Supplementary Fig. S2). Similarly, expected heterozygosity (HE), ranged from 0.4 to 0.6 in the whole dataset and foothill populations from the Tatry exhibited the highest values (0.52–0.6). Populations also varied in gene diversity (HS) ranging from 0.43 to 0.65 with the highest values (0.65–0.61) in one foothill and two subalpine populations from the Tatry. The proportion of rare alleles (DW index)31 exhibited a similar trend as well, with the range from 82.44 in the Jeseníky subalpine population to 411.79 in the Tatry foothill population. Despite this variation, we did not detect consistent differences between the group of foothill and subalpine populations across three mountain ranges in diversity index (non-significant ANOVA with region as a random factor) (Table 2). Instead, each mountain range tended to show a different pattern (Supplementary Fig. S2).
Table 1.
Genetic diversity of 16 populations of Primula elatior from foothill and subalpine habitats in the three mountain ranges investigated.
| Pop. ID | Locality | Range | Group | Elevation | No. Ind. | No. Alleles | Allelic richness | HO | HE | HS | DW index | No. private alleles | HWE testa | MIGRATE-N |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | Žďárský potok u Rýmařova | Jeseníky | foothill | 735 | 18 | 41 | 2.6 | 0.43 ± 0.28 | 0.44 ± 0.26 | 0.43 | 240.76 | 2 | ✓ | |
| F2 | V Mlýnkách | Jeseníky | foothill | 618 | 13 | 44 | 2.71 | 0.35 ± 0.29 | 0.46 ± 0.28 | 0.45 | 121.95 | 1 | PRIV 4**, PACA 78**, PV 4767*, PV 23741* | ✓ |
| S1 | Velká Kotlina | Jeseníky | subalpine | 1397 | 13 | 36 | 2.61 | 0.49 ± 0.36 | 0.46 ± 0.30 | 0.54 | 82.44 | 0 | PACA 78*, PV 23741* | ✓ |
| S2 | Velká Kotlina edge | Jeseníky | subalpine | 1301 | 16 | 39 | 2.6 | 0.46 ± 0.30 | 0.48 ± 0.27 | 0.51 | 84.78 | 0 | PRIV 4*, PACA 78* | ✓ |
| F3 | Strážné | Krkonoše | foothill | 776 | 15 | 45 | 2.75 | 0.41 ± 0.37 | 0.46 ± 0.33 | 0.49 | 175.15 | 2 | PRIV 4***, PACA 38** | ✓ |
| F4 | Spálený mlýn | Krkonoše | foothill | 788 | 10 | 39 | 2.68 | 0.41 ± 0.35 | 0.46 ± 0.30 | 0.49 | 84.45 | 0 | PACA 38* | ✓ |
| F6 | Michlův mlýn | Krkonoše | foothill | 658 | 9 | 35 | 2.52 | 0.39 ± 0.29 | 0.43 ± 0.26 | 0.45 | 150.71 | 1 | PACA 38* | |
| F7 | Svatý Petr | Krkonoše | foothill | 816 | 5 | 34 | 2.82 | 0.43 | 0.5 | 0.57 | 138.8 | 0 | ||
| S3 | Velká Kotelní jáma | Krkonoše | subalpine | 1280 | 20 | 45 | 2.62 | 0.40 ± 0.32 | 0.45 ± 0.31 | 0.53 | 145.06 | 4 | PRIV 4***, PACA 78** | ✓ |
| S4 | Malá Sněžná jáma | Krkonoše | subalpine | 1368 | 20 | 41 | 2.52 | 0.34 ± 0.32 | 0.44 ± 0.32 | 0.52 | 227.05 | 1 | PRIV 4***, PACA 78**, PV 4767*, PACA 38* | ✓ |
| F5 | Poludnica | Tatry | foothill | 1055 | 17 | 68 | 3.72 | 0.57 ± 0.33 | 0.62 ± 0.29 | 0.61 | 411.79 | 9 | PRIV 4***, PV 1973** | ✓ |
| F8 | Ždiar, Tatranská Kotlina | Tatry | foothill | 769 | 15 | 63 | 3.33 | 0.48 ± 0.34 | 0.54 ± 0.33 | 0.58 | 380.05 | 5 | PACA 78*** | |
| S5 | Štefánikova chata | Tatry | subalpine | 1709 | 14 | 33 | 2.29 | 0.39 ± 0.43 | 0.41 ± 0.29 | 0.49 | 167.63 | 1 | PV 279**, PACA 78***, PRIV 7**, PV 4767*, PV 23741* | ✓ |
| S9 | Zamkovského chata | Tatry | subalpine | 1470 | 7 | 34 | 2.66 | 0.54 ± 0.34 | 0.49 ± 0.28 | 0.57 | 314.54 | 1 | ||
| S11 | Kondraczka | Tatry | subalpine | 1950 | 5 | 43 | 3.53 | 0.5 | 0.51 | 0.65 | 227.41 | 1 | ||
| S12 | Tristar | Tatry | subalpine | 1450 | 5 | 34 | 2.82 | 0.43 | 0.44 | 0.62 | 195.04 | 0 |
aOnly loci with significant deviation from HWE are listed; P-values were estimated by 1000 permutations (*P < 0.05, **P < 0.01, ***P < 0.001).
Table 2.
