Abstract
Background
Tibetan pigs, which inhabit the Tibetan Plateau, exhibit distinct phenotypic and physiological characteristics from those of lowland pigs and have adapted well to the extreme conditions at high altitude. However, the genetic and epigenetic mechanisms of hypoxic adaptation in animals remain unclear.
Methods
Whole-genome DNA methylation data were generated for heart tissues of Tibetan pigs grown in the highland (TH, n = 4) and lowland (TL, n = 4), as well as Yorkshire pigs grown in the highland (YH, n = 4) and lowland (YL, n = 4), using methylated DNA immunoprecipitation sequencing.
Results
We obtained 480 million reads and detected 280679, 287224, 259066, and 332078 methylation enrichment peaks in TH, YH, TL, and YL, respectively. Pairwise TH vs. YH, TL vs. YL, TH vs. TL, and YH vs. YL comparisons revealed 6829, 11997, 2828, and 1286 differentially methylated regions (DMRs), respectively. These DMRs contained 384, 619, 192, and 92 differentially methylated genes (DMGs), respectively. DMGs that were enriched in the hypoxia-inducible factor 1 signaling pathway and pathways involved in cancer and hypoxia-related processes were considered to be important candidate genes for high-altitude adaptation in Tibetan pigs.
Conclusions
This study elucidates the molecular and epigenetic mechanisms involved in hypoxic adaptation in pigs and may help further understand human hypoxia-related diseases.
Electronic supplementary material
The online version of this article (10.1186/s40104-019-0316-y) contains supplementary material, which is available to authorized users.
Keywords: DNA methylation, Hypoxic adaptation, MeDIP-seq, Tibetan pig
Background
Epigenetic modification, including DNA methylation and histone modification, results in long-term regulations of gene expression [1]. In mammals, DNA methylation predominantly occurs at the C-5 position of cytosine in CpG dinucleotides, which are sparsely distributed in the genome, with the exception of short genomic regions called CpG islands (CGIs) [2, 3]. CpG methylation stabilizes the chromatin structure and may regulate the accessibility of these DNA regions for the transcriptional machinery [4]. Short-term severe hypoxia results in long-lasting changes in the genome-wide DNA methylation status, some of which may be highly correlated with the transcriptional modulation of several genes involved in functional pathways [5]. DNA methylation also plays an important role in regulating the transcription of hypoxia-response genes [6]. For example, DNA methylation in the promoter region of the estrogen receptor gene has been shown to increase under conditions of chronic hypoxia in the uterine arteries of pregnant sheep, thereby inhibiting gene expression [7]. Another previous study revealed significant genome-wide epigenetic differences between Ethiopians living at high and low altitudes [8]. Therefore, DNA methylation, an important epigenetic mechanism, may allow for the comprehensive profiling of hypoxic responses and adaptation in animals. Furthermore, as hypoxic stress is known to influence gene expression via the modification of DNA methylation, which has been studied with regard to tumor suppressor genes and cancer progression [9]; hypoxia has been shown to promote DNA hypermethylation in tumors [10] and alter gene expression in osteoblasts [11].
An important technical advance for analyzing genome-wide DNA methylation includes immunoprecipitation, which is used to enrich the genome portion containing either 5-methylcytosine or 5-hydroxymethylcytosine, depending on the antibody used, followed by high-throughput sequencing [12–14]. The methylated DNA immunoprecipitation (MeDIP)-based approach results in the enrichment of methylated DNA regions and can be combined with gene-by-gene polymerase chain reaction (PCR) detection and tiling microarrays [15–17]. MeDIP, in conjunction with high-throughput sequencing (MeDIP-seq), provides a genome-wide coverage technique that has successfully been used to profile global DNA methylation patterns in mammalian genomes [13] and in several tissues, including human breast cancer cells [18], peripheral blood mononucleocytes [19], and malignant nerve tumors [20].
Tibetan pigs inhabit the Tibetan Plateau and have a well-developed heart, which is better adapted to hypoxia than that of lowland pigs [21, 22]. A number of rapidly evolving and positively selected genes have been reported for Tibetan pigs based on high-throughput sequencing [23–27], and several important candidate genes for high-altitude adaptation, including genes involved in angiogenesis, ATP binding, and glucose metabolic processes as well as important pathways such as the hypoxia-inducible factor 1 (HIF-1) signaling pathway, have been revealed by comparative transcriptomic and proteomic analyses of Tibetan pig heart tissues [28]. However, the mechanisms involved in the regulation of gene expression at high altitude remain unclear. In the present study, we used MeDIP-seq to examine the genome-wide DNA methylation landscape in the heart tissues of Tibetan and Yorkshire pigs grown in high- and lowland areas to evaluate whether DNA methylation regulates the expression of certain key genes, potentially involved in high-altitude adaptation. Our results may further elucidate the regulation of genes involved in adaptation to high altitude in other species.
Methods
Sample preparation and DNA isolation
Castrated boars from four groups, which were conceived, born, and raised in either highland (Linzhi, 3000 m) or lowland (Beijing, 100 m) areas, including Tibetan highland pigs (TH, n = 9), Yorkshire highland pigs (YH, n = 9), Tibetan lowland pigs (TL, n = 9), and Yorkshire lowland pigs (YL, n = 9), were slaughtered and sampled at six months of age. The immigrant groups (YH and TL) descended from populations that had migrated to their raising places approximately three years earlier and had been bred for one generation. Prior to the experiment, all pigs were vaccinated and regularly dewormed during feeding. The feeding method included free access to drink water and feed. The same feeding method was maintained for different pigs at the same elevation. DNA was isolated from heart tissue samples using the TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China).
Library construction and sequencing
DNA (1 μg) was sonicated using a Covaris sonication system (Covaris, Woburn, MA, USA) according to previously established parameters [19] to obtain approximately 250-bp fragments. To end-repair DNA fragments, “adenine” nucleotides were added to the 3′ end and ligated to Agencourt® AMPure XP beads (Beckman Coulter, Brea, CA, USA). The Magnetic Methylated DNA Immunoprecipitation Kit (Diagenode, Liège, Belgium) was used to perform the methylation analysis by MeDIP. DNA enriched by MeDIP was amplified by PCR using the TruSeq DNA sample preparation kit and PCR Prep Box (Illumina, San Diego, CA, USA). Clustering of the index-coded samples was performed on a cBot Cluster Generation System using the cBot user guide (Illumina). After cluster generation, the libraries were sequenced using a HiSeq 2500 platform (Illumina), and 50 bp long single-end reads were generated. Four biological replicates were included in each pig group examined.
