Abstract
An animal’s directional heading within its environment is encoded by the activity of head direction (HD) cells. In rodents, these neurons are found primarily within the limbic system in the interconnected structures that form the limbic HD circuit. In our accompanying report in this issue, we describe two HD cell populations located outside of this circuit in the medial precentral cortex (PrCM) and dorsal striatum (DS). These extralimbic areas receive their HD signals from the limbic system but do not provide critical input or feedback to limbic HD cells (Mehlman ML, Winter SS, Valerio S, Taube JS. J Neurophysiol 121: 350–370, 2019.). In this report, we complement our previous lesion and recording experiments with a series of neuroanatomical tracing studies in rats designed to examine patterns of connectivity between the PrCM, DS, limbic HD circuit, and related spatial processing circuitry. Retrograde tracing revealed that the DS receives direct input from numerous structures known to contain HD cells and/or other spatially tuned cell types. Importantly, these projections preferentially target and converge within the most medial portion of the DS, the same area in which we previously recorded HD cells. The PrCM receives direct input from a subset of these spatial processing structures. Anterograde tracing identified indirect pathways that could permit the PrCM and DS to convey self-motion information to the limbic HD circuit. These tracing studies reveal the anatomical basis for the functional relationships observed in our lesion and recording experiments. Collectively, these findings expand our understanding of how spatial processing circuitry functionally and anatomically extends beyond the limbic system into the PrCM and DS.
NEW & NOTEWORTHY Head direction (HD) cells are located primarily within the limbic system, but small populations of extralimbic HD cells are found in the medial precentral cortex (PrCM) and dorsal striatum (DS). The neuroanatomical tracing experiments reported here explored the pathways capable of transmitting the HD signal to these extralimbic areas. We found that projections arising from numerous spatial processing structures converge within portions of the PrCM and DS that contain HD cells.
Keywords: dorsal striatum, head direction cell, limbic system, medial precentral cortex, navigation, spatial cognition
INTRODUCTION
Head direction (HD) cells are modulated by the animal’s orientation within its environment, firing only when the animal’s head is pointing in a specific direction. A population of HD cells contains subsets of neurons tuned to each direction and therefore provides a continuous representation of the animal’s current HD, much like a compass (Taube et al. 1990). Ultimately, this HD signal is thought to generate an internal sense of direction used for navigation (Butler et al. 2017; Valerio and Taube 2012; van der Meer et al. 2010).
In the rodent brain, HD cells are found primarily within the limbic system in a group of interconnected structures that form the limbic HD circuit. The HD signal is thought to be generated subcortically within this circuit via the reciprocal connections between the dorsal tegmental nucleus and lateral mammillary nucleus. Subsequently, the HD signal is conveyed to multiple cortical structures via the anterior thalamus (Taube 2007). Interestingly, small populations of HD cells are found outside of the limbic HD circuit in the medial precentral cortex (PrCM), located in the frontal lobe, and in the dorsal striatum (DS), a component of the basal ganglia (Mizumori et al. 2000, 2005; Ragozzino et al. 2001; Wiener 1993). We previously recorded extralimbic HD cells in the PrCM and DS and found that these neurons displayed characteristics nearly identical to those of limbic HD cells recorded in the anterodorsal thalamic nucleus (ADN), suggesting that the extralimbic HD cell activity is driven by output from limbic HD cells (Mehlman et al. 2019). Indeed, disrupting the limbic HD circuit by lesioning the ADN eliminated HD cell activity in the PrCM and DS. Neurons modulated by the animal’s head movements, angular head velocity (AHV) cells, were also recorded in the PrCM and DS. The activity of these AHV cells was relatively unaffected by ADN lesions. Furthermore, we found that combined lesions of the PrCM and DS did not affect limbic HD cell activity recorded in the ADN. Collectively, the results from our previous lesion and recording experiments reveal a unidirectional functional relationship in which the PrCM and DS receive the HD signal from the limbic system but do not provide critical input or feedback to limbic HD cells.
Functionally related structures require underlying anatomical relationships; if a signal is transmitted from one structure to another, the structures must be physically connected via direct and/or indirect anatomical pathways. Therefore, the results of our lesion and recording experiments raise important questions about the patterns of anatomical connectivity between the PrCM, DS, and limbic HD circuit. What are the pathways capable of transmitting the HD signal and other spatial information from the limbic HD circuit to the PrCM and DS? Conversely, what are the pathways that could convey information, such as a self-motion signal, from the PrCM and DS to the limbic HD circuit? Here, we explore these issues in a series of neuroanatomical tracing studies in rats. Retrograde tracing was performed to examine direct projections from the limbic HD circuit to the PrCM and DS. Additionally, an anterograde tracer was used to visualize output pathways arising from the PrCM and DS.
These tracing studies provide a detailed, comprehensive, and novel description of the anatomical connections that permit the PrCM and DS to interact with a wide variety of structures involved in spatial processing (Table 1). We identify numerous projections that arise from the limbic HD circuit and converge within portions of the PrCM and DS that contain HD cells and AHV cells. Interestingly, portions of the DS that do not appear to contain HD cells receive less numerous and less dense spatial inputs. Additionally, we describe indirect pathways capable of conveying information from the PrCM and DS to the limbic HD circuit. These neuroanatomical tracing studies, coupled with our previous lesion and recording experiments, demonstrate that the PrCM and DS are functionally and anatomically integrated into the brain’s spatial processing circuitry to a greater extent than traditionally recognized.
Table 1.
Groups of structures in which labeling patterns were examined following retrograde tracer injections into the DS or PrCM
| Limbic HD Circuit Structures | Vestibular Input Structures | Motor Input Structures | Other Thalamic Nuclei | Other Spatial Processing Structures |
|---|---|---|---|---|
| DTN* | MVe | EPN | AVN* | HPC |
| LMN* | NPH | LHb | AMN* | dSub |
| ADN* | SGN | IPN | LDN* | vSub |
| RSP* | NRe* | PPC* | ||
| PoS* | PT | POR | ||
| PaS* | aCl | |||
| MEC* |
aCl, anterior claustrum; ADN, anterodorsal thalamic nucleus; AMN, anteromedial thalamic nucleus; AVN, anteroventral thalamic nucleus; dSub, dorsal subiculum; DS, dorsal striatum; DTN, dorsal tegmental nucleus; EPN, entopeduncular nucleus; HPC, hippocampus; IPN, interpeduncular nucleus; LDN, laterodorsal thalamic nucleus; LHb, lateral habenula; LMN, lateral mammillary nucleus; MEC, medial entorhinal cortex; MVe, medial vestibular nucleus; NPH, nucleus prepositus hypoglossi; NRe, nucleus reuniens of the thalamus; PaS, parasubiculum; POR, postrhinal cortex; PoS, postsubiculum; PPC, posterior parietal cortex; PrCM, medial precentral cortex; PT, parataenial nucleus of the thalamus; RSP, retrosplenial cortex; SGN, supragenual nucleus; vSub, ventral subiculum.
Brain area contains head direction (HD) cells.
MATERIALS AND METHODS
Subjects
Female Long-Evans rats (n = 29) weighing ~300 g were used in the experiments (Harlan Laboratories, Indianapolis, IN). Animals were pair-housed before surgery and housed individually following surgery. Food and water were available ad libitum and colony rooms were kept on a 12:12-h light-dark cycle at all times. All experimental procedures were approved by the Dartmouth College Institutional Animal Care and Use Committee and conformed to the standards outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Surgical Procedures
Animals were anesthetized with isoflurane (3% vaporized in oxygen) and then placed in a stereotaxic frame (David Kopf Instruments, Tujunga, CA). An incision was made to expose the skull, and small holes were drilled above the structures to be injected with tracer. Injections were delivered through a blunt-tip 33-gauge stainless steel cannula connected via polyethylene tubing to a 10-μl Hamilton syringe (Hamilton, Reno, NV) that was depressed at a constant rate by a syringe pump (0.1 μl/min; Razel Scientific Instruments, Stamford, CT). The cannula was left in place for 6 min following injection delivery to aid diffusion.
Retrograde Tracer Injections
Cholera toxin subunit B (CTB) conjugated to the red fluorescent dye Alexa Fluor (AF) 594 (Thermo Fisher Scientific, Waltham, MA) was used as a retrograde tracer (dissolved in phosphate buffer to a concentration of 1% wt/vol). CTB is taken up by axon terminals located at the injection site and then transported retrogradely within the axons, ultimately accumulating within the cell bodies from which these axons arise. Importantly, CTB does not cross synapses and therefore labels only the neurons that project directly to the injection site (i.e., polysynaptic connections are not labeled) (Conte et al. 2009a). It is possible that CTB is also taken up by fibers of passage that course through the injection site without forming synaptic connections, particularly if the fibers are damaged (Conte et al. 2009b); the protocol we used, however, has been shown to minimize the uptake of CTB by fibers of passage (Conte et al. 2009a). A total of 23 animals each received a 0.15-μl injection of CTB. Importantly, these injections targeted the specific portions of the striatum and cortex in which HD cells and AHV cells have been recorded (Mehlman et al. 2019; Mizumori et al. 2000, 2005; Wiener 1993). Eleven animals received an injection into the dorsomedial striatum (DMS) [1.3 to −0.3 mm anterior/posterior (A/P); 1.5 to 1.8 mm medial/lateral (M/L); −4.5 mm dorsal/ventral (D/V)], and seven animals received an injection into the overlying cortical area corresponding to the lateral portion of the PrCM and the adjacent primary motor cortex (M1) (0.5 to −0.3 mm A/P; 1.5 to 1.7 mm M/L; −1.4 mm D/V). For simplicity and to be consistent with the existing HD cell literature, we refer to this cortical area as the PrCM, although this area includes portions of M1 and has been alternatively defined as the medial agranular cortex (Hoover and Vertes 2007; Ostlund et al. 2009; Reep et al. 1990), anteromedial cortex (Sinnamon and Galer 1984), motor cortex (Condé et al. 1995), secondary motor cortex (Paxinos and Watson 1997, 2009; Swanson 1998; Yin 2009), vibrissa motor cortex (Brecht et al. 2004), and frontal orienting field (Erlich et al. 2011). Finally, five additional animals received a CTB injection into more lateral and ventral portions of the DS, which we refer to as the dorsolateral striatum (DLS) (0.5 to 0.1 mm A/P; 3.5 to 3.8 mm M/L; −5.5 mm D/V). The coordinates listed above, and all that follow, measure A/P and M/L relative to bregma and D/V relative to the cortical surface.
A subset of the animals described above (n = 6) received a second injection consisting of CTB conjugated to the green fluorescent dye AF 488 (Thermo Fisher Scientific); these injections targeted the hemisphere contralateral to the hemisphere injected with red CTB. The green CTB failed to retrogradely label cells with the same efficacy as the red CTB; therefore, green labeling patterns were not analyzed in these animals. When red labeling patterns were examined, there were no differences between animals injected with only red CTB and animals injected with red and green CTB.
Anterograde Tracer Injections
An adeno-associated virus (rAAV5-hSyn-EYFP; UNC Vector Core, Chapel Hill, NC) was used as an anterograde tracer. This virus infects neurons located at the injection site, causing them to express enhanced yellow fluorescent protein (EYFP) within their cell bodies and processes. Importantly, this virus does not cross synapses and therefore infects only the neurons located directly at the injection site (i.e., polysynaptic connections are not labeled) (Nassi et al. 2015). Additionally, this virus does not infect fibers of passage that course through the injection site without forming synaptic connections (Chamberlin et al. 1998). This virus primarily infects cell bodies located at the injection site, resulting in the anterograde transport of EYFP from these cell bodies to their associated processes; however, the rAAV5 virus can also infect axon terminals located at the injection site, resulting in the retrograde transport of the virus from these axons to their associated cell bodies, ultimately producing EYFP expression within these cell bodies (Aschauer et al. 2013; Chamberlin et al. 1998). A total of six animals each received a 0.25-μl injection of the anterograde tracer; these injections targeted the same brain areas as the retrograde tracer injections described above. Three animals received an injection into the DMS (0.4 to −0.2 mm A/P; 1.8 mm M/L; −4.5 mm D/V), two animals received an injection into the PrCM (0.3 to −0.2 mm A/P; 1.5 to 1.6 mm M/L; −1.4 mm D/V), and one animal received an injection into the DLS (0.1 mm A/P; 3.5 mm M/L; −5.5 mm D/V).
Histological Processing and Analyses
Animals injected with the retrograde tracer received a lethal dose of Euthasol (pentobarbital sodium and phenytoin sodium; 250 mg/kg injected intraperitoneally) and were transcardially perfused with 0.9% saline followed by 4% paraformaldehyde; this process occurred 1 wk after surgery. Brains were removed and postfixed in 4% paraformaldehyde for ~7 days and then cryoprotected in 20% sucrose for ~4 days.
Animals injected with the anterograde tracer received a lethal dose of Euthasol (250 mg/kg injected intraperitoneally) and were transcardially perfused with 0.9% saline followed by 10% formalin; this process occurred 9 wk after surgery. Brains were removed and postfixed in 10% formalin for 1 h and then cryoprotected in 20% sucrose for ~4 days.
Following cryoprotection, brains were sectioned on a cryostat. Coronal sections (30 μm thick) were acquired throughout the A/P axis of the brain; three consecutive sections were acquired every ~200 μm, starting at the level of the olfactory bulbs and ending at the level of the posterior cerebellum. The three consecutive sections were mounted on three series of glass slides (each series containing one of the sections). Two series were mounted using a medium containing a fluorescent DAPI stain (Fluoroshield with DAPI; Sigma-Aldrich, St. Louis, MO); these sections were examined under a fluorescent microscope (Olympus, Tokyo, Japan) equipped with filters for viewing red (CTB), yellow-green (EYFP), and blue (DAPI) signals. Representative images of injection sites and fluorescent labeling patterns were acquired using a digital camera (Olympus) connected to the microscope; image processing software was used to 1) uniformly adjust brightness and contrast levels (ImageJ; NIH, Bethesda, MD) and 2) automatically stitch together sequences of separate images into a single composite image covering a large area of tissue (Image Composite Editor; Microsoft, Redmond, WA). The third series of sections was Nissl-stained with thionine and examined under a light microscope (Leitz; Leica, Carnaxide, Portugal). Cytoarchitecture and anatomical landmarks were examined in brain sections using the DAPI and Nissl stains; these features were compared with a rat brain atlas (Paxinos and Watson 2009) to determine the location of structures of interest in each section.
Retrograde Tracing Analyses
The injection site in each animal was examined for the presence of CTB, indicated by a bright red fluorescent signal emitted by the AF 594 dye. For each animal, the extent of CTB diffusion was reconstructed by examining sections containing the injection site and tracing the area of tissue containing CTB onto a series of atlas plates illustrating coronal sections throughout the A/P axis of the DS and PrCM (1.6 to −1.3 mm). Next, image processing software (ImageJ) was used to compute the total area of tissue containing CTB in each animal; using these measurements independent-samples t-tests were performed to determine if the extent of CTB diffusion in the DMS was comparable to that observed in the DLS and PrCM (SPSS; IBM, Armonk, NY). All means are reported with ±SE. Nine animals were excluded from subsequent histological analyses due to extensive diffusion of tracer into unintended areas (e.g., the corpus callosum) or because an insufficient volume of tracer was delivered at the injection site (these animals are also excluded from Fig. 1).
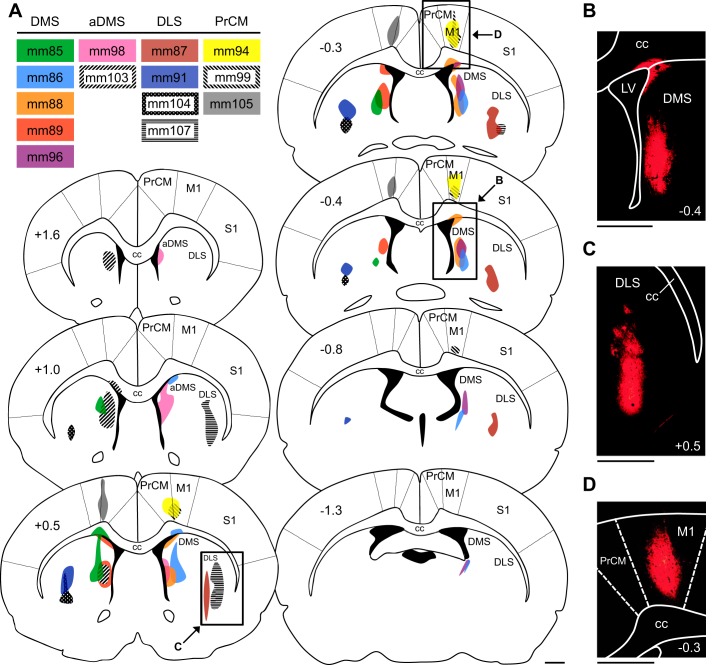
Fig. 1.
