Significance
The mitochondrial uniporter is a calcium channel complex that is tightly regulated by the Ca2+-binding paralogous proteins MICU1 and MICU2, which work together to inhibit the channel. However, MICU1 and MICU2 are nonredundant and how each contributes to channel regulation is unclear. To better understand their mechanism, we solved the structure of MICU2. Based on comparison with MICU1, we propose a model in which the C-terminal helices of the MICU1/MICU2 monomers come together in the middle of the protein complex and may constitute the gating mechanism. The mechanism of uniporter channel regulation will be important for basic ion-channel biology and for elucidating the roles for mitochondrial calcium in disease, including the debilitating neuromuscular disorder of MICU1 deficiency.
Keywords: mitochondria, calcium, MICU2, EF-hand, MR-SAD
Abstract
The mitochondrial uniporter is a Ca2+-channel complex resident within the organelle’s inner membrane. In mammalian cells the uniporter’s activity is regulated by Ca2+ due to concerted action of MICU1 and MICU2, two paralogous, but functionally distinct, EF-hand Ca2+-binding proteins. Here we present the X-ray structure of the apo form of Mus musculus MICU2 at 2.5-Å resolution. The core structure of MICU2 is very similar to that of MICU1. It consists of two lobes, each containing one canonical Ca2+-binding EF-hand (EF1, EF4) and one structural EF-hand (EF2, EF3). Two molecules of MICU2 form a symmetrical dimer stabilized by highly conserved hydrophobic contacts between exposed residues of EF1 of one monomer and EF3 of another. Similar interactions stabilize MICU1 dimers, allowing exchange between homo- and heterodimers. The tight EF1–EF3 interface likely accounts for the structural and functional coupling between the Ca2+-binding sites in MICU1, MICU2, and their complex that leads to the previously reported Ca2+-binding cooperativity and dominant negative effect of mutation of the Ca2+-binding sites in either protein. The N- and C-terminal segments of the two proteins are distinctly different. In MICU2 the C-terminal helix is significantly longer than in MICU1, and it adopts a more rigid structure. MICU2’s C-terminal helix is dispensable in vitro for its interaction with MICU1 but required for MICU2’s function in cells. We propose that in the MICU1–MICU2 oligomeric complex the C-terminal helices of both proteins form a central semiautonomous assembly which contributes to the gating mechanism of the uniporter.
The mitochondrial calcium uniporter is an ancient calcium channel found in all major eukaryotic taxa, with dramatic lineage-specific diversification and losses (1). In animals the uniporter imparts mitochondria with a mechanism for rapid Ca2+ uptake into the matrix and plays a major role in coupling energy metabolism with cellular excitation events (2). The human uniporter is a multisubunit protein complex that consists of a pore-forming component MCU (3, 4), its apparently inactive paralog MCUb (5), a single transmembrane helix-containing subunit called EMRE that activates the channel (6), and two paralogous EF-hand Ca2+-binding proteins, MICU1 and MICU2 (7–11), that reside in the mitochondrial intermembrane space. A third MICU paralog (MICU3) appears to be a part of the uniporter in neuronal cells, where it is specifically expressed (8, 12). Although human MCU is the pore-forming subunit, it is not sufficient in reconstitution studies to transport Ca2+ and has a strict functional requirement for coexpression with the metazoan-specific protein EMRE (6, 13).
Recent studies utilizing a broad variety of techniques have led to significant advances in our understanding of MICU1/MICU2 function. Early seminal studies (14–16), performed before the uniporter components were discovered, demonstrated that the uniporter is regulated by extramitochondrial calcium. We now have strong evidence that MICU1 and MICU2 underlie this calcium regulation (7–10, 17, 18). Specifically, the two proteins work together to permit Ca2+ uptake by the uniporter only at concentrations exceeding a threshold Ca2+ level. Support for the joined function comes from the observation that mitochondrial Ca2+ uptake is blocked completely in cells in which either one of these proteins is locked in the off state by mutations in their EF-hands so they are unable to bind Ca2+ (7). In isolation, both MICU1 and MICU2 bind Ca2+ in a cooperative fashion with submicromolar affinities (17). They form homodimers in solution, which exchange upon mixing to form heterodimers. Curiously, the heterodimer exhibits even higher Ca2+-binding cooperativity, and with submicromolar affinity it is capable of sensing cytosolic Ca2+ signals directly (17).
The current model that emerges from these and other studies postulates that MICU1 and MICU2 inhibit the channel at resting cytosolic Ca2+ levels, and as Ca2+ concentration rises during cellular signaling events Ca2+ binding to the EF-hands of the MICU1/MICU2 heterocomplex relieves this inhibition, allowing Ca2+ uptake through the channel (2). The Ca2+ affinity of MICU1 and MICU2 determines the “threshold” Ca2+ required to allow transport through the uniporter. Due to the cooperativity of Ca2+ binding, the MICU1–MICU2 complex works effectively as the uniporter’s on–off switch (17).
Structures of components of the uniporter complex recently determined have started to hint at the mechanism of mitochondrial calcium uptake. The NMR structure of the pore-forming region of MCU from Caenorhabditis elegans has defined the basic structural features of the channel (19). More recently, several groups reported high-resolution cryo-EM and X-ray structures of MCU from fungi (20–23). In these structures the channel is made of four MCU monomers arranged into a dimer of dimers. The X-ray structure of MICU1 has also been determined, revealing that in the apo state the protein forms a somewhat irregular hexameric assembly that can be viewed as a trimer of homodimers, or, alternatively, as being built of two layers of trimers (24). The C-terminal α-helices of the MICU1 monomers are positioned in close proximity to each other in the center of a semiopen hexameric assembly. However, they do not appear to contribute significantly to the intermolecular contacts and are somewhat disordered in the crystal, having a different orientation for each of the monomers. The monomer of MICU1 is built of two lobes, each harboring one functional and one structural EF-hand. In the Ca2+-bound state MICU1 crystallized as a dimer, albeit the protein was lacking the C-terminal α-helix. Although different protein constructs were used for the apo and Ca2+-bound conditions, a significant change in the conformation upon Ca2+ binding could be inferred from comparison of the two structures (24). The change in conformation can be monitored by fluorescence intensity of Trp-substituted MICU mutants (17). The heterodimeric complex of MICU1 and MICU2 also displays a conformational change upon Ca2+ binding, as shown by monitoring hydrophobic surface area exposure using a fluorescent dye (17).
Although MICU1 and MICU2 are paralogous and reside within a heterodimeric complex, studies to date demonstrate their nonequivalence: The physiological function of MICU2 requires the presence of MICU1, whereas the ability of MICU1 to serve as a “gatekeeper” appears to be independent of MICU2 (7). Biochemical and genetic studies place MICU1 intermediate between MICU2 and the pore, such that loss of MICU2 still leads to a pore guarded by MICU1 (albeit at a different threshold), whereas loss of MICU1 leads to a fully unguarded pore. The amino acid sequence of MICU2 recapitulates several features of MICU1. It has an N-terminal predicted mitochondrial targeting sequence, two predicted functional EF-hand Ca2+-binding sites (8), and two putative structural EF-hand sites like MICU1. Despite only ∼28% sequence identity between MICU1 and MICU2, the two proteins must share some key structural features that allow them to exchange between homodimers and heterodimers and to respond to Ca2+ in a cooperative fashion. At the same time, they are clearly nonredundant functionally. We need more information on the structure of MICU2 to resolve this apparent paradox.
Here we report the X-ray structure of MICU2 in the inhibitory (Ca2+-free) state. The protein forms a dimer in which the monomers are related by approximate twofold rotational symmetry. The core structure of MICU2 is very similar to that of MICU1, both as a monomer and as a dimer. The interaction interface within the dimer involves hydrophobic contacts between the functional EF1 of one monomer and the structural EF3 of another monomer, which are conserved across MICUs and enable exchange of MICU1 and MICU2 homodimers into heterodimers. A distinctive feature of MICU2 is a long C-terminal helix structurally separated from the core domain. We find that this helix is critical for MICU2 function in cellular physiology, yet is not necessary for MICU2–MICU1 interaction in vitro. We show that a computational model of the MICU1–MICU2 complex can be generated upon substituting MICU2 structure for three MICU1 monomers within the previously published structure of the hexameric assembly of the apo MICU1 structure. We propose that C-helices contributed by both proteins form a semiautonomous assembly possibly constituting the gating mechanism of the uniporter complex.
