Significance
Elucidation of the molecular mechanism underlying crop adaptation to local environments, and the expansion of their cultivation area, will contribute to the development of germplasm resilient to climate changes. In this study, we cloned and functionally characterized a chilling tolerant quantitative trait locus (QTL), HAN1, that confers chilling tolerance in temperate japonica rice. In the HAN1 promoter, a functional nucleotide polymorphism results in an increase of putative MYB cis-element in the allele of temperate japonica rice, and enhances its chilling tolerance, allowing it to adapt to a temperate climate during northward expansion. We can now select for the superior allele of HAN1 to improve chilling tolerance in rice, thereby contributing to global food security in response to climate changes.
Keywords: rice, jasmonate, chilling tolerance, temperate adaption
Abstract
Rice (Oryza sativa L.) is a chilling-sensitive staple crop that originated in subtropical regions of Asia. Introduction of the chilling tolerance trait enables the expansion of rice cultivation to temperate regions. Here we report the cloning and characterization of HAN1, a quantitative trait locus (QTL) that confers chilling tolerance on temperate japonica rice. HAN1 encodes an oxidase that catalyzes the conversion of biologically active jasmonoyl-L-isoleucine (JA-Ile) to the inactive form 12-hydroxy-JA-Ile (12OH-JA-Ile) and fine-tunes the JA-mediated chilling response. Natural variants in HAN1 diverged between indica and japonica rice during domestication. A specific allele from temperate japonica rice, which gained a putative MYB cis-element in the promoter of HAN1 during the divergence of the two japonica ecotypes, enhances the chilling tolerance of temperate japonica rice and allows it to adapt to a temperate climate. The results of this study extend our understanding of the northward expansion of rice cultivation and provide a target gene for the improvement of chilling tolerance in rice.
Rice (Oryza sativa L.), a chilling sensitive crop that feeds more than half the world’s population, originated in subtropical regions of Asia and subsequently expanded to a wide range of geographical regions (1). The two subspecies are composed of five ecotypes: indica, aus, aromatic, temperate japonica, and tropical japonica rice. These ecotypes are characterized by specific climate adaptations based on agro-ecological cultivation conditions (2, 3). While indica rice is mainly cultivated in tropical and subtropical regions, japonica rice experienced a wider habitat expansion. A japonica rice subgroup extended to tropical regions of Southeast Asia and evolved into tropical japonica. Another subgroup, termed temperate japonica, expanded to more northern and higher elevation regions and reached the northern limits of its natural cultivation area. Through enhanced chilling tolerance, temperate japonica rice adapted progressively to a temperate climate (4). Tolerance to low temperatures above 0 °C, termed chilling tolerance, was one of the most important factors that ensured the northward expansion of rice (5, 6). Genes involved in chilling adaptation should exhibit allelic differences across latitudinal gradients. The relative frequencies in the allele pool present in different geographical regions may reflect the adaptation mechanism of rice to new climatic conditions (6, 7).
Chilling tolerance in rice is a quantitative trait controlled by multiple genetic factors and the environment. Dissecting the genetic basis of chilling tolerance in rice is still at its infant stage. A number of quantitative trait loci (QTLs) that confer chilling tolerance have been mapped, and some have been genetically characterized; examples are qLTG3-1, Ctb1, qCTS7, COLD1, qCTS-9, CTB4a, and qPSR10 (6–13). In addition to COLD1 and CTB4a, the natural variation of the gene bZIP73 has been identified to enhance rice adaptation to cold habitats (14). However, the natural allele which confers chilling tolerance on temperate japonica rice specifically has been seldom discovered, and thus the molecular mechanisms underlying the adaptation to chilling in temperate japonica rice during its northward expansion are still unknown. In this study we identified a chilling tolerance QTL, dubbed HAN1 (“han” termed “chilling” in Chinese). The natural allele of temperate japonica rice, distinct from other ecotypes and wild rice populations, conferred chilling tolerance and enabled the expansion of temperate japonica rice to temperate regions.
Results
HAN1 Is a Major QTL for Chilling Tolerance in Temperate Japonica Rice.
