Significance
The key epigenetic regulator EZH2 plays a central role in fibrosis and abnormal angiogenesis in scleroderma. EZH2-target genes mediating profibrotic and antiangiogenic effects in scleroderma patients have been identified and characterized. Inhibiting EZH2 by repurposing already-existing EZH2 inhibitors currently being trialed in cancer might provide a therapeutic approach to scleroderma.
Keywords: Scleroderma, epigenetics, EZH2, angiogenesis, fibrosis
Abstract
Scleroderma (SSc) is a complex disease that involves activation of the immune system, vascular complications, and tissue fibrosis. The histone methyltransferase enhancer of zeste homolog 2 (EZH2) mediates trimethylation of lysine 27 of histone 3 (H3K27me3), which acts as a repressive epigenetic mark. Both EZH2 and H3K27me3 were elevated in SSc dermal fibroblasts and endothelial cells compared with healthy controls. EZH2 inhibitor DZNep halted fibrosis both in vitro and in vivo. In SSc fibroblasts, DZNep dose-dependently reduced the expression of profibrotic genes and inhibited migratory activity of SSc fibroblasts. We show that epigenetic dysregulation and overexpression of LRRC16A explains EZH2-mediated fibroblast migration in SSc. In endothelial cells, inhibition of EZH2 restored normal angiogenesis in SSc via activating the Notch pathway, specifically by up-regulating the Notch ligand DLL4. Our results demonstrate that overexpression of EZH2 in SSc fibroblasts and endothelial cells is profibrotic and antiangiogenic. Targeting EZH2 or EZH2-regulated genes might be of therapeutic potential in SSc.
Systemic sclerosis (Scleroderma, SSc) is a systemic disease comprised of blood vascular dysfunction, autoimmunity, and excessive fibroblast activation that ultimately results in tissue damage and organ failure. The underlying cause of SSc appears to be multifactorial as various cell types and pathways have been implicated. Recent studies have provided strong support for the involvement of epigenetic mechanisms in SSc pathogenesis (1–5). Alterations of histone marks, DNA methylation, and noncoding RNAs were observed in immune cells, endothelial cells (ECs), and fibroblasts isolated from patients with SSc, pointing to the importance of examining epigenetic regulation in this disease (6).
The impact of a histone methyltransferase, the enhancer of zeste homolog 2 (EZH2), in SSc pathogenesis was examined in this study. This enzyme is involved in T cell differentiation, EC-mediated angiogenesis, as well as myofibroblast transformation and tissue fibrosis—essentially all three aspects of SSc pathogenesis (7–9). EZH2 induces histone H3 lysine 27 trimethylation (H3K27me3) and is the catalytic component of the highly conserved polycomb-repressive complex 2 (PRC2) that represses transcription. However, in certain circumstances, EZH2 activates transcription, which is PRC2-independent (10). EZH2 is involved in the regulation of EC function, particularly by affecting genes associated with cell adhesion (7, 11). In the tumor microenvironment, EZH2 is regulated by hypoxia, where EZH2 appears to induce angiogenesis (12–14). In contrast, other studies showed that EZH2 silencing or EZH2 inhibitors enhanced migration and angiogenesis of ECs (15). In addition, EZH2 inhibitor DZNep increased angiogenesis in ischemic muscles, the circulating levels of proangiogenic cells, and blood flow recovery in mice (15). EZH2 appears to promote fibrosis. Its expression is increased in patients with idiopathic pulmonary fibrosis and chronic kidney disease (8, 16), as well as in animal models including bleomycin-induced lung fibrosis, carbon tetrachloride liver fibrosis, and unilateral ureteral obstruction kidney fibrosis (8, 16–18). Inhibition of EZH2 reduces TGFβ-Smad signaling pathways and suppresses activation of the EGFR and PDGFR pathways through up-regulation of PTEN (8, 16). In addition, miR-132, MeCP2, and EZH2 operate in regulatory epigenetic relay pathways that result in transcriptional repression of PPARγ, ultimately leading to stimulation of myofibroblast transdifferentiation (17, 19).
In this study, we focused on the involvement of EZH2 in SSc angiogenesis and fibrosis, utilizing dermal ECs and fibroblasts isolated from healthy controls and SSc patients. We first established the expression profile of EZH2 in both cell types in SSc, then assessed its functional roles in SSc ECs and fibroblasts, as well as in vivo in an animal model. Through DNA methylation profiling and literature mining, we were able to identify key genes involved in mediating the fibroblast migratory and antiangiogenic effects of EZH2 in SSc, then validate their roles with functional assays.
Materials and Methods
Patients.
All patients were recruited from the University of Michigan Scleroderma Program and met the American College of Rheumatology/European League Against Rheumatism criteria for the classification of SSc (20). The demographics and clinical characteristics of enrolled patients are summarized in SI Appendix, Table S1. Two 4-mm punch biopsies from the distal forearm of healthy volunteers and diffuse or limited cutaneous SSc patients were obtained for fibroblast and EC isolation. All subjects included in this study signed a written informed consent. All procedures in this study were reviewed and approved by the Institutional Review Board of the University of Michigan.
Cell Culture.
Dermal fibroblasts and ECs were isolated as described (3, 21–23). After digestion, ECs were magnetically labeled with anti-CD31 antibodies (CD31 MicroBead Kit) and added to LS Columns (Miltenyi Biotech) attached to MidiMACS Separator. Negatively selected cells, which were fibroblasts, were cultured in RPMI 1640 with 10% FBS and antibiotics. The positively selected CD31+ ECs were grown in EBM-2 media with growth factors (Lonza). Cells between passages 3 and 6 were used in the experiments.
mRNA Extraction and qRT-PCR.
Total RNA was isolated using Direct-zol RNA MiniPrep Kit (Zymo Research), followed by preparation of cDNA using the Verso cDNA synthesis kit (Thermo Scientific). Primers along with Power SYBR Green PCR master mix (Applied Biosystems) were used for qPCR run by the ViiA 7 Real-Time PCR System. Primer sequences are as follows: VEGF FW: ATGAACTTTCTGCTGTCTTGGGT; RV: TGGCCTTGGTGAGGTTTGATCC; TGFB1 FW: TTGCTTCAGCTCCACAGAGA; RV: GTTGGACAACTGCTCCACCT; DNMT1 FW: CGACTACATCAAAGGCAGCAACCTG; RV: TGGAGTGGACTTGTGGGTGTTCTC; DNMT3A: FW: CGAGTCCAACCCTGTGATGATTG; RV: CGTGGTCTTTGCCCTGCTTTATG; NOTCH1 FW: TCCACCAGTTTGAATGGTCA; RV: AGCTCATCATCTGGGACAGG; NOTCH4 FW: AACTCCTCCCCAGGAATCTG; RV: CCTCCATCCAGCAGAGGTT; JAG2 FW: GGTCGTACTTGCACTCACAATACC; RV: GTAGCAAGGCAGAGGGTTGC; NUMB FW: AGCGCAAGCAGAAGCGGGAG; RV: CGGCGTGGGATGGCATGAGG; FBXW7 FW: GGCGCCGCGGCTCTTTTCTA; RV: GCTGCCCACAGAGAGCAGTTCC; ACTIN FW: GTCAGGCAGCTCGTAGCTCT; RV: GCCATGTACGTTGCTATCCA. The other primers were KiCqStart® SYBR® Green Primers from Sigma or QuantiTect Primer Assays from Qiagen.