Genetic diversity and differentiation of foothill and subalpine populations within the target mountain ranges.
| Group | No. of ind./pop. | No. of alleles | No. of private alleles | Allelic richness | HE | HS | DW index | FST | Among populations variation (%) | Pairwise FST among populations (min-max values) | Among groups variation (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Jeseníky Mts | |||||||||||
| Subalpine populations | 29/2 | 44 | 0 | 2.61 ± 0.005 | 0.47 ± 0.01 | 0.53 ± 0.015 | 83.61 ± 1.17 | 0.03 | 2.76* | 0.035 | |
| Foothill populations | 31/2 | 53 | 3 | 2.66 ± 0.055 | 0.45 ± 0.01 | 0.44 ± 0.01 | 181.36 ± 59.41 | 0.08 | 7.82*** | 0.057 | |
| Subalpine × foothills | 6.11 (p = 0.33) | ||||||||||
| Krkonoše Mts | |||||||||||
| Subalpine populations | 40/2 | 56 | 5 | 2.57 ± 0.05 | 0.45 ± 0.005 | 0.53 ± 0.005 | 186.06 ± 41 | 0.21 | 21.42*** | 0.132 | |
| Foothill populations | 39/4 | 60 | 3 | 2.69 ± 0.093 | 0.46 ± 0.019 | 0.50 ± 0.035 | 137.28 ± 26.41 | 0.05 | 5.47*** | (0.035) 0.059 (0.08) | |
| Subalpine × foothills | 4.26 (p = 0.14) | ||||||||||
| Tatry Mts | |||||||||||
| Subalpine populations | 31/4 | 68 | 3 | 2.83 ± 0.352 | 0.46 ± 0.038 | 0.58 ± 0.053 | 226.16 ± 44.82 | 0.24 | 24.18*** | (0.065) 0.125 (0.174) | |
| Foothill populations | 32/2 | 87 | 14 | 3.53 ± 0.195 | 0.58 ± 0.04 | 0.6 ± 0.015 | 395.92 ± 15.87 | 0.03 | 3.03 (p = 0.1) | 0.06 | |
| Subalpine × foothills | 2.32 (p = 0.25) | ||||||||||
| Significance of differences between groups of foothill and subalpine populations | F1,12 = 0.09 | F1,12 = 0.09 | F1,12 = 0.3 | F1,12 = 0.08 | |||||||
P-values were estimated by 1000 permutations (*P < 0.05, **P < 0.01, ***P < 0.001).
Additionally, we calculated for subalpine pop. S5 from the Nízké Tatry Mts and foothill pop. F5 and F8 among population variation (subalpine × foothill pop.) = 23.82% (p = 0.33), FST = 0.27, and pairwise FST among populations = (0.06) 0.11 (0.137).
Among-population differentiation (pairwise FST) was low, but varied considerably from 0.04 to 0.28, with an average of 0.13 (Supplementary Table S3). In the Krkonoše and the Tatry, we observed lower FST values between foothill populations (average of 0.06 for six comparisons in the Krkonoše, and 0.06 for one comparison in the Tatry) compared to those between subalpine populations (on average 0.14 for six comparisons in the Tatry and 0.13 for one comparison in the Krkonoše). In contrast, two subalpine populations in the Jeseníky, that occupied different parts of one glacial cirque, were less differentiated from each other (0.04) than were their two foothill counterparts (0.06). Between the foothill and subalpine populations, we observed at most a moderate differentiation, with the highest mean pairwise FST values in the Krkonoše (0.10) followed by the Tatry (0.09), and the lowest values in the Jeseníky (0.08).
The results of an AMOVA (analysis of molecular variance) confirmed high intrapopulation variability by assigning the highest proportion of variation always among individuals within populations (from 97.24% to 73.27%). In congruence with pairwise FST values, two subalpine populations in the Krkonoše and four in the Tatry were more differentiated from each other than their foothill counterparts: 21.42% and 24.18% of among-subalpine population variation compared to 5.47% and 3.03% of among-foothill population variation in the Krkonoše (four populations) and the Tatry (two populations), respectively. The lowest variation (2.76%), although still significant, was found between subalpine populations from the Jeseníky. Finally, hierarchical AMOVAs within each mountain range showed that the foothill-subalpine differentiation was non-significant and accounted for little variation (from 2.32% to 6.11%, Table 2). To conclude, even though the structure results revealed some differentiation between foothill and subalpine populations, this differentiation is still markedly lower than differentiation observed between subalpine populations in two of the three mountain ranges.
Gene flow, migration rates and model selections
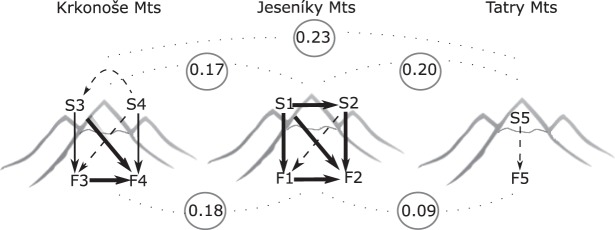
We further addressed the role of migration in foothill-subalpine population differentiation by testing for the presence of gene flow and estimating the strength of gene flow between populations in a coalescent framework using migrate-n. Within each region, we analysed pairs of representatively sampled (≥10 individuals per population) subalpine-subalpine, foothill-subalpine, and foothill-foothill populations; for each pair, we modelled past evolutionary history under Bayesian inference of five migration models differing in presence and directionality of migration. The models with unidirectional migration gained the best support in all population pairs analysed (Table 3, Fig. 3, Supplementary Table S4). In the case of the foothill-subalpine pairs, models of unidirectional migration from subalpine to foothill populations were consistently the best models fit across different population pairs from different mountain ranges. The estimated number of immigrants (Nm) was overall high (>1) (ranging from 5 to 551 with an average of 160), which suggests that migration via ongoing gene flow has probably a greater effect on the extent of differentiation between populations than genetic drift (Nm values >1)32. However, besides migration, the large estimates for the number of immigrants could also at least partly reflect the recent shared ancestry of populations33.
Figure 3.
Migration and differentiation of foothill (F) and subalpine (S) populations of P. elatior in the three mountain ranges of central Europe. The values between mountain ranges are overall mean pairwise FST between foothill and subalpine populations. The arrows indicate preferred migration scenario, their width reflects estimated number of immigrants per generation (Nm): dashed arrow (5–21), standard arrow (68–73), and thick arrow (161–551). Created by V. Konečná.
We observed the lowest estimated migration rates between subalpine populations in the Krkonoše (Nm = 5), where subalpine populations are separated by a mountain ridge. In contrast, in the Jeseníky, a relatively weak barrier to gene flow appears to exist between the two populations residing within one glacial cirque based on the high migration rate estimated (Nm = 266). The highest number of immigrants in the entire dataset was also found between two foothill populations (Krkonoše, F3 and F4; Nm = 551) (Table 3, Fig. 3).