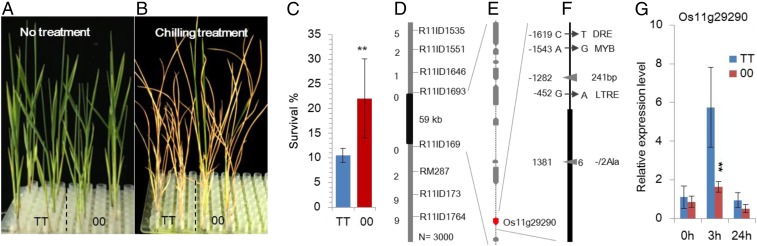
To understand the subspecies divergence of chilling tolerance and to identify the potential genes responsible for adaptation to chilling, a recombinant inbred line (RIL) population derived from a cross between indica cultivar Teqing and temperate japonica cultivar 02428 was used for linkage analysis of chilling tolerance. The indica rice parent Teqing was chilling sensitive, while the japonica rice parent 02428 was highly tolerant to chilling stress (SI Appendix, Fig. S1 A and B). In the RIL population, there was a continuous distribution in chilling tolerance at the seedling stage, and two QTLs were detected on chromosomes 4 and 11 for chilling tolerance in rice (SI Appendix, Fig. S1 C and D, and Table S1). Chilling Tolerance 1 (HAN1) was a major QTL located on chromosome 11, with a logarithm (base 10) of odds (LOD) value of 11.5. The positive allele from 02428 improved chilling tolerance with an increase of 20.4% in seedling survival rate and accounted for 35.8% of the total phenotypic variation under a chilling stress condition (8 °C for 6 d). A near-isogenic line (NIL) population was further developed in the Teqing genetic background. Compared with the indica genotype, HAN1Teqing/Teqing, plants with the japonica genotype, HAN102428/02428, had a higher survival rate following chilling treatment. Furthermore, the chilling tolerance of plants with the heterozygous genotype HAN1Teqing/02428 was close to the midparent value of HAN1Teqing/Teqing and HAN102428/02428, suggesting that HAN1 had an additive effect but no significant dominant effect on chilling tolerance (Fig. 1 A–C and SI Appendix, Fig. S1 E and F).
Fig. 1.
Map-based cloning of HAN1. (A and B) Performance of seedlings with two genotypes in the near-isogenic line (TT, Teqing/Teqing; 00, 02428/02428) before (A) and after chilling treatment (B). (C) Chilling tolerance of two genotypes in the near-isogenic line background (Student’s t test, **P < 0.01, n = 5). (D) Fine mapping of HAN1 on the long arm of chromosome 11. The numbers of recombinants are shown on the left, and the InDel marker loci used for fine-mapping are shown on the right of the linkage map. (E) There were a total of 10 putative genes in the 59-kb chromosomal interval containing HAN1. The candidate gene LOC_Os11g29290 is shown in red. (F) DNA sequence comparison of the HAN1 candidate gene between the two parental rice cultivars. The negative and positive numbers indicate the positions of polymorphic sites in the promoter and coding region of HAN1, respectively, relative to the ATG start codon. The arrows indicate the SNPs between Teqing (Right) and 02428 (Left), and the two triangles show the InDels between Teqing (both insertions) and 02428 (both deletions). The predicted effects of these SNPs/InDels are shown in the rightmost column. (G) Comparison of the expression levels of the candidate gene LOC_Os11g29290 between two genotypes of NIL under low temperature treatment (Reference gene is the Actin gene numbered LOC_Os03g50885; Student’s t test, **P < 0.01, n = 3).
Through high-density molecular markers in the extremely tolerant and highly sensitive RILs, HAN1 was further mapped to an 871-kb interval between the two marker loci R11ID1646 and R11ID1733 (SI Appendix, Table S2). In the NIL population (BC5F2) of HAN1 with 3,000 individuals, 11 recombinants between R11ID1535 and R11ID1764 were selected for progeny tests of chilling tolerance. The progeny tests revealed that HAN1 cosegregated with a 59-kb interval between R11ID1693 to R11ID1699 (Fig. 1D and SI Appendix, Table S3). There are 10 genes annotated in this region based on the Nipponbare reference genome (Fig. 1E). Among them, the gene numbered LOC_Os11g29290 is chilling responsive and exhibits differential expression between the two genotypes of the NIL in response to chilling stress treatment. The Teqing allele displayed a higher expression level than the 02428 allele at the third hour of chilling stress treatment (Fig. 1G). Sequence comparison identified several nucleotide variations in LOC_Os11g29290 between the two parental lines. There was an insertion/deletion (InDel) of 241 bp and several SNPs in cis-elements predicted to be involved in low temperature response in the promoter region (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace). There was also one 6-bp InDel that caused amino acid changes in the coding region of the gene (Fig. 1F). Therefore, we consider LOC_Os11g29290 to be a strong candidate gene for HAN1.
HAN1 Is a Negative Regulator of Chilling Tolerance in Rice.
To confirm that HAN1 is the gene underlying the chilling tolerance QTL, we first analyzed the chilling tolerance of HAN1 overexpression and knockout lines. The overexpression lines consisted of the maize ubiquitin promoter fused with either the Teqing or 02428 allele of HAN1. These overexpression lines withered at the seedling stage and did not survive to reproductive growth. Thus, no progeny were available for further analysis (SI Appendix, Fig. S2). Instead, we used the inducible gene expression system mediated by the estrogen receptor to overexpress HAN1 (15). As shown in Fig. 2 A–E and SI Appendix, Fig. S3, the T2 generation transgenic lines carrying either pER8::HAN102428 or pER8::HAN1Teqing had higher gene expression levels under both normal and chilling treatment conditions, and were more sensitive to low temperature treatment than the control Nipponbare in the presence of estradiol. Comparatively, the chilling tolerance of the transgenic lines carrying pER8::HAN1Teqing was similar to the lines expressing pER8::HAN102428, excluding the functional difference in the coding regions between the two parental alleles (SI Appendix, Fig. S3D). Conversely, two CRISPR/Cas9 knockout lines carrying A or T base insertions in the coding region were more chilling-tolerant than the parental control Nipponbare (Fig. 2 F–J). These findings suggest that HAN1 negatively regulates chilling tolerance.
