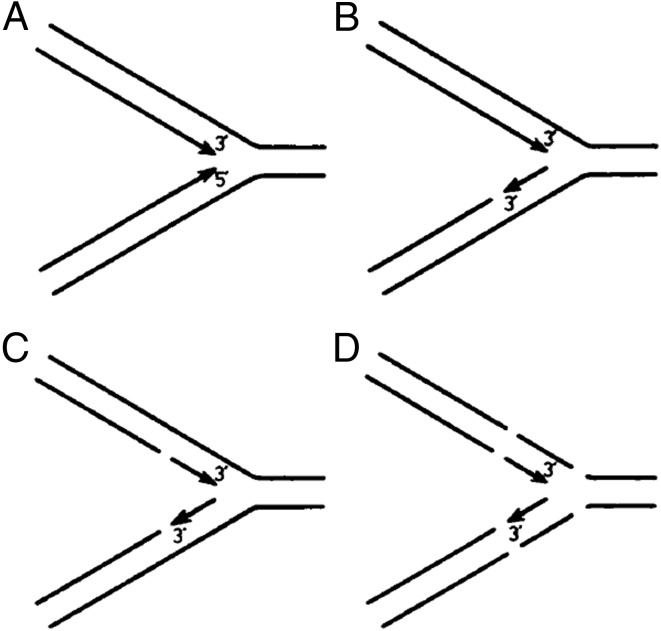
The antiparallel structure of double-stranded DNA, together with the known 5′→3′ directionality of DNA polymerases, necessitates that the two DNA strands are replicated in opposite directions. The leading strand is synthesized in the same direction as the replication fork, whereas the lagging strand is replicated in the opposite direction. In a 1968 paper in PNAS, Reiji and Tsuneko Okazaki and colleagues (1) proposed that the lagging strand is replicated discontinuously in the form of small fragments that subsequently are matured into one continuous strand. A review of the current state of molecular biology published that year (2) named these small fragments “Okazaki fragments,” as they have been called since. The seminal studies of Okazaki et al. (1) generated the textbook model of the semidiscontinuous replication fork, with a continuous leading strand and a discontinuous lagging strand. However, their original experimental results were not in accord with this model, and their studies suggested that all nascent DNA fragments are small. Was the leading strand also synthesized discontinuously, as was depicted in figure 1 of ref. 1 (Fig. 1C)? Fifty years after the landmark paper, this question has been answered by Cronan et al. (3), who show that the leading strand is indeed replicated continuously. However, this strand is fragmented due to ribonucleotide excision repair (RER) (4, 5). RER removes genomic ribonucleotides that are erroneously inserted by replicative DNA polymerases (Fig. 2A).
Fig. 1.
(A–D) Models for the possible structure and reaction in the replicating region of DNA. Reprinted with permission from ref. 1.
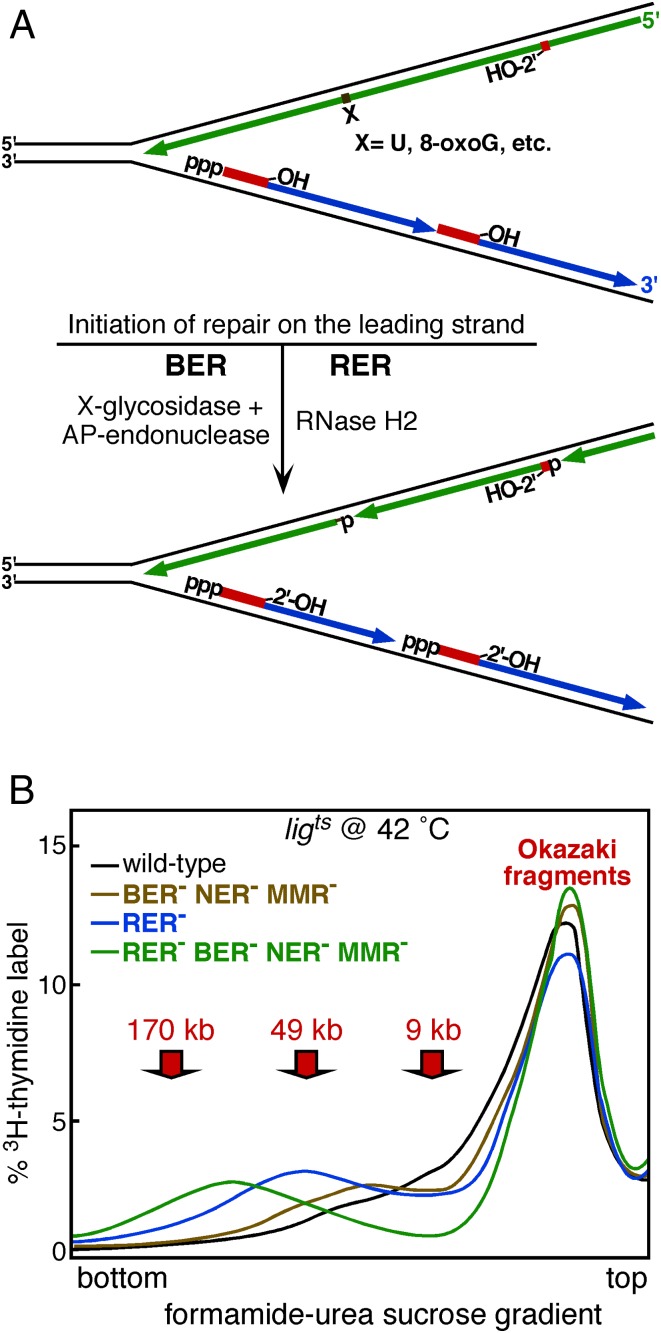
Fig. 2.
(A) DNA incision intermediates in the BER of a damaged or unnatural base (X), or the RER of a genomic ribonucleotide. Only repair on the leading strand, which leads to its fragmentation, is shown. (B) Size distribution, in a formamide-urea-sucrose gradient, of single-stranded replication intermediates. Ligts cells were shifted to 42 °C to inactivate ligase, and cells were pulse-labeled with 3H-thymidine for 2 min. The BER− NER− MMR− defective mutant is mutY mug ung nth nei tag alkA mutM nfi uvrA mutS; the RER− defective mutant is rnhB. Curves redrawn from ref. 3, fig. 3b. DNA size markers are approximate. E. coli Okazaki fragments are 1 to 2 kb.