Results
Structure of MICU2.
We crystallized the Mus musculus MICU2 (UniProt ID Q8CD10) devoid of the approximate predicted N-terminal mitochondrial targeting sequence. The construct expressed in Escherichia coli contains residues 68–432 of MICU2 and, at the N terminus, a short cloning sequence with a (His)6 tag. Similar to the human MICU2 (17), this purified mouse MICU2 protein construct forms a dimer in solution in the presence and absence of Ca2+ and is stabilized by Ca2+ (SI Appendix, Fig. S1 A–C). The Ca2+-free protein produced orthorhombic crystals (space group P21212) that diffracted to 2.5 Å. Based on the Matthews coefficient (3.04 Å3/Da) and the estimated solvent content (59.6%), we determined that the asymmetric unit contains two molecules of MICU2, consistent with our observation that the protein forms a dimer in solution. Our attempts at solving the structure by molecular replacement using the human MICU1 structure as a search model [Protein Data Bank (PDB) ID codes 4NSC and 4NSD], which has 28% sequence identity with MICU2 (SI Appendix, Fig. S2), were unsuccessful. Thus, to solve the structure, we crystallized the selenomethionine (SeMet) substituted variant of MICU2. These crystals diffracted to 4.0 Å (SI Appendix, Table S1). We solved the structure using a modified MR-SAD technique (Methods). Despite the low resolution of the SeMet dataset, the electron density map was of excellent quality, allowing unambiguous sequence assignment and model building (SI Appendix, Fig. S3). The initial model was refined against the 2.5-Å resolution native dataset to Rfree = 25% (Table 1). The final structure spans 92% of the polypeptide chain (PDB ID code 6EAZ) (25).
Table 1.
Data collection and refinement statistics for Ca2+-free MICU2
| Parameter | MICU2 6EAZ |
| Wavelength | 1.00 |
| Resolution range | 83.8–2.50 (2.59–2.50) |
| Space group | P 21 21 2 |
| Unit cell (a, b, c, α, β, γ) | 112.56 125.52 72.15 90 90 90 |
| Total reflections | 113,158 (8,563) |
| Unique reflections | 35,265 (3,319) |
| Multiplicity | 3.2 (2.6) |
| Completeness, % | 98.0 (93.9) |
| Mean I/sigma(I) | 14.9 (2.0) |
| Wilson B-factor | 50.89 |
| R-merge | 0.053 (0.45) |
| R-meas | 0.063 (0.56) |
| R-pim | 0.033 (0.32) |
| CC1/2 | 0.999 (0.872) |
| CC* | 1 (0.965) |
| Reflections used for R-free | 1,769 (155) |
| R-work | 0.20 (0.31) |
| R-free | 0.25 (0.34) |
| No. of nonhydrogen atoms | 5,601 |
| Macromolecules | 5,518 |
| Ligands | 48 |
| Solvent | 35 |
| Protein residues | 663 |
| RMS, bonds | 0.008 |
| RMS, angles | 1.18 |
| Ramachandran favored, % | 95.27 |
| Ramachandran allowed, % | 3.82 |
| Ramachandran outliers, % | 0.92 |
| Clashscore | 5.65 |
| Average B-factor | 67.54 |
| Macromolecules | 67.48 |
| Ligands | 78.45 |
| Solvent | 61.39 |
Statistics for the highest-resolution shell are shown in parentheses.
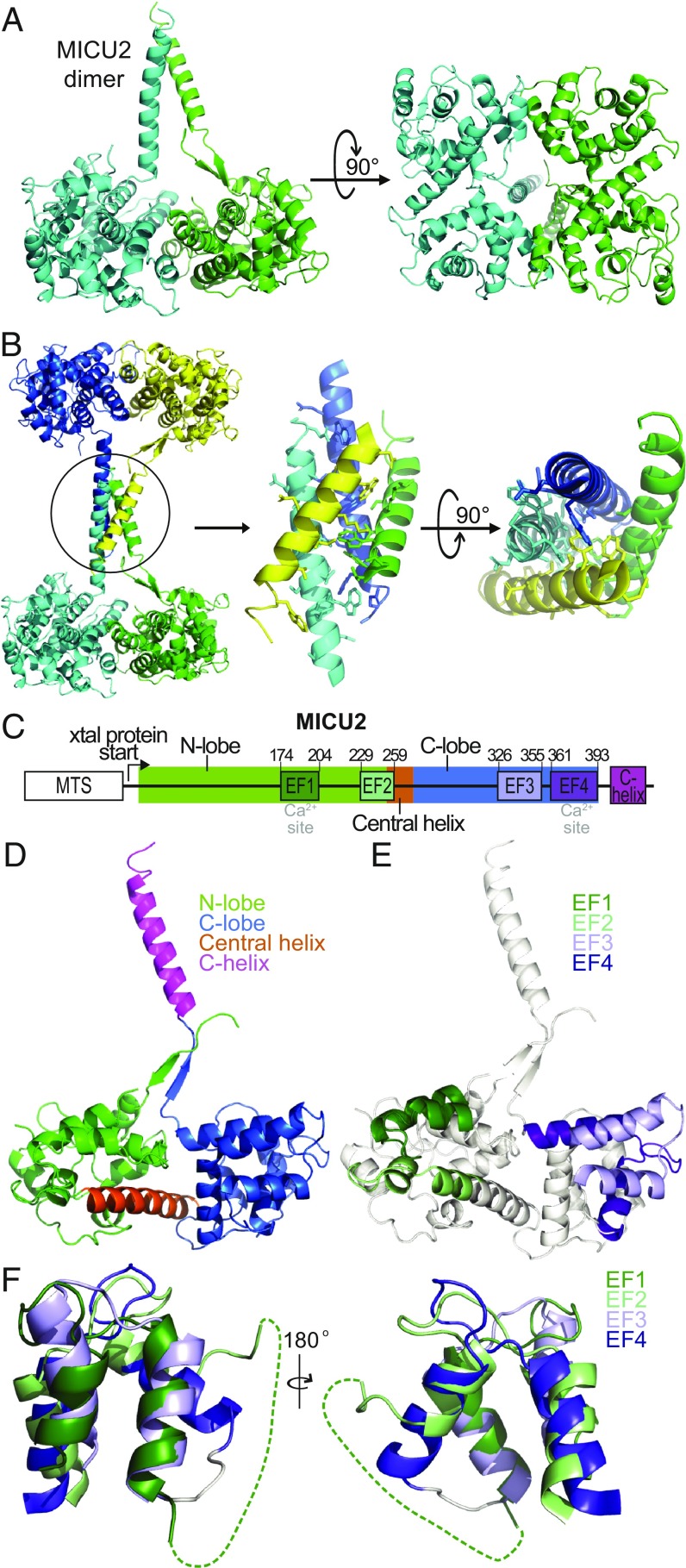
The core structure of the MICU2 monomer is similar to that of MICU1 (24). It consists of two lobes, each containing a pair of EF-hand motifs. There are only two structures in the PDB having significant similarity to MICU2: MICU1 (rmsd = 4.8 Å for 281 residues) (24), followed by citrin (rmsd = 5.2 Å for 151 residues) (26), as calculated by the Dali server (27). The asymmetric unit of the crystal is a dimer in which the monomers are related to each other by noncrystallographic approximate twofold rotational symmetry. The C-terminal α-helices of both monomers in a dimer point in a direction approximately orthogonal to the plane of the dimer and interact with each other (Fig. 1A). This interaction is further stabilized by contacts with another pair of C-helices contributed by an adjacent MICU2 dimer related by a crystallographic symmetry operator. In effect, every MICU2 dimer in the crystal lattice is connected to another dimer by a four-helix bundle (Fig. 1B). This dimer–dimer interaction appears to be the major determinant of the sparse packing of the crystal lattice, which accounts for the high solvent content (SI Appendix, Fig. S4). There are extensive side-chain contacts within the four-helix bundle, with major contribution coming from antiparallel helix–helix interactions (Fig. 1B and SI Appendix, Fig. S5). The striking arrangement of MICU2 molecules underscores the structural independence of the C-helix from the core of the protein and its propensity for self-association. Of importance is also the fact that the orientation and relative position of the C-helices with respect to the protein core are different in each monomer of the dimer. In one monomer (chain A) a short stretch of β-strand with contribution from the N-terminal segment of the polypeptide chain allows the C-helix to extend further away from the core (Fig. 1A). There is no β-strand at a similar position in chain B, the N-terminal segment is not resolved, and the orientation of the C-helix is different. Thus, the C-terminal helix of MICU2 is structurally independent from the core of the protein, and it retains flexibility in position and orientation with respect to the core.
