Significance
Basal-like/BRCA1-associated breast cancer (BC) is a very aggressive form of BC that frequently occurs in young women, with devastating effects. Since tailored therapies are lacking for this type of tumor, scientists and clinicians are searching for weaknesses that can be therapeutically exploited. Here we describe the role of the transcription factor aryl hydrocarbon receptor (AhR) in supporting BC growth by controlling reactive oxygen species (ROS) levels and the tumor-promoting features of the microenvironment. In BC cells, AhR activation mediates the link between intracellular ROS regulation and the protumorigenic functions of the surrounding immune system. We propose that tailored inhibition of AhR-regulated pathways can lead to BC eradication by pushing it beyond its ROS tolerance limit and depriving it of tumor-supporting immune cells.
Keywords: triple-negative breast cancer, aryl hydrocarbon receptor, reactive oxygen species, tumor-associated macrophages, amphiregulin
Abstract
Cancer cells have higher reactive oxygen species (ROS) than normal cells, due to genetic and metabolic alterations. An emerging scenario is that cancer cells increase ROS to activate protumorigenic signaling while activating antioxidant pathways to maintain redox homeostasis. Here we show that, in basal-like and BRCA1-related breast cancer (BC), ROS levels correlate with the expression and activity of the transcription factor aryl hydrocarbon receptor (AhR). Mechanistically, ROS triggers AhR nuclear accumulation and activation to promote the transcription of both antioxidant enzymes and the epidermal growth factor receptor (EGFR) ligand, amphiregulin (AREG). In a mouse model of BRCA1-related BC, cancer-associated AhR and AREG control tumor growth and production of chemokines to attract monocytes and activate proangiogenic function of macrophages in the tumor microenvironment. Interestingly, the expression of these chemokines as well as infiltration of monocyte-lineage cells (monocyte and macrophages) positively correlated with ROS levels in basal-like BC. These data support the existence of a coordinated link between cancer-intrinsic ROS regulation and the features of tumor microenvironment. Therapeutically, chemical inhibition of AhR activity sensitizes human BC models to Erlotinib, a selective EGFR tyrosine kinase inhibitor, suggesting a promising combinatorial anticancer effect of AhR and EGFR pathway inhibition. Thus, AhR represents an attractive target to inhibit redox homeostasis and modulate the tumor promoting microenvironment of basal-like and BRCA1-associated BC.
Cancer cells have a highly dynamic and heterogeneous metabolism that enables them to generate energy, maintain redox homeostasis, and undertake biosynthesis (1, 2). In addition, cancer metabolism has the ability to influence the communication of the tumor cells with nearby immune cells by controlling the nutrient status of the surrounding tumor microenvironment (TME) (3–6). Hence, the study of cancer-associated metabolic alterations has presented attractive therapeutic opportunities in several preclinical models of cancers, including breast, colorectal, and lung cancer (7–9).
Among all forms of breast cancer (BC), the basal-like (commonly being triple-negative based on defined markers; TNBC) is generally more aggressive, is of poor prognosis, and frequently appears in women carriers of mutations in the tumor suppressor BRCA1. More and more evidence supports the idea that the study of TNBC dysregulated metabolism will lead to efficacious therapeutic approaches against this aggressive disease (10). Compared with other BC subtypes, these cancers have increased glutamine consumption and heightened sensitivity to glutamine depletion (11). Moreover, in addition to BRCA1 mutations, this subtype harbors loss-of-function mutations in Tp53 tumor suppressor, which together promote antioxidant responses (12, 13). Therefore, basal-like BC tends to accumulate higher levels of reactive oxygen species (ROS) because of its genetic and metabolic alterations.
Here we found that human BC with low expression or inactivation of BRCA1 specifically expresses aryl hydrocarbon receptor (AhR), a ligand-activated transcription factor that regulates the expression of a large superfamily of antioxidant molecules known as cytochrome p450 proteins (CYP1A1, CYP1A2, and CYP1B1) (14). In normal and malignant mammary cells, AhR activity is triggered by ROS induced by glutathione deprivation or absence of functional NRF2 antioxidant function. In the same conditions, AhR directly promotes the expression of amphiregulin (AREG), a ligand of the epidermal growth factor receptor (EGFR).
Using in vitro and in vivo models of basal-like/TNBC, we demonstrate that AhR−AREG signaling pathway positively supports tumorigenesis by controlling ROS and shaping the protumorigenic functions of TME. Furthermore, chemically and genetically induced AhR loss of function sensitizes tumor cells to Erlotinib, an EGFR inhibitor, thus suggesting a promising combinatorial antitumor strategy for the treatment of TNBC.
Results
AhR Is Activated by ROS in Normal and Malignant Mammary Cells.
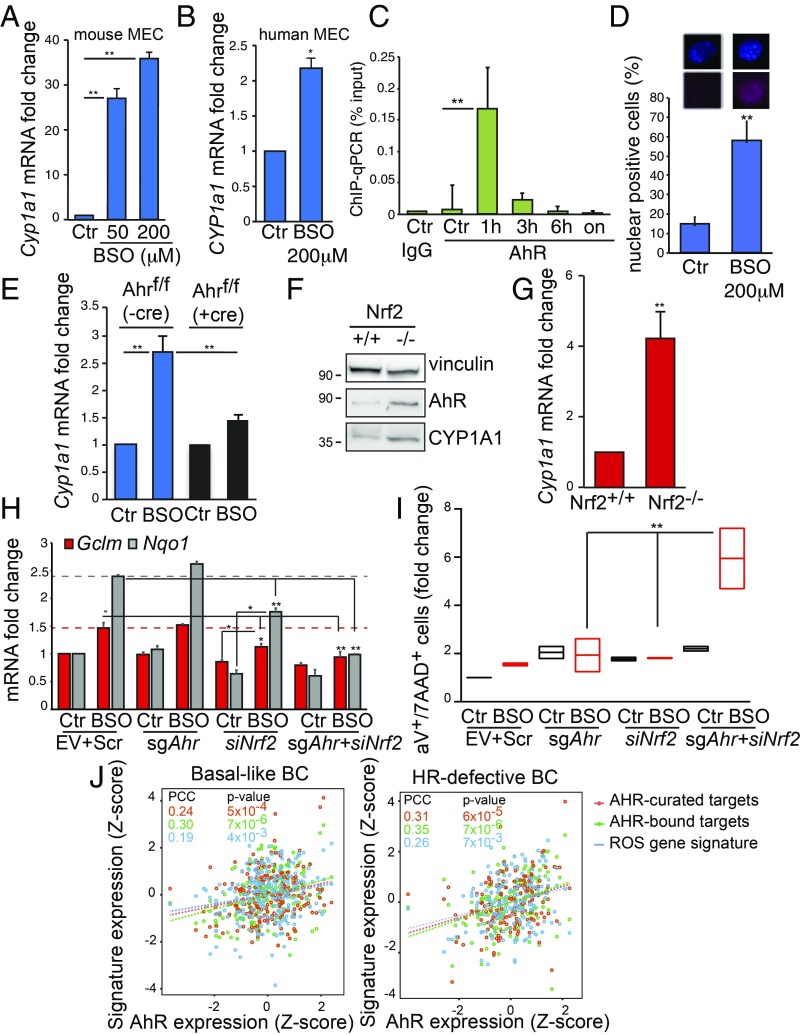
AhR redox activity has been mainly associated with the detoxification of xenobiotics and pollutants (15), while NRF2 has been mainly associated with the regulation of glutathione metabolism (2). However, studies of Ahr or Nrf2 knockout mice suggest a potential cross-talk between these factors in the maintenance of redox homeostasis (16). We found that long-term treatment of mouse and human mammary epithelial cells (MEC) with buthionine sulfoximine (BSO), a glutathione synthesis inhibitor (17), led to increased expression of AhR antioxidant target Cyp1a1 but did not affect Ahr mRNA levels (Fig. 1 A and B and SI Appendix, Fig. S1A). This was due to the ability of AhR to bind the Cyp1a1 promoter as shown by chromatin immunoprecipitation (ChIP) assay followed by qPCR in cells treated with BSO at different time points (Fig. 1C). Compared with IgG antibody control, AhR recruitment peaked at 1 h posttreatment, suggesting that BSO can trigger AhR transcriptional activity as rapidly as the well-characterized AhR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (18, 19). Immunocytochemistry assay showed a high frequency of cells positive for nuclear AhR after 2 h of exposure to BSO compared with control conditions (Fig. 1D and SI Appendix, Fig. S1B). To further test the specificity of AhR activation of Cyp1a1 by BSO, we isolated primary MEC from the mammary glands of female Ahr conditional knock-in mice (Ahrf/f) in which Cre recombinase excises exon_2 encoding the basic domain responsible for DNA binding (20). As expected, Ahr exon_2 expression was found to be relatively lower in Ahrf/f MEC infected with Cre-expressing adenovirus (SI Appendix, Fig. S1C). In these settings, BSO-induced up-regulation of Cyp1a1 was significantly abrogated (Fig. 1D).
Fig. 1.
AhR is activated by ROS in normal and malignant MEC. (A) Cyp1a1 mRNA levels in mouse MEC left untreated (Ctr) or treated with 50 and 200 μM BSO for 24 h. (B) CYP1A1 mRNA levels in human MCF10A cells left untreated (Ctr) or treated with 200 μM BSO for 24 h. (C) ChIP−qPCR assay to detect AhR on Cyp1a1 promoter in COMMA-1D cells treated with 200 µM BSO for indicated time points (n = 3 per group). ChIP with IgG antibody was used as a negative control. (D) Representative images of immunocytochemistry analysis of AhR nuclear staining in cells treated with BSO (200 µM for 2 h) or left untreated (Ctr). Bar graph shows the percentage of nuclear AhR positive cells (n = 100). Additional examples are reported in SI Appendix, Fig. S1B. (E) Cyp1a1 mRNA levels in MEC that were isolated from Ahrf/f mice, infected with Cre-expressing (+cre) or EV control (−cre) adenoviruses, and then treated or not with 50 µM BSO for 24 h (n = 3 per group). (F) Immunoblot showing AhR and CYP1A1 proteins in Nrf2+/+ or Nrf2−/− MEC. Vinculin is loading control. (G) Cyp1a1 mRNA levels in MEC isolated from Nrf2+/+ or Nrf2−/− female mice (n = 5 per genotype). (H) The mRNA analysis of NRF2-targets Gclm and Nqo1 in COMMA-1D cells that were transfected with sgRNA against mouse AhR (sgAhR) and siRNA oligos specific for mouse Nrf2 (siNrf2) and then subjected to BSO (200 µM) for 24 h. Cells manipulated with EV and nontargeting (scramble, scr) siRNA were used as control; n = 3 per group. (I) COMMA-1D cells were treated as in H, harvested 48 h posttreatment, and stained with annexinV/7-AAD apoptosis detection kit. (J) Positive association between AhR expression, AhR-curated targets, AhR-bound targets, and the “ROS gene signature” in basal-like and homologous recombination (HR) defective BC within the TCGA human BC dataset. See Materials and Methods for details. PCC, Pearson’s Correlation Coefficient. *P ≤ 0.05, **P ≤ 0.01.
