Significance
Hypochlorous acid (HOCl) is a powerful oxidant that kills microorganisms. HypT has been identified as a HOCl-sensing transcription factor regulating several genes to enhance survival during HOCl stress in Escherichia coli. However, the structure and a detailed action mechanism of HypT have not yet been reported. In this study, we identified Salmonella Typhimurium HypT as a crucial factor in survival within macrophages and presented structures of HypT. The full-length structure shows interesting features describing a type of tetrameric assembly for the LysR family transcription regulator. The regulatory domain structures at various states give important clues to understanding the HOCl-sensing mechanism. Combining these results, we provided a molecular mechanism for HypT that explains its HOCl-sensing ability and structural changes.
Keywords: hypochlorous acid, LysR-type transcription regulator, HypT, methionine oxidation, repression mechanism
Abstract
Hypochlorous acid (HOCl) is generated in the immune system to kill microorganisms. In Escherichia coli, a hypochlorite-specific transcription regulator, HypT, has been characterized. HypT belongs to the LysR-type transcriptional regulator (LTTR) family that contains a DNA-binding domain (DBD) and a regulatory domain (RD). Here, we identified a hypT gene from Salmonella enterica serovar Typhimurium and determined crystal structures of the full-length HypT protein and the RD. The full-length structure reveals a type of tetrameric assembly in the LTTR family. Based on HOCl-bound and oxidation-mimicking structures, we identified a HOCl-driven methionine oxidation mechanism, in which the bound HOCl oxidizes a conserved methionine residue lining the putative ligand-binding site in the RD. Furthermore, we proposed a molecular model for the oxidized HypT, where methionine oxidation by HOCl results in a conformational change of the RD, inducing a counter rotation of the DBD dimers. Target genes that are regulated by HypT and their roles in Salmonella were also investigated. DNase I footprinting experiments revealed a DNA segment containing two pseudopalindromic motifs that are separated by ∼100 bp, suggesting that only the oxidized structure makes a concomitant binding, forming a DNA loop. An understanding of the HypT-mediated mechanism would be helpful for controlling many pathogenic bacteria by counteracting bacterial HOCl defense mechanisms.
Hypochlorous acid (HOCl) is ubiquitously produced in higher eukaryotes, including humans and Drosophila (1–3). HOCl is also commonly used to kill microorganisms in food industries, hospitals, and households due to its high reactivity toward macromolecules that causes oxidation of proteins and extensive damage to DNA (4–6). HOCl directly oxidizes amino acids containing amines and sulfurs by electrophilic reaction (6, 7). In particular, methionine is oxidized to methionine sulfoxide by HOCl at physiological pH via the chlorinated forms at the reaction rates approaching the diffusion limit (4–6, 8).
Many transcription factors that respond to different oxidative stresses have been identified (9). Among these, OxyR has been well characterized as a major H2O2-specific transcription factor in most Gram-negative bacteria (10–12). OxyR is a typical LysR-type transcriptional regulator (LTTR). These usually function in a tetrameric assembly that involves an N-terminal DNA-binding domain (DBD) and a C-terminal regulatory domain (RD). The crystal structure of Pseudomonas aeruginosa OxyR revealed that OxyR specifically recognizes H2O2 in a small ligand-binding site located in an interface between RD-I and RD-II, the two subdomains of RD (13). OxyR has two redox states for two invariant cysteine residues in RD-II. In the absence of H2O2, the cysteine residues remain as free thiol forms in the conformation representing the inactive state. In the presence of H2O2, the cysteine residues are oxidized to a disulfide, leading to a conformational change to the active state (14).
Recently, a novel HOCl-specific transcription factor (HypT) was identified in Escherichia coli (15). HypT contributes to HOCl resistance in E. coli by up-regulating the biosynthesis of methionine and cysteine, and by down-regulating genes promoting iron acquisition (15–17). E. coli HypT belongs to the LTTR family, but its oligomeric state in the functional form had previously been elusive (15, 18). Biochemical and mutational studies have suggested that three methionine residues (Met123, Met206, and Met230) play an important role in its activity (18, 19). Individual substitution of the methionine residues by isoleucine abolished the activity of HypT to the HOCl resistance of the bacteria (19). However, it remains unclear how HypT specifically and sensitively recognizes HOCl and how it controls regulons at the molecular level. In this study, we identified an ortholog of E. coli HypT in Salmonella enterica serovar Typhimurium (S. Typhimurium) as a crucial factor in survival within macrophages. We determined the crystal structures of HypT from S. Typhimurium at various states, which account for its molecular mechanisms of action and regulation.
Results
Identification of HypT in S. Typhimurium.
For a HypT ortholog candidate, we found an uncharacterized LysR-type transciption regulator gene (WP_000383509) in the S. Typhimurium genome that showed high sequence identity (82%) to E. coli HypT. The gene contained three methionine residues (Met123, Met206, and Met230) that were previously characterized as important HOCl-sensing residues in E. coli HypT (19). However, two cysteine residues (Cys4 and Cys150) in E. coli HypT are replaced like other HypT homologs (19) (SI Appendix, Fig. S1).