Fig. 1.
Structure of the Ca2+-free MICU2. (A) Cartoon representation of the MICU2 dimer (B) MICU2 tetrameric assembly in the crystal lattice arising from crystallographic symmetry (Left). Four C-helices connecting two dimers make extensive parallel and antiparallel interactions (Right). Residues with significant interaction are shown as sticks (see also SI Appendix, Fig. S5). (C) Domain organization of MICU2. (D) View of MICU2 monomer (A chain) with N-lobe, C-lobe, central helix, and C-helix colored differently for emphasis. (E) View of MICU2 monomer (A chain) with each of the four EF-hands colored differently for emphasis. Note that the exiting helix of EF2 is a part of the central helix. (F) The two pairs of EF-hands of MICU2 structurally aligned using the SSM function of Coot. Two different orientations (front and back) are shown.
The core of the MICU2 monomer consists of two lobes arranged around a long central helix (residues 244–268; Fig. 1 C and D). An integral part of each lobe is a pair of EF-hand motifs (Fig. 1 C and E). Only EF1 in the N-lobe and EF4 in the C-lobe are canonical EF-hands that have the capability of binding Ca2+, whereas EF2 and EF3 are the structural EF-hands (SI Appendix, Fig. S2). An unusual feature of the EF-hand pair in the N-lobe is a long loop that connects the exiting helix of EF1 and the entering helix of EF2. This loop is not resolved in the crystal in either monomer of MICU2. A corresponding segment of MICU1 is also not resolved in its crystal structure (24). Sequence alignment shows that this segment is highly variable in the MICU family, particularly in MICU2 (SI Appendix, Fig. S6). The corresponding loop between EF3 and EF4 of MICU2 is much shorter and well-resolved in the crystal. Despite this difference, the relative disposition of the helical segments for both EF-hand pairs is similar and their structures can be aligned with each other (Fig. 1F). The significance of the structural and evolutionary variability of the EF1–EF2 connecting loop is unclear.
Comparison with MICU1.
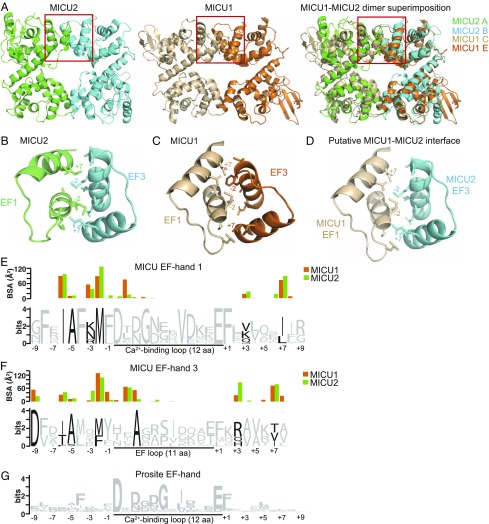
The core structure of mouse MICU2 determined here resembles that of the human MICU1 in the apo form, as determined by Wang et al. (24). These authors identified three domains in the MICU1 structure: the N-domain, the N-lobe, and the C-lobe. All three domains of MICU1 have their equivalent segments in MICU2, although the N-domain of MICU2 is less extensive and more integrated with the N-lobe. Despite that difference the two structures can be superimposed not only at the level of each lobe, but also the entire core domain can be superimposed, and even a dimer of MICU2 can be superimposed with a dimer of MICU1 (Fig. 2A). Such an extensive structural similarity likely contributes to the ability of these proteins to exchange between homo- and heterodimers (17). However, the structural compatibility of the interaction sites that stabilize the monomer–monomer interaction is also necessary. To address this issue, we analyzed the interaction interfaces in MICU1 and MICU2 homodimers in more detail (Fig. 2 B–F).
Fig. 2.
Compatibility of interaction interfaces in MICU2 and MICU1 homodimers. (A) The dimer of MICU2 (6EAZ) is shown on the left, a dimer of MICU1 (4NSC, chains C and E) is shown in the middle, and the superimposition of the MICU1 and MICU2 dimers is shown on the right (only the core domains are shown). Red boxes indicate the EF1–EF3 interaction region, which is shown in more detail in B–D. Note that due to the symmetry the complete interaction interface consists of two such regions. (B) MICU2 interaction interface. Residues for the C-lobe EF3 (chain B) interaction with the N-lobe EF1 (chain A) are shown. Side chains of residues that make significant contribution to the interaction are shown in stick representation. Residues in positions −6, −5, −3, −2, +3, and +7 with respect to the loop and the L2 position within the loop are shown for EF1. Residues in positions −9, −6, −5, −2, +3, and +7 with respect to the loop and the L2 and L3 position within the loop are shown for EF3. These residues are both conserved in the MICU family, in contrast to the typical EF-hand signature, and show significant interaction (D–F). Residues −2 and +7 with respect to the loop are labeled. (C) MICU1 interface residues for the C-lobe EF3 (E chain) interaction with the N-lobe EF1 (C chain) are shown (from 4NSC). Residues in similar positions as in B are shown as sticks and/or emphasized with labeling. (D) The putative MICU1–MICU2 interface is shown from superimposition of the MICU1 and MICU2 dimers. The same residues as in B and C are shown as sticks and/or emphasized with labeling. (E–G) Protein sequence logos are shown for (E) MICU EF-hand-1, (F) MICU EF-hand-3, and (G) an EF-hand motif (PS50222) generated by PROSITE and modified for the purposes of this figure (50). The MICU EF-hand logos were generated using WebLogo (49) from alignments of ∼500 metazoan MICU1/2/3 sequences. The average BSA for each residue, as calculated by the PISA server, is shown above the sequence logo for MICU EF-hand 1 (E) and EF-hand 3 (F), with the orange bar indicating the BSA for the residue in MICU1 (average of 6) and the green bar for the residue in MICU2 (average of 2). Residues that are conserved in the MICU family but not in the PROSITE EF-hand motif and make significant contribution to the interaction are emphasized on the MICU logos in black.
The interaction interface between MICU2 monomers in the homodimer involves the exposed surfaces of EF1 of one monomer and EF3 of another (Fig. 2B). Residues in positions −6, −2, and +7 with respect to the Ca2+-binding loop in both EF1 and EF3, as well as residues −9, L2, and +3 in EF3, make the greatest contribution to the solvent-excluded surface area that constitutes the monomer–monomer interaction interface (see bar graphs in Fig. 2 E and F). As the energy of protein–protein interaction is directly related to the buried surface area (BSA) (28, 29), these residues make a major contribution to the binding energy of MICU2–MICU2 interaction. The contacts between the MICU2 monomers are mostly hydrophobic (Fig. 2 E and F and SI Appendix, Fig. S7). Due to the twofold rotational symmetry of the homodimer, the complete interaction interface comprises two EF1–EF3 interaction sites for the total solvent-excluded surface area of ∼990 Å2 [analyzed by the PISA server (30)]. The MICU1 homodimer is built in a similar way (Fig. 2 C, E, and F and SI Appendix, Fig. S8). Wang et al. (24) pointed to the importance of the salt bridge between Arg221 and Asp376 in MICU1 for homodimer stability. This interaction is not present in MICU2, where the Arg is substituted with Ser (SI Appendix, Fig. S2). However, several other contributing residues are either preserved or substituted with similar residues (SI Appendix, Figs. S7 and S8). Thus, we conclude that the interfaces between the monomers in MICU1 and MICU2 homodimers are structurally compatible with each other (Fig. 2 D–F and SI Appendix, Figs. S7 and S8), permitting the exchange of monomers between homo- and heterodimers.
