Significance
Specific APOL1 variants are a strong risk factor for human kidney disease. Previous reports examining the intracellular localization of the APOL1 protein in kidney and glomerular podocytes have yielded inconsistent results. Here we demonstrate differential localization of wild-type and risk variant APOL1 polypeptides, with the wild type localizing predominantly to lipid droplets and risk variant forms localizing predominantly to the endoplasmic reticulum. We further demonstrate that the localization of risk variant APOL1 modulates cytotoxic effects, and that perturbations increasing lipid droplet localization of risk variant polypeptides decrease this cytotoxicity. These findings have significant implications for understanding the disease mechanism of APOL1-associated kidney disease and for development of new therapeutic approaches.
Keywords: APOL1, kidney, lipid droplet, podocyte, autophagy
Abstract
Two coding variants in the apolipoprotein L1 (APOL1) gene (termed G1 and G2) are strongly associated with increased risk of nondiabetic kidney disease in people of recent African ancestry. The mechanisms by which the risk variants cause kidney damage, although not well-understood, are believed to involve injury to glomerular podocytes. The intracellular localization and function of APOL1 in podocytes remain unclear, with recent studies suggesting possible roles in the endoplasmic reticulum (ER), mitochondria, endosomes, lysosomes, and autophagosomes. Here, we demonstrate that APOL1 also localizes to intracellular lipid droplets (LDs). While a large fraction of risk variant APOL1 (G1 and G2) localizes to the ER, a significant proportion of wild-type APOL1 (G0) localizes to LDs. APOL1 transiently interacts with numerous organelles, including the ER, mitochondria, and endosomes. Treatment of cells that promote LD formation with oleic acid shifted the localization of G1 and G2 from the ER to LDs, with accompanying reduction of autophagic flux and cytotoxicity. Coexpression of G0 APOL1 with risk variant APOL1 enabled recruitment of G1 and G2 from the ER to LDs, accompanied by reduced cell death. The ability of G0 APOL1 to recruit risk variant APOL1 to LDs may help explain the recessive pattern of kidney disease inheritance. These studies establish APOL1 as a bona fide LD-associated protein, and reveal that recruitment of risk variant APOL1 to LDs reduces cell toxicity, autophagic flux, and cell death. Thus, interventions that divert APOL1 risk variants to LDs may serve as a novel therapeutic strategy to alleviate their cytotoxic effects.
People of recent African ancestry have an approximately fivefold increased risk of developing chronic kidney disease compared with other groups, largely attributable to disease-associated risk variants (G1 and G2) in the apolipoprotein L1 gene, APOL1 (1–3). These kidney disease APOL1 variants differ from wild type (G0) by two amino acid substitutions (p.S342G and p.I384M) in the G1 allele, and by a deletion of two amino acids near the C terminus (p.delN388/Y389) in the G2 allele (1). Increased risk of APOL1-associated kidney disease is inherited as a recessive trait (2, 4, 5). The odds ratio of disease in individuals with high-risk versus low-risk APOL1 genotypes varies from ∼5-fold for hypertension-attributed kidney disease to 10- to 20-fold for focal segmental glomerulosclerosis (FSGS) and 29- to 89-fold for HIV-associated nephropathy (1, 2, 6, 7). The mechanisms by which APOL1 risk variants promote progressive end-stage kidney disease remain elusive. Among the proposed pathways contributing to APOL1 variant-associated cellular toxicity and death are stress-activated protein kinases, necrosis, pyroptosis, autophagy, endoplasmic reticulum (ER) stress, and apoptosis (8–13).