The evidence that AhR could respond to the intracellular depletion of reduced glutathione prompted us to test the relationship between AhR and NRF2 in the control of ROS levels in normal and malignant MEC. Compared with MEC isolated from Nrf2 wild-type (Nrf2+/+) female mice, MEC from Nrf2 null (Nrf2−/−) mice did not express Nrf2 mRNA and accumulated both AhR and Cyp1a1 proteins (Fig. 1E and SI Appendix, Fig. S1D). These changes were associated with an increase in Cyp1a1 mRNA, while AhR levels were not affected (Fig. 1F and SI Appendix, Fig. S1E). NRF2 bona fide target Hmox1 was down-regulated while Nqo1 mRNA was unaffected in Nrf2−/− compared with Nrf2+/+ cells (SI Appendix, Fig. S1 F and G). Then, the consequences of down-regulated Ahr and/or Nrf2 (separately or in combination) were assessed. Briefly, first, we deleted Ahr by cell transfection with single-guide (sg)RNA (sgAhr) followed by puromycin selection, and then we applied Nrf2 siRNA (siNrf2) for 1 d prior to BSO treatment. Control cells (Ctr) were left untreated, and additional controls were generated for Ahr and Nrf2 down-regulation by applying an empty sgRNA vector (EV) and a nontargeting (scramble, Scr) siRNA, respectively. Cells were collected at 24 and 48 h for RNA and apoptosis analyses, respectively. Nrf2 mRNA levels were low in siNrf2-transfected cells, compared with Scr control (SI Appendix, Fig. S1H). Cyp1a1 expression was specifically affected by sgAhR in both untreated (Ctr) and BSO-treated cells (SI Appendix, Fig. S1I). In EV+Scr cells, NRF2 targets Nqo1 and Gclm were properly up-regulated by BSO treatment within 24 h, while they were not affected in sgAhr samples and were marginally altered in siNrf2 cells. Low levels of both Ahr and Nrf2 dramatically decreased BSO-induced Nqo1 and Gclm levels (Fig. 1G). This resulted in a significant increase in apoptosis in Ahr/Nrf2-deleted cells as measured by annexinV/7-aminoactinomycin D (7-AAD) staining (Fig. 1H).
We next examined whether AhR activation could also be a marker of oxidative stress in human basal-like/TNBC. The expression of AhR and its canonical targets, CYP1A1 and CYP1B1, were found to be higher in BC with genetic mutations (Van’t Veer dataset) or low expression (TCGA cohort) of BRCA1 gene (SI Appendix, Fig. S1 J and K). Through bioinformatics analysis of TCGA data of basal-like BC and BC with homologous recombination DNA repair defects (HR-defective BC; see Materials and Methods for additional details), we found that expression of AhR gene and two AhR-regulated gene sets positively correlated with an oxidative stress gene expression signature (Fig. 1I) (21). Together, these data indicate that both NRF2 and AhR may act as sensors of oxidative stress in normal and malignant MEC.
ROS-Regulated AhR Controls Expression of the EGFR Ligand AREG.
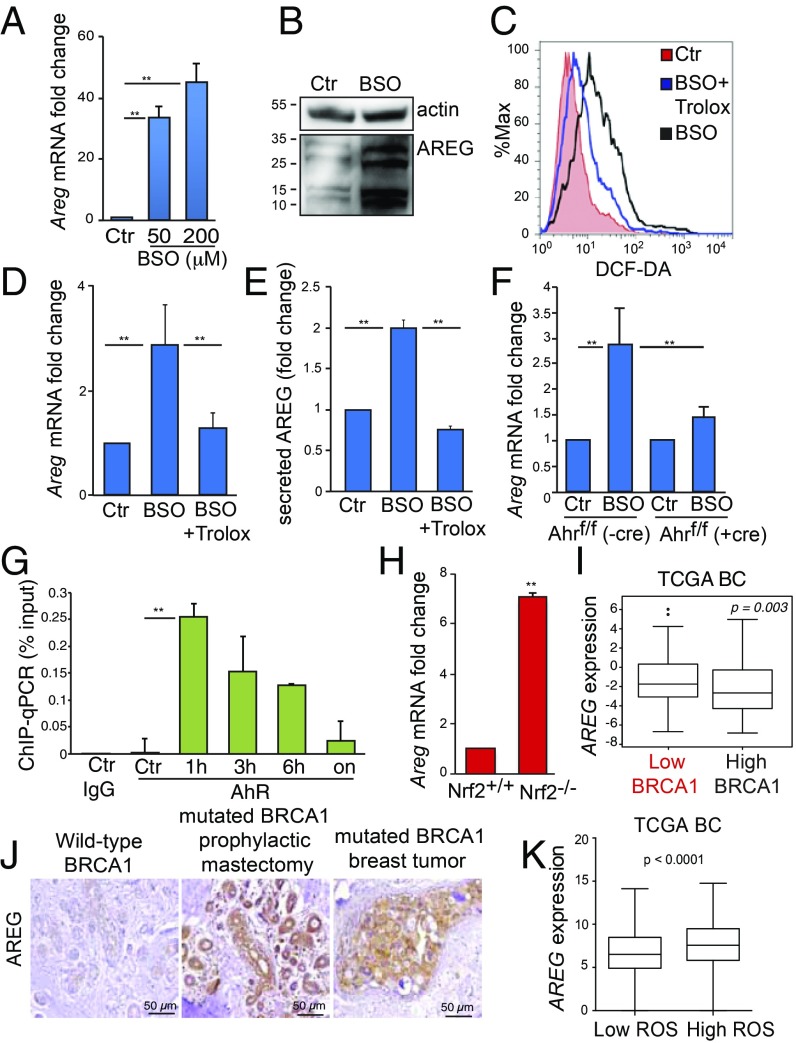
Cells use nontoxic levels of ROS to activate specific signaling pathways that regulate proliferation and malignant transformation (22). Furthermore, some studies have reported a correlation between xenobiotic-induced AhR activation and high levels of the EGFR ligand, AREG (23, 24). Therefore, we tested the hypothesis that AhR could modulate the EGFR pathway in conditions of oxidative stress in addition to an antioxidant response. In primary mouse MEC and in nontumorigenic human breast epithelial cells (MCF10A), BSO greatly induced AREG protein levels (Fig. 2 A and B and SI Appendix, Fig. S2A).
Fig. 2.
AREG expression is regulated by ROS-activated AhR and is elevated in BRCA1-associated BC. (A) Areg mRNA levels in mouse MEC that were left untreated (Ctr) or treated with the indicated doses of BSO for 24 h (n = 3 per group). (B) Immunoblot showing AREG protein in mouse MEC that were left untreated (Ctr) or treated with 200 µM BSO for 24 h. Vinculin is loading control. (C) Representative FACS profile of ROS levels in mouse cells treated with 200 µM BSO, with or without 250 µM Trolox and stained with DCF-DA after 24 h. (D) Areg mRNA in mouse cells treated as in C (n= 3 per group). (E) ELISA measurement of secreted AREG protein in culture medium of mouse cells treated as in C (n = 5 per group). (F) Areg mRNA levels in MEC that were isolated from Ahrf/f mice, infected with Cre-expressing (+cre) or EV control (−cre) adenoviruses, and treated or not with 50 µM BSO for 24 h (n = 3 per group). (G) ChIP−qPCR assay to detect AhR on Areg promoter in COMMA-1D cells treated with 200 µM BSO and harvested at the indicated time points (n = 3 per group). ChIP with IgG antibody was used as a negative control. (H) Areg mRNA in MEC isolated from Nrf2+/+ or Nrf2−/− virgin female mice (n = 5 per genotype). (I) AREG mRNA levels in TCGA BC grouped according to low (bottom tertile) or high (top tertile) BRCA1 expression (n = 1,102). (J) Representative images of IHC to detect AREG protein in samples from a BRCA1 wild-type reduction mammoplasty from healthy women used as controls, a prophylactic mastectomy in a woman heterozygous for a BRCA1 mutation (BRCA1 mutant), and a human BRCA1 mutant basal/TNBC breast tumor. (K) Expression levels of AREG in basal-like BC with low ROS or high ROS based on the ROS gene signature. **P ≤ 0.01.
EGFR is a member of a large family of receptor tyrosine kinases that also includes HER2 (ERBB2/NEU), ERBB3, and ERBB4. All these receptors promote intracellular signaling in the form of homodimers or heterodimers and upon binding to a large spectrum of soluble ligands including EGF, epiregulin, AREG, epigen, neuregulin (NRG1/2/3/4), transforming growth factor alpha, and Heparin-binding EGF-like growth factor (HB-EGF) (25). In different cancer types, Erbb receptors and ligands are differentially regulated during tumorigenesis and influence tumor progression and response to therapies (26, 27).
To verify the specificity of Areg regulation by ROS in MEC, we assessed the expression of different Erbb ligands in mouse MEC treated with BSO. Of note, these cells mainly express Egfr, Erbb2, and Erbb3 receptors (SI Appendix, Fig. S2B). In these cells, among all known Erbb ligands, BSO mainly induced the expression of Areg (SI Appendix, Fig. S2C). Areg mRNA up-regulation by BSO appears to be ROS-mediated, since cotreatment of mouse MEC with the antioxidant Trolox abolished both BSO-induced ROS and the accumulation of this transcript (Fig. 2 C and D). Once translated, AREG is a membrane-bound protein whose activation is regulated by release of its extracellular domain from the membrane (28). Indeed, BSO treatment promoted AREG release into the culture medium of MCF10A cells in a Trolox-sensitive manner (Fig. 2E).