We have further analyzed how well the EF1–EF3 interaction interface is preserved throughout evolution within the MICU family. From comparison of protein logos generated for the EF-hand domains of MICU1/MICU2/MICU3 that are involved in the interaction interface (EF1 and EF3) with the EF-hand domain signature, it is clear that the residues with significant contribution to the monomer interaction interface are highly conserved, despite their lack of conservation in the EF-hand domain generally (Fig. 2 E–G). For example, the hydrophobic residues at positions −6, −2, and +7 with respect to the Ca2+-binding loop in EF1 are highly conserved in the MICU family. Importantly, these are the same positions that contribute the most to the total solvent-excluded surface area in MICU1 and MICU2 (Fig. 2 E and F). A notable exception appears to be the Ala residue at −5 position, which is extremely highly conserved in MICU family, yet owing to its small side chain it makes little contribution to BSA. It seems plausible that a small side chain is required at this position for surface compatibility. Based on the above analysis we conclude that the compatibility of the EF1–EF3 monomer–monomer interaction interfaces is likely a common feature of proteins in the MICU family.
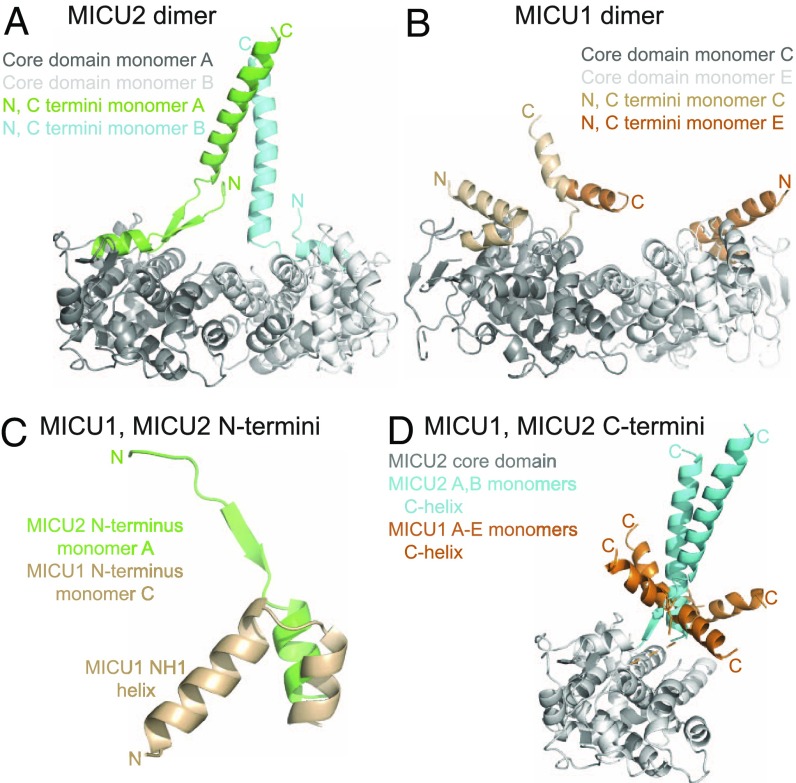
While the core structures of MICU2 and MICU1 are very similar, there are essential differences between the two proteins in the composition and structure of their N-terminal and C-terminal segments (Fig. 3). In one chain of MICU2 a short N-terminal segment of the polypeptide chain stabilizes the extended position of the C-helix via a short stretch of β-strand (Fig. 3A). There is no corresponding β-strand in MICU1. The polypeptide chain of MICU1 extends significantly further toward the N terminus but it folds into an α-helix [helix NH1 in Wang et al. (24)] that packs tightly against the N-lobe of MICU1 (Fig. 3C). The lack of the β-strand is likely the reason for greater mobility of the C-helix in MICU1 compared with MICU2 (Fig. 3D). It is important to note that the NH1 helix of MICU1 is connected at its N terminus directly to a stretch of positively charged residues (KKKKR), which is required for MICU1 function (31). This sequence is highly conserved in MICU1 but absent from MICU2. It appears that the difference in the structure and disposition of MICU1 and MICU2 N termini (Fig. 3C) might be one of the factors defining the unique contribution of each of these proteins to uniporter function.
Fig. 3.
The N termini and C termini differentiate MICU2 from MICU1. (A) The MICU2 dimer (6EAZ) is shown with the N and C termini colored for emphasis, labeled N and C, respectively. (B) The MICU1 dimer (4NSC, C and E monomers) is shown with the N and C termini colored for emphasis. The orientation shown is the same as for the MICU2 dimer in A. (C) The MICU1 and MICU2 N termini are shown highlighting that the MICU1 NH1 helix folds back (toward the core domain), while the MICU2 corresponding region does not. Note that in MICU1 the amino acid sequence immediately preceding NH1 helix is the lysine-rich segment specific for MICU1 which is indispensable for function. (D) MICU1 monomers from the 4NSC hexamer and MICU2 monomers from the 6EAZ dimer are superimposed. The core domain of MICU2 monomer A is shown in gray for reference. The C-helices of the five monomers of MICU1 are colored in orange (note the sixth was not resolved in 4NSC), and the two monomers of MICU2 are colored in cyan.
C-Terminal Helix Is Required for MICU2 Function.
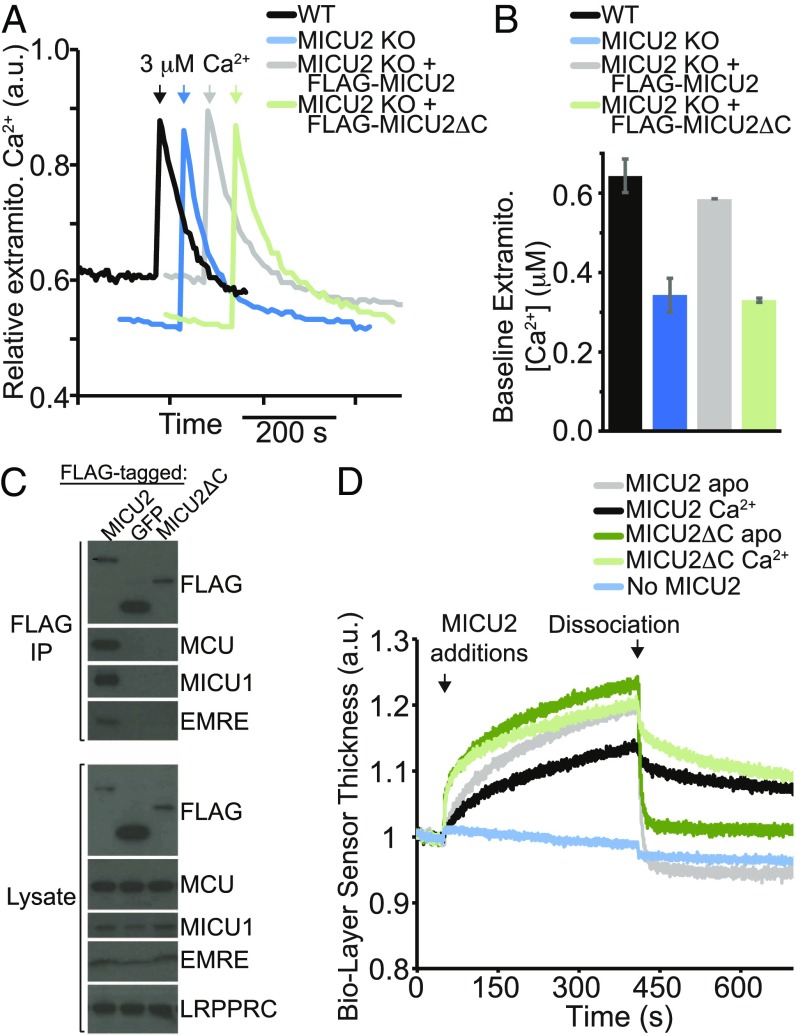
Comparison of MICU2 with MICU1 structure suggests a difference in the position/rigidity of the C-helix in relation to the core of the protein (Fig. 3D). Is that difference functionally important? While it has previously been shown that the C-helix of MICU1 is required for its function and for the interaction with the uniporter complex (7, 24), to our knowledge no one has evaluated the functional significance of the C-terminal helix of MICU2. Therefore, we expressed the full-length MICU2 or MICU2 missing the C-helix (MICU2ΔC) in HEK-293T cells having their native MICU2 knocked out. In these experiments MICU2ΔC was expressed at ∼50% higher level than MICU2, which ensures that any observed difference in function is not caused by reduced expression. MICU2 has been shown to change the threshold for uniporter-mediated calcium uptake (17, 18). We have shown that, in the absence of MICU2, the threshold is ∼400 nM, whereas in its presence the threshold increases to 600 to 800 nM (17). We can take advantage of this difference to dissect the function of MICU1 and MICU2. To assay the threshold-modulating function of MICU2ΔC, we measured the baseline Ca2+ after permeabilizing HEK-293T cells and allowing the mitochondria to take up calcium until a steady state is reached (10 min after permeabilization), an assay which we have previously validated (17). MICU2 KO cells reach a lower baseline Ca2+ level than WT cells (Fig. 4 A and B), suggesting a lower threshold for mitochondrial calcium uptake, as expected. While reintroduction of MICU2 (MICU2 KO+MICU2) restores the threshold of MICU2 KO cells back to the WT level, reintroduction of MICU2ΔC (MICU2 KO+MICU2ΔC) has no effect, suggesting that MICU2ΔC is unable to perform the function of the full-length MICU2 (Fig. 4 A and B). We followed this baseline calcium quantitation with a 3 µM pulse of Ca2+, finding that all four cell lines (WT, MICU2 KO, MICU2 KO+MICU2, and MICU2 KO+MICU2ΔC) take up calcium with similar kinetics (Fig. 4A), suggesting that all are capable of mitochondrial calcium transport. Thus, in contrast to MICU2, MICU2ΔC does not contribute to the regulation of uniporter-mediated calcium transport in this assay in HEK-293T cells.