Western Blots.
Equal amounts of protein from lysed cells were subjected to SDS/PAGE and Western blotting. EZH2 and H3K27me3 were detected using anti-human EZH2 and H3K27me3 antibodies (Cell Signaling) while β-actin and histone H3 were used as loading controls (anti-β-actin antibodies and anti-histone H3 antibodies were from Sigma Aldrich and Cell Signaling, respectively). Band quantification was performed using GelQuant.NET (BiochemLab Solutions) and ImageJ (24).
Manipulation of EZH2 Expression in Fibroblasts and ECs.
DZNep (Cayman Chemicals), an EZH2 inhibitor, was dosed at 0.2–5 μM for fibroblasts and 5 μM for ECs for 48 h, with PBS as a negative control. When indicated, another EZH2 inhibitor, GSK126 (Cayman Chemicals), was dosed at 0.5–10 μM for 72 h in SSc dermal fibroblasts. Cell viability was checked using Trypan blue and was not affected by either EZH2 inhibitor with the dosages used. To evaluate the effect of EZH2 on angiogenesis in ECs, we used 75 nM EZH2 siRNA (Santa Cruz Biotechnology) to transfect ECs for 48 h. Overexpression of EZH2 in ECs was achieved by transfecting 0.33 μg of EZH2 (control vector pCMV6-XL5; Origene) using Lipofectamine 2000 (Invitrogen) in T12.5 flasks. After 5 h, culture media were changed to EGM supplemented with bovine brain extract (Lonza). Matrigel tube formation assay was performed 24 h after transfection. Overexpression of EZH2 was also done in fibroblasts, using 0.1 μg of either control or EZH2 vector in a 12-well plate for 24–72 h. Successful transfection was confirmed by qPCR.
Cell Migration Assay.
To evaluate the effect of EZH2 on cell migration, we performed cell migration assays using SSc fibroblasts treated with DZNep, or normal fibroblasts with EZH2 overexpressed in a 12-well plate. Cells were grown to confluence, and a wound gap was created by a scratch instrument. The media was replaced with RPMI 1640 with 0.1% FBS, and pictures were taken using EVOS XL Core Cell Imaging System (Life Technologies) at 0 h and 48 h after scratch. Quantification of the gap difference was done using ImageJ (24). In a separate set of experiments, SSc dermal fibroblasts were plated in 96 Well Image Lock Microplate and treated with another EZH2 inhibitor GSK126 (0.5–10 µM). Wounds were created using the WoundMaker. The plate was then placed in IncuCyte to acquire data and images. Quantification was done using the Analysis module in the IncuCyte software.
Gel Contraction Assay.
To examine the effect of EZH2 inhibition on gel contraction, we followed the procedure as described (25). SSc dermal fibroblasts were treated with GSK126 (0.5–10 µM) for 72 h before suspension in culture media at 2 × 106 cells/mL. Cells were then mixed with collagen solution from the Cell Contraction Assay kit (Cell Biolabs) and plated in a 24-well plate. Culture media was added after the collagen polymerized. After 1 d, the collagen matrix was released, and the size of the collagen gel was measured and analyzed after 5 h using ImageJ (24).
Matrigel Tube Formation Assay.
ECs were plated in eight-well Lab-Tek chambers coated with growth factor reduced Matrigel (BD Biosciences). The cells were fixed and stained after 8-h incubation. Pictures of each well were taken using EVOS XL Core Cell Imaging System (Life Technologies). Quantitation of the tubes formed by ECs was performed using the Angiogenesis Analyzer function in ImageJ (24).
Bleomycin Skin Fibrosis Model.
A bleomycin-induced skin fibrosis model was used similar to what was described (26, 27). Fifteen-week-old C57BL/6 mice (Jackson Laboratory) were preconditioned on supplemental DietGel 76A (ClearH2O) for 2 wk before starting the experiment. Skin fibrosis was induced by intracutaneous injection of 100 μL of bleomycin (0.5 mg/mL) in PBS, every day for 2 wk in a defined area (∼1 cm2) on the upper back. Intracutaneous injection of 100 μL of PBS was used as control. One group of mice received injections of PBS, and the other two were challenged with bleomycin. Daily oral administration of DZNep (2 mg/kg in 20% DMSO/50% PEG 400/30% PBS) was initiated together with the first challenge of bleomycin and continued for 2 wk. Vehicle control consisting of 20% DMSO/50% PEG 400/30% PBS was used. Oral gavage was performed by the Unit for Laboratory Animal Medicine’s In-Vivo Animal Core. In a separate study, daily i.p. administration of GSK126 (0.5 mg/kg or 5 mg/kg in 20% DMSO/50% PEG 400/30% PBS) or vehicle control (20% DMSO/50% PEG 400/30% PBS) was used in the bleomycin fibrosis model described above. Mice were killed by CO2 inhalation, and the skin from the defined area was harvested at the end of the study. A portion of the skin was fixed in neutral buffered formalin (10%), washed in 70% ethanol, and paraffin embedded. Another portion of the skin was snap frozen for hydroxyproline measurement. All animal protocols used in this study were approved by the Institutional Animal Care & Use Committee at the University of Michigan.
Histological Analysis.
Fixed skin was paraffin embedded and sectioned at the University of Michigan Comprehensive Cancer Center Histology Core. Skin sections were stained with Masson’s trichrome (Sigma-Aldrich). Stained sections were analyzed with an Olympus BX51\DP72 microscope. Dermal thickness was determined by measuring the maximal distance between the epidermal-dermal junction and the dermal-s.c. fat junction. Three measurements were averaged from each skin section. Measurements were performed using the measurement tool in the cellSens imaging software package (Olympus). To examine the effect of EZH2 inhibition on H3K27me3, immunohistochemistry was performed using anti-H3K27me3 (Cell Signaling) at a 1:200 dilution.
Hydroxyproline Assay.
Skin sections were weighed and hydrolyzed in 6 M HCl at 120 °C for 3 h. Hydrolyzed skin supernatant and hydroxyproline standards (Sigma-Aldrich) were transferred to a microplate and dried at 60 °C. Samples and standards were oxidized with Chloramine T oxidation buffer for 5 min at room temperature. 4-(Dimethylamino) benzaldehyde was added to the wells and incubated at 60 °C until the standards were well defined. Absorbance was measured at 560 nm using a Synergy HT microplate reader (BioTek Instruments). Hydroxyproline values were normalized to tissue weight.