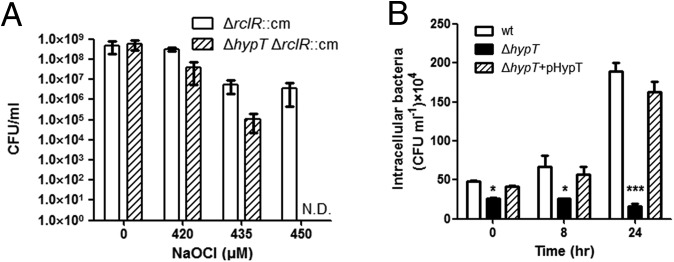
The role of WP_000383509 against HOCl stress was investigated using Salmonella strains cultured in the M9 minimal medium. To generate HOCl stress, NaOCl were treated to the bacteria for 15 min, and then the viabilities were measured. To exclude the effect of another HOCl-sensing transcription factor RclR (20), the susceptibility to HOCl was analyzed using the rclR-deleted strain. Disruption of the gene dropped the viability by 1 or 2 log values under the rclR-deleted background, and the viability was recovered by adding the gene (Fig. 1A and SI Appendix, Fig. S2A). These findings indicate that WP_000383509 is involved in resistance of Salmonella to HOCl stress.
Fig. 1.
Functional importance of hypT (WP_000383509) from S. Typhimurium. (A) Viabilities of the ΔrclR strain and ΔhypT (WP_000383509)/ΔrclR strain depending on the concentration of HOCl. Error bars represent the SD (N.D., not detected). (B) Survival of Salmonella strains in RAW264.7 murine macrophage cell line. Wild-type strain (wt), the hypT-deleted strain (ΔhypT), and the hypT-deleted strain harboring pUHE21-2lacIq::hypT (ΔhypT+pHypT) were added to monolayer of RAW264.7 cells at a multiplicity of infection of 10. After 30 min, 0, 8, or 24 h after infection, intracellular bacteria were plated for enumeration. Each strain was tested in triplicate, and the average cfu·mL−1 was calculated. Asterisks indicate statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001) by unpaired t test comparing the ΔhypT to the wild-type strain.
To further examine the role of the gene in Salmonella pathogenesis, we measured the replication ability of the WP_000383509-deleted S. Typhimurium strain under rclR-intact background within the macrophage vacuoles presumably containing HOCl. Deletion of the gene dramatically decreased the replicative ability of Salmonella within macrophages, and the complementation of the gene fully restored the replicative ability (Fig. 1B). Our results indicate that the WP_000383509 is crucial in the survival of Salmonella in macrophages regardless of the presence of rclR. Taken together, our findings indicate WP_000383509 as HypT of S. Typhimurium (StHypT).
A Configuration in the StHypT Tetramer.
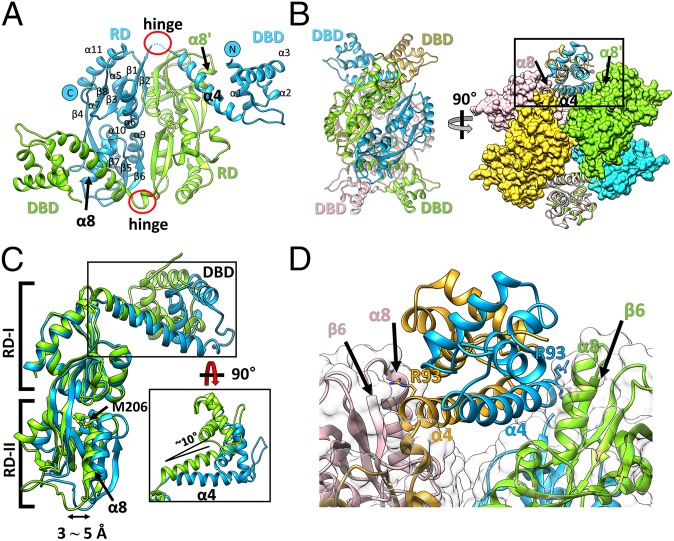
We determined the structure of the full-length StHypT protein (StHypT-FL) at 3.3-Å resolution using crystals grown under reducing conditions. The asymmetric unit contained two StHypT molecules, and each molecule consisted of the N-terminal DBD (residues 1–93) and the RD (residues 100–302), which are linked by a flexible hinge (residues 94–99) (Fig. 2A). A globular homotetramer was built by combining two neighboring asymmetric units, which is consistent with the measured molecular mass using the size exclusion chromatography results (SI Appendix, Fig. S3). These observations strongly suggest that the tetramer is the functional unit of StHypT, as in most LTTR family proteins. The StHypT tetramer can be described as having dimer-of-dimers symmetry, formed by two dimeric interfaces (the DBD-to-DBD in SI Appendix, Fig. S4A and the RD-to-RD in SI Appendix, Fig. S4B) (Fig. 2B). The structure of the DBD dimer is very similar to that of the typical LTTRs (21), and the dimeric interface between the DBDs is also largely stabilized by hydrophobic interactions in the linker helix (α4), as in most LTTRs (SI Appendix, Fig. S4A) (13). Likewise, the overall structure of the RD dimer of StHypT is similar to that of typical LTTRs. The RD dimer of StHypT is stabilized by β/β packing interactions between the β4-β8-β3-β1-β2 sheets of one protomer and the β6-β5-β7-β4-β8 sheets of the other protomer. The angle between each protomer is ∼60° (SI Appendix, Fig. S4B).