APOL1 is expressed only in humans and some higher primates (14, 15). APOL1 circulates in the plasma in association with the densest fraction of the high-density lipoprotein (HDL) (14, 15). Risk variant APOL1 in serum function as trypanolytic factors to protect against infection by the parasite agent of African sleeping sickness, Trypanosoma brucei rhodesiense. However, levels of circulating APOL1 are not correlated with the presence or absence of APOL1-associated kidney disease (16, 17). The causal mediator of kidney disease is believed to be APOL1 expressed in renal parenchymal cells, rather than circulating plasma APOL1 biosynthesized in and secreted from the liver (18). Although APOL1 is expressed in kidney podocytes, endothelial cells, and proximal tubule epithelial cells, podocytes are considered the major target of APOL1-mediated effects. A recent study by Beckerman et al. (10) demonstrated that mice with podocyte-specific overexpression of APOL1 risk variants develop overt kidney disease. Despite the recessive nature of APOL1-associated disease risk, most investigators believe the effect of risk variant (G1 and G2) APOL1 to be mediated by a toxic gain-of-function effect on cells.
Several groups have investigated the intracellular localization and function of APOL1 in kidney cells, with differing conclusions. APOL1 has previously been reported to localize to the ER, partially to mitochondria, as well as to endosomes, lysosomes, and autophagosomes (10, 19, 20). Interestingly, although APOL1 has been well-characterized as an extracellular HDL-associated molecule (21), a hypothesized role as an intracellular lipid-binding protein has not been demonstrated (10, 19). APOL1, the only secreted member of the APOL family, is thought to be mediated by a putative N-terminal signal peptide (22, 23). However, the mechanisms for APOL1 secretion remain ambiguous, with lack of evidence for the involvement of the secretory pathway and an unclear mode of intracellular trafficking. Of interest, several apolipoproteins, including apolipoprotein A-V (ApoA-V) and apolipoprotein B-100 (24, 25), have been characterized as lipid droplet (LD)-associated proteins. In the case of ApoA-V, Shu et al. (24) demonstrated that ApoA-V is present in the cytosol associated with cytosolic lipid droplets and that the C terminus of ApoA-V is essential for LD association. Moreover, ApoA-V was associated with very low density lipoprotein (VLDL) isolated from culture medium, indicating the possibility of a postsecretory interaction between ApoA-V and VLDL (26).
In this study, we demonstrate differential localization of wild-type (G0) APOL1 predominantly to LDs, in contrast to predominant ER localization of risk variant APOL1 (G1 and G2). Using superresolution structured illumination microscopy (SR-SIM) and live-cell imaging, we demonstrate clear differences in localization between wild-type and risk variant APOL1 expressed in various cell types, including human primary podocytes. Shifting APOL1 localization from the ER to LDs through multiple interventions reduced autophagic flux, cellular toxicity, and death. Modulating APOL1 recruitment to lipid droplets may be a potential therapeutic intervention to delay the progression of APOL1-associated kidney disease.
Results
APOL1 Is a Lipid Droplet-Associated Protein.
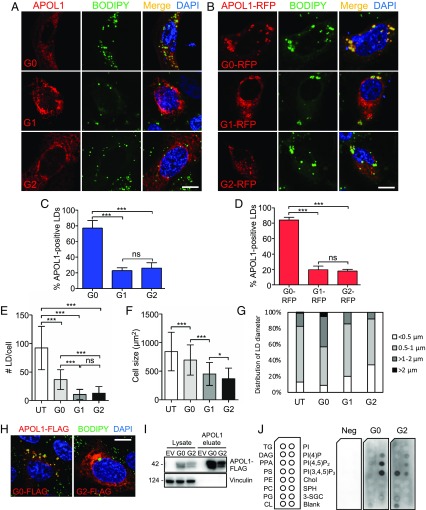
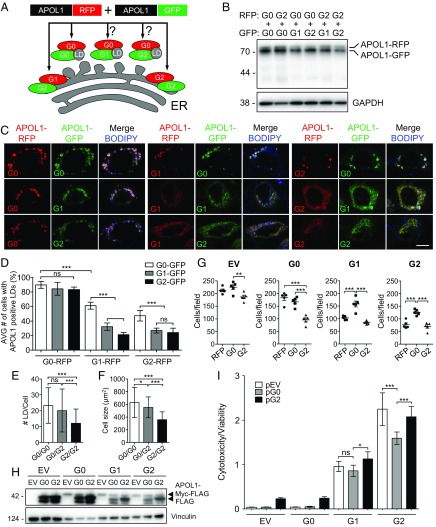