Retrograde tracer injections in the dorsal striatum (DS) and medial precentral cortex (PrCM). A: atlas plates depicting coronal sections throughout the anterior/posterior (A/P) axis of the DS and PrCM. The number at top left of each plate indicates its A/P location in mm relative to bregma. The injection site for each animal is illustrated (color coded by animal, listed in inset), with shaded areas representing the extent of cholera toxin subunit B diffusion. Labeled arrows point to the approximate locations of the images in B–D (the area shown in each image is outlined by a black rectangle). B: image of an injection site in the dorsomedial striatum (DMS; acquired from mm88). C: image of an injection site in the dorsolateral striatum (DLS; acquired from mm107). D: image of an injection site in the PrCM (acquired from mm94). In B–D, the number at bottom right of each image indicates its A/P location in mm relative to bregma. All scale bars are 1 mm. aDMS, anterior dorsomedial striatum; cc, corpus callosum; LV, lateral ventricle; M1, primary motor cortex; S1, primary somatosensory cortex.
Next, structures of interest were examined for the presence of retrogradely labeled cell bodies. CTB remains in vesicles after being transported to cell bodies; therefore, labeled cell bodies display a granular red fluorescent signal emitted by the AF 594 dye (Conte et al. 2009a). Five groups of structures were examined (Table 1). First, each structure in the limbic HD circuit was examined: the dorsal tegmental nucleus (DTN), lateral mammillary nucleus (LMN), anterodorsal thalamic nucleus (ADN), retrosplenial cortex (RSP), postsubiculum (PoS), parasubiculum (PaS), and medial entorhinal cortex (MEC). Each of these structures contains HD cells, and the DTN and LMN also contain AHV cells (Boccara et al. 2010; Taube 2007). Because of the relatively long A/P axis of the RSP, this structure was subdivided into an anterior portion (aRSP; −1.7 to −5.3 mm A/P) and a posterior portion (pRSP; −5.3 to −9.4 mm A/P). Second, three structures thought to convey vestibular signals to the limbic HD circuit were examined: the supragenual nucleus (SGN), nucleus prepositus hypoglossi (NPH), and medial vestibular nucleus (MVe) (Butler and Taube 2015; Clark and Taube 2012; Clark et al. 2012). Each of these structures contains AHV cells (Taube 2007; Winter and Taube 2014). Third, three structures thought to convey motor information to the limbic HD circuit were examined: the entopeduncular nucleus (EPN), lateral habenula (LHb), and interpeduncular nucleus (IPN) (Clark et al. 2009; Taube 2007). The LHb contains AHV cells (Sharp et al. 2006). Fourth, other thalamic nuclei known to contain spatially tuned neurons, including HD cells, were examined: the anteroventral thalamic nucleus (AVN), anteromedial thalamic nucleus (AMN), laterodorsal thalamic nucleus (LDN), nucleus reuniens (NRe), and parataenial nucleus (PT) (Jankowski et al. 2014, 2015; Mizumori and Williams 1993; Tsanov et al. 2011). The final group of brain areas examined included a variety of other structures involved in spatial processing, some of which contain HD cells and AHV cells (Chen et al. 1994; Wilber et al. 2014): the hippocampus (HPC), dorsal subiculum (dSub), ventral subiculum (vSub), posterior parietal cortex (PPC; as defined by Wilber et al. 2014), postrhinal cortex (POR; as defined by Burwell and Amaral 1998), and anterior claustrum (aCl; as defined by Jankowski and O’Mara 2015). Labeling patterns were also examined within the DS and PrCM.
For each structure of interest, labeling was assessed in each animal using a semiquantitative approach in which the average number of labeled cell bodies within a 1-mm2 area was estimated after labeling patterns throughout the structure were examined in sections spanning its entire A/P axis. Estimates of labeling density were scored as follows: −, no labeled cells; +, <5 labeled cells per 1 mm2; ++, 5–10 labeled cells per 1 mm2; and +++, >10 labeled cells per 1 mm2. These estimates were based on labeling observed within the ipsilateral hemisphere (i.e., the hemisphere in which red CTB was injected); labeling density was also examined within the contralateral hemisphere and rated relative to the density of labeling observed within the ipsilateral hemisphere (i.e., if labeled cells were observed within the contralateral hemisphere, the labeling density was rated as comparable to, less than, or much less than the density of labeling observed within the ipsilateral hemisphere).
When examining labeling patterns in cortical areas, we determined the layer(s) in which labeled cell bodies were located. In some cases, the borders between each cortical layer could not be determined, and the location of labeled cells was characterized more generally as superficial (layers I–II/III), intermediate (approximately layer IV), or deep (layers V–VI).
Anterograde Tracing Analyses
The injection site in each animal was examined for the presence of viral infection. Infected neurons accumulate EYFP within their cell bodies and therefore emit a bright yellow-green fluorescent signal. For each animal, the extent of viral infection was reconstructed by examining sections containing the injection site and tracing the area of tissue displaying EYFP expression within cell bodies onto a series of atlas plates illustrating coronal sections throughout the A/P axis of the DS and PrCM (1.6 to −1.3 mm). One animal was excluded from subsequent histological analyses because no EYFP expression was observed at the injection site (this animal is also excluded from Fig. 6).
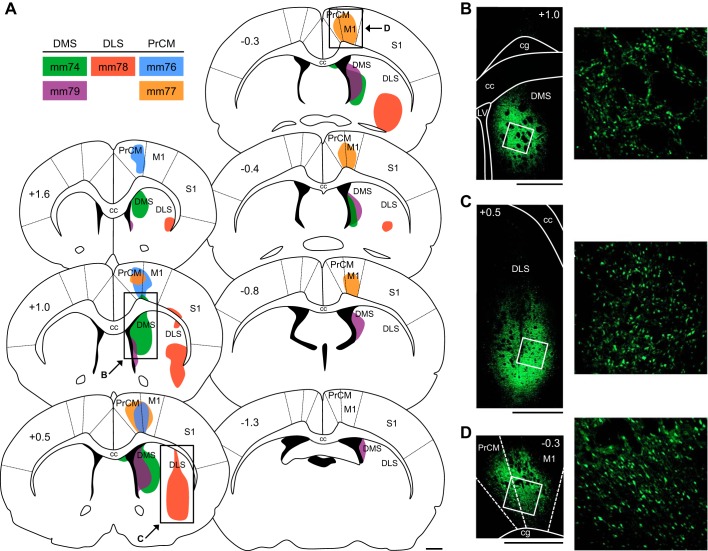
Fig. 6.
Anterograde tracer injections in the dorsal striatum (DS) and medial precentral cortex (PrCM). A: atlas plates depicting coronal sections throughout the anterior/posterior (A/P) axis of the DS and PrCM. The number at top left of each plate indicates its A/P location in mm relative to bregma. The injection site for each animal is illustrated (color coded by animal, listed in inset), with shaded areas representing the portion of tissue infected by the virus. Labeled arrows point to the approximate locations of the images in B–D (the area shown in each image is outlined by a black rectangle). B: image of an injection site in the dorsomedial striatum (DMS; acquired from mm74). C: image of an injection site in the dorsolateral striatum (DLS; acquired from mm78). D: image of an injection site in the PrCM (acquired from mm77). In B–D, right, the image shows the area outlined by the white square (left) at higher magnification. In B–D, left, the number at top right or top left of each image indicates its A/P location in mm relative to bregma. All scale bars are 1 mm. cc, Corpus callosum; cg, cingulum; LV, lateral ventricle; M1, primary motor cortex; S1, primary somatosensory cortex.
In addition to accumulating within cell bodies, EYFP accumulates within the processes of infected neurons; therefore, efferent fibers arising from infected neurons become fluorescently labeled and can be visualized in brain sections. We first examined the primary output pathways arising from the injection site by reconstructing the paths of labeled fiber bundles. Starting at the injection site, we followed the course of labeled fiber bundles across serial coronal sections and identified the locations in which putative terminal fields were formed.
Next, structures of interest were examined for the presence of labeled fibers. These brain areas included 1) the limbic HD circuit structures, 2) the vestibular input structures, and 3) the motor input structures (Table 1). Additionally, the DS and PrCM were examined for fiber labeling.
The atlas plates used in all histological analyses illustrate anatomical borders based on the atlas of Paxinos and Watson (1997, 2009), unless otherwise noted.
RESULTS
Projections to the DS: Retrograde Tracing
Retrograde tracer injections in the DS.
To examine projections from spatial processing circuitry to the DS, we injected the retrograde tracer CTB into different portions of the DS. Figure 1A illustrates the injection site for each animal, with shaded areas representing the extent of CTB diffusion. Importantly, the injections that targeted the DMS were generally generally restricted to the most medial portion of the DS located immediately adjacent to the lateral ventricle. We previously recorded HD cells and AHV cells in this area at A/P levels ranging from 1.6 to −0.4 mm and found that the DS HD signal was eliminated when the limbic HD circuit was disrupted (Mehlman et al. 2019). In two animals, CTB diffusion was restricted to relatively anterior levels of the DMS (aDMS; mm98 and mm103); in the other animals, CTB diffusion was confined to more intermediate and/or posterior levels of the DMS (mm85, mm86, mm88, mm89, and mm96). The injections that targeted the DLS (mm87, mm91, mm104, and mm107) delivered CTB to portions of the DS located lateral and ventral to the primary recording locations in our previous study. Diffusion of tracer into the overlying cortex was not observed in any of the animals described above.
Projections from the limbic HD circuit to the DS.
Figures 2 and 3 illustrate the patterns of retrograde labeling observed in mm88, a representative animal that received a CTB injection into the DMS (Fig. 1, A and B). In this animal, retrogradely labeled cells were observed cortically within the ipsilateral and contralateral RSP and MEC. Within the RSP, labeling was 1) observed within the superficial, intermediate, and deep layers, 2) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, and 3) denser within the pRSP compared with the aRSP. Labeled cells were observed within both the dysgranular and granular subregions of the RSP (Fig. 2, A and B). In general, labeling was slightly denser within the dysgranular subregion (dRSP) compared with the granular subregion (gRSP). Within the MEC, labeling was 1) confined to the deep layers, 2) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, and 3) observed along the entire A/P axis of the MEC, with slightly denser labeling occurring at posterior levels compared with anterior levels (Fig. 2, C and D).
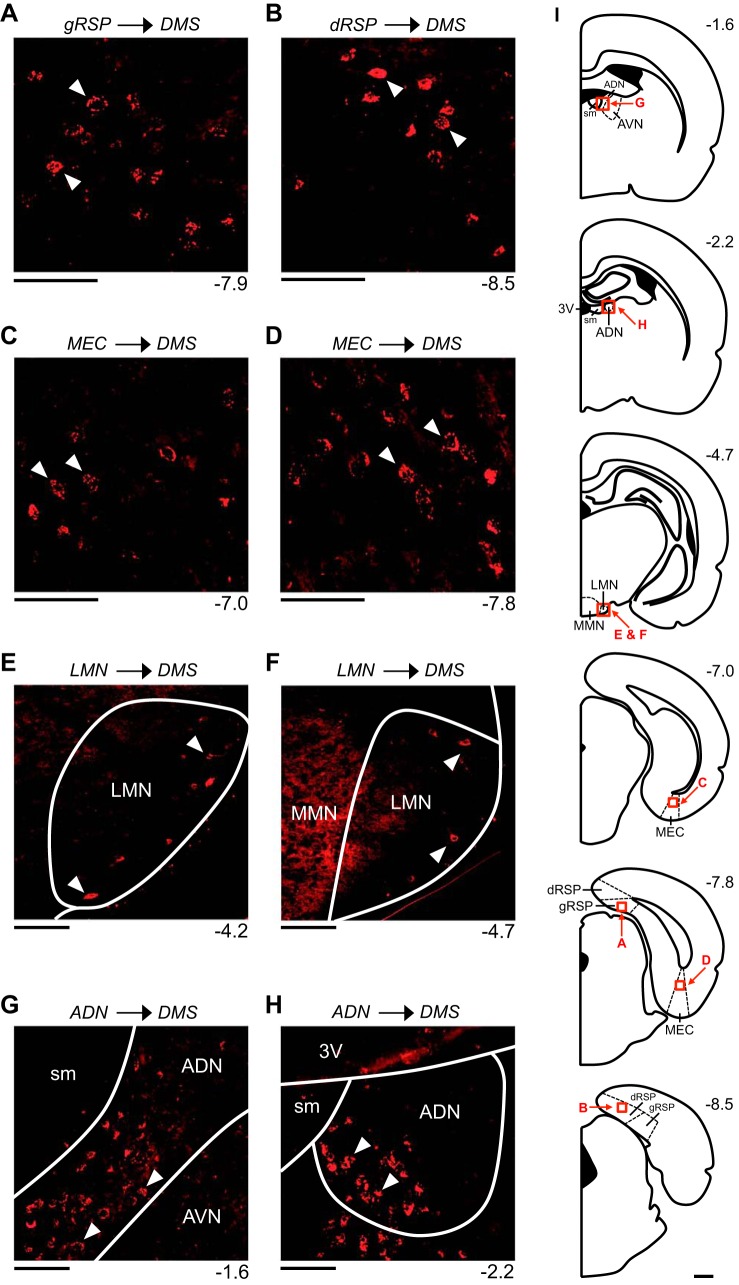
Fig. 2.
Retrograde labeling patterns observed within limbic head direction (HD) circuit structures following an injection into the dorsomedial striatum (DMS). All images are acquired from mm88, a representative animal that received a cholera toxin subunit B injection into the right DMS. A and B: labeled cells within the superficial layers of the granular retrosplenial cortex (gRSP; A) and the deep layers of the dysgranular retrosplenial cortex (dRSP; B). C and D: labeled cells within the deep layers of the medial entorhinal cortex (MEC). E and F: labeled cells within the shell region of the lateral mammillary nucleus (LMN). G and H: labeled cells within the anterodorsal thalamic nucleus (ADN). In A–H, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma, and the white arrowheads in each image indicate examples of labeled cells. All images show labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is medial, and right is lateral. Scale bars in A–H are 100 μm. I: atlas plates depicting coronal sections. The number at top right of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–H (the area shown in each image is outlined by a red square). Scale bar in I is 1 mm. 3V, third ventricle; AVN, anteroventral thalamic nucleus; MMN, medial mammillary nucleus; sm, stria medullaris.
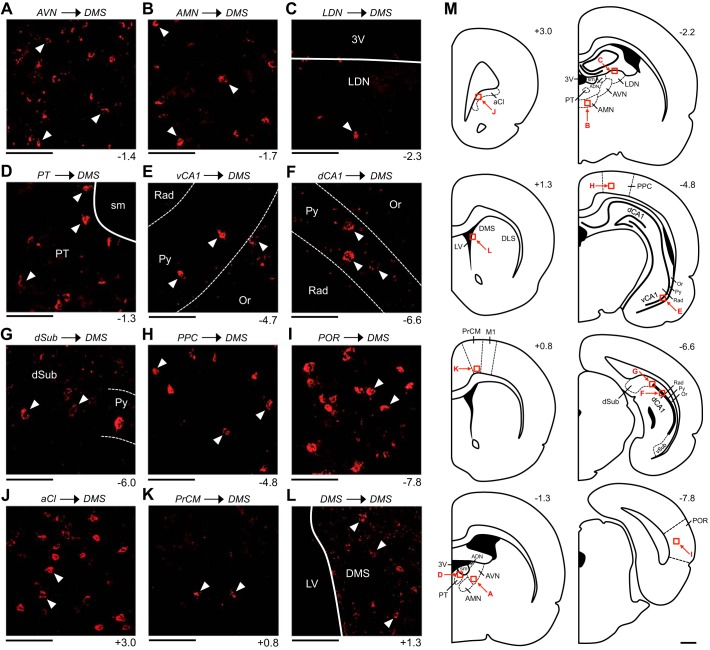
Fig. 3.
Retrograde labeling patterns observed within spatial processing structures following an injection into the dorsomedial striatum (DMS). All images are acquired from mm88, a representative animal that received a cholera toxin subunit B injection into the right DMS. A: labeled cells within the ventral portion of the anteroventral thalamic nucleus (AVN). B: labeled cells within the anteromedial thalamic nucleus (AMN). C: labeled cell within the dorsal portion the laterodorsal thalamic nucleus (LDN). D: labeled cells within the parataenial nucleus of the thalamus (PT). E and F: labeled cells within the pyramidal cell (Py) layer of the ventral (vCA1; E) and the dorsal CA1 area of the hippocampus (dCA1; F). G: labeled cells within the portion of the dorsal subiculum (dSub) located immediately adjacent to the dCA1. H: labeled cells within the deep layers of the posterior parietal cortex (PPC). I: labeled cells within the deep layers of the postrhinal cortex (POR). J: labeled cells within the anterior claustrum (aCl). K: labeled cells within layer VI of the medial precentral cortex (PrCM). L: labeled cells within the portion of the DMS located immediately anterior to the injection site. In A–L, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma, and the white arrowheads in each image indicate examples of labeled cells. All images show labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is medial, and right is lateral. Scale bars in A–L are 100 μm. M: atlas plates depicting coronal sections. The number at top right of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–L (the area shown in each image is outlined by a red square). Scale bar in M is 1 mm. 3V, third ventricle; ADN, anterodorsal thalamic nucleus; DLS, dorsolateral striatum; LV, lateral ventricle; M1, primary motor cortex; Or, oriens layer of the hippocampus; Rad, stratum radiatum layer of the hippocampus; sm, stria medullaris; vSub, ventral subiculum.