Fig. 2.
HAN1 is a negative regulator of chilling tolerance. (A) The DNA construct used for estrogen-inducible overexpression of HAN1 in rice. VEX is the gene coding a chimeric transcriptional activator that binds the pER8 promoter, which consists of eight copies of the LexA operator fused upstream of the −46 35S minimal promoter, to activate transcription of HAN1 in the presence of exogenous estrogen. (B and C) Performance of pER8::HAN102428 transgenic (No. 1–3) and wild-type (WT) lines before (B) and after chilling treatment (C). (D) Expression levels of HAN1 in pER8::HAN102428 transgenic lines and wild type. (E) Survival rate (%) of pER8::HAN102428 transgenic lines and wild type after chilling stress treatment. The DNA construct designed for knockout mutation of HAN1 using the CRISPR/Cas9 system. (G–H) Performance of knockout han1 mutant (han1-1 and han1-2) and wild-type (NP) lines before (G) and after chilling treatment (H). (I) Partial DNA sequences from the HAN1 knockout mutants (han1-1 and han1-2) show one base (A or T) insertion in CDS region of HAN1 for the two alleles, resulting in frameshift mutations. (J) Survival rate (%) of HAN1 knockout mutants and wild-type control after chilling treatment. Significant differences were determined by one-way ANOVA (*P < 0.05, or **P < 0.01, n = 3) in D, E, and G.
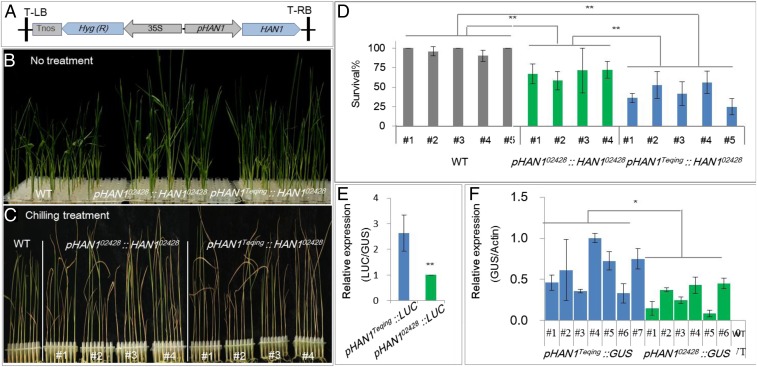
A complementation test was then carried out to determine the allelic variation between the two parental lines. As the coding regions of the two parental alleles of HAN1 are equally functional (SI Appendix, Fig. S3), a 1.8-kb DNA fragment of the HAN1 promoter region from either 02428 or Teqing was fused to the coding region of the HAN1 02428 allele to generate pHAN1Teqing::HAN102428 or pHAN102428::HAN102428 transgenic lines within the Teqing background, respectively. As shown in Fig. 3 A–D, consistent with the observation in the HAN1 overexpression lines, both of the transgenic lines showed increased chilling sensitivity compared with the parental line Teqing. However, pHAN1Teqing::HAN102428 lines were more sensitive to chilling stress than pHAN102428::HAN102428 lines, showing that allelic variations in the HAN1 promoter region confer genetic variation in chilling tolerance.
Fig. 3.
Genetic variations in the parental alleles of HAN1. (A) DNA constructs carrying the HAN102428 coding region driven by either Teqing (pHAN1Teqing) or 02428 (pHAN102428) promoter region. (B and C) Performance of pHAN102428::HAN102428 and pHAN1Teqing::HAN102428 transgenic plants and wild type (Teqing) before (B) and after chilling treatment (C). (D) Relative survival rates of pHAN102428::HAN102428 and pHAN1Teqing::HAN102428 transgenic plants and wild type (Teqing) after chilling treatment. (E) Relative expression levels under chilling treatment (4 °C) in Arabidopsis protoplasts of the LUC reporter genes driven by pHAN102428, and pHAN1Teqing. pHAN102428 and pHAN1Teqing are the promoters from the two parental lines. (F) RT-qPCR analysis of GUS reporter gene expression in pHAN1Teqing::GUS and pHAN102428::GUS transgenic lines in the background of temperate japonica rice cultivar Nipponbare under chilling stress (4 °C for 3 h; WT, Nipponbare; Actin as reference). Significant differences in D–F were determined by one-way ANOVA (*P < 0.05, or **P < 0.01, n ≥ 3).
To confirm functional differences between the HAN1 promoters of Teqing and 02428 alleles, we employed an Arabidopsis protoplast system, in which firefly luciferase (LUC) fused to pHAN1 was expressed to monitor the promoter activity of HAN1 with different variations (7). As shown in Fig. 3E, pHAN1Teqing::LUC protoplasts exhibited higher relative expressions of LUC than pHAN102428::LUC protoplasts under chilling stress conditions. Moreover, expression analysis of β-glucuronidase (GUS) reporter gene in pHAN1Teqing::GUS and pHAN102428::GUS T2 generation transgenic plants also revealed that HAN1 was chilling-induced and HAN1Teqing was higher than that of HAN102428 (Fig. 3F). These results are consistent with the observations of the complementation test and expression analysis (Figs. 1G and 3 A–D). Taken together, all of these results indicate that HAN1 is the causal gene responsible for the chilling tolerance QTL. HAN1 acts as a negative regulator in a dose-dependent manner in rice. Sequence variations in the promoter region result in functional divergence in chilling tolerance between the two parental lines.