DNA Methylation and Bioinformatics Analysis.
Genomic DNA from five patients with diffuse cutaneous SSc was isolated from PBS- and DZNep (5 μM)-treated dermal fibroblasts, and subsequently, bisulfite converted using an EZ DNA Methylation kit (Zymo Research). Genome-wide DNA methylation was evaluated using the Illumina Infinium MethylationEPIC BeadChip Array, which allows measuring DNA methylation levels in over 850,000 CpG sites across the genome, and covers 27,364 annotated genes. The array also covers enhancers, regions of open chromatin identified by ENCODE project, DNase hypersensitive sites, and miRNA promoter regions. Analysis was performed using the Illumina GenomeStudio platform as described (28). The average level of DNA methylation (β) on each CpG site was compared between PBS- and DZNep-treated samples. Differentially methylated CpG sites were defined as those with a differential methylation score ≥|22| (equivalent to P value of less than 0.01 after adjusting for multiple testing) and a mean methylation difference greater than 10% between the two groups. Differentially methylated genes were analyzed for Gene Ontology (GO), network, and pathway enrichments using the Database for Annotation, Visualization, and Integrated Discovery (DAVID V.6.7) (29, 30).
Gene Knockdown Conditions in Fibroblasts and ECs.
To evaluate the involvement of LRRC16A in EZH2-mediated cell migration, we knocked down LRRC16A in EZH2-overexpressing fibroblasts. The knockdown condition for LRRC16A was optimized by transfecting fibroblasts with 100 nM control and LRRC16A siRNA (Dharmacon) for 48 h (70% knockdown of LRRC16A). EZH2 was overexpressed in normal fibroblasts using 0.1 μg of either control or EZH2 vectors. On the second day of transfection, LRRC16A or control siRNA was added to the cells to knock down LRRC16A. After 72 h of EZH2 overexpression (hence 48 h of LRRC16A knockdown), the ability of the cells to migrate was evaluated by a scratch-wound assay.
To investigate whether genes in the Notch signaling pathway are involved in EZH2-mediated tube formation in ECs, we knocked down JAG1, JAG2, or DLL4 in ECs, as these genes were shown to be involved in angiogenesis (31–33). The knockdown condition for JAG1, JAG2, and DLL4 siRNA (all from Dharmacon) was 200 nM, 25 nM, and 25 nM for 48 h, respectively. SSc ECs were simultaneously treated with 5 μM DZNep and control or siRNA of the gene of interest for 48 h before the Matrigel assay was performed.
Statistics.
Results were expressed as mean ± SD. To determine the differences between groups, Mann–Whitney U test, unpaired t test, or Kruskal–Wallis test were performed using GraphPad Prism version 6 (GraphPad Software, Inc). P values of less than 0.05 were considered statistically significant.
Results
EZH2 Expression Was Elevated in both SSc Fibroblasts and ECs.
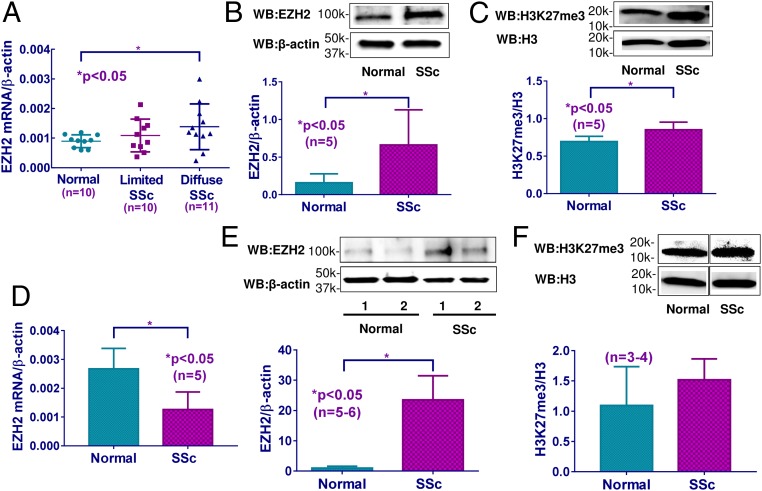
Increased EZH2 mRNA levels were detected in fibroblasts isolated from diffuse cutaneous SSc patients, but not in fibroblasts isolated from healthy controls or limited cutaneous SSc patients (Fig. 1A). The level of EZH2 did not show significant correlation with the modified Rodnan skin score (SI Appendix, Fig. S1). The protein levels of EZH2 in diffuse cutaneous SSc fibroblasts were also significantly elevated compared with healthy controls (Fig. 1B). Since EZH2 catalyzes trimethylation of H3K27, we also measured H3K27me3 levels in these cells. As shown in Fig. 1C, significant increase in H3K27me3 was observed in diffuse cutaneous SSc fibroblasts compared with healthy controls, consistent with increased EZH2 levels observed in these cells. The expression of EZH2 in ECs isolated from skin biopsies was also examined. We observed a significant reduction of EZH2 mRNA in SSc ECs compared with controls (Fig. 1D); however, at the protein level, EZH2 was significantly elevated in SSc ECs (Fig. 1E). The reasons for this discrepancy between the mRNA and protein levels is not clear. We speculate this could be due to posttranscriptional and/or translational modifications, or different RNA and protein degradation rates. Slight elevation of H3K27me3 in SSc ECs was also observed, however, this did not reach statistical significance (Fig. 1F).
Fig. 1.
The expression of EZH2 and H3K27me3 in SSc fibroblasts and ECs. (A) EZH2 mRNA levels were significantly elevated in fibroblasts from diffuse cutaneous SSc patients compared with healthy fibroblasts, but not in limited cutaneous SSc fibroblasts. EZH2 protein levels (B) and H3K27me3 levels (C) were significantly increased in diffuse cutaneous SSc fibroblasts compared with healthy controls. (D) EZH2 mRNA levels were significantly lower in diffuse cutaneous SSc ECs compared with healthy ECs. (E) EZH2 protein levels were significantly higher in diffuse cutaneous SSc ECs compared with healthy ECs. (F) The expression of H3K27me3 was not significantly different between diffuse cutaneous SSc ECs and healthy controls. Results are expressed as mean ± SD. The Mann–Whitney U test was performed, and *P < 0.05 was considered significant.
Inhibition of EZH2 in SSc Dermal Fibroblasts Affected Genes Involved in both Fibrosis and Angiogenesis.