The apparently promiscuous localization of APOL1 suggests it is able to be translocated to various organelles. Since apolipoproteins have an affinity for lipids, we hypothesized possible differences in subcellular organellar localization of APOL1 wild-type (G0) compared with its risk variant forms (G1 and G2). We examined localization of multiple different recombinant constructs of APOL1 (untagged or tagged with FLAG, tagRFPT, and GFP) in primary human podocytes, human and mouse podocyte cell lines, human kidney (HEK-293) cells, human cervical carcinoma (HeLa) cells, and human liver (Huh7) cells (Fig. 1 and SI Appendix, Figs. S1 and S2). Highly overexpressed APOL1 localized throughout cells in a reticular pattern (SI Appendix, Fig. S1). In contrast, wild-type APOL1 G0 in untagged or RFP-tagged form moderately overexpressed in human primary podocytes localized predominantly to LDs (best visualized as RFP-tagged APOL1; Fig. 1B), whereas a smaller fraction of G0 and most of G1 and G2 retained a reticular pattern (Fig. 1); 76.9% of cells expressing untagged APOL1 G0 exhibited APOL1-positive LDs, whereas only 22.8% of G1-expressing cells and 26.0% of G2-expressing cells contained APOL1-positive LDs (Fig. 1C). Similarly, of cells expressing APOL1 G0-RFP, G1-RFP, and G2-RFP, respectively, 84.1, 19.6, and 17.8% exhibited APOL1-positive LDs (Fig. 1D). In primary podocytes, heterologous expression of APOL1 risk variants reduced both numbers of LDs per cell (Fig. 1E) and cell-projection area (Fig. 1F) and the podocytes were more rounded. Human podocytes expressing APOL1 risk variant G1 or G2 exhibited smaller average numbers of LDs (11 and 13, respectively) than podocytes expressing wild-type APOL1 G0 (36 LDs) or untransfected podocytes (92 LDs) (Fig. 1E). In addition, G1-RFP– and G2-RFP–expressing podocytes averaged 434 and 339 µm2, respectively, compared with podocytes expressing G0-RFP (677 µm2) or untransfected podocytes (834 µm2) (Fig. 1F). Moreover, the smaller numbers of LDs in cells expressing APOL1 risk variants G1 and G2 also showed reduced dimension, with none of the cells exhibiting LDs greater than 2 µm in size (Fig. 1G). Our results show reproducible and quantifiable differences in cell size, lipid droplet number, and lipid droplet size between human primary podocytes transiently transfected with G0 and risk variants.
Fig. 1.
APOL1 is a lipid droplet-associated protein that can alter lipid droplet number and size. (A and B) Fluorescence micrographs of human primary podocytes transiently transfected with untagged APOL1 (G0, G1, or G2) (A) or with RFP-tagged APOL1 (B). Lipid droplets were labeled with BODIPY 493/503 (green) and nuclei with DAPI. (Scale bar, 10 µm.) (C and D) Percentage of human primary podocytes containing APOL1-positive LDs (C) (as in A) or APOL1-RFP–positive LDs (D) (as in B). Data from each group represent >50 podocytes from three independent experiments. Data are presented as means ± SD. ***P < 0.001; ns, nonsignificant. (E) Number of LDs per cell in APOL1-RFP–expressing human primary podocytes. Data from three independent experiments, each with at least 30 cells per group, are presented as means ± SD. ***P < 0.001; ns, nonsignificant. (F) Cross-sectional area (number of pixels) of human primary podocytes transfected with APOL1-RFP, quantified from pooled cells from three separate experiments for each group with untransfected (UT) n = 545, G0 n = 131, G1 n = 48, and G2 n = 75 presented as means ± SD. *P < 0.05, ***P < 0.001. (G) Distribution of lipid droplet diameters in untransfected human primary podocytes or podocytes transfected with APOL1-RFP (G0, G1, G2). (H) Indirect immunofluorescence of human primary podocytes transfected with APOL1-FLAG (G0-FLAG and G2-FLAG) and labeled with BODIPY 493/503 and DAPI. (Scale bar, 10 µm.) (I) Immunoblot analysis of purified APOL1-FLAG; 1% Nonidet P-40 lysates from human primary podocytes transfected 16 to 18 h previously with Apol1-FLAG (G0, G2) or empty vector were incubated with anti-FLAG agarose and eluted with 3× FLAG peptide. (J) APOL1-FLAG solid-phase lipid binding. Affinity-purified APOL1-FLAG (G0, G2) or 3× FLAG peptide (negative control) were used in a lipid overlay assay. APOL1-FLAG (G0) bound strongly to PI(4)P, PI(4,5)P2, and PI(3,4,5)P3 and less strongly to PPA and CL. APOL1-FLAG (G2) bound more strongly to PS and CL than to PI(4,5)P2 or PI(3,4,5)P3. One of two experiments with identical results performed with one of two immunopurified preparations of recombinant APOL1. 