HAN1 Is a Chilling-Induced Endoplasmic Reticulum Protein.
The HAN1 protein is predicted to be located in the endoplasmic reticulum (ER) when analyzed with ProtComp 9.0 software (linux1.softberry.com/berry.phtml). To investigate the subcellular localization of HAN1 experimentally, we transiently expressed two fusion genes, HAN1 fused with green fluorescent protein (GFP) (35S::HAN1-GFP), and the ER marker gene HDEL fused with red fluorescent protein (RFP) (35S::HDEL-RFP), in rice protoplasts and tobacco leaves. The HAN1-GFP fluorescence emissions fully overlapped with the HDEL-RFP signals in the cytoplasm of rice protoplasts (SI Appendix, Fig. S4 A–E). In tobacco leaf cells, the overlapping fluorescence signals were presented as a net-like shape, typical of ER subcellular images (SI Appendix, Fig. S4 F–K). Thus, HAN1 is targeted to the ER.
According to gene expression databases (CREP: crep.ncpgr.cn), HAN1 is expressed ubiquitously in all rice tissues (SI Appendix, Fig. S5). During germination, its expression levels are relatively high in radicle and coleoptile tissues, whereas at maturity, these expression levels are high in roots, glumes, and stamens. The transgenic plants that carried the reporter gene GUS, driven by the HAN1 promoter region from either the Teqing or 02428 allele in the Nipponbare background were histologically stained to analyze the expression pattern. Similar spatial-temporal GUS staining patterns were detected in pHAN1Teqing::GUS and pHAN102428::GUS plants. GUS staining was detected in both the radicle and coleoptile, but not in the plumule during germination. During reproductive growth period, GUS was expressed strongly in the roots, hulls, and stamens and weakly in other tissues (SI Appendix, Fig. S6 A–J). The leaves showed very weak staining under normal conditions, but were stained strongly after exposure to chilling stresses (SI Appendix, Fig. S6K).
HAN1 Functions in JA-Mediated Chilling Tolerance.
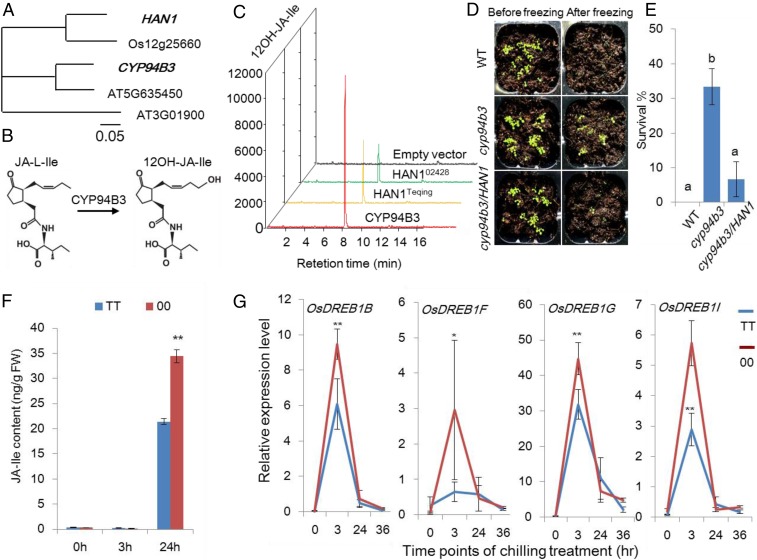
Phylogenetic analysis revealed that HAN1 belonged to the cytochrome P450 superfamily. An Arabidopsis member of this family, CYP94B3 (AT3G48520), has been shown to act as an oxidase to metabolize JA-Ile, the active form of JA, into 12OH-JA-Ile, an inactive form (16) (Fig. 4 A and B). To test the role of HAN1 in JA-Ile catabolism, HAN1 was expressed in yeast (Saccharomyces cerevisiae), and JA-Ile-12-hydroxylase activity was then assayed. Microsomal fractions prepared from yeast cells transformed with an empty vector pYES2NT-C (negative control), CYP94B3 open reading frame (positive control), and either the Teqing or 02428 allele of HAN1, were incubated with JA-Ile, and the reaction products were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS). Similar to microsomes isolated from the CYP94B3-expressing cells, both HAN1Teqing and HAN102428 produced detectable amounts of 12OH-JA-Ile, whereas 12OH-JA-Ile was not produced in the reaction containing microsomes from empty vector-containing control cells (Fig. 4C and SI Appendix, Fig. S7). Therefore, HAN1 has a conserved function to its Arabidopsis homolog CYP94B3, and both the Teqing and 02428 alleles possess JA-Ile-12-hydroxylase activity.