Fig. 2.
Overall structure of wild-type full-length StHypT. (A) The structure of the asymmetric unit containing two protomers (chain A in green, chain B in cyan). The N and C termini and the secondary structural elements are labeled on chain B. (B) Two orthogonal views of the tetramer structure by combining two asymmetric units related to the crystallographic twofold symmetry. (Left) The four promoters are shown in ribbon representation with different colors (green, cyan, yellow, and pink). (Right) RDs are shown in the surface representations, and DBDs are in the ribbon diagram. The contact region between α4 in the DBD and α8 in RD-II is marked as a box, which is enlarged in D. The yellow and pink protomers are symmetry-related molecules to the green and cyan protomers, respectively. (C) Structural superposition of two protomers using the RDs as references. The DBDs have 10° angular variation (black box), and RD-II has a 3∼5 Å translational shift between two protomers at α8. (D) A close-up view of the box region of B. Each linker helix (α4) makes direct contact with α8 in RD-II of the other protomer in the asymmetric unit. Concomitantly, the Arg93 residues in α4 (stick representation) interact with the loop between α8 and β6 in RD-II.
When two protomers in the asymmetric unit were superposed using the RD as a reference, we observed a translational variation (3∼5 Å) in the subdomain RD-II of the RD and a rotational variation (∼10°) of the DBD (Fig. 2C). The two variations are structurally related mainly through the interaction between Arg93 of the DBD and the α8 in RD-II of the other subunit in the dimer (Fig. 2 B and D). The interdomain hinge and the intersubunit interaction between the DBDs and the RDs suggest that the structural changes in the RD result in the structural change of the DBD.
The domain arrangement of StHypT is remarkably different from any other LTTR structure (22–26). To analyze the conformational difference between StHypT and the typical LTTRs, the dimeric units of StHypT and OxyR from P. aeruginosa were taken from the tetramers based on the RD-RD dimeric interface (SI Appendix, Fig. S5A). OxyR, a typical LTTR, displayed an intrinsic asymmetric feature. One subunit exhibited the so-called “extended conformation,” in which the DBD is extended away from the RD, and there is no direct interaction between them (22–25) (SI Appendix, Fig. S5B, Left; yellow). The other subunits exhibited the “compact conformation,” in which a folded-in DBD makes close contact with the RD of the same subunit (SI Appendix, Fig. S5B, Left; tan). In contrast, all subunits in the StHypT tetramer were in a largely single conformation, distinguished from both the extended and the compact conformations (SI Appendix, Fig. S5B, Right). In StHypT, the DBD of one subunit in the dimer was near the RD of the other subunit, with close proximity (albeit no direct interaction) between the linker helix (α4) and the RD-II (SI Appendix, Fig. S5B, Right).
These conformational differences between the subunits of StHypT and OxyR resulted in large differences in the DNA-binding modes. The two DBD dimers in the OxyR tetramer face the same direction (SI Appendix, Fig. S5B, Left), and the OxyR tetramer binds to two adjacent sites of a slightly curved DNA molecule (SI Appendix, Fig. S5C). In contrast, each of the DBD dimer in the StHypT tetramer face opposite sides with a parallel arrangement (Fig. 2B).
HOCl-Binding Site in the RD.
To reveal more detailed features of the StHypT RD, we determined its crystal structure (StHypT-RD) at a higher resolution (2.0 Å; SI Appendix, Table S1). The wild-type StHypT-RD showed a very similar structure as the RD region of HypT-FL (rmsd 0.628 Å for 188 Cα atoms). The typical ligand-binding site cavity of LTTR was well conserved in StHypT-RD, located between RD-I and RD-II (Fig. 3A) (22). Interestingly, Met206, one of three important methionine residues in E. coli HypT (19), was placed near the ligand (or solvent)-accessible cavity (Fig. 3B). The Met206 residue was in the α8 helix of RD-II, and its side chain was inserted into a small hydrophobic pocket composed of Leu174, Tyr200, Tyr205, Leu209, Ile210, Pro248, Tyr250, and Ile273 residues (Fig. 3B). Regarding the other two methionine residues, Met123 was surround by the hydrophobic residues in RD-I, and Met230 was close to the twofold axis of the RD dimer (Fig. 3A and SI Appendix, Fig. S6 A and B).
Fig. 3.
The cavity between the two subdomains of StHypT-RD. (A) A 2.0-Å resolution structure of StHypT-RD, which is drawn in ribbon representation. The dimer, colored in pink and gray, is generated by the crystallographic symmetry (Left). Three methionine residues are in ball-and-stick representation (green). The cavities between the two subdomains (RD-I and RD-II) are indicated by red arrows, which is detailed in B. (B) An enlarged intersection view of the cavity. The Met206 residue (green, ball-and-stick) is located in a solvent-accessible position and is surrounded by the residues shown in brown sticks. (C and D) Structural details of cavities are depicted with the omit maps around water molecules in apo-form (C) and an HOCl (or OCl−) molecule in HOCl-soaked StHypT-RD (D). The Fo-Fc omit maps were contoured at 3.0 σ (mesh). Dotted-lines indicate the interaction between protein and small molecules (water or HOCl). N, O, S, and Cl atoms are colored in blue, red, yellow, and green, respectively.