To detect the earliest lagging-strand pieces synthesized, before they could be matured into continuous DNA, Okazaki et al. (1) used pulse-labeling techniques in Escherichia coli, with 3H-thymidine pulses as brief as 5 s. Given that the DNA replication rate is 500 to 1,000 nt/s and an average Okazaki fragment is 1 to 2 kb in size, it only takes a few seconds to synthesize a single Okazaki fragment. The pulse-labeled DNA was denatured and size-fractionated on an alkaline sucrose gradient. Surprisingly, even though biochemical studies at the time suggested that the leading strand could be synthesized in a continuous fashion (6), virtually all of the labeled DNA fractionated near the top of the gradient, with a DNA length of 1 to 2 kb. Only a small fraction of the label appeared as a shoulder with a size of up to 10 kb, similarly to the result shown in Fig. 2B (black curve). Obviously, the data supported the model in Fig. 1C more than the idealized model in Fig. 1B for semidiscontinuous DNA replication. These curious results were not just an E. coli oddity or artifact; they extended to the gram-positive bacterium Bacillus subtilis and to bacteriophage T4 (7).
Over the next few years, the Okazaki team introduced several improvements and gained more advanced knowledge of the small fragments that were named after them. They showed that the pulse-labeled small fragments could be chased into large fragments upon addition of an excess of cold thymidine, indicating that they were true intermediates in the synthesis of chromosomal DNA. They discovered that the synthesis of Okazaki fragments is primed with a short RNA primer (8). They also showed that the direction of DNA synthesis in vivo was the same as determined in vitro (i.e., 5′→3′) and that the RNA primer indeed was at the 5′ end of an Okazaki fragment. However, despite all these improvements, the fact remained that all nascent DNA was small in size, begging the question regarding the exact nature of leading-strand DNA replication. After the untimely death of Reiji Okazaki in 1975, Tsuneko continued the groundbreaking studies that she and her husband had initiated at Nagoya University. Her team determined the structures of the RNA primers in prokaryotes and eukaryotes (9). Tsuneko Okazaki’s recent account of their studies of replication intermediates can be found in ref. 10.