Table 3.
Summary of the best fitting migration models (based on Bayes factor, see Supplementary Table S4) for foothill and subalpine populations of P. elatior, their differentiation, and numbers of immigrants per generation (Nm) between populations estimated by migrate-n.
| Range | Model | Nm | Pairwise FST |
|---|---|---|---|
| Jeseníky | F1 → F2 | 206 | 0.06 |
| Krkonoše | F3 → F4 | 551 | 0.05 |
| Jeseníky | S1 → F1 | 261 | 0.08 |
| Jeseníky | S2 → F1 | 21 | 0.09 |
| Jeseníky | S1 → F2 | 161 | 0.07 |
| Jeseníky | S2 → F2 | 173 | 0.07 |
| Krkonoše | S3 → F3 | 68 | 0.07 |
| Krkonoše | S4 → F3 | 17 | 0.11 |
| Krkonoše | S3 → F4 | 257 | 0.09 |
| Krkonoše | S4 → F4 | 73 | 0.13 |
| Nízké Tatry | S5 → F5 | 14 | 0.13 |
| Jeseníky | S1 → S2 | 266 | 0.04 |
| Krkonoše | S4 → S3 | 5 | 0.13 |
F = foothill populations, S = subalpine populations.
Discussion
Our population genetic investigation of central European Primula elatior populations provided strong evidence for parallel colonization of subalpine habitats within each of the three mountain ranges that are the focus of our study (Fig. 1). In each case, subalpine populations are more closely related to the geographically closest foothill populations from the same mountain range than to their subalpine counterparts from other mountain ranges. Although island-like distributed subalpine environments are likely to trigger the evolution of recurrently adapted populations, the parallel colonization of subalpine habitats across different mountain ranges in one species has only rarely been documented with genetic data12–14. Future studies on parallel ecotype evolution should also focus on mechanisms behind the genetic parallelism. For instance, whether repeated selection from the standing genetic variation has important role in formation of parallel ecotypes14,34, whether the selection of independent mutations takes place in parallel evolution13, or if adaptive introgression via borrowed alleles from adapted species can facilitate this process35.
The example of the parallel origin of ecotypes along an elevational gradient, although with the opposite direction of colonization than in our study, was recently documented for Heliosperma pusillum, where distinct isolated ecotypes occupying cave entrances in foothills originated from a widespread alpine lineage36. The impact of elevational difference on divergence between foothill (montane) and alpine populations has been documented in Dianthus callizonus37, Primula merrilliana23, and Solidago virgaurea38. The parallel origin of ecotypes has been also documented previously with genetic data in different environments such as sand dune vs. non-dune in Helianthus petiolaris39, sand dune vs. rock in Senecio lautus40, serpentine vs. non-serpentine soils in Solidago virgaurea41, wave vs. crab predation in the mollusc Littorina saxatilis42, and freshwater vs. saltwater in the fish Gasterosteus aculeatus34. The absence of similar studies in subalpine plants might simply reflect less attention to this phenomenon of differentiation in subalpine environment.
In contrast to differentiation among the three distinct mountain ranges (mean pairwise FST = 0.13), foothill and subalpine populations within each region were less differentiated (pairwise FST ranging from 0.08 to 0.10) and accounted for a negligible and non-significant proportion of variation in AMOVA tests (from 2.32% to 6.11% variation). The variation in these values across mountain ranges likely reflects different spatial and elevational distances between subalpine and foothill populations in each region as well as heterogeneity of habitats. In the Jeseníky and northern part of the Tatry (S9, S11; Vysoké/Belanské Tatry) we observed a nearly continuous occurrence of P. elatior along an elevational gradient (corresponding with mean pairwise FST between foothill and subalpine populations of 0.08 and 0.07, respectively)43,44. In contrast, in the Krkonoše and the Nízké Tatry, there was a gap in the occurrence of several hundred meters, which can restrict gene flow (reflected by higher mean pairwise FST of 0.10 and 0.13, respectively), as was also revealed in Knautia in the Krkonoše45. Available population genetic studies have documented that the levels of differentiation among foothill and alpine/subalpine populations may vary considerably among species, from high (mean pairwise FST = 0.19 in Primula merrilliana23) to low (mean pairwise FST = 0.02 in Arabidopsis arenosa46 also from the Tatry).
Our study shows that genetic differentiation along an elevational gradient may vary within a species, from one mountain range to another, although this pattern appears to be less extreme in Primula relative to Heliosperma, where the individual alpine-foothill pairs varied considerably in differentiation (mean FST 0.17–0.43)36. The extent of differentiation likely indicates if populations have an older or younger origin/divergence. Besides elevational differences, lower or comparable differentiation to our study was also observed in two Primula species between grassland and forest habitats in lowland habitats at a fine-scale of several kilometres (mean FST = 0.02 in P. elatior16 and 0.08 in P. veris)47. Our observation in P. elatior thus suggests that elevation is generally a weak isolating barrier whose strength may further vary across independent colonization events in different mountains.
The relatively low foothill-subalpine differentiation that we observed in P. elatior is likely due to high levels of retained ancestral polymorphism and/or persisting gene flow along the elevational gradient. This was suggested by coalescent modelling in which we found consistent support for models with unidirectional migration from subalpine to foothill populations. The unidirectional migration likely reflected gene flow after the colonization of subalpine habitats in postglacial times. This was the best supported model in all three mountain ranges, however, with varying estimated strength across the regions. Such unidirectional downslope migration has been observed also in P. merrilliana23, in which the seed mobility is generally restricted in a similar way to P. elatior due to the lack of any long-distance seed dispersal adaptation48. In the mountains, seed flow may be further enhanced by washing downslope by mountain streams, but pollen flow may be restricted along an elevational gradient due to differences in flowering phenology between populations at different elevations4. In the closely related species P. veris, differences in the flowering phenology affect gene flow even among distinct habitats in the same (lowland) elevation47.
