Significance
Mounting evidence suggests that rare genetic variations contribute to the genetic risk for autism spectrum disorder (ASD). Here, we describe the first ASD-associated in-frame deletion in the human dopamine (DA) transporter (hDAT) gene. This mutation results in the deletion of the conserved Asn336 (∆N336). By integrating molecular insights from X-ray crystallography, electron paramagnetic resonance, and electrophysiology, we uncovered a conformational state of the transporter promoted by ∆N336. It is defined by a “half-open and inward-facing” conformation of the intracellular gate that leads to specific dysfunctions in DA homeostasis as determined in the brain of Drosophila melanogaster expressing hDAT ∆N336. Importantly, hDAT ∆N336 flies display increased fear and impaired social interactions, traits associated with DA dysfunction and ASD.
Keywords: dopamine transporter, leucine transporter, amphetamine, efflux, autism
Abstract
The human dopamine (DA) transporter (hDAT) mediates clearance of DA. Genetic variants in hDAT have been associated with DA dysfunction, a complication associated with several brain disorders, including autism spectrum disorder (ASD). Here, we investigated the structural and behavioral bases of an ASD-associated in-frame deletion in hDAT at N336 (∆N336). We uncovered that the deletion promoted a previously unobserved conformation of the intracellular gate of the transporter, likely representing the rate-limiting step of the transport process. It is defined by a “half-open and inward-facing” state (HOIF) of the intracellular gate that is stabilized by a network of interactions conserved phylogenetically, as we demonstrated in hDAT by Rosetta molecular modeling and fine-grained simulations, as well as in its bacterial homolog leucine transporter by electron paramagnetic resonance analysis and X-ray crystallography. The stabilization of the HOIF state is associated both with DA dysfunctions demonstrated in isolated brains of Drosophila melanogaster expressing hDAT ∆N336 and with abnormal behaviors observed at high-time resolution. These flies display increased fear, impaired social interactions, and locomotion traits we associate with DA dysfunction and the HOIF state. Together, our results describe how a genetic variation causes DA dysfunction and abnormal behaviors by stabilizing a HOIF state of the transporter.
Dopamine (DA), a monoamine neurotransmitter, plays an important role in the central nervous system by regulating a variety of functions, including cognition, emotion, motor activity, and motivation. Altered dopaminergic signaling is linked to multiple neuropsychiatric disorders, such as attention deficit hyperactive disorder; mood disorders; schizophrenia; and, more recently, autism spectrum disorder (ASD) (1–3). The DA transporter (DAT), a presynaptic transmembrane Na+/Cl− symporter, acts by regulating duration of the dopaminergic response by reuptake of released DA. DAT is the primary target site for several psychostimulant drugs, including amphetamine (AMPH) (4). AMPH promotes DAT-mediated DA efflux by inducing human DAT (hDAT) N terminus posttranslational modifications (i.e., phosphorylation), a process regulated by the interactions of the N terminus DAT with the plasma membrane phosphatidylinositol 4,5-bisphosphate (5).
Research into the structural dynamics of DAT suggests that it follows an alternating access model (6–8), wherein the transporter can alternate between an “outward-facing” state and an “inward-facing” state. This mechanism is defined by an extracellular gate as well as an intracellular gate, with the latter known to regulate transport cycle and conformational stability through rearrangements of several structural elements, including the third intracellular loop (IL3), the amino N terminus, and multiple transmembrane helices (9, 10).
ASD is a single condition that comprises a heterogeneous group of developmental disorders manifested by impairments across two core domains: deficits in social communication and interaction and patterns of restricted behaviors, interests, and activities (11). While the causes of ASD are unknown, genetic variants are established as important factors in risk. Analysis of rare variants has provided insight into multiple pathways and systems affected in ASD, and has implicated copy number variations, loss-of-function mutations, and de novo missense mutations as contributing factors (12, 13).
Here, we undertook a closer examination of the structural and functional significance of a rare in-frame deletion of residue N336 (∆N336) in hDAT (hDAT ∆N336) identified in a patient with ASD. The functional perturbations that stem from ∆N336 provided an opportunity to obtain X-ray crystallographic insights as well as molecular dynamics (MD) details into the role of DAT structural domains controlling DA transport. They suggest that the two gates of neurotransmitter transporters can act independently during the transport process and be uncoupled by rare variants. Moreover, these findings have implications of how the DAT and other biogenic amine transporters can operate in an efflux mode (i.e., under the influence of AMPH). They also define how disruption of specific gate conformations of DAT translate into abnormal brain DA function and atypical complex animal behaviors in flies, such as social interactions and fear response.
Methods
Subjects and Clinical Assessment.
Subjects from an ASD family were recruited by the Autism Simon Simplex Collection as described previously (14). The proband was evaluated for ASD with the Autism Diagnostic Interview-Revised and the Autism Diagnostic Observation Schedule, as well as a battery of behavioral and cognitive tests to characterize phenotype. Additional measures captured subclinical ASD-related traits in parents and unaffected siblings, and a comprehensive family medical history was obtained.
hDAT ∆N336 Identification.
Exome capture, sequencing, data processing, and variant calling were conducted as described previously (15, 16). The ∆N336 variant was validated, and inheritance patterns were determined by PCR and Sanger sequencing of all family members. Amplifying primers were designed using Primer3 and subjected to a BLAST-like alignment tool search to ensure specificity. PCR was carried out using 7.1 nmol of amplifying primers and 12 ng of genomic DNA in a final volume of 20 μL. Sequence analysis was performed using Sequencher v5.0.1 (Gene Codes).
Electron Paramagnetic Resonance Protocol.
Cysteine residues were introduced using site-directed mutagenesis into leucine transporter (LeuT), LeuT ∆V269, and LeuT V269N constructs. Experiments were conducted as described by Claxton et al. (17). In Fig. 1 and SI Appendix, Fig. S5, apo refers to Na+ and leucine-free transporter, while the +Na/Leu state was obtained by addition of 200 mM NaCl and fourfold molar excess of Leu relative to LeuT. Double electron-electron resonance (DEER) (18) was performed at 83 K on a Bruker 580 pulsed electron paramagnetic resonance (EPR) spectrometer operating at Q-band frequency using a standard four-pulse sequence (19). DEER echo decays were analyzed to obtain distance distributions.
Fig. 1.
In LeuT, deletion of V269 supports a HOIF conformation of the intracellular gate. Distance distributions of extracellular and intracellular spin-labeled Cys pairs reveal changes in the conformational equilibrium induced by ∆V269 in LeuT. (A) Extracellular reporter pairs (309–480) tagged on a 3D structure of LeuT. (D) Intracellular reporter pairs (7–86) tagged on a 3D structure of LeuT. (B and E) Distance distributions of the extracellular and intracellular reporter pair for LeuT in the apo conformation (black), in the presence of Na+ (red), or in the presence of Na+ plus Leu (blue). (C and F) Distance distributions of the extracellular and intracellular reporter pair for LeuT ∆V269 in the apo conformation (black), in the presence of Na+ (red), or in the presence of Na+ and Leu (blue).
Purification, Crystallization, and Structure Determination.
The purification and crystallization of LeuT ∆V269 were performed as previously reported (20, 21). Briefly, the plasmid encoding Aquifex aeolicus LeuT ∆V269 was transformed into Escherichia coli C41 electrocompetent cells. The transformed cells expressing LeuT ∆V269 were grown in large quantities in Terrific Broth to an absorbance at 600 nm of 0.8. The culture was then induced using 0.1 mM isopropyl-β-d-thiogalactopyranoside and incubated on a shaker for 20 h at 20 °C. The cell membranes were harvested from the bacterial culture by sonication, followed by centrifugation. The harvested membranes were solubilized using a buffer containing 20 mM Tris⋅HCl (pH 8.0), 200 mM NaCl, 40 mM n-Dodecyl-β-D-Maltopyranoside (DDM), and 5 mM leucine. LeuT ∆V269 was purified from this solubilized material using metal affinity chromatography. The partially purified protein was subjected to thrombin digestion overnight to remove the affinity tag. The protein sample was further purified and exchanged into the crystallization buffer [50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 40 mM n-Octyl-β-D-Glucopyranoside (β-OG), and 5 mM leucine] by size exclusion chromatography. The purified protein was concentrated up to 4 mg/mL in the presence of 10% glycerol and 10 μM phospholipids.
Crystallization attempts were performed with the purified protein using the 96-well hanging drop vapor diffusion method at 20 °C. The diffraction data were collected at the Advanced Light Source (beamline 5.0.2; Lawrence Berkeley National Laboratory) using a crystal that was grown in the condition containing 10% PEG 3000, 100 mM sodium cacodylate trihydrate (pH 6.5), and 200 mM magnesium chloride hexahydrate. The diffraction data were indexed, integrated, and scaled using the microdiffraction assembly method in the XDS program (22). The structure was determined at 2.6 Å in the C2 space group by the molecular replacement method using the program Phaser from the Phenix software suite (23). The LeuT structures available in several different conformations were used individually as a search model in the molecular replacement method (7, 24–26). The best solution was obtained using LeuT in the outward-facing occluded conformation [Protein Data Bank (PDB) ID code 2Q72] (SI Appendix, Table S1). The region comprising the V269 deletion in the model was manually built using Coot (27). The structure was refined to reasonable R-factors using Phenix Refine. The data collection, model building, and refinement statistics are tabulated in SI Appendix, Table S2.