Fig. 4.
MICU2 C-terminal helix is required for function and interaction with the uniporter in HEK-293T cells in situ, but not for MICU1–MICU2 interaction in vitro. (A) Extramitochondrial calcium levels in digitonin-permeabilized HEK-293T cells (WT), HEK-293T cells with MICU2 knocked out (MICU2 KO), and MICU2 KO cells with FLAG-MICU2 or FLAG-MICU2 with the C-terminal α-helix deleted (FLAG-MICU2ΔC). At the indicated time, a 3 µM pulse of CaCl2 was given. Fluorescence signal is normalized on a scale of 0–1. (B) The extramitochondrial Ca2+ concentration before the 3 µM pulse of CaCl2 (“baseline”) is quantified 10 min after cell permeabilization across biological replicates and mean ± SEM is reported (n = 3). (C) FLAG coimmunoprecipitation to probe the interaction between FLAG-tagged MICU2, MICU2ΔC, or a negative control mito-targeted GFP and other components of the uniporter complex (MCU, MICU1, and EMRE) in MICU2 KO cells. A representative experiment is shown. (D) Biolayer interferometry traces showing MICU2 or MICU2ΔC binding to the streptavidin-coated ForteBio tips loaded with biotinylated MICU1ΔC in a buffer containing Ca2+ or EGTA. Dissociation begins at the indicated time point, when the well solution for the tips is changed to buffer lacking the MICU2 protein. Signal is corrected for nonspecific binding by subtracting signal obtained from tips lacking the addition of biotinylated MICU1.
If MICU2ΔC is unable to affect the uniporter’s Ca2+ threshold, does it interact with the uniporter complex in situ? To answer this question, we used coimmunoprecipitation with an antibody against the FLAG tag attached to the N terminus of MICU2 or MICU2ΔC. While FLAG-MICU2 coimmunoprecipitates MCU, EMRE, and MICU1, no coimmunoprecipitation of these proteins was observed with FLAG-MICU2ΔC under the same conditions (Fig. 4C). Mitochondria-localized GFP, used here as a negative control, also does not coimmunoprecipitate any of the uniporter components, supporting the view that coimmunoprecipitation requires specific protein–protein interaction under our experimental conditions. Hence, our results suggest that the C-terminal helix of MICU2 contributes to its ability to interact with the uniporter complex in situ.
Next, we tested the role of MICU2’s C-helix in the interaction of MICU2 with MICU1 in vitro. It has been shown that both MICU1ΔC and MICU2ΔC each form homodimers in vitro, suggesting that the C-helix is not required for homodimerization (17, 24), which is consistent with the crystal structures of MICU1 and MICU2. Also, we have shown that MICU1ΔC and MICU2 form a heterodimer even though they do not coimmunoprecipitate with each other in cells (7, 17). Therefore, we wondered if MICU2ΔC and MICU1ΔC can interact in vitro. To this end, we established a biolayer interferometry assay. Biotinylated MICU1ΔC was immobilized on a streptavidin sensor, and the relative biolayer sensor thickness was measured during incubation in buffer, to establish a baseline, followed by incubation with solution containing MICU2 or MICU2ΔC (Fig. 4D). Both proteins showed binding to the immobilized MICU1ΔC on the sensor in both the presence and absence of Ca2+. Subsequent to the binding step, the dissociation of MICU2 and MICU2ΔC was monitored upon switching to a buffer lacking these proteins. These experiments show that, even in the absence of the C-terminal helices of MICU1 and MICU2, these proteins still interact with each other in vitro, even though we do not find evidence of interaction in situ.
Discussion
The 2.5-Å resolution X-ray structure of the inhibited form of MICU2 presented here sheds light on the function of MICU2 and on the mechanism of uniporter regulation by the MICU1–MICU2 complex. MICU1 and MICU2 share 28% sequence identity, with 50% linear sequence similarity. The core parts of the two proteins spanning ∼80% of the structure are very similar in 3D space. The core domain of MICU2, like that of MICU1, consists of two lobes, each built around a pair of EF-hand structural motifs. EF-1 in the N-lobe and EF-4 in the C-lobe are the canonical EF-hands capable of Ca2+ binding with the structural support from their inactive pairmates, EF-2 and EF-3, respectively. Extensive structural similarity between the core domains of MICU1 and MICU2 combined with complementarity of the interaction interfaces explains why the two proteins exchange readily between homo- and heterodimers (17).
While MICU1 and MICU2 are structurally very similar, they also have two distinctly unique features, which may explain why they are not functionally redundant. One is the prominent C-helix extending away from the core of the protein. The second key difference is in the N-terminal segment of the polypeptide chain, which in MICU2 forms a short stretch of β-strand with the base of the C-helix (Fig. 3A). In MICU1 the β-strand is replaced with an N-terminal α-helix (NH1) that interacts with the core domain (Fig. 3B). Importantly, we find that deletion of the C-helix renders MICU2 inactive but does not preclude dimerization in vitro, an effect akin to that reported for MICU1 (Fig. 4). Thus, the structural data highlight the compatibility of the core domains of MICU2 and MICU1 and the divergent structure and autonomous function of their C-terminal helices.
At present, a crystal structure of the MICU1–MICU2 heterodimer is not available, but the availability of the MICU2 structure allows us to begin to model the interaction. We generated a model of the MICU1–MICU2 heterodimer (Fig. 5A) by substituting one of the MICU2 monomers in the homodimer (PDB ID code 6EAZ, ref. 25) with a MICU1 monomer (PDB ID code 4NSC), which is straightforward, considering their structural similarity and compatibility of the monomer–monomer interfaces (Fig. 2). Unfortunately, our model of MICU1–MICU2 heterodimer does not include the C-helices of these proteins due to their mobility (Fig. 3D). An important feature of this model is the direct contact between the functional EF1 in the N-lobe of one monomer and the EF3 in the C-lobe of the other monomer. Although EF3 does not coordinate calcium, it is structurally coupled to EF4. Thus, the monomer–monomer interaction interface in the MICU1–MICU2 heterodimer offers a direct route for structural coupling between the Ca2+-binding sites in the MICU1–MICU2 complex that likely contributes to the cooperativity of the Ca2+ binding (17) and ultimately to the cooperativity of uniporter activation (17). The model also offers a plausible explanation why the mutations of the EF-hand sites in one protein (either MICU1 or MICU2) render the whole complex locked in the off state (7).
Fig. 5.