3-SGC, 3-sulfogalactosylceramide; Blank, solvent blank; Chol, cholesterol; CL, cardiolipin; DAG, diacylglycerol; PPA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI(4)P, phosphatidylinositol (4)-phosphate; PI(4,5)P2, phosphatidylinositol (4,5)-bisphosphate; PI, phosphatidylinositol; PI(3,4,5)P3, phosphatidylinositol (3–5)-trisphosphate; PS, phosphatidylserine; SPH, sphingomyelin; TG, triglyceride.
Because of the differential association of APOL1 variants with LDs, we examined potential differences in lipid binding. Wan et al. (27) previously demonstrated that wild-type APOL1 can bind phosphatidic acid, an anionic phospholipid linked to LD fusion and bound by the LD protein CIDEA (28, 29). Here, APOL1-FLAG fusion proteins (G0 or G2) were immunoprecipitated from human primary podocytes using anti-FLAG antibody conjugated to beads followed by elution with excess 3× FLAG peptide (Fig. 1 H–J). Incubation of the purified APOL1 with solid-phase lipid strips revealed predominant binding of G0-FLAG to PI(4)P, PI(4,5)P2, PI(3,4,5)P3, phosphatidic acid, and, to a lesser degree, cardiolipin, whereas G2-FLAG bound more strongly to phosphatidylserine and cardiolipin (Fig. 1J). These differential lipid binding patterns of APOL1 G0 versus G2 suggest that differential intracellular localization may be mediated in part by differences in lipid binding. Taken together, these results show that APOL1 risk variants have a defect in LD binding which correlates with and may contribute to reduced LD formation in smaller, rounded cells.
Wild-Type APOL1 Localizes Predominantly to LDs, Whereas APOL1 Risk Variants Localize Predominantly to the ER.
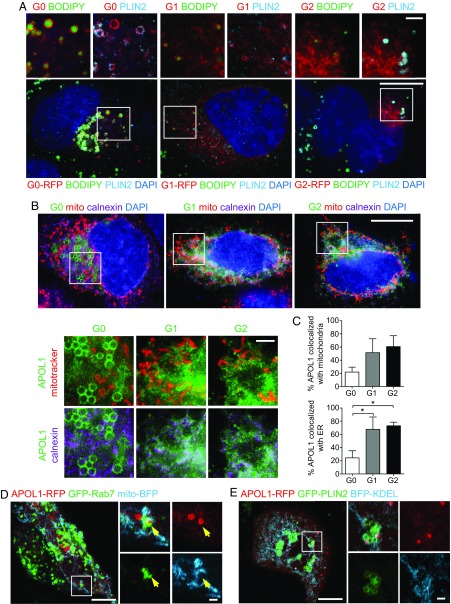
We applied SR-SIM to localize APOL1 relative to known lipid droplet-interacting organelles. In most cells, APOL1 G0 localized to LDs positive for the lipid droplet marker perilipin-2 (PLIN2) (Fig. 2A), whereas G1 and G2 only occasionally were observed at LDs (Fig. 2A). By contrast, most of the APOL1 variants G1 and G2 were found in a reticular pattern colocalizing with the ER marker calnexin (Fig. 2 B and C; 68 and 73% colocalization with calnexin for G1 and G2, respectively), whereas G0 showed significantly less colocalization with calnexin (Fig. 2C; 24%). APOL1-positive LDs derived from the ER partially colocalized with mitochondria (mito-BFP) and occasionally with the endosomal marker GFP-Rab7 (Fig. 2D and Movie S1). Live-cell imaging showed repeated, transient interaction (colocalization) of APOL1 with Rab7-positive structures (SI Appendix, Fig. S3 and Movie S1). APOL1 G0-RFP localized in close proximity to, but rarely completely surrounding, GFP-PLIN2–positive LDs (Fig. 2E and Movie S2), as also shown for endogenous PLIN2. Observed less frequently in still images were smaller, punctate APOL1-positive punctate structures often distinct from but occasionally colocalizing with GFP-PLIN2–positive structures (Fig. 2E, SI Appendix, Fig. S3, and Movie S2). These results reinforce the diversity and plasticity of the previously reported as well as the currently observed multiple sites of intracellular APOL1 localization, including mitochondria and endosomes.
