Abstract
The intestinal epithelium constitutes a crucial defense to the potentially life-threatening effects of gut microbiota. However, due to a complex underlying vasculature, hypoperfusion and resultant tissue ischemia pose a particular risk to function and integrity of the epithelium.
The Small Ubiquitin-like MOdifier (SUMO) conjugation pathway critically regulates adaptive responses to metabolic stress, and is of particular significance in the gut, as inducible knockout of the SUMO-conjugating enzyme Ubc9 results in rapid intestinal epithelial disintegration. Here, we analyzed the pattern of individual SUMO isoforms in intestinal epithelium and investigated their roles in intestinal ischemia/reperfusion (I/R) damage. Immunostaining revealed that epithelial SUMO2/3 expression was almost exclusively limited to crypt epithelial nuclei in unchallenged mice. However, intestinal I/R or overexpression of Ubc9 caused marked enhancement of epithelial SUMO2/3 staining along the crypt-villus axis. Unexpectedly, a similar pattern was found in SUMO1 knockout mice. Ubc9 transgenic mice, but also SUMO1 knockout mice were protected from I/R injury as evidenced by better preserved barrier function and blunted inflammatory responses. PCR array analysis of microdissected villus-tip epithelia revealed a specific epithelial contribution to reduced inflammatory responses in Ubc9 transgenic mice as key chemotactic signaling molecules such as IL17A were significantly downregulated. Together, our data indicate a critical role particularly of the SUMO2/3 isoforms in modulating responses to I/R and provide the first evidence that SUMO1 deletion activates a compensatory process that protects from ischemic damage.
Keywords: inflammation, stress responses, post-translational modifications, SUMOylation
Introduction
The integrity of the intestinal epithelium is crucial for preserving the body’s homeostasis. Maintenance of this barrier generates substantial metabolic demands that arise in part from a need for rapid cell renewal, nutrient, electrolyte, fluid transport processes, and a state of ‘physiologic’ inflammation that ensures containment of bacteria and antigens1. Combined with an underlying vasculature, prone to shunting oxygen-rich blood away from the villus tip, these demands render the intestinal epithelium particularly sensitive to reductions of blood flow and resultant mucosal ischemia/hypoxia2. However, dramatic breakdown of intestinal integrity is generally prevented by extremely effective adaptive processes2–4. Further harnessing these protective responses bears great potential in clinical settings in which mucosal hypoperfusion plays an important role, such as frank vascular occlusion, neonatal necrotizing enterocolitis 5, inflammatory bowel disease2, and systemic inflammatory responses associated with certain surgeries6 and/or different forms of shock7.
Post-translational modification with Small Ubiquitin-like MOdifiers (SUMO) is an effective way to dynamically influence responses to acute states of cellular stress through modification of transcription factors8, enzyme and receptor activity9, proteasomal degradation10, cellular trafficking11, protein aggregation12, chromatin targeting13, and protein-protein interactions14. Much of current literature has focused on SUMO modifications with respect to isolated proteins. This has led to conflicting reports with regard to global outcomes as multiple components of a given pathway may be modified by SUMO in a distinct spatiotemporal sequence. Important new insights on the relevance of the SUMOylation system stem from studies manipulating Ubc9, the sole SUMO E2 conjugating enzyme. Here, rapid epithelial disintegration and death from intestinal failure after inducible global Ubc9 knock-out draws attention to the unique dependency of the intestinal epithelium on a functioning SUMO machinery15.
Vertebrates encode four different SUMO proteins: SUMO1, the highly homologous SUMO2 and 3 (often combined as SUMO2/3), and a still largely enigmatic SUMO4. Determining the specific contribution of these SUMO isoforms for health and disease has been extremely difficult, which is a major obstacle towards harnessing tissue protective effects of SUMOylation. For example, SUMO2/3-conjugated protein levels increase substantially after stimuli such as heat shock, oxidative stress, DNA damage, or hypoxia and ischemia13, 16–19, however, due to rapid cycles of SUMO2/3 conjugation and deconjugation, these modifications are extremely dynamic. In addition, in vivo work is complicated by the redundancy between SUMO paralogs that compensates for the knockout of SUMO1 or SUMO3 and by the fact that global knock-out of SUMO2 is embryonically lethal precluding functional studies of this particular isoform20–22.
In the present work, we characterized the expression patterns of the major SUMO isoforms in the gut under physiological conditions and examined the effect of modulating SUMOylation on maintenance of gut integrity in a murine model of intestinal I/R. To the best of our knowledge, this study provides the first evidence of differential regulation particularly of SUMO2/3 along the crypt villus axis as well as a significant induction of SUMO2/3 modification with I/R challenge. Work with mice overexpressing the sole E2 ligase, Ubc9, as well as from SUMO1 knockout mice provide strong evidence that compensatory mechanisms targeting SUMO2/3 overexpression may be utilized to protect intestinal barrier integrity from I/R injury.
Materials and Methods
Animal experiments
Animal work was approved by the Duke University and the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) Animal Care and Use Committees (ACUC) respectively. Initial studies were performed in C57BL/6 mice obtained from Jackson Laboratories (Bar Harbor, ME). Experiments in the previously established Ubc9 transgenic mice (Ubc9Tg) were performed in the “H3” and “N2” founder lines23. SUMO3 knockout animals had previously been developed in our laboratory.22 SUMO1 knockout animals were graciously made available by Dr. Michael R. Kuehn (NIH)20 and were backcrossed with C57BL/6 mice.
For mesenteric I/R, mice were anesthetized with 1% isoflurane-supplemented by a onetime administration of 80mg/kg ketamine, 5mg/kg xylazine intraperitoneally, and 0.05mg/kg buprenorphine subcutaneously. A midline laparotomy was then made and a 2–3cm ileal loop isolated and rendered ischemic by placing aneurysm clips (Kent Scientific, Torrington, CT) on the peripheral branches of the superior mesenteric artery as well as across the intestine itself in order to block flow through collaterals. Ischemia was maintained for 15, 30, or 45 minutes, after which the clamps were removed and the incision closed. During the procedure, the body temperature was maintained at 37.0±0.3°C rectal temperature using a feedback-controlled heating pad. For the reperfusion phase of injury, mice were allowed to recover in a heated box with supplemental oxygen for a variable amount of time (0.5, 1, 3, or 6 hours). After euthanasia, tissue was rapidly removed and snap frozen in liquid nitrogen for RNA and protein isolation or transferred into 4% paraformaldehyde (PFA) for paraffin embedding. To obtain snapshot images representative of in vivo SUMO expression and distribution, a subset of mice was euthanized, and perfused with 0.9% NaCl (10 ml) followed by 4% PFA (30 ml) via transcardiac puncture. The intestines were then removed, placed in 4% PFA overnight, transferred to phosphate buffered saline (PBS), and embedded in paraffin. A further subset of animals undergoing I/R (n=4 in 3 hour and 6 hour reperfusion groups) received 0.6 mg/g body weight FITC dextran (4,000 MW at a concentration of 80 mg/ml; Sigma-Aldrich, St. Louis, MO) by gavage 3 hours before euthanasia 24. At euthanasia, plasma was harvested and the FITC concentration quantitated by fluorometry against a FITC-dextran standard curve.
Epithelial fractions for western blotting
Intestinal epithelial protein for western blotting was obtained by two methods. We obtained an epithelial enriched fraction by mechanically scraping the superficial cell layer from the opened luminal surface of small intestines. Sequential fractionation of cells along the villus/crypt axis was performed using a modified version of the low-temperature method described previously25. Briefly, ~ 5cm of distal small intestine was swiftly removed after euthanasia and immediately flushed with ice-cold Hank’s Balanced Salt Solution (HBSS) containing 20mM N-ethylmaleimide (NEM) and 0.5mM dithiothreitol (DTT) (Sigma), opened longitudinally, and cut into 10mm pieces. Gut pieces were then brought into ice cold chelating buffer consisting of 27 mM-trisodium citrate, 5 mM-Na2HPO4, 96 mM-NaCl, 8 mM-KH2PO4, 1.5mM-KCl, 0.5mM-DTT, 55mM-D-sorbitol, 44mM-sucrose supplemented with 20mM NEM. Tissue was incubated under agitation and then transferred to a tube with fresh buffer, while the remaining lifted-off epithelia were spun down at 120×G, and the cell pellet frozen in liquid nitrogen and stored at −80°C. This was repeated 6 times to obtain epithelial fractions from most apical (F1) to most basal (F7). PCR for fatty acid-binding protein 2 (FABP2), Leucinerich repeat-containing G-protein coupled receptor 5 (LGR5) and lysozyme transcripts (Supplemental Figure 1A), H&E stain (Supplemental Figure 1B,C) and Western blot for the crypt markers lysozyme and βcatenin (Supplemental Figure 1D) were used to verify that this technique produced a fractionated epithelial cell harvest from villus to crypt.