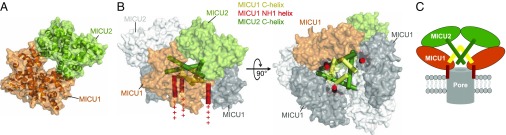
Computational models of the MICU1–MICU2 complex. (A) A model of the MICU1–MICU2 heterodimer built of the core domains of MICU2 and MICU1. The dimer is shown as a semitransparent surface along with a cartoon representation. (B) Two different orientations of the MICU1–MICU2 complex modeled as a trimer of heterodimers based on the published structure of the apo MICU1. The surfaces of all proteins are shown and labeled, and the helices are shown as cylinders for the C-helices and for MICU1’s NH1 helix, which follows the poly-lysine region of MICU1 (SI Appendix, Fig. S6). On the left, the place where the poly-lysine region of MICU1, which precedes NH1, may occupy is labeled as ++++. (C) Schematic representation of the MICU1–MICU2 oligomeric complex. The core domains of MICU1 and MICU2 form the outer shell, with MICU1 between MICU2 and the pore. The C-helices of MICU1 (yellow cylinders) and of MICU2 (dark green cylinders) interact with each other. The NH1 helix of MICU1 (red cylinder) connects the MICU1/MICU2 complex to the uniporter pore (MCU/EMRE) and/or inner mitochondrial membrane lipids.
Previous biochemical data suggest that MICU1 and possibly also the MICU1–MICU2 complex associate into higher oligomers. Functionally competent MICU1 constructs that include the C-helix form a hexamer in solution in the absence of Ca2+ (17, 24). In the presence of Ca2+ variable oligomers of MICU1 were observed, which reduced to a dimeric state when the C-helix was deleted (23). Hence, the C-helix is required for MICU1 oligomerization in vitro and it is also absolutely required for MICU1 function in the cell (7, 24). Vecellio Reane et al. (32) reported that MICU1 and MICU2 when coexpressed form dimers, tetramers, and hexamers, as judged by their mobility on blue native PAGE. These data suggest that oligomerization might be important or even necessary for MICU1 function. At present, the apo MICU1 (PDB ID code 4NSC) is the only available structure that can serve as a template for modeling the MICU1–MICU2 oligomer. The hexameric assembly of MICU1 in the asymmetric unit of the crystal can be viewed as a trimer of homodimers, or, alternatively, as being built of two trimers, one closely packed and one relatively open (cf. supplemental figure 5 in ref. 24). The MICU1 C-helices packed into a bundle are confined in the central cavity of the complex (24) (SI Appendix, Fig. S9). We have found that upon substitution of MICU1–MICU2 heterodimers for the three MICU1 homodimers, and with slight adjustment of the C-helices, a compact bowl-shaped oligomeric structure can be obtained (Fig. 5B). The closely packed trimer in the complex comprises the three MICU2 molecules. The restricted space in the central cavity greatly limits the possible orientations of the C-helices, which facilitates the modeling, yet it is sufficient to accommodate them all without steric clashes. Interestingly, the N-terminal helices (NH1) of the three MICU1 monomers run in parallel and point in the same direction in the opening of the bowl. Since the sequence immediately preceding these helices in MICU1 (not resolved in the structure) is the poly-lysine region (SI Appendix, Fig. S6), then such an arrangement would create a patch of positive charge (marked with “+” signs in Fig. 5B), a plausible site of interaction with the poly-aspartate–containing C-terminal segments of EMRE, the negatively charged MCU residues at the apex of the channel, or negatively charged mitochondrial inner membrane phospholipids such as cardiolipin. This interaction site could allow for a conformational change of MICU1/MICU2 upon Ca2+ binding to be communicated to the pore. Moreover, the centrally located cluster of the C-helices contributed by both MICU1 and MICU2 could block Ca2+ access to the pore and also could be rearranged by Ca2+-induced changes in the core domains of MICU1 and MICU2 to facilitate Ca2+ uptake by the channel. A schematic representation of this model highlighting these key features is shown in Fig. 5C. While it would be premature to speculate about the Ca2+-dependent structural changes in the MICU1–MICU2 complex, two aspects of the proposed model appear relevant. One is the fact that the monomer–monomer interface involves the functional EF1, which in MICU1 changes its conformation on Ca2+ binding and is likely to do so in MICU2. In the context of the proposed model this change might push the monomers slightly apart, causing a shift of the C-helix. Also in EF4, whose exiting helix is connected directly to the C-helix in both MICU1 and MICU2, a structural change induced by Ca2+ is likely to affect the relative disposition of the C-helices in the complex, which as we propose might constitute part of the gating mechanism.
Our computational model of the MICU1–MICU2 complex (Fig. 5 A and B) awaits experimental validation. In particular, the apparent mismatch between the approximate threefold symmetry of the model and the currently available tetrameric structures of MCU (20–23) needs to be resolved. For a structurally competent interaction with the channel’s pore, a match in symmetry would seem to be required, such as two- or fourfold symmetry of the MICU complex or a hexameric composition of the channel subunits [e.g., a trimer of dimers, such as that in the Orai Ca2+ channel (33)]. Future experiments will need to address this issue.
The symmetry notwithstanding, we notice that there are at least three experimental observations reported in the literature that are consistent with our model of the MICU1–MICU2 complex. First, mutation of the poly-lysine region of MICU1 prevents coimmunoprecipitation with MCU (31) and EMRE (34). In our model, the N termini of MICU1 connected to the poly-lysine segments are positioned on one side of the complex, presumably that which faces the mitochondrial inner membrane, making them available for interaction with EMRE, the apex of the MCU pore, or with negatively charged phospholipids of the inner membrane (Fig. 5B). Second, MICU2 does not coimmunoprecipitate with MCU and EMRE in the absence of MICU1 (7). In our model MICU2 is located on the side of the complex opposite to MICU1, away from the putative membrane-associated region, precluding direct contact with MCU or EMRE. Third, it has been reported that MICU1 and MICU2 interactions involve disulfide bond formation between cysteines in the C-terminal helices: MICU1 has been shown to be a specific client of MIA40, an oxidoreductase resident in the mitochondrial intermembrane space, which introduces an intermolecular disulfide bond that links MICU1 and MICU2 into a heterodimer (35, 36). Our model includes the C-helices of MICU1 and MICU2 in close proximity, allowing for the possibility of mixed disulfide formation. While it is reassuring that these observations are consistent with our model, it is important to keep in mind that they do not prove the validity of the model, nor do they exclude other models.
It is important to bear in mind that the physiology and composition of the uniporter varies greatly along the eukaryotic tree of life (SI Appendix, Fig. S10) (1). In fact, the lack of uniporter activity in Saccharomyces cerevisiae was exploited for the initial identification of the uniporter machinery (11). Fungi for which the MCU tetrameric structures have recently been determined (Neurospora crassa, Cyphellophora europaea, Neosartorya fischeri, and Fusarium graminearum) have MCU homologs, but not other components of the uniporter complex. The calcium transport properties of these fungi are highly divergent from the classically defined uniporter activity extensively documented in animal mitochondria (37). EMRE appears to have arisen with metazoa (6). MICU1 is the most ancient MICU paralog, while MICU2 and MICU3 are more recent innovations (8). Due to the large differences in the uniporter’s physiology across organisms, it is conceivable that the metazoan channel itself may be quite different as it evolved from MCU alone to requiring EMRE for function and MICU(s) for regulation (6). The fungal uniporters contain only MCU and appear to transport calcium with very slow kinetics. It is possible that, because the calcium flux is much slower in the fungal MCU-only complexes, MICU-mediated channel regulation is not required. Alternatively, we have shown that in the absence of MICU1 the human uniporter complex can transport Mn2+ (38), raising the hypothesis that fungal MCU channels that naturally lack MICU1 may also transport other ions. Understanding what could be the evolutionary advantage of a transition from MCU alone to the more “complex” mammalian uniporter, including roles for EMRE and MICU1/MICU2, will undoubtedly help in understanding the role of the uniporter in varied cellular environments in the human body.
Methods
Protein Expression.
Competent BL21(DE3) E. coli were transformed with pETDuet-1 vector (EMD Millipore) containing M. musculus MICU2 (residues 68–432) between BamHI and NotI sites with an N-terminal His-tag (such that the N-terminal sequence before the protein is MGSSHHHHHHSQDP). Cells were grown to OD600 = 0.4 shaking at 37 °C in Luria Broth media, then induced with 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma). Cells were pelleted 4 h after induction and bacterial pellet was frozen at −80 °C until purification.
SeMet-MmMICU2 Expression, Purification, and Crystallization.
SeMet-MmMICU2 protein was expressed similarly to the native protein, with a few changes. (i) Competent NEB Xtal-express (methionine auxotroph; New England BioLabs) cells were used and (ii) after OD600 = 0.4, cells were pelleted and resuspended in Selenomethionine Medium Complete from Molecular Dimensions, grown for another 1 h, and the Molecular Dimensions SeMet solution was added, followed by growth for 1 h, followed by induction with 0.3 mM IPTG. Protein was purified and crystallized as the native, except 2 mM DTT was included in all steps.
