Abstract
The role of the N-methyl-d-aspartate receptor (NMDAr) as a contributor to maladaptive neuroplasticity underlying the maintenance of chronic pain is well established. Agmatine, an NMDAr antagonist, has been shown to reverse tactile hypersensitivity in rodent models of neuropathic pain while lacking the side effects characteristic of global NMDAr antagonism, including sedation and motor impairment, indicating a likely subunit specificity of agmatine’s NMDAr inhibition. The present study assessed whether agmatine inhibits subunit-specific NMDAr-mediated current in the dorsal horn of mouse spinal cord slices. We isolated NMDAr-mediated excitatory postsynaptic currents (EPSCs) in small lamina II dorsal horn neurons evoked by optogenetic stimulation of Nav1.8-containing nociceptive afferents. We determined that agmatine abbreviated the amplitude, duration, and decay constant of NMDAr-mediated EPSCs similarly to the application of the GluN2B antagonist ifenprodil. In addition, we developed a site-specific knockdown of the GluN2B subunit of the NMDAr. We assessed whether agmatine and ifenprodil were able to inhibit NMDAr-mediated current in the spinal cord dorsal horn of mice lacking the GluN2B subunit of the NMDAr by analysis of electrically evoked EPSCs. In control mouse spinal cord, agmatine and ifenprodil both inhibited amplitude and accelerated the decay kinetics. However, agmatine and ifenprodil failed to attenuate the decay kinetics of NMDAr-mediated EPSCs in the GluN2B-knockdown mouse spinal cord. The present study indicates that agmatine preferentially antagonizes GluN2B-containing NMDArs in mouse dorsal horn neurons.
NEW & NOTEWORTHY Our study is the first to report that agmatine preferentially antagonizes the GluN2B receptor subunit of the N-methyl-d-aspartate (NMDA) receptor in spinal cord. The preferential targeting of GluN2B receptor is consistent with the pharmacological profile of agmatine in that it reduces chronic pain without the motor side effects commonly seen with non-subunit-selective NMDA receptor antagonists.
Keywords: agmatine, arginine decarboxylase, glutamate, l-arginine, neuroplasticity, polyamine
INTRODUCTION
The decarboxylated form of l-arginine, agmatine, antagonizes the N-methyl-d-aspartate receptor (NMDAr) in a manner that we hypothesize may be GluN2B subunit selective. We and others have shown that agmatine inhibits NMDAr-evoked current (Yang and Reis 1999) and behavior (Fairbanks et al. 2000; Roberts et al. 2005) as well as nitric oxide (NO) production (Galea et al. 1996). Agmatine also inhibits the development of chronic pain (Courteix et al. 2007; Fairbanks et al. 2000; Horváth et al. 1999), but, notably, without the motor toxicity that is often observed with NMDAr antagonists (Fairbanks et al. 2000; Nguyen et al. 2003).
It is well established that the NMDAr plays a critical role in synaptic plasticity throughout the central nervous system (CNS). NMDAr-mediated potentiation of synaptic strength between nociceptive afferents and their targets in the dorsal horn is a central mechanism behind persistent pain following nerve injury (Sandkühler and Gruber-Schoffnegger 2012; Sandkühler and Liu 1998). However, the use of clinically available NMDAr antagonists has not led to effective treatments because of a range of adverse effects. These include, but are not limited to, cognition (Serafini et al. 2013), learning and memory deficits (Butelman 1989; Sanger and Joly 1991), sedation (Nelson et al. 1997), motor dysfunction (Parsons 2001), phencyclidine (PCP)-like stimulatory behavior (Sagratella et al. 1992), and PCP-primed abuse potential (Beardsley et al. 1990). Consequently, many of the NMDAr antagonists were not well tolerated and have not advanced clinically (Chen and Lipton 2006; Nelson et al. 1997). It has been proposed that these side effects are associated with high-affinity NMDAr antagonists (Nelson et al. 1997) or subunit-nonselective antagonists (Sanger and Joly 1991). Interest in development of subunit-selective NMDAr antagonists has increased in recent years for improving treatment of a variety of CNS disorders (Chaki and Fukumoto 2015; Gerhard et al. 2016; Zádori et al. 2014), including pain (Boyce et al. 1999; Zhuo 2017).
The NMDAr is composed of GluN1 (glycine binding) and GluN2 (glutamate binding) subunits. There are four distinct isoforms of the GluN2 subunit (GluN2A–GluN2D); their gene expression ratio has been shown to vary considerably throughout the CNS (Goebel and Poosch 1999). In the spinal cord, GluN2B protein has been shown to have restricted localization in the dorsal horn (Boyce et al. 1999), where sensory processing is governed. Electrophysiological evidence suggests that NMDArs containing GluN2B and GluN2D subunits have a dominant role in lamina I synaptic transmission (Hildebrand et al. 2014). Low-affinity (memantine, amantadine) or NMDA receptor subunit 2B-selective (GluN2B) antagonists such as ifenprodil (Williams et al. 1993) are thought to have a higher therapeutic index (Layton et al. 2006; Qu et al. 2009) due to more limited motor effects, which is consistent with the expression pattern of GluN2B. Development of GluN2B subtype-selective NMDAr antagonists may offer an improved pharmacological option for neuropathic pain (Beinat et al. 2010; Niesters and Dahan 2012; Santangelo et al. 2012).
On the basis of the aforementioned pharmacological profile of agmatine, we have hypothesized that it preferentially antagonizes the GluN2B receptor subunit of the NMDAr. However, such a proposal has not been previously assessed physiologically. The aim of this study was to determine whether agmatine preferentially antagonizes the GluN2B subunit of the NMDAr. We specifically hypothesized that agmatine selectively reduces the long-duration, GluN2B-mediated decay kinetics of NMDAr-mediated evoked excitatory postsynaptic currents (eEPSCs). Using selective optogenetic activation of Nav1.8-expressing nociceptive afferents in dorsal root in an in vitro spinal cord slice preparation, we determined the inhibition characteristics of superfused agmatine, the GluN2B-selective antagonist ifenprodil, and the GluN2A-selective antagonist PEAQX (also referred to as NVP-AAM077). We further hypothesized that the inhibitory characteristics of agmatine on NMDAr-mediated EPSCs would not be evident in spinal cord slices from mice with a conditional knockdown (KD) of the GluN2B subunit. We compared inhibitory responses to both agmatine and ifenprodil between slices generated from GluN2B-KD mice and controls.
MATERIALS AND METHODS
Animals.
Mice were bred for optogenetic or GluN2B conditional knockout features and deeply anesthetized (isoflurane) at 6–8 wk for spinal cord extraction and sectioning. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Spinal cord slice preparation.
Transcardial perfusion was performed on Nav1.8-channelrhodopsin-2 (ChR2) or GluN2B-floxed mice (typically 7 wk old) using oxygenated (95% O2-5% CO2) high-sucrose/kynurenic acid artificial cerebrospinal fluid (aCSF) containing (in mM) 95 NaCl, 1.8 KCl, 1.2 KH2PO4, 0.5 CaCl2, 7 MgSO4, 26 NaHCO3, 15 glucose, 50 sucrose, and 1 kynurenic acid (Doolen et al. 2012). The spinal cord was extracted and sliced (400 µm thick, transverse) using a vibrating microtome (Leica VT1200S; Wetzlar, Germany). Slices were incubated at 37°C for 1 h in oxygenated aCSF containing (in mM) 127 NaCl, 1.8 KCl, 1.2 KH2PO4, 2.4 CaCl2, 1.3 MgSO4, 26 NaHCO3, and 15 glucose (Doolen et al. 2012). Slices were moved to a chamber with oxygenated aCSF at room temperature until recording.
Patch-clamp electrophysiology.
Slices were perfused with oxygenated 30°C aCSF. The patch pipette (5–10 MΩ) was filled with a solution containing (in mM) 110 Cs2SO4, 2 MgCl2, 0.5 CaCl2, 5 HEPES, 5 EGTA, 5 ATP-Mg, 0.5 GTP-Na, and 5 tetraethylammonium. An Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) was used to record membrane currents at a holding potential of +50 mV. Data were acquired using a Digidata 1322A and pCLAMP 8.0 software (Molecular Devices). Strychnine (5 µM; glycine receptor antagonist), picrotoxin (100 µM; GABA receptor, antagonist), and 1,2,3,4-tetrahydro-6-nitro-2,3-dioxobenzo[f]quinoxaline-7-sulfonamide [NBQX, 10 µM; α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, antagonist] were added to the perfusate to isolate NMDAr-mediated currents. Cells were visualized by differential interference contrast optics on an Olympus BX50WI microscope (Tokyo, Japan). All recordings were performed on small neurons located in lamina II of the dorsal horn. The substantia gelatinosa (SG) was located as a translucent band in close proximity to the dorsal surface of the spinal cord.
