Significance
Staphylococcus aureus is one of the most significant human bacterial pathogens that has the capacity to cause serious infections and become highly resistant to antibiotics. In this study, we identified a metabolic adaptation mechanism used by S. aureus to simultaneously circumvent killing by one of the last-line antistaphylococcal antibiotics, daptomycin, and attack from host innate immune cells. This process led to enhanced bacterial survival and was mediated by a change in bacterial membrane phospholipid composition sufficient to impair daptomycin membrane penetration and significantly affect neutrophil chemotactic responses. These results highlight the importance of bacterial membrane lipid adaptation in bacterial pathogenesis and provide crucial insights into potentially novel therapeutic targeting.
Keywords: S. aureus, daptomycin, cardiolipin, phosphatidylglycerol, neutrophils
Abstract
Staphylococcus aureus is a notorious human bacterial pathogen with considerable capacity to develop antibiotic resistance. We have observed that human infections caused by highly drug-resistant S. aureus are more prolonged, complicated, and difficult to eradicate. Here we describe a metabolic adaptation strategy used by clinical S. aureus strains that leads to resistance to the last-line antibiotic, daptomycin, and simultaneously affects host innate immunity. This response was characterized by a change in anionic membrane phospholipid composition induced by point mutations in the phospholipid biosynthesis gene, cls2, encoding cardiolipin synthase. Single cls2 point mutations were sufficient for daptomycin resistance, antibiotic treatment failure, and persistent infection. These phenotypes were mediated by enhanced cardiolipin biosynthesis, leading to increased bacterial membrane cardiolipin and reduced phosphatidylglycerol. The changes in membrane phospholipid profile led to modifications in membrane structure that impaired daptomycin penetration and membrane disruption. The cls2 point mutations also allowed S. aureus to evade neutrophil chemotaxis, mediated by the reduction in bacterial membrane phosphatidylglycerol, a previously undescribed bacterial-driven chemoattractant. Together, these data illustrate a metabolic strategy used by S. aureus to circumvent antibiotic and immune attack and provide crucial insights into membrane-based therapeutic targeting of this troublesome pathogen.
The Gram-positive organism, Staphylococcus aureus, is one of the most important human bacterial pathogens, with a worldwide distribution and an ability to cause infection of almost any human tissue (1). Effective treatment of staphylococcal infections has been hampered by the emergence of antibiotic resistance, leading to increased reliance on last-line antibiotics such as daptomycin (2). Daptomycin is a cyclic lipopeptide antibiotic that interacts with bacterial cell membranes, but the precise mechanisms of action remain elusive (3). Notably, human infections caused by daptomycin-resistant S. aureus have been associated with persistent and complicated infections (2, 4). We and others have recently shown that clinically derived daptomycin-resistant isolates caused persistent infections in nonmammalian and murine septicaemia models (5, 6), raising the question about the correlation between daptomycin resistance, immune evasion, and bacterial survival in vivo (5, 6).
Bacteria have evolved highly conserved mechanisms mediating adaptation and maintenance of membrane integrity to defend against host microbicidal peptides (7, 8). Mutations in genes related to phospholipid biosynthesis are consistently reported in Gram-positive bacteria resistant to daptomycin (4). The most abundant phospholipid found in Gram-positive bacterial membranes, including staphylococcal membranes, is phosphatidylglycerol (PG). PG can be converted to cardiolipin (CL) and lysyl-phosphatidylglycerol (L-PG) by the enzymes cardiolipin synthase (Cls) and multiple peptide resistance factor (MprF), respectively (9–11). Gain-of-function mutations in MprF have been associated with daptomycin resistance (8, 9, 12). Most human bacterial pathogens have a Cls homolog that catalyzes condensation of two PG molecules to yield one CL and one glycerol molecule (13). In S. aureus, there are two cls genes, with cls2 encoding the major CL synthase (10, 11). Thus far, the bacterial membrane adaptation response to antibiotic and innate immune exposure in humans is poorly understood.
Neutrophils form one of the most fundamental host innate immune effectors against bacterial pathogens, including S. aureus (14). The clinical significance of neutrophils is well illustrated by the predisposition to severe and recurrent staphylococcal infections in patients with functional or quantitative neutrophil deficiencies (15). Neutrophil recruitment to the site of infection caused by Gram-positive bacteria is often mediated by bacterial-driven chemoattractants such as formylated peptides and phenol soluble modulins or by endogenous cytokines (e.g., IL-8) released from host cells (16, 17). In response, bacteria have evolved mechanisms that interfere with neutrophil chemotaxis, including chemotaxis inhibitory protein of S. aureus and formyl peptide receptor-like 1 inhibitor (18, 19). Deepening our understanding of how bacteria evolve during human infection to simultaneously circumvent key innate immune effectors and antibiotic selection pressure is crucial in our pursuit of novel therapeutic strategies.
Results
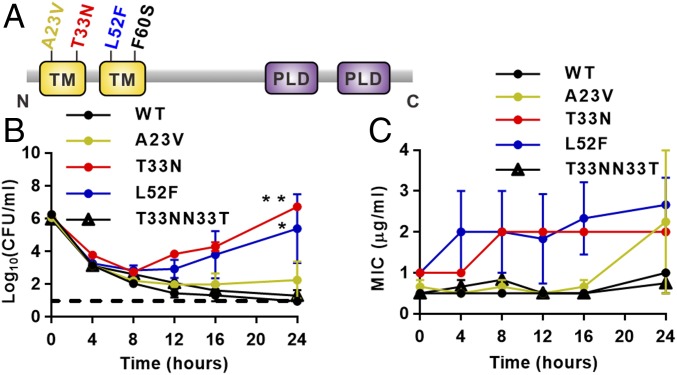
Mutations in S. aureus Cardiolipin Synthase Lead to the Evolution of Daptomycin Resistance and Antimicrobial Failure.