Fig. 4.
HAN1 fine-tunes the JA-mediated chilling tolerance in rice. (A) Phylogenetic tree showing the relationships between HAN1 and the homologous genes from rice and Arabidopsis. (B) Conversion of JA-Ile to 12OH-JA-Ile catalyzed by the Arabidopsis protein CYP94B3 (16). (C) Levels of 12OH-JA-Ile detected by mass spectrometry in the mixture of microsomal proteins from yeast cells incubated with the substrate JA-Ile. (D and E) Survival of the Arabidopsis mutant cyp94b3, the cyp94b3 transgenic line expressing HAN1, and the wild type Col-0 before and after freezing treatment (−10 °C for 1.5 h. There was a significant difference between a and b by Student’s t test with P < 0.01 and n = 3). (F) The levels of JA-Ile in fresh leaves of the two genotypes in rice NIL (TT, Teqing/Teqing; 00, 02428/02428) after 0, 3, and 24 h of chilling treatment (Student’s t test, **P < 0.01, n = 3). (G) Comparison of the relative expression of CBF/DREB1 genes downstream of JA signal pathway between the two genotypes in rice NIL (TT, Teqing/Teqing; 00, 02428/02428) by RT-qPCR after chilling treatment (Student’s t test, **P < 0.01, *P < 0.05, n = 3).
We further confirmed the functional role of HAN1 in planta. The rice HAN1 gene fused with CaMV35S promoter was transformed into the Arabidopsis mutant cyp94b3, which we identified as being more tolerant to freezing stress than wild-type Col-0 plants. HAN1 restored the freezing sensitivity of the cyp94b3 mutant (Fig. 4 D and E). Therefore, HAN1 and CYP94B3 are functionally conserved with respect to JA-Ile catabolism. In rice NILs, HAN1Teqing/Teqing allele was expressed at a significantly higher level than HAN102428/02428 following three hours of chilling treatment, and there was less JA-Ile in fresh plants after a 24-h chilling treatment (Figs. 1G and 4F). This implied that HAN1Teqing/Teqing plants, which exhibited a higher gene expression, catabolized more JA-Ile than HAN102428/02428 plants, resulting in increased sensitivity to low temperature compared with HAN102428/02428 plants.
Jasmonate positively regulates freezing tolerance in Arabidopsis as a critical upstream signal of the CBF/DREB1 pathway, the convergent signaling pathway of cold and chilling responses in plants (17–19). We analyzed the expression of OsDREB1s by real-time quantitative polymerase chain reaction (RT-qPCR) in NILs carrying the two HAN1 genotypes under low temperature conditions. A total of 10 CBF/DREB1s in rice were analyzed (20). As shown in Fig. 4G, the expression levels of four OsDREB1 genes (OsDREB1B, OsDREB1F, OsDREB1G, and OsDREB1I) were relatively higher in NIL-HAN102428/02428. Taken together, these results suggest that HAN1 exerts negative feedback control on JA-Ile levels and performs a key role in attenuating jasmonate over-accumulation. The temperate japonica rice cultivar 02428 has a lower expression of HAN1, which causes it to maintain higher JA-Ile levels and higher gene expressions in the CBF/DREB1-dependent pathway, thus conferring greater chilling tolerance compared with the indica rice cultivar Teqing.
Natural Variation in HAN1 Shows Allelic Differentiation Along Geographic Clines.
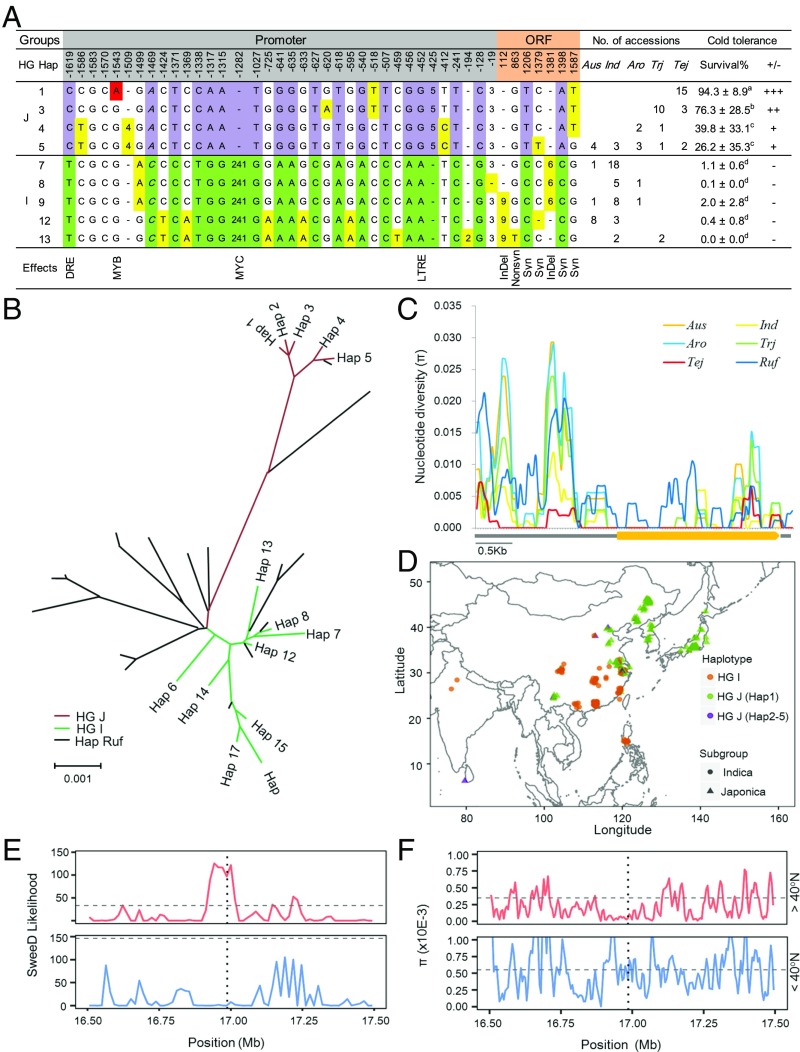