Rosetta Molecular Modeling.
Structural models for hDAT and hDAT ∆N336 were created in parallel using the Rosetta molecular modeling suite (revision 57712; Rosetta Commons). Models were generated based on the hDAT sequence and the Drosophila melanogaster DAT (PDB ID code 4XP9) as a structural template. The sequence identity was 55%. The sequence was threaded onto the template based on a sequence alignment generated by ClustalW2. Backbone coordinated in-gap regions were reconstructed using the cyclical coordinate decent algorithm and refined with the kinematic loop closure algorithm and RosettaMembrane. A total of 1,000 models were generated. The model for analysis is the best-scoring model by the RosettaMembrane energy function that had a root-mean-square deviation (rmsd) to the template of less than 1 Å. The Rosetta energies for hDAT and hDAT ∆N336 were normalized, and the per residue differences between hDAT and hDAT ∆N336 were calculated. These energy differences were mapped onto the structural models, and PyMOL visualization revealed sites where the energy difference was the greatest.
Homology Modeling and Fine-Grained Simulations.
We created 250 homology models of the transmembrane domain of hDAT (residues 60–603) based on the outward-open crystal structure of the D. melanogaster DAT (dDAT) (PDB ID code 4XP1) (28) using MODELLER v.9.16 following the procedure described by Buchmayer et al. (29). The models contained two Na+ ions and one Cl− ion in their respective putative binding sites and a single DA molecule in the central binding site, as observed in the dDAT crystal structure. A previously modeled extracellular loop 2 (EL2) (30) was introduced as a replacement for the crystal structure EL2, due to extensive crystallographic contacts between the EL2 and the cocrystallized antibody, as well as the truncation introduced in the dDAT EL2 sequence for crystallization. The deletion at residue 336 in the hDAT sequence was introduced as either a single deletion of N336 or a deletion of N336, followed by moving K337 to the previous 336 position. The resulting ∆N336 hDAT sequence is the same in each model, but transmembrane 6 (TM6) is shorter by one residue in the former model, while IL2 is shorter by one residue in the latter. The 250 models were scored and sorted by their discrete optimized protein energy score. The best 20 are ranked by their rmsd of the Cα atoms from the template, with secondary ranking of side-chain rmsd to the template. The three models with the lowest Cα atom rmsd to the template were inserted into a preequilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membrane bilayer using the membed procedure (31). The lipids are described by the Berger parameters (32), the protein is described by the AMBER99sb-ildn forcefield (33), and DA is described by the general Amber force field (GAFF) (34) parameters and restrained electrostatic potential (RESP) charges (35). Simulation was performed with GROMACS version 5.1.2. The inserted systems were equilibrated by restraining the protein with 1,000, 100, 10 and 1 kJ⋅mol−1⋅nm−2 restraints, each for 2.5 ns. The equilibrated systems were then put into production and run for 150 ns per model. The temperature was maintained at 310 K using a v-rescale thermostat, and the pressure was maintained semiisotropically with Parrinello–Rahman pressure coupling at 1 bar. The pressure coupling time constant was set to 20 ps, and the compressibility was set to 4.5 × 10−5⋅bar−1. Long-range electrostatic interactions were represented with a particle mesh Ewald method (1 nm cutoff), and Van der Waals interactions were imposed utilizing a Lennard-Jones potential with a 1 nm cutoff.
Cell Culture.
The hDAT-pCIHygro and hDAT-pEGFP expression vectors were engineered to contain hDAT, hDAT ∆N336, or hDAT N336V sequence. Vector sequences were confirmed via Sanger sequence to ensure no off-target mutations were generated. Vector DNA was transiently transfected into cells. The hDAT expression vectors contained GFP for cell selection. Fugene-6 (Promega) in serum-free media was used to transfect CHO cells ∼24 h after plating using a 1:3 DNA/lipid ratio. Cells were maintained in a 5% CO2 incubator at 37 °C in Ham’s F-12 medium supplemented with 10% FBS, 1 mM l-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin. Assays were conducted 24–48 h after transfection.
[3H]DA Uptake.
Saturation kinetics of [3H]DA uptake were performed as described previously (1). DAT-dependent DA uptake was determined using a range of concentrations (10 nM–10 μM) of a mixture of [3H]DA and unlabeled DA. Forty-eight hours before each uptake assay, hDAT, hDAT ∆N336, or hDAT V336N cells were seeded (50,000 cells per well) into 24-well plates. Preceding each uptake experiments, cells were washed in KRH buffer [25 mM Hepes, 120 mM NaCl, 5 mM KCl, 1.2 mM CaCl2, and 1.2 mM MgSO4 supplemented with 5 mM d-glucose, 100 μM ascorbic acid, 1 mM tropolone, and 100 μM pargyline (pH 7.4)]. DA uptake was terminated after 10 min by washing twice in ice-cold KRH buffer. Scintillation fluid was added, and [3H] was quantified in a microplate liquid scintillation counter. Nonspecific binding was calculated in the presence of 10 μM cocaine.
For each data point in the uptake assays in whole Drosophila brain, four brains were dissected and placed in 1 mL of Schneider’s Drosophila Medium with 1.5% BSA. Brains were placed in a 12-μm pore standing well containing 1.4 mL of hemolymph-like solution (HL3) with 1.5% BSA per well. Brains were removed and treated with 50 μM cocaine or vehicle, and incubated for 10 min. Brains were again removed, excess solution was allowed to drain, and brains incubated for 15 min in 0.7 mL HL3 with 1.5% BSA containing 200 nM [3H]DA. Brains were washed with KRH buffer and placed in 100 μL of 0.1% SDS solution in a top count plate. Scintillation fluid was added, and [3H]DA was measured. Apparent affinity (Km) and maximum velocity (Vmax) values were determined in all experiments by nonlinear regression using GraphPad Prism 4.0 (GraphPad Software).
Cell Surface Biotinylation and Immunoblotting.
Cell surface biotinylation and immunoblotting experiments were performed as described previously (1). Cells were seeded in six-well plates (10^6 cells per well) and transfected for 24–48 h before assaying. For each experiment, cells were incubated with sulfo-NHS-S-S-biotin (Pierce Chemical Company) to label surface-localized transporter. Biotinylated protein was pulled down using streptavidin-coupled beads. Protein was separated via SDS/PAGE, and immunoblotting was performed using an anti-DAT monoclonal antibody at 1:1,000 (MAB369; Millipore), β-actin antibody used at 1:5,000 (A5441; Sigma–Aldrich), and goat–anti-rat–HRP-conjugated secondary antibody used at 1:5,000 (sc-2006; Santa Cruz Biotechnology). ECL substrate (170-5061; Bio-Rad) was used for chemiluminescence detection. Protein quantification was performed on an ImageQuant LAS4000 imager (GE Healthcare Life Sciences), and analyzed using ImageJ (NIH).
Amperometry and Patch-Clamp Electrophysiology.
Cells were plated at a density of ∼20,000 per 35-mm culture dish. To intracellularly load DA, a programmable puller (model P-2000; Sutter Instruments) was used to fabricate quartz recording pipettes with a resistance of 3–5 mΩ. Pipettes were filled with 2 mM DA in an internal solution containing 120 mM KCl, 10 mM Hepes, 0.1 mM CaCl2, 2 mM MgCl2, 1.1 mM EGTA, 30 mM dextrose adjusted to pH 7.35, and 275 mOsm. Upon gaining access to the cells, the internal solution was allowed to diffuse into the cell for 10 min. To record DA efflux, a carbon fiber electrode was juxtaposed to the plasma membrane and held at +700 mV (a potential greater than the oxidation potential of DA). Amperometric current was calculated in response to the 10 μM AMPH addition using an Axopatch 200B amplifier and pCLAMP software (Molecular Devices). DA efflux was quantified as the peak value of the amperometric current.
Male flies of the appropriate genotype (five males, 1–3 d old) were selected and treated with standard food supplemented with vehicle (5 mM ascorbic acid and 10 mM sucrose) or l-dopa (l-DOPA; 5 mM). After 24 h, whole brains were manually removed and placed in a mesh holder in Lub’s external solution (130 mM NaCl, 10 mM Hepes, 1.5 mM CaCl2, 0.5 mM MgSO4, 1.3 mM KH2PO4, 34 mM dextrose adjusted to pH 7.35, and 300 mOsm). A carbon fiber electrode was held at +700 mV and positioned in the tyrosine hydroxylase (TH)-positive protocerebral posterior lateral (PPL) 1 DA neuronal region. Amperometric current was calculated in response to 20 μM AMPH addition using an Axopatch 200B amplifier and pCLAMP software (Molecular Devices). DA efflux was quantified as the peak value of the amperometric current.
Steady-State Currents and Current–Voltage Relationships.