Additionally, in mm88, retrogradely labeled cells were observed subcortically within the ipsilateral and contralateral LMN and within the ipsilateral ADN. Within the LMN, labeling was 1) denser within the ipsilateral hemisphere compared with the contralateral hemisphere and 2) observed along the entire A/P axis. The majority of labeled cells were observed at the most anterior and most posterior levels of the LMN, with relatively less labeling occurring at intermediate levels. These labeled cells were confined to the outer portion of the LMN (i.e., the most lateral and ventral portions of the LMN), and labeling was also observed in the portion of tissue immediately surrounding the LMN, sometimes referred to as the ventral tuberomammillary nucleus (Paxinos and Watson 2009); we refer to these regions collectively as the LMN shell (Fig. 2, E and F) (Biazoli et al. 2006). A red fluorescent signal was also observed medial to the LMN within the medial mammillary nucleus (MMN) (Fig. 2F). This signal did not appear to arise from retrogradely labeled cells; rather, this signal appeared to be the result of tissue autofluorescence, a phenomenon observed previously within the MMN (Huang et al. 2017). Within the ADN, the density of labeling was uniform along the entire A/P axis, with labeled cells confined to the medial and ventral portions of the ADN (Fig. 2, G and H).
Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DMS. Interestingly, the two animals that received an injection into the aDMS (i.e., mm98 and mm103) displayed sparser labeling within the LMN and ADN compared with animals that received an injection into intermediate and/or posterior levels of the DMS (Table 2).
Table 2.
Summary of retrograde labeling patterns observed in each animal following a retrograde tracer injection into the DS or PrCM
| Limbic HD Circuit Structures | Other Thalamic Nuclei | Other Spatial Processing Structures | Other | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Animal | Target (area) | LMN | ADN | aRSP | pRSP | MEC | AVN | AMN | LDN | PT | HPC | dSub | vSub | PPC | POR | aCl | DS | PrCM |
| mm85 | DMS (1.90) | + | ++ | + | + | ++ | ++ | +++ | − | + | + | − | − | +++ | +++ | +++ | +++ | + |
| mm86 | DMS (2.05) | ++ | +++ | + | + | +++ | +++ | +++ | + | ++ | + | + | + | +++ | +++ | +++ | +++ | + |
| mm88 | DMS (2.25) | ++ | +++ | + | ++ | +++ | +++ | +++ | + | ++ | + | + | + | +++ | +++ | +++ | +++ | + |
| mm89 | DMS (2.10) | ++ | +++ | + | ++ | +++ | ++ | +++ | + | + | + | + | + | +++ | +++ | +++ | ++ | + |
| mm96 | DMS (1.40) | ++ | +++ | + | ++ | +++ | +++ | +++ | + | ++ | + | + | + | +++ | +++ | +++ | +++ | + |
| mm98 | aDMS (1.30) | + | + | + | ++ | +++ | + | + | − | + | + | + | + | +++ | +++ | +++ | ++ | + |
| mm103 | aDMS (2.30) | + | + | + | + | ++ | + | ++ | − | + | + | + | − | +++ | ++ | +++ | ++ | + |
| mm87 | DLS (1.95) | + | − | − | − | + | − | − | − | − | − | − | − | − | − | +++ | +++ | ++ |
| mm91 | DLS (1.95) | + | − | − | − | + | − | − | − | − | − | − | − | − | + | +++ | +++ | + |
| mm104 | DLS (1.50) | + | − | − | − | + | − | − | − | − | − | − | − | − | − | ++ | +++ | ++ |
| mm107 | DLS (2.45) | + | − | − | − | + | − | − | − | − | − | − | − | − | − | ++ | ++ | +++ |
| mm94 | PrCM (2.05) | + | − | + | − | + | − | − | − | − | − | − | − | ++ | − | +++ | − | +++ |
| mm99 | PrCM (1.35) | + | − | ++ | − | + | − | − | − | − | − | − | − | ++ | − | +++ | − | +++ |
| mm105 | PrCM (1.65) | + | − | + | − | + | − | − | − | − | − | − | − | ++ | − | +++ | − | +++ |
Labeling patterns: −, no labeled cells; +, <5 labeled cells per 1 mm2; ++, 5–10 labeled cells per 1 mm2; +++, >10 labeled cells per 1 mm2. Second column indicates the injection target (DMS, dorsomedial striatum; aDMS, anterior dorsomedial striatum; DLS, dorsolateral striatum; PrCM, medial precentral cortex) and the total area (in mm2) of tissue containing cholera toxin subunit B at the injection site. aCl, anterior claustrum; ADN, anterodorsal thalamic nucleus; AMN, anteromedial thalamic nucleus; AVN, anteroventral thalamic nucleus; DS, dorsal striatum; dSub, dorsal subiculum; HD, head direction; HPC, hippocampus; LDN, laterodorsal thalamic nucleus; LMN, lateral mammillary nucleus; MEC, medial entorhinal cortex; POR, postrhinal cortex; PPC, posterior parietal cortex; PT, parataenial nucleus of the thalamus; aRSP, anterior retrosplenial cortex; pRSP, posterior retrosplenial cortex; vSub, ventral subiculum.
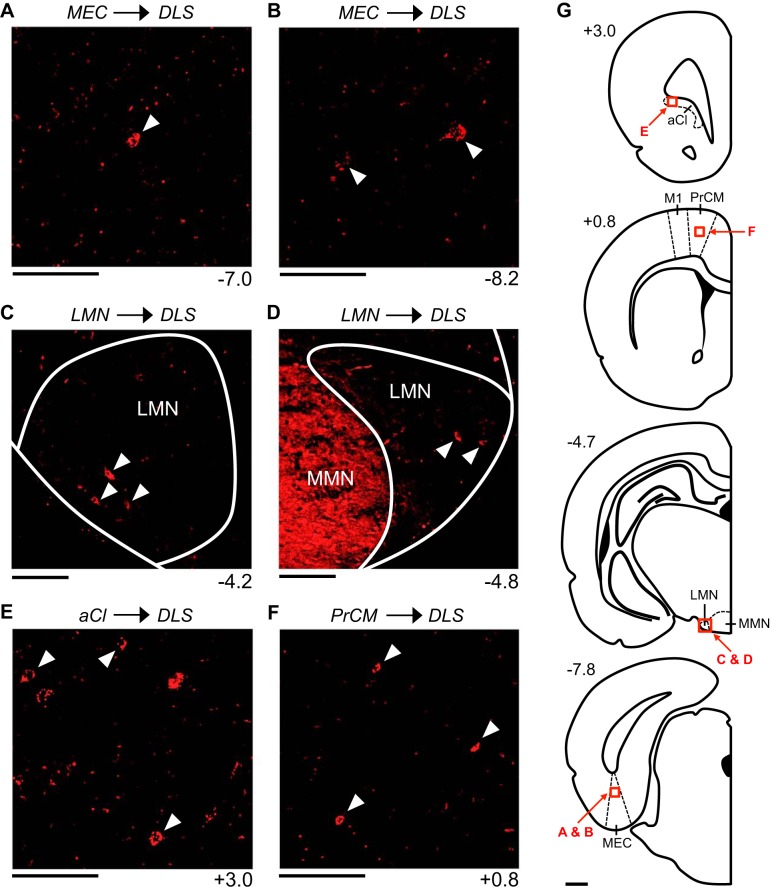
Figure 4 illustrates the patterns of retrograde labeling observed in mm104, a representative animal that received a CTB injection into the DLS (Fig. 1A). In this animal, retrogradely labeled cells were observed cortically within the ipsilateral and contralateral MEC; however, no labeling was observed within the RSP. Within the MEC, labeling was 1) confined to the deep layers, 2) much denser within the ipsilateral hemisphere compared with the contralateral hemisphere, 3) denser at posterior levels compared with anterior levels, and 4) much sparser compared with the MEC labeling observed in animals that received a CTB injection into the DMS (Fig. 4, A and B; Table 2). Additionally, in mm104, retrogradely labeled cells were observed subcortically within the ipsilateral and contralateral LMN; however, no labeling was observed within the ADN. Within the LMN, labeling was 1) comparable within the ipsilateral and contralateral hemispheres, 2) observed along the entire A/P axis, 3) confined to the shell region of the LMN, and 4) sparser compared with the LMN labeling observed in animals that received a CTB injection into the DMS (Fig. 4, C and D; Table 2). Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DLS (Table 2).
Fig. 4.
Retrograde labeling patterns observed within limbic head direction (HD) circuit structures and other spatial processing structures following an injection into the dorsolateral striatum (DLS). All images are acquired from mm104, a representative animal that received a cholera toxin subunit B injection into the left DLS. A and B: labeled cells within the deep layers of the medial entorhinal cortex (MEC). C and D: labeled cells within the shell region of the ipsilateral (C) and contralateral (D) lateral mammillary nucleus (LMN). E: labeled cells within the lateral portion of the anterior claustrum (aCl). F: labeled cells within the intermediate layers of the medial precentral cortex (PrCM). In A–F, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma, and the white arrowheads in each image indicate examples of labeled cells. All images (except D) show labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is lateral, and right is medial (in D, left is medial and right is lateral). Scale bars in A–F are 100 μm. G: atlas plates depicting coronal sections. The number at top left of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–F (the area shown in each image is outlined by a red square). Scale bar in G is 1 mm. M1, primary motor cortex; MMN, medial mammillary nucleus.
None of the animals that received a CTB injection into the DS displayed retrograde labeling within the DTN, PoS, or PaS.
Projections from vestibular and motor structures to the DS.
None of the animals that received a CTB injection into the DS displayed retrograde labeling within the SGN, NPH, MVe, EPN, LHb, or IPN.
Projections from other thalamic nuclei to the DS.
In addition to the ADN, retrograde labeling was observed within other thalamic nuclei involved in spatial processing. In mm88, labeled cells were observed within the ipsilateral AVN, AMN, LDN, and PT. Within the AVN, labeling was confined to the anterior and ventral portions of the structure (Fig. 3A). Within the AMN, labeled cells were observed along the entire A/P axis, with the densest labeling occurring at anterior levels (Fig. 3B). Within the LDN, labeled cells were observed, albeit rarely, along the entire A/P axis (less than 5 labeled cells were observed in total; Fig. 3C). Finally, within the PT, labeled cells were observed along the entire A/P axis, with the densest labeling occurring at anterior levels (Fig. 3D). No labeled cells were observed within the NRe. Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DMS. Compared with animals that received an injection into intermediate and/or posterior levels of the DMS, the two animals that received an injection into the aDMS displayed sparser labeling within the AVN, AMN, and PT and no labeling within the LDN or NRe (Table 2).
Interestingly, none of the animals that received a CTB injection into the DLS displayed retrograde labeling within the AVN, AMN, LDN, PT, or NRe (Table 2).
Projections from other spatial processing structures to the DS.
Retrograde labeling was observed within a variety of other structures involved in spatial processing. In mm88, labeled cells were observed within the ipsilateral HPC, dSub, and vSub and within the ipsilateral and contralateral PPC, POR, and aCl. Within the HPC, labeling was confined to posterior levels (−4.6 to −6.9 mm A/P). These labeled cells were observed exclusively within the CA1 area of the HPC, primarily within the pyramidal cell layer, and occasionally within the adjacent portions of the stratum radiatum and oriens layers. Labeled cells were observed within both the dorsal CA1 (dCA1) and the ventral CA1 (vCA1), with vCA1 labeling occurring at more anterior levels compared with dCA1 labeling (Fig. 3, E and F). Note that labeling within the dCA1 was observed at A/P levels that are posterior to where place cells are typically recorded in the dCA1. Additionally, labeled cells were observed within the dSub and vSub, albeit rarely (in total, less than 5 labeled cells were observed within each area); these cells tended to be located within the portions of the dSub and vSub located immediately adjacent to the dCA1 and vCA1, respectively (Fig. 3G).
Within the PPC, labeling was 1) observed within all the layers, with the deep layers displaying the densest labeling (specifically, layer V), 2) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, 3) observed along the entire A/P axis of the PPC (−4.2 to −4.8 mm), and 4) slightly denser within the medial portion of the PPC compared with the lateral portion (Fig. 3H). Within the POR, labeling was 1) confined to the deep layers, with the densest labeling observed within layer V, 2) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, and 3) observed along the entire A/P axis of the POR (−7.7 to −9.3 mm) (Fig. 3I). Finally, within the aCl, labeling was 1) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, 2) observed along the entire A/P axis of the aCl (3.3 to 2.8 mm), and 3) observed along the entire M/L axis of the aCl (Fig. 3J).
Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DMS. Labeling patterns did not display notable differences when animals that received injections into the aDMS were compared with animals that received injections into intermediate and/or posterior levels of the DMS (Table 2).
Interestingly, none of the animals that received a CTB injection into the DLS displayed retrograde labeling within the HPC, dSub, vSub, or PPC. One of these animals (mm91) displayed sparse labeling within the POR; these labeled cells were observed within the deep layers of the ipsilateral POR. All animals that received a CTB injection into the DLS displayed labeling within the aCl; labeling was much denser within the ipsilateral hemisphere compared with the contralateral hemisphere, and labeled cells were observed exclusively within the lateral portion of the aCl (Fig. 4E; Table 2).
Projections from the DS and PrCM to the DS.
In mm88, retrograde labeling was observed within the PrCM and within the DS itself. Within the PrCM, labeling was observed primarily within layers II/III and VI and was much denser within the ipsilateral hemisphere compared with the contralateral hemisphere (Fig. 3K). The labeled cells typically displayed a pyramidal or irregular morphology. Importantly, PrCM labeling was observed at A/P levels corresponding to our HD cell and AHV cell recording locations in the PrCM (0.7 to −0.8 mm) (Mehlman et al. 2019); labeling was densest at the anterior portion of this A/P range. Within the DS, labeled cells were observed exclusively within the ipsilateral hemisphere, immediately anterior and posterior to the injection site in the DMS; labeling was much denser anterior to the injection site compared with posterior (Fig. 3L). Additionally, sparse labeling was observed lateral to the injection site. Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DMS, and no notable differences were observed when animals that received injections into the aDMS were compared with animals that received injections into intermediate and/or posterior levels of the DMS (Table 2).
In mm104, retrograde labeling was also observed within the PrCM and within the DS itself. Within the PrCM, labeling was 1) observed primarily within the intermediate and deep layers, 2) much denser within the ipsilateral hemisphere compared with the contralateral hemisphere, 3) observed at A/P levels corresponding to our recording locations in the PrCM (0.7 to −0.8 mm) (Mehlman et al. 2019), and 4) denser compared with the PrCM labeling observed in animals that received a CTB injection into the DMS (Fig. 4F; Table 2). Within the DS, labeled cells were observed exclusively within the ipsilateral hemisphere, immediately anterior and posterior to the injection site in the DLS; labeling was much denser anterior to the injection site compared with posterior. Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the DLS (Table 2).
In sum, these retrograde tracing experiments reveal that the DS receives numerous direct projections from the limbic HD circuit and a variety of other structures involved in spatial processing. These projections target and converge within the portion of the DMS in which we previously recorded HD cells and AHV cells (Mehlman et al. 2019). In contrast, the DLS receives less numerous and less dense direct projections from spatial processing structures (Table 2). Importantly, the total area of tissue containing CTB at the injection site was, on average, comparable in the DMS and DLS [DMS, 1.94 ± 0.15 mm2; DLS, 1.97 ± 0.20 mm2; independent-samples t-test, t(7) = −0.145, P = 0.889]. Therefore, the differential labeling patterns observed following CTB injections into the DMS vs. DLS likely reflect true topographic organization in which spatial inputs into the DS preferentially target the most medial portion, rather than resulting from tracer simply diffusing over a larger area within the DMS compared with the DLS. It is possible, however, that the efficacy of CTB uptake and subsequent retrograde transport varies across the M/L axis of the DS, which could contribute to the observed topography.
Projections to the PrCM: Retrograde Tracing
Retrograde tracer injections in the PrCM.
To examine projections from spatial processing circuitry to the PrCM, we injected the retrograde tracer CTB into the PrCM. Figure 1A illustrates the injection site for each animal, with shaded areas representing the extent of CTB diffusion. Importantly, these injections were largely confined to the intermediate and deep layers of the portion of cortex overlying the DMS. As described in materials and methods, this cortical area includes portions of M1 and has been alternatively defined using a variety of nomenclature; for simplicity, we refer to this area as the PrCM. We previously recorded HD cells and AHV cells in this cortical area at A/P levels ranging from 0.7 to −0.8 mm and found that the PrCM HD signal was eliminated when the limbic HD circuit was disrupted (Mehlman et al. 2019). Diffusion of tracer into the underlying striatum was not observed in any of these animals.
Projections from the limbic HD circuit to the PrCM.