Next, we investigated whether AhR was involved in regulating AREG expression. MEC isolated from female Ahrf/f mice were infected with Cre-expressing adenovirus prior exposure to BSO. Thus, BSO-induced up-regulation of Areg was abrogated by loss of transcriptional activity of AhR (Fig. 2F). This suggested that, like Cyp1a1 (Fig. 1D), Areg might be a direct AhR transcriptional target. Indeed, a putative XRE element (5′-G/T N T/G GCGTG A/C-3′) was identified at −260 bp from the ATG start codon. COMMA-1D cells were treated with BSO for different time points before ChIP−qPCR assay. Compared with IgG antibody control, AhR enrichment at Areg promoter started at 1 h posttreatment and gradually declined overtime (Fig. 2G).
It is worth noting that MEC from Nrf2−/− mice accumulated more Areg mRNA than MEC from Nrf2+/+ mice, ruling out that the transcript increase is regulated by NRF2 (Fig. 2H).
In the TCGA dataset, AREG expression was also found to be higher in BC with low levels of BRCA1 (Fig. 2I). Moreover, by immunohistochemistry assay (IHC), AREG protein expression was found to be significantly elevated in mammary preneoplastic tissues of BRCA1 mutation carriers and in the corresponding advanced tumors (Fig. 2J and SI Appendix, Fig. S2D). Consistently, in the TCGA BC dataset, AREG levels were also associated with a high ROS score (Fig. 2K). Therefore, AREG is a transcriptional target of AhR in MEC, and its expression correlates with AhR and ROS levels in human BC.
AhR−AREG Axis Is Required for BRCA1-Associated Mammary Tumorigenesis.
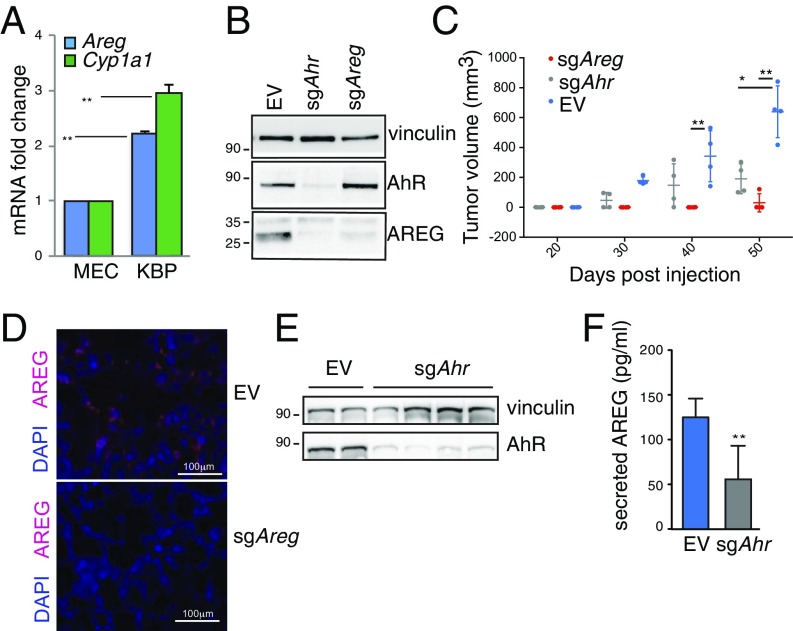
To characterize the functional involvement of the AhR–AREG axis in basal-like and BRCA1-associated tumors, we took advantage of a transplantable mouse primary mammary tumor cell line [K14cre BRCA1f/f p53f/f (KBP)] isolated from a mammary tumor arising in the K14cre Brca1f/f Trp53f/f basal-like/TNBC mouse model (29). Mammary tumors originating from KBP cells resemble spontaneous basal-like/TNBC (30). Compared with normal MEC, the NRF2 target Nqo1 was previously shown to be down-regulated in mouse and human Basal-like/TNBC tumors, as a consequence of defective NRF2 function (12). However, Nqo1 was still induced by exposure to BSO in these cells, suggesting a coordinated transcriptional control of this gene by NRF2 and AhR as shown in Fig. 1 (SI Appendix, Fig. S3A). AhR targets Cyp1a1 and Areg were highly expressed in KBP cells, compared with normal MEC (Fig. 3A). We then used CRISPR/Cas9 gene editing to delete mouse Ahr and Areg in mammary tumor cells by transient transfection. KBP cells with sgRNA against Ahr (sgAhr) or Areg (sgAreg) were maintained under selection for 3 d before analysis in vitro or transplantation in vivo. The sgAreg and sgAhr treatments of KBP cells did not affect their proliferation prior to transplantation (SI Appendix, Fig. S3B) but did induce a significant decrease in AREG and AhR proteins compared with cells transfected with control empty vector (EV) (Fig. 3B). Notably, Ahr deletion also reduced AREG protein, confirming that Areg is an AhR downstream target (Fig. 3B).
Fig. 3.
AhR and AREG support tumorigenesis in a mouse model of BRCA1-associated BC. (A) Levels of Areg and Cyp1a1 mRNAs in mouse MEC and KBP cells (n = 3 per group). (B) Immunoblot showing AhR and AREG proteins in lysates from KBP tumors transfected with EV and sgRNAs against mouse AhR (sgAhr) or Areg (sgAreg) and selected in puromycin-containing media for 3 d. Vinculin is loading control. (C) Representative plot of tumor volume increase over time in FVB female mice transplanted with KBP cells expressing EV, sgAhr, or sgAreg (n = 4 per group). (D) Representative immunofluorescence staining of nuclei (DAPI) and AREG protein (red) in EV and sgAreg KBP tumors. (E) Immunoblot showing AhR protein in lysates from EV and sgAhr KBP tumors. Vinculin is loading control. (F) ELISA measurement of AREG protein in lysates from EV and sgAhr KBP tumors. *P ≤ 0.05, **P ≤ 0.01.
Next, we transplanted EV-, sgAreg-, or sgAhr-transfected KBP cells into the mammary fat pads of virgin female mice and monitored tumor growth until tumors from EV-treated KBP cells reached humane endpoint (tumor volume = 1 cm3). Tumor development in mice receiving either sgAreg- or sgAhr-transfected KBP cells was significantly reduced compared with animals receiving EV-transfected KBP cells (Fig. 3C and SI Appendix, Fig. S3C). Areg deletion almost completely prevented the expansion of KBP tumor cells in vivo, possibly as a result of a cell-autonomous requirement for AREG in these cells. Indeed, Areg deletion in KBP cells impaired cell growth in vitro as shown by Sulforhodamine B (SRB) assay over a period of 12 d (SI Appendix, Fig. S3D).
We confirmed that small tumors growing from sgAreg-treated cells showed a significant reduction in AREG expression measured by IHC (Fig. 3D). The sgAhr-treated cells had almost undetectable expression of AhR that consequently affected AREG secretion (Fig. 3 E and F). Then, we tested whether AREG was the main ERBB ligand to be regulated by AhR in KBP tumors. RNA sequencing (RNA-seq) showed a similar expression profile of ERBB receptors, but higher levels of Areg, Hbegf, and Nrg1 ligands were observed in KBP tumors compared with normal mammary gland tissue (SI Appendix, Fig. S3E). However, neither Hbegf nor Nrg1 expression levels were affected by Ahr deletion compared with Areg, further underlying the existence of a specific AhR−AREG axis in these tumors (SI Appendix, Fig. S3F).
AhR−AREG Axis Regulates the Phenotype and Function of Macrophages in BRCA1-Deleted Mouse Mammary Tumors.
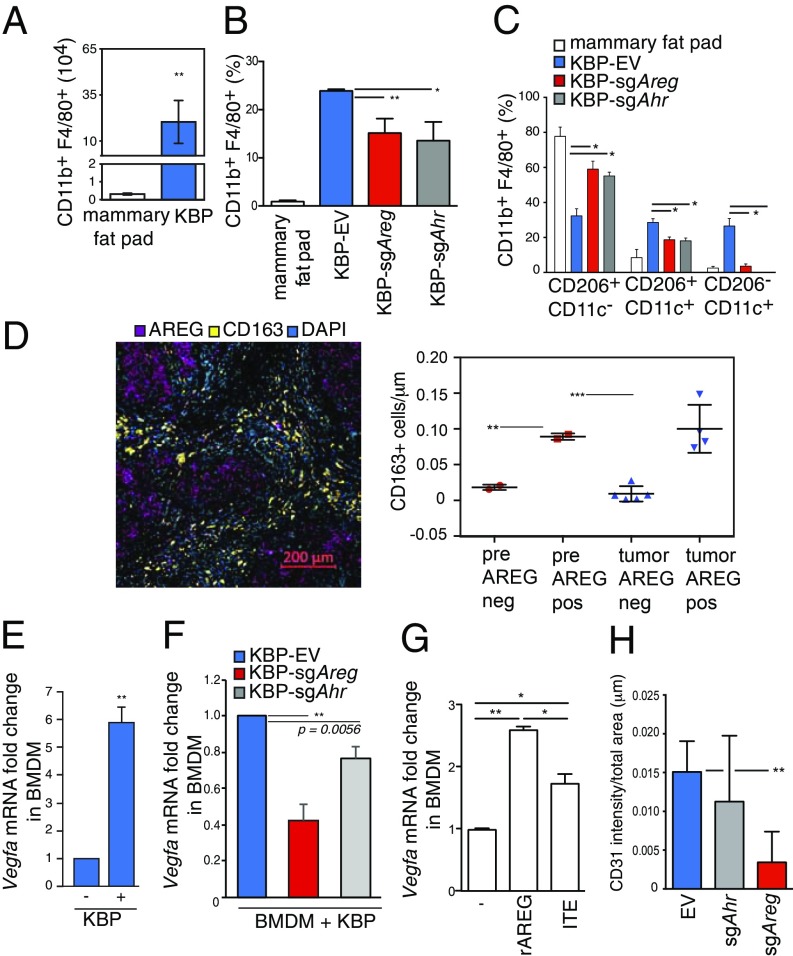
AhR and AREG are both expressed in innate and adaptive immune cell populations to regulate immunity, inflammation, and tissue repair (31, 32). However, apart from a few studies (33–35), the roles of these proteins in the TME are still uncertain. Macrophages are the most abundant immune cells recruited to the breast tumor site, where they become “tumor-associated macrophages” (TAM). TAM have complex genetic and molecular characteristics resulting in extraordinary plasticity and are particularly abundant in BC and present at all stages of progression (36). To examine the characteristics of TAM, we first analyzed macrophages resident in normal mammary tissue of virgin and nulliparous female mice. These cells typically expressed integrin αM chain (CD11b), EGF-like module-containing mucin-like hormone receptor-like 1 (F4/80), MER protooncogene tyrosine kinase (MerTK) and cluster of cifferentiation 64 (CD64) (37), as well as the mannose receptor C type 1 (MRC1/CD206) (SI Appendix, Fig. S4A), which is also expressed by TAM (SI Appendix, Fig. S5B) (38). KBP tumors showed a significant increase in CD11b+F4/80+ macrophages compared with mammary fat pad (Fig. 4A). These macrophages expressed EGFR phosphorylation at tyrosine 106, suggesting activation of EGFR in TAM as previously found in other tumor models (39, 40) (SI Appendix, Fig. S4C). Compared with control tumors, KBP tumors from sgAreg- or sgAhR-treated cells had fewer CD11b+F4/80+ macrophages with a surface marker profile of nontumorigenic, tissue-resident macrophage in the mammary fat pad (Fig. 4 B and C).