Cells were cultured on 12-mm round glass coverslips (CS-12R; Warner) for 24–48 h before recording. Patch-clamp recordings were performed using an Axopatch 200B amplifier, and currents were acquired using pCLAMP software. Rapid application of 10 μM AMPH was achieved by a piezoelectric translator (PZ-150M; Burleigh) moving a theta glass (no. 1407201; Hilgenberg) with vehicle solution flowing continuously through one barrel and AMPH solution through the other barrel. The piezoelectric translator was driven through the pCLAMP software with a voltage step (5–10 ms) filtered at 25–250 Hz. Current traces plotted in all figures represent the mean current for at least 10 sweeps.
Drosophila Genetics.
Drosophila homozygotes for the DAT-null allele DATfmn (dDAT KO) (36) and flies harboring TH-Gal4 (37) were outcrossed to a control line [Bloomington Indiana (BI) 6326)]and selected by PCR or eye color. TH-GAL4 (Bl 8848) and M{vas-int.Dm}ZH-2A, M{3xP3-RFP.attP′} ZH-22A (Bl 24481) were obtained from the BI stock center and outcrossed to dDAT KO flies carrying the white (w1118) mutation (BI stock no. 6236) for five to five to 10 generations. Transgenes (hDAT or hDAT ∆N336) were cloned into pBI-UASC (38), and constructs were injected into embryos from M{vas-int.Dm}ZH-2A, M{3xP3-RFP.attP′}ZH-22A (Bl 24481). Initial potential transformants were isolated and selected.
Drosophila Locomotion Behavioral Analysis.
Three days posteclosion, male flies were collected and placed into tubes with food for 72 h. Locomotion was recorded by beam breaks and analyzed using equipment/software from TriKinetics. For the AMPH-induced locomotion, males were starved for 6 h and then fed sucrose (5 mM) containing either AMPH (10 mM) or vehicle.
Drosophila Escape-Response Behavioral Analysis.
Flies were reared on a 12-h day/night cycle at 25 °C with behavioral experiments performed during the day cycle. Adult male flies (2–5 d old) were selected, manually transferred, and allowed to acclimate to the recording chamber, 2.65 × 1.35 Sylgard silicone testing encapsulant (Dow Corning). The testing chamber was illuminated, and basal locomotion was recorded for 2 s before the auditory stimulus (sound of a predatory wasp) and for 3 s following the auditory stimulus. Each auditory stimulus was presented for 1,000 ms. Drosophila behavior was recorded at 1,000 frames per second using a Phantom v310 (Vision Research). Behaviors were analyzed using custom code written in MATLAB (v. R2016a; MathWorks).
Results
Exome Sequencing of Subjects with ASD Reveals a Rare Inherited In-Frame Deletion of ΔN336 in the hDAT Gene.
Exome capture and sequence analysis identified a rare in-frame deletion at the conserved ∆N336 [hg19 Chr5:1416233-1416236 (CTTG/C)] in the hDAT gene (SLC6A3) in a single ASD family. The family harboring ∆N336 belongs to the Simons Simplex Collection, a well-characterized ASD collection (14). Sanger sequencing evaluation of inheritance and carrier status in available family members confirmed paternal transmission to the proband and absence of this allele in the unaffected sibling (SI Appendix, Fig. S1, Left). The ∆N336 was absent in all other analyzed Simons Simplex Collection families (n = 825) and absent in an independent cohort of nonautistic controls (n = 869) from the American Recovery and Reinvestment Act (ARRA) Autism Sequencing Consortium (39). High cross-species conservation at position 336 (SI Appendix, Fig. S1, Right) suggests a damaging effect of the deletion on transporter function.
Relevant family medical history and multiple assessment scores are provided in SI Appendix, Supplementary Information I. The affected male subject with ASD displayed a normal intelligence quotient (IQ; full scale IQ = 94) and no history of related medical comorbidities. The father of the subject transmitted the variant and tested broader autism phenotype.
Deletion of V269 in LeuT (Which Corresponds to N336 in hDAT) Traps the Intracellular Gate in an Intermediate Open Conformation.
LeuT, a bacterial homolog of hDAT, has been extensively used as a model system for understanding the structural and dynamic bases of Na+-coupled transport in the neurotransmitter/sodium symporter superfamily (20, 40). Models of the conformational changes underlying Na+ and substrate-coupled alternating access have been inferred from crystal structures and investigated spectroscopic analyses using EPR. A common feature of these models is the opening and closing of the intracellular and extracellular gates through the coordination of the N terminus and the extracellular loop (EL4), respectively, although there are substantial differences between the models regarding other structural elements involved in alternating access (8, 17).
To determine the effects of the ∆N336, we introduced the corresponding deletion (20) in LeuT (LeuT ∆V269) and monitored the ligand-dependent conformational dynamics of the extracellular and intracellular gates using spin-label pairs 309/480 and 7/86, respectively. These pairs have been used previously to monitor the isomerization of the transporter between outward-facing, inward-facing, and doubly occluded conformations (8). In LeuT ∆V269, the distance measurements between the spin labels using DEER reveal profound changes in the distance distributions relative to the wild type (Fig. 1). In the absence of Na+ and substrate (apo form), the dominant distance component for the extracellular pair is substantially shorter in the ∆V269 than in the wild type. This suggests collapse of the extracellular gate, as previously observed for the same spin-label pairs in the Y268A mutation, which traps the transporter in an inward-open conformation (8) (Fig. 1 B and C). Consistent with this interpretation, the pair monitoring the intracellular gate shows a predominantly long-distance component (inward-open conformation), with the short-distance component, assigned to the occluded state in the wild-type background, greatly suppressed (Fig. 1 E and F). Remarkably, Na+/Leu has no effect on the probability distribution on the extracellular side of LeuT ∆V269 (Fig. 1 B and C). However, it resets the intracellular gate to a distinct shorter distance component, defined here as “half-open and inward-facing” (HOIF) (Fig. 1F). While this component is observed in the wild-type background (Fig. 1E), its population is minimal compared with the more stable HOIF conformation in LeuT ∆V269.
The DEER distance distributions demonstrate that apo LeuT ∆V269 is predominantly in an outward-closed/inward-open conformation (compare Fig. 1B with Fig. 1C and Fig. 1E with Fig. 1F, black lines), which would be predicted to slow down transport as isomerization to an outward-facing conformation is a central step in symport. Moreover, binding of Na+ alone (compare Fig. 1B with Fig. 1C and Fig. 1E with Fig. 1F, red lines) does not stabilize the outward-facing conformation, which is presumed to enable binding of substrate. Finally, binding of substrate and Na+ (compare Fig. 1B with Fig. 1C and Fig. 1E with Fig. 1F, blue lines) to LeuT ∆V269 reduces the distance of the intracellular spin-label pairs, stabilizing the intracellular gate in a HOIF conformation. This novel HOIF conformation promoted by a single amino acid deletion has a low probability in the wild-type LeuT, where, in the presence of Na+ and substrate, the intracellular gate is closed for the majority of transporters (compare Fig. 1E with Fig. 1F). Therefore, the native valine at position 269 shapes the dynamics of the intracellular gate by impairing the formation of a more open conformation when Na+ and substrate are bound, as well as the efflux of substrate.
Crystal Structure of LeuT ∆V269 Reveals Structural Rearrangements near the Intracellular Gate.
To identify the structural consequences of ∆V269, we determined the crystal structure of LeuT ∆V269 to a resolution of 2.6 Å. The structure of LeuT ∆V269 was solved in an l-leucine– and sodium-bound outward-facing occluded conformation. The asymmetrical unit contained a single protomer, and the structure determined was similar to previously solved structures in an occluded conformation (Cα rmsd ∼ 0.5 Å with PDB ID code 2Q72). The l-leucine could be modeled into the substrate binding site, where F253, a key residue implicated in the extracellular gate, occludes access from the extracellular vestibule, a characteristic of LeuT in the outward-facing occluded conformation. Sodium ions at the Na1 and Na2 sodium binding sites could be modeled unambiguously.