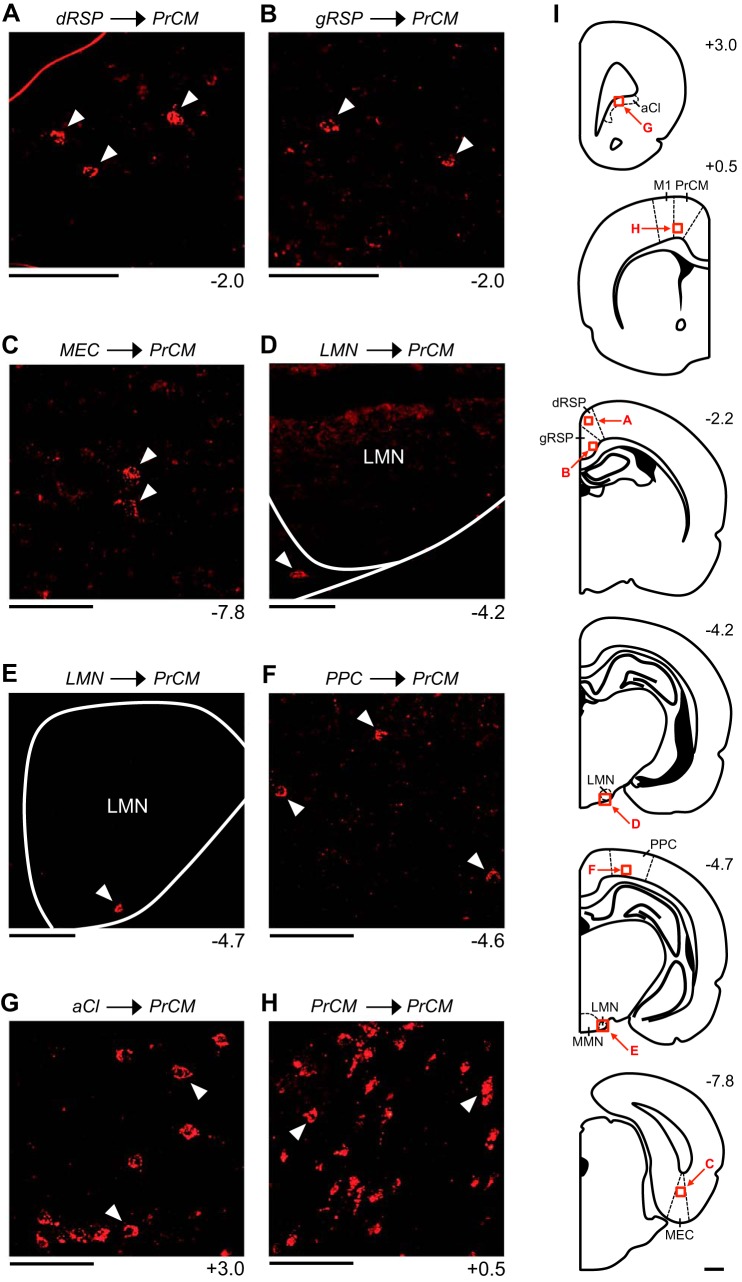
Figure 5 illustrates the patterns of retrograde labeling observed in mm94, a representative animal that received a CTB injection into the PrCM (Fig. 1, A and D). In this animal, retrogradely labeled cells were observed cortically within the ipsilateral and contralateral RSP and MEC. Within the RSP, labeling was 1) observed within the superficial, intermediate, and deep layers, 2) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, and 3) observed within both the dysgranular and granular subregions. Labeled cells were observed exclusively within the aRSP, with the densest labeling occurring at the most anterior levels of the aRSP (Fig. 5, A and B). Within the MEC, labeling was confined to the deep layers and was much denser within the ipsilateral hemisphere compared with the contralateral hemisphere (in total, less than 5 labeled cells were observed within the contralateral hemisphere). Labeled cells were observed along the entire A/P axis of the MEC (Fig. 5C). Additionally, in mm94, labeled cells were observed subcortically within the ipsilateral and contralateral LMN. Within the LMN, labeling was 1) comparable in the ipsilateral and contralateral hemispheres, 2) observed along the entire A/P axis, and 3) confined to the shell region of the LMN (Fig. 5, D and E). No labeling was observed within the DTN, ADN, PoS, or PaS. Similar patterns of retrograde labeling were observed in all other animals that received a CTB injection into the PrCM (Table 2).
Fig. 5.
Retrograde labeling patterns observed within limbic head direction (HD) circuit structures and other spatial processing structures following an injection into the medial precentral cortex (PrCM). All images are acquired from mm94, a representative animal that received a cholera toxin subunit B injection into the right PrCM. A and B: labeled cells within the superficial layers of the dysgranular retrosplenial cortex (dRSP; A) and the deep layers of the granular retrosplenial cortex (gRSP; B). C: labeled cells within the deep layers of the medial entorhinal cortex (MEC). D and E: labeled cells within the shell region of the lateral mammillary nucleus (LMN). F: labeled cells within the deep layers of the posterior parietal cortex (PPC). G: labeled cells within the anterior claustrum (aCl). H: labeled cells within the deep layers of the contralateral PrCM. In A–H, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma, and the white arrowheads in each image indicate examples of labeled cells. All images (except H) show labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is medial, and right is lateral (in H, left is lateral and right is medial). Scale bars in A–H are 100 μm. I: atlas plates depicting coronal sections. The number at top right of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–H (the area shown in each image is outlined by a red square). Scale bar in I is 1 mm. M1, primary motor cortex; MMN, medial mammillary nucleus.
Projections from vestibular and motor structures to the PrCM.
None of the animals that received a CTB injection into the PrCM displayed retrograde labeling within the SGN, NPH, MVe, EPN, LHb, or IPN.
Projections from other thalamic nuclei to the PrCM.
None of the animals that received a CTB injection into the PrCM displayed retrograde labeling within the AVN, AMN, LDN, NRe, or PT (Table 2).
Projections from other spatial processing structures to the PrCM.
In mm94, retrogradely labeled cells were observed within the ipsilateral and contralateral PPC and aCl. Within the PPC, labeling was 1) observed within the superficial, intermediate, and deep layers, 2) much denser within the ipsilateral hemisphere compared with the contralateral hemisphere (in total, less than 5 labeled cells were observed within the contralateral hemisphere), and 3) observed along the entire A/P axis of the PPC (−4.2 to −4.8 mm) (Fig. 5F). The density of labeling was uniform along the entire M/L axis of the PPC. Within the aCl, labeling was 1) denser within the ipsilateral hemisphere compared with the contralateral hemisphere, 2) observed along the entire A/P axis of the aCl (3.3 to 2.8 mm), and 3) observed along the entire M/L axis of the aCl (Fig. 5G). No labeled cells were observed within the HPC, dSub, vSub, or POR. Similar retrograde labeling patterns were observed in all other animals that received a CTB injection into the PrCM (Table 2).
Projections from the DS and PrCM to the PrCM.
In mm94, retrograde labeling was not observed within the DS; however, labeled cells were observed within the PrCM itself, in both the ipsilateral and contralateral hemispheres. Within the ipsilateral PrCM, labeled cells were observed immediately anterior and posterior to the injection site within the superficial, intermediate, and deep layers. Within the contralateral PrCM, labeled cells were observed at the same A/P levels as the injection site within the superficial, intermediate, and deep layers (Fig. 5H). Similar patterns of retrograde labeling were observed in all other animals that received a CTB injection into the PrCM (Table 2).
In sum, these retrograde tracing experiments reveal that the PrCM receives multiple direct projections from the limbic HD circuit and other spatial processing structures. Compared with the DMS, the PrCM receives less numerous and generally less dense projections from structures involved in spatial processing (Table 2). Importantly, the total area of tissue containing CTB at the injection site was, on average, comparable in the DMS and PrCM [DMS, 1.94 ± 0.15 mm2; PrCM, 1.67 ± 0.21 mm2; independent-samples t-test, t(6) = 1.066, P = 0.328]; therefore, the differential labeling patterns observed following CTB injections into the DMS vs. PrCM are unlikely due to a differential extent of CTB diffusion within these two brain areas. However, it is possible that the efficacy of CTB uptake and subsequent retrograde transport varies between the DMS and PrCM, which could contribute to the observed differential labeling patterns.
Projections Arising from the DS: Anterograde Tracing
Anterograde tracer injections in the DS and fiber labeling.
To examine projections from the DS to spatial processing circuitry, we injected the anterograde tracer rAAV5-hSyn-EYFP into different portions of the DS. Figure 6A illustrates the injection site for each animal, with shaded areas representing the portion of tissue infected by the virus, indicated by EYFP expression within cell bodies (Fig. 6, B and C). Importantly, the injections that targeted the DMS (mm74 and mm79) were generally restricted to the most medial portion of the DS located immediately adjacent to the lateral ventricle. We previously lesioned this area and observed no significant changes in limbic HD cell activity (Mehlman et al. 2019). Diffusion of tracer into the overlying cortex was not observed in either of these animals. However, the animal that received an injection into the DLS (mm78) displayed slight diffusion of tracer into the overlying somatosensory cortex (Fig. 6A).
An EYFP signal was also observed within fibers emanating from the infected area; these labeled fibers, putative long-range axonal projections, were visualized throughout the brain in serial coronal sections to reconstruct output pathways. Two general types of labeled fibers were observed, large-diameter fibers and small-diameter fibers, and these fiber types could be readily differentiated on the basis of their appearance and organization. The primary projections arising from the injection site were composed of large-diameter fibers; these smooth, threadlike fibers were uniform in diameter along their length and did not display branching. Large-diameter fibers were typically organized into bundles located within white matter. All fibers within each bundle were oriented similarly (i.e., the fibers ran roughly parallel to each other), and the individual fibers were readily observed (Fig. 7, A and C). Labeled fibers displaying these characteristics are thought to course through the brain area in which they are observed without forming local synaptic connections (Sesack et al. 1989). The second type of labeled fibers displayed a much smaller, more irregular diameter, and varicosities and branch points could often be observed along the length of these fibers (e.g., see Fig. 10B). Typically located outside of white matter and intermixed with cell bodies, these small-diameter fibers often formed densely packed and poorly organized fields lacking consistent fiber orientation (Fig. 7, B and D). These areas were considered putative terminal fields and the sites of potential synaptic connections (Sesack et al. 1989).
Fig. 7.
Appearance of labeled fibers following an injection into the dorsomedial striatum (DMS) or medial precentral cortex (PrCM). A: image of large-diameter fibers within the internal capsule (ic), acquired from mm74, a representative animal that received a virus injection into the right DMS. B: image of small-diameter fibers forming a putative terminal field within the globus pallidus (GP), acquired from mm74. C: image of large-diameter fibers within the ic, acquired from mm77, a representative animal that received a virus injection into the right PrCM. D: image of small-diameter fibers forming a putative terminal field within the entopeduncular nucleus (EPN), acquired from mm77. These small-diameter fibers are located outside of the small, circular fascicles that stipple the EPN; the white arrowhead indicates a single large-diameter fiber located within a fascicle. In A–D, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma. All images show fiber labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is medial, and right is lateral. Scale bars in A–D are 100 μm. E: atlas plates depicting coronal sections. The number at top right of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–D (the area shown in each image is outlined by a red square). Scale bar in E is 1 mm.
Fig. 10.
Fiber labeling patterns observed within spatial processing structures following an injection into the medial precentral cortex (PrCM). All images are acquired from mm77, a representative animal that received a virus injection into the right PrCM. A: small-diameter fibers within the deep layers of the granular retrosplenial cortex (gRSP). B: small-diameter fiber within the intermediate layers of the postsubiculum (PoS). C: small-diameter fibers within the supragenual nucleus (SGN). D: small-diameter fibers within the nucleus prepositus hypoglossi (NPH). E: small-diameter fibers within the medial vestibular nucleus (MVe). F: small-diameter fibers within the interpeduncular nucleus (IPN) and large-diameter fibers within the cerebral peduncle (cp). In A–F, the number at bottom right of each image indicates its anterior/posterior (A/P) location in mm relative to bregma, and the white arrowheads in each image indicate examples of labeled fibers. Images in A, B, and F show fiber labeling within the hemisphere ipsilateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is medial, and right is lateral. Images in C, D, and E show fiber labeling within the hemisphere contralateral to the injection site and are oriented as follows: top is dorsal, bottom is ventral, left is lateral, and right is medial. Scale bars in A–F are 100 μm. G: atlas plates depicting coronal sections. The number at top right of each plate indicates its A/P location in mm relative to bregma. Labeled arrows point to the approximate locations of the images in A–F (the area shown in each image is outlined by a red square). Scale bar in G is 1 mm. 4V, fourth ventricle; dRSP, dysgranular retrosplenial cortex.
Primary DS output pathways.
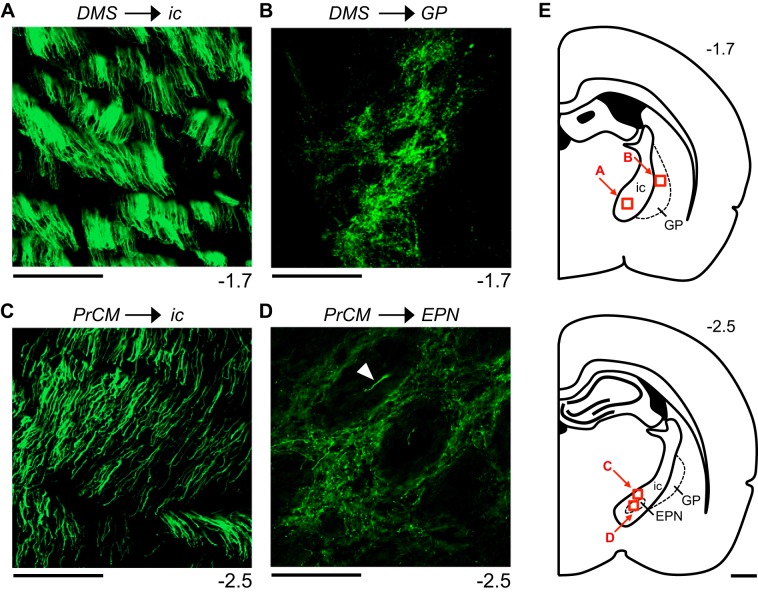
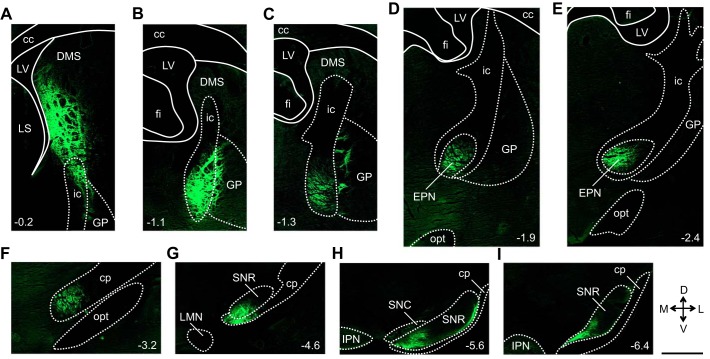
Figure 8 illustrates the reconstructed output pathways observed in mm74, a representative animal that received a virus injection into the DMS (Fig. 6A). EYFP expression was observed within and surrounding cell bodies located at the injection site, with minimal expression observed within the small, circular fascicles that stipple the DS (Fig. 6B). Labeled fibers arising from the injection site, primarily bundles of large-diameter fibers, exited the DMS ventrally to enter the internal capsule (ic) and globus pallidus (GP) (Fig. 8A). Within the ic, these fibers coursed posteriorly and ventrally (Fig. 7A and Fig. 8, B and C). Small-diameter fibers were observed exiting the lateral edge of the ic to enter the adjacent GP where terminal fields were formed (Figs. 7B and 8C). The large-diameter fibers within the ic continued to course posteriorly and ventrally, ultimately entering the EPN (Fig. 8, D and E). Within the EPN, the majority of labeled fibers were large-diameter fibers observed primarily within the small, circular fascicles that stipple the EPN. Small-diameter fibers were also observed within the EPN and were located outside of the fascicles. From the EPN, the large-diameter fibers continued to course posteriorly and ventrally, now traveling within the medial and ventral portions of the cerebral peduncle (cp) (Fig. 8F). At the level of the substantia nigra pars reticulata (SNR), the large-diameter fibers within the cp gave rise to small-diameter fibers that exited the dorsal edge of the cp to enter the medial and ventral portions of the SNR, where large terminal fields were formed (Fig. 8, G, H, and I). At this level, EYFP expression was also observed within cell bodies located in the medial portion of the substantia nigra pars compacta (SNC) (Fig. 8H). This portion of the SNC has been shown to project directly to the DMS (Lerner et al. 2015), and it is likely that EYFP expression in these neurons resulted from the retrograde transport of the virus (Aschauer et al. 2013; Chamberlin et al. 1998). This output pathway was observed exclusively within the hemisphere ipsilateral to the injection site; no labeled fibers were observed within the contralateral hemisphere. Similar patterns of fiber labeling were observed in mm79, an animal that received a virus injection into a more posterior portion of the DMS compared with mm74 (Fig. 6A).
Fig. 8.
Reconstruction of the primary dorsomedial striatum (DMS) output pathways. A–I show a series of images acquired from mm74, a representative animal that received a virus injection into the right DMS. Starting at the injection site (A), this series of images illustrates the location of labeled fibers at progressively more posterior levels, ultimately tracing a pathway from the DMS to the substantia nigra pars reticulata (SNR). The number at bottom left or bottom right of each image indicates its anterior/posterior location in mm relative to bregma. All images show fiber labeling within the hemisphere ipsilateral to the injection site and are oriented as indicated in inset (D, dorsal; V, ventral; M, medial; L, lateral). Scale bar is 1 mm and applies to all panels. cc, Corpus callosum; cp, cerebral peduncle; EPN, entopeduncular nucleus; fi, fimbria of the hippocampus; GP, globus pallidus; ic, internal capsule; IPN, interpeduncular nucleus; LMN, lateral mammillary nucleus; LS, lateral septal nucleus; LV, lateral ventricle; opt, optic tract; SNC, substantia nigra pars compacta.