In contrast to the low foothill-subalpine differentiation that we observed, individual subalpine populations were the most highly differentiated entities within two of the three studied regions (21.42% and 24.18% variation in the Krkonoše and the Tatry, respectively, split into distinct genetic clusters, Fig. 2). For generalization of the differentiation of subalpine populations, however, more populations shall be compared. High differentiation between subalpine populations in the Krkonoše has been documented also in another species growing in the glacial cirques – Gentiana pannonica (14.15%)49. The Jeseníky was the only exception, in which both subalpine populations were restricted to a single glacial cirque with likely high opportunities for gene flow and this is likely responsible for the low among-population differentiation and FST values. These results suggest that mountain ridges may act as relatively strong migration barriers, which can affect gene flow more strongly than elevational differences between subalpine and foothill populations. Although higher differentiation among subalpine populations may also reflect past bottleneck events during their origin from their foothill counterparts, we do not consider this likely. We have not observed significantly reduced levels of diversity in these populations (Table 2) – a clear sign of past bottleneck in populations50.
In contrast to the differentiation observed among subalpine populations, differentiation among foothill populations was low (3.03–7.82% in different regions). This relatively low differentiation is may be the product of a higher density and large population size of foothill populations. Pollination is more efficient in large populations and the larger population size may provide more opportunities for gene flow among populations16,51.
Our results show that subalpine colonization did not appear to lead to a loss of genetic diversity relative to foothill populations. The differences in genetic diversity of subalpine vs. foothill populations were non-significant over the replicated mountain ranges (Table 2). Moreover, neither index of genetic diversity shows any consistent trend across the regions with respect to the elevational groups (Supplementary Fig. S2). On the other hand, foothill populations from the Tatry were together the most genetically diverse of all populations included in our study. This could be a consequence of the foothill habitat16,17,52, which may have experienced the long-term isolation of populations in this area serving as a glacial refugium for temperate species46,53. Generally, subalpine populations of P. elatior appear to be able to maintain genetic diversity equal to foothill populations despite smaller population sizes and the spatial isolation of the glacial cirques in which they occur. This could be due to a large size of the initial colonizing population (ruling out founder effect) and the relative stability of the habitat in postglacial time providing good conditions for the persistence of sufficiently large populations. An additional non-exclusive explanation might be a gradual and relatively slow pace of the shifts of the treeline during the Holocene, which might have maintained sufficient population sizes without bottlenecks, and thus preserve genetic diversity during the colonization process23.
In summary, we describe a case of parallel colonization of subalpine habitats from multiple foothill gene pools. Our results imply that there is a distinct mountain diversity in the subalpine habitats – the subalpine populations are genetically differentiated from their foothill counterparts and from each other. In addition, subalpine populations regularly preserve genetic diversity at similar levels relative to their foothill counterparts. However, ongoing gene flow, in particular from subalpine to foothill habitats together with low levels of differentiation, likely linked to a recent (postglacial) origin of the subalpine populations, seems to prevent any stronger differentiation along the elevational gradient that may lead to speciation.
Methods
Study species and sampling
Primula elatior is an outcrossing perennial plant with a basal rosette of leaves. Flowers are produced in an umbel, characterized by distyly and self-incompatibility25. The main pollinators are Hymenoptera (mostly bumblebees) and Diptera54,55. Seed mobility in this species is generally restricted due to lack of any adaptations for long-distance dispersal, for instance, compared to closely related species P. vulgaris48. Therefore, seeds are dispersed autonomously for short distances, and occasionally they can be washed downslope from streamside habitats.
Plant material was collected in 2015–2016 in the three mountain ranges in central Europe: the Jeseníky, the Krkonoše, and the Tatry. In the Jeseníky and the Krkonoše, we sampled all known populations in subalpine glacial cirques and representative set from distinct valleys in the foothills. In the Tatry, we included populations from two distinct subalpine parts: the southern mountain range of the Nízké Tatry, the northern-eastern mountain range of the Vysoké/Belanské Tatry, and representative foothill populations in the basins between these two mountain ranges (called overall the Tatry). Our dataset comprises in total 202 individuals from 16 populations (Table 1 and Supplementary Table S1). In each mountain range, we sampled multiple foothill (below treeline) and subalpine (above treeline) populations. Individuals in the populations were sampled randomly, but with a minimum distance of approximately 1 m between individuals to avoid collecting clones. Leaf tissue from five to twenty individuals per population was immediately dried in silica gel for subsequent DNA extraction and microsatellite genotyping.
Microsatellite genotyping
Genomic DNA was extracted from dry tissue using a modified NaCl/CTAB protocol56. We employed 12 microsatellite loci developed by Van Geert et al.57, Bickler et al.58, and Seino et al.59 (Supplementary Table S2). From those, seven loci, originally developed for P. veris58, were successfully cross-amplified in P. elatior: PV 23741, PV 21795, PV 27775, PV 4767, PV 23424, PV 19773, and PV 279. The others (Paca 11, Paca 38, Paca 78, Priv 4, and Priv 7) have been already cross-amplified in P. elatior by Seino et al.59 and Van Geert et al.57. Our cross-amplification of the 12 loci was successful, meaning that we consistently amplified variable loci across the sample set, contrary to three additional loci Paca 40459, PV 888058, and PV 2672058, which were not consistently amplified, and therefore not employed in the study. The fluorescently labelled primers (dyes: PET, NED, 6-FAM, and VIC; Applied Biosystems) were designed into two multiplexes based on the results from a complementary threshold analysis in Multiplex manager 1.2. Microsatellite loci were amplified using the QIAGEN Multiplex PCR Kit, with a total reaction volume of 5 μl of QIAGEN mix. The mix contained 0.25 μl of forward primer and 0.25 μl of reverse primer, 1 μl of ddH2O, 2.5 μl of Master Mix, and we added 1 μl (10 ng) of DNA. The PCR amplification was conducted in a thermocycler (Eppendorf Mastercycler Pro) under the following conditions for both multiplexes: 5 min of denaturation at 95 °C, followed by 35 cycles of 95 °C at 30 s, 57 °C for 90 s, 72 °C for 40 s, and a final extension of 68 °C for 30 min. Amplification products were separated using 3130xl Genetic Analyser (DNA laboratory of Faculty of Science, Charles University, Prague) with GeneScan 500 LIZ (Applied Biosystems) as an internal standard.