Protein Purification.
Bacterial cell pellet was thawed and lysed in buffer A containing 500 mM NaCl, 25 mM Hepes, pH 7, 20 mM imidazole, cOmplete protease inhibitors (Roche), lysozyme, and benzonase. Solution was sonicated for 3 min on ice and centrifuged for 1 h at 4 °C at 20,000 × g. Lysate was collected and loaded onto a HisTrap 10 mL column (GE Healthcare) using an AKTAPure. The column was washed with buffer A, followed by elution with an imidazole gradient in buffer B (500 mM imidazole, 150 mM NaCl, and 25 mM Hepes, pH 7). Fractions were collected and concentrated using Millipore 30,000 molecular weight cutoff centrifugal filters to prepare for loading on a size-exclusion chromatography (SEC) column.
SEC.
Protein was loaded onto a Superdex 200 HiLoad column (GE Healthcare) equilibrated with buffer C (150 mM NaCl and 25 mM Hepes, pH 7) with either 5 mM CaCl2 or 1 mM EGTA added. Protein for crystallography was collected from the peak of the sample ran in buffer containing EGTA and was concentrated to 6 mg/mL before crystallization. Purity was assessed by SDS/PAGE with Instant Blue staining (Expedeon).
Differential Scanning Fluorimetry.
Protein was used at 0.2 mg/mL in buffer containing 150 mM NaCl and 50 mM Hepes at pH 7, along with 5× Sypro Orange dye (Life Technologies). Experiments were performed using an Applied Biosystems 7500 Fast Real Time PCR machine with a temperature ramp rate of 2% from 25 °C to 95 °C, along with a melting protocol for differential scanning fluorimetry using SYPRO Orange as described by the manufacturer. Experiments were performed under apo conditions (with 5 mM EGTA in the buffer) or with 5 mM Ca2+. Data are presented as the average ± SD of the melting temperature calculated.
Crystal Growth and Freezing.
Initial screening for crystallization conditions was performed using sparse-matrix screens from Microlytics, Qiagen, and Hampton Research, followed by targeted optimization of crystal growth. Crystals for data collection were grown by the vapor diffusion method. Specifically, protein was used at 6 mg/mL following concentration after SEC. Well solution contained 8% PEG 3350 and 150 mM trilithium citrate, pH 7. Protein and mother liquor were mixed in a 1:1 ratio and pipetted onto a siliconized glass coverslip (HR3-277; Hampton Research), which was sealed onto a 48-well plate (HR3-275; Hampton Research). Needles obtained from initial crystallization were used for microseeding (Hampton Seed Bead HR2-320) in subsequent crystal setups, which allowed for growth of thicker crystals. Crystals from this condition diffracted in the best case to ∼2.9 Å. While seeking a heavy-atom derivative, we were able to attain better resolution as follows. After a week of crystal growth, crystals were incubated overnight with 10 mM ethyl mercuric phosphate (HR2-446; Hampton Research) and were subsequently frozen using 30% glycerol as cryoprotectant by plunging into liquid nitrogen. Although we found no evidence of mercury being incorporated into the crystal, the resolution was slightly improved. It is unclear which aspect of the procedure caused the improvement in resolution. SeMet-MICU2 crystals were prepared in the same way, except with 5 mM DTT in the solution and without incubation with ethyl mercuric phosphate.
X-Ray Diffraction Data.
For the native dataset at 2.5-Å resolution we collected 180° of 1°-oscillation frames from a single crystal at 100 K. The data were collected at Advanced Photon Source (Argonne National Laboratory) beamline 24-ID-C at 1.00-Å wavelength. The data were integrated and scaled using autoPROC (39). For the SeMet derivative, the data were collected at Advanced Light Source (Lawrence Berkeley National Laboratory) beamline 8.2.2 at 0.97 Å. We collected two datasets from the same crystal at 100 K. The data were processed using iMosflm (40) followed by quick scaling with Pointless and Aimless programs [part of the CCP4 package (41)]. Extensive statistics provided by these programs indicated fast deterioration of the crystal due to radiation damage. Upon selecting only the best frames from the two datasets and scaling them together, a 4-Å-resolution dataset of sufficient quality for phase determination by SAD was obtained (SI Appendix, Table S1).
Structure Determination.
All our attempts at solving the structure by molecular replacement using parts of 4NSC, 4NSD, or an unbiased method using BALBES (42) were unsuccessful. We used a modification of the MR-SAD technique that obviates the need for an MR-derived model as input. Briefly, the initial phase information was obtained from the anomalous dispersion of selenium in the 4-Å resolution dataset obtained from crystals of the SeMet substituted protein. Heavy-atom substructure was determined with ShelxD (43) using hkl2map graphic user interface (44). Subsequent density modification with ShelxE returned an electron density map that was too fragmented for main-chain tracing. The atomic model required as input for MR-SAD, as implemented in Phaser-EP, a part of the PHENIX package (45), was obtained by manual [with Coot (46)] or automatic [with ARP/wARP (46)] placement of a few helices in the electron density map produced by ShelxE. Phaser-EP running in the MR-SAD mode refined the positions and occupancies of the heavy-atom sites that were correctly identified by ShelxD, deleted the incorrect sites, and found all of the remaining Se sites. Upon subsequent density modification with AutoBuild [part of the PHENIX package (47)] the resulting electron density map was of excellent quality (SI Appendix, Fig. S3). It was suitable for main-chain tracing and, given the known positions of SeMet, it was sufficient for unambiguous sequence assignment. The resulting model was refined against the 2.5-Å resolution native dataset using several cycles of model building (with Coot) and automatic refinement (with Phaser). The statistics for data collection and model refinement are shown in Table 1 (generated using PHENIX v 1.13).
Cell Culture.
Stable cell lines were made using lentiviral infection followed by selection using 2 μg/mL puromycin. HEK-293T cells were originally obtained from the American Type Culture Collection. MICU2 KO was achieved using transcription activator-like effector nuclease to generate a clonal cell line, which has previously been reported (7).
Coimmunoprecipitation and Western Blot Experiments.
Cells were washed with cold PBS and scraped from a confluent 15-cm dish. Cell pellet was lysed with buffer containing 150 mM NaCl and 25 mM Hepes at pH 7, 1 mM EGTA, 0.2% DDM, and cOmplete protease inhibitors for 30 min at 4 °C and centrifuged for 10 min at 20,000 × g at 4 °C, and the concentrations of the cleared lysates were determined using the Bradford assay. Lysate concentrations were normalized to 2 mg/mL by dilution with the lysis buffer.
For lysates that were used for the coimmunoprecipitation experiments, FLAG M2 affinity gel (Sigma) was added and incubated for 1 h at 4 °C. Beads were washed three times with 600 μL of lysis buffer, followed by addition of SDS sample reducing buffer (Bio-Rad). Western blots were performed on lysates and immunoprecipitates by running the samples on either 4 to 20% gradient or 12% Tris-glycine gels (XP04205BOX or XP00125BOX ; Thermo Fisher), transferring to a PVDF membrane (1704157; Bio-Rad) using a TransBlot Turbo Transfer System (Bio-Rad), blocking with 5% milk in TBST, and immunoblotting using antibodies against MICU1 [homemade (11)], MCU (14997S; Cell Signaling Technologies), FLAG (2368; Cell Signaling Technologies), LRPPRC (SAB2700419; Sigma), EMRE (made in collaboration with Bethyl), and MICU2 (ab101465; Abcam). Experiments were performed three times and representative experiments are presented herein.
Mitochondrial Calcium Physiology Experiments.
Cells were centrifuged at 1,200 × g for 3 min, washed with PBS, and centrifuged for another 3 min at 1,200 × g. The cell pellet was resuspended in buffer containing 125 mM KCl, 2 mM K2HPO4, 1 mM MgCl2, 20 mM Hepes at pH 7.2, 0.005% digitonin, 5 mM glutamate and malate, and 1 μM cell-impermeable Fluo-3 dye (Life Technologies). Cells were used at a density of 5 million cells per mL for each experiment. Green fluorescence of the Fluo-3 dye was monitored using a PerkinElmer LS55 fluorimeter with stirring at room temperature. At the time indicated on the traces shown, a pulse of CaCl2 was given at the final concentration indicated. For absolute quantification of the calcium concentration, the minimum and maximum fluorescence of each solution was determined by ending the experiment with 200 µM EGTA and 2 mM CaCl2. For the quantification of the baseline calcium levels, the baseline reading was determined after stirring for 10 min, at which point the fluorescence signal had stabilized. Experiments shown are representative of at least three replicates. Quantification of baseline calcium levels is reported as the average of three replicates (error bars indicate SEM).