In the apo-structure of StHypT-RD, two water molecules were bound in the cavity around the Met206 residue (Fig. 3C). To better define the HOCl-binding site, we briefly soaked the HypT-RD crystals in HOCl and determined the structure at 1.75-Å resolution (SI Appendix, Table S1). Remarkably, a peanut-shaped electron density map was found in the cavity (consistent with the typical ligand-binding site of LTTRs), and the conserved Met206 residue was placed near the peanut-shaped electron density map (SI Appendix, Fig. S7A). Difference Fourier maps excluded the possibility that the electron density represented two water molecules. Furthermore, we examined a bromine ion in the place of the electron density map in a crystal structure grown in the presence of NaBr (SI Appendix, Fig. S7B). Collectively, we concluded that the electron density map represents an HOCl (or ClO−) molecule. The local environment surrounding the electron density map also likely holds an HOCl molecule (Fig. 3D). The backbone -NH group of Ser110, Tyr205, and Met206 and the side chain hydroxyl group of Ser110 are together involved in its binding. In summary, HypT-RD captures an HOCl molecule near the Met206 residue, which might be important for the selectivity and specificity of StHypT.
To investigate the role of Met206 in the bacteria, we added a plasmid that overexpressed mutant hypT (M206Q or M206L) to the hypT/rclR-disrupted Salmonella strain. Glutamine was chosen to mimic methionine sulfoxide, an oxidized methionine (27, 28), and leucine was chosen to represent the oxidation-free state because leucine is rarely oxidized. The transcriptional impact of the HypT M206Q mutant was similar to the wild-type HypT, while that of the HypT M206L mutant was significantly reduced (SI Appendix, Fig. S2B). We did not see better activity for hypT M206Q, possibly because methionine sulfoxide is bulkier than glutamine, and thus glutamine seems not to fully mimic the oxidized methionine. However, substitution with the leucine residue failed to activate HypT, indicating the importance of the oxidization of Met206 for the activity of HypT. Taken together, our results further confirmed the importance of Met206 in sensing HOCl.
The Roles of Methionine Residues in the Oxidized Conformation.
We determined the structure of a HypT RD M206Q mutant protein at 2.6-Å resolution to understand the HOCl sensing mechanism of HypT. The asymmetric unit of the crystal contained two RD dimers in different conformations (SI Appendix, Fig. S8). Of these, one dimer (chain A:B) showed essentially the same structure as the wild-type RD structure (rmsd 0.802 Å for 367 Cα atoms). The Gln206 in the mutant structure (chain A:B) was in the reduced conformation, since the side chain was inserted into the small hydrophobic pocket (as is Met206 in the wild-type structure) (Figs. 3B and 4A). However, the other dimer (chain C:D) showed substantial structural differences from the RD structure in the reduced conformation (rmsd 2.401 Å for 308 Cα atoms), especially in the active site near the Gln206 residue and in the intersubdomain orientation in the RD (Fig. 4A).
Fig. 4.
Structural comparison between the reduced and oxidized form of StHypT-RD. (A) Superposition of the Met206 (or M206Q) residue and its surrounding local environment in StHypT. The hydrophobic core in the RD is indicated by a red line. The apo-form of wild type, the HOCl-soaked form of wild type, chain B in the M206Q mutant, and chain C in the M206Q mutant of StHypT-RD are in green, cyan, pink, and orange, respectively. All of the structures display the reduced conformation, except for the M206Q StHypT-RD chain C (orange) in the oxidized conformation. (B) The superposition of the reduced form (the apo-form of wild-type StHypT-RD; green) and oxidized form (the M206Q StHypT-RD chain C; orange). RD-I and RD-II subdomains are indicated. The position of the DBD is shown as a dark blue oval. The rotation angle of the β1-β2 sheet and the shift length of α5 are indicated by a red curved arrow and a black arrow, respectively. (C) The intersubunit orientation in the reduced and oxidized conformations. The structures in the reduced conformation are superposed in Left. The representative structure in the reduced conformation (the wild-type StHypT-RD; green) is superposed on the structure in the oxidized conformation (M206Q StHypT-RD chain D; orange). Note the angle of α8 between the two different conformations. For clarity, the subunit on the back side of the RD dimer in Cα is represented in gray.
The Gln206 residue was moved away from the small hydrophobic pocket consisting of α5 in RD-I, resulting in the oxidized conformation (Fig. 4A). This oxidized conformation might result from the polar side chain of glutamine, which is not as suitable for the hydrophobic pocket as is the side chain of methionine. Interestingly, this conformational change in Gln206 induced asymmetric changes between the protomers (chain C:D) in the oxidized conformation. In chain C, the entire RD-I subdomain rotated around RD-II by ∼30° within the RD, as the α5, β1, and β2 in RD-I were moved because the small hydrophobic pocket was collapsed (Fig. 4B). In chain D, RD-II was rotated in the dimer instead of RD-I. The β6-β5 sheet of RD-II of chain D packed together with the β1 and β2 of RD-I in chain C (SI Appendix, Fig. S4B). The rotation of RD-I in chain C caused the rotation of RD-II in chain D. As a result, the RD-II of chain D is rotated by ∼30°, with movement of α8 in the RD-II by 9 Å with respect to RD-I (Fig. 4C). These observations suggest that the oxidation of Met206 leads to a global positional shift of the RD dimers and the DBDs in the full-length HypT tetramer.