MeDIP-seq data analysis
Raw reads obtained by MeDIP-seq were preprocessed using FASTX (version 0.0.13) (http://hannonlab.cshl.edu/fastx_toolkit/index.html) by removing reads containing adapters, low-quality reads, and reads shorter than 20 bp. The remaining (clean reads were then mapped to the pig reference genome of Sus scrofa 10.2.69 (ftp://ftp.ensembl.org/pub/release-69/fasta/sus_scrofa/dna/Sus_scrofa.Sscrofa10.2.69.dna.toplevel.fa.gz) using Bowtie (version 0.12.8). Methylation enrichment peaks were counted (DiffScore ≥50) using MACS (version 1.4) [29]. The MEDIPS package [30] was used to identify differentially-methylated regions [DMRs; window size = 500 bp, fold change ≥1.33 or ≤ 0.75, P ≤ 0.001]. Differentially methylated genes (DMGs) within DMRs were annotated using the UCSC database (https://genome.ucsc.edu/). According to the positions of the DMRs, we annotated related genomic elements as follows: promoter, 5′- untranslated region (UTR), coding sequence (CDS), 3′-UTR, exon, intron, and transcription termination region (TTR). The promoter region was defined as the 5000 bp sequence upstream of the transcription initiation site, and TTR was defined as the 5000 bp downstream of the transcription termination site. The CGI regions included “shores” (2000 bp flanking the CGIs) and “shelves” (2000 bp beyond CpG shores), while “open sea” regions were located outside CGIs. DMGs were classified into functional categories using Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations using DAVID (https://david.ncifcrf.gov/home.jsp) [31].
Bisulfite sequencing PCR to validate MeDIP-seq
Approximately 2 μg of DNA from each sample was subjected to bisulfite conversion with the EpiTect Bisulfite Kit (Qiagen, Frankfurt, Germany) according to the manufacturer’s protocol. Using MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi), we designed five pairs of bisulfite sequencing PCR (BSP) primers for five DMGs, including the branched-chain keto acid dehydrogenase E1 subunit beta (BCKDHB), epoxide hydrolase 2 (EPHX2), glutamic-oxaloacetic transaminase 2 (GOT2), retinoid X receptor gamma (RXRG), and ubiquitin D (UBD) genes, to confirm the results of MeDIP-seq (Additional file 1). All PCR products were gel-purified using a gel purification kit (BioTeke Corporation, Beijing, China), then ligated into the pMD18-T vector (TaKaRa, Kusatsu, Shiga, Japan), and transformed into Escherichia coli DH5α competent cells (Beijing Biotech Co., Ltd., Beijing, China). Eight to twelve monoclones from each individual were sequenced by Sangon Biotech Co., Ltd. (China). The acquired sequences were processed using the online tool QUMA (QUantification tool for Methylation Analysis) [32]. Samples from five individuals (differing from the four individuals whose samples were used for MeDIP-seq) from each group were used for BSP. Differences in DNA methylation for each gene were analyzed using Fisher’s exact test, and differences were considered statistically significant at P < 0.05.
Quantitative real-time PCR
RNA extraction and cDNA synthesis were performed using the RNAprep Pure Kit (for tissue) and FastKing RT Kit (Tiangen Biotech). The hypoxanthine phosphoribosyltransferase (HPRT) gene was used as an internal control. The primers for HPRT and the five DMGs (BCKDHB, EPHX2, GOT2, RXRG, and UBD) are listed in Additional file 2. Quantitative real-time PCR (qPCR) was performed using SuperReal PreMix Plus (Tiangen Biotech) and a CFX96 real-time system (Bio-Rad, CA, USA). A cDNA pool of all samples was used for calibration, and three replications of each sample were performed. Samples from eight individuals from each of the four groups were used for qPCR. Gene expression levels were calculated using the 2−ΔΔCt method [33] and analyzed with one-way analysis of variance using SAS (version 9.1) (SAS Institute, Inc., Cary, NC, USA). Graphs were prepared using SigmaPlot (version 10.0) (Systat Software, San Jose, CA, USA), and data are presented as the mean ± standard error of the mean.
Results
Summary of MeDIP-seq data
Approximately 480 million reads were generated by MeDIP-seq from 16 heart tissue samples (n = 4 per group) of TH, YH, TL, YL (approximately 30 million reads per individual). Approximately 85% of these reads were mapped to the porcine reference genome, and approximately 55% were uniquely mapped to specific regions (Additional file 3). The MeDIP-seq reads were distributed in all chromosomes (1–18 and X), with many peak shapes in the four groups of pigs. The density of normalized reads mapped to the proximal and distal regions of chromosomes was higher than that of reads mapped to other regions in all individuals. Similar uneven distributions have been observed in previous studies [34].
Methylation peaks
Scanning of the MeDIP-seq reads revealed 280679, 287224, 259066, and 332078 highly methylated regions (peaks) in the TH, YH, TL, and YL samples, respectively (Table 1). The lengths of the peaks were 500–1200 bp and averaged 943–1027 bp in the four groups (Additional file 4: Figure S1). The peak regions covered 11.99%, 12.82%, 10.47%, and 15.06% of the genome in TH, YH, TL, and YL, respectively. Most of the methylated peaks mapped to intergenic regions (236438–302779 peaks), followed by intron regions (13951–18130 peaks), while only 3024–3897, 3129–3934, and 2524–3338 peaks mapped to promoters, exons, and TTRs, respectively (Additional file 4: Figure S2).
Table 1.