We have previously collected and reported on S. aureus isolates from nine patients with bloodstream infection who were all treated with daptomycin (4). Samples were collected as soon as the infection was detected and later in infection when resistance to daptomycin and treatment failure was evident (4). All patients had persistent bacteremia and complicated infections involving heart valves, bone and joints, and deep soft tissues (4). Whole-genome sequencing of the nine paired samples identified nonsynonymous point mutations in cls2 in daptomycin-resistant isolates, which were tightly positioned in the two predicted N-terminal transmembrane domains encoded by cls2, resulting in A23V, T33N, L52F, and F60S amino acid substitutions (Fig. 1A) (4). To study these nucleotide changes in cls2, independent of other mutations observed in the clinical daptomycin-resistant strains, allelic replacement experiments were performed to introduce the individual point mutations into a daptomycin-susceptible clinical isolate (A8819) producing A8819Cls2A23V, A8819Cls2T33N, and A8819Cls2L52F (SI Appendix, Table S1). Several attempts to generate the F60S substitution were unsuccessful. Daptomycin susceptibility was most perturbed with the T33N mutation (A8819Cls2T33N), which led to a minimum inhibitory concentration (MIC) of daptomycin similar to that observed in our clinical resistant isolates (MIC 2 μg/mL) compared with the susceptible parent strain (MIC 0.5 μg/mL) (SI Appendix, Table S1). This magnitude rise in MIC of daptomycin has been associated with therapeutic failure and poor patient outcomes (2), confirming the significance of the increase. Chromosomal repair of the T33N mutation (A8819Cls2T33NN33T) restored daptomycin susceptibility back to WT levels (MIC 0.5 μg/mL) (SI Appendix, Table S1). L52F and A23V mutations also increased MICs of daptomycin but to a lesser degree (up to 1 μg/mL) (SI Appendix, Table S1).
Fig. 1.
Effect of Cls2 amino acid substitutions on daptomycin killing and resistance emergence. (A) The predicted amino acid change in Cls2 by individual cls2 point mutations. C, carboxyl terminus; N, amino terminus; PLD, phospholipase D domain; TM, transmembrane domain. (B) Time–kill analyses showing quantitative bacterial counts during exposure to 2 μg/mL daptomycin. Dashed line indicates the detection limit. Comparison of the area under the curve between mutants and WT by one-way ANOVA (*P < 0.05, **P < 0.01). (C) Daptomycin MIC of staphylococcal cells at each time point from the time–kill analysis. Error bars represent mean ± SEM, three independent experiments.
To assess the functional effect of the reduced daptomycin susceptibility with the cls2 mutations, we performed daptomycin treatment assays (Fig. 1B). Daptomycin concentrations (2 and 4 μg/mL) were chosen to mimic free daptomycin concentration observed in deep tissues and bones under standard human dosing (20, 21). Using a concentration of 2 μg/mL, we showed that daptomycin was rapidly bactericidal (≥3 log10 reduction in cfu) against all strains over 8 h; however, significant regrowth occurred back to the starting inocula by 24 h for A8819Cls2T33N and A8819Cls2L52F (Fig. 1B). In contrast, killing to undetectable levels was seen for the daptomycin-susceptible parent strain (A8819) and the T33N repaired strain (A8819Cls2T33NN33T). Similar findings were observed with daptomycin concentrations up to 4 μg/mL (SI Appendix, Fig. S1). Daptomycin treatment failure with A8819Cls2T33N and A8819Cls2L52F directly correlated with progressive resistance, with an MIC of daptomycin increasing up to 4 μg/mL, suggesting that additional adaptation occurred under daptomycin exposure (Fig. 1C). Together, these data show that as seen in patients, the observed cls2 point mutations genetically predisposed S. aureus to resistance evolution and therapeutic failure under antibiotic selection.
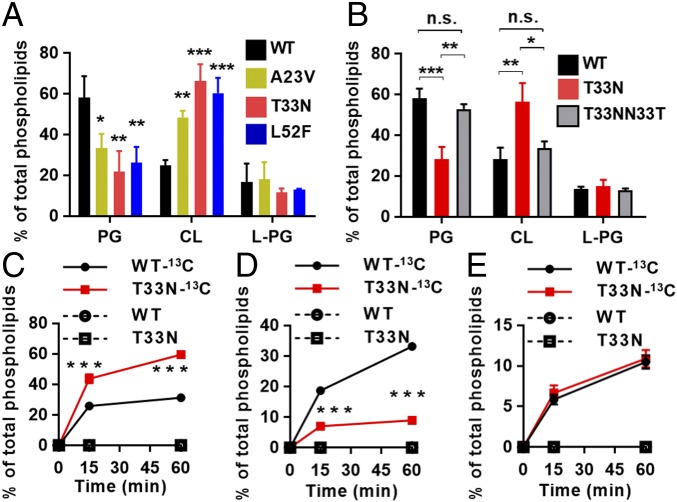
Amino Acid Substitutions in S. aureus Cardiolipin Synthase Enhance Cardiolipin Biosynthesis and Alter Bacterial Membrane Phospholipid Composition.