Pharmacology.
Agmatine (Sigma Aldrich, St. Louis, MO), ifenprodil (Tocris, Bristol, UK), and PEAQX (Tocris) were dissolved in aCSF and individually perfused onto the slice at a rate of 1 ml/min with a peristaltic pump. Baseline eEPSC recordings were taken before drug perfusion. To allow for adequate drug diffusion and equilibration in the recording chamber; drugs were perfused for 10 min before recording. A cumulative concentration response method was implemented with the lowest concentration of the individual drug applied first (agmatine: 300 µM, ifenprodil: 1 µM, or PEAQX: 13.3 nM), followed by the second concentration (agmatine: 1,000 µM, ifenprodil: 10 µM, or PEAQX: 40 nM), and, finally, the highest concentration (agmatine: 3,000 µM, ifenprodil: 100 µM, or PEAQX: 400 nM). Each experimental group included recordings from spinal cord slices collected from a minimum of three individual mice.
Stimulation.
For optogenetic stimulation, blue light (470 nm) pulses (pulse width 8–10 ms) were produced by a 1-W light-emitting diode (LED; Thorlabs, Newton, NJ), collimated, and deflected down the light path of the microscope and through the ×40 objective to activate ChR2-expressing (ChR2+) fibers in the dorsal root (approximate length 3 mm). For electrical stimulation, a suction electrode was placed at the end of the dorsal root. Electrical stimulation pulses, generated by a WPI A310 Accupulser (Sarasota, FL), consisted of square waves with typical pulse widths of 2–5 ms and amplitudes of 3–6 mA. Testing for a monosynaptic excitatory postsynaptic current was performed at the beginning of each neuronal recording. This was done by stimulation of the dorsal root at frequencies of 0.2 and 1 Hz. If no failures occurred with 1-Hz stimulation and no jitter occurred with 0.2-Hz stimulation, the eEPSC was considered to be elicited monosynaptically from Nav1.8+ afferent terminals (Honsek et al. 2015). A similar procedure was conducted for electrical stimulation.
Approximately five eEPSCs (stimulation rate = 0.1 Hz) were averaged for measurement using Clampfit 8.2 software (Molecular Devices). Amplitude was measured from baseline (before the light pulse) to the peak of the eEPSC. Recovery was determined when the eEPSC declined to its baseline amplitude. The decay constant (τdecay) was determined by fitting the peak of the eEPSC to the point of recovery with a standard exponential function.
Photic activation of nociceptors using Nav1.8-ChR2 transgenic mice.
Conditional expression of ChR2 was targeted to Nav1.8+ sensory neurons in mice. The detailed methods have been previously reported (Daou et al. 2013), based on a line developed by Stirling and colleagues (Stirling et al. 2005). Briefly, homozygous mice carrying Nav1.8-cAMP response element (Cre) on a C57B/6 background were crossed with homozygous mice carrying floxed ChR2 in their ROSA26 locus, also on a C57B/6 background (Ai32 mice purchased from The Jackson Laboratory) (Madisen et al. 2012). This cross yielded 100% offspring expressing ChR2 in Nav1.8+ primary afferent sensory neurons, which was evidenced by a nocifensive reaction to illumination of hind paws with 470-nm light from an LED (Plexon) with an attached fiber optic cable. Using teased tibial nerve fiber recordings in these mice, we have demonstrated that 77% of neurons responding to 470-nm light were C polymodal nociceptors (Uhelski et al. 2017). All Nav1.8-ChR2 mice used in these experiments were male.
Generation of GluN2B-KD mice.
The generation of the GluN2B-floxed mouse was initiated by Dr. E. Delpire (Vanderbilt University), as previously described (Brigman et al. 2010). The GluN2B mutant mouse was generated by the Gene-Targeted Mouse Core of the INIAstress consortium. This Integrative Neuroscience Initiative on Alcoholism examines the link between stress and alcohol. A breeding colony of homogenous GluN2B-floxed mice was established. At time of weaning (postnatal day 21), all animals received either an intrathecal injection of 5 µl of 0.9% saline (control) or AAV9.CMV.HI.eGFP-Cre.WPRE.SV40 (GluN2B-KD; Penn Vector Core, University of Pennsylvania). All GluN2B-KD or control mice in these experiments were male and ranged from postnatal day 42–56 at time of experiment. Within GluN2B-KD mice, neurons expressing Cre recombinase were not distinguished from neurons not infected with adeno-associated viral (AAV) vector.
Molecular validation of GluN2B-KD.
Electrophysiological recordings were made on lumbar spinal cord slices collected from mice injected either with saline (control) or AAV9.CMV.HI.eGFP-Cre.WPRE.SV40 (GluN2B-KD). Confirmation of GluN2B knockdown was not performed at the cellular level in the slice preparation due to technical limitations. As such, we cannot rule out the possibility that recordings were made from neurons not transduced with the viral vector and included in the overall analyses. However, molecular validation was conducted on remaining lumbar spinal cord that was subjected to quantitative real-time reverse transcription (RT-qPCR) and Western blot to confirm general GluN2B-KD in the spinal cord of each animal.
RT-qPCR confirmation of GluN2B knockdown.
Remaining lumbar spinal cord tissue remaining was collected in TRIzol reagent (phenol and guanidine isothiocyanate solution) to confirm the genotype of each animal. Total RNA was extracted according to the manufacturer’s instructions. The RNA pellet was dissolved in nuclease-free water, and RNA concentration was estimated by spectrophotometric analysis using the NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). An equal amount of RNA was used for each reaction.
The expression levels of NMDAr subunits GluN2A and GluN2B were determined by estimating the messenger RNA copy number through the RT-qPCR method. All reactions were set up in 96-well format (multiplate 96-well PCR plates; Bio-Rad) and carried out in a CFX96 Touch real-time PCR detection system (Bio-Rad) using an iTaq Universal SYBR green one-step kit (BIO-RAD). The oligonucleotide primers used were mouse GluN2A: F 5′-TCTATGACGCAGCCGTCTTGAACT-3′ and R 5′-TGTGGTAGCAAAGATGTACCCGCT-3′, mouse GluN2B: F 5′-ATG AAGAGGGGCAAGGAGTT-3′ and R 5′-CGATGATGGAGGAGACTTGG-3′, and mouse 18S: F 5′-AAGACGATCAGATACCGTCGTAG-3′ and R 5′-TCCGTCAATTCCTTTAAGTTTCA-3′ (Dhar and Wong-Riley 2009; Tajerian et al. 2015). All reactions were run in triplicate. Each 20 µl of reaction mixture contained 10 μl of 2× master mix, 0.25 μl of enzyme mix, 300 nM of each primer, and 40 ng of RNA. Two no-template control (NTC) wells were included in each run. Wells were sealed with optically clear film. The PCR cycling conditions were 20 min at 50°C, 1 min at 95°C, and then 45 cycles each of denaturation at 95°C for 10 s and annealing and extension at 60°C for 30 s. Melting curve analysis was performed to ensure the amplification of a single product in each reaction. Amplification data were analyzed by CFX Manager software version 3.1 (Bio-Rad). Data [threshold cycle (Ct) values] were analyzed using a comparative Ct method (ΔΔCt method (Schmittgen and Livak 2008). 18S was used as an endogenous control because it has been validated as a stable normalization gene for RT-qPCR (Hunkapiller et al. 1991). To obtain the ΔCt value for each of the samples, the Ct value of 18S was subtracted from the Ct value of target (GluN2A/2B). The ΔΔCt was obtained by using the ΔCt experimental value (AAV9-Cre injected) minus the ΔCt control value (saline injected), and then the fold change (2−ΔΔCt) was calculated.
Western blot confirmation of GluN2B knockdown.