Information on methylation peaks in the four groups of pigs
| Samples | Number of peaks | Mean peak length | Median peak length | Total peak length | Peak covered size in genome, % |
|---|---|---|---|---|---|
| TH | 280678 | 1199.23 | 995 | 336598369 | 11.99 |
| YH | 287224 | 1253.43 | 1027 | 360015941 | 12.82 |
| TL | 259066 | 1135.53 | 943 | 294176561 | 10.47 |
| YL | 332078 | 1273.89 | 1045 | 423031970 | 15.06 |
TH Tibetan highland pig, YH Yorkshire highland pig, TL Tibetan lowland pig, YL Yorkshire lowland pig
Differentially-methylated regions among the four groups
Comparison of TH vs. YH and TL vs. YL resulted in 6829 and 11997 DMRs respectively, which revealed the breed effects in highland and lowland environments. Comparison of TH vs. TL and YH vs. YL resulted in 2826 and 1286 DMRs, respectively, which revealed the effects of hypoxia on DNA methylation (Additional file 5). The number of DMRs between the two breeds (TH vs. YH or TL vs. YL) was higher than that between the two elevations (TH vs. TL or YH vs. YL; Table 2). Most DMRs were distributed in intergenic regions, followed by introns, TTRs, promoters, and CDS regions, while 5′- and 3′- UTRs contained few DMRs (Additional file 4: Figure S3). Furthermore, DMRs were mostly in open sea regions, followed by shelf, shore, and CGI regions (Additional file 4: Figure S4). Of the between-breed DMRs, much fewer (39.45% and 20.54% in high- and lowland comparison groups, respectively) were hypermethylated in Tibetan pig (Table 2). In comparison between the highland and lowland pigs of the same breed showed that Tibetan pigs had a high percentage (68.32%) and Yorkshire pigs had a low percentage (45.65%) of hypermethylated DMRs in the highland groups. Overall, the DNA methylation patterns of the two breeds differed in response to high-altitude hypoxia. We annotated 384, 619, 192, and 92 DMGs from DMRs identified by comparing TH vs. YH, TL vs. YL, TH vs. TL, and YH vs. YL, respectively (Table 2 and Additional file 5). Of these, 14 DMGs were common to the four comparisons groups, 18 were common to the TH vs. TL and YH vs. YL, while 183 DMGs were common to the TH vs. YH and TL vs. YL (Additional file 4: Figure S5).
Table 2.
Numbers of DMRs revealed by four pairwise comparisons
| Comparison groups | Total DMRs | Hypermethylated DMRs | Hypomethylated DMRs | DMGs |
|---|---|---|---|---|
| TH vs. YH | 6829 | 2694 (39.45%) | 4135 (60.55%) | 384 |
| TL vs. YL | 11997 | 2464 (20.54%) | 9533 (79.46%) | 619 |
| TH vs. TL | 2828 | 1932 (68.32%) | 896 (31.68%) | 192 |
| YH vs. YL | 1286 | 587 (45.65%) | 699 (54.35%) | 92 |
TH Tibetan highland pig, YH Yorkshire highland pig, TL Tibetan lowland pig, YL Yorkshire lowland pig
DMGs validation and gene expression
Five DMGs, BCKDHB, EPHX2, GOT2, RXRG, and UBD, which all exhibited higher levels of DNA methylation in TH than in YH, were selected to measure methylation at CpG sites, based on bisulfite sequencing and the expression levels detected by real-time fluorescence qPCR (Additional file 4: Figure S6). The methylated peaks and CpG sites evaluated were all within the intronic regions of the five genes. In the bisulfite sequencing experiment, the methylation of BCKDHB, GOT2, RXRG, and UBD was higher in TH than in YH (P < 0.05), while only EPHX2 showed no significant difference in methylation between TH and YH (P > 0.05; Additional file 4: Figure S6 and Table 3). In the qPCR experiment, the gene expression levels of the five DEGs were lower in TH than in YH, which indicated that up-methylation in the intron regions down-regulated the expression of these genes (Additional file 4: Figure S5 and Additional file 6). The concordance between the data obtained by bisulfite sequencing and qPCR indicated that the DMGs identified by MeDIP-seq were credible.
Table 3.
Methylation ratios of CpG sites and Fisher’s exact P values for the five selected genes
| Gene | TH | YH | TL | YL | P-value in TH/YH | P-value in TL/YL | P-value in TH/TL | P-value in YH/YL | |
|---|---|---|---|---|---|---|---|---|---|
| BCKDHB | Methylated | 113 | 97 | 108 | 96 | 0.0029 | 0.0457 | 0.3329 | 1.0000 |
| Unmethylated | 7 | 23 | 12 | 24 | |||||