Fig. 2.
Differential localization of wild-type (G0) APOL1 to LDs and its risk variants (G1, G2) primarily to the ER. (A) SR-SIM micrographs of representative primary human podocytes transiently transfected with APOL1-RFP, labeled for perilipin-2 (light blue), and with BODIPY 493/503 and DAPI (blue). [Scale bars, 10 µm and 2 µm (Insets).] (B) SR-SIM micrographs of primary human podocytes transiently transfected with APOL1-GFP, labeled for mitochondria (MitoTracker in red), ER (calnexin in magenta), and DAPI (blue). [Scale bars, 10 µm and 2 µm (Insets).] (C) Colocalization analysis of APOL1-GFP with MitoTracker Red (mitochondria) or calnexin (endoplasmic reticulum). Data are presented as means ± SD. *P < 0.05. (D and E) Yellow arrow indicating colocalization of APOL1-RFP with GFP-Rab7. Airyscan micrographs of live primary human podocytes coexpressing APOL1-RFP (G0) with GFP-Rab7 and mito-BFP (D) or with GFP-PLIN2 and BFP-KDEL (E). [Scale bars, 10 µm and 2 µm (Insets).] See SI Appendix, Fig. S3 for time frame still images. See also Movies S1 and S2.
APOL1 Risk Variants Can Translocate from the ER to LDs by Cell Treatment with Oleic Acid.
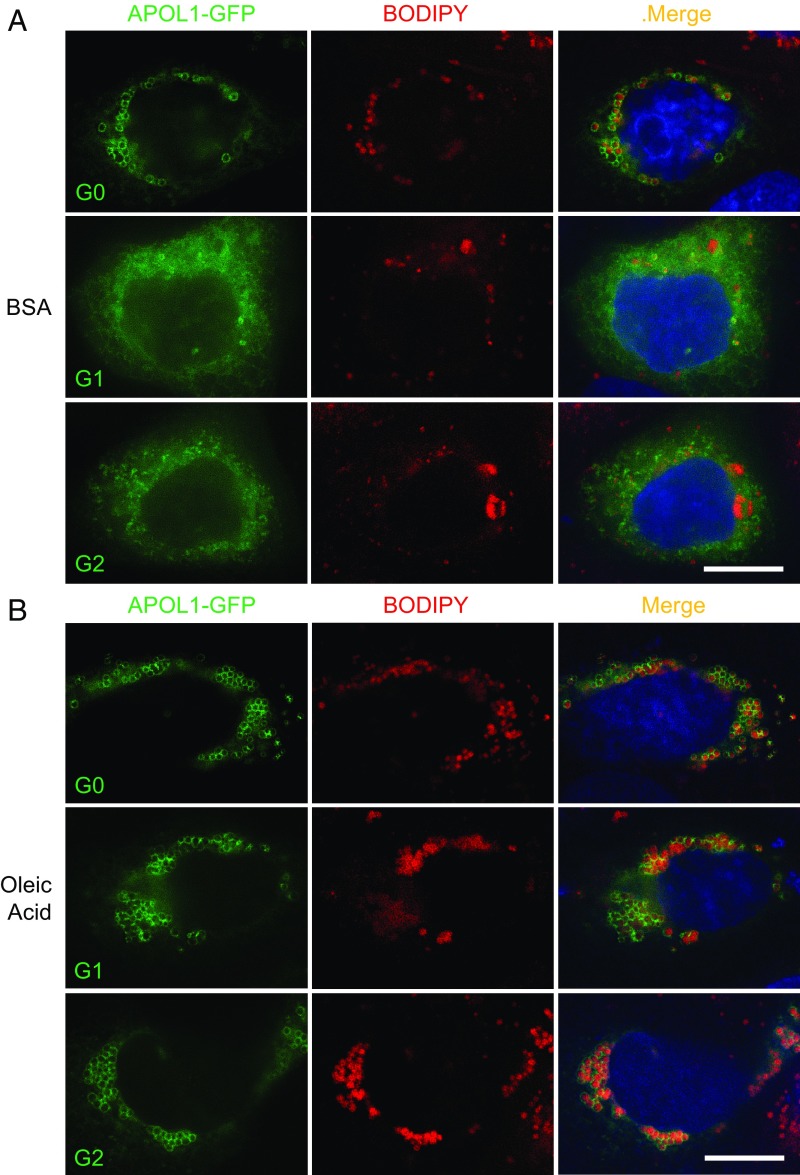
The preferential LD localization of wild-type APOL1 prompted us to hypothesize that the toxic gain-of-function effect of ER-accumulated APOL1 risk variants might be attenuated by forcing the redistribution of APOL1 risk variants to LDs by enhancing LD size and LD number by treatment with oleic acid (OA) (30, 31). OA treatment of human primary podocytes, human podocyte cell lines, or HEK-293 cells transiently transfected with APOL1 G0 led to an increase in LD size and number, as expected (Fig. 3 and SI Appendix, Fig. S4). Moreover, SIM micrographs revealed that OA treatment of podocytes transfected with APOL1 G1 or G2 led to the redistribution of these risk variant polypeptides from an ER-like pattern to LDs (Fig. 3). Thus, OA promotes translocation of APOL1 risk variants from the ER to LDs.