Additional remaining segments of lumbar spinal cords following slice preparation were collected in T-PER protein extraction reagent (catalog no. 78510; Thermo Scientific) to quantify the protein levels of GluN2B following injection of either saline or AAV9-Cre. Tissues were homogenized with one scoop of 0.5-mm glass beads (catalog no. 11079105; BioSpec Products) using a Bullet blender storm homogenizer (catalog no. BBY24M; MIDSCI) for 5 min at speed 9. This suspension was centrifuged at 14,000 rpm for 15 min at 4°C, and supernatants containing EDTA-free protease inhibitor cocktail (catalog no. 87785; Thermo Scientific) were then frozen at −80°C until use. Protein concentration was determined using the Pierce BCA (bicinchoninic acid) protein assay kit (catalog no. 23225; Thermo Scientific). Each sample containing 25 μg of protein was mixed with 4× Laemmli sample buffer (catalog no. 1610747; Bio-Rad) and loaded on 4–15% gradient SDS-PAGE gels. Electrophoresis was carried out for 120 min (PowerPac HC high-current power supply, 300 W; Bio-Rad) set at 85 V in 1× running buffer (10× Tris/glycine/SDS; catalog no. 1610732). The protein was transferred into a polyvinylidene difluoride (PVDF) membrane (catalog no. 05317-10; Sigma). The PVDF membrane was activated in 100% methanol for 2 min (catalog no. JT9830-2; VWR) and then rinsed with transfer buffer (25 mM Tris base, 192 mM glycine, and 20% ethanol). A “sandwich” containing sponge-filter paper, the gel, and the membrane was made, and protein transfer was performed for 60 min at 80 V. The nonspecific binding sites were blocked by incubating the membrane for 60 min at room temperature with Odyssey blocking buffer TBS (catalog no. PN-927-50000). The antibodies were diluted in Odyssey blocking buffer. Membrane was washed four times with TBST (Tris-buffered saline, Tween 20 to 0.01%). Proteins were detected by overnight incubation at 4°C with the primary antibody (rabbit polyclonal anti-NR2B, 1:1,000, catalog no. ab65783; Abcam) followed by incubation with an IRDye 800CW goat anti-rabbit IgG (H + L) at 1:20,000 dilution (catalog no. 925-32211; LI-COR Biosciences). β-Actin was used as an internal control and was detected with the mouse monoclonal anti-β-actin antibody (1:5,000, catalog no. ab6276; Abcam), followed by incubation with an IRDye 680CW goat anti-mouse IgG (H + L) at 1:20,000 dilution (catalog no. 926-32220; LI-COR Biosciences). The signals were detected using Odyssey (LICOR Biosciences) and quantified using the Image J 1.43u software (Bethesda, MD).
Statistical analysis.
Statistical analyses and graphing were performed with SigmaPlot/SigmaStat (Systat Software, Chicago, IL) and Microsoft Excel (Microsoft, Redmond, WA). A two-tailed Student’s t-test was performed to determine differences between individual means. A one-way and/or two-way ANOVA with Kruskal-Wallis post hoc test analysis was conducted to determine differences between multiple means (such as concentration-response relationships) and/or between treatment groups (e.g., control vs. GluN2B-KD). A P value of 0.05 or less was considered significant, and all data are means ± SE.
RESULTS
Isolation of C-fiber-evoked monosynaptic NMDAr-mediated currents in vitro.
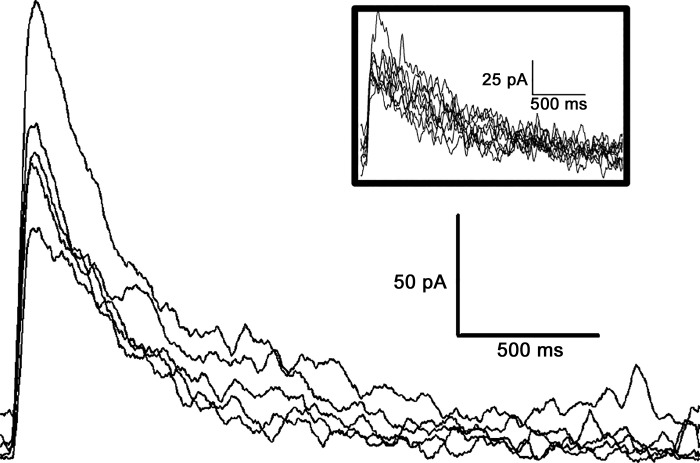
ChR2 expression was driven by the Nav1.8 promoter, which is expressed by almost all nociceptors (Shields et al. 2012). When AMPA, GABA, and glycine receptor antagonists were added to the perfusate and the cell was held at +50 mV, blue light pulsed onto the dorsal root evoked a fast-rising and slow-decaying (decay constant = 290 ± 59 ms) EPSC (Fig. 1). The average eEPSC duration was 2,479 ± 350 ms, and the average amplitude was 81 ± 18 pA. This current was indicative of NMDAr-mediated EPSCs found in the dorsal horn (Hildebrand et al. 2014). The typical conduction distances were ~3 mm and latencies ~6 ms; the estimated 0.5 m/s conduction velocity indicated that the blue light applied to these spinal cord slices activated C-fibers.
Fig. 1.
N-methyl-d-aspartate receptor (NMDAr)-mediated monosynaptic evoked excitatory postsynaptic currents (eEPSCs) from dorsal root fibers. Whole cell voltage-clamp recordings of blue light-evoked NMDAr-mediated eEPSC traces were collected from small lamina II neurons with monosynaptic connections with dorsal root C-fibers. Blue light (470 nm) pulses (8–10 ms) were used to activate Nav1.8+ fibers in the dorsal root at 0.2 Hz or 1 Hz (inset); the absence of jitter and failures suggests a monosynaptic connection.
To determine whether the eEPSCs of each neuron were evoked poly- or monosynaptically, light pulses were delivered to the dorsal root at frequencies of 1 and 0.2 Hz. If there were no failures of eliciting the eEPSC at 1 Hz and no jitter of the eEPSC at 0.2 Hz (example in Fig. 1), the response was considered to be mediated by a monosynaptic connection from a Nav1.8+ terminal to the recorded cell (Honsek et al. 2015). A similar procedure was applied when electrical stimulation was delivered to the dorsal root of slices from GluN2B-knockdown or control mice (data not shown).
Agmatine abbreviated the recovery kinetics of evoked NMDAr currents in Nav1.8-ChR2 mice.
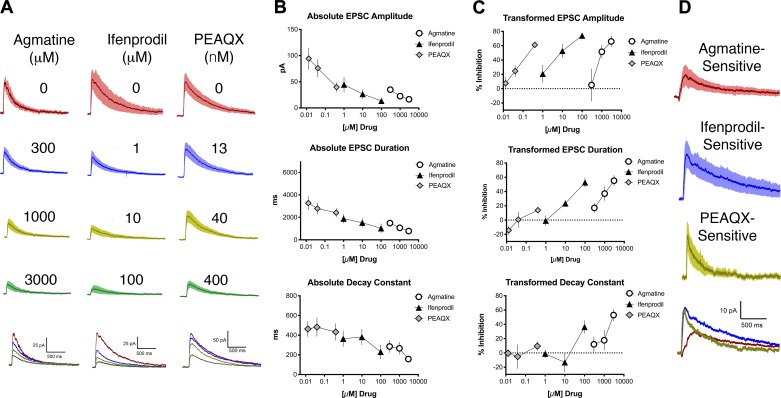
NMDArs containing the GluN2B subunit experience slower closing kinetics (>200 ms) than NMDArs containing GluN2A subunits (<100 ms) (Santucci and Raghavachari 2008). Ifenprodil antagonizes NMDArs containing GluN2B subunits (Williams et al. 1993), thus abbreviating the recovery kinetics of NMDAr-mediated EPSCs. It has been shown that ifenprodil also decreases NMDAr-mediated EPSC amplitude (Lei and McBain 2002). Following the application of agmatine, the eEPSC’s amplitude, duration, and decay constant decreased significantly (Fig. 2A, left) in a concentration-dependent manner (Fig. 2A, bottom left, and Fig. 2, B and C, n = 8; ANOVA, amplitude: F2,18 = 6.1, P < 0.01; duration: F2,18 = 28, P < 0.0001; decay constant: F2,18 = 9.9, P < 0.01). Consistent with the pattern expected of a GluN2B-selective antagonist, the application of ifenprodil also decreased these parameters significantly (Fig. 2A, center) in a concentration-dependent manner (Fig. 2A, bottom center, and Fig. 2, B and C, n = 10; ANOVA, amplitude: F2,16 = 22, P < 0.001; duration: F2,16 = 29, P < 0.001; decay constant: F2,16 = 5.6, P = 0.02), suggesting antagonism of NMDArs containing GluN2B subunits. The application of PEAQX, a selective antagonist of NMDArs containing GluN2A subunits, decreased the amplitude and duration, but not the decay constant, of the eEPSC (Fig. 2A, right, and Fig. 2, B and C, n = 8; ANOVA, amplitude: F2,14 = 31, P < 0.0001; duration: F2,14 = 5.2, P = 0.02; decay constant: F2,14 = 0.7, P = 0.5). Changes in eEPSC amplitude and/or recovery kinetics in response to ifenprodil, agmatine, and PEAQX were observed in SG cells that met the two criteria of receiving monosynaptic input.
Fig. 2.