Western blotting
Cells and tissue samples were lysed in Western blot lysis buffer as described previously26, sonicated and heat inactivated by boiling for 10 minutes. Proteins were electrophoresed on SDS-PAGE 4%–15% gels (Bio-Rad, Hercules, CA) and then transferred to PVDF membranes (Bio-Rad). Membranes were blocked using Tris/HCl-buffered salt solution supplemented with 0.1% Tween 20 and 5% skim milk powder, and incubated with primary antibody overnight at 4°C. Primary antibodies were: polyclonal anti-SUMO1 (Novus Biologicals, Littleton, CO), polyclonal anti-SUMO2/3 (Covance, Denver, PA), monoclonal anti-Ubc9 (BD Bioscience, San Jose, CA), monoclonal anti-β-catenin (Thermo Fisher Scientific, Rockford, IL), polyclonal anti-lysozyme (Thermo), and polyclonal anti-IL17A antibody (Novus). After washing, membranes were incubated with anti-rabbit or mouse horseradish peroxidase conjugates (Bio-Rad) and targets visualized using the Supersignal West Pico Luminol Enhancer solution (Thermo). Membranes were stripped and re-probed with a mouse monoclonal anti-β-actin antibody (Sigma) as loading control. Quantification of band intensity was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Laser microdissection and capture of intestinal villus-tip epithelia
Microdissection was performed from 8μm paraffin sections after deparaffination with CitraSolv (Fisher Scientific, Hanover Park, IL) and sequential ethanol baths. Microdissection was performed on an Axio Observer Z1 microscope equipped with PALM MicroBeam UV cutting beam (Carl Zeiss MicroImaging, München, Germany). Intestinal samples from Ubc9Tg and WT littermates after 45minutes of ischemia and 3 hours of reperfusion were used for these studies. To obtain consistent samples, we limited tissue harvest to villi with no more than an injury score of 2 according to the scoring system of Chiu et al. 27, i.e. to villi with intact epithelial covering but some evidence of a developing sub-epithelial space. Samples were cut using a 20x/0.40, Korr LD PlanNeofluar objective, yielding samples that consisted almost exclusively of villus-tip epithelia (Supplemental Figure 2A-C). A total of 2,000,000μm2 per sample was harvested.
RNA isolation from microdissected tissue and PCR array analysis
RNA from harvested tissue sections was isolated using the RNeasy FFPE kit (Qiagen, Redwood City, CA) according to the manufacturers specifications. Isolated RNA was then pre-amplified using the PreAMPcDNA synthesis kit in combination with PreAMP primer mix specific for the mouse cytokines and chemokines RT2 Profiler PCR array (Catalog number: PAMM-150Z) and RT2 SYBR green qPCR mastermix (all products Qiagen). PCR was performed on a LightCycler 480 (Roche Molecular Systems, Pleasanton, CA) following the RT2 Profiler program recommendations. Data analysis was based on the crossing point (CP) determined by the second derivative maximum analysis method and was performed on the RT2 Profiler PCR array webportal.
Myeloperoxidase (MPO) assay
MPO activity was measured in snap frozen tissue as described previously28. Briefly, tissue was homogenized in potassium phosphate buffer containing 0.5% hexadecyltrimethylammonium bromide (Sigma) and MPO activity was determined by measuring the oxidation of o-dianisidine in presence of hydrogen peroxide (Sigma).
Gene transcription analysis (microarray and RT-qPCR analysis)
RNA was isolated using a commercially available kit (Macherey-Nagel, Bethlehem, PA). Microarray analysis was performed on 4 independent samples for each of 6 groups: sham (WT and Ubc9Tg), 45 minutes ischemia and 3 hour reperfusion (WT and Ubc9Tg), and 45 minutes ischemia and 6 hour of reperfusion (WT and Ubc9Tg). Total RNA was labeled on a sample-by-sample basis according to the manufacturer’s guidelines for use with the Mouse Clariom S GeneChip (Affymetrix, Santa Clara, CA). Labeled cRNA were hybridized to these arrays in blinded, interleaved fashion. The Scanner 3000 (Affymetrix) was used in conjunction with GeneChip Operation Software (Affymetrix) to generate one .CEL file per hybridized cRNA. Next, the Expression Console (Affymetrix) was used to summarize the data contained across all .CEL files and generate 22,206 SST-RMA normalized transcript cluster expression values. Quality of this expression was challenged and assured via Tukey box plot, covariance-based PCA scatter plot, and correlation-based heat map using functions supported in “R” (http://www.cran.r-project.org). To remove noise-biased expression values, lowess modeling was performed using expression values by class (CV~mean expression). Lowess fits were then overplotted to identify the common low-end expression value in which the relationship between mean expression (signal) and CV (noise) deviated from linearity (mean expression value = 3.75). Subsequent flooring of measurements to this value reduced the overall number of gene-level measurements to 21,768. For transcript clusters not discarded, ANOVA testing of the expression values across samples using class as the factor was performed under BH FDR multiple comparison condition resulting in 6,387 genes with a corrected p < 0.05. These genes were consecutively post hoc challenged via TukeyHSD test. The final number of 5,487 differentially expressed genes were selected based on a p value < 0.05 and an absolute difference of means ≥1.5X. Next, annotations for these subset transcript clusters were obtained from NetAffx (Affymetrix), while enriched pathways and functions for the subset transcript clusters by class comparison were identified using Ingenuity Pathway Analysis (IPA) (Qiagen). For RT-qPCR analysis, equal amounts of RNA were reversed-transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). qPCR was performed with the TaqMan® Universal PCR Master Mix (Applied Biosystems) and FAM-labelled TaqMan® Gene Expression Assays (Thermo) for fucosyltransferase 2 (Fut2; Mm00490152_s1), suppressor of cytokine signaling 3 (Socs3; Mm00545913_s1), interleukin 6 (Il6; Mm0046190_m1), chemokine (C-X-C motif) ligand 2 (Cxcl2; Mm00436450_m1), endothelial cell selectin (Sele; Mm00441278_m1), platelet selecting (Selp; Mm01295931_m1), a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif 1 (Adamts1; Mm01344169_m1), matrix metallopeptidase 8 (Mmp8; Mm00439509_m1), or tyrosine aminotransferase (Tat; Mm01244282_m1), each with a VIC-labeled beta-actin primer/probe set (Actb; Mm02619580_g1) for internal calibration. All samples were tested in triplicate on a 7500 Fast Real-Time PCR System (Applied Biosystems) and analysed via the 2-ΔΔCT method.
Tissue staining and injury scoring
Sections were deparaffinized, hematoxylin and eosin-stained (H&E) and coded to conceal both genotype and experimental conditions prior to evaluation. Intestinal injury was quantified using the scoring system established by Chiu et al.27. The highest degree of injury in each field of view using a 200x magnification was noted to calculate average values for the entire length of the specimen. The final injury score resulted from 2 independently-performed examinations of each sample. Naphthol AS-D chloroacetate esterase staining for tissue neutrophils was performed following the manufacturer’s instructions (Sigma). Positively stained cells were quantified microscopically at 200x magnification in 6 unrelated regions of interest per section and reported in relation to the number of villi included in these regions.
Immunofluorescence
Sections were deparaffinized and heat-mediated antigen retrieval was performed in sodium citrate buffer. After blocking at room temperature, sections were incubated with the SUMO1, SUMO2/3 or Ubc9 antibodies specified above plus a monoclonal anti-e-cadherin antibody (BD Bioscience) for anatomical orientation. Labeling was achieved with goat anti-rabbit IgG or goat anti-mouse IgG conjugated to Alexa 488 or Cy3 (Thermo) and slides were mounted with ProLong anti-fade with 4’,6-diamidino-2-phenylindole (DAPI) as counterstain (Thermo).
Statistical Analysis
Statistical significance of differences among groups was determined by the two-sample t-test. Included bar graphs depict means ± SEM. Results were considered statistically significant if p ≤ 0.05.