To investigate the natural variations in HAN1, we sequenced the 1.8-kb promoter and the coding region in 101 accessions from a core germplasm collection, including 42 indica, 16 tropical japonica, 19 temperate japonica, and 24 accessions from other ecotypes. Based on nucleotide polymorphisms, we identified 14 haplotypes (Fig. 5 A and B). Among these haplotypes, Hap1 was found specifically in temperate japonica rice cultivars (including the parental cultivar 02428). Hap2, Hap3, and Hap4 were found in tropical japonica rice cultivars, with Hap3 being the most prevalent. Hap5 was presented in both indica and japonica rice cultivars. Haps 6–14 were mainly presented in the indica rice cultivars, and Hap7 was the dominant haplotype that was found in the parental line Teqing. Based on their phylogenetic relationships, there were two haplotype groups that displayed a clear indica-japonica differentiation in the HAN1 genomic region. Haps 1–5 were assigned to the japonica haplotype group (HG J), while the others were assigned to the indica haplotype group (HG I). Evaluation of chilling tolerance revealed that HG J was significantly more tolerant than HG I (Fig. 5A). In the HG J group, Hap1 in temperate japonica was the most tolerant to low temperatures, followed by Hap3 in tropical japonica (Fig. 5 A and B).
Fig. 5.
The haplotypes and evolutionary analysis of HAN1. (A) Haplotype analysis of HAN1 in 101 rice germplasm accessions. The purple and green shaded SNPs or InDels represent polymorphisms in the japonica and indica types, respectively. The highlighted red A is found only in Hap1 in temperate japonica rice, while yellow polymorphisms are not unique among the rice cultivars (There were significant differences among different letters by Student’s t test with P < 0.01 and n ≥ 3). (B) A phylogenetic tree showing the relationships between HAN1 haplotypes from cultivated rice and O. rufipogon accessions. (C) Nucleotide diversity of HAN1 in the five cultivated rice subgroups of O. sativa and O. rufipogon accessions. The thick bar represents CDS region of HAN1, and the thin bars before or after the CDS are the 1.8-kb promoter or the 0.5-kb downstream region of HAN1, respectively. Aus, Ind, Aro, Trj, Tej, and Ruf represent the five ecotypes of cultivated rice, such as aus, indica, aromatic rice, tropical japonica and temperate japonica rice, and wild rice (O. rufipogon). (D) The geographical distribution of cultivated rice with different HAN1 haplotypes. There are two groups, the indica group HG I and the japonica group HG J. HG J is further divided into two subgroups HG J (Hap 1) and HG J (Haps 2–5) based on presence or absence of the MYB cis element. (E) The likelihood of a selective sweep of the 1.0-Mb region surrounding HAN1 in the rice population cultivated in typical temperate climate areas in high latitude regions (>40° N), or elsewhere in low latitude regions (<40° N). (F) Nucleotide diversity in the 1.0-Mb region flanking HAN1 in the two populations in E. The analyses performed in A, B, and C were based on DNA sequence information generated by Sanger sequencing in a population of 101 rice germplasm accessions, while the analyses in D, E, and F were based on NGS data and geographic information in a collection of 572 rice accessions.
We next examined the evolutionary origin of HAN1. The promoter and coding regions of HAN1 in a total of 21 wild rice accessions (Oryza rufipogon) were sequenced and included in phylogenetic analysis together with the DNA sequences from the 101 landraces. As shown in phylogenetic tree (SI Appendix, Fig. S8), haplotype tree (Fig. 5B), and haplotype network (SI Appendix, Fig. S9) of HAN1, HG I and HG J descended independently from wild rice Or-I and Or-III respectively. It is noteworthy that we did not identify a wild rice accession with the Hap1 haplotype, nor did we identify one that clustered close to it. Hap1 is much closer to the japonica haplotype Hap2, with only a single nucleotide polymorphism at position −1543(G/A). Therefore, the most chilling-tolerant haplotype of HAN1, Hap1, originated directly from japonica rice during the divergence between tropical japonica and temperate japonica rice rather than from O. rufipogon during japonica domestication.