Similar output pathways were also observed in mm78, an animal that received a virus injection into the DLS (Fig. 6, A and C). Labeled fibers arising from the injection site, primarily bundles of large-diameter fibers, coursed posteriorly and medially through the GP to enter the ic. Within the GP, small-diameter fibers formed terminal fields. Within the ic, the large-diameter fibers continued to course posteriorly; at the level of the EPN, large- and small-diameter fibers were observed within this structure. Continuing to course posteriorly via the cp, the large-diameter fibers ultimately gave rise to small-diameter fibers that exited the dorsal edge of the cp, entered the SNR, and formed a large terminal field. Compared with mm74 and mm79, mm78 displayed fiber labeling within more lateral portions of the ic, EPN, cp, and SNR. Therefore, projections arising from the DMS and DLS are topographically organized within a common primary output pathway targeting the SNR; projections arising from the DMS travel within the medial portion of this pathway, whereas those arising from the DLS course through more lateral portions.
Projections from the DS to other brain areas.
In addition to reconstructing the output pathways described above, we examined the limbic HD circuit structures (Table 1) for the presence of fiber labeling; no labeled fibers were observed within any of these structures in mm74, mm79, or mm78. Moreover, none of these animals displayed fiber labeling within any of the vestibular input structures (Table 1) or within the PrCM. As described above, large- and small-diameter fibers were observed within the EPN in mm74, mm79, and mm78; however, none of these animals displayed fiber labeling within the other motor input structures (Table 1).
These anterograde tracing experiments reveal that neither medial nor lateral portions of the DS project directly to structures in the limbic HD circuit or to related vestibular input structures; however, the DS does project directly to the EPN, a motor input structure.
Projections Arising from the PrCM: Anterograde Tracing
Anterograde tracer injections in the PrCM and fiber labeling.
To examine projections from the PrCM to spatial processing circuitry, we injected the anterograde tracer rAAV5-hSyn-EYFP into the PrCM. Figure 6A illustrates the injection site for each animal, with shaded areas representing the portion of tissue infected by the virus, indicated by EYFP expression within cell bodies (Fig. 6D). Importantly, these injections were confined to the portion of cortex overlying the DMS. As described in materials and methods, this cortical area includes portions of M1 and has been alternatively defined using a variety of nomenclature; for simplicity, we refer to this area as the PrCM. We previously lesioned this cortical area and observed no significant changes in limbic HD cell activity (Mehlman et al. 2019). Diffusion of tracer into the underlying striatum was not observed in any of these animals.
An EYFP signal was also observed within fibers emanating from the infected area; these labeled fibers were visualized throughout the brain in serial coronal sections to reconstruct output pathways. As described above, two types of labeled fibers, large-diameter fibers and small-diameter fibers, were observed and readily differentiated on the basis of their appearance and organization (Fig. 7) (Sesack et al. 1989).
Primary PrCM output pathways.
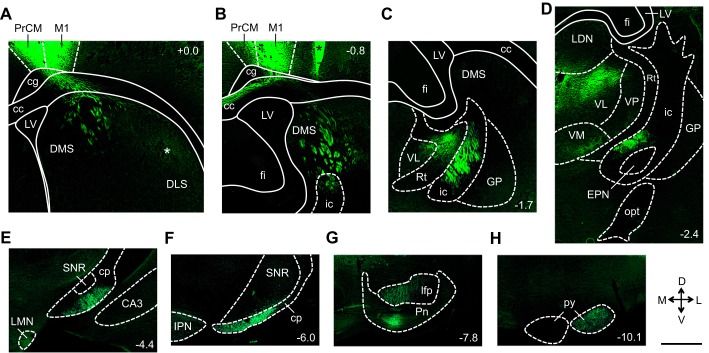
Figure 9 illustrates the reconstructed output pathways observed in mm77, a representative animal that received a virus injection into the PrCM (Fig. 6, A and D). Large-diameter fibers arising from the injection site coursed ventrally through the cingulum (cg) and into the corpus callosum (cc) (Fig. 9A). Within the cc, one group of large-diameter fibers turned medially, traveling through the dorsal portion of the cc into the contralateral hemisphere (Fig. 9B). Here, small-diameter fibers were observed exiting the dorsal edge of the cc to enter all the layers of the contralateral PrCM. Additionally, small-diameter fibers were observed exiting the ventral edge of the cc to enter the contralateral DLS.
Fig. 9.
Reconstruction of the primary medial precentral cortex (PrCM) output pathways. A–H show a series of images acquired from mm77, a representative animal that received a virus injection into the right PrCM. Starting at the injection site (A), this series of images illustrates the location of labeled fibers at progressively more posterior levels, ultimately tracing a pathway from the PrCM to the pyramidal tract (py). White asterisk in A indicates the location of a putative terminal field formed by small-diameter fibers. Black asterisk in B denotes an area not infected by the virus (bright green appearance is due to autofluorescence caused by tissue folding, not enhanced yellow fluorescent protein expression). The number at top right or bottom right of each image indicates its anterior/posterior location in mm relative to bregma. All images show fiber labeling within the hemisphere ipsilateral to the injection site and are oriented as indicated in inset (D, dorsal; V, ventral; M, medial; L, lateral). Scale bar is 1 mm and applies to all panels. CA3, CA3 area of the hippocampus; cc, corpus callosum; cg, cingulum; cp, cerebral peduncle; DLS, dorsolateral striatum; DMS, dorsomedial striatum; EPN, entopeduncular nucleus; fi, fimbria of the hippocampus; GP, globus pallidus; ic, internal capsule; IPN, interpeduncular nucleus; LDN, laterodorsal thalamic nucleus; lfp, longitudinal fasciculus of the pons; LMN, lateral mammillary nucleus; LV, lateral ventricle; M1, primary motor cortex; opt, optic tract; Pn, pontine nuclei; Rt, reticular thalamic nucleus; SNR, substantia nigra pars reticulata; VL, ventrolateral thalamic nucleus; VM, ventromedial thalamic nucleus; VP, ventral posterolateral thalamic nucleus.
A second group of large-diameter fibers traveled laterally and ventrally within the cc; these fibers exited the ventral edge of the cc to enter the medial and central portions of the ipsilateral DS (Fig. 9A). Within the DS, these large-diameter fibers coursed posteriorly and ventrally within the small, circular fascicles that stipple the DS (Fig. 9, A and B). Additionally, small-diameter fibers were observed in more lateral portions of the DS; these fibers were located outside of the fascicles, forming a terminal field in the dorsal portion of the DLS (Fig. 9A). The large-diameter fiber bundles in the medial and central portions of the DS entered the ic, where these fibers continued to travel posteriorly and ventrally (Fig. 7C and Fig. 9, B, C, and D). From the ic, these large-diameter fibers continued to course posteriorly within the cp (Fig. 9, E and F), then within the longitudinal fasciculus of the pons (lfp) (Fig. 9G) and ultimately within the pyramidal tract (py) (Fig. 9H).
Small-diameter fibers were observed arising from this ipsilateral output pathway at multiple levels. At the level of the thalamus, small-diameter fibers coursed medially to exit the ic and enter the lateral edge of the thalamus (Fig. 9C). These small-diameter fibers appeared to terminate primarily within the ventral anterior thalamic nucleus, ventrolateral thalamic nucleus, ventromedial thalamic nucleus, posterior thalamic nuclear group, mediodorsal thalamic nucleus, centrolateral thalamic nucleus, and lateral posterior thalamic nucleus (Fig. 9D). At the level of the EPN, small-diameter fibers arising from the ic traveled ventrally to exit the ic and entered the EPN, forming a terminal field located outside of the small, circular fascicles that stipple the EPN (Figs. 7D and 9D). At the level of the pontine nuclei (Pn), small-diameter fibers exited the ventral edge of the lfp to enter the Pn and form a terminal field (Fig. 9G). Additionally, at these posterior levels, small-diameter fibers were observed exiting the dorsal edge of the lfp and py, with some fibers traveling into the contralateral hemisphere.
Similar patterns of fiber labeling were observed in mm76, an animal that received a virus injection into a more anterior portion of the PrCM compared with mm77 (Fig. 6A).
Projections from the PrCM to other brain areas.
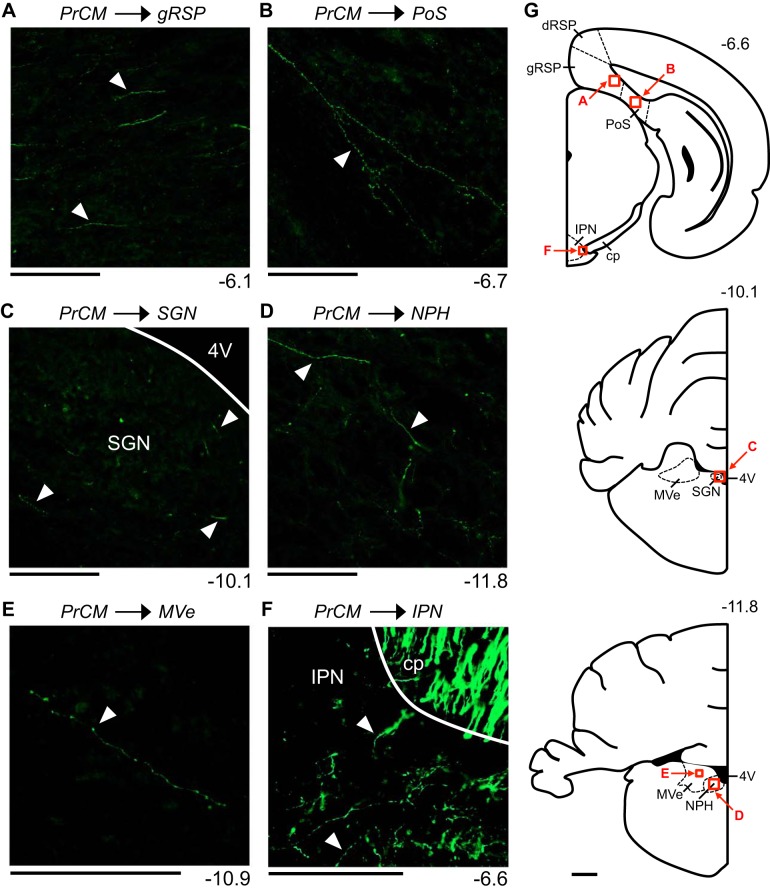
In addition to reconstructing the output pathways described above, we examined the limbic HD circuit structures (Table 1) for the presence of fiber labeling. Small-diameter fibers were observed within the ipsilateral and contralateral aRSP, and to a lesser extent, within the ipsilateral pRSP. These labeled fibers were located within the superficial, intermediate, and deep layers, and within both the dysgranular and granular subregions (Fig. 10A). Additionally, small-diameter fibers were observed within the intermediate and deep layers of the ipsilateral PoS (Fig. 10B). No labeled fibers were observed within the DTN, LMN, ADN, PaS, or MEC.
Small diameter fibers were also observed within the ipsilateral and contralateral SGN (Fig. 10C), NPH (Fig. 10D), and MVe (Fig. 10E), the three vestibular input structures located in the brain stem. Regarding the motor input structures (Table 1), small-diameter fibers were observed within the ipsilateral EPN (Fig. 7D), as described above, as well as within the lateral portion of the ipsilateral IPN (Fig. 10F). No labeled fibers were observed within the LHb. These patterns of fiber labeling were observed in both mm77 and mm76 (fiber labeling within the MVe was observed only in mm77). Compared with the fiber labeling observed within the primary output pathways, fiber labeling within the RSP, PoS, SGN, NPH, MVe, and IPN was relatively sparse. Within these structures, small numbers of individual labeled fibers were typically observed, rather than terminal fields formed by large numbers of overlapping labeled fibers.
These anterograde tracing experiments reveal sparse but direct projections from the PrCM to limbic HD circuit structures, vestibular input structures, and motor input structures.
DISCUSSION
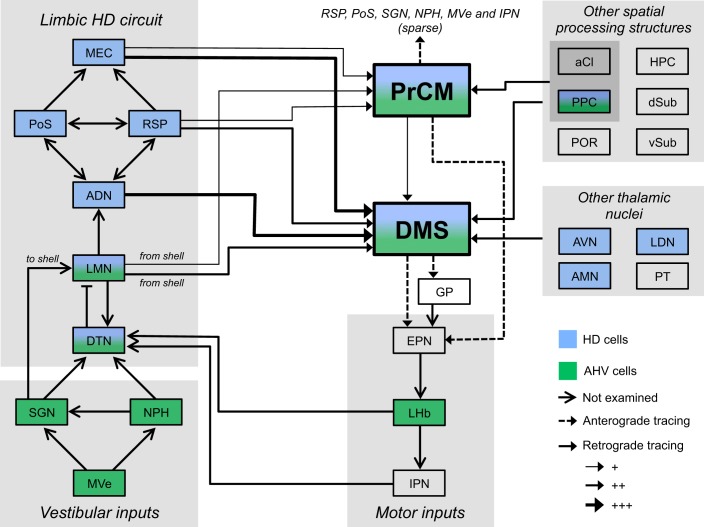
Our accompanying report showed that lesioning the ADN to disrupt limbic HD circuit output eliminated PrCM and DS HD cell activity, demonstrating a functional relationship in which the PrCM and DS receive the HD signal from the limbic system (Mehlman et al. 2019). Here, we examined the anatomical basis for this functional relationship by performing a series of retrograde tracing experiments designed to examine the pathways that could convey the HD signal from the limbic HD circuit to the PrCM and DS. Furthermore, we previously found that combined PrCM and DS lesions did not significantly affect limbic HD cell activity in the ADN. In the present study, we explored the anatomical pathways that could permit the PrCM and DS to convey information to the limbic HD circuit in a series of anterograde tracing experiments. Importantly, our neuroanatomical studies were designed to examine the afferent and efferent connectivity of the specific portions of the frontal cortex and striatum in which HD cell and AHV cell activity has been recorded (Mehlman et al. 2019; Mizumori et al. 2000, 2005; Wiener 1993). No previous study has systematically described how these areas are anatomically related to the limbic HD circuit and other spatial processing structures. Our anatomical characterization of the PrCM is particularly valuable given that it is a relatively ambiguous brain area associated with a variety of nomenclature and inconsistently defined borders. These experiments reveal extensive patterns of anatomical connectivity between the PrCM, DS, and spatial processing circuitry; these connections are summarized in the circuit diagram in Fig. 11. To the best of our knowledge, we observed multiple connections that have not been described previously in the rat, including projections from the LMN and aCl to the DS and PrCM and projections from the AMN, LDN, and PT to the DMS. An important caveat, although we report numerous projections that arise from spatial processing structures and terminate within the PrCM and DS, it is unclear if these projections arise directly from spatially tuned neurons.
Fig. 11.
Circuit diagram summarizing the patterns of connectivity between the medial precentral cortex (PrCM), dorsomedial striatum (DMS), and spatial processing circuitry. Left portion of the diagram depicts the limbic head direction (HD) circuit; the reciprocally connected dorsal tegmental nucleus (DTN) and lateral mammillary nucleus (LMN) are thought to generate the HD signal, which is serially transmitted from the LMN to the anterodorsal thalamic nucleus (ADN) and, in turn, to the retrosplenial cortex (RSP), postsubiculum (PoS), and medial entorhinal cortex (MEC) (Taube 2007). The limbic HD circuit receives critical inputs conveying self-motion information from vestibular structures (bottom left) and motor structures (bottom center) (Clark et al. 2009; Clark and Taube 2012). Other spatial processing structures are listed on the right portion of the diagram; arrows arising from the gray shaded boxes indicate projections from all structures contained within the shaded area. Blue shading denotes structures containing HD cells, and green shading indicates structures containing angular head velocity (AHV) cells (Jankowski et al. 2015; Mizumori and Williams 1993; Sharp et al. 2006; Taube 2007; Tsanov et al. 2011; Wilber et al. 2014; Winter and Taube 2014). Arrows with solid lines indicate connections observed in retrograde tracing experiments, with the weight of each line representing the relative density of the projection. Arrows with dashed lines indicate connections observed in anterograde tracing experiments. Solid lines terminating in an open arrowhead denote connections reported previously but not examined here. Note that numerous projections arising from limbic HD circuit structures and other spatial processing structures, many of which contain HD cells, converge within the DMS, and to a lesser extent, within the PrCM. These two extralimbic areas project to the limbic HD circuit indirectly via the entopeduncular nucleus (EPN); additionally, the PrCM sends sparse but direct projections to structures in the limbic HD circuit, vestibular input structures, and a motor input structure. +, sparse; ++, intermediate; +++, dense; aCl, anterior claustrum; AMN, anteromedial thalamic nucleus; AVN, anteroventral thalamic nucleus; dSub, dorsal subiculum; GP, globus pallidus; HPC, hippocampus; IPN, interpeduncular nucleus; LDN, laterodorsal thalamic nucleus; LHb, lateral habenula; MVe, medial vestibular nucleus; NPH, nucleus prepositus hypoglossi; POR, postrhinal cortex; PPC, posterior parietal cortex; PT, parataenial nucleus of the thalamus; SGN, supragenual nucleus; vSub, ventral subiculum.