Fig. 4.
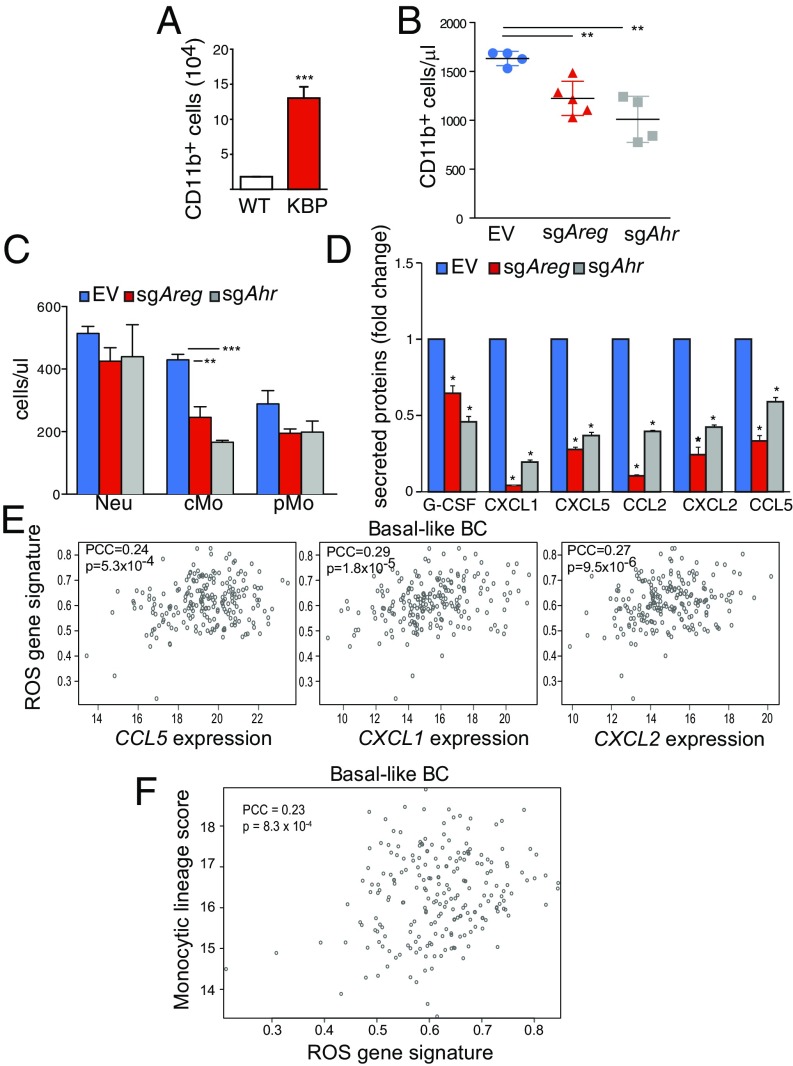
AhR−AREG axis influences the number and function of TAM. (A) Absolute numbers of CD11b+ F4/80+ cells in normal mammary fat pad from wild-type virgin and nulliparous mouse females and KBP tumor-bearing female mice (n ≥ 5 per group). (B) Percentages of CD11b+ F4/80+ cells in the mammary fat pad and EV, sgAhr, or sgAreg KBP tumor-bearing mice (n ≥ 5 per group). (C) Percentages of the indicated subpopulations of CD11b+ F4/80+ macrophages in the mammary fat pad and EV, sgAhr, or sgAreg KBP tumor-bearing mice (n ≥ 5 per group). (D) (Left) Representative immunohistochemistry staining of CD163 (surface marker for human macrophages) and AREG staining in a human primary BRCA1-mutated BC. Nuclei were stained with DAPI. (Right) Total count of CD163-positive (CD163+) macrophages in AREG positive (AREG+) or negative (AREG−) areas in reduction mammoplastic (pre) or tumor tissues from BRCA1 mutant carriers. See Materials and Methods and SI Appendix, Fig. S4D for additional details. (E) Vegfa mRNA levels in BMDM cultured alone (−), or after coculture with KBP cells (+) for 24 h (n = 3 per group). (F) Vegfa mRNA levels in BMDM after coculture with KBP cells expressing EV, sgAhr, or sgAreg for 24 h (n = 3 per group). (G) Vegfa mRNA levels in BMDM that were left untreated (−) or treated with rAREG (50 ng/mL) or ITE (10 μM) for 24 h (n = 3 per group). (H) Quantification of CD31 staining intensity as an indicator of angiogenesis in EV, sgAhr, or sgAreg tumors (n ≥ 5 per group). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
Next, the relevance of TAM in BRCA1-deficient human BC was quantified. IHC staining for CD163 showed that tumor-associated AREG expression correlated with high density and close proximity of macrophages in breast preneoplastic tissues and tumors from BRCA1 mutant carriers (Fig. 4D and SI Appendix, Fig. S4D). Collectively, these data postulate a role of AhR−AREG signaling in attracting TAM into the breast TME.
One well-described function of TAM is to produce vascular endothelial growth factor A (VEGF-A), which facilitates angiogenesis and metastasis (41). In vitro coculture systems between KBP cells and bone marrow-derived macrophages (BMDM) showed that BMDM had a dramatic increase in Vegfa mRNA expression after being in contact with tumor cells (Fig. 4E). These changes contributed to an overall increase in the level of secreted VEGF-A protein in the culture medium of KBP−BMDM cocultures (SI Appendix, Fig. S4E). In contrast to BMDM, KBP cells maintained a high basal level of Vegfa mRNA that did not change after coculture (SI Appendix, Fig. S4F). VEGF-A production by BMDM was mainly AREG-dependent, since deletion of KBP-associated Areg strongly reduced Vegfa expression in BMDM in coculture systems (Fig. 4F). Furthermore, recombinant AREG (rAREG) significantly increased Vegfa mRNA levels in BMDM to a higher extent than AhR activation by 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) (Fig. 4G). We also discovered a significant increase in EGFR expression in BMDM after coculture with KBP cells, further supporting the ability of BMDM to respond to AREG-mediated signaling (SI Appendix, Fig. S4G). Collectively, these data support the ability of cancer-associated AhR and AREG expression to affect the density and tumor-supporting properties of TAM within mammary TME. Corroborating these in vitro findings, we found that sgAreg tumors had less CD31-positive endothelial cells, indicating a low degree of tumor vasculature (Fig. 4H).
AhR−AREG Axis Influences Myeloid Cell Recruitment in BRCA1-Deleted Mouse Mammary Tumors.
Normal mammary ductal genesis is characterized by the epithelial cell-dependent recruitment of monocytes, which mature in situ into macrophages that provide critical support for developing tissue (42). Similarly, during mouse BC tumorigenesis, monocytes can be recruited by developing mammary tumors, where they mature into protumoral TAM characterized by high CD11c expression (43). Furthermore, increase in peripheral blood monocytes is a key feature of human and mouse malignancies, which correlates positively with TAM density in human cancer (44). We found that KBP tumor-bearing animals contained increased numbers of CD11b+ monocytes in the peripheral blood (Fig. 5A). In contrast, monocytes were significantly reduced in the peripheral blood of sgAreg and sgAhr tumor-bearing mice (Fig. 5B). Further stratification of these cells revealed significantly reduced numbers of classical monocytes (CD11b+Ly6-ChiLy6-G−CX3CR1+) in the blood of sgAreg and sgAhr tumor mice, with no significant changes in neutrophils (CD11b+Ly6-CintLy6-G+CX3CR1−) or patrolling monocytes (CD11b+Ly6-Cint/loLy6-G− CX3CR1+) (Fig. 5C).
Fig. 5.
AhR−AREG axis regulates peripheral monocyte count in mammary tumor-bearing animals. (A) Absolute number of CD11b positive (CD11b+) monocytes in the peripheral blood of wild-type (WT) and KBP tumor-bearing mice (n = 5 per group). (B) Absolute number of CD11b+ monocytes in mice bearing mammary tumors derived from EV-, sgAhr-, or sgAreg-transfected KBP cells (n ≥ 5 per group). (C) Absolute numbers of the indicated subpopulations among CD11b+ cells in mice bearing EV, sgAhr, or sgAreg KBP tumors (n ≥ 5 per group). (D) Levels of indicated chemokines in cultured KBP cells expressing EV, sgAhr, or sgAreg vectors (n = 4 per group). (E) Positive correlation between CCL5, CXCL1, and CXCL2 mRNA levels and the ROS gene signature in the TCGA human basal-like BC dataset. See Materials and Methods for details. (F) Positive correlation between monocytic lineage cell count and the ROS gene signature in the TCGA human basal-like BC dataset. See Materials and Methods for details. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
The recruitment and activation of monocytes and macrophages are regulated by specific chemokines and cytokines in the TME (45). Compared with MEC, KBP cells released significantly higher levels of chemokines important for the recruitment and activation of monocytes and macrophages such as granulocyte-colony stimulating factor (G-CSF), chemokine (C-X-C motif) ligand 1 (CXCL1), C-X-C motif ligand 5 (CXCL5), C-C motif chemokine ligand 2 (CCL2), C-X-C motif ligand 2 (CXCL2), and C-C motif chemokine ligand 2 (CCL5) (SI Appendix, Fig. S5A) (46). Interestingly, sgAreg and sgAhr KBP tumor cells displayed significantly lower production of all chemokines elevated in KBP Ctr (Fig. 5D). Treatment of sgAhr tumor cells with rAREG considerably rescued the levels of G-CSF, CXCL1, CXCL5, CCL2, CXCL2, and CCL5 chemokines (SI Appendix, Fig. S6B). This result suggests that AhR expression affects chemokine production through AREG modulation. Mouse data were validated by analysis of human BC in the TCGA cohort. Expression of CXCL1, CXCL2, and CCL5 chemokines correlated positively with AhR and AREG expression and was high in BC with low BRCA1 levels (SI Appendix, Fig. S5C). Interestingly, these chemokines were also increased in basal-like BC with high ROS content (Fig. 5E), along with an elevated infiltration of monocytic lineage cells (monocytes and macrophages) in the TME (Fig. 5F). Our data show that AhR−AREG pathway stimulates the recruitment of monocytic cells in the TME, and these changes correlate with high levels of cancer-associated ROS in basal-like BC.