Results
SUMO1 and SUMO2/3 are differentially expressed along crypt villus axis of small and large intestine
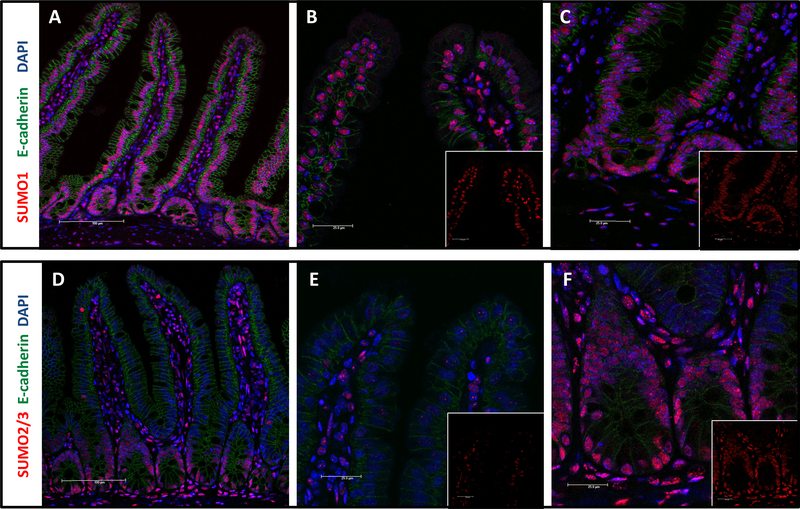
Immunofluorescence staining of intestinal samples revealed a distinct epithelial gradient of SUMO2/3 staining, with very low signal in villus tip but strong nuclear staining outlining the crypt epithelia (Figure 1D-F). In contrast, SUMO1 exhibited some degree of gradient in epithelial cells, but this was less pronounced, and SUMO1 positive epithelial cell nuclei were evident throughout the villus crypt axis (Figure 1A-C and Supplemental Figure 1D). Furthermore, submucosal cells both in the villus core as well as in the intestinal wall were robustly and uniformly positive for both SUMO1 and SUMO2/3. This general staining pattern for SUMO1 and SUMO2/3 was found also in colonic samples, which indicates that a crypt-pronounced SUMO2/3 distribution exists in other regions of the intestine too (Supplemental Figure 3).
Figure 1: SUMO1 and SUMO2/3 distribution in murine small intestine.
Immunostaining of small intestinal sections from animals that were paraformaldehyde-perfused via transcardiac puncture. Sections were stained as indicated with e-cadherin (green), DAPI (blue), and either SUMO1 (red: A-C) or SUMO2/3 (red: D-F). A and D: small intestinal section, 200x; B and E: villus tip, 600x; C and F: crypt, 600x.
We had previously shown that active SUMOylation processes typically increase particularly the nuclear SUMO signal vs. a more diffuse cytoplasmic stain of the free SUMO pool.19 To verify that the observed, increased nuclear staining pattern indeed signified increased SUMO conjugation of proteins, we performed Western blot analysis of epithelial fractions from small intestine (Supplemental Figure 1D). Here the high molecular smear representative of SUMOylated proteins was largely limited to crypt epithelial fractions when probing for SUMO2/3 and, to a lesser extent, for SUMO1. Interestingly, the sole SUMO ligase Ubc9 was expressed at higher levels in epithelial crypt cells too. Together these data suggest that SUMO conjugation is highly regulated under baseline conditions in intestinal epithelia where it defines the crypt epithelial compartment.
Enhanced SUMO2/3 signal in the small intestinal villus tip area after ischemia/reperfusion (I/R) injury
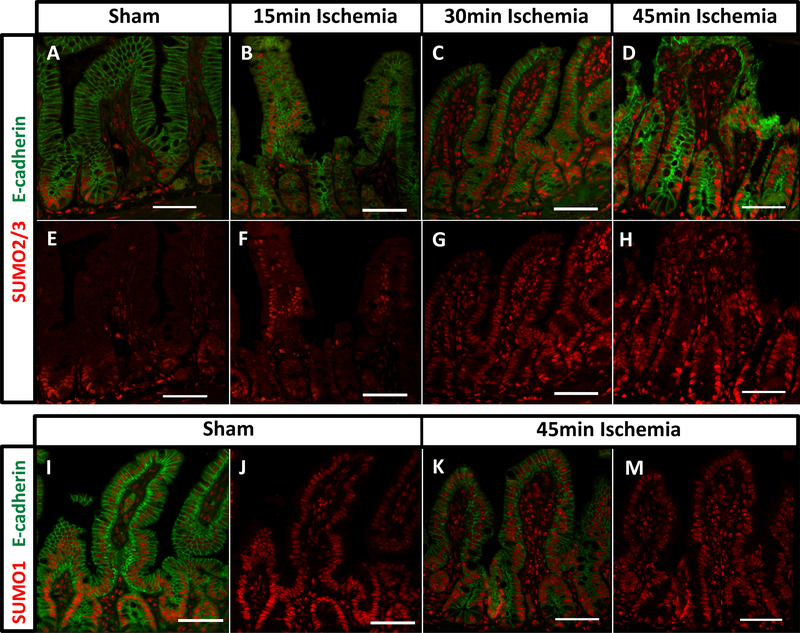
We have previously shown that SUMO2/3 conjugation is rapidly induced in states of ischemia.17, 19 To investigate whether intestinal epithelial SUMO2/3 signal was responsive to ischemic stress despite its tight restriction under baseline conditions, we subjected mice to mesenteric I/R injury. For this, we examined ischemic challenges that ranged from 15 to 45 minutes followed by a consistent reperfusion time of 30 minutes. As outlined in Figure 2, I/R caused a rapid expansion of SUMO2/3 staining, initially as a spotty nuclear signal in more apical villus regions but with ischemic times longer than 30 minutes, as a robust epithelial SUMO2/3 staining within the villus tip. In contrast, differences in SUMO1 staining were much subtler due to the relatively high baseline villus tip signal (Figure 2 I-M).
Figure 2: SUMO2/3 staining expands into the villus-tip region after I/R.
Sections were stained as indicated with e-cadherin (green: A-D) and SUMO2/3 (red: A-H) in animals subjected to sham procedure (A,E), or 15 (B,F), 30 (C,G), and 45 minutes (D,H) of ischemia. Additional sections were stained for e-cadherin (green: I, K) and SUMO1 (red: I-M) after sham procedure (I, J) or 45 minutes of ischemia (K, M). Reperfusion period was 30 minutes in all I/R animals. Magnification 200x, size bar denotes 50μm.
Ubc9 overexpression leads to enhanced nuclear SUMO2/3 staining in intestinal villus epithelia of Ubc9Tg animals
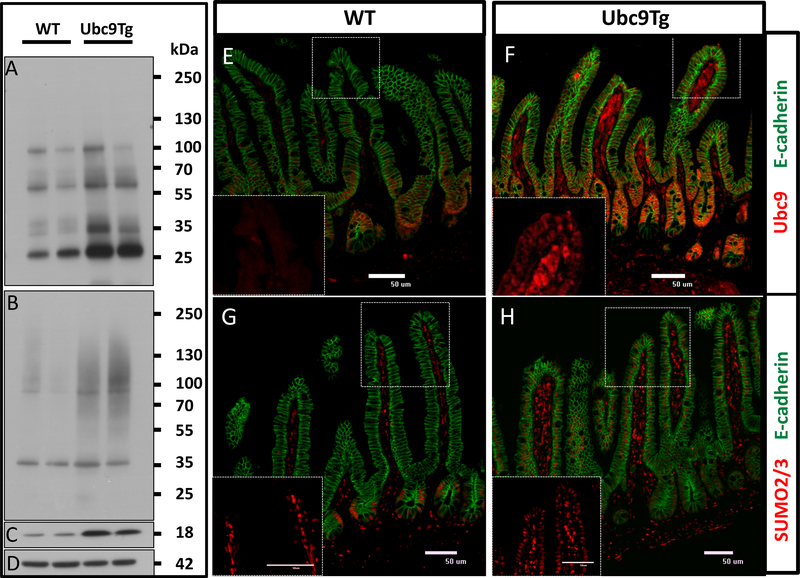
Our previous work had suggested the Ubc9 may be a rate limiting step in the SUMO conjugation process29. In agreement, elevated Ubc9 levels in Ubc9 transgenic (Ubc9Tg) animals translated into elevated baseline levels of SUMO1 and SUMO2/3 modified proteins in epithelial enriched fractions taken from the small intestine (Figure 3A-D). In accordance, immunostaining of small intestinal sections documented a noticeable extension of both Ubc9 (Figure 3 E,F) and nuclear epithelial SUMO2/3 (Figure 3 G,H) staining into the villus tip in Ubc9Tg mice as compared to WT controls.
