Significance
Human gut microbes form a complex community with vast biosynthetic potential. Microbial products and metabolites released in the gut impact human health and disease. However, defining causative relationships between specific bacterial products and disease initiation and progression remains an immense challenge. This study advances understanding of the functional capacity of the gut microbiota by determining the presence, concentration, and spatial and temporal variability of two enterotoxic metabolites produced by the gut-resident Klebsiella oxytoca. We present a detailed mode of action for the cytotoxins and recapitulate their functionalities in disease models in vivo. The findings provide distinct molecular mechanisms for the enterotoxicity of the metabolites allowing them to act in tandem to damage the intestinal epithelium and cause colitis.
Keywords: intestinal microbiota, antibiotic-induced diarrhea, DNA damage, tubulin inhibitor, dysbiosis
Abstract
Establishing causal links between bacterial metabolites and human intestinal disease is a significant challenge. This study reveals the molecular basis of antibiotic-associated hemorrhagic colitis (AAHC) caused by intestinal resident Klebsiella oxytoca. Colitogenic strains produce the nonribosomal peptides tilivalline and tilimycin. Here, we verify that these enterotoxins are present in the human intestine during active colitis and determine their concentrations in a murine disease model. Although both toxins share a pyrrolobenzodiazepine structure, they have distinct molecular targets. Tilimycin acts as a genotoxin. Its interaction with DNA activates damage repair mechanisms in cultured cells and causes DNA strand breakage and an increased lesion burden in cecal enterocytes of colonized mice. In contrast, tilivalline binds tubulin and stabilizes microtubules leading to mitotic arrest. To our knowledge, this activity is unique for microbiota-derived metabolites of the human intestine. The capacity of both toxins to induce apoptosis in intestinal epithelial cells—a hallmark feature of AAHC—by independent modes of action, strengthens our proposal that these metabolites act collectively in the pathogenicity of colitis.
The gastrointestinal tract provides an enormous interface to mediate interactions with the resident microbial community (1). Nutrients, metabolites, cellular components, and virulence factors derived from trillions of gut microbes influence human health and disease (2–4). During homeostasis, the stable microbial community also resists invasion of nonnative bacteria and expansion of low-abundance, potentially harmful microbes known as pathobionts (5–7). Disruption of microbial community structure and function through factors such as inflammation, diet, or medication leads to dysbiosis, characterized by decreased diversity, loss of beneficial microbes, and the expansion of pathobionts (7–9). Given the complexity of the gut ecosystem, we currently understand little about conditions and mechanisms enabling commensal microbes to become pathobionts.
Secreted microbial products are likely to perform key functions in the transition from normal to disease-causing activities. The gut microbiota harbors a vast biosynthetic capacity to generate natural products with remarkably diverse chemistries (10–12). These have affinities for equally diverse targets and the potential to mediate both interactions between microbes and the host (1, 13). Identifying causative relationships between microbial products and host phenotypes, however, remains an immense challenge (14). Despite the difficulty of finding specificity in this complex ecosystem, studies of enterotoxin-producing Klebsiella oxytoca have revealed a surprisingly simple model of pathobiont activity.
K. oxytoca is a resident of the human gut, yet in some patients taking penicillin, expansion of this pathobiont results in antibiotic-associated hemorrhagic colitis (AAHC) (15, 16), a right-sided segmental colitis characterized by bloody diarrhea and severe cramps. Colitogenic strains of K. oxytoca carry a secondary metabolite biosynthetic gene cluster that is critical to cause disease in an animal model (17). The cluster encodes a nonribosomal peptide assembly pathway similar to the pyrrolobenzodiazepine (PBD)-producing synthetases of actinomycetes (18). We isolated and determined the structure of tilivalline (TV), the pyrrolo[2,1-c][1,4]benzodiazepine product of K. oxytoca (17). TV induced apoptotic cell death and loss of barrier integrity in polarized human epithelial cells in vitro, suggesting that these activities are key to K. oxytoca pathogenicity in AAHC (17).
Recent elucidation of the biosynthesis of TV, however, revealed that the enterotoxin gene cluster produces three distinct secondary metabolites, two of which exhibit cytotoxicity (19–21). These studies showed that the nonribosomal peptide synthetase (NRPS) platform does not yield TV directly, but instead an N-acylprolinal, which reacts spontaneously to form two further secondary metabolites we named tilimycin (TM) and culdesacin (19). The intrinsic reactivity of TM with indole yields TV. All three substances are stable in vitro, and importantly, although culdesacin has no obvious bioactivity, both TM and TV are cytotoxic to human cells (19, 20).
Insights into the cytotoxic functionalities of these substances in human cells are provided by their structures. TM and TV belong to the PBD family of natural products, which exhibit antibacterial and anticancer activity by alkylating DNA (22). This family of potent cytotoxic agents has been extensively investigated for use in systemic chemotherapy (23, 24). Structure–activity data for PBDs imply that TM will form a similar PBD–DNA adduct (25). By contrast, presence of an indole substituent on the diazepine ring of TV should block this activity. The molecular basis of TV cytotoxicity is thus an open question.
Here, we establish the causal links between K. oxytoca metabolites and disease. We first demonstrate that both cytotoxins are produced in the human body and use a murine model to determine their concentrations during an active phase of AAHC. We identify the different molecular targets of TM and TV and present a detailed mode of action study. Remarkably, the data show that the enterotoxin gene cluster produces distinct DNA-damaging (TM) and microtubule-stabilizing (TV) secondary metabolites. Although the functionalities of the enterotoxins differ, each substance triggered the apoptotic cell death characteristic for the colonic epithelium in AAHC (16, 17). These findings illustrate the versatility of bacterial host interactions mediated by a single secondary metabolite biosynthesis pathway and provide insights into the molecular mechanisms of pathobiont activity.
Results
K. oxytoca Enterotoxins Are Produced in the Human Intestine.
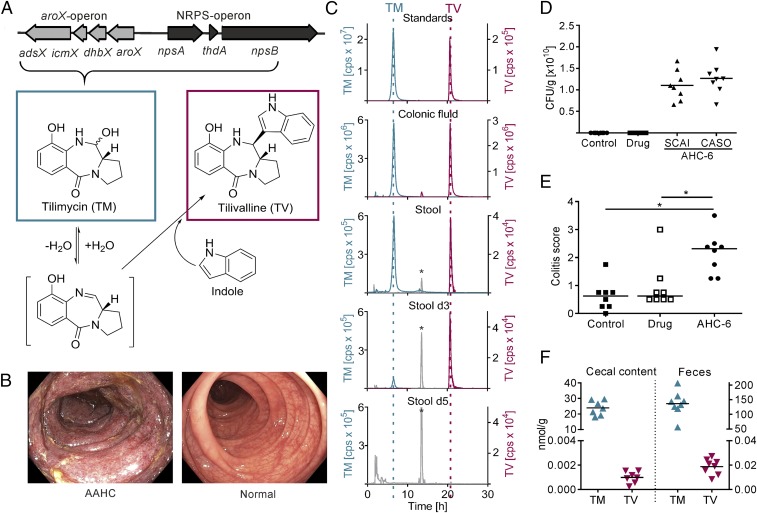
The enterotoxin gene cluster (Fig. 1A) encodes two NRPS modules, NpsA and NpsB, and additional enzymes required to generate an anthranilate precursor (19). Genetic inactivation of the NRPS-operon eliminates cytotoxicity in vitro and pathology in vivo (17). Two end products of this secondary metabolite pathway are cytotoxic: TM and TV (19, 20). In the report of Tse et al. (20), the alternative name, “kleboxymycin,” was proposed for the substance we call TM. TV is formed by the intrinsic reactivity of an imine intermediate of TM with indole. Since stool of healthy humans typically contain millimolar concentrations of indole (26), concomitant production of both cytotoxins TV and TM in the gut is expected. To test this prediction, we analyzed colonic fluid and stool samples of AAHC patients. Presence of both enterotoxins in human samples taken during the active phase of disease was confirmed (Fig. 1 B and C and SI Appendix, Table S1). The triggering antibiotic therapy was terminated at diagnosis since patient conditions improve by halting selective growth of K. oxytoca (16). During the active phase of AAHC, stool of patient A contained both enterotoxins and 107 colony-forming units (cfu) K. oxytoca⋅g−1 in marked contrast to the 102 cfu⋅g−1 feces typically cultured from asymptomatic intestinal carriers of K. oxytoca (27). Follow-up stool samples of this patient also contained TM and TV 3 d later, but after 5 d the metabolites were no longer detectable (Fig. 1C) consistent with reduced abundance of the pathobiont in stool (104 cfu⋅g−1). The incidence of AAHC is very low; thus, the number of patient samples available for analyses is quite limited. Nevertheless, presence of both enterotoxins in human disease as well as the temporal loss and elimination of the substances once therapy is started are compelling observations linking the microbial products to active colitis.
Fig. 1.
Enterotoxins are present in vivo during colitis. (A) K. oxytoca aroX- and NRPS-operons are required to produce tilimycin (TM), which reacts spontaneously with indole to form tilivalline (TV). (B) Endoscopic images of the transverse colons of AAHC patient A with severe edema and a diffuse hemorrhagic mucosa with erosions (Left) and a healthy subject (Right). (C) Representative HPLC-ESMS chromatograms; TM (m/z 235.1004, ±1 ppm) and TV (m/z 334.1477, ±1 ppm) detected in colonic luminal fluid obtained at colonoscopy (patient C) and stool (patient A) during acute AAHC and at day 3 (d3) but not day 5 (d5) after cessation of antibiotics. Ten micromolar TM and 10 nM TV in n-butanol were used as standards. *Peaks at retention time of 13 min (m/z 334.1477, ±1 ppm) are interferences caused by sample matrix and the applied gradient. (D) K. oxytoca colony-forming units per gram of cecal content of mice from control, drug, and K. oxytoca AHC-6 infection group (each n = 8) determined with indicated selection agar as means. (E) Colitis scores of colonized mice compared with controls. Bars indicate medians (n = 8). Kruskal–Wallis test followed by Dunn’s multiple comparison (*P ≤ 0.05). (F) TM and TV concentrations detected in cecal content (n = 7) and feces (n = 8) of K. oxytoca colonized mice. Bars indicate means.