Genetic structure and diversity
Allele sizes were determined in genemarker 2.6 (SoftGenetics). We checked possible presence of null alleles, stuttering, large allele dropout by the program micro-checker 2.2.360. None of the loci showed presence of null alleles in more than half of the populations (maximum seven for Priv 4 and six for Paca 78), and we thus retained all 12 microsatellite loci in analyses. We tested if loci significantly deviated from HWE in each population in R, package pegas61. None of the populations had significant deviation from HWE in more than half of the loci (maximum seven in S5) (Table 1).
First, we explored population structure in the entire dataset as well as separately for each region using Bayesian clustering in structure 2.3.362 employing Abel HPC cluster of the University of Oslo. We used independent allele frequencies model with admixture, which allows for mixed ancestry of individuals. The number of clusters was set from K = 1 to K = 10 for entire dataset and from K = 1 to K = max, in which “max” equalled number of sampled populations in particular regions (K = 6 for each of the Krkonoše and the Tatry datasets, K = 4 for the Jeseníky dataset). Analysis for each K was performed with 20 replicates, the initial length of burn-in period 100,000 and 1,000,000 of Markov chain Monte Carlo (MCMC) replicates after burn-in. Similarity coefficients among runs of the same K63 were calculated using Structure-sum-2009 script31 in R 3.3.264. For an optimal number of clusters (K), we considered the partition, where the rising likelihood of K values started to flatten and which also exhibited high similarity among replicated runs for that particular K (Supplementary Fig. S1). Some analyses allowed several possibilities for the optimal number of K, due to the hierarchical genetic structure of populations65, in that case, we presented several partitions. Outputs from structure analyses were graphically visualized in Structure Plot V2.066.
Further, we visualized relationships among individuals and populations using distance-based approaches. Firstly, genetic relationships among individuals were plotted in centred principal component analysis (PCA) calculated in R, package adegenet67. Secondly, networks of pairwise FST distance among populations were created based on neighbor-net algorithm in SplitsTree 4.13.168. Nei’s pairwise FSTs69, in which heterozygosities are weighted by group sizes, and therefore comparison between populations with different sizes of individuals is possible, were calculated in R, packages adegenet67 and hierfstat70.
In order to test differences in population genetic properties, we calculated descriptive statistics of all populations with respect to small number of individuals and imbalance sampling. Due to varying number of individuals samples per populations (from five to 20 individuals) we employed following subsampling strategy to rule out the effect of varying sample size: five individuals per population were randomly selected, we calculated the corresponding statistics and repeated the process 100 times and we further presented mean value with standard deviation from 100 replicates (https://github.com/MarekLipan/Population-subsampling/blob/master/Genind_subsampling_func.R). We calculated observed heterozygosity (HO) and expected heterozygosity (HE) by subsampling in R, packages adegenet67 and hierfstat70. Further, we calculated the numbers of alleles, allelic richness (with reference population size of five individuals per population, 1,000 permutations), and Nei’s unbiased estimator for gene diversity (HS)71, which is corrected for small sample size, in Microsatellite analyser (MSA) 4.0572. Furthermore, we quantified the proportion of rare alleles using the frequency of down-weighted marker index (DW index), calculated as a ratio of means from the presence-absence matrix of alleles, which makes the measure less sensitive to different number of individuals per population31. DW index was calculated using R-script AFLPdat31. Finally, a number of private alleles was counted in R, package PopGenKit73. Differences in allelic richness, HE, HS, and in DW index between foothill and subalpine group were tested by hierarchical ANOVA with region as a random effect factor in R, package stats. Further, hierarchical structuring of genetic variation among populations within foothill vs. subalpine group in each region was revealed by analysis of molecular variance (AMOVA) in Arlequin 3.174. AMOVAs were calculated by the method of the number of different alleles (FST-like) with 1,000 permutations. Finally, the structuring of variation among foothill and subalpine populations in each region was also explored by hierarchical AMOVAs.
Estimation of gene flow direction and migration rates
To identify the direction and intensity of migration among the foothill and subalpine populations, we searched for optimal models of migrations between populations within each target mountain range in a coalescent framework using migrate-n version 3.6.1175. We have chosen two pairs of foothill and subalpine populations from the Jeseníky, the Krkonoše, and one pair from sub-range of the Tatry (Nízké Tatry), focusing on populations with the maximum number of genotyped individuals (n ≥ 10). To keep the simulation scenarios feasible, we worked with two-population models that were iterated among all possible population combinations within each mountain range: we analysed pairs of subalpine-subalpine, foothill-subalpine, and foothill-foothill populations. For each pair, we modelled past evolutionary history under Bayesian inference of five migration models differing in presence and directionality of migration. We allowed bidirectional migration between two populations (model 1), unidirectional migration (models 2 and 3), panmixia (model 4 assuming that two populations belong to one panmictic population), and zero migration between two separate populations (model 5). In case of foothill-subalpine comparisons, the model 2 allowed migration from foothill to subalpine while the model 3 assumed only migration from subalpine to foothill populations.