To determine the effect of the clinically derived cls2 point mutations on membrane phospholipid composition, we extracted total lipids from the daptomycin-susceptible clinical strain (A8819), the three cls2 point mutants (A8819Cls2T33N, A8819Cls2A23V, and A8819Cls2L52F), and the repaired strain (A8819Cls2T33NN33T). We first assessed lipid profiles using TLC (SI Appendix, Fig. S2A), which showed that relative to A8819, the three cls2 point mutants had an increase in membrane CL content and a reduction in PG, with no change in L-PG. A time-course lipid analysis over bacterial growth phases supported these findings (SI Appendix, Fig. S2 B and C). The concentration of individual phospholipid species was then quantified using liquid chromatography coupled with mass spectrometry (LC-MS). Consistent with the TLC analysis, the percentage of CL among total phospholipids increased significantly in the three cls2 point mutants compared with A8819, whereas the PG content decreased significantly (Fig. 2A). None of the cls2 mutations had an effect on L-PG. Overall, the membrane phospholipid changes were most pronounced for A8819Cls2T33N (Fig. 2A), which paralleled the magnitude of daptomycin resistance. As seen with daptomycin susceptibility, repair of the T33N mutation restored the membrane phospholipid profile back to WT levels (Fig. 2B and SI Appendix, Fig. S2D). The increase in CL in the cls2 point mutants was not secondary to an increase in cls2 transcription or Cls2 membrane quantity, and Cls2 membrane localization was unchanged compared with A8819 (SI Appendix, Fig. S3).
Fig. 2.
Effect of cls2 point mutations on membrane phospholipid profiles. (A) The molar ratio of PG, CL, and L-PG among total phospholipids determined by LC-MS. (B) Repair of Cls2 T33N (T33NN33T) restored membrane phospholipid profiles to WT levels. Phospholipid biosynthesis was assessed after treatment with 13C-glycerol. The ratios of (C) 13C-CL, (D) 13C-PG, and (E) 13C-L-PG (all isotopomers) among total lipid content were determined by LC-MS. Mean ± SD, three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001, one-way ANOVA. n.s., nonsignificant. Student’s t test was used to compare T33N to WT (C–E).
To test whether the cls2 mutations increased cardiolipin synthesis activity in vivo, bacteria were metabolically labeled with 13C-glycerol, which is incorporated into the glycerol backbone and head group of PG and CL (SI Appendix, Fig. S4). The formation of complex PG or CL mass isotopomers (+3, +6, and +9) by LC-MS can then be used to measure de novo biosynthesis of these phospholipids (SI Appendix, Fig. S4). Compared with WT, we showed that CL biosynthesis significantly increased over time in our T33N mutant strain (Fig. 2C), and this was mirrored by a reduction in PG biosynthesis (Fig. 2D). Given that PG is both a metabolic end-product and precursor for CL, these results suggest that CL is synthesized from a subpool of PG in the mutant to account for the different degree of 13C-labeling. No difference in biosynthesis of L-PG was observed (Fig. 2E). Together, these data confirm that clinically relevant Cls2 amino acid substitutions caused enhanced membrane cardiolipin biosynthesis that was proportional to the degree of antibiotic resistance.
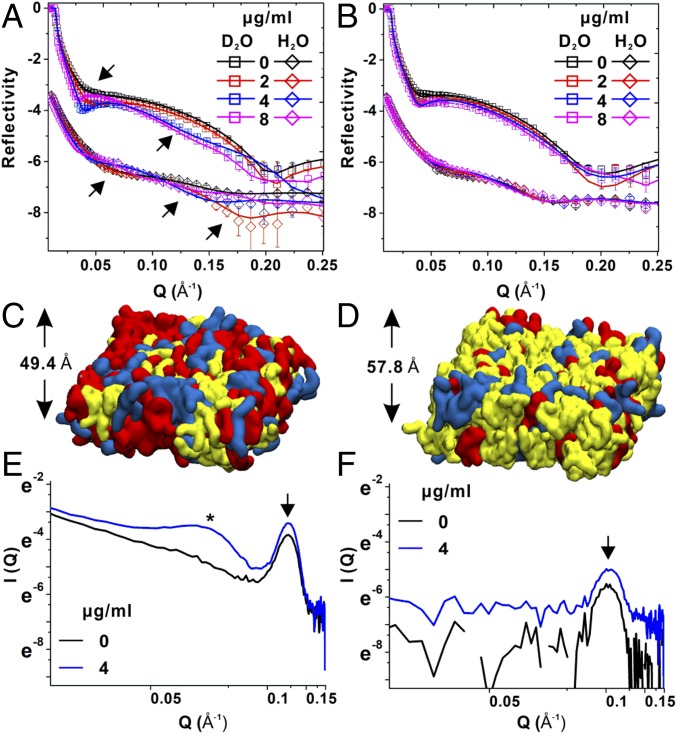
Changes in S. aureus Membrane Composition Block Daptomycin Mediated Lipid Extraction, Membrane Penetration, and Disruption.
To assess the effect of Cls2-mediated lipid changes on daptomycin–membrane interactions, we reconstituted Gram-positive bacterial bilayer membranes immobilized on a planar surface for characterization by neutron reflectometry (NR) (22, 23) (SI Appendix, Fig. S5A). The bilayers were incubated with isotopic solvents (D2O and H2O), and the NR profiles of WT and A8819Cls2T33N were modeled simultaneously to analyze the bilayer structures (Fig. 3 A and B). The cardiolipin-rich membrane caused by the Cls2 T33N amino acid substitution was thicker compared with the WT membrane (57.8 ± 3.7 Å versus 49.4 ± 3.3 Å, respectively) (Fig. 3 C and D and SI Appendix, Fig. S5B and Tables S2 and S3). Despite the overall differences in membrane thickness, the cell surface charge was similar, and using superresolution microscopy of cell surface-bound fluorescent daptomycin, we showed that the level and distribution of daptomycin binding on A8819Cls2T33N cell surface was similar to A8819 (SI Appendix, Fig. S6). These data support the hypothesis that structural modification of the membrane impairs daptomycin penetration and membrane disruption rather than initial binding.
Fig. 3.
Daptomycin–membrane interactions. The neutron reflectivity profiles (symbols) and fits (lines) of (A) A8819 and (B) A8819Cls2T33N membrane models in D2O and H2O are shown. The NR profiles in H2O are offset for clarity. The arrows indicate fringe shifts of NR profiles. The molecular models of reconstituted (C) A8819 and (D) A8819Cls2T33N membranes show a thicker but more CL-rich membrane structure (with CL in yellow, PG in red, and L-PG in blue). Small-angle neutron scattering profiles measured for (E) A8819 and (F) A8819Cls2T33N membranes treated with and without daptomycin (4 μg/mL). The arrow indicates the lipid bilayer, whereas the star indicates the Bragg peak as the sign of micelle formation.