We then asked what concentrations of TM and TV are relevant to disease. Endoscopy performed on AAHC patients is typically preceded by acute diarrhea and colonic lavage; thus, physiologically relevant concentrations of the enterotoxins cannot be determined from the clinical samples. To address this key point quantitatively, we developed analytic methods using a murine disease model. Cecal contents and feces of mice colonized with K. oxytoca AHC-6 were collected during an active phase of AAHC (Fig. 1 D and E). Samples of diseased animals (n = 8) contained TM and TV, but both enterotoxins were absent in control mice (SI Appendix, Fig. S1). TM was more abundant in cecal content (24 ± 4 nmol⋅g−1) and feces (136 ± 40 nmol⋅g−1) of infected mice compared with TV (1 ± 0.4 and 19 ± 6 pmol⋅g−1, respectively) (Fig. 1F). Enterotoxin concentrations were also higher in feces compared with cecal contents. This finding might reflect the consistency of samples (liquid/solid ratio) or, possibly, continued production of toxins during intestinal passage. We conclude that TM and lower amounts of TV are produced in the human and murine intestine. The level of K. oxytoca colonization in experimental animals is much higher than in patients during active AAHC (1010 vs. 107 cfu⋅g−1 stool); thus, we expect that the quantities of K. oxytoca enterotoxins sufficient to cause colitis in patients are lower than the concentrations determined in the murine model.
TM and TV Disrupt Cell Cycle Progression.
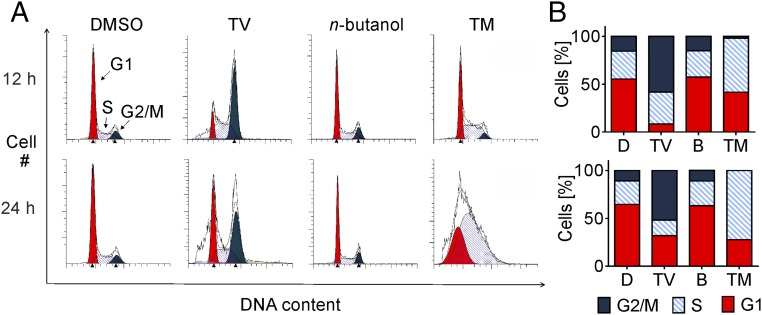
Growth-inhibitory activities of TM and TV were determined in a variety of human tumor cell lines and nontransformed vascular endothelial cells revealing 50% inhibitory concentrations (IC50) in the (sub)micromolar range (SI Appendix, Table S2). In contrast to TV, TM also exhibited antibacterial activity (SI Appendix, Table S3). To gain insights into the cellular processes affected by these generally toxic compounds, we next tested their effects on cell cycle progression. Using flow cytometry, distinct profiles of cell cycle disruption were observed for populations of HeLa cells treated with TV, TM, or solvents. TV treatment of HeLa cells led to an accumulation of cells in the G2/M phase (Fig. 2). TM-treated cells were markedly arrested in G1 or S phase.
Fig. 2.
TV arrests cells at G2/M phase and TM extends S phase. HeLa cells were treated with TV (10 µM), TM (2.5 µM), DMSO (D), or n-butanol (B) for 12 and 24 h. DNA was quantified by flow cytometry of PI-stained cells. (A) One representative cell cycle profile per treatment is shown. (B) Relative proportion of cells in phases are indicated as means (n = 3).
TM Is a DNA-Damaging Agent.
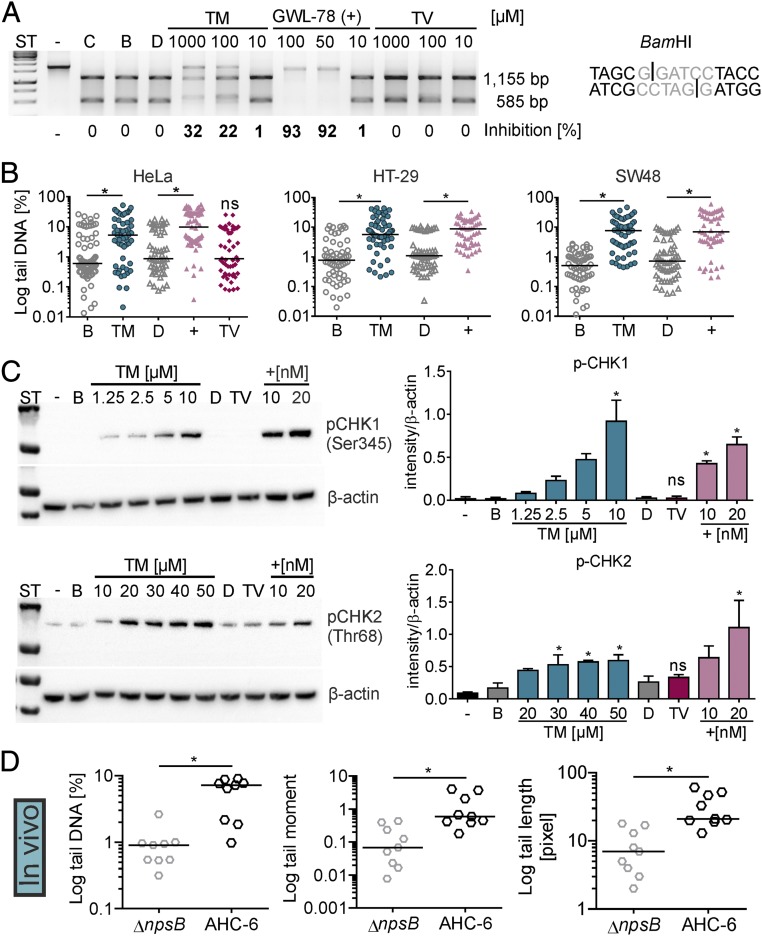
Accumulation of a large fraction of cells at G1/S phase following exposure to TM is consistent with its predicted DNA-alkylating activity. We used biochemical and cellular assays to test this possibility. Structure–activity relationship data have shown that the diazepine ring system of PBDs interacts with the minor groove and stabilizes double-stranded DNA (dsDNA) to thermal denaturation in vitro (25). We determined a 0.5 °C higher melting temperature (Tm) for a dsDNA containing a putative PBD binding site after reaction with an equimolar amount of TM compared with solvent (SI Appendix, Fig. S2A). This value is in good agreement with the 0.7 °C ΔTm measured for the closest structurally related natural product analog DC81 using calf thymus DNA (28) and less than GWL-78 (SI Appendix, Fig. S2B), a PBD-poly(N-methylpyrrole) conjugate engineered to strengthen minor groove contacts (29). We then asked whether the sequence selectivity predicted for TM (30) blocks site-specific endonuclease activity. Indeed, cleavage of a BamHI recognition site was inhibited in a concentration-dependent manner by TM and control GWL-78, but not by TV or buffer (Fig. 3A). By contrast, TM did not inhibit an endonuclease with an A-T–rich binding site (SI Appendix, Fig. S2C).
Fig. 3.
TM interacts with DNA and induces cellular DNA damage in vitro and in vivo. (A) DNA substrate with a BamHI site (Right) was incubated with buffer (C), solvents n-butanol (B) or DMSO (D), different concentrations of TM and TV, or the positive control GWL-78 (+). Nuclease activity on treated DNA compared with solvent and uncut control (−) was visualized by electrophoresis. Percent inhibition is shown. (B) Tail DNA (in percentage) for comet of HeLa treated 4 h with 10 µM TM, 10 µM GWL-78 (+), 20 µM TV, or solvents. HT-29 and SW48 cells were treated with 1 mM TM or controls. Bars represent medians of each dataset (n ≥ 50 cells). Kruskal–Wallis test followed by Dunn’s multiple comparison (*P ≤ 0.05). (C) Phosphorylated (p)-CHK1 in lysates of HT-29 cells treated 4 h with increasing concentrations of TM, GWL-78 (+), 20 µM TV, or solvents. p-CHK2 detected in HT-29 lysates after 8-h treatment (Left). Means ± SEM of p-CHK1/2 signals obtained from three independent cell lysates normalized to β-actin are shown (Right). One-way ANOVA followed by Sidak’s multiple comparison (*P ≤ 0.05) (ns = not significant). (D) Comet of cecal enterocytes of infected mice (24 h) showed tail DNA, tail length, and tail moment were significantly different when mice were colonized with K. oxytoca AHC-6 (WT) compared with the ∆npsB-mutant. Bars represent medians of each dataset (n = 9 mice, with ≥50 cells per mouse), and significance was determined with Mann–Whitney test (*P ≤ 0.05).