Figure 3: Overexpression of Ubc9 enhances baseline small intestinal epithelial SUMO2/3 expression.
Mucosal scrapings from small intestines of WT or Ubc9Tg animals were analyzed by Western blot for SUMO1 (A), SUMO2/3 (B), Ubc9 (C), and bActin (D) protein levels. Markers denote molecular weight in kilo Dalton. Small intestinal sections from wild type (E,G) or Ubc9Tg animals (H,F) were stained for Ubc9 (red: E,F) or SUMO2/3 (red: G,H) as well as e-cadherin (green: E-H). Magnification 200x. Inserts detail the villus tip staining for Ubc9 or SUMO2/3 respectively.
Ubc9Tg animals are protected from intestinal ischemia/reperfusion (I/R) injury
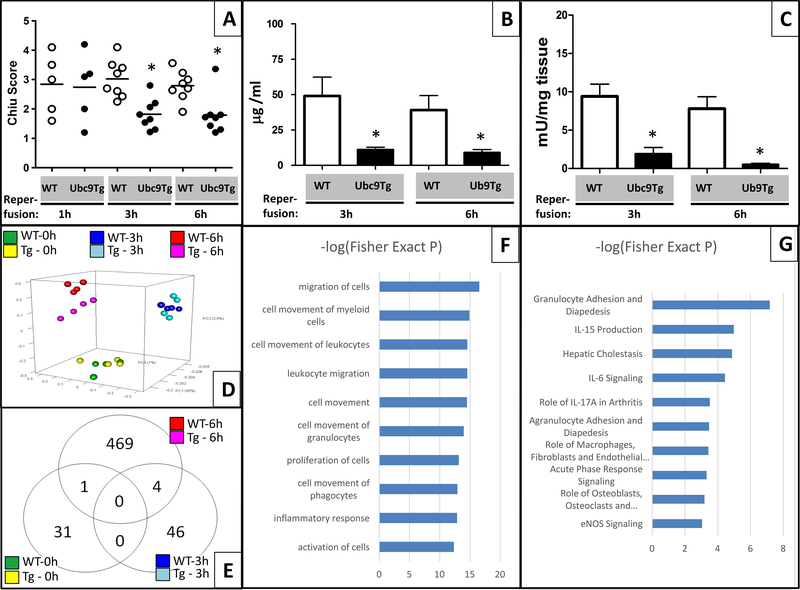
Next, we investigated, whether enhanced SUMO conjugation was a protective response by using Ubc9Tg animals and their wild-type (WT) littermates in the mesenteric I/R model. We examined a consistent ischemic stimulus (45 minutes) with multiple reperfusion times to have endpoints at the height of injury (1-hour reperfusion) and different stages of inflammation and healing (3- and 6-hour reperfusion). Histological scoring indicated that the severity of damage inflicted after 45 minutes of ischemia was comparable between WT and Ubc9Tg animals (Chiu score after 1 hour reperfusion; Ubc9Tg: 2.9±1.5 vs. WT: 2.7±1.0). However, Ubc9Tg animals exhibited a significantly enhanced recovery 3 hours (Chiu score 1.9±0.6 vs. WT: 2.7±0.4) and 6 hours (Chiu score 1.7±0.8 vs. WT: 2.7±0.6) after reperfusion (Figure 4A). In agreement, we observed significantly less leakage of gavaged FITC dextran into the serum in Ubc9Tg animals both 3 hours (11±4μg/ml vs. WT: 49±30μg/ml) and 6 hours (9±5μg/ml vs. WT: 39±23μg/ml) after reperfusion. (Figure 4B). In addition, a marked reduction in the infiltration of inflammatory cells was seen in Ubc9Tg as compared to WT animals as evidenced by reduced myeloperoxidase (MPO) activity in Ubc9Tg 3 hours (Ubc9Tg: 19±19mU/mg tissue vs. WT: 94±35mU/mg tissue) and 6 hours (Ubc9Tg: 5±4mU/mg tissue vs. WT: 78±35mU/mg tissue) after reperfusion (Figure 4C). A similar trend was observed when counting Naphthol AS-D Chloroacetate esterase-stained neutrophils. Few positive cells were encountered 1 hour after re-perfusion in either mouse strain, but in Ubc9Tg animals these numbers remained relatively low (3-hour reperfusion: 0.6±0.3 neutrophils/crypt; 6-hour reperfusion: 0.5±0.4 neutrophils/crypt) while we observed significant inflammatory infiltration in WT animals (3-hour reperfusion: 7.4±3.0 neutrophils/crypt; 6-hour reperfusion: 6.2±1.7 neutrophils/crypt (data not shown). Therefore, while initial injury appeared comparable, Ubc9Tg recovered faster than their littermate controls, likely due to a blunted inflammatory response in the reperfusion period.
Figure 4: Ubc9 Tg animals are protected from injury due to blunted inflammatory responses after intestinal I/R.
(A) Sections from wild type or Ubc9Tg animals undergoing 45 minutes of ischemia and either 1, 3 or 6 hours of reperfusion were stained by H&E and scored using the Chiu intestinal I/R score. n=6/condition. (B) Plasma levels of gavaged FITC dextran (in μg FITC dextran/ml plasma) as a measure of intestinal permeability from wild type or Ubc9Tg animals after 45 minutes of ischemia and either 3 or 6 hours of reperfusion. n=4/condition. (C) Myeloperoxidase assay of intestinal samples harvested after 45 minutes and 3 or 6 hours of reperfusion. n=6/condition, *p≤0.05. Microarray analysis of transcriptional responses in Ubc9Tg (Tg) vs. wild type littermates after I/R. (D) Covariance-based Principal Component Analysis (PCA) scatter plot depicts the Ubc9Tg vs. wild type sample-to-sample relationships by time using the union set of 5,487 genes identified differentially expressed for at least one possible sample type comparison. (E) Venn diagram summarizes the number of differential expressed genes observed between Ubc9Tg vs wild type that are either exclusive by time or shared across time. The top 10 significantly enriched (F) functions and (G) pathways returned from the Ingenuity Pathway Analysis (IPA) tool when provided the list of 474 genes differentially expressed between Ubc9Tg and wild type subjected to 45minutes of ischemia and 6 hours of reperfusion. n=4/condition.
Reduced inflammatory gene expression in Ubc9Tg vs. WT animals after intestinal I/R.
As a significant portion of SUMO target proteins are transcription factors, we next aimed to define the protective phenotype seen in Ubc9 animals using RNA microarray analysis. In this analysis, we tested whole-tissue intestinal samples of Ubc9Tg vs. WT animals undergoing sham procedure or 45 minutes of ischemia with either 3 or 6 hours of reperfusion (raw data has been deposited in NCBI’s Gene Expression Omnibus [http://www.ncbi.nlm.nih.gov/geo/)], GEO Series accession number GSE96733). A total of 22,206 gene-level expression measurements were generated per sample from which our analysis identified a final set of 5,487 as being differentially expressed (for approach and selection criteria, see Materials and Method section of the manuscript). Principal components analysis (Figure 4D) demonstrated separation between sham and the different I/R treatment groups but differences between respective WT and Ubc9Tg animals only in the 6-hour reperfusion cohort (distribution of differentially regulated genes is shown in the Venn diagram, Figure 4E). From this group, we selected 9 differentially regulated genes for validation by qPCR (Table 1) and found the same overall trend, although differences in fold changes were noted. IPA pathway and function analysis of differentially regulated genes in this group revealed a strong enrichment in functions associated with inflammatory cell recruitment such as “migration of cells”, “cell movement of myeloid cells/leukocytes/granulocytes/phagocytes”, or “inflammatory response” (Figure 4F). Consequently, the most significantly enriched pathways when comparing Ubc9Tg and WT animals after 6 hours of reperfusion, were “granulocyte adhesion and diapedesis” as well as pathways associated with cytokines such as IL15, IL6, and IL17A (Figure 4G).
Table 1: Verification of microarray data by qPCR analysis.
Relative expression of 9 selected genes in Ubc9Tg vs. WT animals. n=6 for qPCR and n=4 for microarray.
| Gene symbol | Gene ID | qPCR | Microarray |
|---|---|---|---|
| Fut2 | 14344 | −3.30 | −18.31 |
| Socs3 | 12702 | −3.89 | −4.04 |
| Il6 | 16193 | −3.64 | −7.15 |
| Cxcl2 | 20310 | −3.16 | −3.33 |
| Selp | 20344 | −3.89 | −4.99 |
| Sele | 20339 | −2.88 | −3.26 |
| Adamtsl | 11504 | −2.01 | −2.88 |
| Mmp8 | 17394 | −3.82 | −2.57 |
| Tat | 234724 | −5.04 | −16.27 |
These results demonstrate that Ubc9 overexpression is associated with a complex reprogramming of the inflammatory response after I/R.