Fig. 3.
Treatment with oleic acid can enrich APOL1(G0)-RFP on LDs and redistribute risk variants G1 and G2 from a reticular pattern to LDs. SR-SIM micrographs of representative human primary podocytes transiently transfected with APOL1-GFP and treated with vehicle (fatty acid-free BSA) (A) or oleic acid (0.8 mM) (B), and then labeled with BODIPY 665/676 and DAPI. (Scale bars, 10 µm.)
Redistribution of APOL1 Risk Variants to Lipid Droplets Reduces Cell Toxicity.
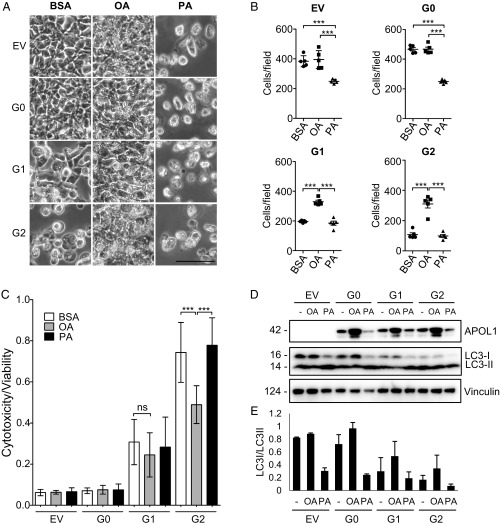
Our observation that APOL1 risk variant polypeptides can translocate from the ER to LDs prompted us to hypothesize that this APOL1 G1/G2 redistribution to LDs might also reduce cell toxicity and death. To determine the effect of promoting LD formation on cytotoxicity, we turned to our well-established HEK-T-Rex cell lines stably expressing tetracycline-inducible wild-type APOL1 and its risk variants (8). As expected, palmitic acid (PA) treatment of HEK-T-Rex cells during induction of APOL1 expression reduced the number of adherent cells regardless of APOL1 genotype (Fig. 4 A and B). In contrast, OA treatment during induction increased the number of adherent cells more than twofold and reduced cytotoxicity (Fig. 4 A and B). Although the cytotoxicity/viability assay results were consistent with bright-field microscopy observations, only G2-overexpressing cells had statistically significant reduction in cytotoxicity following OA treatment (Fig. 4C). OA treatment-associated increased APOL1 and reduced cytotoxicity were accompanied by reduced autophagic flux (diminished LC3-I to LC3-II conversion), suggesting that redistribution of APOL1 risk variants to or near LDs diminished their cytotoxicity, perhaps by autophagic flux inhibition (Fig. 4 D and E). These results demonstrate that the redistribution of APOL1 risk variants to LDs in response to OA is accompanied by reduced autophagic flux and improved cell survival.