The special SNP in the temperate japonica rice at -1543(G/A) site is predicted to be located within the MYB cis-element (WAACCA in Hap1, WAACCG in others). When the base at this site was mutated from 02428 type (A) to Teqing type (G), pHAN102428m::LUC had higher expression levels of LUC than pHAN102428::LUC in the transient expression system of protoplasts (SI Appendix, Fig. S10A). In the transgenic plant han1ΔMYB, with the knockout of this MYB element which was generated by CRISPR/Cas9 in the genetic background of temperate japonica rice cultivar Zhonghua 11, the expression level of HAN1 was higher under chilling stress treatment. Consequently, the mutant showed stronger chilling tolerance than the wild type (SI Appendix, Fig. S10 B–G). Taken together, the temperate japonica rice has a special cis-element MYB which negatively regulates the expression of HAN1 but positively regulates chilling tolerance.
The nucleotide diversity of HAN1 varies among the rice ecotypes. As shown in Fig. 5C, temperate japonica exhibits lower nucleotide diversity compared with other ecotypes, suggesting that adaptive selection or a genetic bottleneck played a role during the northward expansion of rice. An examination of the geographical distribution in our previously described collection of 572 resequenced rice accessions with known geographical information revealed that the enrichment of HAN1 haplotypes differed across latitudinal clines (Fig. 5D). Hap1 is restricted to accessions of temperate japonica from higher latitude regions (a few come from a higher altitude region, the Yunnan-Guizhou Plateau in China) where the temperature is relatively low during the rice planting season. Other rice cultivars belonging to HG J are distributed around the world. However, most HG I cultivars are grown in lower latitude regions where the temperature is relatively high during the rice growing season (Fig. 5D). In a sweep-D scan of the 572 rice accessions (Fig. 5E), a selective signal was detected in a rice population cultivated in a typical temperate climate region (>40° N). This signal was not present in other regions. Compared with the average genomic level, a significantly lower π value was detected only in HAN1, and not in the flanking regions in this population. Therefore, a bottleneck effect was excluded (Fig. 5F). Taken together, the most chilling-tolerant haplotype of HAN1, Hap1, arose in parallel with the northward expansion of temperate japonica rice, and has been fixed due to adaptive selection to high latitude growth conditions.
Discussion
HAN1 Functions to Balance Chilling Tolerance and Growth in Plants.
Jasmonate is a plant hormone involved in stress responses (21). When plants encounter low temperatures, expression of JA biosynthesis genes is induced and the level of active JA-Ile increases, thus activating downstream CBF/DREB1-dependent and/or independent signaling to enhance the cold stress response (17, 18, 22–27). However, JA antagonizes GA in regulating plant growth via the interaction between JAZ proteins and DELLA (28). Over-accumulation of JA will also hinder plant growth and development. As a result, a fine-tuned system of JA metabolism is required to balance its effects on stress response and plant growth. HAN1 plays such a role through metabolizing the active form of jasmonic acid, JA-Ile. HAN1 prevents over-accumulation of active JA under chilling stress conditions and maintains growth potential upon removal of stresses. In HAN1-knockout mutant rice plants, the chilling tolerance increased as expected but growth was severely retarded (Fig. 2H and SI Appendix, Fig. S11A). However, overexpression of HAN1 led to plant death at the seedling stage (SI Appendix, Fig. S2), and introduction of another copy of HAN1 in transgenic plants led to a decrease in grain production (SI Appendix, Fig. S11B). These observations suggest that the expression of HAN1 should be fine-tuned. In natural variants, expression of the temperate japonica allele of HAN1 is induced to a relatively low level and maintains more JA-Ile under chilling stress compared with the indica allele, which consequently confers increased chilling tolerance. Because the expression level of this allele only varies during exposure to chilling stress, introduction of HAN102428 into indica rice cultivars can enhance chilling tolerance. Furthermore, it has no negative effects on components of plant growth such as plant height, tiller number, spikelet number, and grain weight (SI Appendix, Fig. S11C). Therefore, Hap1 of HAN1 is an excellent allele for improving chilling tolerance in rice breeding. Through these aspects, the expression of HAN1 is fine-tuned, and acts to balance stress tolerance and plant growth by regulating the metabolism of JA-Ile.
Natural Variation in HAN1 Underlies the Adaptation to a Temperate Climate During the Northward Expansion of Japonica Rice.