Agmatine decreases the amplitude, duration, and decay constant of blue light-evoked N-methyl-d-aspartate receptor (NMDAr)-mediated excitatory postsynaptic currents (EPSCs) in a concentration-dependent manner. A: averaged blue light-evoked NMDAr-mediated EPSCs, with SE (shading), before (baseline, top traces for each drug) and after application of various concentrations of drug. For each set of traces, drug concentrations from top to bottom are as follows: agmatine 300, 1,000, and 3,000 µM; ifenprodil 1, 10, and 100 µM; PEAQX 13.3, 40, and 400 nM. Bottom traces show averaged EPSCs for all 3 concentrations with the baseline (no drug) represented for each treatment group by a red line, the lowest concentration for each drug by a blue line, the intermediate concentration by a yellow-green line, and the highest concentration by a green line. B: concentration-response relationships for absolute values of amplitude, duration, and decay constant of blue light-evoked NMDAr-mediated EPSCs for agmatine (circles), ifenprodil (triangles), and PEAQX (diamonds). C: transformed concentration-response relationships for amplitude, duration, and decay constant of blue light-evoked NMDAr-mediated EPSCs for agmatine (circles), ifenprodil (triangles), and PEAQX (diamonds) from the data shown in B. PEAQX did not decrease the evoked EPSC (eEPSC) decay constant, but it did decrease the eEPSC amplitude, suggesting antagonism of GluN2A-containing NMDArs. D: plots of EPSC difference currents (baseline trace minus highest drug concentration trace) for agmatine-, ifenprodil-, and PEAQX-sensitive components of NMDAr EPSCs based on the data shown in A, with agmatine represented by the red line, ifenprodil by the blue line, and PEAQX by the yellow-green line.
We next subtracted average eEPSCs recorded in the presence of the highest concentration of each drug (3,000 µM agmatine, 100 µM ifenprodil, and 400 nM PEAQX, Fig. 2D) from the baseline eEPSC to further interrogate the contribution of NMDAr subunits. Decay constants of difference currents were then calculated with an exponential function similar to the method of Hildebrand et al. (2014). The decay constants for the agmatine- and ifenprodil-sensitive EPSC currents in the Nav1.8-ChR2 mice were similar and higher (335 ± 81 and 381 ± 99 ms, respectively) than for PEAQX (204 ± 29 ms).
Effect of GluN2B-KD on agmatine inhibition of EPSCs.
As a second approach to determine whether agmatine depends on NMDArs that contain the GluN2B subunit, we applied agmatine to slices from GluN2B-floxed mice that at time of weaning received an intrathecal injection of either saline (control mice) or AAV9-Cre to knockdown the amount of GluN2B in spinal cord (GluN2B-KD).
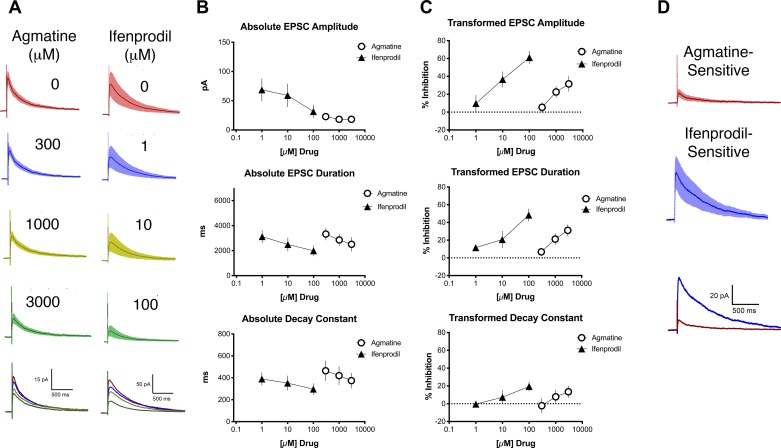
As a wild-type control, GluN2B-floxed mice were injected with 0.9% saline instead of AAV9-Cre at the time of weaning. NMDAr-mediated EPSCs were evoked by electrical stimulation of the dorsal root (typical duration 2 ms, typical amplitude 5 mA, typical conduction distance 3 mm, typical conduction latency 5 ms). The NMDAr-mediated eEPSCs from slices from control mice had an average amplitude of 46 ± 12 pA, a duration of 3,573 ± 441 ms, and a decay constant of 423 ± 52 ms (Fig. 3). All evoked NMDAr-mediated EPSCs studied were determined to be from a neuron that had a monosynaptic connection with the dorsal root. In these control slices, agmatine concentration-dependently reduced the amplitude of the eEPSC, the eEPSC duration, and the decay constant (Fig. 3A, left, and Fig. 3, B and C, n = 14; ANOVA, amplitude: F2,36 = 5.4, P < 0.01; duration: F2,34 = 7.9, P < 0.01; decay constant: F2,31 = 3.4, P < 0.05). It is noteworthy that the magnitude of effect of agmatine is smaller in the experiments presented in Fig. 3 compared with those in Fig. 2. Strain differences may account for these changes and/or difference in population of stimulated afferents, C polymodal nociceptors in the case of Fig. 2 and all sensory afferents in the case of Fig. 3, which may dilute the effect of agmatine.
Fig. 3.
Agmatine dose-dependently inhibits evoked N-methyl-d-aspartate receptor (NMDAr)-mediated excitatory postsynaptic currents (eEPSC) amplitude, duration, and decay constant in GluN2B-floxed control mice. A: electrically evoked NMDAr-mediated averaged EPSCs, with SE (shading), before (baseline, top trace for each drug) and after application of various concentrations of drug. For each set of traces, drug concentrations from top to bottom are as follows: agmatine 300, 1,000, and 3,000 µM; ifenprodil 1, 10, and 100 µM. Bottom traces show averaged EPSCs for all 3 concentrations. The baseline (no drug) is represented for each treatment group by a red line, the lowest concentration for each drug by a blue line, the intermediate concentration by a yellow-green line, and the highest concentration by a green line. B: concentration-response relationships for absolute values of EPSC amplitude, duration, and decay constant of NMDAR-mediated electrically evoked EPSCs for agmatine (circles) and ifenprodil (triangles). C: transformed concentration-response relationships for different parameters of NMDAr-mediated electrically evoked EPSCs for agmatine (circles) and ifenprodil (triangles) from the data shown in B. D: plots of EPSC difference current traces (baseline trace minus highest drug concentration trace) for agmatine- or ifenprodil-sensitive components of NMDAr-EPSCs based on the data shown in A.
Likewise, ifenprodil decreased all of these parameters of the eEPSC in a concentration-dependent manner (Fig. 3A, right, and Fig. 3, B and C, n = 10; ANOVA, amplitude: F2,15 = 6.4, P = 0.01; duration: F2,14 = 5.3, P = 0.02; decay constant: F2,22 = 4.0, P = 0.03). As expected, these results were consistent with the results from the slices Nav1.8-Ch2 mice activated by light stimulation that were featured in Fig. 1. We subtracted average EPSCs recorded in the presence of the highest concentration of each drug (3,000 µM agmatine, 100 µM ifenprodil) from the baseline EPSC to determine the contribution of NMDAr subunits. In these control mice, the decay constant for agmatine-sensitive currents was 430 ± 112 ms and that for ifenprodil-sensitive currents was 474 ± 105 ms (Fig. 3D).