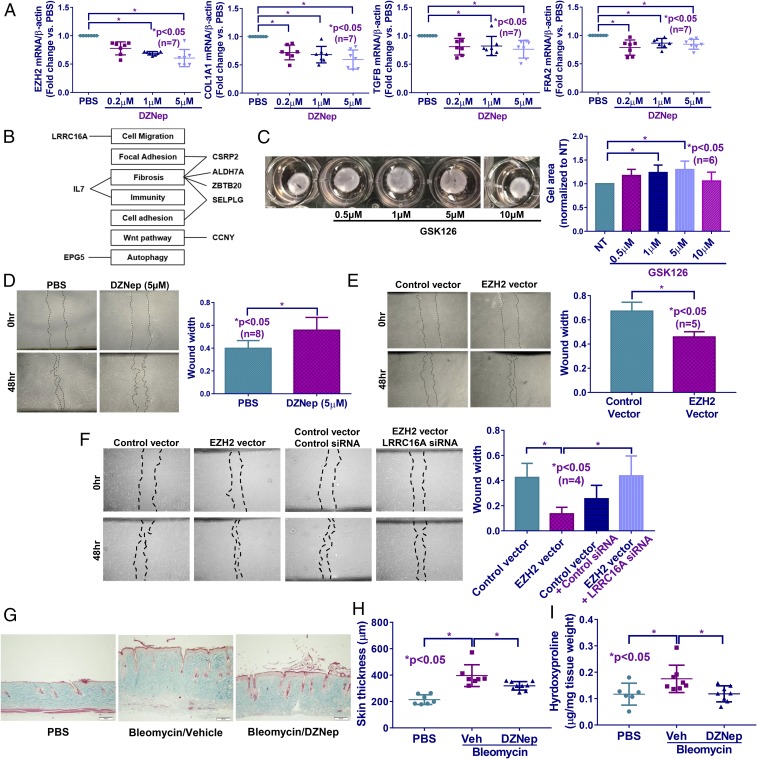
To investigate whether EZH2 plays a role in SSc fibrosis, SSc dermal fibroblasts were treated with different doses of DZNep, which is an EZH2 inhibitor. As expected, DZNep dose-dependently decreased EZH2 expression (Fig. 2A and SI Appendix, Fig. S2). In addition, DZNep also dose-dependently decreased H3K27me3, COL1A1, TGFB, as well as the profibrotic transcription factor FRA2 (Fig. 2A and SI Appendix, Fig. S2). Other fibrosis-related genes were also examined; DZNep (5 μM) did not alter the myofibroblast marker ACTA2 or the antifibrotic gene PPARG, while it decreased the expression of the antifibrotic transcription factor FLI1 significantly (Table 1).
Fig. 2.
DZNep treatment in diffuse cutaneous SSc fibroblasts and animal model led to an antifibrotic phenotype. (A) Inhibiting EZH2 by DZNep in SSc dermal fibroblasts resulted in a dose-dependent reduction in COL1A1, TGFB, and FRA2. (B) Selected hypomethylated and hypermethylated genes affected by EZH2 inhibition that were implicated in fibrosis or fibrosis-related processes. (C) Inhibition of EZH2 by GSK126 dose-dependently relaxed gel contraction in SSc fibroblasts. (D) Scratch-wound assay showing reduced wound closure (cell migration) at 48 h in SSc fibroblasts treated with DZNep (5 μM) compared with PBS control. (E) EZH2 overexpression in fibroblasts from healthy individuals increased cell migration at 48 h. (F) Increased cell migration by EZH2 overexpression in normal fibroblasts was reversed by knocking down LRRC16A simultaneously, implicating LRRC16A in EZH2-mediated cell migration. (G and H) Skin fibrosis was induced in mice by daily bleomycin injection. Mice treated with DZNep showed a significant reduction in dermal thickness compared with vehicle-treated mice. (I) Hydroxyproline content was significantly reduced in DZNep-treated mice. Results are expressed as mean ± SD. Kruskal–Wallis test or Mann–Whitney U test was performed, and *P < 0.05 was considered significant. (Scale bars: 100 μm.)
Table 1.
The expression of genes after EZH2 inhibition by DZNep (5 μM, 48 h) in diffuse cutaneous SSc fibroblasts
| Gene | Expression fold change vs. control (n = 7 patient pairs) | P value |
| ACTA2 | 0.88 ± 0.21 | 0.18 |
| PPARG | 1.09 ± 0.40 | 0.58 |
| FLI1 | 0.72 ± 0.25 | 0.01 |
| DNMT1 | 0.83 ± 0.16 | 0.01 |
| DNMT3A | 0.70 ± 0.25 | 0.01 |
| MECP2 | 0.78 ± 0.22 | 0.02 |
| ALDH7A1 | 0.91 ± 0.27 | 0.36 |
| CCNY | 0.69 ± 0.14 | 0.002 |
| CSRP2 | 0.73 ± 0.25 | 0.02 |
| EPG5 | 0.80 ± 0.19 | 0.048 |
| ZBTB20 | 0.70 ± 0.31 | 0.048 |
| IL7 | 0.66 ± 0.11 | 0.001 |
| LRRC16A | 0.79 ± 0.16 | 0.005 |
| SELPLG | 1.70 ± 0.57 | 0.007 |
Data are expressed as fold change vs. PBS-treated cells (mean ± SD).
EZH2 Inhibition in SSc Fibroblasts Led to Genome-Wide Changes in DNA Methylation.
Although EZH2 preferentially targets histones, it has been shown to serve as a recruitment platform for DNA methyltransferases (DNMTs) (34). To examine how EZH2 affects DNA methylation, fibroblasts from diffuse cutaneous SSc patients were treated with 5 μM DZNep or PBS (n = 5 patient pairs) and the isolated DNA was subjected to the Illumina Infinium MethylationEPIC BeadChip Array. A total of 37 differentially methylated CpG sites were observed (SI Appendix, Table S2). Of these, 17 (11 genes) were hypomethylated, and the remaining 20 sites (13 genes) were hypermethylated. After GO analysis and literature mining, we selected several genes that are relevant to fibrosis or fibrosis-related processes such as cell migration and adhesion for further analysis (Fig. 2B). The mRNA expression of the genes in Fig. 2B was measured by qPCR. Only SELPLG, which was hypermethylated after DZNep treatment, was significantly elevated compared with PBS controls in SSc fibroblasts (Table 1). While the hypomethylated ALDH7A1 was unaltered, the rest of the genes, including hypomethylated CCNY and EPG5, as well as hypermethylated CSRP2, ZBTB20, IL7, and LRRC16A, decreased significantly after DZNep treatment in SSc fibroblasts (Table 1).
Since EZH2 affects DNA methylation via DNMTs and methyl CpG binding protein 2 (MECP2) (17, 19, 35), the expression of DNMT1, DNMT3A, and MECP2 was examined in SSc fibroblasts; all three decreased after DZNep treatment (Table 1).
We further confirmed the expression changes of the aforementioned genes in EZH2-overexpressing normal fibroblasts (Table 2). Successful EZH2 overexpression was confirmed by qPCR (Table 2). Under this condition, significant elevation of COL1A1, TGFB (at 72 h after transfection), FRA2, and FLI1 was observed (Table 2). Interestingly, significant reduction of ACTA2 as well as elevation of PPARG was observed when EZH2 was overexpressed in normal dermal fibroblasts (Table 2). Overexpression of EZH2 also led to significant increase in DNMT1, DNMT3A, and MECP2.