Laser microdissection and capture reveals modulated chemokine responses in villus tip epithelial cells of Ubc9Tg animals.
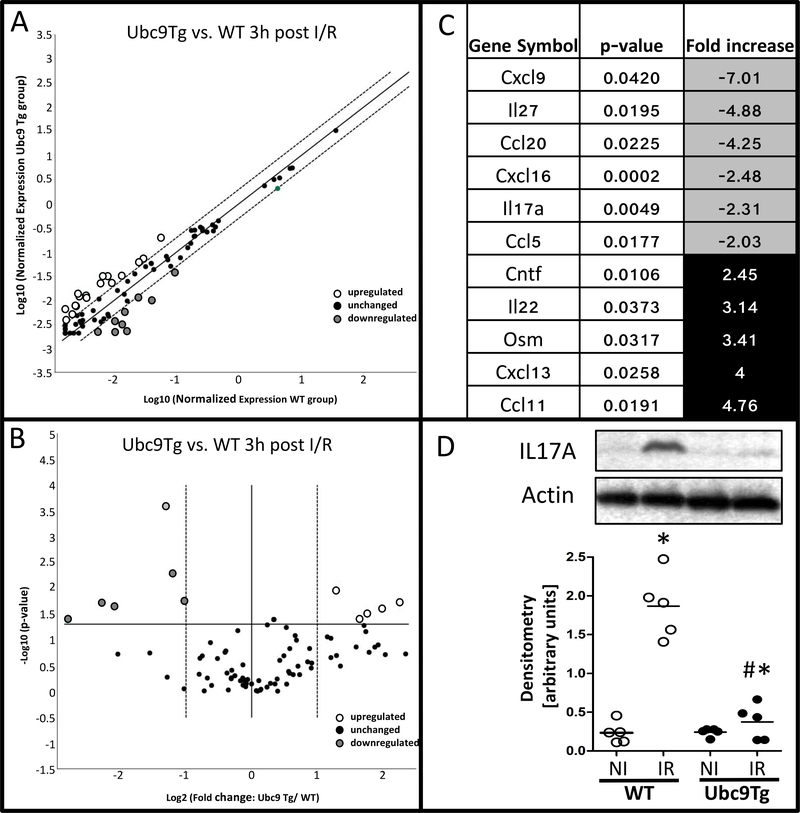
Given the fact that histological changes were evident 3 hours after reperfusion, whereas our microarray from whole tissue identified the most relevant changes only 6 hours after reperfusion, we hypothesized that earlier responses in villus tip epithelia may have been underrepresented in the whole tissue expression analysis. The villus tip not only constitutes the location of the most pronounced injury but also that of the most apparent increase in SUMO expression with either I/R or Ubc9 overexpression. Therefore, we obtained tissue highly enriched for villus tip epithelia by laser microdissection and capture and we examined the expression of major chemokine and cytokines via PCR array. Figure 5A and B show scatter and volcano plots which represent significantly regulated genes in villus tip samples from Ubc9Tg animals vs. WT littermates 3 hours after reperfusion. Five genes (out of 84) were induced at least twofold (p-value < 0.05), whereas 6 genes were downregulated at least twofold (p-value < 0.05) (Figure 5C). These results show that SUMO-overexpression modulates early epithelial inflammatory signaling with a marked suppression of chemotactic factors such as Cxcl9, Cxcl16, Ccl5, Ccl20, Il17a and Il27 in line with the reduced inflammatory infiltration observed in these animals. Because IL17A constitutes a major determinant of chemotactic responses, we verified IL17A protein levels in these animals. Notably, we found that 6 hours after reperfusion, IL17A protein was significantly induced in ischemic vs. non-ischemic small intestinal loops, and confirmed that this response was blunted in Ubc9Tg animals (Figure 5D). Together, these findings further support that SUMOylation controls inflammation after I/R injury and identify a specific epithelial contribution to inflammatory signaling in this context.
Figure 5: Modulation of villus-tip epithelial responses in Ubc9Tg animals after I/R.
Intestinal epithelial villus-tip epithelia from ischemic regions of wild type and Ubc9Tg (45 minutes of ischemia and 3 hours of reperfusion) were isolated by laser microdissection and capture and RNA was isolated to perform a cytokine targeted PCR array. A) Scatter and B) Volcano- blot outline the distribution of regulated genes, significantly up- and downregulated genes are listed in C). D) IL17A protein levels in non-ischemic small intestinal loops (NI) and loops subjected to ischemia/reperfusion (IR; 45minutes ischemia and 6 hours or reperfusion). Quantification was performed by densitometry and is represented as the ratio of IL17A to bActin band intensity. n=5; *p≤0.05
SUMO1 knockout (SUMO1-KO) animals display compensatory overexpression of SUMO2/3 in intestinal epithelia and are partially protected from I/R injury.
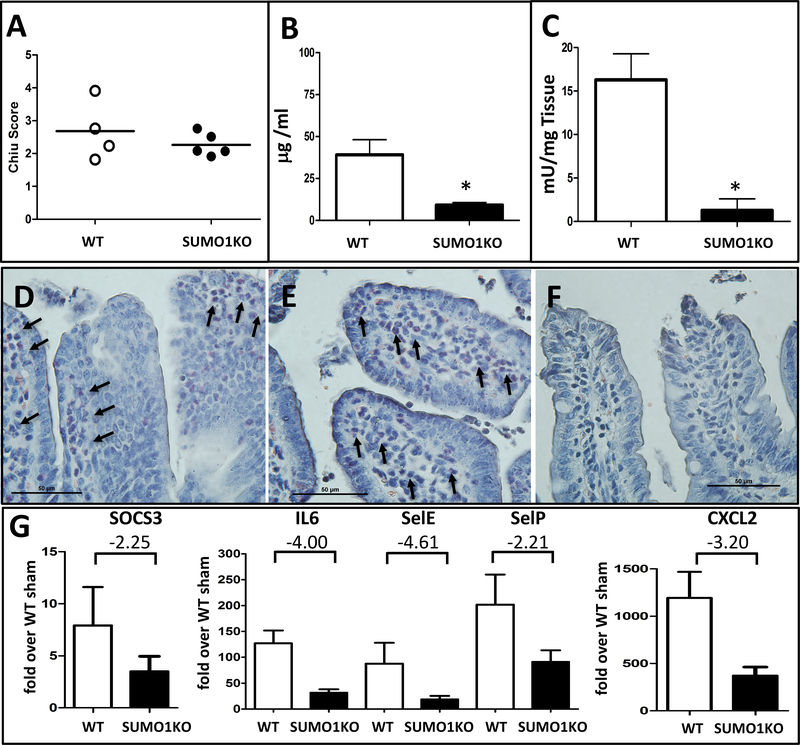
The specific contribution of SUMO2/3 vs. SUMO1 in adaptive responses to injury has never been examined in vivo. To address this, we first defined the relative distribution of the major SUMO isoforms in the intestines of SUMO1 and SUMO3 knock-out animals. No phenotypical abnormalities have been described in these animals20–22 and indeed, when we stained SUMO3 knock-out animals under baseline conditions, we found no differences in either SUMO1 or SUMO2/3 distribution compared to WT littermates (Supplemental Figure 4). However, unchallenged SUMO1-KO animals unexpectedly displayed an enhanced nuclear SUMO2/3 expression that clearly extended into the epithelium of the villus tip and thus mirrored findings from Ubc9Tg animals or after I/R injury (Supplemental Figure 5). When exposed to the mesenteric I/R model (45minutes of ischemia and 6 hours of reperfusion), minor differences in overall injury severity were observed between SUMO1-KO vs. WT littermates (Figure 6A). However, SUMO1-KO animals displayed better intestinal barrier preservation after FITC dextran gavage (SUMO1-KO:9.2±3.2μg FITC dextran/ml plasma vs. WT 39.1μ±18.1 μg FITC dextran/ml plasma) (Figure 6B), together with a blunted inflammatory response as evidenced by the significantly reduced myeloperoxidase (MPO) activity (SUMO1-KO: 1.35±2.91mU/mg vs. WT 16.26±5.97mU/mg) (Figure 6C). In agreement, staining of cells from the granulocyte lineage with Naphthol AS-D Chloroacetate, revealed a numerous infiltration of polymorphnuclear cells in seemingly well-preserved villi adjacent to sites of epithelial injury in WT mice (Figure 6D,E) but only isolated neutrophils in SUMO1-KO mice (Figure 6F). Lastly, and consistent with the transcriptional pattern observed in Ubc9Tg animals, we observed a blunted response in the selected group of genes involved in inflammatory cell recruitment (Figure 6G). Together, this indicates that compensatory SUMO2/3 overexpression in SUMO1-KO is sufficient to regulate important inflammatory responses in I/R injury. However, the more complete protective phenotype in Ubc9Tg animals suggests that SUMO1 and/or SUMO-independent effects of Ubc9 may also contribute to adaptive responses.