AhR−AREG Axis Is a Promising Therapeutic Target for Human BRCA1-Associated BC.
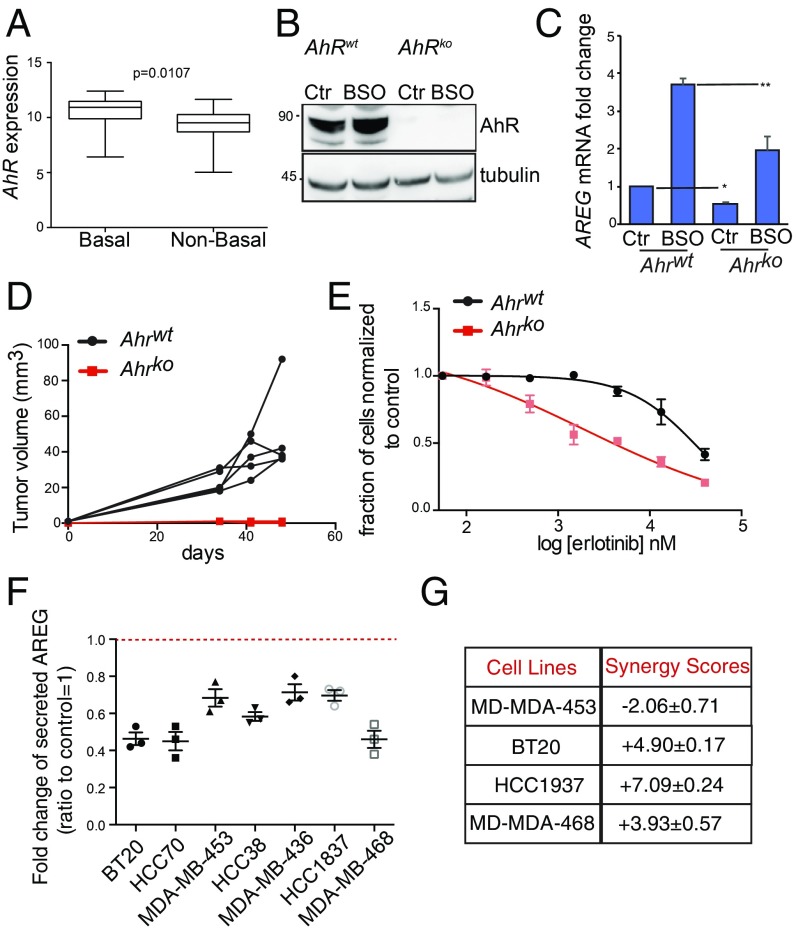
Then, we sought to test the oncogenic role of AhR in human BC as previously performed in the mouse model. As found in TCGA data analysis, AhR mRNA expression was more elevated in basal-like versus nonbasal-like BC cell lines (Fig. 6A). Next, MDA-MB-468 cells were CRISPR/Cas9-edited to obtain stable isogenic cell lines that were proficient (AhR wild-type, AhRwt) or deficient (AhR knock-out, AhRko) for AhR. Compared with AhRwt, AhRko cells did not express AhR, and, consequently, they had a significant low level of AREG and CYP1A1 expression both at basal level and upon exposure to BSO (Fig. 6 B and C and SI Appendix, Fig. S6A). In these cells, AhR deletion dramatically impaired their ability to both grow in vitro and form tumor in the fat pad of Nonobese diabetic/severe combined immunodeficiency (NOD/SCID) female mice (Fig. 6D and SI Appendix, Fig. S6B). Given the low levels of AREG mRNA in AhRko cells, we tested their sensitivity to Erlotinib, an EGFR tyrosine kinase inhibitor (47). AREG expression has been associated with resistance to EGFR inhibitors in breast, lung, and colon cancers (48–51). Compared with AhRwt tumor cells, AhRko cells were highly sensitive to EGFR inhibition as measured by a standard 5-d SRB assay (Fig. 6E).
Fig. 6.
AhR−AREG axis is a promising therapeutic target in basal-like and BRCA1-associated BC. (A) AhR expression levels in basal-like versus non−basal-like BC cell lines included in the Cancer Cell Line Encyclopedia. See Materials and Methods for details. (B) Immunoblot of MDA-MB-468 cell line carrying a wild-type (AhRwt) or deleted form of AhR (AhRko). Cells were left untreated (Ctr) or exposed to 500 μM BSO for 24 h. (C) AREG mRNA levels in cells treated as in B. (D) Representative plot of tumor volume increase over time after transplantation of MDA-MB-468 AhRwt and AhRko cells in the fat pad of immune-compromised NOD-SCID female mice. (E) Sensitivity of MDA-MB-468 AhRwt and AhRko cells to increasing doses of EGFR inhibitor, Erlotibin, as measured by SRB growth assay. (F) Levels of secreted AREG in the media of the indicated cell lines after treatment with AhRi (CH-223191) for 24 h and represented as ratio to their respective AREG levels in control (untreated) conditions; n = 3. (G) Synergy scores for CH-223191 (AhRi), and Erlotinib combinatorial treatment in the indicated BC cell lines as calculated using the SynergyFinder web application (see Materials and Methods for details, and see SI Appendix, Fig. S7). *P ≤ 0.05, **P ≤ 0.01.
The above results prompted us to evaluate the therapeutic value of interfering with AhR oncogenic function through the use of a potent and specific AhR inhibitor (AhRi), namely CH-223191 (52). Treatment of a subset of basal-like BC cell lines with 1 μM CH-223191 for 24 h significantly reduced secretion of AREG, independently of the variability of AREG basal level in these cells (Fig. 6F and SI Appendix, Fig. S6C). In both MDA-MB-468 and HCC1937, treatment with AhRi affected AREG secretion in a dose-dependent manner and in the absence of any external stimulus (SI Appendix, Fig. S6 D and E). Consistent with this finding, CH-223191-treated HCC-1937 cells showed a defect in the EGFR phosphorylation normally induced by incubation in nutrient-rich culture medium compared with untreated cells or cells exposed to Erlotinib (SI Appendix, Fig. S6F). These data suggest that AhR inhibition by a chemical compound can affect EGFR activation.
Given the effect of AhR inhibition on AREG levels and EGFR phosphorylation, we investigated whether the targeting of AhR might synergize with Erlotinib treatment to curtail BC cell growth. We seeded MDA-MB-453, BT20, MDA-MB-468, and HCC1937 cells in 96-well plates and treated them with various combinations of Erlotinib and CH-223191. Erlotinib was used at threefold serial dilution starting at 25 μM (five dilutions total), while CH-223191 was used at threefold serial dilution starting at 50 μM (nine dilutions total). First, we scored drug toxicity by calculating cell density using the SRB colorimetric assay. Then, we determined whether there was any synergy in therapeutic activity between CH-223191 and Erlotinib, using SynergyFinder (53). This analysis revealed a high degree of therapeutic synergy between Erlotinib and CH-223191 in cell lines with high EGFR expression (BT20, MDA-MB-468, and HCC1937) but an antagonistic effect where EGFR expression was low (MDA-MB-453) (Fig. 6G and SI Appendix, Fig. S7). These findings demonstrate a therapeutic potential of targeting both the cell-extrinsic (secreted AREG) and cell-intrinsic (intracellular AhR activation) components of the AhR−AREG axis for the treatment of BRCA1-associated BC.
Discussion
To date, most of the studies focused on AhR are linked to its role as environmental sensor for dioxins and xenobiotics. Recent work has elucidated a multitasking role of AhR in the control of cancer cell survival and tumor-associated immune system functions (54). The fact that AhR is chronically activated in many tumor types, including BC, supports the premise that AhR might be a promising drug target for anticancer therapies. However, the benefits of targeting AhR are still under debate given the contradictory observations that AhR is both protumorigenic and a tumor suppressor, and the multitude of activities elicited by different AhR ligands (14). In our work, analysis of human BC data and the use of human and mouse BC models supports an oncogenic role of AhR in basal-like and BRCA1-associated BC. In addition to controlling ROS, AhR stimulates transcription of the EGFR ligand AREG and thereby activates EGFR signaling in both normal and malignant MEC. Interestingly, among all known Erbb ligands, AREG is the main target of ROS-activated AhR pathway. Thus, AhR ensures cell survival and proliferation by coordinating an antioxidant response and activating the potent tumor-promoting signaling pathway mediated by EGFR. Indeed, AhR deletion by CRISPR/Cas9 gene editing dramatically impaired the in vivo growth of mouse and human TNBC cells.
Besides controlling cell intrinsic functions, AhR signaling influences the infiltration and phenotypic properties of macrophages in the TME. Tumor-infiltrating macrophages take on a trophic role that facilitates angiogenesis, extracellular matrix breakdown, and tumor cell motility, particularly in BC (41). Conversely, human BC cells can educate macrophages to adopt a tumorigenic and immunosuppressive phenotype that allows the BC cells to avoid immune surveillance and continue their invasion and growth (55). In our tumor model, we have found that AhR and its downstream effector AREG regulate a cluster of monocyte/macrophage-related chemokines that shape the immune landscape of BRCA1-deficient mammary tumors, resulting in an increase in CD11b+F4/80+CD206+ TAM with a tumorigenic phenotype. Indeed, our coculture of mouse BRCA1-deficient tumor cells with BMDM provides a clear demonstration of the mutual communication between macrophages and tumor cells in the expression of VEGF-A and the control of tumor angiogenesis. Analyses of TCGA human BC dataset have corroborated the correlation between myeloid-related chemokines (CCL5 and CXCL1/2) and expression levels of BRCA1, AhR, and AREG. Strikingly, the expression of these chemokines and the presence of myeloid populations are associated with basal-like BC with high ROS content.