DNA alkylation at guanine bases by a PBD or other agents should trigger a host cellular DNA damage response and activate multiple DNA repair enzymes including the base- and nucleotide-excision repair pathways. Incomplete excision removal of the PBD adduct may also lead to DNA single- and double-strand breaks (31). To test whether the K. oxytoca enterotoxins exert DNA damage, HeLa cells were treated with TM, TV, or the DNA-alkylating control GWL-78, and then subjected to comet analysis, a gel electrophoresis-based method to measure DNA damage in individual cells. Significantly increased DNA fragmentation was observed with HeLa cells after TM or GWL-78 treatment, compared with TV or solvents. Similar results were obtained with the colon cancer cell lines HT-29 and SW48 (Fig. 3B and SI Appendix, Fig. S2D). Lysates of the TM- and GWL-78–treated HT-29 cells also exhibited increased phosphorylation of the cell cycle checkpoint kinases CHK1 and CHK2 (Fig. 3C). Thus, the effects of TM on DNA alert master regulators of cellular responses to DNA damage and replication stress, and lead to accumulation of DNA single-strand and double-strand breaks.
To assess whether the intestinal epithelium exhibits genomic instability when exposed to TM, we chose to analyze tissues before day 5 of infection when apoptosis and exfoliation of the lining are excessive (Fig. 1E; see Fig. 6A). We used a pilot study to monitor the temporal increase in TM and TV concentrations in stool during the first 72 h of colonization (SI Appendix, Fig. S2E). TM was detected after 12 h and increased thereafter. TV production was comparatively delayed. To focus on the bioactivity of TM, we colonized additional animals with K. oxytoca AHC-6 or the toxin-deficient npsB mutant for 24 h when TV levels in stool should still be low (SI Appendix, Fig. S2E). Upon killing, we determined cells per gram of stool and quantified TM and TV in cecal contents and feces (SI Appendix, Fig. S2 F and G). Trace amounts of TM were also detected in pooled blood samples and in some cases kidney of infected animals. Comet analysis revealed significant DNA fragmentation in enterocytes isolated from ceca of mice colonized by wild-type K. oxytoca, but not the toxin-deficient mutant (Fig. 3D).
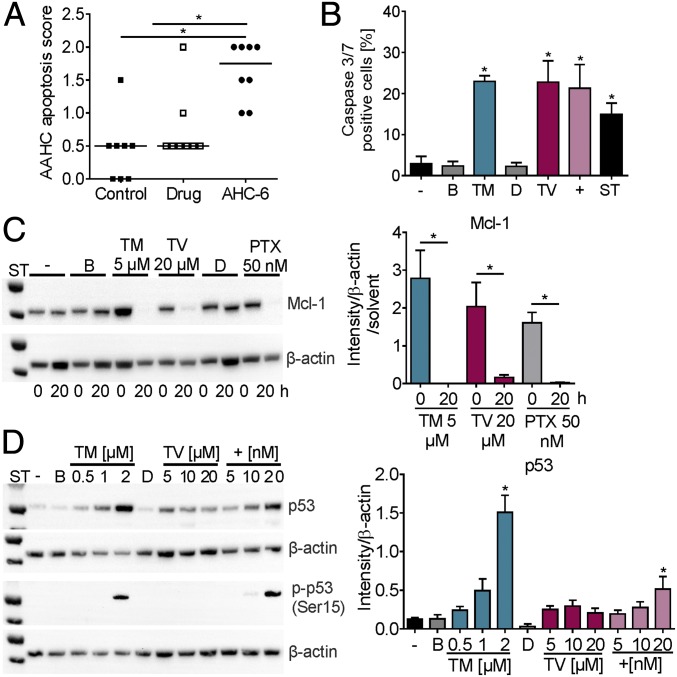
Fig. 6.
In vitro effects of TM and TV match features of AAHC. (A) Histopathological apoptosis scores of colons of untreated (control), amoxicillin/clavulanate treated (drug), and mice colonized with K. oxytoca (AHC-6). Bars indicate medians (n = 8). Kruskal–Wallis test followed by Dunn’s multiple comparison (*P ≤ 0.05). (B) Percent caspase 3/7-positive SW48 cells without (−) or after 24-h treatment with 2 µM TM, 60 µM TV, 20 nM GWL-78 (+), 2 µM staurosporin (ST), and solvents (B and D). Values are means ± SD (n = 3). (C) Total Mcl-1 protein in HT-29 cells treated 0 and 20 h with TM, TV, PTX, or solvents. Means ± SEM of signal for total Mcl-1 normalized to β-actin and solvents obtained from three independent cell lysates are shown (Right). (D) Total p53 protein and Ser15 phosphorylation (p-p53) detected in SW48 lysates treated 12 h with TM, TV, and GWL-78 (+) as indicated (Left). Means ± SEM of signal for total p53 normalized to β-actin obtained from three independent cell lysates are shown (Right). One-way ANOVA followed by Sidak’s multiple comparison (*P ≤ 0.05).
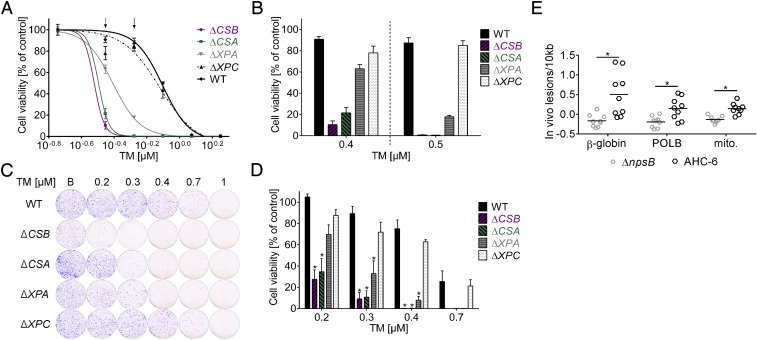
To gain insights into how TM-induced DNA damage is sensed and repaired in host cells, we next asked whether mutation of genes encoding key repair factors would render cells hypersensitive to TM. Inactivating mutations were generated in the human haploid cell line HAP1 (32, 33). Equal numbers of cells for each mutant line were cultured with increasing concentrations of TM, TV, or controls, and cell viability was measured (Fig. 4 A and B and SI Appendix, Fig. S3 A and B). Illudin S was chosen as the DNA-alkylating control because it is well characterized in this assay and is closer in size to TM than GWL-78 (SI Appendix, Fig. S3A). We observed pronounced hypersensitivity to TM with mutant cells lacking the Cockayne syndrome group A or B (CSA or CSB) proteins compared with wild-type survival. These factors mediate transcription-coupled repair (TCR), a subpathway of nucleotide excision repair (NER) that targets DNA alterations blocking translocation of RNA polymerase through expressed genes. Cells lacking the NER factor xeroderma pigmentosum protein A (XPA), which functions downstream of CSA/B, also showed significantly increased susceptibility to multiple concentrations of TM. Lower viability was not detected for any mutant cell line tested with TV compared with wild type, but mutant cells exposed to control substance illudin S exhibited a pattern of sensitivity similar to TM (SI Appendix, Fig. S3 A and B). DNA lesions caused by illudin S are efficiently repaired by TCR, but poorly recognized by the global genome branch of nucleotide excision repair (GG-NER) in which XPC functions (34). The results of the short-term dose–response assay were confirmed with a long-term colony formation assay (Fig. 4 C and D and SI Appendix, Fig. S3 C and D). The results of these experiments verify that HAP1 cells lacking CSA, CSB, and XPA were significantly more sensitive to TM, revealing that TCR proteins can recognize the DNA alterations caused by TM. If this mechanism of DNA damage recognition is also valid in vivo, we would expect a higher incidence of lesions to be detected in nontranscribed regions of the genome compared with highly expressed genes. To test this hypothesis, we performed a long amplicon analyses on genomic DNA isolated from cecal enterocytes of infected mice. Indeed, the lesion burden measured for a 8.7-kb region of the β-globin gene exceeded the frequency of lesions scored in a 6.6-kb region of the heavily transcribed DNA-polymerase β gene and a 10-kb segment of the mitochondrial genome. The lesion burden in all fragments was significantly higher in the DNA of mice infected with wild-type bacteria compared with animals colonized with the mutant (Fig. 4E). We conclude that TM is a DNA-binding and -damaging agent with genotoxic effects on host cells in vitro and in vivo.
Fig. 4.
TM induces hypersensitivity in human DNA repair-deficient cells and causes DNA lesions in vivo. (A) Dose–response survival curves of TM-treated mutants deficient in CSA, CSB, and XPA, but not XPC, show TM hypersensitivity compared with wild type (WT). Values are normalized to solvent controls and represent means ± SEM of three technical replicates. Data from one of three biological replicates are shown. (B) Cell viability shown as means ± SEM at two assay concentrations (indicated with arrows in A). (C) Colony formation by cells treated with n-butanol (B) or the indicated concentrations of TM before recovery in drug-free medium. Macroscopic colonies were stained with crystal violet. (D) Values of C normalized to solvent. Means ± SEM are shown (n = 3). Significance of results for mutants compared with WT was determined with one-way ANOVA followed by Sidak’s multiple comparison (*P ≤ 0.05). (E) Higher lesion burden in genomic DNA isolated from cecal enterocytes of K. oxytoca AHC-6 (WT) colonized mice (24 h) compared with mice infected with the ∆npsB-mutant was detected with long amplicon quantitative PCR. Lesions/10 kb in β-globin, DNA-polymerase β (POLB) and in a region of the mitochondrial genome (mito.) are shown. Bars indicate means (n = 9), and significance was determined with unpaired t test (*P ≤ 0.05).
Tubulin Is the Molecular Target of TV.