Figure 6: SUMO1 knock-out (SUMO1-KO) animals display a blunted inflammatory response to intestinal I/R injury.
(A) H&E stained section from wild type and SUMO1-KO animals were scored using the Chiu intestinal I/R score. (B) Plasma levels of gavaged FITC dextran (in μg FITC dextran/ml plasma) from wild type or SUMO1-KO animals. (C) Myeloperoxidase assay. (D-F) Representative images from Naphthol AS-D Chloroacetate esterase staining of intestinal sections from wild type (D,E) or SUMO1-KO animals (F) after I/R injury. Images were taken from relatively intact sections adjacent to areas of significant injury. Arrows denote infiltrating neutrophils. Magnification 400x. (G) qPCR from post-ischemic small intestines in SUMO-1 KO and WT mice. Graphs represent fold change over sham treated wild type animals and notes the fold difference of response between SUMO1-KO and WT, consistent with Table 1. Assays were performed on wild type (n=4) or SUMO1-KO (n=5) animals subjected 45 minutes of ischemia and 6 hours of reperfusion. *p≤0.05.
Discussion
SUMO conjugation allows for rapid cellular reprogramming in states of severe metabolic stress such as I/R. Under such conditions, especially SUMO2/3-conjugation rapidly increases29 and evidence suggests an important adaptive and cell-protective role through regulation of metabolism, hypoxia adaptive responses, inflammation, RNA processing, and SUMOylation-dependent ubiquitin conjugation10, 30–38. Our present study adds important knowledge to the field by showing a distinct SUMO2/3, and to a lesser degree SUMO1, crypt-to-villus gradient under physiologic conditions and, importantly, a marked expansion of epithelial SUMOylation in an I/R injury model. Mice overexpressing Ubc9, and – unexpectedly – mice lacking SUMO-1, demonstrated enhanced nuclear SUMO2/3 signal in intestinal epithelial cells and improved outcomes after mesenteric I/R, primarily through blunting of inflammatory responses. This data provides powerful support that SUMOylation particularly with the SUMO2/3 isoforms, is a crucial protective response after intestinal I/R injury.
While SUMOylation is clearly essential for intestinal homeostasis15 as well as for barrier integrity and inflammatory control during bacterial invasion39, the intestinal expression pattern of the SUMO paralogs has never been studied in detail. Hypothesizing that SUMOylation events may perform a protective role in this metabolically challenged tissue, we expected a strong SUMO signal in the apical epithelium in parallel with the steep oxygen gradient that exists along the crypt/villus axis2. However, the exact opposite was the case, with strong SUMO2/3 signal in the connective tissue and the epithelial crypt but little staining of villus-tip epithelia. Results from Demarque et al. bear impressive evidence of the functional importance of our findings. Here, the induced loss of Ubc9 in adult animals led to the depletion of the small intestinal proliferative compartment, to massive disturbances of intestinal epithelial architecture and ultimately, to rapid demise from intestinal failure15. Therefore, SUMOylation is an indispensable and, as shown by our data, a highly-regulated mechanism for small intestinal epithelial organization. What remains unknown is how SUMOylation is regulated in this setting, and what its specific functions are. It will be extremely interesting to integrate the observed SUMO2/3 conjugation pattern with existing knowledge on crypt-villus gradients of regulators of intestinal epithelial renewal and differentiation40.
Interestingly, the physiologic restriction of SUMO2/3 signal was rapidly lifted after I/R, which led to a severity-dependent expansion of SUMOylation into the apical epithelium. The villus tip is particularly exposed to metabolic challenges such as hypoxia2 and free radical generation41, and therefore injury following I/R is typically most pronounced here27. Increased global SUMOylation, particularly of SUMO2/3 has been reported in different organs after I/R including kidney, heart, and brain17, 19, 29, 42, 43, and is thought to be part of an overall cytoprotective response. However, tools capable of defining the specific contribution of SUMOylation and of the individual SUMO isoforms to protective responses in this context have been lacking. The SUMO E2 conjugase Ubc9 is considered a gatekeeper within the SUMOylation process, as evidenced by the fact that SUMO1/2/3 overexpression alters free SUMO levels but not those of SUMO-modified proteins29. In our current work, we now observed that Ubc9 overexpression induced SUMO1 and SUMO2/3 signal particularly in the villus tip and that it conveyed significant protection in a mesenteric I/R model. Together with previous work that correlated expression levels of Ubc9 with SUMO conjugation levels in the brain and, importantly, with increased tolerance to focal cerebral ischemia23, this data strongly support the idea that global SUMOylation levels may be harnessed to protect against I/R injury and related inflammation.
SUMOylation prominently modifies components of the transcriptional apparatus (recently reviewed in 44). However, so far only one work has attempted to define the impact of altered SUMOylation on gene expression in vivo. In a neuron-specific SUMO1/2/3 knock-down model45, SUMO served as a transcriptional repressor under physiologic conditions but had decidedly gene-activating effects following transient cerebral ischemia. Unlike our study, pro-growth and anti-apoptotic pathways were prominently enhanced. While direct comparisons between the very distinct approaches are of course problematic, the divergent findings may highlight that SUMOylation plays substantially different roles in a largely differentiated, non-replicating tissue displaying very low SUMO conjugation under physiologic conditions (brain) vs. a rapidly regenerating tissue with relatively high baseline SUMO signal (intestine). Importantly, the gut is exposed to trillions of bacteria and milligrams of lipopolysaccharide (LPS) within its lumen. Following intestinal injury, it is therefore imperative to prevent dysregulated inflammatory activation and to rapidly re-institute barrier integrity. In Ubc9Tg mice, this appears to be achieved at least in part by regulating influx of inflammatory cells as evidenced both by our pathological findings and by the prominent regulation of genes involved in inflammatory cell infiltration, such as that of adhesion proteins (p-selectin46, e-selectin47), of chemotactic factors (Cxcl148, 249, 3, 550), of inflammatory regulators (Il651, Il15, and Il17A52) and proteases (e.g. AdamTs 1, 4, 14, Adam8, Mmp8)53. Upstream analysis of the differentially expressed genes provided for a candidate network of 8 major regulators of stress responses (Supplemental Figure 6), most of which are known to be modulated by SUMOylation31, 32, 54–58. It is noteworthy, that these findings show significant overlap with the work of Fritah et al.39, which identifies SUMOylation as an important host-protective mechanism against gut-invasive pathogens through reduction of inflammatory epithelial destruction and which implicates, by PCR array analysis and SILAC-based quantitative proteomics, many of the same responses (Il6, Cxcl3, Cxcl1, Nfkbia and Xbp1).
Similar to our results in Ubc9Tg animals, Fritah et al.39 observed that pro-inflammatory gene expression was not significantly altered under baseline conditions in mice, haploinsufficient for Ubc9. However, the lack of a signal in early post-ischemic time-points also raised the question of whether responses of the villus-tip epithelium, i.e. the tissue with most dramatic changes of SUMOylation, were adequately represented in our whole tissue microarray study. An important limitation of published genetic approaches15, 39 including our own, is the inability to dissect cell type-specific responses to changes in SUMOylation. We aimed to address this by a focused analysis of apical epithelial transcriptional responses from samples obtained through laser microdissection and capture. Ensuing cytokine and chemokine-targeted analysis supported observations from our microarray data that SUMOylation modulates Il17a expression, a key cytokine linking T-cell activation to neutrophil mobilization and activation59 and one of great importance in intestinal inflammation60, 61. The significance of these results is supported by the parallel decrease of the Il17a target gene Ccl2059, 61 and the induction of Il22, a member of the IL10 family known to inhibit production of Il17a62. Interestingly, in addition to Il17a, other factors downregulated in Ubc9Tg animals also shared T-cell chemotactic and regulatory functions (Cxcl963, Il2764, Ccl2065, Cxcl1666, Ccl567) in parallel with a growing appreciation both of T-cell responses during early ischemia-reperfusion events68 and of the role of SUMO in regulating these69.