Fig. 4.
Treatment of T-Rex-293 cells with oleic acid reduces cellular toxicity and autophagic flux. (A) Bright-field phase-contrast micrographs of representative APOL1 T-Rex-293 cells stably expressing empty vector (EV), G0, G1, or G2 pretreated with vehicle (fatty acid-free BSA), 1 mM oleic acid, or 1 mM palmitic acid for 3 h and then induced for 18.5 h with tetracycline (10 ng/mL) to express APOL1. (Scale bar, 50 µm.) (B) Quantitation of T-Rex-293 cells stably expressing an empty vector or APOL1 variants G0, G1, or G2 treated with BSA, 1 mM OA, or 1 mM PA and induced to express APOL1 with tetracycline (10 ng/mL) for 16 h. ***P < 0.001. (C) Cell cytotoxicity/viability after 22-h treatment with tetracycline (10 ng/mL). Data from two independent experiments, each with eight experimental replicates, are presented as means ± SD. ***P < 0.001; ns, nonsignificant. (D) Immunoblot of APOL1-FLAG, LC3, and vinculin from whole-cell lysates prepared after 22-h induction with tetracycline (10 ng/mL) and treatment with 1 mM OA or 1 mM PA. (E) Quantitation of band intensities by densitometry from two independent experiments for LC3I and LC3II mean densities (from C) displayed as a ratio (means ± SD).
Wild-Type APOL1 Can Recruit Toxic APOL1 Risk Variants to LDs, Reducing Cytotoxicity.
Risk of APOL1-associated kidney disease is inherited as an autosomal recessive trait, despite the gain-of-function toxicity observed in the presence of risk variant polypeptides. We therefore hypothesized that APOL1 G0 coexpression might attenuate toxic effects of APOL1 variants G1 and G2 in podocytes by redirecting localization of G1 and G2 to LDs (Fig. 5A). We used equivalent amounts of APOL1-RFP with APOL1-GFP for each combination of G0 expressed with its risk variants (Fig. 5B). When all nine combinations of RFP-tagged APOL1 (G0, G1, or G2) were coexpressed with GFP-tagged APOL1 (G0, G1, and G2), we observed recruitment of G1-RFP or G2-RFP to the LDs by wild-type APOL1 (G0) (Fig. 5C). G0-GFP coexpression doubled the respective proportions of G1-RFP and G2-RFP localizing at LDs from 32.4 to 61.3% and 24.2 to 47.3% (Fig. 5D). Coexpression of wild-type APOL1 with G2-RFP increased the average number of lipid droplets per cell from 12 to 20 (Fig. 5E) and average cell size from 359 to 546 μm2 (Fig. 5F).
Fig. 5.