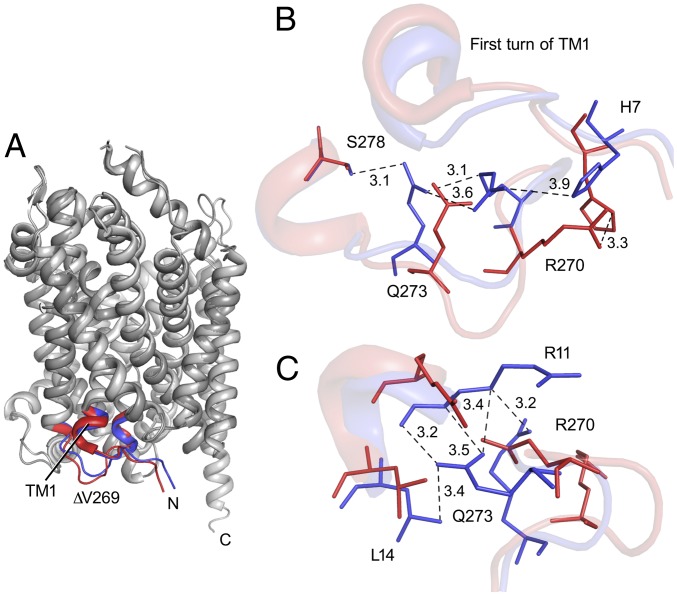
Differences in the ∆V269 and wild-type (PDB ID code 2A65) structures were observed at the intracellular regions that interact with the loop harboring V269 (Fig. 2A). The N terminus and the first helical turn of TM1 showed significant side-chain reorientations in the ∆V269 structure. The distance between the side chains of residues H7 and R270, which are within hydrogen bonding distance in PDB ID code 2A65, is greater in the ∆V269 structure. Furthermore, the deletion of V269 resulted in the translocation of the subsequent five residues (270–274) in the structure, such that the R270 side chain has shifted toward the first helical turn of TM1 and Q273 and away from H7. The deletion also results in a change in the side-chain position of Q273, which moves closer to the first turn of TM1 and now exists within hydrogen bonding distance of S278 (Fig. 2B), which corresponds to D345 in hDAT (discussed below) (20). The movement of the first helical turn of TM1 nearer to the V269 deletion region is facilitated by a series of interactions between the L14 and R11 of TM1 and Q273 (Fig. 2C).
Fig. 2.
Structure of LeuT ∆V269. (A) Superposition of LeuT ∆V269 with PDB ID code 2A65. The primary differences in the intracellular region are highlighted in red (PDB ID code 2A65) and blue (∆V269). (B) Reorientation of the residues immediately following the ∆V269 is displayed. The movement of the first helical turn of the TM1 region is displayed in cartoon representation in the background. (C) Residues responsible for movements in the first turn of TM1 are highlighted (also SI Appendix, Fig. S2).
The differences in the regions other than the N termini are highlighted in red (PDB ID code 2A65) and blue (∆V269) (SI Appendix, Fig. S2A). On the extracellular side, the differences in the structures are minimal. The loop region (153–162) present at the entrance of the extracellular vestibule moves toward the central transmembrane axis in the ∆V269 structure compared with PDB ID code 2A65, and occupies the position similar to the loop in the Y268A mutant structure (PDB ID code 3TT3), which is inward-facing (SI Appendix, Fig. S2B). The deletion of V269 also resulted in a side-chain flip of residue W63. The superposition of the ∆V269 structure with PDB ID code 2A65, and PDB ID code 3TT3 (inward-open LeuT), shows the progression of the indole ring flip of W63 (SI Appendix, Fig. S2B). A water molecule now resides in the space that was occupied by the side chain of V269 in PDB ID code 2A65. This water molecule is coordinated by the indole ring of W63 (SI Appendix, Fig. S2C). The region 438–444 in the ∆V269 structure has also moved away from the deletion region compared with PDB ID code 2A65. The side chain of N274 has shifted “outwards,” and Ile441 has moved away from the V269 deletion region, while also displacing the 438–444 loop. A similar reorientation is also observed in PDB ID code 3TT3, but the loop region slides “up” toward the membrane instead of moving out away from the transmembrane core (SI Appendix, Fig. S2D).
These differences and similarities between the ∆V269 structure, PDB ID code 2A65, and PDB ID code 3TT3 suggest that the deletion of V269 in LeuT could favor a conformation that is neither completely outward-facing nor inward-facing, as observed in EPR experiments. However, usage of β-OG, l-leucine, and sodium in the purification and crystallization conditions favored crystallization of ∆V269 in an outward-facing occluded conformation, which accounts for diminished changes in the EL4 region compared with the DEER data.
Modeling Predicts hDAT ∆N336 Disrupts Coordination Between the N Terminus, IL3, and TM7 Through the Formation of a New K337–D345 Interaction.
To extend our understanding of the consequences of the mutation to hDAT, we carried out homology modeling of hDAT ∆N336 using the Rosetta algorithm. We found that the deletion of N336 leads to a change in orientation of K337. The ∆N336 positions K337 in an orientation that enables an interaction linking IL3 to TM7 through D345 (Fig. 3A, red), while in the wild type (Fig. 3A, blue), IL3 does not interact with TM7 through D345 since K337 faces the intracellular side. Of note, in the ∆V269 structure, the R270 side chain (K337 in hDAT) has shifted toward the first helical turn of TM1 and Q273. The deletion also results in a change in the side-chain position of Q273, which now exists within hydrogen bonding distance of S278 (D345 in hDAT).
Fig. 3.
Modeling of hDAT ∆N336 predicts the formation of a new K337-D345 hydrogen bond. (A) Modeling using Rosetta shows that hDAT (blue) positions a free residue K337 toward the intracellular space. Modeling hDAT ∆N336 (red) reveals a repositioning of K337 that, in turn, forms a new K337-D345 hydrogen bond that is absent in hDAT. This K337-D345 bond may stabilize IL3 by linking it to TM7. (B) Cartoon representation of a homology model of hDAT embedded in a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer. The chloride (cyan) and Na+ (blue) ions; DA (orange); and side chains of K66 (green), K337 (yellow), and D345 (red) are highlighted. (C) hDAT at 50 ns: salt bridge between K66 and D345. (D) hDAT ∆N336 at 50 ns: salt bridge between K66 and D345 (replica 2). (E) hDAT ∆N336 at 100 ns: residue D345 forms two salt bridges, one to K66 and a second to K337 (replica 2) (also SI Appendix, Fig. S3).
Our finding of a potential Q273–S278 hydrogen bond interaction in LeuT and K337–D345 hydrogen bond interaction in hDAT ∆N336 (Q273–S278 interaction in LeuT ∆V269) is mechanistically significant in light of recent MD simulations suggesting that the Na2 is regulated by the K66–D345 interaction (41). According to these simulations, impairments of the K66–D345 interaction affect the kinetics of an inward-open to inward-closed transition of the intracellular gate (41, 42).
Next, we probed the stability of the K337–D345 interaction in hDAT ∆N336 by MD simulations. In our simulations, the K66–D345 salt bridge in wild-type hDAT strengthens the interactions between TM1a and TM7 (Fig. 3B) and stabilizes the bundle domain (TM1, TM2, TM6, and TM7) at the intracellular side (Fig. 3C). Simulations of hDAT showed a stable salt bridge between the side chains of K66 and D345 over the 150-ns time course (SI Appendix, Fig. S3A). Rare occasional short breaking of the salt bridge was observed in all three replicas, but it reformed after a few nanoseconds. No direct interaction between the side chain of K337 and D345 was seen (SI Appendix, Fig. S3B). In contrast, the simulations suggest a very different scenario for hDAT ∆N336 (Fig. 3 D and E, representations of the hDAT ∆N336 bundle at two different times in the simulation). The K66–D345 salt bridge is much less stable, with the distance increasing substantially for long time periods, indicative of a much weaker interaction (SI Appendix, Fig. S3C). The reorientation of the side chain of K337 promoted by ∆N336 shortens the IL3 loop, as observed in the crystal structure of the LeuT Δ269, and consequently weakens the interaction of K66 with D345. The new orientation of K337 favors an increased interaction with D345 (SI Appendix, Fig. S3D), effectively promoting a competition between the side chains of K337 and K66 and, as a consequence, weakening the interaction between TM7 and TM1a. The new K337–D345 interactions, as noted in Fig. 3A, may promote a less dynamic intracellular gate, thereby establishing the conserved N336 as a pivotal residue to the dynamics of the intracellular gate of hDAT, and possibly to transport function.
∆N336 Impairs hDAT Function.
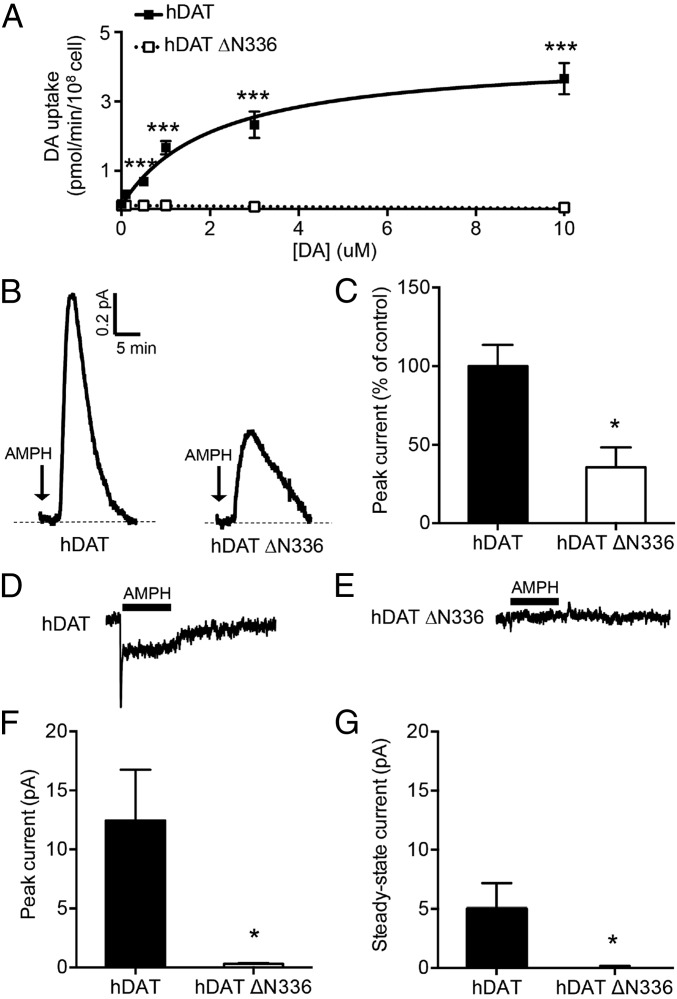
EPR analysis, X-ray crystallography, and hDAT modeling predict that ∆N336 disrupts intracellular gating dynamics and potentially compromises function. Therefore, we asked whether hDAT ∆N336 is capable of DA transport. We found that hDAT ∆N336 cells display a significant reduction in DA uptake. The hDAT ∆N336-expressing cells, compared with hDAT, were below measurable DA uptake levels to calculate the Vmax of DA transport and Km of DA. Fig. 4A show representative plots of DA uptake kinetics for hDAT (Vmax = 4.33 ± 0.44 pmol⋅min−1 per 108 cells; Km = 2.10 ± 0.59 μM) and hDAT ∆N336 cells. The reduced DA transport was not associated with a reduction in either total or surface expression of hDAT ∆N336 (SI Appendix, Fig. S4, Top). Surface fractions were quantitated, normalized to total hDAT, and expressed as a percent of hDAT (SI Appendix, Fig. S4, Bottom) (P = 0.781 by t test; n = 4, in duplicate).