| Methylated ratio | 0.9417 | 0.8083 | 0.9000 | 0.8000 | |||||
| EPHX2 | Methylated | 133 | 146 | 129 | 148 | 0.3200 | 0.0500 | 0.5200 | 0.9100 |
| Unmethylated | 62 | 54 | 70 | 52 | |||||
| Methylated ratio | 0.6821 | 0.7300 | 0.6482 | 0.7400 | |||||
| GOT2 | Methylated | 104 | 108 | 96 | 96 | 0.0300 | 0.2463 | 1.0000 | 0.5085 |
| Unmethylated | 0 | 6 | 0 | 3 | |||||
| Methylated ratio | 1.0000 | 0.9474 | 1.0000 | 0.9697 | |||||
| RXRG | Methylated | 279 | 295 | 268 | 244 | 0.0160 | 0.7000 | 1.05E-05 | 0.0970 |
| Unmethylated | 27 | 52 | 73 | 62 | |||||
| Methylated ratio | 0.9118 | 0.8501 | 0.7859 | 0.7974 | |||||
| UBD | Methylated | 75 | 66 | 62 | 71 | 0.0483 | 0.1301 | 0.0098 | 0.3677 |
| Unmethylated | 5 | 14 | 16 | 9 | |||||
| Methylated ratio | 0.9375 | 0.8250 | 0.7949 | 0.8875 |
Functional annotation of DMGs
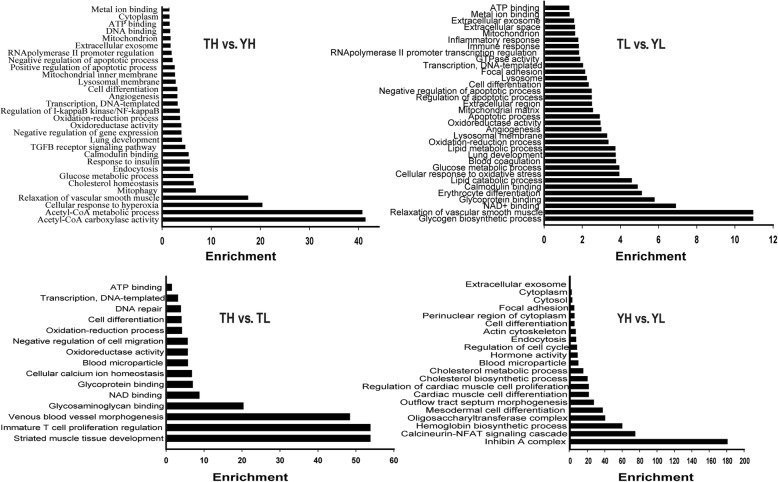
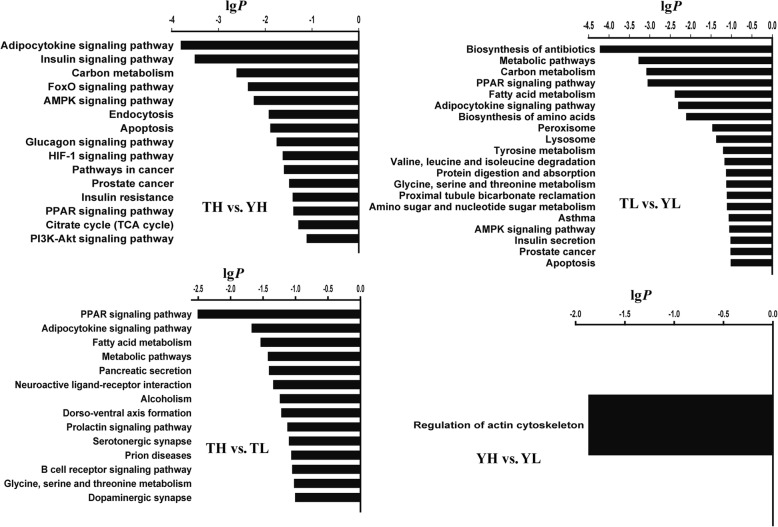
The main GO terms enriched in the 384 DMGs that were identified by TH vs. YH comparison and might be related to hypoxic adaptation were acetyl-CoA carboxylase activity, cellular response to hyperoxia, and relaxation of vascular smooth muscle, while the KEGG pathways included adipocytokine, insulin, forkhead box protein O (FoxO), AMP-activated protein kinase (AMPK) signaling, etc. The main GO terms enriched in the 619 DMGs that were identified by TL vs. YL comparison and that could be related to inherent genetic differences between the two breeds were glycogen biosynthetic process, relaxation of vascular smooth muscle, and NAD+ binding, while the KEGG pathways included biosynthesis of antibiotics and metabolic and peroxisome proliferator-activated receptor (PPAR) signaling pathways. The main GO terms enriched in the 192 DMGs that were identified by TH vs. TL comparison and might be related to hypoxic responses in Tibetan pigs were striated muscle tissue development, venous blood vessel morphogenesis, and glycosaminoglycan binding, while the KEGG pathways included PPAR and adipocytokine signaling and fatty acid metabolism. The main GO terms enriched in the 92 DMGs that were identified by YH vs. YL comparison and that could be related to responses to hypoxia in Yorkshire pigs were inhibin A complex, calcineurin–nuclear factor of activated T cells (NFAT) signaling cascade, and hemoglobin biosynthetic process, while the main KEGG pathway category was regulation of actin cytoskeleton (Additional file 7, Additional file 8, Fig. 1, and Fig. 2).
Fig. 1.
Gene Ontology (GO) terms enriched in DMGs identified by comparison of TH vs. YH, TL vs. YL, TH vs. TL and YH vs. YL. The X-axis indicates GO enrichment, and Y-axis indicates GO terms
Fig. 2.
KyotoEncyclopedia of Genes and Genomes (KEGG) pathways enriched in DMGs identified by comparison of TH vs. YH, TL vs. YL, TH vs. TL and YH vs. YL. The X-axis indicates the pathway terms and Y-axis indicates KEGG pathway enrichment
We observed the overrepresentation of several GO terms and KEGG pathways (common in the TH vs. YH and TL vs. YL), which included angiogenesis (enriched with FGFR2, EPAS1, and ANGPT2), the AMPK signaling pathway (enriched with IGF1R and FOXO1), and cell differentiation (enriched by ERG, GRB2, FOXO1, and ANGPT2). DMGs between TH and YH were also enriched in several interesting pathways, which included HIF-1 signaling (AKT3, ANGPT2, EIF4E2, HK2, ICA, IGF1R, IL6R, and MAP2K1), pathways in cancer (AKT3, EPAS1, FADD, FGFR2, GRB2, KIT, MAP2K1, STAT5B, FOXO1, and SMAD2), phosphoinositide 3-kinase (PI3K/protein kinase B (also known as AKT) signaling (FGFR2, MAP2K1, IL6R, KIT, IGF1R, ANGPT2, AKT3, and EIF4E2), and insulin signaling (MAP2K1, INPPL1, GRB2, HK2, ACACA, FOXO1, ACACB, PCK2, PPP1CB, PRKAR2A, PTPN1, AKT3, and EIF4E2) (Additional file 8). The aforementioned pathways were all connected to the HIF-1 signaling pathway in common (https://www.kegg.jp/pathway/hsa04066) and could be involved in the regulation of oxygen balance in the tissues and cells of animals. Based on the functions of DMGs found among the four groups, 19 DMGs were identified, which were possibly related to high-altitude adaptation in Tibetan pigs (Table 4).
Table 4.