The first indication that DNA repair might be responsible for some of the small fragments detected in pulse-labeling studies came from the study of an E. coli sof (for short Okazaki fragment) mutant (11). The sof mutant is defective for dUTPase, which sanitizes the dNTP pool by hydrolyzing erroneously synthesized dUTP. This analog is readily incorporated into DNA during DNA replication, but the uracil base is subsequently excised by uracil-DNA-glycosylase, followed by base excision repair (BER) of the abasic moiety. In the sof mutant, the high level of dUTP resulted in frequent dUMP misincorporation and hence frequent BER, which caused severe fragmentation of newly synthesized DNA. However, it was also clear from these studies that uracil excision did not contribute to the generation of short fragments in wild-type cells (11). Nevertheless, they reinforced the notion that DNA repair might contribute to the generation of short fragments on the leading strand. But which form of DNA repair? A mutant defective for all forms of BER, nucleotide excision repair (NER), and mismatch repair (MMR) still failed to show the expected bimodal distribution of sizes for a fully semidiscontinuous DNA replication model (12).
Even before Okazaki et al. (1) published their seminal studies, it was known that DNA polymerases could use ribonucleoside triphosphates (rNTPs) as substrates, albeit poorly (13). Critical insights into the nature and abundance of genomic ribonucleotides came from studies in yeast. The replicative DNA polymerases show a robust discrimination of 104 to 105 for dNTP versus rNTP incorporation (14). At first glance, this low frequency of misincorporation should not pose a serious problem. However, the intracellular concentrations of the four rNTPs are 30- to 200-fold higher than those of the corresponding dNTPs. Therefore, in a simple competition assay, when rNTPs and dNTPs were present at their physiological concentrations, a much more frequent misincorporation occurred, about one ribonucleotide per kilobase (14). These genomic ribonucleotides are excised by the RER pathway, which is initiated by RNase H2-catalyzed incision at the 5′-ribonucleotide position (Fig. 2A) (4). The RER pathway is specific for RNase H2 (RnhB in E. coli), because RNase H1 does not cut at single genomic ribonucleotides. In E.coli, the dNTP pools are not as low as they are in yeast; nevertheless, the dATP concentration is one-twentieth that of ATP (15). It appears that ribonucleotide misinsertion could be the source of the half-century-old leading-strand fragmentation problem.
To carry out their analysis, Cronan et al. (3) make use of a temperature-sensitive ligase strain in which DNA ligase activity is shut down rapidly upon a shift from 28 to 42 °C, preventing ligation of processed Okazaki fragments and the ligation of excision repair events. In addition, they make an important modification to the original Okazaki protocol. Anticipating that ribonucleotides would remain in the genome in an E. coli rnhB mutant that is defective for RER, they could no longer rely on alkaline sucrose gradients for the size separation of nascent single-stranded DNA. RNA is sensitive to alkali, and the DNA could be fragmented at ribonucleotide insertion sites during centrifugation under alkaline conditions. Therefore, they turned to formamide-urea-sucrose gradients carried out at neutral pH, which showed a similar separatory power of denatured DNA. With this improvement, they first reinvestigated the size distribution of labeled DNA from a mutant defective in BER, NER, and MMR (Fig. 2B, brown curve). Compared with the isogenic wild-type strain, the higher-molecular-weight shoulder on the peak of Okazaki-size products had shifted slightly to the left, indicative that some repair of newly replicated DNA occurred. However, products larger than ∼50 kb in size were absent. Remarkably, the profile from an rnhB mutant showed a bimodal distribution of product sizes, with the high-molecular-weight products centered around 50 kb (Fig. 2B, blue curve). Finally, when all repair mutations were combined, a robust high-molecular-weight distribution was observed, centered around 80 kb (Fig. 2B, green curve). Importantly, >90% of the small fragments mapped to the lagging strand, and 90% of the large DNA mapped to the leading strand. These data are consistent with an earlier study by Okazaki et al. (16), in which a mutation in DNA polymerase I caused a defect in the joining of all small nascent fragments, because E. coli DNA polymerase I is required for RER as well as Okazaki fragment maturation (5).
The new study by Cronan et al. (3) shows that the incorporation of ribonucleotides and their subsequent excision by RER is a frequent event. Cronan et al. assess the frequency of this event by measuring the number of ribonucleotides present in a plasmid propagated in an rnhB strain. From the fraction of the isolated plasmid that was nicked by RNase H2 and using the reasonable assumption that ribonucleotide insertion is random, Cronan et al. use the Poisson equation to calculate the misinsertion frequency at one ribonucleotide per 8 to 9 kb, or about 500 per genome. By 1 order of magnitude, RER in bacteria appears to be the most frequently employed DNA repair pathway, as it is in eukaryotes.