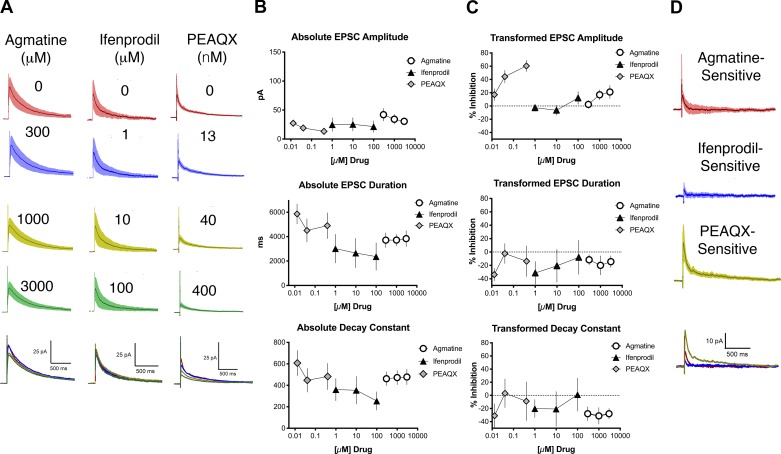
We next injected GluN2B-floxed mice with AAV9-Cre (termed GluN2B-KD mice). The eEPSCs from the slices harvested from GluN2B-KD mice had an average amplitude of 37 ± 6.6 pA, a duration of 3560 ± 456 ms, and a decay constant of 403 ± 46 ms (Fig. 4). There were no significant differences in any of these parameters between KD and control mice from Fig. 3. The long-duration eEPSC and decay constant were likely due to NMDArs containing GluN2D subunits (Hildebrand et al. 2014). All evoked NMDAr-mediated EPSCs studied were determined to be from a neuron that had a monosynaptic connection with dorsal root afferents. In contrast to control mice, the application of 300, 1,000, and 3,000 µM agmatine elicited no significant concentration-dependent change in eEPSC duration or decay constant; however, a concentration-dependent change in eEPSC amplitude was evident (Fig. 4A, left, and Fig. 4, B and C, n = 10; ANOVA, amplitude: F2,18 = 5.4, P = 0.02; duration: F2,18 = 0.16, P = 0.85; decay constant: F2,18 = 0.085, P = 0.92). The transformed values following agmatine application were compared between control (Fig. 3C) and GluN2B-KD (Fig. 4C) and were confirmed to be statistically different between the treatment groups for eEPSC duration and decay constant values (duration: F1,72 = 29, P < 0.0001; decay constant: F1,69 = 24, P < 0.0001), but not for amplitude (amplitude: F1,67 = 1.1, P = 0.29). These results were similar to those seen following application of ifenprodil (1, 10, and 100 µM) to spinal cord slices from GluN2B-KD mice, except that no significant concentration-dependent change in eEPSC amplitude was evident (Fig. 4A, middle, and Fig. 4, B and C, n = 5; ANOVA, amplitude: F2,12 = 3, P = 0.24; duration: F2,12 = 0.31, P = 0.74; decay constant: F2,12 = 0.27, P = 0.77). The transformed values following ifenprodil application were compared between control (Fig. 3C) and GluN2B-KD (Fig. 4C) and were confirmed to be statistically different between the treatment groups for eEPSC duration, decay constant values, and amplitude (duration: F1,33 = 18, P < 0.0002; decay constant: F1,34 = 4.4, P < 0.04; amplitude: F1,29 = 25, P < 0.0001). Various concentrations of PEAQX were also applied (13.3, 40, and 400 nM) to slices from GluN2B-KD mice. PEAQX elicited a significant concentration-dependent change in eEPSC amplitude, consistent with antagonism of NMDArs containing GluN2A subunits. However, the eEPSC duration and the decay constant did not change in a concentration-dependent manner (Fig. 4A, right, and Fig. 4, B and C, n = 7; ANOVA, amplitude: F2,18 = 5.9, P = 0.01; duration: F2,18 = 0.92, P = 0.42; decay constant: F2,18 = 0.54, P = 0.59).
Fig. 4.
Agmatine and ifenprodil did not decrease the N-methyl-d-aspartate receptor (NMDAr)-mediated excitatory postsynaptic currents (EPSC) duration or decay constant in GluN2B-knockdown (KD) mice. A: traces of NMDAr-mediated electrically evoked EPSCs (eEPSCs), with SE (shading), before (baseline, top traces for each drug) and after application of increasing concentrations of drug. For each set of traces, drug concentrations from top to bottom are as follows: agmatine 300, 1,000, and 3000 µM; ifenprodil 1, 10, and 100 µM; PEAQX 13.3, 40, and 400 nM. Bottom traces show averaged EPSCs for all 3 concentrations. The baseline (no drug) is represented for each treatment group by a red line, the lowest concentration for each drug by a blue line, the intermediate concentration by a yellow-green line, and the highest concentration by a green line. There was no significant change in the NMDAr-mediated eEPSC duration or decay constant with increasing concentrations of agmatine and ifenprodil. There was a concentration-dependent decrease in the amplitude of the eEPSC following application of PEAQX. B: concentration-response relationships for absolute values of EPSC amplitude, duration, and decay constant of NMDAr-mediated eEPSCs for agmatine (circles), ifenprodil (triangles), and PEAQX (diamonds). C: transformed concentration-response relationships for different parameters of NMDAr-mediated eEPSCs for agmatine (circles), ifenprodil (triangles), and PEAQX (diamonds) based on the data shown in B. PEAQX alone decreased NMDAr-mediated eEPSC amplitude. D: plots of EPSC subtraction traces (baseline trace minus highest drug concentration) for agmatine-, ifenprodil-, and PEAQX-sensitive components of NMDAr-EPSCs based on the data shown in A, with agmatine represented by the red line, ifenprodil by the blue line, and PEAQX by the yellow-green line.
Difference currents demonstrated agmatine’s lack of NMDA antagonism in GluN2B-KD mice.
Agmatine- and ifenprodil-sensitive currents were calculated in GluN2B-KD mice by subtracting EPSCs following application of the highest concentration of drug (3000 µM for agmatine, 100 µM for ifenprodil, and 400 nM for PEAQX) from their baseline EPSC (Fig. 4D). In these GluN2B-KD mice, the decay constant for agmatine was 121 ± 55 ms, that for ifenprodil was 175 ± 64 ms, and that for PEAQX was 195 ± 75 ms. These agmatine- and ifenprodil-sensitive decay constants from GluN2B-KD currents were compared with the agmatine- and ifenprodil-sensitive decay constants from control mice in Fig. 3. There was a significant decrease in the decay constant of agmatine- and ifenprodil- sensitive currents of the GluN2B-KD mice compared with the control mice, supporting agmatine’s antagonism of GluN2B subunits (Student’s t-test, P = 0.04 for agmatine and P = 0.05 for ifenprodil).
Post hoc tissue analysis.
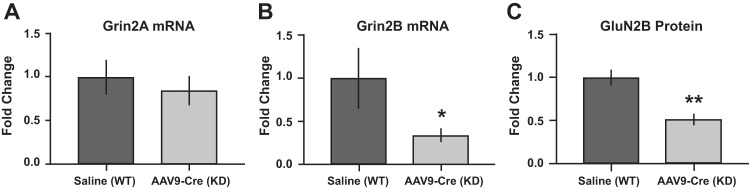
Lumbar spinal cord sections were collected from GluN2B-floxed mice and processed for verification of either presence of GluN2B (control) or reduction of GluN2B (GluN2B-KD). Levels of GluN2A mRNA were also evaluated to determine the potential for compensatory changes. The mRNA expression level for GluN2A and GluN2B in spinal cord tissues was evaluated using RT-qPCR in both control and GluN2B-KD mice. No change was observed in the expression level of GluN2A between saline controls and AAV9-Cre injected mice, as expected (Fig. 5A). However, GluN2B mRNA expression was downregulated in AAV9-Cre-injected mice compared with saline-injected controls (Fig. 5B). Consistently, GluN2B protein expression was also reduced in AAV9-Cre-injected mice compared with saline-injected controls (Fig. 5C). These results confirmed reduction of spinal GluN2B in the AAV9-Cre-injected mice.
Fig. 5.
Confirmation of GluN2B-knockdown (KD) in mouse spinal cord. GluN2B-floxed mice were injected at time of weaning with saline (control) or adeno-associated virus expressing Cre recombinase (AAV9-Cre; conditional knockdown). Quantitative real-time reverse transcription (RT-qPCR) of spinal cord tissue indicates no changes in Grin2A mRNA levels (A) but a significant decrease in Grin2B mRNA (B) in GluN2B-KD mice compared with controls. C: GluN2B protein was also reduced in spinal cord from GluN2B-KD mice relative to controls. *P < 0.05; **P < 0.01, significant difference from saline control (unpaired Student’s t-test).
DISCUSSION
This study was conducted to determine whether agmatine selectively antagonizes NMDArs containing GluN2B subunits in the substantia gelatinosa (SG) of the spinal cord. To assess this question, in vitro patch-clamp experiments were performed by selective photic activation of nociceptive afferents innervating the SG of transverse spinal cord slices from Nav1.8-ChR2 mice (Daou et al. 2013) and dorsal root electrical stimulation in GluN2B-KD or control mice. First, agmatine inhibited the NMDAr-mediated eEPSC amplitude, demonstrating NMDAr antagonism. It is well established that NMDArs containing the GluN2B subunit exhibit slow recovery kinetics (decay constant ~280 ms) (Arrigoni and Greene 2004; Hildebrand et al. 2014; Tong and MacDermott 2014). Agmatine significantly decreased the duration and decay constant of NMDAr-mediated eEPSCs of slices from Nav1.8-ChR2 mice. Ifenprodil was applied as a positive control for GluN2B inhibition at concentrations within a range that has been shown to effectively antagonize GluN2B subunits (Arrigoni and Greene 2004; Kumar and Huguenard 2003; Li and Clark 2002; Williams et al. 1993). Ifenprodil had an effect similar to that of agmatine in decreasing the eEPSC recovery time and decay constant. This result suggests that agmatine antagonizes NMDArs containing GluN2B subunits.