Using NR, we then characterized the effect of the observed membrane changes on daptomycin interactions. Daptomycin treatment at 2, 4, and 8 μg/mL significantly shifted the fringe of the NR curves of the WT membranes in a concentration-dependent manner (Fig. 3A, arrows), indicating extraction and solubilization of the membrane by daptomycin. In contrast, there was a less pronounced change in reflectivity for the cardiolipin-rich, daptomycin-resistant A8819Cls2T33N membrane (Fig. 3B). Analysis of the NR curves showed that at a daptomycin concentration of 2 μg/mL, the lipid volume fraction of the WT membrane was reduced, and no daptomycin was seen within the membrane consistent with a lipid extraction mechanism (24) (Table 1 and SI Appendix, Table S4). At a higher daptomycin concentration (4 μg/mL), antibiotic membrane penetration was evident, extending through the bilayer and causing dislocation of acyl-chains (Table 1 and SI Appendix, Table S4). At the highest concentration (8 μg/mL), the membrane was completely solvated by daptomycin, with a substantial decrease in the lipid volume fraction from 86.5 to 26.6% (Table 1 and SI Appendix, Table S4). For the thicker, cardiolipin-rich A8819Cls2T33N membrane, only mild lipid extraction was observed at 2, 4, and 8 μg/mL with the lipid volume fraction only reducing from 78.5 to 62.8% (Table1 and SI Appendix, Table S5). The A8819Cls2T33N membrane resisted daptomycin penetration and remained intact (Table 1 and SI Appendix, Table S5).
Table 1.
Summary of bilayer coverage and contents for the symmetric membrane bilayer deposited on silicon oxide surface
| Layer | A8819 | A8819Cls2T33N | ||||||
| 0 μg/mL Dp* | 2 μg/mL Dp* | 4 μg/mL Dp* | 8 μg/mL Dp* | 0 μg/mL Dp* | 2 μg/mL Dp* | 4 μg/mL Dp* | 8 μg/mL Dp* | |
| HG1† | 60.5 ± 4.8‡ | 31 ± 10 | 30.3 ± 6.4 (HG), 4.5 ± 0.1 (Dp) | 10.5 ± 1.5 (HG + Dp) | 52.5 ± 8.3 | 23.7 ± 7.9 | 26.8 ± 5.9 | 23.7 ± 8.2 |
| Tails§ | 86.5 ± 0.8 | 71.7 ± 1.5 | 52.8 ± 1.3 (Tails), 4.5 ± 0.1 (Dp) | 26.6 ± 1.6 (Tails), 12.5 ± 1 (Dp) | 78.5 ± 1.3 | 66.3 ± 1.4 | 62 ± 1.1 | 62.8 ± 1.1 |
| HG2¶ | 46 ± 5.4 | 33.6 ± 3.8 | 15.8 ± 2.9 (HG), 17.2 ± 1.2 (Tails), 4.5 ± 0.1 (Dp) | 14.7 ± 0.2 (HG), 11.9 ± 3.1 (Tails), 12.5 ± 1 (Dp) | 27.7 ± 2.1 | 16 ± 2.8 | 16.8 ± 2.8 | 17.9 ± 2.8 |
| Dp layer | — | — | 17.4 ± 1.9 | 12.6 ± 1.3 | — | — | 14.9 ± 1.4 | 14.1 ± 0.9 |
Dp, daptomycin.
HG1, head groups of the inner leaflet of the membrane.
Values are volume fractions (%) with two SDs.
Tails, acyl-chains of the membrane bilayer.
HG2, head groups of the outer leaflet of the membrane.
To further define the daptomycin–membrane interaction, we then used small-angle neutron scattering (SANS), which provided information on the daptomycin aggregation characteristics within the bilayer membranes under physiologically relevant conditions (25). Consistent with our NR data, at a clinically relevant daptomycin concentration of 4 μg/mL, we observed daptomycin penetrate and form aggregates straddling the bilayer membrane for WT (A8819) but not for the A8819Cls2T33N membranes (Fig. 3 E and F). These data also concur with our bacterial killing data, which showed complete bacterial killing for WT A8819 but therapeutic failure toward A8819Cls2T33N (SI Appendix, Fig. S1). Using established core-shell and hollow-cylinder models to characterize the daptomycin aggregates (25), it was estimated that daptomycin constituted a 28 ± 0.86 Å radius spherical micelle structure within the membrane bilayer (SI Appendix, Fig. S7). Taken together, these results show that S. aureus adapts during treatment with daptomycin in human infections by increasing its cardiolipin membrane content leading to a thicker membrane that resists daptomycin lipid extraction, and membrane penetration and disruption, promoting bacterial survival.
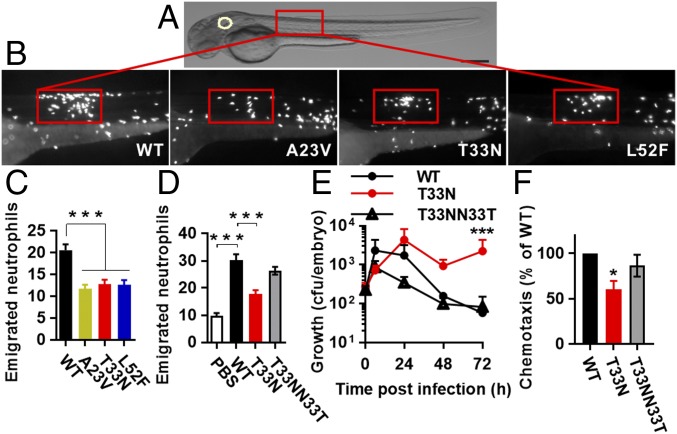
Cls2 Amino Acid Substitutions Impair Neutrophil Recruitment in Vivo and Promote Bacterial Survival.