Comparison with Previous Neuroanatomical Tracing Experiments
Many of our results replicate the general findings of prior neuroanatomical studies. Previous work examining striatal connectivity has described direct projections to the DS arising from thalamic nuclei involved in spatial processing (Cheatwood et al. 2003; Erro et al. 2002; Van Groen and Wyss 1995) as well as the RSP (McGeorge and Faull 1989; van Groen and Wyss 1992; Van Groen and Wyss 2003), entorhinal cortex (McGeorge and Faull 1989; Sørensen and Witter 1983; Swanson and Köhler 1986), PPC (McGeorge and Faull 1989; Reep et al. 2003), HPC and Sub (Groenewegen et al. 1987; Kelley and Domesick 1982), POR (Agster et al. 2016; McGeorge and Faull 1989), and PrCM (Reep et al. 1987, 2003; Reep and Corwin 1999). Efferent projections from the DS to the GP (Nagy et al. 1978a; Rajakumar et al. 1993) and EPN (Fink-Jensen and Mikkelsen 1989; Nagy et al. 1978a; Rajakumar et al. 1993; van der Kooy and Carter 1981) have also been reported. Furthermore, previous experiments investigating the connectivity of the PrCM and surrounding portions of the frontal cortex have described direct projections to these areas arising from the RSP (Hoover and Vertes 2007; Reep et al. 1990; Shibata et al. 2004), MEC (Hoover and Vertes 2007; Reep et al. 1990), and PPC (Kolb and Walkey 1987; Reep et al. 1990, 1994), as well as efferent projections from this cortical area to the RSP (Reep et al. 1987; Van Groen and Wyss 2003), PoS (Reep et al. 1987), EPN, cp, Pn, and py (Reep et al. 1987). Importantly, these previous neuroanatomical tracing studies 1) did not focus on the specific portions of the PrCM and DS that were targeted in our previous lesion and recording studies (Mehlman et al. 2019), 2) did not systematically examine patterns of connectivity with spatial processing circuitry (Table 1), and 3) used types of tracers that differ from those utilized in our studies.
Our findings also address a number of inconsistencies in the existing neuroanatomical literature. Previous retrograde tracing experiments examining projections from the DS to the EPN have reported that these projections arise from either the entire M/L axis of the DS (Fink-Jensen and Mikkelsen 1989, using wheatgerm agglutinin-horseradish peroxidase) or only the central and lateral portions of the DS (Rajakumar et al. 1993, using fluorogold). Previous anterograde tracing studies using [3H]leucine have reported direct projections from the DS to the EPN, but these studies do not indicate the M/L location of the tracer injections in the DS (Nagy et al. 1978a; van der Kooy and Carter 1981). We address these inconsistent and ambiguous findings by showing that anterograde tracer injections confined to either the DMS or DLS similarly label terminal fields within the EPN, replicating the results of Fink-Jensen and Mikkelsen (1989) using an anterograde tracing approach. Previous anterograde tracing studies exploring projections from the frontal cortex to the EPN also report inconsistent and somewhat ambiguous findings; van der Kooy and Carter (1981) observed that [3H]leucine injections in the “cortex dorsal to the head of the caudate putamen” did not produce fiber labeling within the EPN, whereas Reep et al. (1987) showed that [3H]leucine injections in medial portions of the frontal cortex labeled fibers in what appears to be the EPN. In the current study, we observed labeled terminal fields within the EPN following anterograde tracer injections into the PrCM (Fig. 7D), demonstrating that this cortical area indeed projects to the EPN. Finally, the presence of direct projections from the ADN and AVN to the DMS has been unclear. Anterograde tracing experiments have reported fiber labeling in the DMS following Phaseolus vulgaris leucoagglutinin injections into the ADN or AVN (Van Groen and Wyss 1995). However, in a later study examining thalamostriatal projections, Erro et al. (2002) injected the retrograde tracer fluorogold into the DMS and did not report any labeling in the ADN and AVN; either there was an absence of retrogradely labeled cells in these structures, or they simply were not examined. In the current study, we replicate the findings of Van Groen and Wyss (1995) and expand on them by using a retrograde tracing approach to demonstrate that the ADN and AVN, as well as other thalamic nuclei involved in spatial processing, project directly to the DMS but not the DLS (Table 2).
Importantly, the patterns of striatal afferent connectivity we observed in rats are consistent with studies in mice demonstrating that the medial portion of the DS uniquely receives numerous spatial inputs from structures including the anterior thalamic nuclei, entorhinal cortex, RSP, and PPC (Hintiryan et al. 2016; Hunnicutt et al. 2016; Pan et al. 2010; Wall et al. 2013).
In sum, our tracing studies replicate and expand on previous neuroanatomical work to provide a detailed, comprehensive, and novel circuit diagram (Fig. 11) describing how canonical spatial processing structures project to and receive input from the PrCM and DS, two brain areas that contain HD cells and AHV cells but are traditionally excluded from diagrams illustrating the brain’s spatial processing circuitry.
Propagation of the HD Signal to the DS
The DS receives direct input from a variety of structures that contain HD cells (Table 2; Fig. 11), and it is likely that one or more of these monosynaptic connections are responsible for conveying the HD signal to the DS. A large proportion of neurons (>50%) in the ADN are thought to be HD cells (Mehlman et al. 2019; Taube 1995). The ADN projects densely to the DS, and these projections are exclusively ipsilateral. This anatomical finding is consistent with the results of our lesion and recording studies. HD cell activity in the DS was 1) consistently eliminated in animals with complete ipsilateral ADN lesions, 2) consistently spared in animals with incomplete ipsilateral ADN lesions, and 3) unrelated to the size of the contralateral ADN lesion (Mehlman et al. 2019). Therefore, the ADN may convey the HD signal directly to the DS within, but not across, hemispheres. Interestingly, projections from the ADN to the DS arise exclusively from the medial and ventral portions of the ADN (Fig. 2, G and H). In contrast, projections from the ADN to the RSP and PoS arise primarily from the lateral and dorsal portions of the ADN, respectively (Van Groen and Wyss 1995).
Alternatively, DS HD cells could be driven by output from a structure other than the ADN. However, given that HD cell activity in the DS depends on the ADN (Mehlman et al. 2019), DS HD cells must be driven by output from a structure in which HD cell activity is also dependent on the ADN (i.e., the ADN could convey the HD signal to the DS indirectly via an intermediate structure, such as the RSP). In addition to the ADN, the DS receives direct input from other anterior thalamic nuclei involved in spatial processing. These nuclei include the AVN, AMN, and LDN, all of which contain HD cells; compared with the ADN, a smaller proportion of neurons in these nuclei are thought to be HD cells (Jankowski et al. 2015; Mizumori and Williams 1993; Tsanov et al. 2011). The DS also receives direct input from the PT, which contains neurons modulated by the animal’s location (Jankowski et al. 2015). It is not known if HD cell activity in these brain areas depends on the ADN; if so, then it is possible that the AVN, AMN, and/or LDN could transmit the HD signal to the DS. Given that projections from the LDN to the DS are extremely sparse, they are unlikely to solely support DS HD cell activity. In addition to their spatial processing functions, the anterior thalamic nuclei are involved in a variety of mnemonic processes (Aggleton et al. 2010; Mizumori and Williams 1993).
Furthermore, the DS receives direct input from multiple cortical areas containing HD cells: the RSP, MEC, and PPC (Chen et al. 1994; Cho and Sharp, 2001; Sargolini et al. 2006; Wilber et al. 2014). It is possible that one of these projections conveys the HD signal to the DS. Lesioning the ADN eliminates cortical HD cell activity in the PoS and MEC (Goodridge and Taube 1997; Winter et al. 2015), and the HD signals in the RSP and PPC likely depend on ADN output in a similar manner, although these functional relationships have not been demonstrated.
Finally, the DS receives direct input from the LMN, a structure that contains HD cells and is thought to drive HD cell activity in the ADN (Bassett et al. 2007; Blair et al. 1998; Stackman and Taube 1998). Although not demonstrated, it is likely that the LMN HD signal remains intact in animals with ADN lesions; therefore, if output from LMN HD cells directly drives DS HD cells, we would not expect ADN lesions to eliminate DS HD cell activity. However, because we observed an elimination of DS HD cell activity following complete ADN lesions (Mehlman et al. 2019), it is unlikely that the DS HD signal originates directly and exclusively from the LMN. In addition to HD cells, the LMN contains AHV cells similar to those we observed in the DS (Mehlman et al. 2019; Stackman and Taube 1998). It is possible that projections from the LMN transmit the AHV signal to the DS. In the LMN, the AHV signal is thought to originate from vestibular inputs, which include direct projections from the SGN to the LMN (Clark and Taube 2012). Interestingly, these projections terminate within the shell region of the LMN (Biazoli et al. 2006), the same area in which we observed labeled cells following retrograde tracer injections into the DMS or DLS (Fig. 2, E and F, and Fig. 4, C and D). Therefore, the SGN could drive AHV cell activity in the LMN shell, which in turn drives the small population of AHV cells we observed in the DS of both control and lesioned animals. AHV cells in the DS may also be driven or influenced by inputs from the PPC, which contains AHV cells (Wilber et al. 2014) and neurons tuned to other self-motion parameters (Whitlock et al. 2012). It is unclear how AHV cells in the LMN and PPC would be affected in animals with ADN lesions. If one or both of these structures indeed drive DS AHV cells, then we would expect the LMN and/or PPC AHV signals to remain intact following ADN lesions, given that ADN lesions did not disrupt AHV cell activity in the DS.
In addition to receiving input from HD cell-containing structures, the DS also receives direct projections from other brain areas involved in spatial processing, including the HPC, dSub, vSub, POR, and aCl (Table 2; Fig. 3). These structures process information about the animal’s location and environmental context (Bucci et al. 2000; Jankowski and O’Mara 2015; Kim and Fanselow 1992; O’Keefe 1976; Sharp and Green 1994). Projections from these structures may therefore support the activity of DS neurons shown to encode the animal’s location within a maze (Mizumori et al. 2000; see also Berke et al. 2009; Schmitzer-Torbert and Redish 2004) or the identity of the animal’s environmental context (Yamin et al. 2013).
DS Topography
Projections arising from the limbic HD circuit and other brain areas involved in spatial processing display a pronounced topography along the M/L axis of the DS; these projections all converge within the most medial portion of the DS located immediately adjacent to the lateral ventricle (i.e., the DMS), with little to no termination in more lateral and ventral areas (i.e., the DLS) (Table 2). Critically, these anatomical findings parallel the results of all DS HD cell recording studies; HD cells have been recorded exclusively within the DMS, and when recording electrodes target the DLS, no HD cell activity is observed (Mehlman et al. 2019; Mizumori et al. 2000; Ragozzino et al. 2001; Wiener 1993). Therefore, the distribution of HD cells within the DS and the organization of spatial inputs into the DS display a similar topography, suggesting that the anatomical projections we observed indeed drive DS HD cell activity. Although HD cells appear confined to the DMS, we recorded AHV cell activity in both the DMS and DLS (Mehlman et al. 2019). As described above, this AHV cell activity could be driven by projections from the LMN, and it is therefore noteworthy that these projections terminate within both the DMS and DLS (Table 2). It is important to note that we did not examine anatomical connections across the entire M/L axis of the DS in a continuous manner; rather, we described the connectivity of the most medial portion of the DMS and compared it with the connectivity of the medial and ventral portions of the DLS.
The DMS and DLS display differences not only in terms of HD cell activity and spatial inputs but also with regard to their contributions to behavior. The DMS is associated with guiding cognitive, goal-directed behaviors, whereas the DLS is thought to facilitate habitual, stimulus-response behaviors (Devan et al. 2011). Lesions of the DMS disrupt behaviors involving spatial processing and flexible navigation strategies (Devan and White 1999; Devan et al. 1999; Lee et al. 2014; Whishaw et al. 1987; Yin and Knowlton 2004), and the DMS may contribute to these behaviors in a manner dependent on the spatial information provided by local HD cell activity. In contrast, DLS lesions largely preserve spatially guided behaviors (i.e., place learning) while disrupting behaviors mediated by stimulus-response strategies (i.e., response learning) (Devan et al. 1999; Packard and McGaugh 1996; Yin and Knowlton 2004). Navigation associated with response learning is thought to be guided by egocentric spatial information (e.g., “turn left”) (Devan et al. 2011; Packard and McGaugh 1996); this may relate to why the DLS does not appear to contain HD cells, because these neurons encode allocentric, rather than egocentric, spatial information.
It is unclear why the DMS may require the animal’s HD to be represented locally by DS HD cells, since an identical directional signal may be available via afferent fibers arising from HD cells located in the limbic system. DS HD cells may serve to relay the HD signal to other brain areas that do not receive direct input from the limbic HD circuit. PrCM HD cells may serve a similar function. It is interesting that HD cells are found throughout the brain in numerous and diverse subcortical and cortical structures. The ubiquity of these neurons suggests that information about the animal’s orientation within the world is critical for a variety of neural, behavioral, and cognitive functions.
In addition to receiving numerous spatial inputs, the DMS also receives direct and relatively dense input from the SNC (Lerner et al. 2015), mediodorsal thalamic nucleus (Erro et al. 2002), centromedial thalamic nucleus (Erro et al. 2002), centrolateral thalamic nucleus (Cheatwood et al. 2003; Erro et al. 2002), ventrolateral thalamic nucleus (Cheatwood et al. 2003), cingulate cortex (Cheatwood et al. 2003; McGeorge and Faull 1989), auditory cortex (McGeorge and Faull 1989), and visual cortex (Cheatwood et al. 2003; Khibnik et al. 2014; McGeorge and Faull 1989). The extent to which these connections contribute to spatial processing vs. other functions performed by the DMS is unclear.
Propagation of the HD Signal to the PrCM
The PrCM receives direct input from a variety of spatial processing structures. These projections arise from a subset of the structures that project to the DS and are generally less dense compared with the projections targeting the DS (Table 2; Fig. 11). Interestingly, although spatial inputs into the PrCM are relatively less numerous and less dense, we observed a larger percentage of HD cells in the PrCM compared with the DS when recording single neurons in these brain areas (Mehlman et al. 2019).
The RSP, MEC, and PPC each contain HD cells and project directly to the PrCM (Chen et al. 1994; Cho and Sharp 2001; Sargolini et al. 2006; Wilber et al. 2014). It is likely that one or more of these monosynaptic connections are responsible for conveying the HD signal to the PrCM. Importantly, projections from the RSP to the PrCM arise exclusively from the anterior portion of the RSP; it is unclear if this anterior portion contains HD cells because most reports of RSP HD cell activity come from recordings performed in the posterior portion of the RSP (Alexander and Nitz 2015, 2017; Cho and Sharp 2001; Knight et al. 2013; see also Chen et al. 1994). The HD signal in the PrCM must be transmitted from a structure in which HD cell activity is dependent on the ADN, because ADN lesions eliminated PrCM HD cell activity (Mehlman et al. 2019). As described above, ADN lesions abolish the HD signal in the PoS and MEC (Goodridge and Taube 1997; Winter et al. 2015), and the same is likely true for other cortical areas (i.e., the RSP and PPC).
Additionally, the PrCM receives direct input from the LMN shell. As described above, it is unlikely that these projections are solely responsible for transmitting the HD signal, although they could be involved in driving the population of AHV cells we observed in the PrCM of both control and lesioned animals (Mehlman et al. 2019; Stackman and Taube 1998). PrCM AHV cells may also be driven or influenced by inputs from the PPC, as described above with regard to the DS (Whitlock et al. 2012; Wilber et al. 2014). Finally, direct projections from the aCl to the PrCM may convey information pertaining to the animal’s location and environmental features (Jankowski and O’Mara 2015).
Both the retrograde and anterograde tracing experiments revealed projections from the PrCM to the DS, suggesting that the HD signal could be serially transmitted first to the PrCM and in turn to the DMS. This scenario seems unlikely given that these corticostriatal projections terminate primarily within the DLS (Table 2; Fig. 9A). Moreover, in our lesion and recording experiments, we found that incomplete ADN lesions spared HD cell activity in the DS, but not in the PrCM, suggesting that the HD signal can persist in the DS despite being eliminated in the PrCM (Mehlman et al. 2019). Therefore, we conclude that a parallel propagation scenario is more likely, in which the HD signal is directly and independently transmitted to both the PrCM and DS, either from a common structure or from different structures.
What functions could the HD signal serve in the PrCM? The PrCM and adjacent portions of cortex are involved in generating head movements associated with orienting responses (Erlich et al. 2011; Kanki et al. 1983; Sinnamon and Charman 1988; Sinnamon and Galer 1984), and lesioning this cortical area unilaterally produces attentional deficits referred to as hemispatial neglect (Crowne et al. 1983; King and Corwin 1990; Vargo et al. 1988). It is unclear if the HD signal contributes to these functions, which may relate more to egocentric spatial processing compared with allocentric spatial processing. It has been proposed that the PrCM is particularly involved in orienting the head to locations defined by internal signals, rather than to locations defined by the presence of salient stimuli (Erlich et al. 2011; Sinnamon and Charman 1988). This idea suggests that PrCM HD cells may provide spatial information used to form internal representations of the animal’s orientation relative to behaviorally relevant locations within the environment. Other studies have implicated the PrCM in action selection and sequencing related to goal-directed behavior (Erlich et al. 2015; Hanks et al. 2015; Kargo et al. 2007; Ostlund et al. 2009; Yin 2009); it is unclear if these processes utilize the directional signal provided by PrCM HD cells. It is noteworthy that HD cells in the PrCM are uniquely situated to influence motor output, and therefore behavior, because these cells appear to intermix with corticospinal neurons. Our anterograde tracing studies demonstrate that the area in which we previously recorded PrCM HD cell activity (Mehlman et al. 2019) gives rise to descending projections that ultimately travel within the pyramidal tract (Fig. 9H).