Table 2.
The expression of genes after EZH2 overexpression in normal fibroblasts
| Gene | Expression fold change vs. control vector after 24-h transfection (n = 7 patient pairs) | P value |
| EZH2 | 2,468 ± 1,397 | 0.001 |
| COL1A1 | 1.84 ± 0.66 | 0.01 |
| ACTA2 | 0.62 ± 0.20 | 0.001 |
| TGFB | 1.15 ± 0.31 (1.67 ± 0.71) | 0.21 (0.04) |
| FRA2 | 1.52 ± 0.53 | 0.02 |
| PPARG | 1.37 ± 0.28 | 0.005 |
| FLI1 | 1.49 ± 0.36 | 0.004 |
| DNMT1 | 1.18 ± 0.23 (2.75 ± 1.61) | 0.06 (0.04) |
| DNMT3A | 1.38 ± 0.35 | 0.01 |
| MECP2 | 1.93 ± 0.75 | 0.01 |
| IL7 | 8.03 ± 4.95 | 0.003 |
| LRRC16A | 1.26 ± 0.15 | 0.001 |
| SELPLG | 1.25 ± 0.24 | 0.02 |
Data are expressed as fold change vs. control-transfected cells (mean ± SD). Values between parentheses represent expression changes at 72 h after transfection.
Alteration of EZH2 Expression in Dermal Fibroblasts Led to Changes in Gel Contraction and Cell Migration.
SSc fibroblasts possess increased migratory and contractility abilities compared with normal fibroblasts, indicative of an activated phenotype (36). We first performed a gel contraction assay to examine the effect of EZH2 on myofibroblast-mediated contractility. By treating SSc dermal fibroblasts with another EZH2 inhibitor, GSK126, we found that GSK126 dose-dependently inhibited gel contraction (Fig. 2C). Interestingly, an effect of GSK126 on gel contraction is not observed at the highest dose used, which is consistent with a U-shaped dose–response curve and might suggest triggering compensatory mechanisms in SSc fibroblasts to counteract the effects of GSK126 at higher doses. To examine whether EZH2 affects myofibroblast function, a scratch-wound assay was performed. As shown in Fig. 2D, treating SSc dermal fibroblasts with DZNep (5 μM) resulted in a decrease in cell migration at 48 h after scratch, as indicated by the significant increase in wound width. To further confirm our results, a scratch-wound assay was performed using EZH2-overexpressing normal fibroblasts. We showed that overexpression of EZH2 resulted in an increase in wound closure, as indicated by the smaller wound width after EZH2 was overexpressed (Fig. 2E). We also examined the effect of GSK126 on wound healing. As shown in SI Appendix, Fig. S3, at the highest dose 10 µM, GSK126 significantly inhibited wound closure in SSc fibroblasts.
We further tested the involvement of LRRC16A, which was hypermethylated and down-regulated after DZNep treatment in SSc fibroblasts, in cell migration (Fig. 2B, Table 1, and SI Appendix, Table S1). Since this gene was overexpressed in EZH2-overexpressing normal fibroblasts (Table 2), we knocked it down and performed a scratch-wound assay. EZH2-overexpressing cells with LRRC16A knocked down migrated significantly slower compared with EZH2 overexpression alone (Fig. 2F), suggesting that LRRC16A is involved in EZH2-mediated cell migration.
These data suggest that elevated levels of EZH2 result in a profibrotic phenotype in SSc dermal fibroblasts, as indicated by the effect of EZH2 on COL1A1, TGFB, and FRA2 and further supported by its effect on cell migration and gel contraction.
Inhibition of EZH2 Prevented Bleomycin-Induced Skin Fibrosis in Mice.
To further examine the role of EZH2 in fibrosis, we tested the effect of EZH2 inhibitors in the bleomycin-induced skin fibrosis mouse model. Daily dosing of DZNep or GSK126 prevented bleomycin-induced skin fibrosis, as shown by reduction of skin thickness, hydroxyproline content, and H3K27me3 levels in the skin (Fig. 2 G–I and SI Appendix, Figs. S4 and S5).
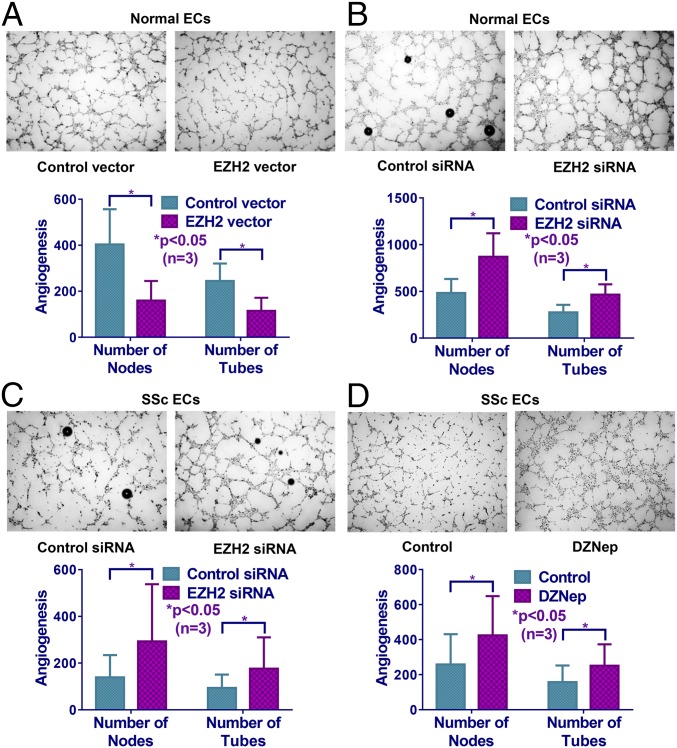
Modulation of EZH2 Expression Affected Tube Formation in ECs.
Because EZH2 affects the regulation of genes involved in EC function, in particular genes associated with cell adhesion and migration (7, 11, 15), we performed Matrigel tube formation assays after EZH2 overexpression or siRNA knockdown in normal dermal ECs (Fig. 3 A and B). Overexpression of EZH2 (18-fold increase in EZH2 mRNA) inhibited angiogenesis, represented by the number of nodes or tubes, while knockdown of EZH2 (88% decrease in EZH2 mRNA) resulted in an increase in angiogenesis (Fig. 3 A and B). Since we showed that EZH2 was overexpressed in SSc ECs (Fig. 1), we knocked down EZH2 in these cells and observed a significant increase in angiogenesis compared with cells transfected with control siRNA (Fig. 3C). Similarly, treating SSc ECs with DZNep, which led to an ∼50% reduction in EZH2 expression in SSc ECs, significantly enhanced angiogenesis (Fig. 3D). These data suggest that EZH2, possibly by repressing proangiogenic genes or activating antiangiogenic genes, inhibits angiogenesis in SSc ECs.