Key differentially methylated genes possibly related to hypoxic adaption in Tibetan pigs
| Gene | Description | TH vs. YH | TL vs. YL | TH vs. TL | YH vs. YL | Functional annotation |
|---|---|---|---|---|---|---|
| AKT3 | AKT serine/threonine kinase 3 | + | – | – | – | HIF-1 signaling pathway, VEGF signaling pathway, pathways in cancer |
| ANGPT2 | Angiopoietin 2 | + | + | + | – | Angiogenesis, HIF-1 signaling pathway |
| CAT | Catalase | + | – | – | – | Oxygen species, apoptosis process, mitochondrion |
| CD38 | Cluster of differentiation 38 | + | + | + | + | Calcium signaling pathway |
| EIF4E2 | Eukaryotic translation initiation factor 4E family member 2 | + | – | – | – | HIF-1 signaling pathway, mTOR signaling pathway, insulin signaling pathway |
| EPAS1 | Endothelial PAS domain protein 1 | + | + | – | – | Angiogenesis, mitochondrion, response to hypoxia |
| ERG | ETS transcription factor-related gene | + | + | + | + | Cell differentiation |
| FADD | Fas associated via death domain | + | – | – | – | Apoptosis, pathways in cancer |
| FGFR2 | Fibroblast growth factor receptor 2 | + | + | – | + | Angiogenesis, Pathways in cancer |
| FOXO1 | Forkhead box O1 | + | + | – | – | Apoptotic process, response to hyperoxia |
| GRB2 | Growth factor receptor bound protein 2 | + | – | – | – | MAPK signaling pathway, insulin signaling pathway, pathways in cancer |
| HK2 | Hexokinase 2 | + | + | – | – | HIF-1 signaling pathway |
| ICA | Porcine inhibitor of carbonic anhydrase | + | + | + | – | HIF-1 signaling pathway |
| IGF1R | Insulin-like growth factor 1 receptor | + | + | – | – | Apoptosis process, HIF-1 signaling pathway |
| IL6R | Interleukin 6 receptor | + | – | – | – | HIF-1 signaling pathway, PI3K-Akt signaling pathway |
| KIT | KIT proto-oncogene receptor tyrosine kinase | + | + | – | – | Inflammatory response, ATP binding, pathways in cancer |
| MAP2K1 | Mitogen-activated protein kinase 1 | + | – | + | – | HIF-1 signaling pathway, VEGF signaling pathway |
| SMAD2 | SMAD family member 2 | + | – | – | – | TGF-beta signaling pathway |
| STAT5B | Signal transducer and activator of transcription 5B | + | – | – | – | Apoptosis process, erythrocyte differentiation, pathways in cancer |
Note: “+” represents differentially methylated genes in some comparisons. “-” represents no differentially methylated genes. TH, Tibetan highland pig; YH, Yorkshire highland pig; TL, Tibetan lowland pig; YL, Yorkshire lowland pig
Discussion
DNA methylation, an epigenetic modification, is an essential regulator of gene expression. Patterns of DNA methylation are determined in early development and are heritable [35, 36]. Although DNA methylation is stably maintained during cell division, it can be dynamically changed in response to the environment [37]. A complete characterization of the methylome and its dynamic changes may serve for an accurate disease prognosis [38]. In the present study, we found that there were differences in DNA methylation between Tibetan and Yorkshire pigs as well as between highland and lowland pigs within the same breed. The patterns of methylation in highland Tibetan pigs may represent regulatory mechanisms for adaptation to high-altitude hypoxia.
Tibetan pigs, which have a long history of living at high altitude and have experienced strong selection, have a well-developed heart, and are well adapted to low oxygen conditions [22]. Immigrant mammals that migrate to high altitudes are exposed to chronic hypoxia, which would result in cardiac hypertrophy and eventual heart failure [39, 40]. We believe that the 384 DMGs identified by comparing TH vs. YH, were of the most interest as potential candidate genes for high-altitude adaptation in Tibetan pigs. Of these 384 DMR, 183 were also identified by comparing TL vs. YL and thus represented stable differences in methylation between the two breeds. However, the 80 DMGs identified by comparing TH vs. TL, could be hypoxia-responsive genes in Tibetan pigs, while the 40 DMGs identified by comparing YH vs. YL could be hypoxia-responsive genes in Yorkshire pigs.
Adaptation to hypoxia is a complex process that involves multiple genes and various pathways [28]. Previous research has demonstrated that intermittent hypoxia may initiate epigenetic changes leading to long-lasting increases in oxidative stress and eventually to manifestations of cardiovascular disease in adult rats, while excessive hypoxic exposure in adult rats may induce the early onset of autonomic dysfunction, caused by DNA hypermethylation [41]. Additionally, DNA methylation has been shown to be involved in hypoxic adaptation in Tibetan chicken embryos [42].
HIF-1, which is a pivotal transcription factor, has numerous target genes that regulate the processes of cell proliferation, angiogenesis, glucose metabolism, and apoptosis in response to hypoxia [43–45]. In this study, eight DMGs, including IGF1R and AKT3, were enriched in the HIF-1 signaling pathway. The IGF1R gene is responsible for cell proliferation and survival and is expressed in many types of cancer cells [46]; enhanced activation of IGF1R is also implicated in the resistance to chemotherapy in a hypoxic microenvironment [47–49]. Previous studies have shown that AKT3 plays an important role in cell proliferation, migration, and survival [50–53] and have revealed that it is also involved in the regulation of estrogen receptor-binding fragment-associated antigen 9 (EBAG9) and vascular endothelial growth factor (VEGF) secretion in ovarian cancer cells [54]. The AKT3 expression could prevent the impaired angiogenesis in endothelial progenitor cells [55], which suggested that AKT3 played a functional role in promoting angiogenesis and vascular permeability by regulating VEGF [56]. Therefore, the high levels of expression and low levels of methylation of IGF1R and AKT3 in the Tibetan pig may trigger changes in their expression, potentially leading to adaptation to hypoxic environments.
Some other noteworthy DMGs, including EPAS1, FGFR2, FOXO1, and SMAD2, were found in this study. A genome-wide scanning revealed that EPAS1 and the other genes had strong selection signals in Tibetans [57–62]. Depletion of FGFR2 in cancer cells attenuated the hypoxia-mediated cell invasion [63], while forkhead box O1 (FOXO1) expression increased in smooth muscle cells and cardiomyocytes under hypoxia [64, 65].
Conclusions
Our findings provide a comprehensive, detailed picture of DNA methylation patterns and distribution under hypoxic adaptation. The MeDIP-seq data identified, with sufficient depth and high resolution, genome- wide spanned DMRs, suggesting that this technique represents an effective approach for DNA methylome analysis. The genes modified by DNA methylation for adaptation to high-altitude in Tibetan pig may be involved in the HIF-1, insulin, and VEGF signaling pathways. We identified 19 DMGs that are potentially related to hypoxic adaptation in Tibetan pig. The present study provides new insights into hypoxic adaptation and its relationship to DNA methylation.