Fig. 4.
hDAT ∆N336 displays impaired DA transport, reduced AMPH-induced DA efflux, and diminished AMPH-induced currents. (A) Representative plot of [3H]DA uptake kinetics in hDAT (■) or hDAT ∆N336 (□) cells (***P ≤ 0.0001 by two-way ANOVA, followed by Bonferroni posttest; n = 6, performed in triplicate). (B) Representative AMPH-induced amperometric currents recorded from hDAT and hDAT ∆N336 cells. Cells were patch-loaded with DA. Arrows indicate application of 10 μM AMPH. (C) Quantification of AMPH-induced DA efflux. Data are represented as the peak amperometric current ± SEM (*P ≤ 0.01 by t test; n = 5). (D and E) Mean AMPH (10 μM)-induced whole-cell current of 10 consecutive sweeps obtained from hDAT and hDAT ∆N336 cells is plotted. The hDAT and hDAT ∆N336 cells were kept under voltage clamp (−60 mV), and AMPH was rapidly applied to the cells for 1 s by a piezoelectric translator (Methods). In contrast to hDAT ∆N336 cells, hDAT cells display a peak and a steady-state current. (F and G) Quantification of AMPH-induced peak current and steady-state current for hDAT- and hDAT ∆N336-expressing cells. Data are represented as amperometric current ± SEM (*P ≤ 0.05 by t test; n = 9) (also SI Appendix, Figs. S4 and S5).
∆N336 Supports Reverse Transport of DA.
AMPH is a psychostimulant that promotes DAT-mediated DA efflux (reverse transport of DA). To measure DA efflux, we adopted the patch-clamp technique in whole-cell patch configuration to allow intracellular perfusion of an internal solution containing 2 mM DA. An amperometric probe was juxtaposed to the plasma membrane of the cell and used to measure reverse transport of DA via oxidation/reduction reactions. The whole-cell electrode, in current-clamp configuration, allows the cell to control its membrane voltage and ensures that cells were equally loaded with intracellular DA (hDAT ∆N336 cells are incapable of DA uptake). Fig. 4B show representative traces of DA efflux in hDAT and hDAT ∆N336 cells. Quantitation of the peak amperometric currents shows reduced DA efflux in hDAT ∆N336 cells compared with hDAT cells (P ≤ 0.01 by t test; n = 5; Fig. 4C). Considering that hDAT ∆N336 has impaired DA uptake while retaining DA efflux, these data support a model where the N336 and the intracellular gate of hDAT differentially regulate inward versus outward transport of DA.
∆N336 Exhibits Impaired Conformational Changes Defining the Transport Cycle.
Rapid (millisecond) application of AMPH induces an hDAT-mediated fast-activating transient current that represents the synchronized inward reorientation of hDAT with the substrate bound (43). This transient current is followed by a cocaine-sensitive inward steady-state current that represents substrate (i.e., AMPH) uptake (43). Therefore, these two currents define, at least in part, the hDAT transport cycle. We voltage-clamped hDAT or hDAT ∆N336 cells using the whole-cell patch configuration at −60 mV while applying 10 μM AMPH. As expected, hDAT cells displayed a fast-activating transient current, followed by a steady-state current (Fig. 4D). Upon washout of AMPH, the steady-state current decayed to baseline levels (Fig. 4D). These characteristic fast-activating transient currents and steady-state currents were notably absent in hDAT ∆N336 cells (Fig. 4E). Quantitation of the peak current (Fig. 4F) and steady-state currents (Fig. 4G) shows that ∆N336 significantly impairs synchronized inward reorientation of hDAT with the substrate bound, as well as substrate-induced transporter function (P ≤ 0.05 by unpaired t test; n = 9). These data further suggest that in hDAT, ∆N336 substrate uptake and efflux are uncoordinated and independent processes.
A HOIF Conformation Is Critical for Transport.
Our results point to a significant role for N336 in hDAT as well as V269 in LeuT for the dynamics of the intracellular gate. However, the question remains of whether the HOIF conformation stems exclusively from a deletion of V269 in LeuT or is a conformation that is regulated by that specific amino acid residue. In other words, what role does the N336 in hDAT have in the HOIF conformation? Furthermore, is the HOIF conformation important for the dynamics of substrate uptake and efflux? To begin to address these questions, we substituted the native LeuT amino acid V269 with the equivalent hDAT ΔN336. For this purpose, we analyzed the distance distributions of reporter pairs on the intracellular and extracellular gates. In the presence of Na+ and substrate, the distance distributions of spin-label pairs 309/480 monitoring the extracellular side (SI Appendix, Fig. S5A) were comparable for LeuT and LeuT V269N (compare SI Appendix, Fig. S5B with SI Appendix, Fig. S5C, blue lines), despite an observed increase of a short-distance component in the apo state and in the presence of Na+ in LeuT V269N (compare SI Appendix, Fig. S5B with SI Appendix, Fig. S5C, black and red lines, respectively).
LeuT V269N has considerable effects on the distribution of spin-label pairs 7/86 monitoring the intracellular gate (SI Appendix, Fig. S5D). In the apo state, LeuT V269N shows a dramatic shift in the transporter distributions favoring an inward-facing conformation (compare SI Appendix, Fig. S5E with SI Appendix, Fig. S5F, black lines). Binding of Na+ alone (SI Appendix, Fig. S5F, red line) reduces the population of inward-facing transporters and exposes a population of transporters wherein the intracellular gate is in a HOIF. Binding of substrate and Na+ also promotes a conformation wherein the intracellular gate may reside in a HOIF conformation (compare SI Appendix, Fig. S5E with SI Appendix, Fig. S5F, blue lines). However, the population of the HOIF conformation in LeuT V269N is greatly reduced compared with LeuT ∆V269, while it remains increased with respect to the wild-type LeuT. These data strongly suggest that the residue at position 269 in LeuT, and possibly at position 336 in hDAT, regulates the mobility of the intracellular gate and its ability to occupy an inward-closed conformation. They also suggest that an asparagine at position 269 promotes a HOIF conformation that allows an occluded conformation in the presence of substrate and Na+, and similar to the low probability conformation noticed in LeuT. These data imply that the probability distribution of the HOIF conformation and the mobility of the intracellular gate are regulated (from bacteria to higher organisms) by amino acid conservation in this loop, including position 269 (i.e., V vs. N). Our data suggest that valine at position 269 inhibits the HOIF conformation, while an asparagine partially stabilizes the HOIF conformation, also allowing for an occluded conformation in the presence of substrate and Na+.
To further explore the importance of the HOIF conformation and the role of N336 in transporter function and substrate uptake, we substituted N336 in hDAT with the corresponding valine residue in LeuT (hDAT N336V). We expected the hDAT N336V cells to display reduced Vmax of DA uptake. SI Appendix, Fig. S5G shows that hDAT N336V cells do exhibit a significant reduction (∼85%) in Vmax and a significant decrease in DA Km compared with hDAT cells (SI Appendix, Fig. S5G, Top). A representative plot of DA uptake kinetics for hDAT and hDAT N336V is shown in SI Appendix, Fig. S5G, Bottom. These data suggest that N336 is pivotal for proper DA transport in addition to the dynamics of the intracellular gate.
Isolated Brains from hDAT ∆N336 Flies Display Decreased DA Uptake and AMPH-Induced DA Efflux.