Fig. 3.
Manipulation of EZH2 expression affected EC angiogenesis in vitro. (A) Overexpression of EZH2 in normal ECs led to a decreased number of nodes and tubes in Matrigel tube formation assay. Knockdown of EZH2 in normal ECs (B) and diffuse cutaneous SSc ECs (C) resulted in increased angiogenesis. (D) Inhibition of EZH2 by DZNep similarly increased angiogenesis in diffuse cutaneous SSc ECs compared with cells treated with PBS control. Results are expressed as mean ± SD. Mann–Whitney U test was performed, and *P < 0.05 was considered significant.
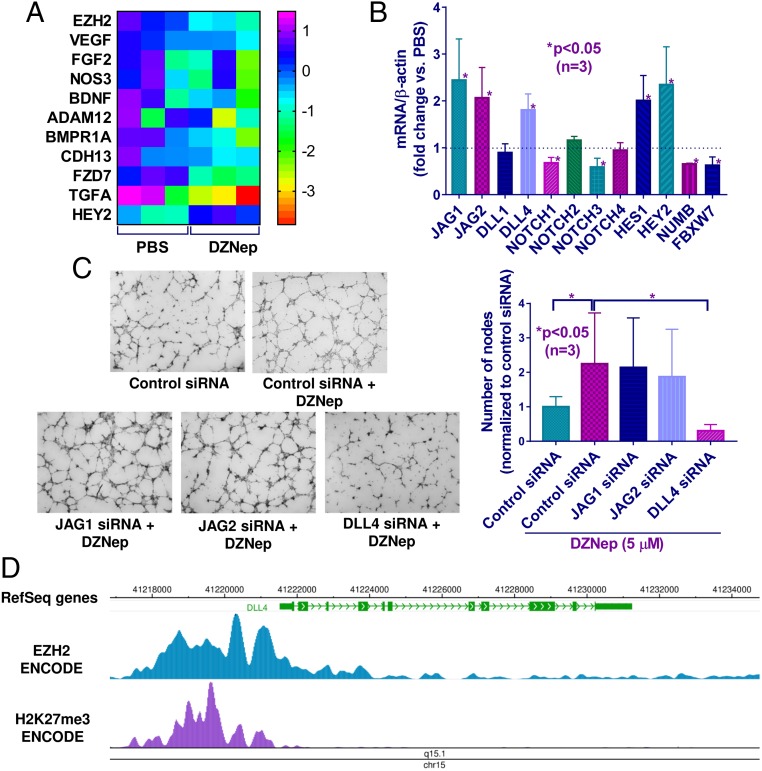
EZH2 Inhibits Angiogenesis in SSc by Repressing the Notch Signaling Pathway.
To identify the mechanism involved in EZH2-mediated inhibition of angiogenesis in SSc ECs, we compiled a list of angiogenesis-associated genes affected by EZH2 in ECs from several publications (7, 15, 37) after thorough literature search in PubMed using the keywords “endothelial cells,” “angiogenesis,” and “EZH2.” We then measured the expression of these genes in SSc ECs after DZNep or PBS treatment. Of all of the proangiogenic genes examined, only HEY2, a Notch target gene, was significantly up-regulated after EZH2 inhibition (Fig. 4A). This prompted us to examine whether other members of the Notch pathway were affected by DZNep. Notch ligands JAG1, JAG2, DLL4, and Notch receptor NOTCH2, as well as Notch target gene HES1, were all up-regulated after EZH2 inhibition (Fig. 4B). In contrast, NOTCH1 and NOTCH3, as well as NUMB and FBXW7, which are Notch signaling inhibitors, were significantly down-regulated (Fig. 4B). These results suggest that EZH2 acts as a repressor for certain Notch genes and activator for others.
Fig. 4.
EZH2 affected diffuse cutaneous SSc EC angiogenesis through the Notch signaling pathway. (A) Heat map of angiogenesis-associated genes in SSc ECs after PBS or DZNep treatment. Only HEY2, a Notch target gene, was significantly up-regulated after DZNep treatment. (B) Relative mRNA expression of genes involved in Notch signaling after EZH2 inhibition by DZNep (5 μM, 48 h) in SSc ECs. Data are expressed as fold change versus PBS-treated cells. (C) Increased angiogenesis mediated by DZNep in SSc ECs was mediated by increased expression of Notch ligand DLL4, as DLL4 knockdown led to significant decrease in angiogenesis when cells were treated simultaneously with DZNep. (D) Genome browser tracks of the DLL4 locus along with ChIP-seq data for EZH2 and H3K27me3 in human umbilical endothelial cells generated by the ENCODE project. Results are expressed as mean ± SD. Unpaired t test or Kruskal–Wallis test were performed, and *P < 0.05 was considered significant.
To gain more insight into Notch signaling in SSc ECs, we measured mRNA expression levels of Notch-related genes in normal and SSc ECs. A significant increase in JAG2 and NUMB was observed, while DLL4, HES1, and HEY2 were decreased in SSc ECs compared with normal ECs (SI Appendix, Fig. S6). The significant increase of Notch inhibitor NUMB and decrease of Notch-target genes HES1 and HEY2 suggest that the Notch signaling pathway is impaired in SSc ECs.
We next examined whether DZNep affects the mRNA expression of Notch related genes in SSc fibroblasts, and did not observe that DZNep significantly affected the expression of Notch ligands and receptors (SI Appendix, Fig. S7). Although down-regulation of NOTCH3 and up-regulation of Notch-regulated gene HES1 were observed after DZNep treatment in SSc fibroblasts, the magnitude was smaller compared with that observed in SSc ECs.
Since Notch pathway, specifically the Notch ligands, affects angiogenesis (31–33), we examined whether the up-regulated Notch ligands JAG1, JAG2, and DLL4 were involved in EZH2-mediated angiogenesis in SSc ECs. As shown in Fig. 4C, the increase in tube formation on Matrigel gel after DZNep treatment in SSc ECs was inhibited by DLL4 knockdown, while no change was observed by JAG1 or JAG2 siRNAs. This suggests that EZH2 inhibits SSc EC tube formation through repressing DLL4, and that inhibition of EZH2 increases tube formation through up-regulation of DLL4. Indeed, ChIP‐seq data demonstrate enrichment of EZH2 binding and H3K27me3 marks at the promoter region of DLL4 in ECs (Fig. 4D) (38), supporting a role for EZH2 and EZH2-mediated trimethylation of H3K27 in regulating DLL4 in ECs.