Additional files
Primers for bisulfite sequencing PCR. (XLSX 11 kb)
Primers for qPCR. (XLSX 11 kb)
Summary of sequencing reads obtained by MeDIP-seq. (XLSX 11 kb)
Figure S1. Distribution of peaks varied with peak length in the four groups of pigs. Figure S2. Distribution of peaks in different gene elements in the four comparsion groups. The X-axis indicates different gene elements, and the Y-axis indicates the number of peaks. Figure S3. Distribution of DMRs in different gene elements in the four groups of pigs. Figure S4. Distribution of DMRs in CpG island, open sea, shelf, and shore regions. Figure S5. Venn diagram of differentially methylated genes (DMGs) among the four comparison groups. TH, Tibetan highland pig; TL, Tibetan lowland pig; YH, Yorkshire highland pig; YL, Yorkshire lowland pig. Figure S6. Validation of differentially methylated or differentially expressed genes by bisulfite sequencing and qPCR, respectively. (a–e) Methylation of CpG dinucleotide in BCKDHB, EPHX2, GOT2, RXRG, and UBD. (f) qPCR results for the five DMGs. (PDF 1380 kb)
DMRs and DMGs in the four groups of pigs. (XLSX 3191 kb)
qPCR and MeDIP-seq results. (XLSX 9 kb)
GO functional analysis of DMGs in the four groups of pigs. (XLSX 24 kb)
KEGG pathway analysis of DMGs in the four groups of pigs. (XLSX 13 kb)
Acknowledgments
We would like to thank Ying Zhang, Liyuan Wang, Yu Fu, and Xuyuan Zhang for assistance with sample collection.
Funding
This study was supported by the National Natural Science Foundation of China (No. 31560615), the National Key Technology Research and Development Program (No. 2015BAD03B02), the Program for Changjiang Scholars and Innovation Research Team in University (No. IRT_15R62), and the Innovation Base Cultivation and Development Project (No. Z171100002217072).
Availability of data and materials
The datasets supporting the conclusions of this article (raw datasets of MeDIP-seq) are available at the National Center for Biotechnology Information under accession No. GSE114779 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114779).
Abbreviations
- AKT
Protein kinase B
- AKT3
AKT serine/threonine kinase 3
- AMPK
AMP-activated protein kinase
- ANGPT2
Angiopoietin 2
- BCKDHB
Branched-chain keto acid dehydrogenase E1 subunit beta
- BSP
Bisulfite sequencing polymerase chain reaction
- CAT
Catalase
- CD38
Cluster of differentiation 38
- CDS
Coding sequence
- CGI
CpG island
- DMG
Differentially methylated gene
- DMR
Differentially methylated region
- EBAG9
Estrogen receptor-binding fragment-associated antigen 9
- EIF4E2
Eukaryotic translation initiation factor 4E family member 2
- EPAS1
Endothelial PAS domain protein 1
- EPHX2
Epoxide hydrolase 2
- ERG
ETS transcription factor-related gene
- FADD
Fas associated via death domain
- FGFR2
Fibroblast growth factor receptor 2
- FOXO1
Forkhead box O1
- GO
Gene Ontology
- GOT2
Glutamic-oxaloacetic transaminase 2
- GRB2
Growth factor receptor bound protein 2
- HIF-1
Hypoxia-inducible factor 1
- HK2
Hexokinase 2
- HPRT
Hypoxanthine phosphoribosyltransferase
- ICA
Inhibitor of carbonic anhydrase
- IGF1R
Insulin-like growth factor 1 receptor
- IL6R
Interleukin-6 receptor
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KIT
KIT proto-oncogene receptor tyrosine kinase
- MAP2K1
Mitogen-activated protein kinase 1
- MeDIP-seq
Methylated DNA immunoprecipitation sequencing
- NFAT
Nuclear factor of activated T cells
- PI3K
Phosphoinositide 3-kinase
- PPAR
Peroxisome proliferator-activated receptor
- qPCR
Real-time fluorescence quantitative polymerase chain reaction
- RXRG
Retinoid X receptor gamma
- SMAD2
SMAD family member 2
- STAT5B
Signal transducer and activator of transcription 5B
- TH
Tibetan highland pig
- TL
Tibetan lowland pig
- TTR
Transcriptional termination region
- UBD
Ubiquitin D
- UTR
Untranslated region
- VEGF
Vascular endothelial growth factor
- YH
Yorkshire highland pig
- YL
Yorkshire lowland pig
Authors’ contributions
HZ conceived and designed the experiments. BZ and DB performed the experiments and analyzed the data. BZ, DB, YC, XG, YZ, and LY contributed reagents/materials/analysis tools. BZ wrote the paper. HZ, YC, XG, and BZ revised the paper. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The experimental procedures were approved by the animal welfare committee of the State Key Laboratory for Agro Biotechnology of China Agricultural University (Approval number XK257). Pig farming is permitted in Linzhi in Tibet, and the field study did not involve endangered or protected species.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Bo Zhang, Email: bozhang06@126.com.
Dongmei Ban, Email: 1903156665@qq.com.
Xiao Gou, Email: gouxiaosa@163.com.
Yawen Zhang, Email: zhuantouliuzi@163.com.
Lin Yang, Email: 18811073189@163.com.
Yangzom Chamba, Email: qbyz628@126.com.
Hao Zhang, Email: zhanghao827@163.com.