Our observation that global SUMO1 deletion results in better outcome following I/R injury was a completely unexpected finding, because it is generally believed that in organs under stress, increased SUMOylation improves outcome and decreased SUMOylation impairs it. For example, increased SUMOylation by overexpression of Ubc9 improves outcome following small intestine I/R (this study) or in experimental stroke23, while silencing SUMO2/3 or SUMO1,2,3 expression worsens outcomes.45, 70 As overexpression of SUMO1 is also reported to support protective responses in a heart failure model,71 we therefore expected outcomes following small intestine I/R injury to be worse in SUMO1-KO mice. SUMO1 deficient animals have been studied and apart from abnormalities in RanGAP1 localization and in formation of PML nuclear bodies20, no phenotype has been attested in these animals20, 21. It was also noted that in SUMO1-KO mice most, if not all, SUMO-1 functions are compensated for by SUMO2/3, albeit with no evidence of increased SUMO2 or SUMO3 mRNA expression.21 Our study confirmed this compensation of SUMO1 by SUMO2/3-overexpression, and, most importantly, showed that such induction of SUMO2/3 blunts intestinal inflammatory responses after I/R. However, this protection from I/R injury is not as effective as in Ubc9Tg animals, which indicates independent effects of SUMO1 and/or of Ubc9 itself. Here, we have to acknowledge that conclusions from our work may be limited by the difficulties we encountered in the quantification of SUMO isoform levels. On the whole-tissue level, high background signal in the connective tissue made it difficult to quantify changes that occurred in epithelial cells and methods to isolate epithelial cells were too complex and lengthy to produce faithful snapshot images of the extremely dynamic SUMOylation process. Therefore, while data from SUMO1 deficient mice underline the importance of SUMO2/3 in the regulation of intestinal protective programs, our approach may have underappreciated changes of SUMO1 levels after I/R and/or in Ubc9 animals. Together, results reported here describe a unique pattern of nuclear SUMO2/3 staining in epithelial cells of the small intestine along the crypt-villus axis; they demonstrate that improved outcome following I/R in Ubc9-overexpressing mice is associated with preserved barrier function and blunted inflammatory responses; and they identify a previously undocumented process, whereby in SUMO1-KO mice, the compensatory overexpression of SUMO2/3 increases tissue I/R resistance. The tremendous protective power of SUMO conjugation reported here further highlights the importance of our ongoing work aimed at boosting the SUMOylation pathway to protect organs from damage induced by metabolic stress.72–75
Supplementary Material
Acknowledgements
The work was supported by a Dream Innovation Grant (DIG), made possible by Duke Anesthesiology’s DREAM Campaign to J.K. and NIH R01 NS081299 and NS097554 to W.P., NIH R01 NS099590 to W.Y. and NIH R56 HL126891 to J.K. as well as a Duke School of Medicine Health Scholar award and a Scientist Development Grant (15SDG25080046) from the American Heart Association to J.K.. J.D.B. received support from a NIH-OxCam Fellowship and the UAB Medical Scientist Training Award and J.D.B. and J.M.H. were supported by the Intramural Research Program of the NINDS/NIH. The authors thank Soman N. Abraham, PhD for discussions and support, Laura Mitrescu, B.S., Jasmine Fowler, B.S., Yang-ja Lee, Ph.D., Pei Miao, B.S. and Daniel Ye, B.S. for technical assistance and Sara Galletti, Ph.D. for help with manuscript preparation. Also, we thank Yasheng Gao, Ph.D. from the Duke light microscopy core facility for helping with the laser microdissection and capture procedures.
Non-standard abbreviations:
- DTT
dithiothreitol
- FITC
fluorescein isothiocyanate
- H+E
hematoxylin + eosin
- I/R
ischemia/reperfusion
- MPO
myeloperoxidase
- NEM
Nethylmaleimide
- PFA
paraformaldehyde
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- SUMO
Small Ubiquitin-like MOdifer
- Tg
transgenic
- WT
wild type
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest related to the work.
References
- 1.Glover LE, Lee JS, Colgan SP. Oxygen metabolism and barrier regulation in the intestinal mucosa. The Journal of clinical investigation 2016;126:3680–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karhausen J, Furuta GT, Tomaszewski JE, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest 2004;114:1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaser A, Flak MB, Tomczak MF, et al. The unfolded protein response and its role in intestinal homeostasis and inflammation. Exp Cell Res 2011;317:2772–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature 2016;529:307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogberg N, Carlsson PO, Hillered L, et al. Intraluminal intestinal microdialysis detects markers of hypoxia and cell damage in experimental necrotizing enterocolitis. J Pediatr Surg 2012;47:1646–1651. [DOI] [PubMed] [Google Scholar]
- 6.Karhausen J, Qing M, Gibson A, et al. Intestinal mast cells mediate gut injury and systemic inflammation in a rat model of deep hypothermic circulatory arrest. Crit Care Med 2013;41:e200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark JA, Coopersmith CM. Intestinal crosstalk: a new paradigm for understanding the gut as the “motor” of critical illness. Shock 2007;28:384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang J, Valin A, Gill G. Regulation of transcription factor activity by SUMO modification. Methods in molecular biology 2009;497:141–152. [DOI] [PubMed] [Google Scholar]
- 9.Wasik U, Filipek A. Non-nuclear function of sumoylated proteins. Biochim Biophys Acta 2014;1843:2878–2885. [DOI] [PubMed] [Google Scholar]
- 10.Praefcke GJ, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin. Trends Biochem Sci 2012;37:23–31. [DOI] [PubMed] [Google Scholar]
- 11.Raman N, Nayak A, Muller S. The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 2013;122:475–485. [DOI] [PubMed] [Google Scholar]
- 12.Krumova P, Meulmeester E, Garrido M, et al. Sumoylation inhibits alphasynuclein aggregation and toxicity. J Cell Biol 2011;194:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendriks IA, Treffers LW, Verlaan-de Vries M, et al. SUMO-2 Orchestrates Chromatin Modifiers in Response to DNA Damage. Cell Rep 2015;10:1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J, Durrin LK, Wilkinson TA, et al. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 2004;101:14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demarque MD, Nacerddine K, Neyret-Kahn H, et al. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology 2011;140:286–296. [DOI] [PubMed] [Google Scholar]
- 16.Golebiowski F, Matic I, Tatham MH, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2009;2:ra24. [DOI] [PubMed] [Google Scholar]
- 17.Yang W, Sheng H, Warner DS, et al. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab 2008;28:269–279. [DOI] [PubMed] [Google Scholar]
- 18.Nunez-O’Mara A, Berra E. Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biological chemistry 2013;394:459–469. [DOI] [PubMed] [Google Scholar]
- 19.Yang W, Sheng H, Warner DS, et al. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab 2008;28:892–896. [DOI] [PubMed] [Google Scholar]
- 20.Evdokimov E, Sharma P, Lockett SJ, et al. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci 2008;121:4106–4113. [DOI] [PubMed] [Google Scholar]
- 21.Zhang FP, Mikkonen L, Toppari J, et al. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol 2008;28:5381–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Wansleeben C, Zhao S, et al. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep 2014;15:878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YJ, Mou Y, Maric D, et al. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS One 2011;6:e25852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Napolitano LM, Koruda MJ, Meyer AA, et al. The impact of femur fracture with associated soft tissue injury on immune function and intestinal permeability. Shock 1996;5:202–207. [DOI] [PubMed] [Google Scholar]
- 25.Flint N, Cove FL, Evans GS. A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. Biochem J 1991;280 ( Pt 2):331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Ma Q, Yang W, et al. Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J Neurochem 2012;123:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu CJ, McArdle AH, Brown R, et al. Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal. Arch Surg 1970;101:478–483. [DOI] [PubMed] [Google Scholar]