We used microsatellite data type with Brownian motion microsatellite model. The mutation rate was set constant over all loci. migrate-n estimated two parameters Theta - θ (mutation scaled population size) and M (mutation scaled immigration rate). The starting values of θ and M were calculated from Wright’s FST using prior values (for θ: minimum = 0.004, delta ranged from 0.9 to 2.0, maximum ranged from 9.0 to 20, bins = 500; for M: minimum = 0, delta ranged from 80 to 100, maximum ranged from 800 to 1000, bins = 500). Prior distributions of population sizes and migration rates were set based on the personal knowledge of the populations from the field with a broader range of both parameters with the aim to achieve the best searching of the space. To reach the stable states we ran each model for each pair multiple times (at least five). We have also checked the effective sample size, which was well over 500 (approximately 3000–5000). After burn-in of 20,000, we sampled 500,000 states from a single Markov chain, one every 5,000 steps. Four chains were run in parallel with heating static scheme (temperatures: 1.0, 1.5, 3.0, 10,000). According to Hodel et al.32, based on θ and M parameters we calculated a number of immigrants per generation following the formula: Nm = [(θx ∗ My→x)/4] (for nuclear loci). For calculating Nm values, we used median values for both θ and M parameters.
To select the most likely model among the five models for each population pair, we used Bayes factors comparison. Bayes factor allows comparing nested and non-nested models, without assuming normality, or large samples76,77. We calculated natural log Bayes factors following the formula: LBF = ln [mL (model1)] − ln [mL (model2)], in which model1 is a model with the highest marginal likelihood and model2 is each of the other models. We used “Bezier” approximated marginal likelihood calculated using the thermodynamic integration with the heating scheme described above). Marginal likelihood is the integral of the likelihood function over the complete parameter range. Afterwards, we calculated the probability of each model following the formula:
in which mLmodeli is the marginal likelihood of modeli and is the sum of the marginal likelihoods of all other models.
Supplementary information
Acknowledgements
The computations were performed using resources provided by UNINETT Sigma2 - the National Infrastructure for High Performance Computing and Data Storage in Norway (http://www.hpc.uio.no/). Access to computing and storage facilities provided under the programme “Projects of Large Research, Development, and Innovations Infrastructures” (CESNET LM2015042), was greatly appreciated. We are grateful to Kristýna Šemberová, who helped us with the maps. This study was supported by Grant No. 925916 from the Charles University Grant Agency. Additional support was provided by Charles University, Primus-SCI-35.
Author Contributions
V.K., M.D.N. and F.K. designed research; V.K. performed research; V.K. and F.K. analysed data; V.K. and F.K. wrote the paper and all authors revised the paper.
Data Availability
All data generated or analysed during this study are included in this article (and its Supplementary Information Files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39669-2.
References
- 1.Körner, C. Alpine plant life: functional plant ecology of high mountain ecosystems. (Springer Science & Business Media, 2003).
- 2.Flatscher, R., Frajman, B., Schönswetter, P. & Paun, O. Environmental heterogeneity and phenotypic divergence: Can heritable epigenetic variation aid speciation? Genet. Res. Int. 1–9, 10.1155/2012/698421 (2012). [DOI] [PMC free article] [PubMed]
- 3.Pfennig DW, et al. Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol. Evol. 2010;25:459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Bertel C, Hülber K, Frajman B, Schönswetter P. No evidence of intrinsic reproductive isolation between two reciprocally non-monophyletic, ecologically differentiated mountain plants at an early stage of speciation. Evol. Ecol. 2016;30:1031–1042. doi: 10.1007/s10682-016-9867-y. [DOI] [Google Scholar]
- 5.Clausen J, Keck DD, Hiesey WM. The Concept of Species Based on Experiment. Am. J. Bot. 1939;26:103–106. doi: 10.1002/j.1537-2197.1939.tb12875.x. [DOI] [Google Scholar]
- 6.Lowry DB. Ecotypes and the controversy over stages in the formation of new species. Biol. J. Linn. Soc. 2012;106:241–257. doi: 10.1111/j.1095-8312.2012.01867.x. [DOI] [Google Scholar]
- 7.Clausen J, Keck DD, Hiesey WM. Regional Differentiation in Plant Species. Am. Nat. 1941;75:231–250. doi: 10.1086/280955. [DOI] [Google Scholar]
- 8.Hiesey WM, Clausen J, Keck DD. Relations between Climate and Intraspecific Variation in Plants. Am. Nat. 1942;76:5–22. doi: 10.1086/281009. [DOI] [Google Scholar]
- 9.Funk WC, Blouin MS, Corn PS, Maxell BA, Pilliod DS. Population structure of Columbia spotted frogs (Rana lueteiventris) is strongly affected by the landscape. Mol. Ecol. 2005;14:1–14. doi: 10.1111/j.1365-294X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- 10.Ohsawa T, Ide Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Glob. Ecol. Biogeogr. 2008;17:152–163. doi: 10.1111/j.1466-8238.2007.00357.x. [DOI] [Google Scholar]
- 11.Levin D. The recurrent origin of plant races and species. Syst. Bot. 2001;26:197–204. [Google Scholar]
- 12.Šrámková-Fuxová G, et al. Range-wide genetic structure of Arabidopsis halleri (Brassicaceae): glacial persistence in multiple refugia and origin of the Northern Hemisphere disjunction. Bot. J. Linn. Soc. 2017;185:321–342. doi: 10.1093/botlinnean/box064. [DOI] [Google Scholar]
- 13.Fustier M, Brandenburg J, Boitard S, Lapeyronnie J, Eguiarte LE. Signatures of local adaptation in lowland and highland teosintes from whole-genome sequencing of pooled samples. Mol. Ecol. 2017;26:2738–2756. doi: 10.1111/mec.14082. [DOI] [PubMed] [Google Scholar]
- 14.Holliday JA, Zhou L, Bawa R, Zhang M, Oubida RW. Evidence for extensive parallelism but divergent genomic architecture of adaptation along altitudinal and latitudinal gradients in Populus trichocarpa. New Phytol. 2016;209:1240–1251. doi: 10.1111/nph.13643. [DOI] [PubMed] [Google Scholar]