Structural Modeling of the Oxidized HypT.
We built a structural model of the full-length HypT in the oxidized state. The RD structures of the M206Q mutant in the oxidized conformation was superposed onto the full-length structure of the HypT tetramer in the reduced conformation. Given that the molecular contact between α4 of the DBD and α8 in the RD governed the angle of the DBD with respect to the RD (Fig. 2D), the rotation of the DBD dimers was expected in response to the HOCl-driven oxidation in the RD. The movement of β1 and α8 of the other subunit resulted in the clockwise rotation of the DBD dimers by ∼20° and a total ∼40° rotation between the DBD dimers in the tetrameric assembly (SI Appendix, Fig. S9).
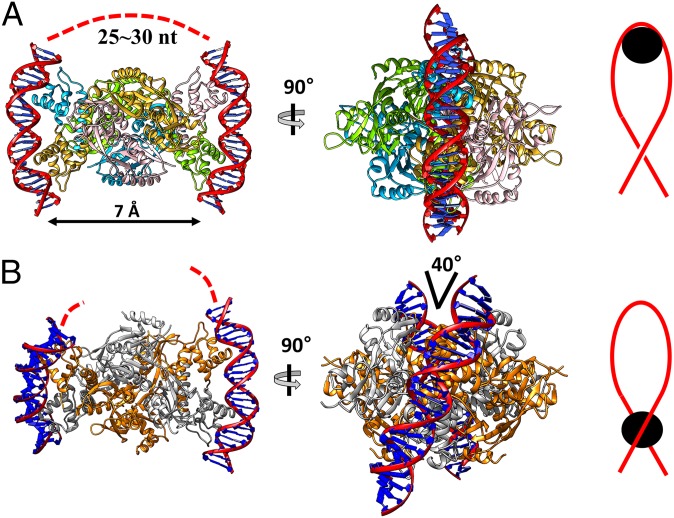
We next compared the possible DNA-binding modes between the reduced and the oxidized structures of the HypT tetramer. The reduced form of the StHypT tetramer could bind to two parallel DNA segments that are curved by interaction with the DBD dimer (Fig. 5A, Left and Center). The RD surfaces of StHypT between the two DBD dimers are highly basic, indicating that the side surface can bind a DNA segment (25–30 nt) (SI Appendix, Fig. S5D) and, thus, connect the two curved bound DNA segments (Fig. 5A, Left). This DNA-winding mode is comparable to the histone octamer that scaffolds the DNA in eukaryotic cells (29).
Fig. 5.
DNA docking models of the reduced and oxidized HypT. Two DNA fragments (red double helices) are docked on the reduced structure (A) and the oxidized structure (B) using the BenM DBD–DNA complex structure (PDB ID code: 4IHS). The flanking DNA between the two fragments is depicted by the dotted red lines. Red lines represent the negatively supercoiled DNA, and black circles indicate the site of binding of the HypT tetramer.
In contrast to two parallel DNA segments bound to the reduced HypT, two DNA segments adopting a crossing angle of ∼40° could be docked onto the DBD dimers of the oxidized HypT tetramer model (Fig. 5B). The crossing angle between the bound DNA segments did not allow the wrapping around the HypT tetramer in the oxidized conformation. Instead, the oxidized HypT could be bound to a negative-supercoiled DNA loop (Fig. 5B).
StHypT Target Genes and StHypT Binding DNA Segments.
We compared the transcription levels of putative target genes (feoA, feoB, fhuA, fhuC, sitC, sitD, metN, metB, and cysH) in S. Typhimurium SL1344 cultured in the M9 minimal media. In the absence of HOCl, the transcription levels of the putative target genes were not influenced by a deletion of hypT (SI Appendix, Fig. S10A). In the presence of HOCl stress, however, the hypT-deletion strain showed a twofold higher transcription level of iron acquisition-related genes (feoA, feoB, fhuA, fhuC, sitC, and sitD) than the wild-type strain. The increased transcription levels in the hypT-deleted mutant returned to the levels of the wild type by adding a hypT-expressing plasmid (SI Appendix, Fig. S10B). Consistently, intracellular iron level of the hypT-deleted strain cultured in the LB medium in the presence of HOCl stress were substantially lower than that of the wild-type strain (SI Appendix, Fig. S10C). However, genes related to the biosynthesis of cysteine and methionine (metN, metB, and cysH) in the hypT-deleted S. Typhimurium showed no significant differences in their transcript levels, unlike E. coli cultured in the LB medium (15) (SI Appendix, Fig. S10 A and B). These discrepancies might result from the different growth media and lower levels of HOCl treatment than for HypT of E. coli.