We utilized D. melanogaster as an animal model to determine, functionally and behaviorally, DA dysfunction caused by ∆N336. We expressed hDAT or hDAT ∆N336 in flies homozygous for the Drosophila DAT-null allele, DATfmn (dDAT KO), adopting the Gal4/UAS system to express a single copy of hDAT or hDAT ∆N336 in a dDATfmn mutant background, selectively in DA neurons. The transgenic flies were generated by using phiC31-based integration, which should lead to the expression of comparable levels of mRNA for the relevant transgenes (hDAT or hDAT ∆N336).
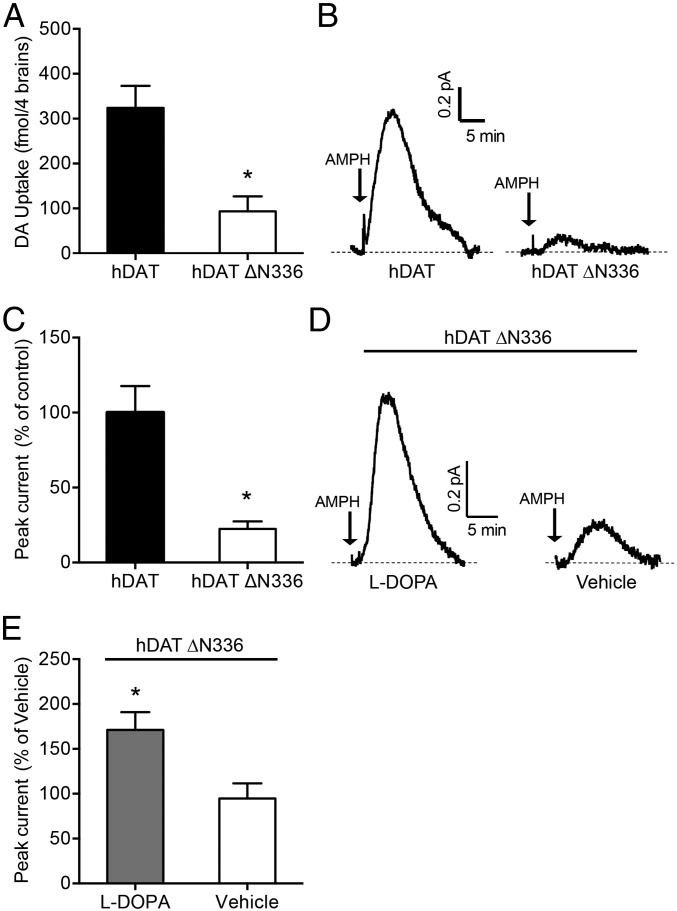
We isolated the brains of flies expressing either hDAT or hDAT ∆N336 to determine possible changes in brain DA uptake (200 nM [3H]DA). We detected a significant reduction in DA uptake in the flies expressing hDAT ∆N336 (93.7 ± 33.22 fmol in four brains) compared with the flies expressing hDAT (324.1 ± 49.14 fmol in four brains) (P ≤ 0.01 by t test; n = 4) (Fig. 5A).
Fig. 5.
In Drosophila brain, hDAT ∆N336 cells display reduced DA uptake and AMPH-induced efflux. (A) Flies expressing hDAT ∆N336 display reduced DA uptake compared with hDAT-expressing brains. Data are represented as DA uptake (200 nM DA) using four Drosophila brains per well ± SEM (*P ≤ 0.01 by t test; n = 4). (B) Representative AMPH-induced (20 μM) amperometric currents recorded from fly brains expressing hDAT or hDAT ∆N336. Arrows indicate application of AMPH. (C) Quantitation of DA efflux. Data are presented as the peak amperometric current ± SEM (*P ≤ 0.01 by t test; n = 4). (D) Flies expressing hDAT ∆N336, when fed an l-DOPA (5 mM) diet for 24 h, display increased DA efflux compared with vehicle-fed flies. Representative AMPH-induced (20 μM) amperometric currents recorded from hDAT ∆N336 fly brains. Arrows indicate application of AMPH. (E) Quantitation of DA efflux. Data are presented as the peak amperometric current ± SEM (*P ≤ 0.05 by t test; n = 4).
To determine the ability of hDAT ∆N336 to support reverse transport of DA in brain, we measured DA efflux from isolated Drosophila brains. We inserted a carbon fiber electrode into isolated Drosophila brains in close proximity to the mCherry-tagged DA-specific neurons. Fig. 5B shows representative traces of DA efflux in brains of flies expressing either hDAT or hDAT ∆N336. AMPH-induced DA efflux recorded in the brain of hDAT ∆N336 flies is significantly lower than that seen in hDAT flies (P ≤ 0.01 by t test; n = 6) (Fig. 5C).
DA Synthesis Enhances Reverse Transport of DA in Flies Expressing hDAT ∆N336.
Deficient DA uptake causes a decrease in neuronal DA content (44). Therefore, it is possible that the reduced ability of AMPH to induce reverse transport of DA in hDAT ∆N336 flies is due to low levels of intracellular DA. Thus, we used l-DOPA, the direct precursor to DA, as a tool for increasing DA content in dopaminergic neurons of hDAT ∆N336 flies. We treated hDAT ∆N336 flies with l-DOPA or vehicle for 24 h and then recorded AMPH-induced amperometric signals from isolated brains. Representative amperometric traces of l-DOPA and vehicle-treated hDAT ∆N336 files are shown in Fig. 5D. Quantitation of peak amperometric currents show that DA efflux in l-DOPA–fed hDAT ∆N336 flies is elevated (182 ± 6.3% increase) compared with vehicle-treated flies (P ≤ 0.05 by t test; n = 4 l-DOPA, n = 5 vehicle) (Fig. 5E). These data demonstrate that hDAT ∆N336 can support reverse transport of DA in intact brains.
hDAT ∆N336 Supports Hyperlocomotion and Increased Grooming.
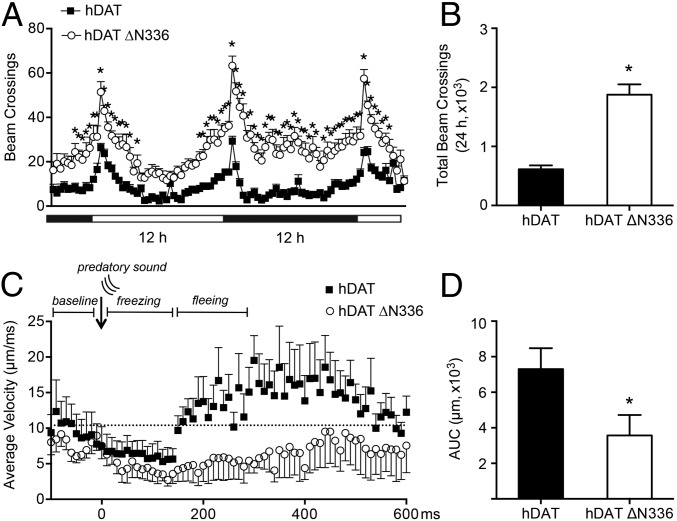
Across many species, locomotion is a fundamental behavior regulated by DA. In Drosophila, we and others have established that locomotion is a DA-associated and DAT-dependent behavior (1, 36). Therefore, measuring changes in locomotion offers a powerful behavioral indicator for probing DA dysfunction in vivo. To this end, we measured locomotion in flies expressing either hDAT or hDAT ∆N336. Fig. 6A shows locomotor activity of these flies by beam crosses over a 36-h period during both the light (horizontal white bar) and dark (horizontal black bar) conditions. Although the hDAT ∆N336 flies are clearly hyperactive, we observed no changes in the locomotive circadian rhythms. Quantitation of total beam crossings in hDAT ∆N336 flies over 24 h is significantly elevated compared with hDAT flies (*P ≤ 0.0001 by Mann–Whitney test; n = 30–31) (Fig. 6B). These data support the notion that hDAT ∆N336 results in impaired DA neurotransmission and increased locomotion.
Fig. 6.
hDAT ∆N336 flies are hyperactive and show prolonged freezing and reduced fleeing. (A) Locomotor activity was assayed over 30 h during the light (horizontal white bars) and dark (horizontal dark bars) cycle. Flies expressing hDAT ∆N336 (○) were hyperactive compared with flies expressing hDAT (■) (*P < 0.05 by two-way ANOVA with Sidak’s post hoc test; n = 31–32). Beam crossings were binned into 20-min intervals. (B) Quantitation of total beam crossings over 24 h. Data are presented as the total beam crossing ± SEM (*P ≤ 0.0001 by Mann–Whitney test; n = 30–31). (C) Average velocity (μm⋅ms−1) was assayed over 600 ms following an auditory stimulus for flies expressing hDAT (■) or hDAT ∆N336 (○). The hashed line indicates preaudible baseline velocity. (D) AUC is calculated as the cumulative velocity from the onset to the end of the predatory sound. Quantitation of AUC over 600 ms for hDAT and hDAT ∆N336 flies. Data are presented as the total AUC ± SEM (*P ≤ 0.05 by t test; n = 8).
hDAT ∆N336 Flies Exhibit Increased Grooming Behavior.