While it is evident from the current study that the leading strand is synthesized largely continuously, the question still remains as to how continuous. Replication forks tend to stall for a variety of reasons, and cells have several mechanisms to restart replication forks (17). Some of these restart mechanisms use the available 3′ terminus for restart, after recombination or fork remodeling, but a de novo restart by RNA priming is also a possible mechanism. The latter would show up as a fragmentation event on the leading strand. In their study of the size distributions of leading strands in the multiple-pathway excision repair-defective strain (RER− BER− NER− MMR−), Cronan et al. (3) provide evidence that, indeed, the leading strand is not completely continuous. Whether these discontinuities are the result of still another repair pathway, the result of replication-fork restart, or the result of the stochastic, unprovoked activity of DNA primase on the leading strand needs further investigation.
Acknowledgments
I thank Andrei Kuzminov and colleagues for sharing their results prior to publication. This research is supported by the National Institutes of Health Grant R35-GM118129.
Footnotes
The author declares no conflict of interest.
See companion article on page 1251 in issue 4 of volume 116.
References
- 1.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc Natl Acad Sci USA. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous Molecular biology comes of age. Nature. 1968;219:825–829. doi: 10.1038/219825a0. [DOI] [PubMed] [Google Scholar]
- 3.Cronan GE, Kouzminova EA, Kuzminov A. Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision. Proc Natl Acad Sci USA. 2019;116:1251–1260. doi: 10.1073/pnas.1814512116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sparks JL, et al. RNase H2-initiated ribonucleotide excision repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaisman A, et al. Investigating the mechanisms of ribonucleotide excision repair in Escherichia coli. Mutat Res. 2014;761:21–33. doi: 10.1016/j.mrfmmm.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goulian M, Kornberg A, Sinsheimer RL. Enzymatic synthesis of DNA, XXIV. Synthesis of infectious phage phi-X174 DNA. Proc Natl Acad Sci USA. 1967;58:2321–2328. doi: 10.1073/pnas.58.6.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okazaki R, et al. In vivo mechanism of DNA chain growth. Cold Spring Harb Symp Quant Biol. 1968;33:129–143. [Google Scholar]
- 8.Sugino A, Hirose S, Okazaki R. RNA-linked nascent DNA fragments in Escherichia coli. Proc Natl Acad Sci USA. 1972;69:1863–1867. doi: 10.1073/pnas.69.7.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitani T, Yoda K, Okazaki T. Discontinuous DNA replication of Drosophila melanogaster is primed by octaribonucleotide primer. Mol Cell Biol. 1984;4:1591–1596. doi: 10.1128/mcb.4.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okazaki T. Days weaving the lagging strand synthesis of DNA—A personal recollection of the discovery of Okazaki fragments and studies on discontinuous replication mechanism. Proc Jpn Acad, Ser B, Phys Biol Sci. 2017;93:322–338. doi: 10.2183/pjab.93.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tye BK, Chien J, Lehman IR, Duncan BK, Warner HR. Uracil incorporation: A source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1978;75:233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amado L, Kuzminov A. Low-molecular-weight DNA replication intermediates in Escherichia coli: Mechanism of formation and strand specificity. J Mol Biol. 2013;425:4177–4191. doi: 10.1016/j.jmb.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berg P, Fancher H, Chamberlin M. The synthesis of mixed polynucleotides containing ribo-and deoxyribonucleotides by purified preparations of DNA polymerase from Escherichia coli. In: Vogel HJ, Bryson V, Lampen JO, editors. Informational Macromolecules. Academic Press; New York: 1963. pp. 467–483. [Google Scholar]
- 14.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okazaki R, Arisawa M, Sugino A. Slow joining of newly replicated DNA chains in DNA polymerase I-deficient Escherichia coli mutants. Proc Natl Acad Sci USA. 1971;68:2954–2957. doi: 10.1073/pnas.68.12.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marians KJ. Lesion bypass and the reactivation of stalled replication forks. Annu Rev Biochem. 2018;87:217–238. doi: 10.1146/annurev-biochem-062917-011921. [DOI] [PMC free article] [PubMed] [Google Scholar]