Application of the GluN2A subunit-selective antagonist PEAQX was used as a negative control for GluN2B-mediated effects on slices from Nav1.8-ChR2 mice. At the concentrations used in this study, PEAQX has been shown to preferentially antagonize GluN2A subunits (Auberson et al. 2002), which do not contribute to the slow GluN2B-mediated decay kinetics (Hildebrand et al. 2014). As expected, PEAQX did not decrease the NMDAr-mediated eEPSC slow recovery kinetics but induced a decrease in eEPSC amplitude.
To confirm that agmatine acts on GluN2B-containing NMDArs, NMDAr-mediated EPSCs evoked by electrical stimulation of the dorsal root were investigated from slices of mice with a conditional knockdown of GluN2B subunits. At time of weaning, GluN2B-floxed mice were intrathecally injected with either saline (wild-type control) or AAV9-Cre (GluN2B-KD). RT-qPCR confirmed that injection of AAV9-Cre significantly decreased Grin2B mRNA expression in spinal cord tissue as compared with mice that received the control injection of saline. Grin2A mRNA levels were unaffected. GluN2B protein was also significantly decreased in AAV9-Cre injected GluN2B-floxed mice. In spinal cord slices of GluN2B-KD mice, agmatine (as well as ifenprodil) did not decrease the duration or decay constant of NMDAr EPSCs, indicating antagonism of NMDArs containing the GluN2B subunit. Agmatine did decrease the NMDAr eEPSC amplitude, but not to the same degree as PEAQX. This result suggests that agmatine may have some activity at the GluN2A subunit. However, at a high concentration (3,000 µM) that induced, on average, a greater than 50% decrease of NMDAr eEPSC duration and decay constant in slices from normal mice, eEPSC amplitude in GluN2B-KD slices following the application of agmatine decreased only an average of ~20%. Therefore, the contribution of GluN2A to the effects of agmatine is likely to be minor.
Difference currents were determined by subtracting postdrug from predrug eEPSCs for Nav1.8-ChR2, GluN2B-KD, and control mice. These difference currents were used to calculate agmatine-, ifenprodil- and PEAQX-sensitive current decay constants. This analysis was used as an additional method to test for the drugs’ antagonism of GluN2B subunit-containing NMDARs. In Nav1.8-ChR2 and control mice, the agmatine- and ifenprodil-sensitive currents were similar and longer than the PEAQX-sensitive currents, suggesting that agmatine has a similar effect on NMDArs as the known GluN2B antagonist, ifenprodil (Williams 1993). Agmatine- and ifenprodil-sensitive current decay constants were also similar in wild-type mice and significantly longer than in GluN2B-KD mice. These difference current calculations support agmatine’s antagonism of long-duration, GluN2B-mediated currents.
This investigation provides strong evidence in support of the GluN2B subunit of the NMDAr as a key neuronal target of agmatine in the spinal cord dorsal horn. Agmatine affected eEPSCs from SG receiving monosynaptic connections from afferents (Honsek et al. 2015; Luz et al. 2010). In spinal cord slice recordings, primary afferents were activated by blue light pulsed onto the dorsal roots of Nav1.8-ChR2 mice. The Nav1.8 channel is expressed in almost all nociceptors (Amaya et al. 2000; Shields et al. 2012; Uhelski et al. 2017), and the estimated conduction velocities of blue light-activated eEPSCs were indicative of activation of C-fibers (conduction velocities <1 m/s). These results suggest that agmatine modulates GluN2B-containing NMDArs that are expressed on secondary neurons receiving monosynaptic input from nociceptive primary afferent C-fibers. We have previously demonstrated that agmatine is released from spinal synaptosomes in a calcium-dependent manner (Goracke-Postle et al. 2007a) and actively transported into the same preparation (Goracke-Postle et al. 2007b), both of which are signature aspects of neurotransmission. It has been long estimated that synaptic transmitter concentrations for typical neurotransmitters, such as glutamate, are in the millimolar range (Riveros et al. 1986; Scimemi and Beato 2009). It is conceivable that synaptic release of multiple vesicles of agmatine could similarly produce transient, subsynaptic agmatine concentrations in the micromolar to millimolar range featured in these in vitro experiments.
Agmatine attenuates hyperalgesia generated by neuropathy, inflammation, and spinal cord injury (Fairbanks et al. 2000). Enhancement in the synaptic strength of nociceptors following injury is believed to occur during the development of hyperalgesia (Sandkühler 2007). Drugs that attenuate activity-dependent synaptic potentiation between nociceptors and their targets in the dorsal horn have been shown to have antihyperalgesic effects (Boyce et al. 1999; Drdla-Schutting et al. 2012). Application of NMDAr antagonists to the spinal cord has been shown to inhibit long-term potentiation evoked by C-fiber stimulation; agmatine’s antagonism of the NMDAr may be a mechanism by which it reduces hypersensitivity accompanying neuropathy and inflammation.
There is emerging evidence for the use of agmatine as a treatment for persistent pain in a clinical setting. In a human trial on patients suffering from sciatica, agmatine demonstrated superiority of pain relief compared with placebo (Keynan et al. 2010). Our study suggests that an important site of agmatine’s action is the GluN2B subunit of the NMDAr. Drugs that target this subunit have demonstrated a good tolerability profile in humans (Marquis et al. 1998; Merchant et al. 1999; Patat et al. 1994). Agmatine may also have beneficial effects when used in combination with opioids in terms of lowering opioid dosage, decreasing opioid withdrawal effects, and attenuating opioid dependence (Kolesnikov et al. 1996; Li et al. 1999; Regunathan 2006).
Summary.
The results of this study provide insight into the cellular mechanism of agmatine’s well-established effects on chronic pain and opioid analgesic tolerance. We provide physiological evidence that agmatine antagonizes NMDArs containing GluN2B subunits, which are expressed on second-order neurons in the SG receiving monosynaptic input from nociceptors. Increasing agmatine levels in the dorsal horn of the spinal cord may have therapeutic value for the treatment of chronic pain and/or reducing opioid dose requirements.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant R01 DA035931 (to C. A. Fairbanks), and NIH Training Grant T32 DA007097 supported both C. D. Peterson and J. J. Waataja. The Gene-Targeted Mouse Core of the INIAstress consortium is supported by NIH Grant AA013514 (to E. Delpire).
Present contact for J. J. Waataja: ReShape Lifesciences Inc. (e-mail: jwaataja@reshapelifesci.com).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
J.J.W., C.D.P., C.A.F., and G.L.W. conceived and designed research; J.J.W., H.V., and C.J.G.-P. performed experiments; J.J.W., H.V., and C.D.P. analyzed data; J.J.W., C.D.P., H.V., C.A.F., and G.L.W. interpreted results of experiments; J.J.W., C.D.P., and C.A.F. prepared figures; J.J.W., C.D.P. and C.A.F. drafted manuscript; J.J.W., C.D.P., C.A.F., G.L.W., C.J.G.-P., P.S., and E.D. edited and revised manuscript; J.J.W., C.D.P., G.L.W., and C.A.F. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Hai Truong for technical assistance in spinal cord slice preparation. E. Delpire and P. Séguéla contributed essential specialized mouse strains.