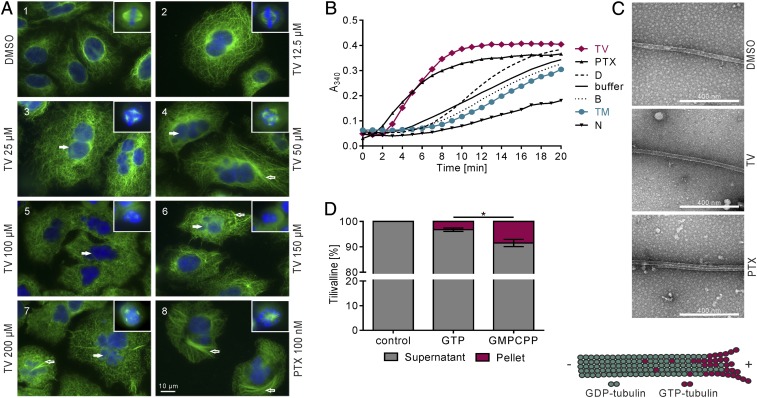
The results above indicate that TV has neither DNA-binding nor genotoxic activity, as expected. Exposure to TV blocked cell cycle progression at G2/M phase (Fig. 2). TV also impeded closure of a scratch created in a monolayer of HeLa cells (SI Appendix, Fig. S4). Considering that microtubules form the mitotic spindle, constitute the cytoskeleton, and facilitate cellular movement, we asked whether TV inhibits microtubule-dependent processes. To observe the effects of TV on the microtubule network, A549 lung carcinoma cells and HT-29 colon cancer cells were exposed to TV and stained with antibodies to tubulin or a spindle pole marker (Fig. 5A and SI Appendix, Fig. S5 A and B). Microscopy revealed aberrant spindle morphologies and micronucleation in A549 cells treated with TV, compared with solvent, in a manner resembling the effects of the microtubule-stabilizing drug paclitaxel (PTX) (Fig. 5A, 1–8). TV induced formation of abnormal type II bipolar spindles with atypically short distances between poles and poor DNA alignment at the metaphase plate (Inset 2). With increasing concentrations of TV, type III tripolar and multipolar spindles (small star-shaped aggregates of microtubules and a ball of DNA) appeared (Insets 3–7) and the number of micronucleated cells increased (3–7, filled arrows). PTX-induced type III spindles are mostly monopolar and with more dense star-shaped microtubules (inset 8). Interphase microtubules of TV treated cells acquired a straight, parallel orientation (4, 5) different from that of control cells. Loosely packed bundles appeared in some cells (4, 6, and 7, open arrows) in contrast to more abundant and compact bundles in cells treated with PTX (8, open arrow). Similar results were obtained for HT-29 cells (SI Appendix, Fig. S5 A and B). These results imply that TV is a microtubule-stabilizing agent.
Fig. 5.
TV targets tubulin. (A) A549 cells were treated with DMSO, increasing doses of TV and PTX for 24 h. DNA was visualized with Hoechst 33342 (blue) and α-tubulin with antibody (green). Insets are mitotic spindles from the same preparation. (B) Tubulin polymerization in the presence of effectors TV (100 µM), PTX (10 µM), TM (10 µM), and nocodazole (N) (10 µM) was monitored via A340 over time. Shown are means (n = 4). (C) Electron micrographs confirm that turbidity reflects microtubule formation in reactions with DMSO, TV (75 µM), and PTX (10 µM). (D) TV present in polymers or supernatant was analyzed via HPLC-UV/VIS. Bars indicate distribution of TV following reactions without or with tubulin plus nucleotide. Shown are means ± SEM (n = 3), and significance was determined with one-way ANOVA followed by a Bonferroni post hoc test (*P ≤ 0.05).
To test whether TV binds tubulin directly, we assayed polymerization of purified αβ-tubulin heterodimers in vitro in the presence of TV, or control substances: PTX, the destabilizing drug nocodazole, and TM. TV but not TM stimulated the formation of polymers in a manner resembling PTX. This activity for TV was confirmed with independent protein preparations and different assay conditions (Fig. 5B and SI Appendix, Fig. S5C). Taken together, the data indicate that the TV effect on polymerization manifests at the nucleation phase, which occurs faster with TV than in buffer alone, and in the total amount of microtubules accumulated at the plateau phase. Transmission electron micrographs of reaction products verified that the increased absorbance we measured was due to formation of microtubules, not other polymers or aggregates (Fig. 5C). We conclude that TV increases polymerization of tubulin into microtubules.
To explore the mechanism underlying this activity, we investigated the stoichiometry of TV in the polymer. Polymerization products were fractionated by centrifugation, and the proportion of TV cosedimenting with microtubules vs. the fraction remaining in the supernatant was determined by HPLC-UV/VIS analysis. In this experiment, we also varied the nucleotide content of tubulin. The protein is active for microtubule assembly in the GTP-bound state, but hydrolysis to GDP results in an inactive conformation; thus, GTP hydrolysis in the polymers and GDP/GTP exchange control the assembled state of tubulin (cartoon, Fig. 5C). We compared cofractionation after polymerization reactions in the presence of GTP or guanosine-5′-[(α,β)-methyleno]triphosphate (GMPCPP), a GTP derivative that hydrolyzes slowly, mimics the GTP-like state of tubulin, and extends it for a longer period of time. HPLC-UV/VIS analysis (Fig. 5D) showed a ∼2.6-fold increase in the proportion of TV recovered in the microtubule-containing pellet for the GMPCPP reaction products compared with the GTP-containing products. This finding indicates that TV interacts better with tubulin loaded with GMPCPP than with GTP. Given that GMPCPP withstands hydrolysis in the microtubule longer than GTP, these results suggest that the GTP-like state of tubulin favors the binding of TV. By contrast, PTX drives inactive GDP-tubulin into microtubules by replacing the requirement for the γ-phosphate of GTP to activate the protein. This interaction stabilizes microtubules by preferential binding of PTX to assembled tubulin with a 1:1 stoichiometry. The properties of TV–tubulin interactions observed here provide strong evidence that TV and PTX stabilize microtubules by different mechanisms. If so, we would also expect tumor cells that have acquired resistance to PTX, due to mutations in β-tubulin affecting drug binding, to remain susceptible to TV. Indeed, two PTX-resistant ovarian cancer lines (35), 1A9PTX10 and 1A9PTX22, were equally sensitive to TV (IC50 = 4.5 and 1.9 µM, respectively) as the parental line 1A9 (IC50 = 3.6 µM), whereas higher doses of PTX were required to inhibit growth of 1A9PTX10 (IC50 = 29.9 nM) and 1A9PTX22 (IC50 = 30.9 nM) compared with 1A9 (IC50 = 2.7 nM).
Cytotoxicity and AAHC.
The predominant histopathological feature of AAHC in humans and animal models are epithelial alterations characterized by increased apoptosis (Fig. 6A) and mucosal hemorrhage. Our earlier work with TV showed that cellular exposure induces apoptosis (17). Relative abundances of the enterotoxins measured in patient and mouse samples in this study imply a predominant role for TM in pathogenicity. We therefore measured caspase 3/7 activity in SW48 cells treated with TM or TV (Fig. 6B), confirming that the metabolites act independently to induce apoptotic cell death. Induction of cellular apoptosis involved loss of the prosurvival Mcl-1 protein (Fig. 6C). Degradation of Mcl-1 was observed following exposure to each enterotoxin as well as the apoptosis inducer PTX. Nonetheless, the molecular targets we identified for the enterotoxins support the prediction that the mechanisms leading to apoptosis will differ.
To assess whether tumor suppressor p53 (TP53) gene expression was up-regulated upon exposure to the cytotoxins, we measured total p53 protein with SW48 cells, since these carry the wild-type TP53 gene. Total p53 protein was elevated through treatment with DNA-reacting agents TM and GWL-78 (Fig. 6D). In a cascade of downstream reactions, p53 can activate a large number of genes including genes encoding DNA damage recognition components of NER. Activation of p53 for a DNA damage response involves phosphorylation of Ser15. Activation at this specific residue was indeed detected in response to both DNA-reactive substances TM and GWL-78, but not by the tubulin inhibitor TV (Fig. 6D). This result strengthens the notion that the cellular responses to the K. oxytoca enterotoxins trigger distinct pathways leading to cell death.
Discussion
Functional characterization of intestinal metabolites and signals in host–microbiota–pathogen interactions is a difficult challenge, but also indispensable in understanding host–microbe interactions relevant for the development of intestinal disease. This study shows that the distinct chemistries of the K. oxytoca enterotoxins alter their molecular targets and confer different functionalities.
The PBD ring system of TM meditates interactions with DNA, which are blocked by the indole substituent in TV. TM showed activities in vitro characteristic for the DNA-reactive PBD family: duplex stabilization and endonuclease protection, induced DNA damage signals, G1/S cell cycle arrest, and DNA fragmentation in cellular model systems. Enterotoxin production in vivo increased genome instability and lesion burden in ceca of infected mice, thus validating the genotoxic mode of action for TM. Whether these effects are due to the predicted TM–DNA adduct formation remains to be answered by structural analysis of the complex. Previous work has shown that the twist of the PBD ring system allows the molecule to fit in the DNA minor groove, causing very little distortion of the overall DNA structure (36). Accordingly, DNA repair mechanisms have difficulty recognizing this DNA adduct (31). We identified TM hypersensitivity in TCR mutants. TCR preferentially removes DNA lesions that block translocation of RNA polymerase II (Pol II) (37). CSB (also known as ERCC6) recognizes and binds lesion-arrested Pol II to initiate TCR. The recent structures determined for the Pol II–CSB complex from yeast (38) revealed that CSB (Rad26) promotes Pol II bypass of modestly bulky lesions to rescue transcription arrest, or when Pol II persists at bulkier sites, to initiate TCR. CSB is thought to recruit additional repair factors such as CSA (also known as ERCC8), UVSSA, XPG, and TFIIH in human cells (37). CSA and CSB have a further regulatory role in general transcription restart following genotoxic stress (39). In GG-NER, the lesion site and helix distortions are initially recognized by the heterotrimeric XPC protein complex or the UV-damaged DNA-binding protein. After lesion detection, both TCR-NER and GG-NER rely on the same machineries to remove the DNA lesion, which includes the lesion binding and verifying factor XPA (40). In good agreement with these findings, mouse cecal cells exposed to the enterotoxins in vivo carried a higher abundance of lesions in a nontranscribed region of the genome compared with a highly expressed gene. Altogether, these data indicate that TM acts as a genotoxin and specifically requires TCR for its removal from DNA.