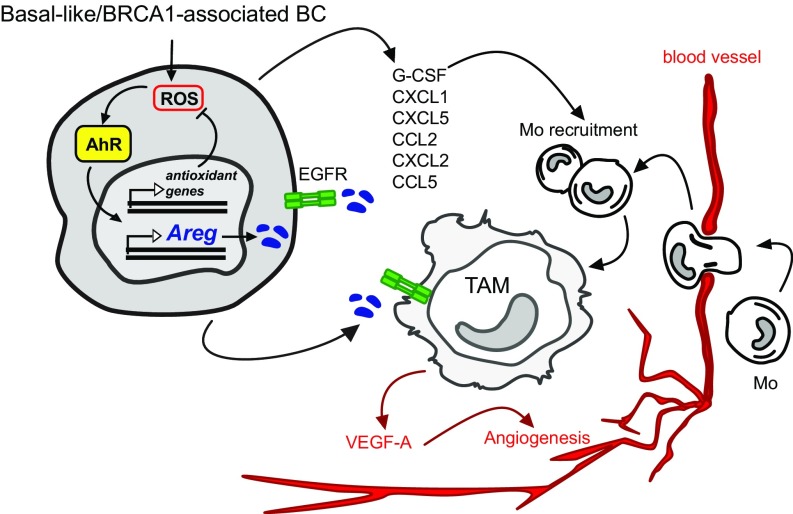
Overall, our observations suggest the following model (graphically summarized in Fig. 7). In basal-like/TNBC or BRCA1-associated BC, genetic and metabolic alterations may lead to chronic high ROS levels that trigger an increase in AhR protein levels and transcriptional activity. In these conditions, AhR activation counters ROS by promoting expression of antioxidant genes, but it also induces the expression of EGFR ligand AREG. Through the release of AREG and specific chemokines (G-CSF, CXCL1/2/5, and CCL2/5) in the TME, AhR activation axis may facilitate the recruitment of monocytes from blood vessels and the activation of protumorigenic and angiogenic TAM.
Fig. 7.
AhR−AREG axis defines a signaling pathway between cellular intrinsic redox mechanisms and surrounding TME. See Discussion for detailed description.
In conclusion, we have established a connection between tumor-intrinsic redox mechanisms and TME composition in BC. Our in vivo work using a Brca1/Trp53-deleted mouse model reveals how both these aspects are prerequisites for tumor progression and maintenance. These observations provide valuable insights into the multifactorial oncogenic activity of AhR and may form the basis of a better-tailored future drug development against one of the most aggressive and challenging type of BC.
Materials and Methods
Mice.
KBP mice were provided by J. Jonkers, Netherlands Cancer Institute (NKI), Amsterdam, The Netherlands, and were on the Friend Virus B/NIH Jackson (FVB/NJ) background. KBP tumor cells were obtained and used for in vivo transplantation studies as described (30). NRF2−/− mice were kindly provided by P. Ohashi, Princess Margaret Cancer Centre, Toronto, and were on the C57/B6 background. AhRf/f mice were purchased from The Jackson Laboratory (stock no. 006203) and were on a mixed background. For mouse and human tumor transplantation studies, FVB and immune-deficient NOD-SCID recipient female mice were 8 to 10 wk old and were purchased from The Jackson Laboratory. All mice were maintained and handled according to Animal Use Protocols (AUP) 4599 and 985 that were approved and regularly revised by the Animal Care and Use Committee of the University Health Network, Toronto.
Human Samples.
Human breast tissue samples for the analysis of AREG expression and infiltration of macrophages were obtained from the Catalan Institute of Oncology, Barcelona. Women carrying BRCA1/2 mutations were recruited by the Genetic Counseling Unit at the Catalan Institute of Oncology, Barcelona. The collection of human tissue samples was approved by the Ethics Committee at Institut d’Investigació Biomédica de Bellvitge (IDIBELL) Institute after obtaining written consent from all participants. See SI Appendix for further details regarding the use of these specimens.
Cell Lines and Treatments.
Mouse COMMA-1D cells (originally provided by S. Muthuswamy, Ontario Cancer Institute, Toronto), primary mouse MEC, and KBP cells were cultured in DMEM/F12 medium containing 10% FBS (Thermo Fisher Scientific), l-glutamine, 1 µg/mL of hydrocortisone (Sigma), 5 µg/mL of insulin (Sigma), and 5 ng/mL of epidermal growth factor (EGF; Sigma). Human MCF10A cells (ATCC) were cultured in DMEM/F12 medium supplemented with 5% horse serum (Thermo Fisher Scientific), 20 ng/mL of EGF, 0.5 µg/mL of hydrocortisone, 100 ng/mL of cholera toxin (Sigma), 10 µg/mL of insulin, and penicillin−streptomycin (Pen-Strep; Thermo Fisher Scientific). Human BC cell lines (ATCC) were cultured under strain-specific conditions according to ATCC recommendations.
Oxidative stress was induced for various times by exposing cells to medium containing 50 µM or 200 µM BSO (Sigma). For ROS scavenging, BSO-exposed cells were cotreated with 250 µM Trolox (EMD Millipore). MEC were starved in 0.5% FBS and nutrient-free medium for 24 h and then treated in the same medium with 50 ng/mL of rAREG from R&D Systems (262-AR-100) and harvested after 24 h. The AhR antagonist CH-223191 (Sigma) was applied to cell cultures at 1, 5, and 10 μM for 24 h (AREG measurement by ELISA), and 900 nM (analysis of EGF receptor phosphorylation in HCC1937) or different doses for 5 d (drug screening). The tyrosine kinase inhibitor, Erlotinib, was administrated to HCC1937 cell line at 600 nM for analysis of EGF receptor phosphorylation by Western blot.
Isolation of Primary Murine MEC.
Primary murine MEC were isolated from 8- to 10-wk-old virgin female mice as previously described (12). MEC were cultured in serum-free medium for 48 h to kill stromal fibroblasts and seeded (5 × 105) in six-well plates for experiments. In the case of AhRf/f mice, MEC (1 × 105) were seeded in six-well plates and infected overnight with prepackaged, ready-to-use adenovirus expressing Cre recombinase (Vector BioLabs). Cells were processed for analysis 48 h after infection.
Preparation of Murine BMDM.
Whole bone marrow was harvested from 10- to 12-wk-old female mice by flushing Hanks’ Balanced Salt Solution through femurs and tibias using a 27-gauge needle (BD Biosciences). Following red blood cell lysis, cells were cultured in 10% RPMI in 10-cm plates overnight. Nonadherent cells were collected and reseeded in Petri dishes in medium containing 20 ng/mL of murine macrophage colony-stimulating factor (M-CSF; Peprotech). After 3 d of culture, cells were provided with fresh medium containing 20 ng/mL of M-CSF. Macrophages were harvested on day 4.
For coculture experiments, BMDM (1 × 106) were seeded in triplicate in six-well plates and incubated with or without KBP cells (2.5 × 105 cells per well). Cells were harvested 48 h later using enzyme-free cell dissociation medium (Millipore) and either processed for flow cytometric analysis or sorted as described in Flow Cytometry and Cell Sorting.
Mouse and Human Tumor Induction and Treatment.
KBP (3 × 105) or MDA-MB-468 (0.5 × 106) cells were transplanted into #4 mammary gland fat pads of syngeneic FVB or NOD-SCID female mice (10 wk old). Diameters of developing tumors were measured in duplicate using digital calipers starting on day 14 (KBP) or day 30 (MDA-MB-468) posttransplantation when tumors became palpable. Tumor volume (in cubic millimeters) was calculated as 1/2(width2 × height). Tumor diameters were measured and volumes calculated as above two times per week.
Mouse Mammary Tumor Dissociation for FACS Analysis.
Tumors were resected from #4 mammary fat pads of transplanted mice, cut into 2- to 3-mm2 pieces, and placed into a C-tube (Miltenyi Biotech) containing 5 mL of Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS, 2 mM l-glutamine, 100 U/mL of penicillin, 100 μg/mL of streptomycin, 0.05 mM β-mercaptoethanol, 0.26 U/mL of Liberase TM (Sigma), and 20 U/mL of DNase I (Sigma). Tumors were mechanically processed using a gentleMACS Octo Dissociator with Heaters (Miltenyi Biotech). Processed samples were filtered once through a 100-µm cell strainer (Falcon), and the corresponding C tubes were rinsed with 5 mL of cold IMDM and passed through the same strainer. Cells were filtered once using a 70-µm strainer (Falcon), followed by a 40-µm strainer (Falcon). Filtered samples were collected in 15-mL Falcon tubes and centrifuged at 1,250 RPM for 8 min at 4 °C. Pellets were incubated with red blood cell lysis buffer for 7 min at room temperature (RT), and then centrifuged at 278 × g for 8 min at 4 °C before resuspension in PBS−/− containing 1% BSA plus 2 mM EDTA. Cell suspensions were subjected to fluorescence-activated cell sorting (FACS)/flow cytometry as described in Flow Cytometry and Cell Sorting.
Flow Cytometry and Cell Sorting.
Flow cytometric analyses of TAM and BMDM were performed using the following Abs: anti-CD49f-AF488 GoH3, anti-CD45.1-AF700 A20, anti-CD11b-Pacific Blue M1/70, anti-F4/80-PE BM8, anti-CD206-APC C068C2, anti-CD11c-APCCy7 N418, and anti-MHCII-PECy7 M5/114.15.2 (all from BioLegend). For FACS experiments, macrophages were identified as CD49flo/−CD45+CD11b+F4/80hi and sorted to >95% purity into 10% RPMI at 4 °C to 8 °C. BMDM were further processed for RNA extraction (as describe in RT-PCR), while TAM were seeded into 96-well flat-bottom plates at 2 × 105 cells per well. For flow cytometric analysis of resident macrophages in mammary glands of naïve FVB mice, the additional Abs anti-MerTk-PE 108928 (R&D Systems) and anti-CD64-PE X54-5/7.1 (BioLegend) were applied.