Biolayer Interferometry.
ForteBio Octet was used to measure the interaction of MICU1 and MICU2. N-terminally His-tagged and Avi-tagged MICU1ΔC was biotinylated. Biotinylation of MICU1ΔC was facilitated by the BirA kit from Avidity (BirA500). One milliliter of 40 μM MICU1ΔC protein was mixed with 9 mL of buffer containing 50 mM Bicine (pH 8.3) and 50 mM NaCl and concentrated to a volume of 900 μL. One hundred microliters of BiomixB [100 mM ATP, 100 mM Mg(OAc)2, and 500 μM d-biotin] and 20 μL of 1 mg/mL BirA biotin-protein ligase were added to initiate the reaction. It was incubated at room temperature for 2 h. Intact mass monitored by Q Exactive mass spectrometer indicated a +225 mass shift and the reaction was completed. Extra biotin and ATP was removed through SEC using a Superdex 75 column. Biotinylated MICU1ΔC was loaded onto streptavidin-coated ForteBio tips at 20 nM. MICU2 was allowed to associate with the bound MICU1 at 1 μM concentration in PBS with 2 mM DTT and 0.015% P20. Dissociation was monitored in the same buffer. Measurements of the nonspecific binding to tips without MICU1 bound were subtracted.
Generation of Sequence Logos.
Sequences of MICU1 (200), MICU2 (100), and MICU3 (200) from different metazoan organisms were compiled, chosen as best bidirectional BLAST hits to the human MICU proteins. The sequences were aligned using Clustal Omega (48). This alignment was used to generate protein logos using the WebLogo server (49). The EF-hand logo shown was generated by PROSITE from 4,641 sequences (PS50222) (50).
Supplementary Material
Acknowledgments
We thank Sarah Calvo, Luke Chao, Jason McCoy, and Scott Vafai for constructive feedback and Piotr Sliz for crystallography advice. This research used resources of the Advanced Light Source, a Department of Energy Office of Science User Facility under Contract DE-AC02-05CH11231 for anomalous data collection (beamline 8.2.2), and of the Advanced Photon Source for the native dataset (Northeastern Collaborative Access Team beamlines). This work was supported in part by National Institutes of Health Grant R01AR071942 (to Z.G. and V.K.M.) and funding from Calico (to W.J. and V.K.K.). V.K.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: V.K.M. is a paid advisor to Janssen Pharmaceuticals, Raze Therapeutics, and 5AM Ventures.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6EAZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817759116/-/DCSupplemental.
References
- 1.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336:886. doi: 10.1126/science.1214977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamer KJ, Mootha VK. The molecular era of the mitochondrial calcium uniporter. Nat Rev Mol Cell Biol. 2015;16:545–553. doi: 10.1038/nrm4039. [DOI] [PubMed] [Google Scholar]
- 3.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raffaello A, et al. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32:2362–2376. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sancak Y, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamer KJ, Mootha VK. MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 2014;15:299–307. doi: 10.1002/embr.201337946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plovanich M, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Csordás G, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallilankaraman K, et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perocchi F, et al. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patron M, Granatiero V, Espino J, Rizzuto R, De Stefani D. MICU3 is a tissue-specific enhancer of mitochondrial calcium uptake. Cell Death Differ. 2019;26:179–195. doi: 10.1038/s41418-018-0113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovács-Bogdán E, et al. Reconstitution of the mitochondrial calcium uniporter in yeast. Proc Natl Acad Sci USA. 2014;111:8985–8990. doi: 10.1073/pnas.1400514111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreau B, Nelson C, Parekh AB. Biphasic regulation of mitochondrial Ca2+ uptake by cytosolic Ca2+ concentration. Curr Biol. 2006;16:1672–1677. doi: 10.1016/j.cub.2006.06.059. [DOI] [PubMed] [Google Scholar]
- 15.Igbavboa U, Pfeiffer DR. EGTA inhibits reverse uniport-dependent Ca2+ release from uncoupled mitochondria. Possible regulation of the Ca2+ uniporter by a Ca2+ binding site on the cytoplasmic side of the inner membrane. J Biol Chem. 1988;263:1405–1412. [PubMed] [Google Scholar]
- 16.Bragadin M, Pozzan T, Azzone GF. Kinetics of Ca2+ carrier in rat liver mitochondria. Biochemistry. 1979;18:5972–5978. doi: 10.1021/bi00593a033. [DOI] [PubMed] [Google Scholar]
- 17.Kamer KJ, Grabarek Z, Mootha VK. High-affinity cooperative Ca2+ binding by MICU1-MICU2 serves as an on-off switch for the uniporter. EMBO Rep. 2017;18:1397–1411. doi: 10.15252/embr.201643748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Payne R, Hoff H, Roskowski A, Foskett JK. MICU2 restricts spatial crosstalk between InsP3R and MCU channels by regulating threshold and gain of MICU1-mediated inhibition and activation of MCU. Cell Rep. 2017;21:3141–3154. doi: 10.1016/j.celrep.2017.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oxenoid K, et al. Architecture of the mitochondrial calcium uniporter. Nature. 2016;533:269–273. doi: 10.1038/nature17656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo J, et al. Cryo-EM structure of a mitochondrial calcium uniporter. Science. 2018;361:506–511. doi: 10.1126/science.aar4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen NX, et al. Cryo-EM structure of a fungal mitochondrial calcium uniporter. Nature. 2018;559:570–574. doi: 10.1038/s41586-018-0333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan C, et al. X-ray and cryo-EM structures of the mitochondrial calcium uniporter. Nature. 2018;559:575–579. doi: 10.1038/s41586-018-0330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baradaran R, Wang C, Siliciano AF, Long SB. Cryo-EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature. 2018;559:580–584. doi: 10.1038/s41586-018-0331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, et al. Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake. EMBO J. 2014;33:594–604. doi: 10.1002/embj.201386523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamer KJ, Grabarek Z. 2018 Apo structure of the mitochondrial calcium uniporter protein MICU2. Protein Data Bank. Available at https://www.rcsb.org/structure/6EAZ. Deposited August 3, 2018.
- 26.Thangaratnarajah C, Ruprecht JJ, Kunji ER. Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat Commun. 2014;5:5491. doi: 10.1038/ncomms6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 28.Chen J, Sawyer N, Regan L. Protein-protein interactions: General trends in the relationship between binding affinity and interfacial buried surface area. Protein Sci. 2013;22:510–515. doi: 10.1002/pro.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gromiha MM, Yugandhar K, Jemimah S. Protein-protein interactions: Scoring schemes and binding affinity. Curr Opin Struct Biol. 2017;44:31–38. doi: 10.1016/j.sbi.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman NE, et al. MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell Rep. 2013;5:1576–1588. doi: 10.1016/j.celrep.2013.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vecellio Reane D, et al. A MICU1 splice variant confers high sensitivity to the mitochondrial Ca2+ uptake machinery of skeletal muscle. Mol Cell. 2016;64:760–773. doi: 10.1016/j.molcel.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai MF, et al. Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. eLife. 2016;5:e15545. doi: 10.7554/eLife.15545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrungaro C, et al. The Ca(2+)-dependent release of the mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca(2+) uptake. Cell Metab. 2015;22:721–733. doi: 10.1016/j.cmet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Patron M, et al. MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol Cell. 2014;53:726–737. doi: 10.1016/j.molcel.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carafoli E, Lehninger AL. A survey of the interaction of calcium ions with mitochondria from different tissues and species. Biochem J. 1971;122:681–690. doi: 10.1042/bj1220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamer KJ, et al. MICU1 imparts the mitochondrial uniporter with the ability to discriminate between Ca2+ and Mn2+ Proc Natl Acad Sci USA. 2018;115:E7960–E7969. doi: 10.1073/pnas.1807811115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 44.Pape T, Schneider TR. HKL2MAP: A graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 48.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hulo N, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.