- 15.Bettin O, Cornejo C, Edwards PJ, Holderegger R. Phylogeography of the high alpine plant Senecio halleri (Asteraceae) in the European Alps: In situ glacial survival with postglacial stepwise dispersal into peripheral areas. Mol. Ecol. 2007;16:2517–2524. doi: 10.1111/j.1365-294X.2007.03273.x. [DOI] [PubMed] [Google Scholar]
- 16.Jacquemyn H, Honnay O, Galsbusera P, Roldán-Ruiz I. Genetic structure of the forest herb Primula elatior in a changing landscape. Mol. Ecol. 2004;13:211–219. doi: 10.1046/j.1365-294X.2003.02033.x. [DOI] [PubMed] [Google Scholar]
- 17.Odat N, Jetschke G, Hellwig FH. Genetic diversity of Ranunculus acris L. (Ranunculaceae) populations in relation to species diversity and habitat type in grassland communities. Mol. Ecol. 2004;13:1251–1257. doi: 10.1111/j.1365-294X.2004.02115.x. [DOI] [PubMed] [Google Scholar]
- 18.Ortego J, Riordan EC, Gugger PF, Sork VL. Influence of environmental heterogeneity on genetic diversity and structure in an endemic southern Californian oak. Mol. Ecol. 2012;21:3210–3223. doi: 10.1111/j.1365-294X.2012.05591.x. [DOI] [PubMed] [Google Scholar]
- 19.Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
- 20.Ellstrand NC, Elam DR. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 1993;24:217–242. doi: 10.1146/annurev.es.24.110193.001245. [DOI] [Google Scholar]
- 21.Günther T, Lampei C, Barilar I, Schmid KJ. Genomic and phenotypic differentiation of Arabidopsis thaliana along altitudinal gradients in the North Italian Alps. Mol. Ecol. 2016;25:3574–3592. doi: 10.1111/mec.13705. [DOI] [PubMed] [Google Scholar]
- 22.Luo Y, Widmer A, Karrenberg S. The roles of genetic drift and natural selection in quantitative trait divergence along an altitudinal gradient in Arabidopsis thaliana. Heredity (Edinb). 2015;114:220–228. doi: 10.1038/hdy.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao J, et al. Genetic diversity and gene flow within and between two different habitats of Primula merrilliana (Primulaceae), an endangered distylous forest herb in eastern China. Bot. J. Linn. Soc. 2015;179:172–189. doi: 10.1111/boj.12305. [DOI] [Google Scholar]
- 24.Valentine, D. H. & Kress, A. In Flora Europaea 3 (ed. Tutin, T.) (Cambridge University Press, 1972).
- 25.Richards, J. Primula. (Timber Press, 2003).
- 26.Konečná, V. Phenotypic variability and evolutionary relationships among populations of Primula elatior along an altitudinal gradient (Master’s thesis). (Charles University, Prague, Czech Republic, 2017).
- 27.Jacquemyn H, Brys R, Hermy M. Patch occupancy, population size and reproductive success of a forest herb (Primula elatior) in a fragmented landscape. Oecologia. 2002;130:617–625. doi: 10.1007/s00442-001-0833-0. [DOI] [PubMed] [Google Scholar]
- 28.Jacquemyn H, Vandepitte K, Roldán-Ruiz I, Honnay O. Rapid loss of genetic variation in a founding population of Primula elatior (Primulaceae) after colonization. Ann. Bot. 2008;103:777–783. doi: 10.1093/aob/mcn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Rossum F, Triest L. Fine-scale genetic structure of the common Primula elatior (Primulaceae) at an early stage of population fragmentation. Am. J. Bot. 2006;93:1281–1288. doi: 10.3732/ajb.93.9.1281. [DOI] [PubMed] [Google Scholar]
- 30.Krahulec F. Species of vascular plants endemic to the Krkonoše Mts (Western Sudetes) Preslia. 2006;78:503–516. [Google Scholar]
- 31.Ehrich D. AFLPDAT: A collection of R functions for convenient handling of AFLP data. Mol. Ecol. Notes. 2006;6:603–604. doi: 10.1111/j.1471-8286.2006.01380.x. [DOI] [Google Scholar]
- 32.Hodel RGJ, D S Cortez MB, Soltis PS, Soltis DE. Comparative phylogeography of black mangroves (Avicennia germinans) and red mangroves (Rhizophora mangle) in Florida: Testing the maritime discontinuity in coastal plants. Am. J. Bot. 2016;103:730–739. doi: 10.3732/ajb.1500260. [DOI] [PubMed] [Google Scholar]
- 33.Beerli P, Felsenstein J. Maximum-Likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics. 1999;152:763–773. doi: 10.1093/genetics/152.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hufford, M. B. et al. The Genomic Signature of Crop-Wild Introgression in Maize. PLoS Genet. 9, (2013). [DOI] [PMC free article] [PubMed]
- 36.Trucchi E, Frajman B, Haverkamp THA, Schönswetter P, Paun O. Genomic analyses suggest parallel ecological divergence in Heliosperma pusillum (Caryophyllaceae) New Phytol. 2017;216:267–278. doi: 10.1111/nph.14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabel A, Sattler J. & Reisch Christoph. Genetic variation and performance of the alpine plant species Dianthus callizonus differ in two elevational zones of the Carpathians. Alp. Bot. 2017;127:65–74. doi: 10.1007/s00035-016-0177-3. [DOI] [Google Scholar]
- 38.Sakaguchi S, et al. Phylogeographic analysis of the East Asian goldenrod (Solidago virgaurea complex, Asteraceae) reveals hidden ecological diversification with recurrent formation of ecotypes. Ann. Bot. 2018;121:489–500. doi: 10.1093/aob/mcx182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrew RL, Ostevik KL, Ebert DP, Rieseberg LH. Adaptation with gene flow across the landscape in a dune sunflower. Mol. Ecol. 2012;21:2078–2091. doi: 10.1111/j.1365-294X.2012.05454.x. [DOI] [PubMed] [Google Scholar]
- 40.Roda, F. et al. Genomic evidence for the parallel evolution of coastal forms in the Senecio lautus complex. 22, 2941–2952 (2013). [DOI] [PubMed]
- 41.Sakaguchi S, et al. Simultaneous evaluation of the effects of geographic, environmental and temporal isolation in ecotypic populations of Solidago virgaurea. New Phytol. 2017;216:1268–1280. doi: 10.1111/nph.14744. [DOI] [PubMed] [Google Scholar]
- 42.Butlin RK, et al. Parallel evolution of local adaptation and reproductive isolation in the face of gene flow. Evolution (N. Y). 2014;68:935–949. doi: 10.1111/evo.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kochjarová, J. In Flóra Slovenska VI/4 (VEDA, 2017).