- 28.Weiss SJ, Slivka A. Monocyte and granulocyte-mediated tumor cell destruction. A role for the hydrogen peroxide-myeloperoxidase-chloride system. J Clin Invest 1982;69:255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Sheng H, Thompson JW, et al. Small ubiquitin-like modifier 3-modified proteome regulated by brain ischemia in novel small ubiquitin-like modifier transgenic mice: putative protective proteins/pathways. Stroke 2014;45:1115–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agbor TA, Cheong A, Comerford KM, et al. Small ubiquitin-related modifier (SUMO)-1 promotes glycolysis in hypoxia. J Biol Chem 2011;286:4718–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comerford KM, Leonard MO, Karhausen J, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A 2003;100:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carbia-Nagashima A, Gerez J, Perez-Castro C, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 2007;131:309–323. [DOI] [PubMed] [Google Scholar]
- 33.Kunz K, Wagner K, Mendler L, et al. SUMO Signaling by Hypoxic Inactivation of SUMO-Specific Isopeptidases. Cell Rep 2016;16:3075–3086. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh YL, Kuo HY, Chang CC, et al. Ubc9 acetylation modulates distinct SUMO target modification and hypoxia response. EMBO J 2013;32:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua G, Ganti KP, Chambon P. Glucocorticoid-induced tethered transrepression requires SUMOylation of GR and formation of a SUMO-SMRT/NCoR1-HDAC3 repressing complex. Proc Natl Acad Sci U S A 2016;113:E635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decque A, Joffre O, Magalhaes JG, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol 2016;17:140–149. [DOI] [PubMed] [Google Scholar]
- 37.Jennewein C, Kuhn AM, Schmidt MV, et al. Sumoylation of peroxisome proliferator-activated receptor gamma by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of proinflammatory cytokines. J Immunol 2008;181:5646–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuro-Gyina PK, Parvin JD. Roles for SUMO in pre-mRNA processing. Wiley Interdiscip Rev RNA 2016;7:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritah S, Lhocine N, Golebiowski F, et al. Sumoylation controls host antibacterial response to the gut invasive pathogen Shigella flexneri. EMBO Rep 2014;15:965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeung TM, Chia LA, Kosinski CM, et al. Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell Mol Life Sci 2011;68:2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turan A, Mahmood A. The profile of antioxidant systems and lipid peroxidation across the crypt-villus axis in rat intestine. Dig Dis Sci 2007;52:1840–1844. [DOI] [PubMed] [Google Scholar]
- 42.Guo C, Wei Q, Su Y, et al. SUMOylation occurs in acute kidney injury and plays a cytoprotective role. Biochim Biophys Acta 2015;1852:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu Y, Lambert JP, Nicholson CK, et al. DJ-1 protects the heart against ischemia-reperfusion injury by regulating mitochondrial fission. J Mol Cell Cardiol 2016;97:56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chymkowitch P, Nguea PA, Enserink JM. SUMO-regulated transcription: challenging the dogma. Bioessays 2015;37:1095–1105. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Liu X, Sheng H, et al. Neuron-specific SUMO knockdown suppresses global gene expression response and worsens functional outcome after transient forebrain ischemia in mice. Neuroscience 2017;343:190–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massberg S, Enders G, Leiderer R, et al. Platelet-endothelial cell interactions during ischemia/reperfusion: the role of P-selectin. Blood 1998;92:507–515. [PubMed] [Google Scholar]
- 47.Russell J, Epstein CJ, Grisham MB, et al. Regulation of E-selectin expression in postischemic intestinal microvasculature. Am J Physiol Gastrointest Liver Physiol 2000;278:G878–885. [DOI] [PubMed] [Google Scholar]
- 48.Mersmann J, Berkels R, Zacharowski P, et al. Preconditioning by toll-like receptor 2 agonist Pam3CSK4 reduces CXCL1-dependent leukocyte recruitment in murine myocardial ischemia/reperfusion injury. Crit Care Med 2010;38:903–909. [DOI] [PubMed] [Google Scholar]
- 49.De Filippo K, Dudeck A, Hasenberg M, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood 2013;121:4930–4937. [DOI] [PubMed] [Google Scholar]
- 50.Li W, Hsiao HM, Higashikubo R, et al. Heart-resident CCR2+ macrophages promote neutrophil extravasation through TLR9/MyD88/CXCL5 signaling. JCI Insight 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fielding CA, McLoughlin RM, McLeod L, et al. IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J Immunol 2008;181:2189–2195. [DOI] [PubMed] [Google Scholar]
- 52.Ferretti S, Bonneau O, Dubois GR, et al. IL-17, produced by lymphocytes and neutrophils, is necessary for lipopolysaccharide-induced airway neutrophilia: IL-15 as a possible trigger. J Immunol 2003;170:2106–2112. [DOI] [PubMed] [Google Scholar]
- 53.Daly MC, Atkinson SJ, Varisco BM, et al. Role of matrix metalloproteinase-8 as a mediator of injury in intestinal ischemia and reperfusion. FASEB J 2016;30:3453–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ungureanu D, Vanhatupa S, Kotaja N, et al. PIAS proteins promote SUMO-1 conjugation to STAT1. Blood 2003;102:3311–3313. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Z, Wang M, Li J, et al. SUMOylation and SENP3 regulate STAT3 activation in head and neck cancer. Oncogene 2016;35:5826–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pascual G, Fong AL, Ogawa S, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature 2005;437:759–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B, Yang Y, Chernishof V, et al. Proinflammatory stimuli induce IKKalphamediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell 2007;129:903–914. [DOI] [PubMed] [Google Scholar]
- 58.Kim TH, Hwang SB, Jeong PY, et al. Requirement of tyrosylprotein sulfotransferase-A for proper cuticle formation in the nematode C. elegans. FEBS Lett 2005;579:53–58. [DOI] [PubMed] [Google Scholar]
- 59.Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology 2010;129:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xin L, Jiang TT, Chaturvedi V, et al. Commensal microbes drive intestinal inflammation by IL-17-producing CD4+ T cells through ICOSL and OX40L costimulation in the absence of B7–1 and B7–2. Proc Natl Acad Sci U S A 2014;111:10672–10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee JW, Wang P, Kattah MG, et al. Differential regulation of chemokines by IL-17 in colonic epithelial cells. J Immunol 2008;181:6536–6545. [DOI] [PubMed] [Google Scholar]
- 62.Myles IA, Fontecilla NM, Valdez PA, et al. Signaling via the IL-20 receptor inhibits cutaneous production of IL-1beta and IL-17A to promote infection with methicillin-resistant Staphylococcus aureus. Nat Immunol 2013;14:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lacotte S, Brun S, Muller S, et al. CXCR3, inflammation, and autoimmune diseases. Ann N Y Acad Sci 2009;1173:310–317. [DOI] [PubMed] [Google Scholar]
- 64.Iwasaki Y, Fujio K, Okamura T, et al. Interleukin-27 in T cell immunity. Int J Mol Sci 2015;16:2851–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ito T, Carson WFt, Cavassani KA, et al. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res 2011;317:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hase K, Murakami T, Takatsu H, et al. The membrane-bound chemokine CXCL16 expressed on follicle-associated epithelium and M cells mediates lympho-epithelial interaction in GALT. J Immunol 2006;176:43–51. [DOI] [PubMed] [Google Scholar]
- 67.Murooka TT, Rahbar R, Platanias LC, et al. CCL5-mediated T-cell chemotaxis involves the initiation of mRNA translation through mTOR/4E-BP1. Blood 2008;111:4892–4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Rabb H, Womer KL. Ischemia-reperfusion and immediate T cell responses. Cell Immunol 2007;248:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ding X, Wang A, Ma X, et al. Protein SUMOylation Is Required for Regulatory T Cell Expansion and Function. Cell Rep 2016;16:1055–1066. [DOI] [PubMed] [Google Scholar]
- 70.Datwyler AL, Lattig-Tunnemann G, Yang W, et al. SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab 2011;31:2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tilemann L, Lee A, Ishikawa K, et al. SUMO-1 gene transfer improves cardiac function in a large-animal model of heart failure. Sci Transl Med 2013;5:211ra159. [DOI] [PubMed] [Google Scholar]
- 72.Bernstock JD, Lee YJ, Peruzzotti-Jametti L, et al. A novel quantitative high-throughput screen identifies drugs that both activate SUMO conjugation via the inhibition of microRNAs 182 and 183 and facilitate neuroprotection in a model of oxygen and glucose deprivation. J Cereb Blood Flow Metab 2016;36:426–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W, Wang L, Paschen W. Development of a high-throughput screening assay for inhibitors of small ubiquitin-like modifier proteases. J Biomol Screen 2013;18:621–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang W, Sheng H, Wang H. Targeting the SUMO pathway for neuroprotection in brain ischaemia. Stroke and Vascular Neurology 2016;1:101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernstock JD, Ye D, Smith JA, et al. Quantitative high-throughput screening identifies cytoprotective molecules that enhance SUMO conjugation via the inhibition of SUMO-specific protease (SENP)2. FASEB J 2017;Epub ahead of print doi: 10.1096/fj.201700711R. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.