For analysis of peripheral blood monocytes, blood (15 μL) was collected from the tail veins of live mice into a heparinized capillary tube, followed by transfer into a 5-mL polystyrene tube containing 100 μL of PBS−/− plus 20 mM EDTA. Peripheral blood samples were stained directly with anti-Ly6C-PE HK1.4, anti-Ly6G-APCCy7 1A8, and anti-F4/80-FITC BM8 (BioLegend) Abs in combination with the Abs described above.
All flow cytometry samples were blocked for a minimum of 10 min in 1:100 anti-CD16/CD32 2.4G2 (eBioscience) containing 1:200 DNase I (protease-free; Roche) before staining in PBS−/− containing 1% BSA plus 2 mM EDTA. After blocking, Abs were added at appropriate dilutions, and cells were stained for 30 min on ice. Dead cells were excluded by adding 5 μL of 7-AAD (BioLegend) during the last 10 min of surface staining. Cells were then washed and either sorted on an Astrios FACS Instrument (Beckman Coulter) or analyzed using a Fortessa Instrument (BD Bioscience) and FlowJo software (Tree Star, Inc.).
For flow analysis of phosphorylated EGFR, tumors were dissociated according to mouse mammary tumor dissociation method. Then 106 cells were suspended in 0.5 mL of PBS−/− and fixed with 0.5 mL of 4% formaldehyde (final concentration 2%) for 10 min at 37 °C. Cells were washed by centrifugation with PBS−/− containing 1% BSA and 2 mM EDTA prior staining with anti-CD49f (AF488 GoH3; 1/200), anti-CD45.1 (AF700 A20; 1/400), anti-CD11b (Pacific Blue M1/70; 1/400), and anti-F4/80-PE (BM8; 1/400) for 30 min on ice. Cells were then washed twice and permeabilized by adding ice-cold Perm Buffer II (BD) with gentle vortexing. Cells were incubated for 30 min on ice and washed twice. Cells were then suspended in 100 μL of primary phospho-EGF Receptor (Tyr1068) antibody (D7A5, 1:1,600; cell signaling) and incubated for 1 h on ice. Cells were washed twice and then resuspended in 100 μL of secondary antibody (goat anti-rabbit APC conjugated, 1/1000; Thermo Fisher Scientific) and incubated for 1 h on ice. Cells were washed twice and analyzed at Fortessa Instrument (BD Bioscience), and data were processed with FlowJo software (Tree Star, Inc.).
CRISPR/Cas9 Gene Editing.
For CRISPR/CAS9 gene-editing studies in mouse and human cells, sgRNAs were evaluated using two different online CRISPR design tools: Zhang Lab online CRISPR design tool (https://zlab.bio/guide-design-resources) and the Zinc Finger Consortium Tool (zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx). To minimize potential off-target mutations, we selected highly specific guide RNA sequences which were predicted to have zero potential off-targets, even after three mismatches in the 20 nucleotide sgRNA sequence. The following oligos were used to synthetize mouse guide target sequences: mouse AhR, forward primer 5′-CACCGCTAGCGTCAGCTACCTGA-3′ and reverse primer 5′-AAACRCAGGTAGCTGACGCTGAGC-3′; and mouse AREG, forward primer 5′-CACCGGTGGACTTGAGCTTTCTGT-3′ and reverse primer 5′-AAACACAGAAAGCTCAAGTCCACC-3′. For CRISPR/CAS9 gene editing of human AhR in MDA-MB-468, an sgRNA targeting human AhR exon1 (NC_000007.14) was designed. The following guide oligos were designed to express the sgRNA: forward primer 5′- CACCGTCACCTACGCCAGTCGCAAG -3′ and reverse primer 5′- AAACCTTGCGACTGGCGTAGGTGAC -3′. In all cases, the annealed double-stranded guide oligo was cloned into the BbsI cut puromycin-modified version of vector pX330 (Addgene plasmid #42230). To obtain a stable MDA-MB-468 line carrying AhR deletion, the px330-PURO hAhR-sgRNA plasmid vector was transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific), followed by brief selection pressure in 1 μg/mL of puromycin for 48 h, and isolation of resistant individual clonal cell lines after 2 wk. A 748-bp genomic PCR amplicon spanning the human AhR exon1 CRISPR/CAS9 sgRNA target sequence was amplified using the forward primer 5′-CACGCCACTGTCCCGAGAGGACGCAGGTG- 3′ and reverse primer 5′-TATGAGCGCAACACAAAGCCAGTTGGTGG- 3′. Direct DNA sequencing of the human AhR exon1-spanning genomic DNA PCR amplicon was performed using the sequencing primers forward 5′- AGTGGTCCCAGCCTACAC -3′ and reverse 5′-GCTGTCAACAAATCAGGACC- 3′. Human AhR exon1 CRISPR/CAS9 frame-shift modifications on each allele were verified by analysis of DNA sequence chromatograms. Human and mouse AhR knock-out was further verified and validated using RT-PCR of downstream targets or Western blotting with AhR-specific antibody (BML-SA210-0025; Enzo Life Sciences).
The siRNA and sgRNA Cell Transfection.
COMMA-1D, KBP, and MDA-MB-468 cells (1 × 105) were seeded into six-well plates and transfected overnight with specific plasmids plus Lipofectamine 3000 (Life Technologies). For studies of AhR and NRF2 combinatorial down-regulation, COMMA-1D cells were first transfected with EV or EV containing AhR sgRNA, then kept in medium with 1 µg/mL of puromycin (Wisent Bio Products) for 2 d prior to transfection with 50 picomoles (pmol) mouse NRF2 siRNA (Thermo Fisher Scientific) KBP transfected cells were cultured in medium with 1 µg/mL of puromycin for 72 h before injection into #4 mammary fat pads of syngeneic female FVB mice. MDA-MB-468 cells were cultured and maintained in medium containing 1 µg/mL of puromycin until expansion of stable resistant clones.
Cell Proliferation Measurement.
KBP cells expressing EV and sgRNA against mouse Areg and AhR were analyzed for proliferation by using 488 EdU Click Proliferation Kit (BD Biosciences) according to manufacture’s guidelines.
Apoptosis Measurement.
Apoptosis was evaluated by Annexin V/7-AAD staining. In brief, cells were collected and stained with FITC-conjugated Annexin V and 7-AAD for 15 min at room temperature in 10× binding buffer. All reagents were purchased from BD Biosciences. Cells were analyzed by a FACSCalibur flow cytometer immediately after staining.
Cell Growth Measurement.
KBP cells were transfected with sgAhr, sgAreg, or EV as described in The siRNA and sgRNA Cell Transfection. Positive selection was applied using 1 μg/mL of puromycin for 48 h. Cells were then resuspended, counted, and plated in six-well plates. Cells were fixed at the indicated time points for subsequent SRB assay analysis. Time points were seeded in triplicate. Cell number was assessed indirectly by using the SRB colorimetric assay (Sigma) according to the manufacturer’s recommendations.
Drug Sensitivity Screening.
Human BC cell lines were seeded at different concentrations accordingly to the cell line to obtain 30% confluency in 96-well plates in triplicate. After 24 h, cells were treated with Erlotinib at threefold serial dilution starting at 25 μM (five dilutions total) and/or CH-223191 at threefold serial dilution starting at 50 μM (nine dilutions total). Cells were maintained in culture for 5 d before calculating cell density using the SRB colorimetric assay (Sigma) according to the manufacturer’s recommendations. Cell density was calculated using the SoftMax Pro software (Molecular Devices).
Immunoblotting.
Mouse and human MEC were collected posttreatment and lysed in radioimmunoprecipitation assay (RIPA) buffer. Protein content was measured using the Bio-Rad Protein Assay (Bio-Rad). Samples were resuspended in 4× Bolt LDS Sample Buffer (Life Technologies) and incubated at 70 °C for 5 min before loading on precast Bolt 4–12% Bis-Tris Plus gels (Life Technologies). Immunoblotting was performed using a standard protocol and primary Abs recognizing the following proteins: AhR (BML-SA210-0025; Enzo Life Sciences), vinculin (SPM227; Abcam), AREG (16036-1-IP; Proteintech), total EGF receptor (#4267; Cell Signaling), phospho-EGF receptor (Tyr1068) (#2234; Cell Signaling), alpha-tubulin (T5168; Sigma), and actin (A2066; Sigma-Aldrich). Primary Abs were visualized using anti-mouse and anti-rabbit ECL HRP-conjugated secondary Abs (Amersham). Membranes were developed for chemiluminescent detection, and images were acquired with GelCapture Software using MicroChemi 2.0/4.2 (FroggaBio).
ELISA and Cytokine Profiling.
Detection of mouse and human AREG in culture supernatants of mouse and human MEC and human BC cell lines was performed using the Mouse and Human Amphiregulin DuoSet ELISA Kit (R&D) according to manufacturer’s protocol. Absorbance was determined at 450 nm on a FlexStation 3 plate reader (Molecular Devices). Cytokine profiling was conducted using the Cytokine Array-Mouse Cytokine Antibody Array kit (Abcam) according to manufacturer’s instructions. Membranes were developed for chemiluminescent detection and images were acquired with GelCapture Software using MicroChemi 2.0/4.2 (FroggaBio).
RT-PCR.
RNA was isolated using the Nucleospin RNA Plus kit (Macherey-Nagel) and reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad) according to manufacturers’ instructions. Quantitative RT-PCR was performed using SYBR Green primers (Applied Biosystems). Mouse ribosomal protein S9 (rps9) and human ribosomal protein S18 (rps18) were used as housekeeping genes to determine relative mRNA expression. All primer sequences are described in SI Appendix, Table S1.
RNA-Seq.
Total RNA was isolated from MEC and KBP mammary tumors using the Nucleospin RNA Plus kit (Macherey-Nagel). Two micrograms of RNA were assessed for quality control using the Agilent Bioanalyzer before library construction. RNA deep sequencing was performed with the Illumina HiSeq 2000 sequencing system at Princess Margaret Genomic Centre, Toronto. Processed sequence data were obtained as .fastq files along with FASTQC data. The regularized log-normalized (rlog) expression values were plotted by transforming the count data to the log2 scale according to the method previously described by Love et al. (56).
ChIP.