- 44.Bureš, L. Chráněné a ohrožené rostliny CHKO Jeseníky. (Agentura Rubico, s.r.o., 2013).
- 45.Kolář F, Kaplan Z, Suda J, Štech M. Populations of Knautia in ecologically distinct refugia on the Hercynian massif belong to two endemic species. Preslia. 2015;87:363–386. [Google Scholar]
- 46.Kolář F, et al. Northern glacial refugia and altitudinal niche divergence shape genome-wide differentiation in the emerging plant model Arabidopsis arenosa. Mol. Ecol. 2016;25:3929–3949. doi: 10.1111/mec.13721. [DOI] [PubMed] [Google Scholar]
- 47.Deschepper, P., Brys, R., Fortuna, M. A. & Jacquemyn, H. Analysis of spatial genetic variation reveals genetic divergence among populations of Primula veris associated to contrasting habitats. Sci. Rep. 1–12, 10.1038/s41598-017-09154-9 (2017). [DOI] [PMC free article] [PubMed]
- 48.Taylor K, Woodell SRJ. Biological Flora of the British Isles: Primula elatior (L.) Hill. J. Ecol. 2008;96:1098–1116. doi: 10.1111/j.1365-2745.2008.01418.x. [DOI] [Google Scholar]
- 49.Ekrtová E, Štech M, Fér T. Pattern of genetic differentiation in Gentiana pannonica Scop.: did subalpine plants survive glacial events at low altitudes in Central Europe? Plant Syst. Evol. 2012;298:1383–1397. doi: 10.1007/s00606-012-0644-2. [DOI] [Google Scholar]
- 50.England PR, et al. Effects of intense versus diffuse population bottlenecks on microsatellite genetic diversity and evolutionary potential. Conserv. Genet. 2003;4:595–604. doi: 10.1023/A:1025639811865. [DOI] [Google Scholar]
- 51.Shao J, Zhang X, Zhang Z, Zhu G. Identification of effective pollinators of Primula merrilliana and effects of flower density and population size on pollination efficiency. J. Syst. Evol. 2008;46:537–544. [Google Scholar]
- 52.Lowry DB, Rockwood RC, Willis JH. Ecological reproductive isolation of coast an inland races. Evolution (N. Y). 2008;62:2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juřičková L, Horáčková J, Ložek V. Direct evidence of central European forest refugia during the last glacial period based on mollusc fossils. Quat. Res. 2014;82:222–228. doi: 10.1016/j.yqres.2014.01.015. [DOI] [Google Scholar]
- 54.Schou O. The distyly in Primula elatior (L.) Hill (Primulaceae), with a study of flowering phenology and pollen flow. Bot. J. Linn. Soc. 1983;86:261–274. doi: 10.1111/j.1095-8339.1983.tb00973.x. [DOI] [Google Scholar]
- 55.Proctor, M., Yeo, P. & Lack, A. The natural history of pollination. (HarperCollins Publishers, 1996).
- 56.Štorchová H, et al. An improved method of DNA isolation from plants collected in the field and conserved in saturated NaCl/CTAB solution. Taxon. 2000;49:79–84. doi: 10.2307/1223934. [DOI] [Google Scholar]
- 57.Van Geert A, et al. Isolation and characterization of microsatellite loci in primrose (Primula vulgaris) Belgian J. Bot. 2006;139:261–264. [Google Scholar]
- 58.Bickler C, A’Hara S, Cottrell J, Rogers L, Bridle J. Characterisation of thirteen polymorphic microsatellite markers for cowslip (Primula veris L.) developed using a 454 sequencing approach. Conserv. Genet. Resour. 2013;5:1185–1187. doi: 10.1007/s12686-013-9997-2. [DOI] [Google Scholar]
- 59.Seino, M. M., de Vega, C., Bazaga, P., Jacquemyn, H. & Herrera, C. M. Development and characterization of microsatellite loci for the primrose Primula vulgaris and successful cross-amplification in the congeneric P. elatior and P. veris. Conserv. Genet. Resour. 653–655, 10.1007/s12686-014-0171-2 (2014).
- 60.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. doi: 10.1111/j.1471-8286.2004.00684.x. [DOI] [Google Scholar]
- 61.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 62.Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. Plos Biol. 2005;3:1289–1299. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.R Core Team. R: a language and environment for statistical computing, Vienna, Austria, https://www.r-project.org/ (2016).
- 65.Ehrich D, et al. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Mol. Ecol. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- 66.Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG. Structure plot: a program for drawing elegant STRUCTURE bar plots in user friendly interface. Springerplus. 2014;3:431. doi: 10.1186/2193-1801-3-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 68.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 69.Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes. 2005;2:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- 71.Nei, M. Molecular evolutionary genetics. (Columbia university press, 1987).
- 72.Dieringer D, Schlötterer C. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes. 2003;3:167–169. doi: 10.1046/j.1471-8286.2003.00351.x. [DOI] [Google Scholar]
- 73.Rioux Paquette, S. PopGenKit: useful functions for (batch) file conversion and data resampling in microsatellite datasets. R package version 1.0. (2012).
- 74.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinformatic. 2005;1:47–50. doi: 10.1177/117693430500100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beerli, P. In Population Genetics for Animal Conservation (eds Bertorelle, G., Bruford, M. W., Hauffe, H. C., Rizzoli, A. & Vernesi, C.) (Cambridge University Press, 2009).
- 76.Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185:313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kass RE, Raftery AE. Bayes factors. J. Am. Stat. Assoc. 1995;90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article (and its Supplementary Information Files).