In contrast, the pentacyclic PBD TV is a tubulin-targeting compound. Biochemical experiments indicate that TV exerts a stabilizing effect on microtubules: enhancing both nucleation and elongation phases of polymerization. This finding is surprising in that, although plants and marine sponges are a rich source of microtubule-stabilizing compounds, bacteria are less common producers (41). The vast majority of microtubule-stabilizing agents bind within one of two distinct nonoverlapping sites on tubulin: the taxane or peloruside site (41). Microtubules assembled in the presence of TV contain a small fraction of the compound bound to the polymer. This quantitative relationship differs strikingly from the taxane and peloruside site ligands, which bind stochiometrically to the microtubules (42). Notably, the bound fraction of TV increased when microtubules were assembled in the presence of the slowly hydrolyzable nucleotide GMPCPP. The longitudinal contacts between GDP tubulin subunits in microtubules are more compressed than those of GMPCPP microtubules (43). It is also known that this compression can alter binding properties of drugs with an interfacial binding site (44).
From a structural point of view, a drug able to bind into the GMPCPP longitudinal interface and not in the GDP longitudinal interface will keep the interface in the expanded conformation, thus mimicking the γ-phosphate effect and stabilizing the microtubule cap. As a result, assembly would be favored, as we observed in the presence of TV. This finding points to a mechanistic hypothesis by which TV could be an interfacial ligand of the longitudinal interface that acts as a matchmaker (for review, see ref. 42). In this model, TV would only fit if the interface is in the expanded GTP/GMPCPP state and not in the compressed GDP state. This tubulin–TV interaction would stabilize the GTP/GMPCPP state, thus preventing microtubule disassembly and enhancing its polymerization, as observed in the biochemical assays (Fig. 5 and SI Appendix, Fig. S5). Based on our results, we propose the longitudinal interface of tubulin dimer association as the binding site for TV, with the requirement that the interface is in the expanded GTP/GMPCPP conformation. This mode of binding would stabilize the active interface preventing depolymerization and stabilizing microtubules. In conclusion, TV is a microbiota-derived tubulin inhibitor, which is now linked to human intestinal disease.
Our findings showing that TM and TV are produced in the human body combined with evidence for the cellular processes they disrupt raise several important questions. Many successful antiproliferative drugs bind to tubulin and suppress microtubule dynamics. The potential importance of TV in cancer therapeutics needs to be explored. We ask therefore, whether—despite low potency—detailed knowledge of TV–tubulin interactions could be valuable given the unusual mode of longitudinal association suggested by this study. This is particularly pertinent considering the problem of innate and acquired resistance to tubulin inhibitors and their side effects (45) that ultimately limits their clinical success (41).
Second, is TM tumorigenic? Colibactins are another group of microbiota-derived small-molecule genotoxins produced by select members of the Enterobacteriaceae (46–49). In contrast to TM’s DNA-damaging activity, alkylation of DNA by colibactin induces acute DNA double-strand breaks and megalocytosis in eukaryotic cells (50). Incomplete repair of host DNA damage following infection by colibactin-proficient Escherichia coli leads to chronic mitotic and chromosomal aberrations as well as increased frequency of gene mutation (51). This contribution to cellular transformation is thought to promote tumorigenesis in colorectal cancer (CRC) (52). Healthy humans (2–9%) carry K. oxytoca in their intestine and ∼50% of isolates are toxin producers (15, 16, 53, 54). Continuous secretion of TM over a lifetime of colonization may cause low-grade DNA damage that could trigger chromosomal instability and contribute to cellular transformation. Thus, the risk of TM toxicity in sporadic and hereditary CRC will be important to assess.
Third, given the potential harm done to the host, we also wonder why gut residents produce genotoxins. Nougayrède et al. (50) proposed that colonizing E. coli may use colibactin to slow the renewal of enterocytes by blocking the cell cycle, thereby prolonging bacterial persistence. Unlike colibactin, TM release does not require host cell contact. K. oxytoca that secrete TM did not show a colonization advantage over the mutant strain during antibiotic induced dysbiosis (17). However, the antibacterial activity toward other gut residents shown in this study implies that TM-mediated antagonism of microbial competitors may confer the real advantage to K. oxytoca during homeostasis.
Finally, the intrinsic reactivity of TM with available indole to produce TV is an unusual feature of this system. Conversion of TM to TV repurposes the molecule, alters its cellular effects on the host, and presumably mediates a different spectrum of microbe–microbe interactions within this niche. In conclusion, the K. oxytoca enterotoxin system contributes remarkable functional versatility to this organism’s activities as a pathobiont of the human intestine.
Methods
Human Colonic Luminal Fluid and Stool Samples.
A 29-y-old female patient (A) suffering from systemic lupus erythematosus was treated with ampicillin/sulbactam for pneumonia and dexibuprofen for arthralgia. After 12 d of antibiotic therapy, she developed severe abdominal pain and hemorrhagic diarrhea. Intestinal ultrasound and colonoscopy showed severe segmental hemorrhagic colitis starting in the descending colon. Symptoms improved 2 d after stopping the antibiotic therapy, and the colitis subsided completely after 10 d without additional specific therapy. Stool culture was negative for Clostridium difficile, Yersinia, EHEC, Shigella, Campylobacter, and Salmonella species. Patient B, a 32-y-old female, and patient C, a 35-y-old female, were treated with amoxicillin/clavulanate in addition to nonsteroidal antiinflammatory drugs for sinusitis and as antibiotic prophylaxis after orthognathic surgery, respectively. Both patients developed bloody diarrhea with abdominal cramps and were diagnosed with acute AAHC by colonoscopy. Colitis subsided after discontinuation of the triggering antibiotic therapy. K. oxytoca isolates from patients A, B, and C carry the enterotoxin gene cluster as determined by PCR and were positive for cytotoxicity. Colonic luminal fluids aspirated during diagnostic colonoscopy and stool samples were stored at −20 °C. The protocol for sample acquisition was approved by the ethics committee of the Medical University of Graz (17-199 ex 05/06) and patients’ informed consent was obtained.
Mouse Infection Models.
Animal experiments were performed as previously described (17). Adult female C57BL/6NCrl mice with SOPF status (Charles River; Janvier Labs) were housed in individually ventilated cages. Studies were performed in accordance with the Commission for Animal Experiments of the Austrian Ministry of Science (GZ 66.007/0006-II/3b/2011 and GZ: BMWFW-66.007/0002-WF/V/3b/2017) and the local ethics committee.
For the AAHC model, 8-wk-old mice of treatment groups received Curam (amoxicillin/clavulanate, 2,000/200 mg; Sandoz) 100 mg/kg/treatment intraperitoneally twice daily at t = 0, 8, 24, 32, 48, 56, 72, and 80 h. Mice were infected intragastrically three times (t = 0, 24, and 48 h) with 1 × 109 cfu of K. oxytoca AHC-6 (with chromosomally integrated kanamycin resistance marker) resuspended in 100 µL of LB broth. At day 5, the mice were killed by cervical dislocation, and the entire intestinal tract was removed.
For short-term colonization experiments, 8- to 13-wk-old mice were administered amoxicillin (0.4 mg⋅mL−1; Genaxxon Bioscience) in drinking water and mice of treatment groups were infected intragastrically once with 1 × 108 cfu of K. oxytoca AHC-6 or ∆npsB.
Colonization by K. oxytoca was quantified via plating 0.02–0.3 g of cecal content homogenized in LB broth on SCAI agar and selective CASO agar (50 μg⋅mL−1 kanamycin). Cecal content was stored at −80 °C for HPLC–electrospray mass spectrometry (ESMS) measurements, and cecal tissue was stored in formalin for histopathological analysis as previously described (17).
Cell Culture and Test Substances.
The HAP1 cell line is a derivative of near-haploid leukemia cell line KBM7 that was reprogrammed to adherent growth (55). HAP1 and mutant derivatives were grown in Iscove’s modified Dulbecco’s medium containing l-glutamine and 25 mM Hepes, pH 7. HeLa cells were cultured in DMEM and T84, HT-29 (CLS Cell Lines Service), and A549 cells in a 1:1 mixture of Ham’s F-12 and DMEM supplemented with 2 mM glutamine. 1A9, PTX10, PTX22 (35), and LNCaP cells were cultured in RPMI medium 1640 supplemented with 1 mM sodium pyruvate. MCF7 were cultured in minimum essential Eagle medium supplemented with 2 mM glutamine and 1 mM sodium pyruvate. Endothelial cell growth medium was used for the cultivation of human umbilical vein endothelial cells (PromoCell). SW48 were grown in McCoy’s 5A (modified) medium. All media were supplemented with 10% FBS and 100 µg⋅mL−1 penicillin/streptomycin (Gibco). Cell lines were obtained from ATCC (if not stated otherwise) and incubated at 37 °C with 5% CO2 in 95% humidity.
n-Butanol solutions of synthetic TM, and DMSO solutions of synthetic TV, GWL-78, PTX, nocodazole, staurosporin, and illudin S were stored at −20 °C.