Apart from impairing daptomycin–membrane interactions, we hypothesized that the altered bacterial membrane phospholipid profile caused by cls2 point mutations may have an effect on S. aureus–host interactions. Neutrophils are the dominant innate immune cell for controlling S. aureus infection (14, 15). To interrogate neutrophil behavior in vivo and in real time with high resolution, we utilized the vertebrate zebrafish (Danio rerio) model system (26). In common with humans, zebrafish have cellular and soluble immune arms and complex tissue environments that enable a mechanistic understanding of human infectious diseases (26, 27). To assess neutrophil trafficking and recruitment to a localized soft tissue staphylococcal infection, we used transgenic embryos with red fluorescent neutrophils (Tg[lyz:dsRed]) (28) that were infected into the somatic muscle with GFP-expressing S. aureus (Fig. 4A). Notably, neutrophil recruitment to the localized infection site was significantly compromised for infection with the three cls2 point mutants compared with infection by A8819 after 6 h postinfection (Fig. 4 B and C). Mutation repair for the T33N strain (A8819Cls2T33NN33T) was sufficient to restore neutrophil recruitment back to WT (A8819) infection levels (Fig. 4D). This attenuated neutrophil response affected bacterial clearance, with more persistent infection observed with A8819Cls2T33N compared with the WT and repaired strains (Fig. 4E). To further validate the observed neutrophil migration findings in zebrafish, we assessed human neutrophil migration within an ex vivo assay. As shown in Fig. 4F, significantly less human neutrophil recruitment was observed with A8819Cls2T33N compared with the WT strain. Mutation repair of the T33N mutant (A8819Cls2T33NN33T) caused similar neutrophil recruitment as WT (Fig. 4F). These results indicated that the Cls2 amino acid substitutions equipped S. aureus with the ability to circumvent neutrophil recognition and response, leading to a persistent infection as observed in patients and animal models (2, 5, 6).
Fig. 4.
Effect of Cls2 amino acid substitutions on neutrophil recruitment. (A) Schematic of a 48-h postfertilization (hpf) zebrafish showing the S. aureus injection sites. Rectangle, somatic muscle. White circle, the otic vesicle. (Scale bar: 250 μm.) (B) Representative images of localized sites of infection with S. aureus strains at 6 h postinfection. (C) The number of emigrated neutrophils to the localized site of infection. For WT, n = 22; A23V, n = 29; T33N, n = 34; L52F, n = 27, pooled from four independent experiments. (D) Neutrophil recruitment was restored to WT levels by the repaired strain A8819Cls2T33NN33T. For PBS, n = 12; WT, n = 21; T33N, n = 22; T33NN33T, n = 23, pooled from three independent experiments. (E) The bacterial burden in zebrafish after a somatic muscle infection. Error bars represent mean + SEM, three independent experiments (***P < 0.001 compared with WT and T33NN33T, χ2 test for trend). (F) Human neutrophil recruitment was assessed using a Transwell system, with neutrophils and bacterial cells in the top and bottom wells, respectively. Five independent experiments. For C, D, and F, error bars represent mean ± SEM. *P < 0.05, ***P < 0.001 compared with WT, Kruskal–Wallis test.
Compositional Changes of Membrane Phospholipids Resulted in Reduced Neutrophil Recruitment in Zebrafish.
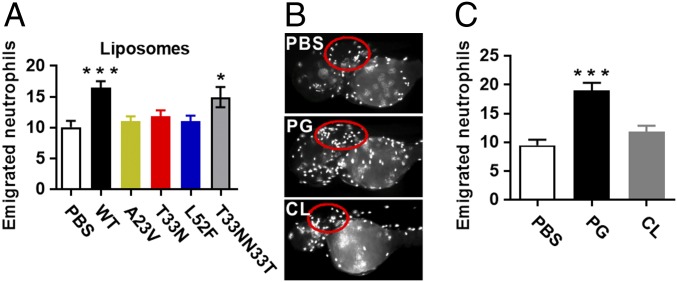
To investigate the contribution of the S. aureus bacterial membrane to neutrophil evasion, total lipids were extracted from each of the cls2 mutant and the daptomycin-susceptible parent (A8819) and repaired strains. Liposomes were then processed and injected into the otic vesicle of zebrafish (Fig. 4A, white circle). The otic vesicle is normally devoid of leukocytes and is relatively confined, preventing dispersion of the liposomes (29). As shown in Fig. 5A, and similar to that seen with infection of live, whole bacterial cells (Fig. 4 B–D), the liposomes from the three cls2 point mutants were significantly attenuated in inducing neutrophil recruitment compared with liposomes from A8819 and the T33N repaired strain (A8819Cls2T33NN33T), suggesting that altered anionic phospholipid composition may be responsible for the immune evasion. To determine the phospholipid driver of this S. aureus evasion response, purified PG and CL that were free of DNA, peptidoglycan, wall-teichoic acid, and lipoteichoic acid from A8819 cells (SI Appendix, Fig. S8) were processed to form liposomes. Injection of the PG- and CL-specific liposomes into the otic vesicle of zebrafish embryos showed that PG liposomes induced substantial neutrophil recruitment, whereas the recruitment of neutrophils by CL liposomes was comparable to the PBS control (Fig. 5 B and C). These data suggest that PG is the major bacterial-mediated phospholipid driver of neutrophil chemoattraction, and the S. aureus adaptation response to daptomycin represents a bacterial membrane-based stealth strategy to simultaneously evade an antibiotic and a key innate immune effector cell to promote survival within a host.
Fig. 5.