ChIP was performed in COMMA-1D cells as previously described (57), with the following modifications: AhR protein (4 µg; Enzo Life Sciences) was prebound to Protein A and G Dynabeads (Life Technologies) for 6 h. DNA fragments were purified with the MinElute PCR purification kit (Qiagen) and processed for qPCR analysis according to the manufacturer’s protocol. Fold enrichment was calculated over input. The statistical significance of differences in enrichment was calculated using the unpaired Student t test. A complete list of PCR primers appears in SI Appendix, Table S2.
ROS Measurement.
To measure intracellular ROS, cells were incubated with 300 nM CM-H2DCFDA (DCF-DA, C6827; Invitrogen) for 10 min at 37 °C. DCF-DA fluorescence was analyzed by flow cytometry using a FACS Canto instrument (BD Biosciences) and FlowJo software.
Immunocytochemistry.
COMMA-1D were seeded onto glass coverslips in 12-well plates in triplicates (1 × 105 per well). Cells were treated with BSO at 200 μM for 1 h. Cells were fixed with paraformaldehyde (2% PAF) at room temperature for 10 min and then washed with PBS 1× twice. Cells were incubated with Blocking buffer containing 10% FBS and 0.05% Triton in PBS 1× at room temperature for 1 h. Primary antibody AhR (BML-SA210-0025; Enzo Life Sciences) was diluted at 1/100 in Blocking buffer and applied at room temperature for 1 h. Cells were washed with 2% FBS Blocking buffer three times prior to incubation with secondary antibody (goat anti-rabbit Alexa Fluor 568; Thermo Fisher Scientific) at 1/1,000 at room temperature for 1 h. Cells were washed with 2% FBS Blocking buffer three times and then stained with DAPI (0.5 μg/mL in PBS 1×) at room temperature for 5 min. Coverslips were rinsed in water and mounted with VectaShield (50 uL/coverslip; Vector Laboratories). Images were acquired with a fluorescence microscope (Zeiss AxioImager M1 equipped with Hamamatsu ORCA Flash4 camera) using Zeiss Zen software and processed with Adobe Photoshop CS5. Data were reported as percentage of cells with AhR positive nuclear staining per total number of cells (n = 100).
Statistical Analyses of Mouse and Human Cell Line Data.
Data were reported in bar graphs as the mean or median ± SEM, with P values calculated using Student t test (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001). The mean was calculated based on a minimum of n = 3 replicates in each experiment, and each experiment was performed at least three times. Data were analyzed by either Microsoft Excel or GraphPad Prism 7.
Supplementary Material
Acknowledgments
We thank Mary Saunders for scientific editing of the manuscript; Rob Cairns for helpful discussions; Thales Papagiannakopoulos for assistance with CRISPR/Cas9 gene editing experiments; and Arianna Sabo, Bruno Amati, and Mathieu Lupien for assistance in performing ChIP assays. This study was supported by Susan G. Komen Career Catalyst Research Grant 410005437 (to C. Gorrini), Canadian Institutes of Health Research (CIHR) Grant MOP-86707 (to T.W.M.), Foundation CIHR Grant FDN-143268 (to T.W.M.), Spanish Ministry of Health Institute of Health Carlos III (ISCIII) Grants PI15/00854 and PI18/01029 (to M.A.P.), Spanish Ministry of Science and Innovation Fondo Europeo de Desarrollo Regional (M.A.P.), Generalitat de Catalunya SGR-2017-449 and Centres de Recerca de Catalunya (CERCA) Program (M.A.P.), Asociación Española Contra el Cáncer Hereditary Cancer Grant (to C.L. and M.A.P.), University of Turin-Compagnia di San Paolo [Pancreatic Cancer Therapy (PANTHER) grant] (to P.C.), Cancer Research Institute (CRI) Irvington Postdoctoral Fellowship (to C.B.), and Wellcome Trust 101067/Z/13/Z (to J.W.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815126116/-/DCSupplemental.
References
- 1.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 2.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 3.Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munn DH, Mellor AL. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013;34:137–143. doi: 10.1016/j.it.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi N, et al. Cancer cells co-opt the neuronal redox-sensing channel TRPA1 to promote oxidative-stress tolerance. Cancer Cell. 2018;33:985–1003.e7. doi: 10.1016/j.ccell.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Port J, et al. Colorectal tumors require NUAK1 for protection from oxidative stress. Cancer Discov. 2018;8:632–647. doi: 10.1158/2159-8290.CD-17-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross MI, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 10.Long JP, Li XN, Zhang F. Targeting metabolism in breast cancer: How far we can go? World J Clin Oncol. 2016;7:122–130. doi: 10.5306/wjco.v7.i1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timmerman LA, et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorrini C, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med. 2013;210:1529–1544. doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks OD, Vousden KH. Metabolic regulation by p53. J Mol Med (Berl) 2011;89:237–245. doi: 10.1007/s00109-011-0735-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: Friend and foe. Nat Rev Cancer. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 16.Hayes JD, Dinkova-Kostova AT, McMahon M. Cross-talk between transcription factors AhR and Nrf2: Lessons for cancer chemoprevention from dioxin. Toxicol Sci. 2009;111:199–201. doi: 10.1093/toxsci/kfp168. [DOI] [PubMed] [Google Scholar]
- 17.Griffith OW, Meister A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (S-n-butyl homocysteine sulfoximine) J Biol Chem. 1979;254:7558–7560. [PubMed] [Google Scholar]
- 18.Luecke-Johansson S, et al. A molecular mechanism to switch the aryl hydrocarbon receptor from a transcription factor to an E3 ubiquitin ligase. Mol Cell Biol. 2017;37:e00630-16. doi: 10.1128/MCB.00630-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, et al. Constitutive regulation of CYP1B1 by the aryl hydrocarbon receptor (AhR) in pre-malignant and malignant mammary tissue. J Cell Biochem. 2008;104:402–417. doi: 10.1002/jcb.21630. [DOI] [PubMed] [Google Scholar]
- 20.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci USA. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang YY, et al. Gene expression after treatment with hydrogen peroxide, menadione, or t-butyl hydroperoxide in breast cancer cells. Cancer Res. 2002;62:6246–6254. [PubMed] [Google Scholar]
- 22.Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du B, Altorki NK, Kopelovich L, Subbaramaiah K, Dannenberg AJ. Tobacco smoke stimulates the transcription of amphiregulin in human oral epithelial cells: Evidence of a cyclic AMP-responsive element binding protein-dependent mechanism. Cancer Res. 2005;65:5982–5988. doi: 10.1158/0008-5472.CAN-05-0628. [DOI] [PubMed] [Google Scholar]
- 24.John K, Lahoti TS, Wagner K, Hughes JM, Perdew GH. The Ah receptor regulates growth factor expression in head and neck squamous cell carcinoma cell lines. Mol Carcinog. 2014;53:765–776. doi: 10.1002/mc.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roskoski R., Jr The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. doi: 10.1016/j.phrs.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Bublil EM, Yarden Y. The EGF receptor family: Spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol. 2007;19:124–134. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Kruspig B, et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci Transl Med. 2018;10:eaao2565, and erratum (2018) 10:eaav9152. doi: 10.1126/scitranslmed.aao2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sternlicht MD, Sunnarborg SW. The ADAM17-amphiregulin-EGFR axis in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2008;13:181–194. doi: 10.1007/s10911-008-9084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorrini C, et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci USA. 2014;111:4472–4477. doi: 10.1073/pnas.1324136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julliard W, Fechner JH, Mezrich JD. The aryl hydrocarbon receptor meets immunology: Friend or foe? A little of both. Front Immunol. 2014;5:458. doi: 10.3389/fimmu.2014.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaiss DMW, Gause WC, Osborne LC, Artis D. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 2015;42:216–226. doi: 10.1016/j.immuni.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 34.Platten M, von Knebel Doeberitz N, Oezen I, Wick W, Ochs K. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front Immunol. 2015;5:673. doi: 10.3389/fimmu.2014.00673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaiss DM, et al. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity. 2013;38:275–284. doi: 10.1016/j.immuni.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noy R, Pollard JW. Tumor-associated macrophages: From mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakubzick C, et al. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Lanaya H, et al. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol. 2014;16:972–977. doi: 10.1038/ncb3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivatsa S, et al. EGFR in tumor-associated myeloid cells promotes development of colorectal cancer in mice and associates with outcomes of patients. Gastroenterology. 2017;153:178–190.e10. doi: 10.1053/j.gastro.2017.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 42.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 43.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: Decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 46.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 47.Ali R, Wendt MK. The paradoxical functions of EGFR during breast cancer progression. Signal Transduct Target Ther. 2017;2:16042. doi: 10.1038/sigtrans.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kappler CS, et al. Oncogenic signaling in amphiregulin and EGFR-expressing PTEN-null human breast cancer. Mol Oncol. 2015;9:527–543. doi: 10.1016/j.molonc.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hobor S, et al. TGFα and amphiregulin paracrine network promotes resistance to EGFR blockade in colorectal cancer cells. Clin Cancer Res. 2014;20:6429–6438. doi: 10.1158/1078-0432.CCR-14-0774. [DOI] [PubMed] [Google Scholar]
- 50.Busser B, et al. Amphiregulin promotes BAX inhibition and resistance to gefitinib in non-small-cell lung cancers. Mol Ther. 2010;18:528–535. doi: 10.1038/mt.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa N, et al. Increases of amphiregulin and transforming growth factor-alpha in serum as predictors of poor response to gefitinib among patients with advanced non-small cell lung cancers. Cancer Res. 2005;65:9176–9184. doi: 10.1158/0008-5472.CAN-05-1556. [DOI] [PubMed] [Google Scholar]
- 52.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ianevski A, He L, Aittokallio T, Tang J. SynergyFinder: A web application for analyzing drug combination dose-response matrix data. Bioinformatics. 2017;33:2413–2415. doi: 10.1093/bioinformatics/btx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stockinger B, Di Meglio P, Gialitakis M, Duarte JH. The aryl hydrocarbon receptor: Multitasking in the immune system. Annu Rev Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 55.Sousa S, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015;17:101. doi: 10.1186/s13058-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bailey SD, et al. Noncoding somatic and inherited single-nucleotide variants converge to promote ESR1 expression in breast cancer. Nat Genet. 2016;48:1260–1266. doi: 10.1038/ng.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.