Discussion
In this study, we provided evidence that overexpression of EZH2 plays a critical role in SSc pathogenesis, specifically in fibrosis and angiogenesis. EZH2 inhibitors appear to be antifibrotic both in vitro and in vivo. Inhibiting EZH2 decreases the expression of profibrotic genes including COL1A1, FRA2, and TGFB in SSc dermal fibroblasts and decreases migratory abilities of these cells. Through examining genome-wide changes in DNA methylation after DZNep treatment in SSc fibroblasts, we identified LRRC16A as a key mediator for the inhibitory effect of DZNep on cell migration. In addition to its effect in fibrosis, we also showed that elevated levels of EZH2 in SSc ECs inhibit angiogenesis. DZNep treatment improves tube formation in SSc ECs by derepressing key genes of the Notch pathway, specifically through the Notch ligand DLL4.
The role of EZH2 in SSc has been examined in a previous study (39). Krämer et al. showed that DZNep exacerbated fibrosis in SSc dermal fibroblasts through up-regulation of profibrotic FRA2. However, neither EZH2 nor H3K27me3 levels were determined after DZNep treatment, therefore whether EZH2 was inhibited is not known. Although the effect of DZNep in the dermal bleomycin model has also been previously evaluated (39) and did not show a favorable outcome, the 2 mg/kg twice a week dosing regimen adopted might not have been sufficient to achieve EZH2 inhibition, due to the unfavorable pharmacokinetic properties of DZNep (half-life in mice is 0.9 h) (40). Therefore, we chose daily administration of DZNep to achieve the optimal therapeutic effect. Indeed, under this therapeutic regimen, DZNep prevented bleomycin-induced skin fibrosis. Similarly, in the bleomycin-induced pulmonary fibrosis model and the unilateral ureteral obstruction renal fibrosis model, daily DZNep administration ameliorated tissue fibrosis effectively (8, 16). In our study, we confirmed the profibrotic effect of EZH2 in SSc fibrosis using two EZH2 inhibitors, DZNep and GSK126.
By examining the effect of DZNep on H3K27me3 expression in SSc fibroblasts, we showed that DZNep inhibited both the expression and the methyltransferase activity of EZH2. DZNep dose-dependently reduced both EZH2 and H3K27me3 in SSc fibroblasts. These results suggest that the profibrotic effect of EZH2 is dependent on both its expression and enzymatic activity. EZH2 can act as a transcription activator (10) as well as a repressor. In fact, the majority of the genes we studied in SSc fibroblast were down-regulated by DZNep, suggesting that EZH2-mediated gene activation might play a more significant role in mediating its profibrotic effects.
To regulate gene expression, EZH2 cooperates with other epigenetic silencing machineries, including DNMTs (34) and histone deacetylases (41). In this study, we showed that EZH2 also affects the expression of DNMT1, DNMT3A, and MECP2, ultimately resulting in genome-wide changes in DNA methylation after DZNep treatment in SSc fibroblasts. Since we showed that DZNep inhibits the invasiveness of SSc fibroblasts in the scratch-wound assay, we were intrigued to see that LRRC16A which is involved in cell migration (42, 43), was hypermethylated and down-regulated following DZNep treatment in SSc fibroblasts. Recent data suggest that loss of EZH2 can lead to hypermethylation in specific loci, suggesting that alternative repressive mechanisms trigger increased DNA methylation during the loss of EZH2, although the exact mechanism behind this observation is still not clear (44). We speculate that EZH2 might protect against inappropriate DNA methylation in specific loci in physiologic conditions, and that increased EZH2 in SSc might impair this DNA methylation balance in the LRRC16A locus.
LRRC16A encodes a cell membrane-cytoskeleton protein that is localized to lamellipodia and macropinosomes and regulates actin polymerization via affecting Rac1 and Trio (42, 43). Knocking down LRRC16A significantly reduced migration of EZH2-overexpressing fibroblasts, providing a functional role of LRRC16A in the profibrotic properties of EZH2 in SSc.
EZH2 affects angiogenesis by controlling gene expression in ECs. In cancer, VEGF promotes angiogenesis by inducing EZH2, which, in turn, silences vasoinhibin-1, an antiangiogenic mediator (45). In addition, migratory properties of human brain microvascular ECs in the tumor microenvironment are altered by miR101-mediated inhibition of EZH2 (12). We showed that EZH2 expression was significantly higher in SSc ECs, and that increased expression of EZH2 impairs angiogenesis in SSc ECs by modulating genes associated with this process. Through overexpression and knockdown of EZH2 we were able to modulate EC tube formation. Furthermore, we showed that DLL4 mediates DZNep-promoted angiogenesis in SSc ECs, suggesting that up-regulated EZH2 represses proangiogenic DLL4, leading to inhibition of angiogenesis in SSc ECs. The unique property of EZH2 in promoting tumor angiogenesis while inhibiting SSc angiogenesis suggests that EZH2 inhibitors have the therapeutic advantage to be used in cancer and autoimmune fibrosing diseases.
We showed that DZNep activates Notch signaling in SSc ECs by up-regulation of Notch ligands, Notch receptors, and Notch-regulated genes. This activation is possibly mediated by down-regulation of Notch inhibitors NUMB and FBXW7. Our results agree with other studies that point to a regulatory role of EZH2 in Notch signaling (46, 47). Interestingly, the expression of the Notch ligand DLL4 was significantly reduced in SSc ECs, and increasing DLL4 by inhibiting EZH2 restored angiogenesis in these cells. This suggests that DLL4 plays a significant role in promoting angiogenesis.
In conclusion, we identified EZH2 as a key epigenetic regulator that promotes fibrosis and inhibits angiogenesis in SSc. We identified LRRC16A as a mediator for EZH2-induced cell migration in SSc fibroblasts. In addition, we revealed that EZH2-Notch axis controls angiogenesis through the Notch ligand DLL4 in SSc. These findings suggest a role for utilizing EZH2 inhibitors to treat SSc.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant T32AR007080 (to P.-S.T.). P.-S.T. is also supported by the Scleroderma Foundation and the Arthritis National Research Foundation. A.H.S. is supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health Grants R01AI097134 and U19AI110502, National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health Grant R01AR070148, and the Lupus Research Alliance. D.K. is supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant K24AR063120 and the National Institute of Allergy and Infectious Diseases Grant UM1AI110557. D.A.F. is supported by the National Institute of Allergy and Infectious Diseases Grant UM1AI110557. The University of Michigan Comprehensive Cancer Center Histology/Immunohistochemistry Core (performed the embedding and cutting of skin samples from the bleomycin study) is supported by the National Cancer Institute of the National Institutes of Health under Award P30CA046592.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813006116/-/DCSupplemental.