Effect of bacterial membrane phospholipids on neutrophil migration in vivo. (A) Quantification of emigrated neutrophils into the zebrafish otic vesicle at 6 h after injection of liposomes constituted from S. aureus membrane phospholipids. For PBS, n = 14; WT, n = 28; A23V, n = 26; T33N, n = 24; L52F, n = 24; T33NN33T, n = 17, pooled from four independent experiments. (B) Representative images of zebrafish otic vesicles (red circles) injected with purified PG or CL liposomes, with (C) quantification of emigrated neutrophils. For PBS, n = 10; PG, n = 17; CL, n = 17, pooled from three independent experiments. Error bars represent the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with PBS for A and C, one-way ANOVA.
Discussion
S. aureus has evolved a wealth of strategies to optimize its survival in various host niches and under nocuous selection pressures (30). Here we report a metabolic resistance mechanism used by S. aureus to prevent membrane lysis by the last line antistaphylococcal antibiotic, daptomycin. Single amino acid substitutions in cardiolipin synthase 2 led to a significant increase in the bacterial membrane CL/PG ratios due to enhanced cardiolipin biosynthesis, which led to a thicker membrane that resisted daptomycin penetration and membrane disruption. This adaption also led to immune evasion. Specifically, we showed that membrane PG acted as a bacterial-driven neutrophil chemoattractant, and in the context of cls2 point mutations, reduced PG membrane content led to less neutrophil trafficking to a localized site of infection and prolonged bacterial survival.
Recent studies on the interactions between daptomycin and bacterial membranes have used indirect methodological approaches such as measuring permeability loss of liposomes exposed to daptomycin using a fluorescence assay and membrane systems that are less relevant to Gram-positive bacteria, particularly using phosphatidylcholine (PC) and not L-PG (31). It was shown that the addition of cardiolipin to 10% of total lipids (using a PC/PG membrane system) was sufficient to prevent daptomycin interaction with membranes (31); however, we have clearly shown that our daptomycin-susceptible clinical strain (A8819) already has a greater percent of cardiolipin than 10% (Fig. 2A), suggesting the possibility of discordance between clinically relevant membranes. More recently, alteration of membrane curvature and diverting daptomycin binding or interference with fluid membrane microdomains have been proposed using the model organism Bacillus subtilis or enterococcal strains (7, 32, 33). However, the phospholipid membrane composition of these organisms is different to S. aureus, and daptomycin–membrane interactions appear to be bacterial species dependent. For example, daptomycin has an irregular membrane binding pattern in B. subtilis and Enterococcus, with a particular predilection for the division septum (7, 32, 33), whereas we have observed universal distribution of daptomycin on S. aureus membranes (SI Appendix, Fig. S6 C and D).
Here we have reconstituted clinically relevant S. aureus membranes and have provided direct structural analysis of daptomycin–membrane interactions. The scattering of free neutrons by matter provides excellent structural detail, and this experimental approach has only recently been used to study biological membranes (25, 34). We used unsaturated phospholipids with complex head group composition to reconstitute the membranes, and the surface coverage achieved for the WT membranes was 86.5% (Table 1), which is higher than what has been previously reported using unsaturated phospholipids with similar techniques (35). Using SANS and NR, we identified three distinct modes of action of daptomycin on the membrane. At lower concentrations (2 μg/mL), lipid extraction occurred and caused lesions in the bilayer membrane, whereas at higher concentrations (4 μg/mL), daptomycin molecules penetrated into the bilayer and formed organized micelles. At the highest concentrations (8 μg/mL), the bilayer was completely solvated by daptomycin. The process of lipid extraction, penetration, micelle formation, and membrane lysis was inhibited in the context of the clinically derived Cls2 amino acid substitution T33N, which coincided with daptomycin resistance. The daptomycin molecules still attached to the outer leaflet of the membrane in the A8819Cls2T33N strain, and this was corroborated with superresolution imaging of fluorescent daptomycin binding to bacterial cells, but the daptomycin was functionally impaired due to the barrier created by the thicker CL-rich membrane. This is an anionic phospholipid driven mechanism of daptomycin resistance and is independent of the charge-based repulsion theory proposed for mprF mutations that alter L-PG membrane content (4, 12).
Subversion of host immune surveillance contributes to persistent bacterial infections (36). Given the importance of neutrophils in eliminating S. aureus (15), we utilized a transgenic zebrafish line carrying red fluorescent neutrophils to characterize the interactions between S. aureus and neutrophils in vivo and in real time during acute infection. We found that infection with the S. aureus cls2 point mutants compromised neutrophil recruitment, and this was associated with a more prolonged bacterial burden in the host. This compromised neutrophil migration was further confirmed using human neutrophils. The purified PG and CL liposomes used for the neutrophil migration studies were found to be free of DNA, peptidoglycan, lipoteichoic acids, and wall-teichoic acids (SI Appendix, Fig. S8), suggesting that established bacterial-driven neutrophil chemoattractants for compromised neutrophil migration were not at play (37). We then focused on the phospholipid membrane components and through injection of purified CL and PG liposomes into zebrafish, established that PG was a driver for neutrophil chemotaxis. This provided a mechanistic model whereby Cls2 amino acid substitutions led to increased cardiolipin synthase activity and increased CL production that was subsequently causing a reduction in PG. This change in lipid profile not only disturbed daptomycin–membrane interactions but also reduced neutrophil chemoattraction to a localized site of infection, finally resulting in a more persistent infection, which has been observed in patients and animal models without a clear explanation (2, 5, 6). Future analyses are still required to investigate the generalization of bacterial PG in inducing neutrophil migration and if PG acts directly as a chemotactic or indirectly via activation of endogenous cells. In conclusion, we have characterized a bacterial metabolic adaptation process that leads to simultaneous evasion of host immune and antimicrobial attack, providing important insights for future therapeutic targeting for this troublesome pathogen.
Materials and Methods
Media and Reagents.