When the wild-type strain was exposed to HOCl, the transcription levels of iron acquisition-related genes were decreased (SI Appendix, Fig. S10D) without changing the mRNA levels of hypT itself (SI Appendix, Fig. S10E). Furthermore, the electrophoretic mobility shift assay (EMSA) with the sit, feo, and fhu operons revealed that the purified HypT protein directly bound to their possible promoter regions (SI Appendix, Fig. S11). Based on these results, we concluded that in Salmonella, HypT directly represses these genes in response to HOCl stress. It is interesting that the iron acquisition-related genes are repressed even in an iron-limited condition. The M9 media used in this study contained a very limited concentration of iron, where the master iron regulator Fur is in the inactive apo-forms and thus cannot repress the iron acquisition genes (30, 31) (SI Appendix, Fig. S10F). We further observed the recovery of the bacterial viability at a lethal dose of HOCl by addition of the membrane permeable the iron chelator dipyridyl (SI Appendix, Fig. S12), indicating that the toxicity of HOCl is mainly mediated by iron in Salmonella.
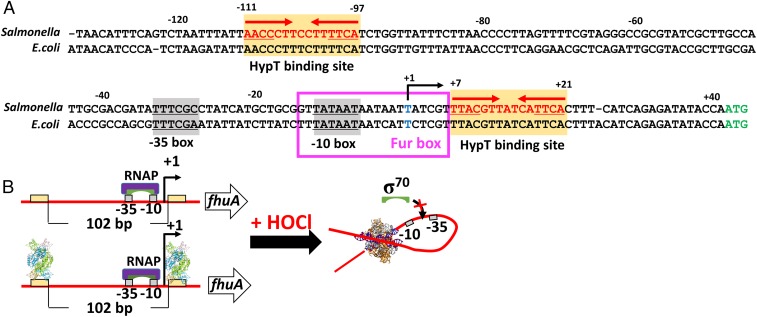
To investigate the HypT–DNA interaction, the fhu operon was selected as a model promoter. Since S. Typhimurium and E. coli have nearly identical nucleotide sequences in the promoter region of the fhu operon, the transcription start site (TSS) and σ70 binding sites (−10 and −35 boxes) in the fhuA promoter were adopted from E. coli (32) (Fig. 6A). Multiple DNase I footprinting experiments revealed two HypT-binding sites: TTACGTTATCATTCA, +7 ∼ +21 from the TSS and AACCCTTCCTTTTCA, −111 ∼ −97 from the TSS (SI Appendix, Fig. S13). The DNA region between the two binding sites contains the −10 box and −35 box, the TSS, and Fur binding site (Fur box). The HypT-binding sites exhibit pseudopalindromic patterns, as is observed in other LTTRs (33, 34) (Fig. 6A). However, the spacing (102 nt) between the two binding sites in StHypT are far different from those of typical LTTRs (7 nt in the case of OxyR; ref. 33). The EMSA, using the various DNA fragments, confirmed the results from DNase I footprinting experiments and further suggested that the reduced and the oxidized HypT tetramers share the binding sites in the fhu promoter (SI Appendix, Fig. S13C).
Fig. 6.
A fhuA promoter and its repression mechanism by HypT. (A) The structure of the fhuA promoter. The fhuA promoter from Salmonella was analyzed by comparing with the E. coli fhuA promoter. The −10 and −35 boxes, recognized by σ70, are highlighted by gray boxes. The StHypT binding sequences (red letters on yellow boxes) were determined by DNase I footprinting assay (SI Appendix, Fig. S13). The pseudopalindromic sites are indicated with underlines and red arrows. The transcription start site and translation initiation codon are shown in blue and green letters, respectively. Fur box is indicated by a magenta box. (B) A proposed repression mechanism of StHypT. Red lines, yellow boxes, and gray boxes represent DNA, the StHypT binding site, and the σ factor binding site (−35 and −10 box), respectively. The combination of purple and green illustrations presents the RNA polymerase complex (RNAP), and among them, the green shapes represent σ70.
Discussion
Given the high sequence similarity between E. coli HypT and StHypT, the structure in the functional states and the regulation mechanisms would be shared. However, previous studies on E. coli HypT have presented a ring-like dodecameric assembly by low-resolution negative-staining electron microscopy (15). Functions of five cysteine residues of E. coli HypT were investigated (35). Only Cys4 in E. coli HypT, which was replaced with threonine in StHypT, was implicated as an oxidation-sensitive residue that could make an intermolecular disulfide bond (35). This difference at the site of Cys4 might account for the higher oligomeric assembly of HypT, which is confined in E. coli HypT (SI Appendix, Fig. S1).