References
- 1.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki MM. Bird a. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 3.Mason K, Liu Z, Aguirre-Lavin T, Beaujean N. Chromatin and epigenetic modifications during early mammalian development. Anim Reprod Sci. 2012;134(1–2):45–55. doi: 10.1016/j.anireprosci.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Susanj C. John M. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21(35):5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 5.Hartley I, Elkhoury FF, Shin JH, Xie B, Gu X, Gao Y, et al. Long-lasting changes in DNA methylation following short-term hypoxic exposure in primary hippocampal neuronal cultures. PLoS One. 2013;8(10):e77859. doi: 10.1371/journal.pone.0077859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson JA, Watson CJ, McCann A, Baugh J. Epigenetics, the epicenter of the hypoxic response. Epigenetics. 2010;5:293–296. doi: 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- 7.Dasgupta C, Chen M, Zhang H, Yang S, Zhang L. Chronic hypoxia during gestation causes epigenetic repression of the estrogen receptor-alpha gene in ovine uterine arteries via heightened promoter methylation. Hypertension. 2012;60:697–704. doi: 10.1161/HYPERTENSIONAHA.112.198242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alkorta-Aranburu G, Beall CM, Witonsky DB, Gebremedhin A, Pritchard JK, Di Rienzo A. The genetic architecture of adaptations to high altitude in Ethiopia. PLOS Genet. 2012;8:e1003110. doi: 10.1371/journal.pgen.1003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31(16):3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thienpont B, Steinbacher J, Zhao H, D’Anna F, Kuchnio A, Ploumakis A, et al. Tumor hypoxia causes DNA hypermethylation by reducing TET activity. Nature. 2016;537(7618):63. doi: 10.1038/nature19081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peter VN, Janja M, Barbara O. Hypoxia mimetic deferoxamine influences the expression of histone acetylation- and DNA methylation-associated genes in osteoblasts. Connect Tissue Res. 2015;56(3):1–24. doi: 10.3109/03008207.2015.1017573. [DOI] [PubMed] [Google Scholar]
- 12.Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. BioTechniques. 2008;44(1):35. doi: 10.2144/000112708. [DOI] [PubMed] [Google Scholar]
- 13.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hume S, Feng S, Shannon MK, Sriharsa P, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12(6):1–8. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komashko VM, Acevedo LG, Squazzo SL, Iyengar SS, Rabinovich A, O'Geen H, et al. Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 2008;18(4):521. doi: 10.1101/gr.074609.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pomraning K, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47(3):142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 17.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18(5):780. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G. Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics. 2010;11(1):137. doi: 10.1186/1471-2164-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Ye M, Li Y, Yan Z, Butcher LM, Sun J, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52(3):203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Feber A, Wilson GA, Zhang L, Presneau N, Idowu B. Down TA, et al. comparative methylome analysis of benign and malignant peripheral nerve sheath tumors. Genome Res. 2011;21(4):515–524. doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Qiangba Y, Shang P, Wang Z, Ma J, Wang L, et al. A comprehensive MicroRNA expression profile related to hypoxia adaptation in the Tibetan pig. PLoS One. 2015;10(11):e143260. doi: 10.1371/journal.pone.0143260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ai H, Yang B, Li J, Xie X, Chen H, Ren J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genomics. 2014;15:834. doi: 10.1186/1471-2164-15-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 2013;45(12):1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Chen L, Long KR. Mu ZP. Hypoxia-related gene expression in porcine skeletal muscle tissues at different altitude. Genet Mol Res. 2015;14(3):11587–11593. doi: 10.4238/2015.September.28.10. [DOI] [PubMed] [Google Scholar]
- 25.Jia C, Kong X, Koltes JE, Gou X, Yang S, Yan D, et al. Gene co-expression network analysis unraveling transcriptional regulation of high-altitude adaptation of Tibetan pig. PLoS One. 2016;11(12):e168161. doi: 10.1371/journal.pone.0168161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Q, Gu Y, Zhou X, Jin L, Guan J, Liu R, et al. Comparative transcriptomics of 5 high-altitude vertebrates and their low-altitude relatives. GigaScience. 2017;6(12):1–9. doi: 10.1093/gigascience/gix105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin L, Mao K, Li J, Huang W, Che T, Fu Y, et al. Genome-wide profiling of gene expression and DNA methylation provides insight into low-altitude acclimation in Tibetan pigs. Gene. 2018;642:522–532. doi: 10.1016/j.gene.2017.11.074. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Chamba Y, Peng S, Wang Z, Ma J, Wang L, et al. Comparative transcriptomic and proteomic analyses provide insights into the key genes involved in high-altitude adaptation in the Tibetan pig. Sci Rep. 2017;7(1):3654. doi: 10.1038/s41598-017-03976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Jérôme E, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):1–9. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chavez L, Jozefczuk J, Grimm C, Dietrich J, Timmermann B, Lehrach H, et al. Computational analysis of genome-wide DNA methylation during the differentiation of human embryonic stem cells along the endodermal lineage. Genome Res. 2010;20(10):1441. doi: 10.1101/gr.110114.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36(Web Server issue):170–175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Zou C, Fu Y, Li C, Liu H, Li G, et al. Genome-wide gene expression and DNA methylation differences in abnormally cloned and normally natural mating piglets. Anim Genet. 2016;47(4):436–450. doi: 10.1111/age.12436. [DOI] [PubMed] [Google Scholar]
- 35.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 36.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 37.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 38.Bock C. Epigenetic biomarker development. Epigenomics-Uk. 2009;1(1):99–110. doi: 10.2217/epi.09.6. [DOI] [PubMed] [Google Scholar]
- 39.Penaloza D, Arias-Stella J. The heart and pulmonary circulation at high altitudes: healthy highlanders and chronic mountain sickness. Circulation. 2007;115:1132–1146. doi: 10.1161/CIRCULATIONAHA.106.624544. [DOI] [PubMed] [Google Scholar]
- 40.Lankford HV, Swenson ER. Dialated hearts at high altitude: words from on high. High Alt Med Biol. 2014;15:511–519. doi: 10.1089/ham.2014.1047. [DOI] [PubMed] [Google Scholar]