Metabolite Analysis.
Human stool samples and cecal contents and feces of mice and control mice spiked with TM and TV were mixed 1:1 (wt/vol) with n-butanol for 5 min, centrifuged [16,000 × g, 15 min, room temperature (RT)] and filtered (nylon, 0.2 µm). Butanol extracts of mouse cecal contents from the AAHC model were mixed with “Silica Gel 60” (particle size, 40–63 µm; Merck) and loaded into a Pasteur pipette containing glass wool. All samples were prepared with MeOH (HPLC grade) and CHCl3 (99.2%, stabilized with 0.6% EtOH). The crude extracts were treated with 30 µL of MeOH, mixed thoroughly by vortexing (20 s), and ultrasonicated (3 min, 22 °C) until homogeneity. The mixtures were applied on prepared silica columns [height, 2 cm; conditioned with CHCl3/MeOH, 1:1 (vol/vol)] and allowed to enter the pad via gravity. The pad was carefully eluted with portions of CHCl3/MeOH (1:1) (vol/vol; 5 × 500 µL), the combined extracts were concentrated by removing the solvents (22 °C, 60 min) and dried on the rotary evaporator (8 mbar, 40 °C, 8 min). The samples were stored at −18 °C.
Human colonic luminal fluid from patient A was centrifuged (4,000 × g, 30 min, RT). Supernatant was filtered (0.45 and 0.22 µm), mixed 1:1 (vol/vol) with n-butanol by vortexing (30 s), and centrifuged (10,000 × g, 5 min, RT). Colonic luminal fluids of patients B and C were mixed 1:1 (vol/vol) with n-butanol, vortexed for 5 min, and centrifuged (16,000 × g, 15 min, RT). Supernatants were filtered (0.22 µm). The organic phase was concentrated to dryness via vacuum centrifugation at 45 °C. Dried extracts were stored at −20 °C. Colonic fluid extract of patient A was purified before measurement. The crude extract was dissolved in 200 µL of MeOH and applied to a preparative TLC plate (silica gel; #Z513040; 1,500 μm; 20 × 20 cm). The plate was evolved using CHCl3/MeOH (=20:1) (vol/vol), and then air-dried. The silica gel in the region Rf = 0.1–0.5 was scraped into 150 mL of MeOH. The mixture was stirred for 20 min at 22 °C and filtered through a cellulose filter. The pad was rinsed with 100 mL of MeOH, and the solvent was removed under reduced pressure.
HPLC–High-Resolution-ESMS.
For HPLC–high-resolution (HR)-ESMS analysis, samples were re-extracted in MeOH (300 µL), transferred to sterile autosampler vials (polypropylene, 250 µL), and held at 20 °C in the autosampler before measurement. HR-ESMS measurements were performed on a Q-Exactive Hybrid Quadrupole-Orbitrap MS after HPLC on a Dionex Ultimate 3000 series instrument (Thermo Fisher Scientific). The HR-MS applies an atmospheric pressure electrospray ionization source (ES) using nitrogen as nebulizer and drying gas. Measurements were performed in positive ionization mode, with a drying gas temperature of 440 °C, spray voltage of 3.5 kV, and a resolution of 70,000 (FWHM). The HPLC system was equipped with a Shodex Asahipak ODP-50 column (4.0 × 125 mm; 5-µm particle size) using mobile phases A [water including 0.1% formic acid (vol/vol)] and B (acetonitrile) under gradient elution conditions: 0–10 min, 10–12.5% B; 10–10.5 min, 12.5–30% B; 10.5–15 min, 30% B; 15–25 min, 30–50% B; 25–30 min, 50% B; 30–30.5 min, 50–10% B; and 30.5–35 min, 10% B. Ions were recorded as protonated [M+H]+ form in single-ion monitoring mode and masses set to m/z: 235.1077 (TM) and 334.1550 (TV). Column temperature was 30 °C; flow rate, 0.5 mL⋅min−1; and injection volume, 10 µL.
Cytotoxicity Assay.
Cell survival was measured via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) staining after 48-h incubation with TM and TV, as previously described (17).
Agar Diffusion Assay.
Cell suspensions (0.5 McFarland) were spread on Columbia Blood Agar and dried for 15 min. Antimicrobial discs were soaked with 10 µL (40 μg) of TM, TV, or solvents, dried, and transferred to the inoculated plates. Depending on the bacteria tested, plates were incubated at 37 °C for 24–48 h under aerobic or anaerobic conditions.
Cell Cycle Analysis.
Cells were harvested and washed twice with ice-cold PBS. The pellet was resuspended in 500 μL of PBS, and 5 mL of 70% ethanol (−20 °C) was added dropwise under constant shaking. Fixed and permeable cells were centrifuged (720 × g, 4 min), washed with PBS containing 0.5% FBS, and then resuspended in 200 μL of PI-hypotonic lysis buffer [0.1% sodium citrate, 0.1% Triton X-100, 100 µg⋅mL−1 RNase A (Fermentas), 50 µg⋅mL−1 PI (Sigma Aldrich)] and incubated for 20 min at RT in the dark. Equivalent numbers of cells (events) were then sorted according to the PI signal strength via FACS (BD LSR II Flow Cytometer; BD Biosciences). BD FACSDIVA software (version 8) and ModFit LT software (version 4 and 5) were used to analyze and plot the data.
Thermal Denaturation.
To generate a DNA template, complementary primers (5′-CGATAACATCTTTTTCATTTGCAAACGCATTTGCAATAGCATGTCCGCAAATGGTAGAT-3′) were heated 10 min at 90 °C, and then slowly cooled (1 h) to RT. Ten micromolar dsDNA in 10 mM Tris⋅HCl, pH 7.5, 10 mM NaCl, and 1 mM MgCl2 was incubated with 10 µM TM, TV, GWL-78, and solvents at 37 °C for 18 h. One hour before analysis, DNA was incubated with SYBR Green at 37 °C. Thermal denaturation was performed from 20 to 90 °C with a ramping of 0.5 °C. Absorbance was measured at 260 nm on a CFX Real-Time PCR Detection System (Bio-Rad) in quadruplicates in three independent experiments.
Endonuclease Inhibition.
DNA substrate (1,740 bp) containing a single BamHI and SspI recognition sequence was generated by PCR. Five hundred nanograms of DNA were incubated with test substances or solvents for 16 h at 37 °C in 10 mM Tris⋅HCl, pH 7.5, 1 mM EDTA. Treated DNA was incubated with 20 U of endonuclease for 3 h at 37 °C, and then resolved electrophoretically. Nuclease activity was calculated as intensity of the cut DNA fragment normalized to the intensity of all bands visible and expressed as inhibition percentage.
Isolation of Enterocytes.
Mice were colonized with K. oxytoca for 24 h, and ceca were extracted, washed with ice-cold PBS, and inverted. Epithelial cell suspension was obtained after 30-min incubation in PBS with 5 mM EDTA. Cells were dissociated by incubation at 37 °C for 20 min in TrypLE Express Enzyme (Thermo Fisher Scientific). In total, 1 × 106 cells were pelleted for genomic DNA (gDNA) isolation, and the remaining cells were kept in DMEM/F12 media with 10% FBS for comet analysis.
Single-Cell Electrophoresis—Comet Assay.
After 4-h treatment with TM, TV, or control substances, cells were harvested by gentle scraping, then pelleted (700 × g, 2 min, 4 °C), washed, and resuspended in ice-cold PBS to obtain 1 × 105 cells⋅mL−1. Cells were carefully mixed with agarose at a 1:10 ratio (vol/vol) and 75 μL/well were immediately transferred onto the comet slide and analyzed following the manufacturer’s instructions (OxiSelect comet assay kit; Cell Biolabs). Tail length, tail DNA, and tail moment, defined as tail length times tail DNA, were calculated with OpenComet software.
Sensitivity of DNA Repair-Deficient Mutants.
Cells were treated with TM, TV, illudin S, or solvent controls. For dose–response curves, cell viability after 48 h was determined using CellTiter-Glo (Promega). After medium was replaced with 50 µL of reagent, plates were shaken for 30–45 min in darkness, and luminescence was detected on a plate reader. Values were plotted by sigmoidal curve fitting. For colony-forming assays, cells were treated for 48 h, followed by incubation in drug-free medium for 72 h. Cells were washed with PBS, fixed in 3.7% formaldehyde for ≥1 h, stained with 0.1% crystal violet in 10% ethanol for ≥1 h, washed, and air-dried. Images were analyzed using CellProfiler.
Long Amplicon PCR.
gDNA was isolated from 1 × 106 cecum cells with the Qiagen Genomic-tip 20/G Kit using the Qiagen Genomic DNA buffer set applying the protocol for tissues. TE buffer (20×, 200 mM Tris⋅HCl, 20 mM EDTA, pH 8.0) was used in all steps. gDNA was quantified via Pico Green standard curve (Quant-iT PicoGreen dsDNA reagent; Invitrogen) (56) using Lambda (λ)/HindIII DNA (Thermo Fisher Scientific). Fluorescence was measured in a plate reader (20-s shaking, 485-nm excitation, 520-nm emission), and DNA quality was assessed with electrophoresis.