Repetitive behaviors, including self-grooming, have been observed in animal models of neuropsychiatric disorders (45). Grooming behavior in Drosophila is a stereotyped sequence of leg and body movements (46). In Drosophila, grooming requires functional DA neurotransmission (47, 48). Therefore, it is possible that ∆N336 flies display altered grooming because of impaired DA clearance. We quantified the percent of time spent grooming in flies expressing ∆N336 compared with wild type. The ∆N336 flies displayed enhanced time grooming (22.9 ± 3.4% time grooming) compared with wild-type flies (6.3 ± 4.2% time grooming) (P ≤ 0.05 by t test; n = 3–5).
hDAT ∆N336 Flies Exhibit Increased Freezing Following an Audible Fear Stimulus.
A fear stimulus assay offers an opportunity to probe an evolutionarily conserved construct. A complex pattern of animal behaviors is involved in the detection and analysis of threat stimuli, and in the situations in which the threat is encountered. Risk assessment is therefore pivotal for establishing an appropriate response to threatening situations, such as the presence of a predator. Risk assessment evokes an evolutionarily conserved brain state, fear, that triggers defensive behaviors. Risk assessment is also important for determining one specific behavioral defense over another, for example, flight over hiding or freezing. The appropriate behavioral choice in threating situations is controlled by specific neurocircuits and determines the ability to survive (49). In humans, dysregulation of these evolutionary conserved circuits is implicated in anxiety-related disorders (49, 50), including ASD.
To evaluate if impairments in DA clearance from the variant ∆N336 alters defensive response (i.e., freezing, flight), we exposed either hDAT or hDAT ∆N336 flies to an auditory fear stimulus, the sound of a predatory wasp. Freezing behavior in Drosophila in response to a threatening stimulus has been previously observed by Zacarias et al. (51) and Gibson et al. (52). Furthermore, the sound of the predatory wasp specifically evokes a fear response in Drosophila larvae (53). We adapted this assay in adult Drosophila and recorded a behavior in which adult flies freeze upon hearing the threatening sound of a predatory wasp. Using 3D printed recording chambers and a high-speed camera that can record a single organism’s movement at a rate of 1,000 frames per second, we analyzed freezing behavior in freely moving hDAT or hDAT ∆N336 flies in response to a threatening auditory stimulus. In freely moving hDAT flies, the sound of a predatory wasp (Fig. 6C, arrow, predatory sound) elicits a short freezing period, followed by a distinctive and rapid increase in average velocity, fleeing (Fig. 6C and Movie S1) (hDAT WT; SI Appendix). In contrast, when exposed to the sound of the predatory wasp, hDAT ∆N336 flies display prolonged freezing and delayed fleeing (Fig. 6C and Movie S2) (hDAT ∆N336; SI Appendix).
The deficient fleeing response of hDAT ∆N336 flies was quantified by calculating the area under the curve (AUC) of the average velocity (total distance traveled in micrometers) from the onset of the predatory sound until flies returned to prestimulus velocity levels (∼600 ms). Quantitation of the AUC reveals flies expressing hDAT ∆N336 have a markedly reduced escape response (3,574 ± 1,147 μm) compared with hDAT flies (7,302 ± 1,176 μm) (P ≤ 0.05 by t test; n = 8) (Fig. 6D). One possible explanation for this deficit in fleeing response in hDAT ∆N336 flies is an increase in the freezing period.
hDAT ∆N336 Flies Show Impairments in Social Interactions.
Grouping is a widespread behavior observed across many animal populations that is often advantageous for survival and is defined as a temporary or permanent aggregation of animals (54). Grouping defines a wide variety of interactions that range from coincidental encountering to social gathering, and can lead to complex behaviors, such as fleeing in response to a threat. In this context, fleeing is an escaping behavior of a prey population. During fleeing, the compression and expansion of the flock size are regulated by complex social interactions among the animals in the flock, as well as by prey–predator interactions (54).
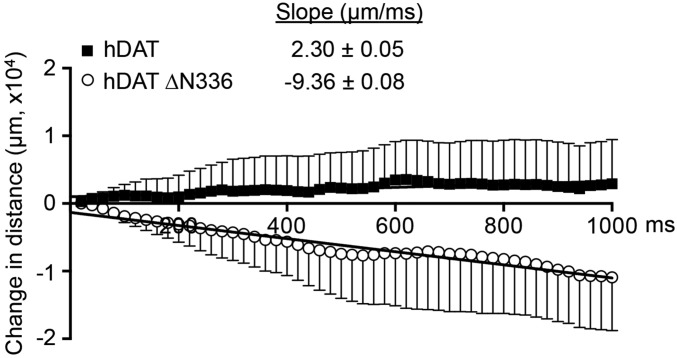
Here, we explored whether hDAT dysfunction induced by ∆N336 influences flock size during an escape response induced by a fear stimulus. We measured the interfly distance across four flies before and after an escape response and calculated their change in social space (sum of distances between a fly and its three neighbors) over time. A positive social space value indicates an expansion in flock size [a normal response under predatory pressure (54)] over time, while a negative social space value indicates a compression of flock size. Fig. 7 illustrates that flies expressing hDAT have a preference for increasing the flock size following an auditory fear response, while flies expressing hDAT ∆N336 have a propensity for a time-dependent aggregation. This significant inverse social space relationship (P < 0.001) was calculated using the slope of the sum of interfly distances in hDAT (2.30 ± 0.05 μm⋅ms−1) and hDAT ∆N336 (−9.36 ± 0.08 μm⋅ms−1) flies over time (Fig. 7). These slopes were calculated over 1,000 ms of recordings (data binned in 20-ms intervals) and normalized to the respective sum of interfly distances calculated at 700 ms after the start of the predatory stimulus. This time point (time 0) was chosen as it occurs immediately after the freezing behavior.
Fig. 7.
hDAT ∆N336 flies show social impairments as measured by proximity to their neighbors during the escape response. The sum of distances (μm) across four flies (social space) was calculated and assayed over 1,000 ms and normalized to the respective social space calculated at time 0. Over time, hDAT flies (■) display increased flock size, as determined by the slope of the fitting of the sum of distances over time (black dotted line: 2.30 ± 0.05 μm⋅ms−1). In contrast, hDAT ∆N336 flies (○) reveal decreased flock size over time (gray dotted line: −9.36 ± 0.08 μm⋅ms−1).
Discussion
ASD is a phenotypically and etiologically complex disease, yet significant evidence for disease risk resides in an individual’s genetics (12, 13). Rare variations represent an important class of this genetic risk, and sequencing efforts have identified new risk variants associated with specific signaling pathways/molecules, including DA (1–3). However, a limitation of these efforts is the difficulty of achieving genetic power to detect rare variant enrichment. Clearly, the exact nature of the genetic risk for ASD conferred by ∆N336 is limited. However, this occurring variant provided a blueprint to gain valuable insights into the mechanisms of transporter function and gating, and how dysfunction of these processes translates in specific abnormal animal behaviors and DA neurotransmission.
We demonstrated that hDAT ∆N336 cells have impaired DA transport and DAT-mediated electrical currents, while partially supporting DA efflux. Using Drosophila as an animal model, we discovered that expression of hDAT ∆N336 reduces DA uptake in whole brain. These data, combined with previous published data (1–3), support the idea that hDAT dysfunction in ASD stems from specific and yet distinct mechanisms. Interestingly, deficits in DA efflux in Drosophila brain can be rescued by oral administration of l-DOPA to the ∆N336 transgenic flies. These data suggest a possible avenue for restoring DA dynamics and efflux function in ASD.
The question remains as to whether a specific impairment in DAT function causes disruptions of defined behaviors and whether these disruptions reveal new mechanisms/behaviors underlying aberrant DA neurotransmission. We found that hDAT ∆N336 flies are hyperlocomotive, associating ∆N336 with an increase in extracellular DA levels. DA dysfunction in hDAT ∆N336 flies is also reflected in increased grooming. This is important since repetitive behaviors have been associated with dysregulated DA homeostasis.
To uncover whether ∆N336, as well as impaired DAT function, disrupts complex behaviors, we developed paradigms to explore “fear” and social interactions in flies. Neuron-specific expression of ∆N336 disrupted flies’ defensive behaviors. Our data demonstrate that hDAT ∆N336 flies display prolonged freezing and delayed fleeing upon an auditory fear stimulus, further supporting the role of DA in specific fear responses. Our results link DAT dysfunction associated with ∆N336 to impaired defensive behaviors and to increased fear.
ASD is also characterized by impairments in social interactions and communications. Therefore, we measured the compression/expansion of a fly flock size under predatory pressure. As expected, flies expressing hDAT have a preference for increasing the flock size following an audible fear response. Instead, flies expressing hDAT ∆N336 have a propensity for a time-dependent aggregation. These data define how specific impairments in hDAT function translate into disrupted social interactions.
The functional and behavioral characterization of variants is essential to provide insights into their impact on neurocircuits and signaling pathways. However, their structural characterization may shed light on the underpinnings of hDAT function. Using LeuT as a template, we show that ∆V269 stabilizes a previously unappreciated conformation of the intracellular gate, where it resides in a HOIF conformation. The loss of uptake function is a possible consequence of a locked conformation of the extracellular gate. In contrast, the unique conformation of the intracellular gate, favored by binding of ions and substrate, allows for reverse transport of DA. Therefore, the intracellular gate differentially regulates inward versus outward transport of DA. These results also suggest that coordination of the extracellular and intracellular gates is a requirement for DA uptake, but not DA efflux, which is not impaired by the stabilization of the HOIF conformation.