Several homologs of HypT based on the sequence similarity have been found in many Gram-negative bacteria (15). However, to date, most of these genes have not been functionally characterized, except for E. coli HypT and StHypT. Although further functional studies are required, Met206 is the hallmark for the HypT orthologs, since the methionine residue is directly involved in HOCl sensing, according to our results. A recent study on a HypT homolog (VV2_1132) from Vibrio vulnificus, in which Met206 is replaced with leucine, confirmed that it did not function as a HOCl-responsive factor (36, 37). Two other methionine residues (Met123 and Met230) were characterized in E. coli HypT (19). Both methionine residues are in the moving parts and have less hydrophobic interactions in the oxidized conformation (SI Appendix, Fig. S6 C and D). Given the greater polarity of the oxidized methionine, the oxidized methionine residues appear to be more stabilized in the oxidized conformation. Thus, our findings suggest that oxidation of the two methionine residues (Met123 and Met230) render the oxidized conformation of HypT to be more favorable. Why was HOCl recognized by a methionine residue in HypT? This question could be simply explained by the preferential reactivity of methionine with HOCl comparted to H2O2, as discussed in ref. 19. Methionine reacts with H2O2 much more slowly than with HOCl. Therefore, HypT has more selectivity for HOCl by adopting methionine residues for sensing oxidative stress, unlike cysteine-based sensors.
Like H2O2, HOCl generates hydroxyl radicals that are more reactive and induce damage to DNA or other essential cellular molecules in the presence of Fe2+ (7, 38, 39). Indeed, the reaction of Fe2+ with HOCl is much faster than with H2O2 (7). Therefore, it could be more important to reduce the cellular levels of Fe2+ to block the reaction in the bacterial cytosol maybe for the survival within macrophages. To better understand the HOCl-resistance mechanisms of Salmonella, further studies would be required on the interplay or the functional division between HypT and the HOCl-responsive cysteine-based transcriptional regulator RclR.
This study revealed that StHypT in the oxidized state is bound to a DNA segment that contains two pseudopalindromic sequences separated by ∼100 bp in the promoter region of the fhuA promoter. A DNA docking model of the StHypT tetramer in the reduced state suggested a 25- to 30-nt spacing between two DNA motifs, as shown in Fig. 5A. Thus, the concomitant binding of the reduced StHypT tetramer to the long fhuA promoter DNA segment would not be allowed. Only separate binding of the StHypT tetramers on one (or both) of the two HypT-binding sites are expected. In contrast, the oxidized StHypT tetramer would fit and be able to bind to the long DNA segment by looping in a negatively supercoiling manner. From these findings, we propose a regulation mechanism for the gene expression of the fhu operon by HypT. In the absence of HOCl stress, the HypT tetramer is bound to one or two HypT-binding motifs, but it cannot repress the target gene because the σ factor is accessible to −35 and −10 boxes (Fig. 6B). When a HOCl triggers the conformational change of HypT, the concomitant binding of the two HypT-binding motifs is allowed, which ultimately form a DNA loop. Because the RNA polymerase-binding sites (−35 and −10 boxes) are in the loop, gene expression would be strongly repressed (Fig. 6B). In addition, the regulatory mechanism would be changed if the spacing between the two HypT-binding motifs were different.
We observed that the HypT-mediated defense system of Salmonella increases the lethal dose of HOCl and is important in survival of the bacteria within macrophages. An understanding of the HOCl defense mechanism at the molecular level would be helpful for controlling many pathogenic bacteria by counteracting them.
Experimental Procedures
Expression and Purification of HypT for Crystallization.
To overexpress the full-length (1–302) and regulatory domain (RD, 97–302) of HypT (WP_000383509), the genes encoding each protein were amplified and cloned into the pProEX-HTa expression vector (Invitrogen). The resulting plasmid encodes the hexa-histidine tag and TEV protease cleavage site at the N terminus of each protein. For the HypT (M206Q) mutant protein, site-directed mutagenesis was performed. The expression and purification methods are provided in SI Appendix. The purified proteins were concentrated to 20 mg/mL and stored at −80 °C until used for crystallization.
Crystallization, Data Collection, and Structural Determination.
The proteins were crystallized at 14 °C using the hanging-drop vapor diffusion method. The structure of StHypT-FL was determined by the single anomalous dispersion method in the Pohang Accelerator Laboratory Beamline 5C (Pohang, Republic of Korea) (40, 41). The detailed procedures are described in SI Appendix and SI Appendix, Table S1.
The other experimental procedures are described in more detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank Pohang Accelerator Laboratory for the use of the beamline 5C. This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Grant 710012-03-1-HD120) (to N.-C.H.), and from the National Research Foundation of Korea (Grant NRF-2017M3A9F6029755) (to N.-C.H). This study was also supported by 2018 Grant 14162MFDS972 from the Ministry of Food and Drug Safety, Republic of Korea (to S.R.). I.J. and D.K. were supported by the BK21 Plus Program of Department of Agricultural Biotechnology, Seoul National University, Seoul, Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5YDW, 5YDO, 5YDV, 5YER, and 5YEZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811509116/-/DCSupplemental.