In total, 3.5 ng of gDNA was used for Long Amplicon PCR (56, 57). PCR was performed in 25-µL reactions containing 1× LongAmp Hot Start Taq2× Master Mix (New England Biolabs), 0.1 µM primer, and nuclease-free water. Temperature profile was used as follows: 94 °C, 2 min; 94 °C, 15 s; x °C (individual), 12 min; 72 °C, 10 min; 4 °C, forever. Annealing temperatures and cycle numbers were adjusted for 8.7-kb β-globin fragment (accession number X14061) to 64 °C and 25 cycles; for 6.6-kb DNA-polymerase β (accession number AA79582) to 65 °C and 24 cycles; for 10-kb long mitochondrial fragment to 64 °C and 17 cycles; for 117-bp short mitochondrial fragment to 60 °C and 18 cycles. Ten microliters of PCR product were quantified in duplicate using Pico Green and lesion burden was calculated according to Furda et al. (56).
Monolayer Reconstitution Assay.
Confluent HeLa cell monolayers were wounded with a pipet tip to generate a “scratch,” washed, and treated with substances. Images were taken with a Nikon Eclipse TE 300 microscope. Phase contrast with a 10× magnification applied to the same xyz coordinates enabled an identical position of each scratch to be monitored over time, and gap closure was calculated (CorelDraw).
Immunofluorescence Microscopy.
A549 cells on coverslips were treated with TV or solvent for 24 h. Cells were washed with PEMP buffer [PEM, 100 mM piperazine-N,N′-bis(2-ethanosulfonic acid) (PIPES) containing 4% polyethylene glycol 8000], permeabilized with 0.5% (vol/vol) Triton X-100 in PEM buffer for 90 s at RT, and fixed with 3.7% (vol/vol) formaldehyde for 30 min at RT. After incubation with α-tubulin (DM1A) mouse mAb (Sigma Aldrich), cells were washed twice and incubated with FITC goat anti-mouse immunoglobulins. DNA was stained with 1 µg⋅mL−1 Hoechst 33342. A Zeiss Axioplan epifluorescence microscope with an ORCA-FLASH 4.0 cooled CCD camera was used to analyze the samples.
HT-29 cells grown on coverslips were fixed with methanol-free 4% formaldehyde for 15 min at RT. Cells were washed 3 × 5 min in PBS and blocked (5% goat serum, 0.3% Triton X-100 in PBS) for 1 h at RT. After overnight incubation at 4 °C with antibodies to β-tubulin or NuMA (Cell Signaling Technology), cells were washed 3 × 5 min in PBS, and then incubated with anti-rabbit IgG (H+L), F(ab′)2 fragment (Alexa Fluor 488 conjugate) for 2 h in the dark. After washing 2 × 5 min in PBS, DAPI staining followed (15 min). The washed coverslips were embedded in Mounting Medium (Roth). A Nikon Inverted Microscope Eclipse Ti-E/B with confocal laser scanning was used to analyze the samples at different magnifications, and images were edited with the Fiji software (ImageJ).
Tubulin Polymerization Assays.
TM, TV, and control substances were diluted in general tubulin buffer (GTB) [80 mM piperazine-N,N'-bis(2-ethanesulfonic acid) sequisodium salt; 2.0 mM MgCl2; 0.5 mM EGTA, pH 6.9]. Tubulin polymerization buffer (TPB) contained GTB with 1 mM GTP and 15% glycerol. Lyophilized porcine α/β-tubulin (tebu-bio), reconstituted at 10 mg⋅mL−1 in ice-cold GTB was mixed with TPB to a final concentration of 3 mg⋅mL−1 tubulin in GTB with 1 mM GTP and 10% glycerol. One hundred micromolar TV, 10 µM TM, 10 µM PTX, and 10 µM nocodazole were added to 100 µL of this suspension in a 96-well plate. The reaction was started immediately in a spectrophotometer at 37 °C. After initial orbital shaking for 5 s, absorbance at 340 nm was measured every min for 1 h under static conditions. Substances diluted in GTB without tubulin served as blanks. To verify these results, tubulin isolated from bovine brain as described (58) was polymerized in the presence of compounds either in glycerol-assembling buffer (GAB) [3.4 M glycerol, 10 mM sodium phosphate (NaPi), 1 mM EGTA, 6 mM MgCl2, 1 mM GTP, pH 6.7] or GTB with 1 mM GTP and 10% glycerol, pH 6.9. Turbidity at 340 nm was measured using a Varioskan Flash multimode microplate reader (Thermo Fisher Scientific) at 37 °C.
Tubulin Pelleting Assay.
To produce GDP-tubulin, purified tubulin was first prepared by three buffer exchanges (500 µL) through 50-kDa Amicon Ultra-4 (Merck) with 10 mM GTP in NaPi buffer [10 mM sodium phosphate (NaPi), 1 mM EGTA, 6 mM MgCl2, 0.1 mM GDP, pH 6.7] followed by a column (Sephadex G-25) buffer exchange to NaPi. Polymerization assay reactions in GAB (200 µL), were performed as described in Methods, Tubulin Polymerization Assays, except the nucleotide was either 1 mM GTP or GMPCPP. A control without tubulin was performed to exclude the possibility that the TV recovered in the pellet had sedimented due to a loss of solubility. Reactions were centrifuged (10,000 × g, 20 min, 37 °C). Supernatants were recovered and pellets were diluted in 200 µL of NaPi buffer (with 1 mM GTP, pH 6.7). Ten micromolar PTX was added as internal standard to both supernatant and pellet to determine the loss of TV during extraction and measurement. Samples were subjected to three rounds of organic extraction with dichloromethane. The recovered dichloromethane was evaporated and dried pellets containing the substances were resuspended in 50% acetonitrile in H2O. Samples were loaded for HPLC-UV/VIS (Agilent 1100 series) and run in a gradient of 2–40% of acetonitrile toward 0.001% formic acid in H2O (1 mL⋅min−1, 25 min). TV values were normalized to the standard PTX. For each type of reaction (without tubulin, GTP to GMCPP), the TV concentration in either pellet or supernatant was normalized to the additive value of both (shown in percentage).
Transmission Electron Microscopy.
Tubulin assembly was performed in GTB (1 mM GTP, 10% glycerol, pH 6.9) with 25 µM tubulin and DMSO, 75 μM TV, or 10 µM PTX. Samples were fixed with 0.1% glutaraldehyde and the reaction was stopped with 100 mM glycine. Four microliters of sample, diluted 1:10 in water, were mounted onto a carbon-coated 400 mesh copper grid. Samples were counterstained with 2% (wt/vol) uranyl acetate before transmission electron microscopy. Micrographs were obtained using a JEOL 1230 transmission electron microscope at 100 keV and a 16 megapixel TemCam-F416 camera from TVIPS.
Immunoblot.
Cells were treated with TM, TV, or control substances (two wells per treatment) and harvested. The pellet was washed with PBS at 4 °C, resuspended in 200 μL of lysis buffer (PBS with protease and phosphatase inhibitor; Roche), lysed via ultrasonication twice for 15 s, and cleared by centrifugation (10,000 × g, 20 min, 4 °C). For immunoblot analysis, 5–10 μg of protein was denatured in SDS sample buffer for 10 min at 100 °C before loading onto a 12% NuPAGE Bis-Tris-protein gel (Thermo Fisher Scientific). Proteins were transferred to PVDF membranes via electroblotting for 2 h at 220 mA, and membranes were blocked overnight in TBS-T buffer (20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1 mL of Tween 20) with 5% BSA or milk powder. All mAbs were purchased from Cell Signaling Technology and used according to the supplier’s recommendations. We used Phospho-Chk1 (Ser345) (133D3) rabbit mAb and phospho-Chk2 (Thr68) (C13C1) rabbit mAb. P53 (DO-7) mouse mAb, phospho-p53 (Ser15) (16G8) mouse mAb, and Mcl-1 (D35A5) rabbit mAb were used to measure apoptosis induction. The secondary antibodies used were anti-rabbit IgG, HRP-linked antibody, and anti-mouse IgG, HRP-linked antibody. Equal loading was confirmed with β-Actin (13E5) rabbit mAb. Detection was achieved with Clarity Western ECL Blotting Substrate (Bio-Rad).
Caspase 3/7 Flow Cytometry.
The CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit containing the SYTOX AADvanced dead cell stain (Thermo Fisher Scientific) was used to determine Caspase-3– or -7–positive cells and exclude necrotic cells. Cells were grown and harvested as described in Methods, Cell Cycle Analysis, washed with ice-cold PBS, and incubated with CellEvent Caspase-3/7 Green Detection Reagent for 25 min at 37 °C. Staining with SYTOX AADvanced dead cell stain solution for 5 min at 37 °C followed. Cells were analyzed using 488-nm excitation and applied standard fluorescence compensation on a BD LSR II Flow Cytometer. Fluorescence emission with 530/30 BP (Caspase-3/7) and 690/50 BP (dead cell stain) filters or their equivalents was used.
Quantification and Statistical Analysis.
Protein and DNA signals were quantified with the Image Lab Software (Bio-Rad). Significance (*P ≤ 0.05) was determined with statistical tests (in GraphPad Prism) specified in figure legends. Values for substances were compared with respective solvents, and n represents the number of independent experiments performed, if not stated otherwise.
Supplementary Material
Acknowledgments
We thank D. E. Thurston and K. M. Rahman for GWL-78, P. Giannakakou for 1A9 cells and the mutant subline PTX10, E. Leitner for bacterial strains, and M. Owusu for assistance in quantifying the clonogenic assays. We also thank NAWI Graz for supporting the Graz Central Laboratory Environmental Metabolomics. We acknowledge networking contributions from European Cooperation in Science and Technology Action CM1407, “Challenging Organic Syntheses Inspired by Nature—From Natural Products Chemistry to Drug Discovery.” Research was funded by the Austrian Science Fund W901 Doktoratskolleg Molecular Enzymology (to E.L.Z., S.S., W.K., and R.B.), BioTechMed-Graz Secretome Flagship (to S.S., E.L.Z., G.G., C.H., and R.B.), and Grant BFU2016-75319-R (Agencia Española de Investigacion/Fondo Europeo de desarrollo regional, Unión Europea) (to J.F.D.) from Ministerio de Economica y Competitividad.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819154116/-/DCSupplemental.