The LeuT ∆V269 crystal structure reveals, in atomic detail, the structural rearrangements induced by the mutation. Substantial repacking is observed for IL3, which changes the side-chain orientation of the highly conserved R270 (K377 in hDAT). The consequences of this repacking are a profound shift in the structure and ligand-dependent dynamics of the N-terminal loop, which acts as an intracellular gate, and the EL4 lid, which blocks the permeation pathway on the extracellular side. While the crystal structure hints at movement of the intracellular gate that extends to the first turn of TM1, the extracellular vestibule adopts an outward-occluded conformation. Previous reports indicated the tendency of solutes, including detergent molecules, to bind in this vestibule and “wedge it” in a more open configuration (25). The stabilization of this conformation may have limited the extent of the repacking on the intracellular side in the crystal structure.
De novo modeling and MD simulations extend these conclusions to hDAT by demonstrating a repacking of the side chain of K377 in hDAT ∆N336, which disrupts the interaction between K66 and D345. In light of recent MD simulations invoking a role for this hydrogen bond in intracellular gating, it is plausible that the weakening of this interaction leads to a more dynamic N-terminal domain supporting specific functions of hDAT (i.e., DA efflux).
Our results uncover how specific hDAT molecular impairments promoted by genetic variants are linked to altered social behaviors. We translated the consequences of the relative changes in the conformational stability of transporter intermediates, both at the brain level and behaviorally. This entailed the development of behavioral assays supported by recent technological advances, including high-speed video recording of predatory and social behaviors in flies. The experimental paradigms we describe here provide a framework for molecular and behavioral analysis of novel DAT variants that are discovered by genetic analyses of individuals with ASD or related neuropsychiatric illness, as well as other disease-linked mutants that are emerging from precision medicine initiatives.
Supplementary Material
Acknowledgments
N.G.C. was supported by an NIH T32 Fellowship from the Ion Channel, and Transporter Biology Training Grant (T32 NS007491-14). This work was supported by NIH Grants MH070039 (to E.G.), GM080403 and HL122010 (to J.M.), U54-GM087519 and S10 RR027091 (to H.S.M.), and DA35263 and DA38058 (to A.G.). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors for data have been deposited in Protein Data Bank, www.wwpdb.org (PDB ID code 6NLE).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816247116/-/DCSupplemental.
References
- 1.Hamilton PJ, et al. NIH ARRA Autism Sequencing Consortium De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder. Mol Psychiatry. 2013;18:1315–1323. doi: 10.1038/mp.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowton E, et al. SLC6A3 coding variant Ala559Val found in two autism probands alters dopamine transporter function and trafficking. Transl Psychiatry. 2014;4:e464. doi: 10.1038/tp.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartier E, et al. Rare autism-associated variants implicate syntaxin 1 (STX1 R26Q) phosphorylation and the dopamine transporter (hDAT R51W) in dopamine neurotransmission and behaviors. EBioMedicine. 2015;2:135–146. doi: 10.1016/j.ebiom.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koob GF, Bloom FE. Cellular and molecular mechanisms of drug dependence. Science. 1988;242:715–723. doi: 10.1126/science.2903550. [DOI] [PubMed] [Google Scholar]
- 5.Hamilton PJ, et al. PIP2 regulates psychostimulant behaviors through its interaction with a membrane protein. Nat Chem Biol. 2014;10:582–589. doi: 10.1038/nchembio.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forrest LR, et al. Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA. 2008;105:10338–10343. doi: 10.1073/pnas.0804659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kazmier K, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21:472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen N, Rickey J, Berfield JL, Reith ME. Aspartate 345 of the dopamine transporter is critical for conformational changes in substrate translocation and cocaine binding. J Biol Chem. 2004;279:5508–5519. doi: 10.1074/jbc.M306294200. [DOI] [PubMed] [Google Scholar]
- 10.Khelashvili G, et al. Spontaneous inward opening of the dopamine transporter is triggered by PIP2-regulated dynamics of the N-terminus. ACS Chem Neurosci. 2015;6:1825–1837. doi: 10.1021/acschemneuro.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Psychiatric Association; DSM-5 Task Force . Diagnostic and Statistical Manual of Mental disorders: DSM-5. 5th Ed American Psychiatric Association; Washington, DC: 2013. [Google Scholar]
- 12.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: On the path to precision medicine. Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Rubeis S, Buxbaum JD. Recent advances in the genetics of autism spectrum disorder. Curr Neurol Neurosci Rep. 2015;15:36. doi: 10.1007/s11910-015-0553-1. [DOI] [PubMed] [Google Scholar]
- 14.Fischbach GD, Lord C. The Simons Simplex collection: A resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 15.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claxton DP, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat Struct Mol Biol. 2010;17:822–829. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 19.Zou P, McHaourab HS. Increased sensitivity and extended range of distance measurements in spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophys J. 2010;98:L18–L20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 21.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol. 2012;19:212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malinauskaite L, et al. A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state. Nat Commun. 2016;7:11673. doi: 10.1038/ncomms11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang KH, Penmatsa A, Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015;521:322–327. doi: 10.1038/nature14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchmayer F, et al. Amphetamine actions at the serotonin transporter rely on the availability of phosphatidylinositol-4,5-bisphosphate. Proc Natl Acad Sci USA. 2013;110:11642–11647. doi: 10.1073/pnas.1220552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stockner T, et al. Mutational analysis of the high-affinity zinc binding site validates a refined human dopamine transporter homology model. PLoS Comput Biol. 2013;9:e1002909. doi: 10.1371/journal.pcbi.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf MG, Hoefling M, Aponte-Santamaría C, Grubmüller H, Groenhof G. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J Comput Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 32.Berger O, Edholm O, Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 35.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- 36.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005;25:7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friggi-Grelin F, et al. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 38.Wang JW, Beck ES, McCabe BD. A modular toolset for recombination transgenesis and neurogenetic analysis of Drosophila. PLoS One. 2012;7:e42102. doi: 10.1371/journal.pone.0042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70:1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 41.Razavi AM, Khelashvili G, Weinstein H. A Markov state-based quantitative kinetic model of sodium release from the dopamine transporter. Sci Rep. 2017;7:40076. doi: 10.1038/srep40076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng MH, Bahar I. Molecular mechanism of dopamine transport by human dopamine transporter. Structure. 2015;23:2171–2181. doi: 10.1016/j.str.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erreger K, Grewer C, Javitch JA, Galli A. Currents in response to rapid concentration jumps of amphetamine uncover novel aspects of human dopamine transporter function. J Neurosci. 2008;28:976–989. doi: 10.1523/JNEUROSCI.2796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giros B, Caron MG. Molecular characterization of the dopamine transporter. Trends Pharmacol Sci. 1993;14:43–49. doi: 10.1016/0165-6147(93)90029-j. [DOI] [PubMed] [Google Scholar]
- 45.Silverman JL, Tolu SS, Barkan CL, Crawley JN. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology. 2010;35:976–989. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connolly K. The social facilitation of preening behaviour in Drosophila melanogaster. Anim Behav. 1968;16:385–391. doi: 10.1016/0003-3472(68)90023-7. [DOI] [PubMed] [Google Scholar]
- 47.Yellman C, Tao H, He B, Hirsh J. Conserved and sexually dimorphic behavioral responses to biogenic amines in decapitated Drosophila. Proc Natl Acad Sci USA. 1997;94:4131–4136. doi: 10.1073/pnas.94.8.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang HY, et al. Overexpression of the Drosophila vesicular monoamine transporter increases motor activity and courtship but decreases the behavioral response to cocaine. Mol Psychiatry. 2006;11:99–113. doi: 10.1038/sj.mp.4001742. [DOI] [PubMed] [Google Scholar]
- 49.Steimer T. The biology of fear- and anxiety-related behaviors. Dialogues Clin Neurosci. 2002;4:231–249. doi: 10.31887/DCNS.2002.4.3/tsteimer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeDoux J. The emotional brain, fear, and the amygdala. Cell Mol Neurobiol. 2003;23:727–738. doi: 10.1023/A:1025048802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zacarias R, Namiki S, Card GM, Vasconcelos ML, Moita MA. Speed dependent descending control of freezing behavior in Drosophila melanogaster. Nat Commun. 2018;9:3697. doi: 10.1038/s41467-018-05875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson WT, et al. Behavioral responses to a repetitive visual threat stimulus express a persistent state of defensive arousal in Drosophila. Curr Biol. 2015;25:1401–1415. doi: 10.1016/j.cub.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W, Yan Z, Jan LY, Jan YN. Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc Natl Acad Sci USA. 2013;110:13612–13617. doi: 10.1073/pnas.1312477110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SH, Pak HK, Chon TS. Dynamics of prey-flock escaping behavior in response to predator’s attack. J Theor Biol. 2006;240:250–259. doi: 10.1016/j.jtbi.2005.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.