REFERENCES
- Amaya F, Decosterd I, Samad TA, Plumpton C, Tate S, Mannion RJ, Costigan M, Woolf CJ. Diversity of expression of the sensory neuron-specific TTX-resistant voltage-gated sodium ion channels SNS and SNS2. Mol Cell Neurosci 15: 331–342, 2000. doi: 10.1006/mcne.1999.0828. [DOI] [PubMed] [Google Scholar]
- Arrigoni E, Greene RW. Schaffer collateral and perforant path inputs activate different subtypes of NMDA receptors on the same CA1 pyramidal cell. Br J Pharmacol 142: 317–322, 2004. doi: 10.1038/sj.bjp.0705744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M. 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett 12: 1099–1102, 2002. doi: 10.1016/S0960-894X(02)00074-4. [DOI] [PubMed] [Google Scholar]
- Beardsley PM, Hayes BA, Balster RL. The self-administration of MK-801 can depend upon drug-reinforcement history, and its discriminative stimulus properties are phencyclidine-like in rhesus monkeys. J Pharmacol Exp Ther 252: 953–959, 1990. [PubMed] [Google Scholar]
- Beinat C, Banister S, Moussa I, Reynolds AJ, McErlean CS, Kassiou M. Insights into structure-activity relationships and CNS therapeutic applications of NR2B selective antagonists. Curr Med Chem 17: 4166–4190, 2010. doi: 10.2174/092986710793348572. [DOI] [PubMed] [Google Scholar]
- Boyce S, Wyatt A, Webb JK, O’Donnell R, Mason G, Rigby M, Sirinathsinghji D, Hill RG, Rupniak NM. Selective NMDA NR2B antagonists induce antinociception without motor dysfunction: correlation with restricted localisation of NR2B subunit in dorsal horn. Neuropharmacology 38: 611–623, 1999. doi: 10.1016/S0028-3908(98)00218-4. [DOI] [PubMed] [Google Scholar]
- Brigman JL, Wright T, Talani G, Prasad-Mulcare S, Jinde S, Seabold GK, Mathur P, Davis MI, Bock R, Gustin RM, Colbran RJ, Alvarez VA, Nakazawa K, Delpire E, Lovinger DM, Holmes A. Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning. J Neurosci 30: 4590–4600, 2010. doi: 10.1523/JNEUROSCI.0640-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butelman ER. A novel NMDA antagonist, MK-801, impairs performance in a hippocampal-dependent spatial learning task. Pharmacol Biochem Behav 34: 13–16, 1989. doi: 10.1016/0091-3057(89)90345-6. [DOI] [PubMed] [Google Scholar]
- Chaki S, Fukumoto K. Potential of glutamate-based drug discovery for next generation antidepressants. Pharmaceuticals (Basel) 8: 590–606, 2015. doi: 10.3390/ph8030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Lipton SA. The chemical biology of clinically tolerated NMDA receptor antagonists. J Neurochem 97: 1611–1626, 2006. doi: 10.1111/j.1471-4159.2006.03991.x. [DOI] [PubMed] [Google Scholar]
- Courteix C, Privat AM, Pélissier T, Hernandez A, Eschalier A, Fialip J. Agmatine induces antihyperalgesic effects in diabetic rats and a superadditive interaction with R(−)-3-(2-carboxypiperazine-4-yl)-propyl-1-phosphonic acid, a N-methyl-d-aspartate-receptor antagonist. J Pharmacol Exp Ther 322: 1237–1245, 2007. doi: 10.1124/jpet.107.123018. [DOI] [PubMed] [Google Scholar]
- Daou I, Tuttle AH, Longo G, Wieskopf JS, Bonin RP, Ase AR, Wood JN, De Koninck Y, Ribeiro-da-Silva A, Mogil JS, Séguéla P. Remote optogenetic activation and sensitization of pain pathways in freely moving mice. J Neurosci 33: 18631–18640, 2013. doi: 10.1523/JNEUROSCI.2424-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci 29: 483–492, 2009. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolen S, Blake CB, Smith BN, Taylor BK. Peripheral nerve injury increases glutamate-evoked calcium mobilization in adult spinal cord neurons. Mol Pain 8: 56, 2012. doi: 10.1186/1744-8069-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drdla-Schutting R, Benrath J, Wunderbaldinger G, Sandkühler J. Erasure of a spinal memory trace of pain by a brief, high-dose opioid administration. Science 335: 235–238, 2012. doi: 10.1126/science.1211726. [DOI] [PubMed] [Google Scholar]
- Fairbanks CA, Schreiber KL, Brewer KL, Yu CG, Stone LS, Kitto KF, Nguyen HO, Grocholski BM, Shoeman DW, Kehl LJ, Regunathan S, Reis DJ, Yezierski RP, Wilcox GL. Agmatine reverses pain induced by inflammation, neuropathy, and spinal cord injury. Proc Natl Acad Sci USA 97: 10584–10589, 2000a. doi: 10.1073/pnas.97.19.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea E, Regunathan S, Eliopoulos V, Feinstein DL, Reis DJ. Inhibition of mammalian nitric oxide synthases by agmatine, an endogenous polyamine formed by decarboxylation of arginine. Biochem J 316: 247–249, 1996. doi: 10.1042/bj3160247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DM, Wohleb ES, Duman RS. Emerging treatment mechanisms for depression: focus on glutamate and synaptic plasticity. Drug Discov Today 21: 454–464, 2016. doi: 10.1016/j.drudis.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel DJ, Poosch MS. NMDA receptor subunit gene expression in the rat brain: a quantitative analysis of endogenous mRNA levels of NR1Com, NR2A, NR2B, NR2C, NR2D and NR3A. Brain Res Mol Brain Res 69: 164–170, 1999. doi: 10.1016/S0169-328X(99)00100-X. [DOI] [PubMed] [Google Scholar]
- Goracke-Postle CJ, Overland AC, Riedl MS, Stone LS, Fairbanks CA. Potassium- and capsaicin-induced release of agmatine from spinal nerve terminals. J Neurochem 102: 1738–1748, 2007a. doi: 10.1111/j.1471-4159.2007.04647.x. [DOI] [PubMed] [Google Scholar]
- Goracke-Postle CJ, Overland AC, Stone LS, Fairbanks CA. Agmatine transport into spinal nerve terminals is modulated by polyamine analogs. J Neurochem 100: 132–141, 2007b. doi: 10.1111/j.1471-4159.2006.04193.x. [DOI] [PubMed] [Google Scholar]
- Hildebrand ME, Pitcher GM, Harding EK, Li H, Beggs S, Salter MW. GluN2B and GluN2D NMDARs dominate synaptic responses in the adult spinal cord. Sci Rep 4: 4094, 2014. doi: 10.1038/srep04094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsek SD, Seal RP, Sandkühler J. Presynaptic inhibition of optogenetically identified VGluT3+ sensory fibres by opioids and baclofen. Pain 156: 243–251, 2015. doi: 10.1097/01.j.pain.0000460304.63948.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth G, Kékesi G, Dobos I, Szikszay M, Klimscha W, Benedek G. Effect of intrathecal agmatine on inflammation-induced thermal hyperalgesia in rats. Eur J Pharmacol 368: 197–204, 1999. doi: 10.1016/S0014-2999(99)00060-6. [DOI] [PubMed] [Google Scholar]
- Hunkapiller T, Kaiser RJ, Koop BF, Hood L. Large-scale and automated DNA sequence determination. Science 254: 59–67, 1991. doi: 10.1126/science.1925562. [DOI] [PubMed] [Google Scholar]
- Keynan O, Mirovsky Y, Dekel S, Gilad VH, Gilad GM. Safety and efficacy of dietary agmatine sulfate in lumbar disc-associated radiculopathy. An open-label, dose-escalating study followed by a randomized, double-blind, placebo-controlled trial. Pain Med 11: 356–368, 2010. doi: 10.1111/j.1526-4637.2010.00808.x. [DOI] [PubMed] [Google Scholar]
- Kolesnikov Y, Jain S, Pasternak GW. Modulation of opioid analgesia by agmatine. Eur J Pharmacol 296: 17–22, 1996. doi: 10.1016/0014-2999(95)00669-9. [DOI] [PubMed] [Google Scholar]
- Kumar SS, Huguenard JR. Pathway-specific differences in subunit composition of synaptic NMDA receptors on pyramidal neurons in neocortex. J Neurosci 23: 10074–10083, 2003. doi: 10.1523/JNEUROSCI.23-31-10074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton ME, Kelly MJ 3rd, Rodzinak KJ. Recent advances in the development of NR2B subtype-selective NMDA receptor antagonists. Curr Top Med Chem 6: 697–709, 2006. doi: 10.2174/156802606776894447. [DOI] [PubMed] [Google Scholar]
- Lei S, McBain CJ. Distinct NMDA receptors provide differential modes of transmission at mossy fiber-interneuron synapses. Neuron 33: 921–933, 2002. doi: 10.1016/S0896-6273(02)00608-6. [DOI] [PubMed] [Google Scholar]
- Li J, Li X, Pei G, Qin BY. Effects of agmatine on tolerance to and substance dependence on morphine in mice. Zhongguo Yao Li Xue Bao 20: 232–238, 1999. [PubMed] [Google Scholar]
- Li X, Clark JD. Hyperalgesia during opioid abstinence: mediation by glutamate and substance P. Anesth Analg 95: 979–984, 2002. [DOI] [PubMed] [Google Scholar]