Rice is a chilling sensitive crop. Low temperature in regions with temperate climates is the main factor in limiting the northward expansion of rice cultivation. It is widely accepted that indica rice and japonica rice originated from different ancestors of O. rufipogon, though the domestication process is still actively debated (3, 29–31). Further divergence occurred within the japonica subspecies; temperate japonica rice separated from tropical japonica rice with distinct geographical adaptation. It is thought that two sequential rounds of northward expansion occurred in temperate japonica rice. The first was from South China to Korea and Japan at the beginning of the first century, and the second was from Korea and Japan to Northeast China in the early 19th century (4). HAN1 is the QTL gene on chromosome 11 for chilling tolerance of temperate japonica rice. A phylogenetic tree constructed from HAN1 sequences reflects the demographic history of rice domestication and expansion. Variations in HAN1 diverged during rice domestication, followed by artificial selection during the northward expansion. We identified two haplotype groups, HG I and HG J, which originated independently from Or-I and Or-III. In the japonica HAN1 haplotype group HG J, the most chilling-tolerant haplotype, Hap1, is mainly represented in temperate japonica, the dominant group of rice accessions from Japan, Korea, and Northeast China. Hap1 differs from other japonica haplotypes in one special functional polymorphism, and no haplotype from wild rice is closely related, indicating that Hap1 originated in japonica rice rather than in wild rice. Therefore, the natural variation at the MYB motif in the promoter of HAN1 provided an adaptive advantage to the high latitude environment during the northward expansion of japonica rice cultivation. To date, several members belonging to the MYB family in plants, including OsMYB4, MYB15, OsMYB3R-2, MYBS3, OsMYB2, MYB96, and OsMYB30, were reported for their roles in cold stress regulation (32–40), but their molecular mechanisms have not been clearly revealed. Therefore, further studies should be conducted to test whether these MYB transcription factors or their homologous proteins bind to this MYB cis-element to regulate cold response of HAN1 in rice.
Materials and Methods
Materials and Methods are described in SI Appendix, Materials and Methods. The original sequencing datasets have been deposited in the Genome Sequence Archive of Bejing Institute of Genomics, Chinese Academy of Sciences (bigd.big.ac.cn/gsa) under Accession numbers: CRA000778, CRA000779, and CRA000995.
Supplementary Material
Acknowledgments
We thank Dr. E Gonzales at the International Rice Research Institute for kindly providing cultivated rice and wild rice seeds. We thank Dr. David Zaitlin for language improvement. This work was supported by the National Natural Science Foundation of China (Grant 31371603) and partly supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant 2018398).
Footnotes
The authors declare no conflict of interest.
Data deposition: The original sequencing datasets have been deposited in the Genome Sequence Archive of Bejing Institute of Genomics, Chinese Academy of Sciences, bigd.big.ac.cn/gsa (accession nos. CRA000778, CRA000779, and CRA000995).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819769116/-/DCSupplemental.
References
- 1.Chang TT. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices. Euphytica. 1976;25:425–441. [Google Scholar]
- 2.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- 5.Wu W, et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc Natl Acad Sci USA. 2013;110:2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Y, et al. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat Commun. 2017;8:14788. doi: 10.1038/ncomms14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujino K, et al. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA. 2008;105:12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito K, Hayano-Saito Y, Kuroki M, Sato Y. Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 2010;179:97–102. [Google Scholar]
- 10.Liu F, et al. Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol Plant. 2013;6:757–767. doi: 10.1093/mp/sss161. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura J, et al. Rice homologs of inducer of CBF expression (OsICE) are involved in cold acclimation. Plant Biotechnol. 2011;28:303–309. [Google Scholar]
- 12.Zhao J, et al. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol J. 2017;15:1141–1148. doi: 10.1111/pbi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao N, et al. Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 2018;177:1108–1123. doi: 10.1104/pp.18.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C, et al. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun. 2018;9:3302. doi: 10.1038/s41467-018-05753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo J, Niu QW, Chua NH. Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]
- 16.Koo AJ, Cooke TF, Howe GA. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA. 2011;108:9298–9303. doi: 10.1073/pnas.1103542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Jiang L, Wang F, Yu D. Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell. 2013;25:2907–2924. doi: 10.1105/tpc.113.112631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, et al. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J Exp Bot. 2017;68:1361–1369. doi: 10.1093/jxb/erx004. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Ding Y, Yang S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23:623–637. doi: 10.1016/j.tplants.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Mao D, Chen C. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS One. 2012;7:e47275. doi: 10.1371/journal.pone.0047275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santino A, et al. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 2013;32:1085–1098. doi: 10.1007/s00299-013-1441-2. [DOI] [PubMed] [Google Scholar]
- 22.Chinnusamy V, et al. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, et al. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 25.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 26.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang DL, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Civáň P, Craig H, Cox CJ, Brown TA. Three geographically separate domestications of Asian rice. Nat Plants. 2015;1:15164. doi: 10.1038/nplants.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vannini C, et al. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004;37:115–127. doi: 10.1046/j.1365-313x.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- 33.Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008;27:1677–1686. doi: 10.1007/s00299-008-0587-9. [DOI] [PubMed] [Google Scholar]
- 34.Park MR, et al. Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ. 2010;33:2209–2230. doi: 10.1111/j.1365-3040.2010.02221.x. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal M, et al. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- 36.Ma Q, et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su CF, et al. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010;153:145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot. 2012;63:2541–2556. doi: 10.1093/jxb/err431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HG, Seo PJ. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015;82:962–977. doi: 10.1111/tpj.12866. [DOI] [PubMed] [Google Scholar]
- 40.Lv Y, et al. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-Amylase expression. Plant Physiol. 2017;173:1475–1491. doi: 10.1104/pp.16.01725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.