Bacterial strains, plasmids, and oligonucleotides used in this study are described in SI Appendix, Table S6. S. aureus cells were cultured in heart infusion broth (HI) (Oxoid) with constant shaking at 37 °C.
Genetic Manipulation.
The vector pIMAY and the Escherichia coli strain DC10B were used to genetically manipulate S. aureus isolate following the published protocol (SI Appendix, SI Materials and Methods) (38).
Daptomycin Susceptibility Testing.
Broth microdilution MIC testing was performed based on guidelines by the Clinical and Laboratory Standards Institute. Daptomycin MIC > 1 μg/mL is officially termed daptomycin-nonsusceptible but was termed daptomycin-resistant throughout the manuscript for clarity. Time–kill assays were performed with an initial bacterial inoculum of 106 cfu/mL in Mueller–Hinton broth supplemented with 50 mg/L calcium and daptomycin. (SI Appendix, SI Materials and Methods).
Lipid Analysis by Mass Spectrometry.
Lipids were extracted following the published protocol (10). Lipid species were processed and quantified according to standard procedures (39). For de novo phospholipid biosynthesis, the metabolic labeling was initiated by the addition of glycerol or [13C]-glycerol (Sigma-Aldrich) to 1 mM after bacterial growth from optical density 600 nm of 0.4 for 30 min at 37 °C. Data collected for lipid composition of PG, CL, and L-PG with 13C-glycerol incorporation were analyzed using Metaboanalyst (https://www.metaboanalyst.ca) (SI Appendix, SI Materials and Methods).
S. aureus Bilayer Membrane Formation and Neutron Reflectometry.
Deposition of the model membrane on the top of a SiO2 surface was performed using a custom-built Langmuir–Blodgett trough (Nima Technology) following published Langmuir–Blodgett and Langmuir–Schaefer procedures (22) (SI Appendix, SI Materials and Methods). Synthetic PG (18:1), CL (18:1), and L-PG (18:1) (Avanti Polar Lipids, Inc.) were mixed at the molar ratios of 69:12:19 and 23:60:17 to create A8819 and A8819Cls2T33N symmetric membrane bilayers, respectively. Specular neutron reflection at solid–liquid interface was carried out on the Platypus time-of-flight neutron reflectometer at the OPAL 20 MW Multipurpose Research Reactor, Lucas Heights, Australia. The final reflectivity (reflected intensity/incident intensity) is presented as a function of momentum transfer. Analysis of the NR profiles was performed using MOTOFIT followed by Monte Carlo Analysis to determine 95% confidence intervals (40). (SI Appendix, SI Materials and Methods).
Small-Angle Neuron Scattering.
Synthetic PG, CL, and L-PG were mixed at the molar ratios of 69:12:19 and 23:60:17 for producing A8819 and A8819Cls2T33N membrane vesicles, respectively. The samples were measured using the small-angle neutron scattering instrument, Quokka, at Australian Nuclear Science and Technology Organization (SI Appendix, SI Materials and Methods).
Zebrafish Strains, Maintenance, and Leukocyte Enumeration.
WT Tübingen and Tg(lyz:DsRed)nz50 zebrafish (28) embryos were maintained in the Monash University Fish Core facility and infected with S. aureus cells according to standard protocols (27) (SI Appendix, SI Materials and Methods). Animal protocols were approved by the Monash University Animal Ethics Committee.
In Vitro Transwell Chemotaxis Assay.
Human venous blood was collected from healthy volunteers for isolating neutrophils with the approval of the Monash University Human Research Ethics Committee and the informed consents from the volunteers were obtained. Chemotaxis assays were performed according to a previously published protocol (41) (SI Appendix, SI Materials and Methods).
Liposome Preparation.
Staphylococcal PG or CL were extracted from TLC plates using chloroform/methanol/water (5:5:1) followed by centrifugation. Liposomes were prepared following an established protocol (42) (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Monash Micro Imaging for the scientific and technical assistance and the Australian Center for Neutron Scattering, Australian Nuclear Science and Technology Organization, for providing the neutron research facilities. We also acknowledge Professors George Eliopoulos and the late Robert C. Moellering, Jr., for their initial contribution of strains and mentorship. We acknowledge funding for this work from Australian National Health and Medical Research Council (NHMRC) Project Grant APP1144303 (to A.Y.P.), and CASS Foundation Grant SM/12/4276 (to A.Y.P.). A.Y.P. and B.P.H. were funded by an NHMRC Practitioner Fellowship, and H.-H.S. was funded by an NHMRC Career Development Fellowship. G.J.L. is an NHMRC Senior Research Fellow. M.J.M. is an NHMRC Principal Research Fellow. The Abberior stimulated emission depletion microscope was funded by Australian Research Council Linkage Infrastructure, Equipment and Facilities Grant LE150100110, 2015. The Australian Regenerative Medicine Institute is supported by grants from the State Government of Victoria and the Australian Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812066116/-/DCSupplemental.