- Luz LL, Szucs P, Pinho R, Safronov BV. Monosynaptic excitatory inputs to spinal lamina I anterolateral-tract-projecting neurons from neighbouring lamina I neurons. J Physiol 588: 4489–4505, 2010. doi: 10.1113/jphysiol.2010.197012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, Hsu YW, Garcia AJ 3rd, Gu X, Zanella S, Kidney J, Gu H, Mao Y, Hooks BM, Boyden ES, Buzsáki G, Ramirez JM, Jones AR, Svoboda K, Han X, Turner EE, Zeng H. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15: 793–802, 2012. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis P, Lecasble M, Passa P. [Quality of life of patient with peripheral arterial obliterative disease treated with ifenprodil tartrate. Results of an ARTEMIS study]. Drugs 56, Suppl 3: 37–48, 1998. doi: 10.2165/00003495-199856003-00005. [DOI] [PubMed] [Google Scholar]
- Merchant RE, Bullock MR, Carmack CA, Shah AK, Wilner KD, Ko G, Williams SA. A double-blind, placebo-controlled study of the safety, tolerability and pharmacokinetics of CP-101,606 in patients with a mild or moderate traumatic brain injury. Ann N Y Acad Sci 890: 42–50, 1999. doi: 10.1111/j.1749-6632.1999.tb07979.x. [DOI] [PubMed] [Google Scholar]
- Nelson KA, Park KM, Robinovitz E, Tsigos C, Max MB. High-dose oral dextromethorphan versus placebo in painful diabetic neuropathy and postherpetic neuralgia. Neurology 48: 1212–1218, 1997. doi: 10.1212/WNL.48.5.1212. [DOI] [PubMed] [Google Scholar]
- Nguyen HO, Goracke-Postle CJ, Kaminski LL, Overland AC, Morgan AD, Fairbanks CA. Neuropharmacokinetic and dynamic studies of agmatine (decarboxylated arginine). Ann N Y Acad Sci 1009: 82–105, 2003. doi: 10.1196/annals.1304.009. [DOI] [PubMed] [Google Scholar]
- Niesters M, Dahan A. Pharmacokinetic and pharmacodynamic considerations for NMDA receptor antagonists in the treatment of chronic neuropathic pain. Expert Opin Drug Metab Toxicol 8: 1409–1417, 2012. doi: 10.1517/17425255.2012.712686. [DOI] [PubMed] [Google Scholar]
- Parsons CG. NMDA receptors as targets for drug action in neuropathic pain. Eur J Pharmacol 429: 71–78, 2001. doi: 10.1016/S0014-2999(01)01307-3. [DOI] [PubMed] [Google Scholar]
- Patat A, Molinier P, Hergueta T, Brohier S, Zieleniuk I, Danjou P, Warot D, Puech A. Lack of amnestic, psychotomimetic or impairing effect on psychomotor performance of eliprodil, a new NMDA antagonist. Int Clin Psychopharmacol 9: 155–162, 1994. doi: 10.1097/00004850-199409000-00003. [DOI] [PubMed] [Google Scholar]
- Qu XX, Cai J, Li MJ, Chi YN, Liao FF, Liu FY, Wan Y, Han JS, Xing GG. Role of the spinal cord NR2B-containing NMDA receptors in the development of neuropathic pain. Exp Neurol 215: 298–307, 2009. doi: 10.1016/j.expneurol.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Regunathan S. Agmatine: biological role and therapeutic potentials in morphine analgesia and dependence. AAPS J 8: E479–E484, 2006. doi: 10.1208/aapsj080356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveros N, Fiedler J, Lagos N, Muñoz C, Orrego F. Glutamate in rat brain cortex synaptic vesicles: influence of the vesicle isolation procedure. Brain Res 386: 405–408, 1986. doi: 10.1016/0006-8993(86)90181-2. [DOI] [PubMed] [Google Scholar]
- Roberts JC, Grocholski BM, Kitto KF, Fairbanks CA. Pharmacodynamic and pharmacokinetic studies of agmatine after spinal administration in the mouse. J Pharmacol Exp Ther 314: 1226–1233, 2005. doi: 10.1124/jpet.105.086173. [DOI] [PubMed] [Google Scholar]
- Sagratella S, Pezzola A, Popoli P, Scotti de Carolis AS. Different capability of N-methyl-d-aspartate antagonists to elicit EEG and behavioural phencyclidine-like effects in rats. Psychopharmacology (Berl) 109: 277–282, 1992. doi: 10.1007/BF02245874. [DOI] [PubMed] [Google Scholar]
- Sandkühler J. Understanding LTP in pain pathways. Mol Pain 3: 9, 2007. doi: 10.1186/1744-8069-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkühler J, Gruber-Schoffnegger D. Hyperalgesia by synaptic long-term potentiation (LTP): an update. Curr Opin Pharmacol 12: 18–27, 2012. doi: 10.1016/j.coph.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkühler J, Liu X. Induction of long-term potentiation at spinal synapses by noxious stimulation or nerve injury. Eur J Neurosci 10: 2476–2480, 1998. doi: 10.1046/j.1460-9568.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Sanger DJ, Joly D. Effects of NMDA receptor antagonists and sigma ligands on the acquisition of conditioned fear in mice. Psychopharmacology (Berl) 104: 27–34, 1991. doi: 10.1007/BF02244550. [DOI] [PubMed] [Google Scholar]
- Santangelo RM, Acker TM, Zimmerman SS, Katzman BM, Strong KL, Traynelis SF, Liotta DC. Novel NMDA receptor modulators: an update. Expert Opin Ther Pat 22: 1337–1352, 2012. doi: 10.1517/13543776.2012.728587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santucci DM, Raghavachari S. The effects of NR2 subunit-dependent NMDA receptor kinetics on synaptic transmission and CaMKII activation. PLOS Comput Biol 4: e1000208, 2008. doi: 10.1371/journal.pcbi.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108, 2008. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Scimemi A, Beato M. Determining the neurotransmitter concentration profile at active synapses. Mol Neurobiol 40: 289–306, 2009. doi: 10.1007/s12035-009-8087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini G, Pompili M, Innamorati M, Dwivedi Y, Brahmachari G, Girardi P. Pharmacological properties of glutamatergic drugs targeting NMDA receptors and their application in major depression. Curr Pharm Des 19: 1898–1922, 2013. doi: 10.2174/13816128113199990293. [DOI] [PubMed] [Google Scholar]
- Shields SD, Ahn HS, Yang Y, Han C, Seal RP, Wood JN, Waxman SG, Dib-Hajj SD. Nav1.8 expression is not restricted to nociceptors in mouse peripheral nervous system. Pain 153: 2017–2030, 2012. doi: 10.1016/j.pain.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Stirling LC, Forlani G, Baker MD, Wood JN, Matthews EA, Dickenson AH, Nassar MA. Nociceptor-specific gene deletion using heterozygous NaV1.8-Cre recombinase mice. Pain 113: 27–36, 2005. doi: 10.1016/j.pain.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Tajerian M, Sahbaie P, Sun Y, Leu D, Yang HY, Li W, Huang TT, Kingery W, David Clark J. Sex differences in a murine model of complex regional pain syndrome. Neurobiol Learn Mem 123: 100–109, 2015. doi: 10.1016/j.nlm.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong CK, MacDermott AB. Synaptic GluN2A and GluN2B containing NMDA receptors within the superficial dorsal horn activated following primary afferent stimulation. J Neurosci 34: 10808–10820, 2014. doi: 10.1523/JNEUROSCI.0145-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhelski ML, Bruce DJ, Séguéla P, Wilcox GL, Simone DA. In vivo optogenetic activation of Nav1.8+ cutaneous nociceptors and their responses to natural stimuli. J Neurophysiol 117: 2218–2223, 2017. doi: 10.1152/jn.00083.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JH, Li YG, Nayak A, Errington ML, Murphy KP, Bliss TV. The suppression of long-term potentiation in rat hippocampus by inhibitors of nitric oxide synthase is temperature and age dependent. Neuron 11: 877–884, 1993. doi: 10.1016/0896-6273(93)90117-A. [DOI] [PubMed] [Google Scholar]
- Williams K. Ifenprodil discriminates subtypes of the N-methyl-d-aspartate receptor: selectivity and mechanisms at recombinant heteromeric receptors. Mol Pharmacol 44: 851–859, 1993. [PubMed] [Google Scholar]
- Yang XC, Reis DJ. Agmatine selectively blocks the N-methyl-d-aspartate subclass of glutamate receptor channels in rat hippocampal neurons. J Pharmacol Exp Ther 288: 544–549, 1999. [PubMed] [Google Scholar]
- Zádori D, Veres G, Szalárdy L, Klivényi P, Toldi J, Vécsei L. Glutamatergic dysfunctioning in Alzheimer’s disease and related therapeutic targets. J Alzheimers Dis 42, Suppl 3: S177–S187, 2014. doi: 10.3233/JAD-132621. [DOI] [PubMed] [Google Scholar]
- Zhuo M. Ionotropic glutamate receptors contribute to pain transmission and chronic pain. Neuropharmacology 112: 228–234, 2017. doi: 10.1016/j.neuropharm.2016.08.014. [DOI] [PubMed] [Google Scholar]