In addition to receiving input from a number of spatial processing structures, the PrCM also receives direct and relatively dense input from the auditory and visual cortices (Reep et al. 1990); it is unclear if these connections contribute to the spatial processing functions performed by the PrCM.
Anatomical Pathways from the PrCM and DS to the Limbic HD Circuit
We previously found that ADN HD cell activity was largely unaffected following combined lesions of the PrCM and DMS (Mehlman et al. 2019). Lesioning these extralimbic areas may have failed to influence activity within the ADN simply because of the absence of anatomical pathways capable of conveying information from the PrCM and DMS to the limbic HD circuit. However, the anterograde tracing studies reported here demonstrate that such pathways indeed exist: the PrCM and DS project indirectly to the limbic HD circuit via the EPN. We observed direct projections from the PrCM and DS to the EPN. We also observed direct projections from the DS to the GP, which in turn projects directly to the EPN (Kincaid et al. 1991; Rajakumar et al. 1993; see also van der Kooy and Carter 1981). The EPN serves as a gateway to the limbic HD circuit; the EPN projects directly to the LHb (Nagy et al. 1978b; Rajakumar et al. 1993; van der Kooy and Carter 1981), which in turn projects directly to the DTN (Liu et al. 1984) and indirectly to the DTN via the IPN (Contestabile and Flumerfelt 1981; Groenewegen et al. 1986; Liu et al. 1984). Therefore, signals from the PrCM and DS could be conveyed to the limbic HD circuit via two routes: 1) EPN → LHb → DTN and 2) EPN → LHb → IPN → DTN. These pathways are illustrated in Fig. 11. Interestingly, a recent neuroanatomical study in mice reported a direct projection from the EPN to the ADN (Wallace et al. 2017), revealing an additional route that could potentially convey information from the PrCM and DS to the limbic HD circuit; it is unclear if such connectivity is also present in rats.
Although the results of our lesion and recording studies suggest that the limbic HD circuit does not receive critical information arising from the PrCM or DMS (Mehlman et al. 2019), projections from the IPN to the DTN appear to convey necessary self-motion signals arising from elsewhere, likely the LHb. Indeed, lesions of the IPN disrupt HD cell activity in the ADN (Clark et al. 2009), and lesions of either the LHb or IPN produce navigation deficits (Clark and Taube 2009; Lecourtier et al. 2004). The LHb contains neurons modulated by the animal’s running speed and AHV (Sharp et al. 2006), and the IPN is thought to convey these self-motion signals to the DTN (Clark et al. 2009). Although AHV cells in the LHb display properties similar to those of the AHV cells we recorded in the PrCM and DMS (Mehlman et al. 2019; Sharp et al. 2006), LHb AHV cells may not depend on output from the PrCM and DMS. If these LHb neurons were influenced by the PrCM and DMS, then we would expect combined PrCM and DMS lesions to disrupt LHb AHV cell activity and, in turn, ADN HD cell activity (due to the transmission of self-motion signals from the LHb to the DTN). However, we did not observe this pattern of results following combined PrCM and DMS lesions (Mehlman et al. 2019), although it is possible that we failed to observe a disruption of ADN HD cell activity due to compensatory mechanisms. Importantly, our lesions were confined to the DMS, targeting the portion of the DS in which we recorded HD cell activity. It is possible that lesioning the DLS would affect the limbic HD signal more significantly than DMS lesions. The DLS may be more likely to provide self-motion information given its general engagement with sensorimotor functions (Devan et al. 2011).
Additionally, we observed sparse but direct projections from the PrCM to structures in the limbic HD circuit (the RSP and PoS), vestibular input structures (the SGN, NPH, and MVe), and a motor input structure (the IPN) (Fig. 10). The functions that these projections serve are unclear, but they are unlikely to convey information critical for the generation and maintenance of the limbic HD signal, given that combined PrCM and DMS lesions did not disrupt ADN HD cell activity (Mehlman et al. 2019). Importantly, the PrCM projects indirectly to the ADN via its direct projections to the RSP and PoS, which both project directly to the ADN (van Groen and Wyss 1990, 1992; Van Groen and Wyss 2003).
Conclusion
The majority of research in the field of navigation and spatial cognition has focused on a relatively small number of structures in the limbic system. Importantly, limbic spatial processing circuitry does not exist in isolation, but rather interacts with other networks in the brain. We first performed a series of lesion and recording experiments (Mehlman et al. 2019) and then a series of companion neuroanatomical tracing studies. Collectively, our results provide a detailed description of how spatial processing circuitry functionally and anatomically extends into two extralimbic areas, the PrCM and DS. Each of these structures contains HD cells, which appear to be driven by direct projections arising from cortical and subcortical structures in the limbic HD circuit. These projections preferentially target and converge within portions of the PrCM and DS that contain HD cells and AHV cells. Given that the PrCM and DS are involved in controlling motor output and behavior, it is possible that the HD signal is conveyed to these brain areas to guide navigation. Our findings provide valuable insights into the spatial processing circuitry beyond the limbic system and inform future research examining the PrCM and DS in the context of navigation and spatial cognition.
GRANTS
This work was supported by National Institute of Neurological Disorders and Stroke Grants NS053907 (to J. S. Taube) and NS096888 (to M. L. Mehlman).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.L.M. and J.S.T. conceived and designed research; M.L.M. and S.S.W. performed experiments; M.L.M. analyzed data; M.L.M. and J.S.T. interpreted results of experiments; M.L.M. prepared figures; M.L.M. drafted manuscript; M.L.M. and J.S.T. edited and revised manuscript; M.L.M. and J.S.T. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Jennifer Marcroft for assisting with data collection and Kyle Smith for providing the anterograde tracer and access to imaging equipment.
Present address of M. L. Mehlman: Department of Neuroscience, University of Rochester School of Medicine and Dentistry, Rochester, NY.
REFERENCES
- Aggleton JP, O’Mara SM, Vann SD, Wright NF, Tsanov M, Erichsen JT. Hippocampal-anterior thalamic pathways for memory: uncovering a network of direct and indirect actions. Eur J Neurosci 31: 2292–2307, 2010. doi: 10.1111/j.1460-9568.2010.07251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agster KL, Tomás Pereira I, Saddoris MP, Burwell RD. Subcortical connections of the perirhinal, postrhinal, and entorhinal cortices of the rat. II. Efferents. Hippocampus 26: 1213–1230, 2016. doi: 10.1002/hipo.22600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander AS, Nitz DA. Retrosplenial cortex maps the conjunction of internal and external spaces. Nat Neurosci 18: 1143–1151, 2015. doi: 10.1038/nn.4058. [DOI] [PubMed] [Google Scholar]
- Alexander AS, Nitz DA. Spatially periodic activation patterns of retrosplenial cortex encode route sub-spaces and distance traveled. Curr Biol 27: 1551–1560.e4, 2017. doi: 10.1016/j.cub.2017.04.036. [DOI] [PubMed] [Google Scholar]
- Aschauer DF, Kreuz S, Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS One 8: e76310, 2013. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett JP, Tullman ML, Taube JS. Lesions of the tegmentomammillary circuit in the head direction system disrupt the head direction signal in the anterior thalamus. J Neurosci 27: 7564–7577, 2007. doi: 10.1523/JNEUROSCI.0268-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke JD, Breck JT, Eichenbaum H. Striatal versus hippocampal representations during win-stay maze performance. J Neurophysiol 101: 1575–1587, 2009. doi: 10.1152/jn.91106.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazoli CE Jr, Goto M, Campos AM, Canteras NS. The supragenual nucleus: a putative relay station for ascending vestibular signs to head direction cells. Brain Res 1094: 138–148, 2006. doi: 10.1016/j.brainres.2006.03.101. [DOI] [PubMed] [Google Scholar]
- Blair HT, Cho J, Sharp PE. Role of the lateral mammillary nucleus in the rat head direction circuit: a combined single unit recording and lesion study. Neuron 21: 1387–1397, 1998. doi: 10.1016/S0896-6273(00)80657-1. [DOI] [PubMed] [Google Scholar]
- Boccara CN, Sargolini F, Thoresen VH, Solstad T, Witter MP, Moser EI, Moser MB. Grid cells in pre- and parasubiculum. Nat Neurosci 13: 987–994, 2010. doi: 10.1038/nn.2602. [DOI] [PubMed] [Google Scholar]
- Brecht M, Krauss A, Muhammad S, Sinai-Esfahani L, Bellanca S, Margrie TW. Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J Comp Neurol 479: 360–373, 2004. doi: 10.1002/cne.20306. [DOI] [PubMed] [Google Scholar]
- Bucci DJ, Phillips RG, Burwell RD. Contributions of postrhinal and perirhinal cortex to contextual information processing. Behav Neurosci 114: 882–894, 2000. doi: 10.1037/0735-7044.114.5.882. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG. Cortical afferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. J Comp Neurol 398: 179–205, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- Butler WN, Smith KS, van der Meer MAA, Taube JS. The head direction signal plays a functional role as a neural compass during navigation. Curr Biol 27: 1259–1267, 2017. doi: 10.1016/j.cub.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler WN, Taube JS. The nucleus prepositus hypoglossi contributes to head direction cell stability in rats. J Neurosci 35: 2547–2558, 2015. doi: 10.1523/JNEUROSCI.3254-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin NL, Du B, de Lacalle S, Saper CB. Recombinant adeno-associated virus vector: use for transgene expression and anterograde tract tracing in the CNS. Brain Res 793: 169–175, 1998. doi: 10.1016/S0006-8993(98)00169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatwood JL, Reep RL, Corwin JV. The associative striatum: cortical and thalamic projections to the dorsocentral striatum in rats. Brain Res 968: 1–14, 2003. doi: 10.1016/S0006-8993(02)04212-9. [DOI] [PubMed] [Google Scholar]
- Chen LL, Lin LH, Green EJ, Barnes CA, McNaughton BL. Head-direction cells in the rat posterior cortex. I. Anatomical distribution and behavioral modulation. Exp Brain Res 101: 8–23, 1994. doi: 10.1007/BF00243212. [DOI] [PubMed] [Google Scholar]
- Cho J, Sharp PE. Head direction, place, and movement correlates for cells in the rat retrosplenial cortex. Behav Neurosci 115: 3–25, 2001. doi: 10.1037/0735-7044.115.1.3. [DOI] [PubMed] [Google Scholar]
- Clark BJ, Brown JE, Taube JS. Head direction cell activity in the anterodorsal thalamus requires intact supragenual nuclei. J Neurophysiol 108: 2767–2784, 2012. doi: 10.1152/jn.00295.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BJ, Sarma A, Taube JS. Head direction cell instability in the anterior dorsal thalamus after lesions of the interpeduncular nucleus. J Neurosci 29: 493–507, 2009. doi: 10.1523/JNEUROSCI.2811-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BJ, Taube JS. Deficits in landmark navigation and path integration after lesions of the interpeduncular nucleus. Behav Neurosci 123: 490–503, 2009. doi: 10.1037/a0015477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BJ, Taube JS. Vestibular and attractor network basis of the head direction cell signal in subcortical circuits. Front Neural Circuits 6: 7, 2012. doi: 10.3389/fncir.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condé F, Maire-Lepoivre E, Audinat E, Crépel F. Afferent connections of the medial frontal cortex of the rat. II. Cortical and subcortical afferents. J Comp Neurol 352: 567–593, 1995. doi: 10.1002/cne.903520407. [DOI] [PubMed] [Google Scholar]
- Conte WL, Kamishina H, Reep RL. Multiple neuroanatomical tract-tracing using fluorescent Alexa Fluor conjugates of cholera toxin subunit B in rats. Nat Protoc 4: 1157–1166, 2009a. doi: 10.1038/nprot.2009.93. [DOI] [PubMed] [Google Scholar]
- Conte WL, Kamishina H, Reep RL. The efficacy of the fluorescent conjugates of cholera toxin subunit B for multiple retrograde tract tracing in the central nervous system. Brain Struct Funct 213: 367–373, 2009b. doi: 10.1007/s00429-009-0212-x. [DOI] [PubMed] [Google Scholar]
- Contestabile A, Flumerfelt BA. Afferent connections of the interpeduncular nucleus and the topographic organization of the habenulo-interpeduncular pathway: an HRP study in the rat. J Comp Neurol 196: 253–270, 1981. doi: 10.1002/cne.901960206. [DOI] [PubMed] [Google Scholar]
- Crowne DP, Richardson CM, Ward G. Brief deprivation of vision after unilateral lesions of the frontal eye field prevents contralateral inattention. Science 220: 527–530, 1983. doi: 10.1126/science.6836298. [DOI] [PubMed] [Google Scholar]
- Devan BD, Hong NS, McDonald RJ. Parallel associative processing in the dorsal striatum: segregation of stimulus-response and cognitive control subregions. Neurobiol Learn Mem 96: 95–120, 2011. doi: 10.1016/j.nlm.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Devan BD, McDonald RJ, White NM. Effects of medial and lateral caudate-putamen lesions on place- and cue-guided behaviors in the water maze: relation to thigmotaxis. Behav Brain Res 100: 5–14, 1999. doi: 10.1016/S0166-4328(98)00107-7. [DOI] [PubMed] [Google Scholar]
- Devan BD, White NM. Parallel information processing in the dorsal striatum: relation to hippocampal function. J Neurosci 19: 2789–2798, 1999. doi: 10.1523/JNEUROSCI.19-07-02789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich JC, Bialek M, Brody CD. A cortical substrate for memory-guided orienting in the rat. Neuron 72: 330–343, 2011. doi: 10.1016/j.neuron.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich JC, Brunton BW, Duan CA, Hanks TD, Brody CD. Distinct effects of prefrontal and parietal cortex inactivations on an accumulation of evidence task in the rat. eLife 4: e05457, 2015. doi: 10.7554/eLife.05457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erro EM, Lanciego JL, Gimenez-Amaya JM. Re-examination of the thalamostriatal projections in the rat with retrograde tracers. Neurosci Res 42: 45–55, 2002. doi: 10.1016/S0168-0102(01)00302-9. [DOI] [PubMed] [Google Scholar]
- Fink-Jensen A, Mikkelsen JD. The striato-entopeduncular pathway in the rat. A retrograde transport study with wheatgerm-agglutinin-horseradish peroxidase. Brain Res 476: 194–198, 1989. doi: 10.1016/0006-8993(89)91558-8. [DOI] [PubMed] [Google Scholar]
- Goodridge JP, Taube JS. Interaction between the postsubiculum and anterior thalamus in the generation of head direction cell activity. J Neurosci 17: 9315–9330, 1997. doi: 10.1523/JNEUROSCI.17-23-09315.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewegen HJ, Ahlenius S, Haber SN, Kowall NW, Nauta WJ. Cytoarchitecture, fiber connections, and some histochemical aspects of the interpeduncular nucleus in the rat. J Comp Neurol 249: 65–102, 1986. doi: 10.1002/cne.902490107. [DOI] [PubMed] [Google Scholar]
- Groenewegen HJ, Vermeulen-Van der Zee E, te Kortschot A, Witter MP. Organization of the projections from the subiculum to the ventral striatum in the rat. A study using anterograde transport of Phaseolus vulgaris leucoagglutinin. Neuroscience 23: 103–120, 1987. doi: 10.1016/0306-4522(87)90275-2. [DOI] [PubMed] [Google Scholar]
- Hanks TD, Kopec CD, Brunton BW, Duan CA, Erlich JC, Brody CD. Distinct relationships of parietal and prefrontal cortices to evidence accumulation. Nature 520: 220–223, 2015. doi: 10.1038/nature14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintiryan H, Foster NN, Bowman I, Bay M, Song MY, Gou L, Yamashita S, Bienkowski MS, Zingg B, Zhu M, Yang XW, Shih JC, Toga AW, Dong HW. The mouse cortico-striatal projectome. Nat Neurosci 19: 1100–1114, 2016. doi: 10.1038/nn.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover WB, Vertes RP. Anatomical analysis of afferent projections to the medial prefrontal cortex in the rat. Brain Struct Funct 212: 149–179, 2007. doi: 10.1007/s00429-007-0150-4. [DOI] [PubMed] [Google Scholar]