References
- 1.Lee AS, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4:18033. doi: 10.1038/nrdp.2018.33. [DOI] [PubMed] [Google Scholar]
- 2.Fowler VG, Jr, et al. S. aureus Endocarditis and Bacteremia Study Group Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–665. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 3.Tran TT, Munita JM, Arias CA. Mechanisms of drug resistance: Daptomycin resistance. Ann N Y Acad Sci. 2015;1354:32–53. doi: 10.1111/nyas.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peleg AY, et al. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One. 2012;7:e28316. doi: 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron DR, et al. Impact of daptomycin resistance on Staphylococcus aureus virulence. Virulence. 2015;6:127–131. doi: 10.1080/21505594.2015.1011532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards RL, et al. Persistent Staphylococcus aureus isolates from two independent cases of bacteremia display increased bacterial fitness and novel immune evasion phenotypes. Infect Immun. 2015;83:3311–3324. doi: 10.1128/IAI.00255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran TT, et al. Daptomycin-resistant Enterococcus faecalis diverts the antibiotic molecule from the division septum and remodels cell membrane phospholipids. MBio. 2013;4:e00281-13. doi: 10.1128/mBio.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst CM, et al. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peschel A, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai M, et al. Staphylococcus aureus requires cardiolipin for survival under conditions of high salinity. BMC Microbiol. 2011;11:13. doi: 10.1186/1471-2180-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koprivnjak T, et al. Characterization of Staphylococcus aureus cardiolipin synthases 1 and 2 and their contribution to accumulation of cardiolipin in stationary phase and within phagocytes. J Bacteriol. 2011;193:4134–4142. doi: 10.1128/JB.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang SJ, Mishra NN, Rubio A, Bayer AS. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother. 2013;57:5658–5664. doi: 10.1128/AAC.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Short SA, White DC. Biosynthesis of cardiolipin from phosphatidylglycerol in Staphylococcus aureus. J Bacteriol. 1972;109:820–826. doi: 10.1128/jb.109.2.820-826.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spaan AN, Surewaard BG, Nijland R, van Strijp JA. Neutrophils versus Staphylococcus aureus: A biological tug of war. Annu Rev Microbiol. 2013;67:629–650. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- 15.Bogomolski-Yahalom V, Matzner Y. Disorders of neutrophil function. Blood Rev. 1995;9:183–190. doi: 10.1016/0268-960x(95)90024-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang R, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 17.Schiffmann E, Corcoran BA, Wahl SM. N-formylmethionyl peptides as chemoattractants for leucocytes. Proc Natl Acad Sci USA. 1975;72:1059–1062. doi: 10.1073/pnas.72.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prat C, Bestebroer J, de Haas CJ, van Strijp JA, van Kessel KP. A new staphylococcal anti-inflammatory protein that antagonizes the formyl peptide receptor-like 1. J Immunol. 2006;177:8017–8026. doi: 10.4049/jimmunol.177.11.8017. [DOI] [PubMed] [Google Scholar]
- 19.de Haas CJ, et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montange D, et al. Penetration of daptomycin into bone and synovial fluid in joint replacement. Antimicrob Agents Chemother. 2014;58:3991–3996. doi: 10.1128/AAC.02344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traunmüller F, et al. Soft tissue and bone penetration abilities of daptomycin in diabetic patients with bacterial foot infections. J Antimicrob Chemother. 2010;65:1252–1257. doi: 10.1093/jac/dkq109. [DOI] [PubMed] [Google Scholar]
- 22.Clifton LA, et al. Asymmetric phospholipid: Lipopolysaccharide bilayers; a Gram-negative bacterial outer membrane mimic. J R Soc Interface. 2013;10:20130810. doi: 10.1098/rsif.2013.0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clifton LA, et al. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir. 2015;31:404–412. doi: 10.1021/la504407v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YF, Sun TL, Sun Y, Huang HW. Interaction of daptomycin with lipid bilayers: A lipid extracting effect. Biochemistry. 2014;53:5384–5392. doi: 10.1021/bi500779g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen HH, Thomas RK, Penfold J, Fragneto G. Destruction and solubilization of supported phospholipid bilayers on silica by the biosurfactant surfactin. Langmuir. 2010;26:7334–7342. doi: 10.1021/la904212x. [DOI] [PubMed] [Google Scholar]
- 26.Hepburn L, et al. Innate immunity. A spaetzle-like role for nerve growth factor β in vertebrate immunity to Staphylococcus aureus. Science. 2014;346:641–646. doi: 10.1126/science.1258705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhuiyan MS, et al. Acinetobacter baumannii phenylacetic acid metabolism influences infection outcome through a direct effect on neutrophil chemotaxis. Proc Natl Acad Sci USA. 2016;113:9599–9604. doi: 10.1073/pnas.1523116113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levraud JP, Colucci-Guyon E, Redd MJ, Lutfalla G, Herbomel P. In vivo analysis of zebrafish innate immunity. Methods Mol Biol. 2008;415:337–363. doi: 10.1007/978-1-59745-570-1_20. [DOI] [PubMed] [Google Scholar]
- 30.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T, et al. Cardiolipin prevents membrane translocation and permeabilization by daptomycin. J Biol Chem. 2014;289:11584–11591. doi: 10.1074/jbc.M114.554444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller A, et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci USA. 2016;113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clifton LA, et al. An accurate in vitro model of the E. coli envelope. Angew Chem Weinheim Bergstr Ger. 2015;127:12120–12123. doi: 10.1002/ange.201504287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes AV, et al. High coverage fluid-phase floating lipid bilayers supported by omega-thiolipid self-assembled monolayers. J R Soc Interface. 2014;11:20140245. doi: 10.1098/rsif.2014.0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monack DM, Mueller A, Falkow S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 37.Schmeling DJ, et al. Chemotaxigenesis by cell surface components of Staphylococcus aureus. Infect Immun. 1979;26:57–63. doi: 10.1128/iai.26.1.57-63.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012;3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu C, et al. RPLC-ion-trap-FTMS method for lipid profiling of plasma: Method validation and application to p53 mutant mouse model. J Proteome Res. 2008;7:4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- 40.Nelson A. Co-refinement of multiple-contrast neutron/X-ray reflectivity data using MOTOFIT. J Appl Cryst. 2006;39:273–276. [Google Scholar]
- 41.Corriden R, et al. Ecto-nucleoside triphosphate diphosphohydrolase 1 (E-NTPDase1/CD39) regulates neutrophil chemotaxis by hydrolyzing released ATP to adenosine. J Biol Chem. 2008;283:28480–28486. doi: 10.1074/jbc.M800039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hope MJ, Bally MB, Webb G, Cullis PR. Production of large unilamellar vesicles by a rapid extrusion procedure: Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.