References
- 1.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 2.Dees C, et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis. 2013;73:1232–1239. doi: 10.1136/annrheumdis-2012-203194. [DOI] [PubMed] [Google Scholar]
- 3.Tsou PS, et al. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis Rheumatol. 2016;68:2975–2985. doi: 10.1002/art.39828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YY, et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+ T cells of patients with systemic sclerosis. Br J Dermatol. 2014;171:39–47. doi: 10.1111/bjd.12913. [DOI] [PubMed] [Google Scholar]
- 5.Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis. 2015;74:1612–1620. doi: 10.1136/annrheumdis-2014-205303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsou PS, Sawalha AH. Unfolding the pathogenesis of scleroderma through genomics and epigenomics. J Autoimmun. 2017;83:73–94. doi: 10.1016/j.jaut.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreger H, et al. Epigenetic regulation of cell adhesion and communication by enhancer of zeste homolog 2 in human endothelial cells. Hypertension. 2012;60:1176–1183. doi: 10.1161/HYPERTENSIONAHA.112.191098. [DOI] [PubMed] [Google Scholar]
- 8.Xiao X, et al. EZH2 enhances the differentiation of fibroblasts into myofibroblasts in idiopathic pulmonary fibrosis. Physiol Rep. 2016;4:e12915. doi: 10.14814/phy2.12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tumes DJ, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity. 2013;39:819–832. doi: 10.1016/j.immuni.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Yan J, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood. 2016;128:948–958. doi: 10.1182/blood-2016-01-690701. [DOI] [PubMed] [Google Scholar]
- 11.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis. 2016;19:9–24. doi: 10.1007/s10456-015-9485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smits M, et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PLoS One. 2011;6:e16282. doi: 10.1371/journal.pone.0016282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smits M, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He M, et al. Cancer angiogenesis induced by Kaposi sarcoma-associated herpesvirus is mediated by EZH2. Cancer Res. 2012;72:3582–3592. doi: 10.1158/0008-5472.CAN-11-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitić T, et al. EZH2 modulates angiogenesis in vitro and in a mouse model of limb ischemia. Mol Ther. 2015;23:32–42. doi: 10.1038/mt.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, et al. Enhancer of zeste homolog 2 inhibition attenuates renal fibrosis by maintaining Smad7 and phosphatase and tensin homolog expression. J Am Soc Nephrol. 2016;27:2092–2108. doi: 10.1681/ASN.2015040457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann J, et al. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714, 714.e1–714.e4. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atta H, et al. Mutant MMP-9 and HGF gene transfer enhance resolution of CCl4-induced liver fibrosis in rats: Role of ASH1 and EZH2 methyltransferases repression. PLoS One. 2014;9:e112384. doi: 10.1371/journal.pone.0112384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang MD, et al. Rosmarinic acid and baicalin epigenetically derepress peroxisomal proliferator-activated receptor γ in hepatic stellate cells for their antifibrotic effect. Hepatology. 2012;55:1271–1281. doi: 10.1002/hep.24792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Hoogen F, et al. 2013 classification criteria for systemic sclerosis: An American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2013;72:1747–1755. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- 21.Tsou PS, et al. Activation of the thromboxane A2 receptor by 8-isoprostane inhibits the pro-angiogenic effect of vascular endothelial growth factor in scleroderma. J Invest Dermatol. 2015;135:3153–3162. doi: 10.1038/jid.2015.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsou PS, et al. Scleroderma dermal microvascular endothelial cells exhibit defective response to pro-angiogenic chemokines. Rheumatology (Oxford) 2015;55:kev399. doi: 10.1093/rheumatology/kev399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsou PS, et al. Lipoic acid plays a role in scleroderma: Insights obtained from scleroderma dermal fibroblasts. Arthritis Res Ther. 2014;16:411. doi: 10.1186/s13075-014-0411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Tsou P-S, Khanna D, Sawalha AH. Methyl-CpG-binding protein 2 mediates antifibrotic effects in scleroderma fibroblasts. Ann Rheum Dis. 2018;77:1208–1218. doi: 10.1136/annrheumdis-2018-213022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haak AJ, et al. Targeting the myofibroblast genetic switch: Inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J Pharmacol Exp Ther. 2014;349:480–486. doi: 10.1124/jpet.114.213520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchings KM, et al. Pharmacokinetic optimitzation of CCG-203971: Novel inhibitors of the Rho/MRTF/SRF transcriptional pathway as potential antifibrotic therapeutics for systemic scleroderma. Bioorg Med Chem Lett. 2017;27:1744–1749. doi: 10.1016/j.bmcl.2017.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coit P, et al. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietras A, von Stedingk K, Lindgren D, Påhlman S, Axelson H. JAG2 induction in hypoxic tumor cells alters Notch signaling and enhances endothelial cell tube formation. Mol Cancer Res. 2011;9:626–636. doi: 10.1158/1541-7786.MCR-10-0508. [DOI] [PubMed] [Google Scholar]
- 32.Oon CE, et al. Role of Delta-like 4 in Jagged1-induced tumour angiogenesis and tumour growth. Oncotarget. 2017;8:40115–40131. doi: 10.18632/oncotarget.16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu C, et al. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer Res. 2007;67:1757–1768. doi: 10.1158/0008-5472.CAN-06-3700. [DOI] [PubMed] [Google Scholar]
- 34.Viré E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 35.Lv YC, et al. Histone methyltransferase enhancer of zeste homolog 2-mediated ABCA1 promoter DNA methylation contributes to the progression of atherosclerosis. PLoS One. 2016;11:e0157265. doi: 10.1371/journal.pone.0157265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi-Wen X, et al. Focal adhesion kinase and reactive oxygen species contribute to the persistent fibrotic phenotype of lesional scleroderma fibroblasts. Rheumatology (Oxford) 2012;51:2146–2154. doi: 10.1093/rheumatology/kes234. [DOI] [PubMed] [Google Scholar]
- 37.Kottakis F, et al. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell. 2011;43:285–298. doi: 10.1016/j.molcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ram O, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krämer M, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis. 2013;72:614–620. doi: 10.1136/annrheumdis-2012-201615. [DOI] [PubMed] [Google Scholar]
- 40.Peer CJ, et al. A rapid ultra HPLC-MS/MS method for the quantitation and pharmacokinetic analysis of 3-deazaneplanocin A in mice. J Chromatogr B Analyt Technol Biomed Life Sci. 2013;927:142–146. doi: 10.1016/j.jchromb.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y, Niederstrasser H, Edwards M, Jackson CE, Cooper JA. Distinct roles for CARMIL isoforms in cell migration. Mol Biol Cell. 2009;20:5290–5305. doi: 10.1091/mbc.E08-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang C, et al. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. Ezh2 loss propagates hypermethylation at T cell differentiation-regulating genes to promote leukemic transformation. J Clin Invest. 2018;128:3872–3886. doi: 10.1172/JCI94645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–197. doi: 10.1016/j.ccr.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao E, et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwon H, et al. Epigenetic silencing of miRNA-34a in human cholangiocarcinoma via EZH2 and DNA methylation: Impact on regulation of Notch pathway. Am J Pathol. 2017;187:2288–2299. doi: 10.1016/j.ajpath.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.