References
- 1.Weiss SJ, LoBuglio AF. Phagocyte-generated oxygen metabolites and cellular injury. Lab Invest. 1982;47:5–18. [PubMed] [Google Scholar]
- 2.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 3.Winterbourn CC, Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins CL, Pattison DI, Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids. 2003;25:259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 5.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Gray MJ, Wholey WY, Jakob U. Bacterial responses to reactive chlorine species. Annu Rev Microbiol. 2013;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folkes LK, Candeias LP, Wardman P. Kinetics and mechanisms of hypochlorous acid reactions. Arch Biochem Biophys. 1995;323:120–126. doi: 10.1006/abbi.1995.0017. [DOI] [PubMed] [Google Scholar]
- 8.Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 9.Hillion M, Antelmann H. Thiol-based redox switches in prokaryotes. Biol Chem. 2015;396:415–444. doi: 10.1515/hsz-2015-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storz G, Tartaglia LA, Ames BN. Transcriptional regulator of oxidative stress-inducible genes: Direct activation by oxidation. Science. 1990;248:189–194. doi: 10.1126/science.2183352. [DOI] [PubMed] [Google Scholar]
- 11.Storz G, Tartaglia LA. OxyR: A regulator of antioxidant genes. J Nutr. 1992;122(Suppl 3):627–630. doi: 10.1093/jn/122.suppl_3.627. [DOI] [PubMed] [Google Scholar]
- 12.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 13.Jo I, et al. Structural details of the OxyR peroxide-sensing mechanism. Proc Natl Acad Sci USA. 2015;112:6443–6448. doi: 10.1073/pnas.1424495112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi H, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 15.Gebendorfer KM, et al. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem. 2012;287:6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candeias LP, Stratford MR, Wardman P. Formation of hydroxyl radicals on reaction of hypochlorous acid with ferrocyanide, a model iron(II) complex. Free Radic Res. 1994;20:241–249. doi: 10.3109/10715769409147520. [DOI] [PubMed] [Google Scholar]
- 17.Dukan S, Touati D. Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drazic A, et al. Tetramers are the activation-competent species of the HOCl-specific transcription factor HypT. J Biol Chem. 2014;289:977–986. doi: 10.1074/jbc.M113.521401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drazic A, et al. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc Natl Acad Sci USA. 2013;110:9493–9498. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker BW, Schwessinger EA, Jakob U, Gray MJ. The RclR protein is a reactive chlorine-specific transcription factor in Escherichia coli. J Biol Chem. 2013;288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alanazi AM, Neidle EL, Momany C. The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr D Biol Crystallogr. 2013;69:1995–2007. doi: 10.1107/S0907444913017320. [DOI] [PubMed] [Google Scholar]
- 22.Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 23.Jo I, et al. The hydrogen peroxide hypersensitivity of OxyR2 in Vibrio vulnificus depends on conformational constraints. J Biol Chem. 2017;292:7223–7232. doi: 10.1074/jbc.M116.743765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muraoka S, et al. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: Unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J Mol Biol. 2003;328:555–566. doi: 10.1016/s0022-2836(03)00312-7. [DOI] [PubMed] [Google Scholar]
- 25.Monferrer D, et al. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol Microbiol. 2010;75:1199–1214. doi: 10.1111/j.1365-2958.2010.07043.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, et al. Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J Mol Biol. 2010;396:1012–1024. doi: 10.1016/j.jmb.2009.12.033. [DOI] [PubMed] [Google Scholar]
- 27.Vogt W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy MR, et al. Impact of methionine oxidation on calmodulin structural dynamics. Biochem Biophys Res Commun. 2015;456:567–572. doi: 10.1016/j.bbrc.2014.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 30.Seo SW, et al. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun. 2014;5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjarnason J, Southward CM, Surette MG. Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J Bacteriol. 2003;185:4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: Specific transcription in dense regions of overlapping promoter-like signals. J Mol Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Heo YJ, et al. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J Bacteriol. 2010;192:381–390. doi: 10.1128/JB.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koentjoro MP, Adachi N, Senda M, Ogawa N, Senda T. Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in complex with a DNA fragment of the recognition-binding site in the promoter region. FEBS J. 2018;285:977–989. doi: 10.1111/febs.14380. [DOI] [PubMed] [Google Scholar]
- 35.Drazic A, et al. Role of cysteines in the stability and DNA-binding activity of the hypochlorite-specific transcription factor HypT. PLoS One. 2013;8:e75683. doi: 10.1371/journal.pone.0075683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang Y, et al. A novel tetrameric assembly configuration in VV2_1132, a LysR-type transcriptional regulator in Vibrio vulnificus. Mol Cells. 2018;41:301–310. doi: 10.14348/molcells.2018.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jang Y, Choi G, Jo I, Choi S, Ha N. Purification, crystallization, and preliminary X-ray crystallographic analysis of VV2_1132, a LysR-type transcriptional regulator from Vibrio vulnificus. Biodesign. 2017;5:44–48. [Google Scholar]
- 38.Pryor WA. Oxy-radicals and related species: Their formation, lifetimes, and reactions. Annu Rev Physiol. 1986;48:657–667. doi: 10.1146/annurev.ph.48.030186.003301. [DOI] [PubMed] [Google Scholar]
- 39.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice LM, Earnest TN, Brunger AT. Single-wavelength anomalous diffraction phasing revisited. Acta Crystallogr D Biol Crystallogr. 2000;56:1413–1420. doi: 10.1107/s0907444900010039. [DOI] [PubMed] [Google Scholar]
- 41.Park SY, Ha SC, Kim YG. The protein Crystallography beamlines at the Pohang light source II. Biodesign. 2017;5:30–34. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.