References
- 1.Fischbach MA, Segre JA. Signaling in host-associated microbial communities. Cell. 2016;164:1288–1300. doi: 10.1016/j.cell.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, et al. Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 5.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. 2017;279:70–89. doi: 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 8.Hajishengallis G, Lamont RJ. Dancing with the stars: How choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–1033. doi: 10.1111/cmi.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joice R, Yasuda K, Shafquat A, Morgan XC, Huttenhower C. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014;20:731–741. doi: 10.1016/j.cmet.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppel N, Balskus EP. Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem Biol. 2016;23:18–30. doi: 10.1016/j.chembiol.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Donia MS, Fischbach MA. HUMAN MICROBIOTA. Small molecules from the human microbiota. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg N, et al. Natural products as mediators of disease. Nat Prod Rep. 2017;34:194–219. doi: 10.1039/c6np00063k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaugerie L, et al. Infectious Colitis Study Group Klebsiella oxytoca as an agent of antibiotic-associated hemorrhagic colitis. Clin Gastroenterol Hepatol. 2003;1:370–376. doi: 10.1053/s1542-3565(03)00183-6. [DOI] [PubMed] [Google Scholar]
- 16.Högenauer C, et al. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med. 2006;355:2418–2426. doi: 10.1056/NEJMoa054765. [DOI] [PubMed] [Google Scholar]
- 17.Schneditz G, et al. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc Natl Acad Sci USA. 2014;111:13181–13186. doi: 10.1073/pnas.1403274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlikova M, et al. Novel pathway of 3-hydroxyanthranilic acid formation in limazepine biosynthesis reveals evolutionary relation between phenazines and pyrrolobenzodiazepines. Sci Rep. 2018;8:7810. doi: 10.1038/s41598-018-26179-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dornisch E, et al. Biosynthesis of the enterotoxic pyrrolobenzodiazepine natural product tilivalline. Angew Chem Int Ed Engl. 2017;56:14753–14757. doi: 10.1002/anie.201707737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse H, et al. A tricyclic pyrrolobenzodiazepine produced by Klebsiella oxytoca is associated with cytotoxicity in antibiotic-associated hemorrhagic colitis. J Biol Chem. 2017;292:19503–19520. doi: 10.1074/jbc.M117.791558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Tesmar A, et al. Biosynthesis of the Klebsiella oxytoca pathogenicity factor tilivalline: Heterologous expression, in vitro biosynthesis, and inhibitor development. ACS Chem Biol. 2018;13:812–819. doi: 10.1021/acschembio.7b00990. [DOI] [PubMed] [Google Scholar]
- 22.Gerratana B. Biosynthesis, synthesis, and biological activities of pyrrolobenzodiazepines. Med Res Rev. 2012;32:254–293. doi: 10.1002/med.20212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartley JA. The development of pyrrolobenzodiazepines as antitumour agents. Expert Opin Investig Drugs. 2011;20:733–744. doi: 10.1517/13543784.2011.573477. [DOI] [PubMed] [Google Scholar]
- 24.Mantaj J, Jackson PJ, Rahman KM, Thurston DE. From anthramycin to pyrrolobenzodiazepine (PBD)-containing antibody-drug conjugates (ADCs) Angew Chem Int Ed Engl. 2017;56:462–488. doi: 10.1002/anie.201510610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurston DE. Advances in the study of pyrrolo[2,1-c][l,4]benzodiazepine (PBD) antitumour antibiotics. In: Neidle S, Waring MJ, editors. Molecular Aspects of Anticancer Drug-DNA Interactions. Vol 1. Macmillan; London: 1993. pp. 54–88. [Google Scholar]
- 26.Darkoh C, Chappell C, Gonzales C, Okhuysen P. A rapid and specific method for the detection of indole in complex biological samples. Appl Environ Microbiol. 2015;81:8093–8097. doi: 10.1128/AEM.02787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zollner-Schwetz I, et al. Role of Klebsiella oxytoca in antibiotic-associated diarrhea. Clin Infect Dis. 2008;47:e74–e78. doi: 10.1086/592074. [DOI] [PubMed] [Google Scholar]
- 28.Thurston DE, et al. Effect of A-ring modifications on the DNA-binding behavior and cytotoxicity of pyrrolo[2,1-c][1,4]benzodiazepines. J Med Chem. 1999;42:1951–1964. doi: 10.1021/jm981117p. [DOI] [PubMed] [Google Scholar]
- 29.Wells G, et al. Design, synthesis, and biophysical and biological evaluation of a series of pyrrolobenzodiazepine-poly(N-methylpyrrole) conjugates. J Med Chem. 2006;49:5442–5461. doi: 10.1021/jm051199z. [DOI] [PubMed] [Google Scholar]
- 30.Basher MA, Rahman KM, Jackson PJM, Thurston DE, Fox KR. Sequence-selective binding of C8-conjugated pyrrolobenzodiazepines (PBDs) to DNA. Biophys Chem. 2017;230:53–61. doi: 10.1016/j.bpc.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Petrusek RL, Uhlenhopp EL, Duteau N, Hurley LH. Reaction of anthramycin with DNA. Biological consequences of DNA damage in normal and xeroderma pigmentosum cell. J Biol Chem. 1982;257:6207–6216. [PubMed] [Google Scholar]
- 32.Mazouzi A, et al. Repair of UV-induced DNA damage independent of nucleotide excision repair is masked by MUTYH. Mol Cell. 2017;68:797–807.e7. doi: 10.1016/j.molcel.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Moder M, et al. Parallel genome-wide screens identify synthetic viable interactions between the BLM helicase complex and Fanconi anemia. Nat Commun. 2017;8:1238. doi: 10.1038/s41467-017-01439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaspers NG, et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair (Amst) 2002;1:1027–1038. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 35.Giannakakou P, et al. Paclitaxel-resistant human ovarian cancer cells have mutant beta-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem. 1997;272:17118–17125. doi: 10.1074/jbc.272.27.17118. [DOI] [PubMed] [Google Scholar]
- 36.Kopka ML, et al. Crystal structure of a covalent DNA-drug adduct: Anthramycin bound to C-C-A-A-C-G-T-T-G-G and a molecular explanation of specificity. Biochemistry. 1994;33:13593–13610. doi: 10.1021/bi00250a011. [DOI] [PubMed] [Google Scholar]
- 37.Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst) 2015;36:13–18. doi: 10.1016/j.dnarep.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Epanchintsev A, et al. Cockayne’s syndrome A and B proteins regulate transcription arrest after genotoxic stress by promoting ATF3 degradation. Mol Cell. 2017;68:1054–1066.e6. doi: 10.1016/j.molcel.2017.11.009. [DOI] [PubMed] [Google Scholar]
- 40.Li CL, et al. Tripartite DNA lesion recognition and verification by XPC, TFIIH, and XPA in nucleotide excision repair. Mol Cell. 2015;59:1025–1034. doi: 10.1016/j.molcel.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohena CC, Mooberry SL. Recent progress with microtubule stabilizers: New compounds, binding modes and cellular activities. Nat Prod Rep. 2014;31:335–355. doi: 10.1039/c3np70092e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field JJ, Díaz JF, Miller JH. The binding sites of microtubule-stabilizing agents. Chem Biol. 2013;20:301–315. doi: 10.1016/j.chembiol.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 43.Alushin GM, et al. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saez-Calvo G, et al. Triazolopyrimidines are microtubule-stabilizing agents that bind the Vinca inhibitor site of tubulin. Cell Chem Biol. 2017;24:737–750.e6. doi: 10.1016/j.chembiol.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 45.Carrion AF, et al. Severe colitis associated with docetaxel use: A report of four cases. World J Gastrointest Oncol. 2010;2:390–394. doi: 10.4251/wjgo.v2.i10.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008;46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowrouzian FL, Oswald E. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog. 2012;53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Payros D, et al. Maternally acquired genotoxic Escherichia coli alters offspring’s intestinal homeostasis. Gut Microbes. 2014;5:313–325. doi: 10.4161/gmic.28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putze J, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nougayrède JP, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 51.Cuevas-Ramos G, et al. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joainig MM, et al. Cytotoxic effects of Klebsiella oxytoca strains isolated from patients with antibiotic-associated hemorrhagic colitis or other diseases caused by infections and from healthy subjects. J Clin Microbiol. 2010;48:817–824. doi: 10.1128/JCM.01741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herzog KA, et al. Genotypes of Klebsiella oxytoca isolates from patients with nosocomial pneumonia are distinct from those of isolates from patients with antibiotic-associated hemorrhagic colitis. J Clin Microbiol. 2014;52:1607–1616. doi: 10.1128/JCM.03373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furda AM, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. Methods Mol Biol. 2012;920:111–132. doi: 10.1007/978-1-61779-998-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Hunt CP, et al. PCR-based analysis of mitochondrial DNA copy number, mitochondrial DNA damage, and nuclear DNA damage. Curr Protoc Toxicol. 2016;67:20.11.1–20.11.25. doi: 10.1002/0471140856.tx2011s67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreu JM. Large scale purification of brain tubulin with the modified Weisenberg procedure. Methods Mol Med. 2007;137:17–28. doi: 10.1007/978-1-59745-442-1_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.