- Huang LW, Simonnet J, Nassar M, Richevaux L, Lofredi R, Fricker D. Laminar localization and projection-specific properties of presubiculuar neurons targeting the lateral mammillary nucleus, thalamus, or medial entorhinal cortex. eNeuro 4: ENEURO.0370-16.2017, 2017. doi: 10.1523/ENEURO.0370-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunnicutt BJ, Jongbloets BC, Birdsong WT, Gertz KJ, Zhong H, Mao T. A comprehensive excitatory input map of the striatum reveals novel functional organization. eLife 5: e19103, 2016. doi: 10.7554/eLife.19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MM, Islam MN, Wright NF, Vann SD, Erichsen JT, Aggleton JP, O’Mara SM. Nucleus reuniens of the thalamus contains head direction cells. eLife 3: e03075, 2014. doi: 10.7554/eLife.03075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MM, O’Mara SM. Dynamics of place, boundary and object encoding in rat anterior claustrum. Front Behav Neurosci 9: 250, 2015. doi: 10.3389/fnbeh.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski MM, Passecker J, Islam MN, Vann S, Erichsen JT, Aggleton JP, O’Mara SM. Evidence for spatially-responsive neurons in the rostral thalamus. Front Behav Neurosci 9: 256, 2015. doi: 10.3389/fnbeh.2015.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki JP, Martin TL, Sinnamon HM. Activity of neurons in the anteromedial cortex during rewarding brain stimulation, saccharin consumption and orienting behavior. Behav Brain Res 8: 69–84, 1983. doi: 10.1016/0166-4328(83)90172-9. [DOI] [PubMed] [Google Scholar]
- Kargo WJ, Szatmary B, Nitz DA. Adaptation of prefrontal cortical firing patterns and their fidelity to changes in action-reward contingencies. J Neurosci 27: 3548–3559, 2007. doi: 10.1523/JNEUROSCI.3604-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley AE, Domesick VB. The distribution of the projection from the hippocampal formation to the nucleus accumbens in the rat: an anterograde- and retrograde-horseradish peroxidase study. Neuroscience 7: 2321–2335, 1982. doi: 10.1016/0306-4522(82)90198-1. [DOI] [PubMed] [Google Scholar]
- Khibnik LA, Tritsch NX, Sabatini BL. A direct projection from mouse primary visual cortex to dorsomedial striatum. PLoS One 9: e104501, 2014. doi: 10.1371/journal.pone.0104501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science 256: 675–677, 1992. doi: 10.1126/science.1585183. [DOI] [PubMed] [Google Scholar]
- Kincaid AE, Penney JB Jr, Young AB, Newman SW. Evidence for a projection from the globus pallidus to the entopeduncular nucleus in the rat. Neurosci Lett 128: 121–125, 1991. doi: 10.1016/0304-3940(91)90774-N. [DOI] [PubMed] [Google Scholar]
- King V, Corwin JV. Neglect following unilateral ablation of the caudal but not the rostral portion of medial agranular cortex of the rat and the therapeutic effect of apomorphine. Behav Brain Res 37: 169–184, 1990. doi: 10.1016/0166-4328(90)90092-S. [DOI] [PubMed] [Google Scholar]
- Knight R, Piette CE, Page H, Walters D, Marozzi E, Nardini M, Stringer S, Jeffery KJ. Weighted cue integration in the rodent head direction system. Philos Trans R Soc Lond B Biol Sci 369: 20120512, 2013. doi: 10.1098/rstb.2012.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb B, Walkey J. Behavioural and anatomical studies of the posterior parietal cortex in the rat. Behav Brain Res 23: 127–145, 1987. doi: 10.1016/0166-4328(87)90050-7. [DOI] [PubMed] [Google Scholar]
- Lecourtier L, Neijt HC, Kelly PH. Habenula lesions cause impaired cognitive performance in rats: implications for schizophrenia. Eur J Neurosci 19: 2551–2560, 2004. doi: 10.1111/j.0953-816X.2004.03356.x. [DOI] [PubMed] [Google Scholar]
- Lee AS, André JM, Pittenger C. Lesions of the dorsomedial striatum delay spatial learning and render cue-based navigation inflexible in a water maze task in mice. Front Behav Neurosci 8: 42, 2014. doi: 10.3389/fnbeh.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, Deisseroth K. Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162: 635–647, 2015. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chang L, Wickern G. The dorsal tegmental nucleus: an axoplasmic transport study. Brain Res 310: 123–132, 1984. doi: 10.1016/0006-8993(84)90015-5. [DOI] [PubMed] [Google Scholar]
- McGeorge AJ, Faull RL. The organization of the projection from the cerebral cortex to the striatum in the rat. Neuroscience 29: 503–537, 1989. doi: 10.1016/0306-4522(89)90128-0. [DOI] [PubMed] [Google Scholar]
- Mehlman ML, Winter SS, Valerio S, Taube JS. Functional and anatomical relationships between the medial precentral cortex, dorsal striatum, and head direction cell circuitry. I: Recording studies. J Neurophysiol 121: 350–370, 2019. doi: 10.1152/jn.00143.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumori SJ, Puryear CB, Gill KM, Guazzelli A. Head direction codes in hippocampal afferent and efferent systems: what functions do they serve? In: Head Direction Cells and the Neural Mechanisms of Spatial Orientation, edited by Wiener SI, Taube JS. Cambridge, MA: The MIT Press, 2005. [Google Scholar]
- Mizumori SJ, Ragozzino KE, Cooper BG. Location and head direction representation in the dorsal striatum of rats. Psychobiology (Austin Tex) 28: 441–462, 2000. [Google Scholar]
- Mizumori SJ, Williams JD. Directionally selective mnemonic properties of neurons in the lateral dorsal nucleus of the thalamus of rats. J Neurosci 13: 4015–4028, 1993. doi: 10.1523/JNEUROSCI.13-09-04015.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JI, Carter DA, Fibiger HC. Anterior striatal projections to the globus pallidus, entopeduncular nucleus and substantia nigra in the rat: the GABA connection. Brain Res 158: 15–29, 1978a. doi: 10.1016/0006-8993(78)90003-3. [DOI] [PubMed] [Google Scholar]
- Nagy JI, Carter DA, Lehmann J, Fibiger HC. Evidence for a GABA-containing projection from the entopeduncular nucleus to the lateral habenula in the rat. Brain Res 145: 360–364, 1978b. doi: 10.1016/0006-8993(78)90869-7. [DOI] [PubMed] [Google Scholar]
- Nassi JJ, Cepko CL, Born RT, Beier KT. Neuroanatomy goes viral! Front Neuroanat 9: 80, 2015. doi: 10.3389/fnana.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe J. Place units in the hippocampus of the freely moving rat. Exp Neurol 51: 78–109, 1976. doi: 10.1016/0014-4886(76)90055-8. [DOI] [PubMed] [Google Scholar]
- Ostlund SB, Winterbauer NE, Balleine BW. Evidence of action sequence chunking in goal-directed instrumental conditioning and its dependence on the dorsomedial prefrontal cortex. J Neurosci 29: 8280–8287, 2009. doi: 10.1523/JNEUROSCI.1176-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard MG, McGaugh JL. Inactivation of hippocampus or caudate nucleus with lidocaine differentially affects expression of place and response learning. Neurobiol Learn Mem 65: 65–72, 1996. doi: 10.1006/nlme.1996.0007. [DOI] [PubMed] [Google Scholar]
- Pan WX, Mao T, Dudman JT. Inputs to the dorsal striatum of the mouse reflect the parallel circuit architecture of the forebrain. Front Neuroanat 4: 147, 2010. doi: 10.3389/fnana.2010.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates: Compact (3rd ed.). New York: Academic, 1997. [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates: Compact (6th ed.). New York: Academic, 2009. [Google Scholar]
- Ragozzino KE, Leutgeb S, Mizumori SJY. Dorsal striatal head direction and hippocampal place representations during spatial navigation. Exp Brain Res 139: 372–376, 2001. doi: 10.1007/s002210100795. [DOI] [PubMed] [Google Scholar]
- Rajakumar N, Elisevich K, Flumerfelt BA. Compartmental origin of the striato-entopeduncular projection in the rat. J Comp Neurol 331: 286–296, 1993. doi: 10.1002/cne.903310210. [DOI] [PubMed] [Google Scholar]
- Reep RL, Chandler HC, King V, Corwin JV. Rat posterior parietal cortex: topography of corticocortical and thalamic connections. Exp Brain Res 100: 67–84, 1994. doi: 10.1007/BF00227280. [DOI] [PubMed] [Google Scholar]
- Reep RL, Cheatwood JL, Corwin JV. The associative striatum: organization of cortical projections to the dorsocentral striatum in rats. J Comp Neurol 467: 271–292, 2003. doi: 10.1002/cne.10868. [DOI] [PubMed] [Google Scholar]
- Reep RL, Corwin JV. Topographic organization of the striatal and thalamic connections of rat medial agranular cortex. Brain Res 841: 43–52, 1999. doi: 10.1016/S0006-8993(99)01779-5. [DOI] [PubMed] [Google Scholar]
- Reep RL, Corwin JV, Hashimoto A, Watson RT. Efferent connections of the rostral portion of medial agranular cortex in rats. Brain Res Bull 19: 203–221, 1987. doi: 10.1016/0361-9230(87)90086-4. [DOI] [PubMed] [Google Scholar]
- Reep RL, Goodwin GS, Corwin JV. Topographic organization in the corticocortical connections of medial agranular cortex in rats. J Comp Neurol 294: 262–280, 1990. doi: 10.1002/cne.902940210. [DOI] [PubMed] [Google Scholar]
- Sargolini F, Fyhn M, Hafting T, McNaughton BL, Witter MP, Moser MB, Moser EI. Conjunctive representation of position, direction, and velocity in entorhinal cortex. Science 312: 758–762, 2006. doi: 10.1126/science.1125572. [DOI] [PubMed] [Google Scholar]
- Schmitzer-Torbert N, Redish AD. Neuronal activity in the rodent dorsal striatum in sequential navigation: separation of spatial and reward responses on the multiple T task. J Neurophysiol 91: 2259–2272, 2004. doi: 10.1152/jn.00687.2003. [DOI] [PubMed] [Google Scholar]
- Sesack SR, Deutch AY, Roth RH, Bunney BS. Topographical organization of the efferent projections of the medial prefrontal cortex in the rat: an anterograde tract-tracing study with Phaseolus vulgaris leucoagglutinin. J Comp Neurol 290: 213–242, 1989. doi: 10.1002/cne.902900205. [DOI] [PubMed] [Google Scholar]
- Sharp PE, Green C. Spatial correlates of firing patterns of single cells in the subiculum of the freely moving rat. J Neurosci 14: 2339–2356, 1994. doi: 10.1523/JNEUROSCI.14-04-02339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PE, Turner-Williams S, Tuttle S. Movement-related correlates of single cell activity in the interpeduncular nucleus and habenula of the rat during a pellet-chasing task. Behav Brain Res 166: 55–70, 2006. doi: 10.1016/j.bbr.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Shibata H, Kondo S, Naito J. Organization of retrosplenial cortical projections to the anterior cingulate, motor, and prefrontal cortices in the rat. Neurosci Res 49: 1–11, 2004. doi: 10.1016/j.neures.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Sinnamon HM, Charman CS. Unilateral and bilateral lesions of the anteromedial cortex increase perseverative head movements of the rat. Behav Brain Res 27: 145–160, 1988. doi: 10.1016/0166-4328(88)90040-X. [DOI] [PubMed] [Google Scholar]
- Sinnamon HM, Galer BS. Head movements elicited by electrical stimulation of the anteromedial cortex of the rat. Physiol Behav 33: 185–190, 1984. doi: 10.1016/0031-9384(84)90098-2. [DOI] [PubMed] [Google Scholar]
- Sørensen KE, Witter MP. Entorhinal efferents reach the caudato-putamen. Neurosci Lett 35: 259–264, 1983. doi: 10.1016/0304-3940(83)90327-0. [DOI] [PubMed] [Google Scholar]
- Stackman RW, Taube JS. Firing properties of rat lateral mammillary single units: head direction, head pitch, and angular head velocity. J Neurosci 18: 9020–9037, 1998. doi: 10.1523/JNEUROSCI.18-21-09020.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW. Brain Maps: Structure of the Rat Brain (2nd ed). New York: Elsevier, 1998. [Google Scholar]
- Swanson LW, Köhler C. Anatomical evidence for direct projections from the entorhinal area to the entire cortical mantle in the rat. J Neurosci 6: 3010–3023, 1986. doi: 10.1523/JNEUROSCI.06-10-03010.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JS. Head direction cells recorded in the anterior thalamic nuclei of freely moving rats. J Neurosci 15: 70–86, 1995. doi: 10.1523/JNEUROSCI.15-01-00070.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taube JS. The head direction signal: origins and sensory-motor integration. Annu Rev Neurosci 30: 181–207, 2007. doi: 10.1146/annurev.neuro.29.051605.112854. [DOI] [PubMed] [Google Scholar]
- Taube JS, Muller RU, Ranck JB Jr. Head-direction cells recorded from the postsubiculum in freely moving rats. I. Description and quantitative analysis. J Neurosci 10: 420–435, 1990. doi: 10.1523/JNEUROSCI.10-02-00420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov M, Chah E, Vann SD, Reilly RB, Erichsen JT, Aggleton JP, O’Mara SM. Theta-modulated head direction cells in the rat anterior thalamus. J Neurosci 31: 9489–9502, 2011. doi: 10.1523/JNEUROSCI.0353-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerio S, Taube JS. Path integration: how the head direction signal maintains and corrects spatial orientation. Nat Neurosci 15: 1445–1453, 2012. doi: 10.1038/nn.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooy D, Carter DA. The organization of the efferent projections and striatal afferents of the entopeduncular nucleus and adjacent areas in the rat. Brain Res 211: 15–36, 1981. doi: 10.1016/0006-8993(81)90064-0. [DOI] [PubMed] [Google Scholar]
- van der Meer MAA, Richmond Z, Braga RM, Wood ER, Dudchenko PA. Evidence for the use of an internal sense of direction in homing. Behav Neurosci 124: 164–169, 2010. doi: 10.1037/a0018446. [DOI] [PubMed] [Google Scholar]
- van Groen T, Wyss JM. The postsubicular cortex in the rat: characterization of the fourth region of the subicular cortex and its connections. Brain Res 529: 165–177, 1990. doi: 10.1016/0006-8993(90)90824-U. [DOI] [PubMed] [Google Scholar]
- van Groen T, Wyss JM. Connections of the retrosplenial dysgranular cortex in the rat. J Comp Neurol 315: 200–216, 1992. doi: 10.1002/cne.903150207. [DOI] [PubMed] [Google Scholar]
- Van Groen T, Wyss JM. Projections from the anterodorsal and anteroventral nucleus of the thalamus to the limbic cortex in the rat. J Comp Neurol 358: 584–604, 1995. doi: 10.1002/cne.903580411. [DOI] [PubMed] [Google Scholar]
- Van Groen T, Wyss JM. Connections of the retrosplenial granular b cortex in the rat. J Comp Neurol 463: 249–263, 2003. doi: 10.1002/cne.10757. [DOI] [PubMed] [Google Scholar]
- Vargo JM, Corwin JV, King V, Reep RL. Hemispheric asymmetry in neglect produced by unilateral lesions of dorsomedial prefrontal cortex in rats. Exp Neurol 102: 199–209, 1988. doi: 10.1016/0014-4886(88)90094-5. [DOI] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Callaway EM, Kreitzer AC. Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron 79: 347–360, 2013. doi: 10.1016/j.neuron.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Saunders A, Huang KW, Philson AC, Goldman M, Macosko EZ, McCarroll SA, Sabatini BL. Genetically distinct parallel pathways in the entopeduncular nucleus for limbic and sensorimotor output of the basal ganglia. Neuron 94: 138–152.e5, 2017. doi: 10.1016/j.neuron.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whishaw IQ, Mittleman G, Bunch ST, Dunnett SB. Impairments in the acquisition, retention and selection of spatial navigation strategies after medial caudate-putamen lesions in rats. Behav Brain Res 24: 125–138, 1987. doi: 10.1016/0166-4328(87)90250-6. [DOI] [PubMed] [Google Scholar]
- Whitlock JR, Pfuhl G, Dagslott N, Moser MB, Moser EI. Functional split between parietal and entorhinal cortices in the rat. Neuron 73: 789–802, 2012. doi: 10.1016/j.neuron.2011.12.028. [DOI] [PubMed] [Google Scholar]
- Wiener SI. Spatial and behavioral correlates of striatal neurons in rats performing a self-initiated navigation task. J Neurosci 13: 3802–3817, 1993. doi: 10.1523/JNEUROSCI.13-09-03802.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber AA, Clark BJ, Forster TC, Tatsuno M, McNaughton BL. Interaction of egocentric and world-centered reference frames in the rat posterior parietal cortex. J Neurosci 34: 5431–5446, 2014. doi: 10.1523/JNEUROSCI.0511-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SS, Clark BJ, Taube JS. Spatial navigation. Disruption of the head direction cell network impairs the parahippocampal grid cell signal. Science 347: 870–874, 2015. doi: 10.1126/science.1259591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SS, Taube JS. Head direction cells: from generation to integration. In: Space, Time and Memory in the Hippocampal Formation, edited by Derdikman D, Knierim JJ. New York: Springer, 2014. doi: 10.1007/978-3-7091-1292-2_4. [DOI] [Google Scholar]
- Yamin HG, Stern EA, Cohen D. Parallel processing of environmental recognition and locomotion in the mouse striatum. J Neurosci 33: 473–484, 2013. doi: 10.1523/JNEUROSCI.4474-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HH. The role of the murine motor cortex in action duration and order. Front Integr Neurosci 3: 23, 2009. doi: 10.3389/neuro.07.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin HH, Knowlton BJ. Contributions of striatal subregions to place and response learning. Learn Mem 11: 459–463, 2004. doi: 10.1101/lm.81004. [DOI] [PMC free article] [PubMed] [Google Scholar]