- 41.Nanduri J, Makarenko V, Reddy VD, Yuan G, Pawar A, Wang N, et al. Epigenetic regulation of hypoxic sensing disrupts cardiorespiratory homeostasis. P Natl Acad Sci USA. 2012;109(7):2515–2520. doi: 10.1073/pnas.1120600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Gou W, Ma J, Zhang H, Zhang Y, Zhang H. Genome methylation and regulatory functions for hypoxic adaptation in Tibetan chicken embryos. Peer J. 2017;5:e3891. doi: 10.7717/peerj.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 44.Xia Y, Choi HK, Lee K. Recent advances in hypoxia-inducible factor (HIF)-1 inhibitors. Eur J Med Chem. 2012;49:24–40. doi: 10.1016/j.ejmech.2012.01.033. [DOI] [PubMed] [Google Scholar]
- 45.Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5(5):378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8(12):915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 47.Eckstein N, Servan K, Hildebrandt B, Pölitz A, Von JG, Wolf-Kümmeth S, et al. Hyperactivation of the insulin-like growth factor receptor I signaling pathway is an essential event for cisplatin resistance of ovarian cancer cells. Cancer Res. 2009;69(7):2996. doi: 10.1158/0008-5472.CAN-08-3153. [DOI] [PubMed] [Google Scholar]
- 48.Jameson MJ, Beckler AD, Taniguchi LE, Allak A, Wagner LBV, Lee NG, et al. Activation of the insulin-like growth factor-1 receptor induces resistance to epidermal growth factor receptor antagonism in head and neck squamous carcinoma cells. Mol Cancer Ther. 2011;10(11):2124. doi: 10.1158/1535-7163.MCT-11-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murakami A, Takahashi F, Nurwidya F, Kobayashi I, Minakata K, Hashimoto M, et al. Hypoxia increases gefitinib-resistant lung cancer stem cells through the activation of insulin-like growth factor 1 receptor. PLoS One. 2014;9(1):e86459. doi: 10.1371/journal.pone.0086459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Littlewood T, Bennett M. Akt isoforms in vascular disease. Vasc Pharmacol. 2015;71:57. doi: 10.1016/j.vph.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin HP, Lin CY, Huo C, Jan YJ, Tseng JC, Jiang SS, et al. AKT3 promotes prostate cancer proliferation cells through regulation of AKT, B-Raf, and TSC1/TSC2. Oncotarget. 2015;6(29):27097–27112. doi: 10.18632/oncotarget.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jie L, Wei H, Ren C, Wen Q, Liu W, Yang X, et al. Flotillin-2 promotes metastasis of nasopharyngeal carcinoma by activating NF-κB and PI3K/Akt3 signaling pathways. Sci Rep. 2015;5:11614. doi: 10.1038/srep11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, Zheng W, Shuai X, Chang RM, Yu L, Fang F, et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6(29):27736–27750. doi: 10.18632/oncotarget.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liby TA, Spyropoulos P, Buff LH, Eldridge J, Beeson C, Hsu T, et al. Akt3 controls vascular endothelial growth factor secretion and angiogenesis in ovarian cancer cells. Int J Cancer. 2012;130(3):532. doi: 10.1002/ijc.26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zheng Y, Xu Z. MicroRNA-22 induces endothelial progenitor cell senescence by targeting AKT3. Cell Physiol Biochem. 2014;34(5):1547–1555. doi: 10.1159/000366358. [DOI] [PubMed] [Google Scholar]
- 56.Qin B, Liu J, Liu S, Li B, Ren J. MiR-20b targets AKT3 and modulates vascular endothelial growth factor-mediated changes in diabetic retinopathy. Acta Bioch Bioph Sin. 2016;48(8):732–740. doi: 10.1093/abbs/gmw065. [DOI] [PubMed] [Google Scholar]
- 57.Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc Natl Acad Sci U S A. 2010;107(25):11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bigham A, Bauchet M, Pinto D, Mao X, Akey JM, Mei R, et al. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 2010;6(9):e1001116. doi: 10.1371/journal.pgen.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329(5987):72. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 60.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329(5987):75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 2011;28(2):1075–1081. doi: 10.1093/molbev/msq290. [DOI] [PubMed] [Google Scholar]
- 62.Xu S, Li S, Yang Y, Tan J, Lou H, Jin W, et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol. 2011;28(2):1003. doi: 10.1093/molbev/msq277. [DOI] [PubMed] [Google Scholar]
- 63.Khurana A, Liu P, Mellone P, Lorenzon L, Vincenzi B, Datta K, et al. HSulf-1 modulates FGF-2 and hypoxia mediated migration and invasion of breast Cancer cells. Cancer Res. 2011;71(6):2152–2161. doi: 10.1158/0008-5472.CAN-10-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Awad H, Nolette N, Hinton M, Dakshinamurti S. AMPK and FoxO1 regulate catalase expression in hypoxic pulmonary arterial smooth muscle. Pediatr Pulm. 2014;49(9):885. doi: 10.1002/ppul.22919. [DOI] [PubMed] [Google Scholar]
- 65.Raeis V, Philip-Couderc P, Roatti A, Habre W, Sierra J, Kalangos A, et al. Central venous hypoxemia is a determinant of human atrial ATP-sensitive potassium channel expression: evidence for a novel hypoxia-inducible factor 1alpha-Forkhead box class O signaling pathway. Hypertension. 2010;55(5):1186–1192. doi: 10.1161/HYPERTENSIONAHA.109.148767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for bisulfite sequencing PCR. (XLSX 11 kb)
Primers for qPCR. (XLSX 11 kb)
Summary of sequencing reads obtained by MeDIP-seq. (XLSX 11 kb)
Figure S1. Distribution of peaks varied with peak length in the four groups of pigs. Figure S2. Distribution of peaks in different gene elements in the four comparsion groups. The X-axis indicates different gene elements, and the Y-axis indicates the number of peaks. Figure S3. Distribution of DMRs in different gene elements in the four groups of pigs. Figure S4. Distribution of DMRs in CpG island, open sea, shelf, and shore regions. Figure S5. Venn diagram of differentially methylated genes (DMGs) among the four comparison groups. TH, Tibetan highland pig; TL, Tibetan lowland pig; YH, Yorkshire highland pig; YL, Yorkshire lowland pig. Figure S6. Validation of differentially methylated or differentially expressed genes by bisulfite sequencing and qPCR, respectively. (a–e) Methylation of CpG dinucleotide in BCKDHB, EPHX2, GOT2, RXRG, and UBD. (f) qPCR results for the five DMGs. (PDF 1380 kb)
DMRs and DMGs in the four groups of pigs. (XLSX 3191 kb)
qPCR and MeDIP-seq results. (XLSX 9 kb)
GO functional analysis of DMGs in the four groups of pigs. (XLSX 24 kb)
KEGG pathway analysis of DMGs in the four groups of pigs. (XLSX 13 kb)
Data Availability Statement
The datasets supporting the conclusions of this article (raw datasets of MeDIP-seq) are available at the National Center for Biotechnology Information under accession No. GSE114779 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE114779).