Coexpression of APOL1 (G0) with its risk variants (G1 or G2) promotes G1/G2 accumulation on LDs and reduces cell death. (A) Diagram illustrating predicted localization of APOL1-RFP (red) or APOL1-GFP (green) variants to the LD or ER. (B) Immunoblot of whole-cell lysates from human primary podocytes transiently cotransfected with the indicated APOL1-RFP and APOL1-GFP variant DNAs in equal amounts. (C) Airyscan micrographs of human primary podocytes cotransfected with the indicated variants of APOL1-RFP (red), APOL1-GFP (green), and BODIPY 665/676 (blue). Merged images in magenta when overlapped with APOL1-RFP, APOL1-GFP, and BODIPY 665/676. (Scale bar, 10 µm.) (D) Quantitation of human primary podocytes containing detectable LDs positive for APOL1-RFP (G0, G1, G2) when cotransfected with the indicated APOL1-GFP variants. Data from three independent experiments per group, each with ≥50 cells analyzed, are presented as means ± SD. ***P < 0.001; ns, nonsignificant. (E) Number of LDs per cell in human primary podocytes cotransfected with APOL1-RFP and APOL1-GFP. Data presented from at least 25 cells per group are presented as means ± SD. ***P < 0.001; ns, nonsignificant. (F) Cross-sectional area of human primary podocytes cotransfected with APOL1-RFP and APOL1-GFP; quantified for each group, G0-RFP/G0-GFP n = 56, G0-RFP/G2-GFP n = 46, and G2-RFP/G2-GFP n = 65, presented as means ± SD. *P < 0.05, ***P < 0.001. (G) Quantitation from bright-field images using five fields from two representative experiments of tetracycline-inducible transgenic APOL1 T-Rex-293 cells stably expressing empty vector (C-terminal MYC and FLAG tags), G0 (APOL1-G0-T-Rex-293), G1 (APOL1-G1-T-Rex-293), or G2 (APOL1-G2-T-Rex-293) cells transfected with plasmids encoding RFP (control plasmid), G0-RFP (G0), or G2-RFP (G2) 4 h before induction of tetracycline (10 ng/mL)-inducible APOL1 for 16 h. **P < 0.01, ***P < 0.001. (H) Immunoblot of APOL1 T-Rex-293 whole-cell lysates after transient transfection with APOL1-FLAG (G0, G2) or empty vector 4 h before tetracycline (10 ng/mL) induction for 18 h and cell collection at 22 h. (I) Quantitation of cytotoxicity/viability of APOL1 T-Rex-293 cells transiently transfected with APOL1-FLAG (G0, G2) or empty vector for 4 h before tetracycline induction for 18 h. Data from three independent experiments, each with eight experimental replicates, are presented as means ± SD. ***P < 0.001, *P < 0.05; ns, nonsignificant.
We next predicted that wild-type APOL1 G0 expression could also reduce the cytotoxicity of APOL1 risk variant polypeptides, and tested this hypothesis in APOL1 T-Rex cells (8). When each APOL1 T-Rex line was transfected with either G0-RFP or G2-RFP, there was a reduced number of cells for the empty vector and G0 lines following transfection with G2-RFP, suggesting augmentation of the toxic effect of the risk variants (Fig. 5G). In contrast, when the G1 and G2 lines were transfected with G0-RFP, there was almost double the number of cells compared with the control RFP transfection (Fig. 5G). Cytotoxicity was reduced postinduction of FLAG-tagged APOL1 risk variants after subsequent transient transfection of untagged G0, but not G2 (Fig. 5I). Consistent with the effects of OA, APOL1 risk variant recruitment to LDs by overexpressed wild-type APOL1 reduced cytotoxicity and cell death. These results suggest that APOL1 risk variant polypeptide accumulation in the ER promotes cytotoxicity that can be alleviated by wild-type APOL1 recruitment of APOL1 risk variants to LDs.
Discussion
APOL1 is a component of high-density lipoprotein circulating in human blood. Its association with HDL and its well-established intracellular endosomal–lysosomal trafficking in trypanosomes prompted us to hypothesize a role for APOL1 in lipid transport and/or metabolism (14, 21, 32). Previous groups have suggested that APOL1 might associate with lipid droplets. Here, we provide direct evidence for APOL1 localization at and recruitment to lipid droplets. We have also demonstrated clear differences in the intracellular localization of wild-type (G0) and risk variant (G1 and G2) APOL1 polypeptides. In resting cells, the G0 form localizes predominantly to LDs, while the G1 and G2 risk variant forms localize preferentially to the ER (Fig. 6). LDs are dynamically synthesized from the ER and directly contact other organelles, including mitochondria, endosomes, Golgi complex, and peroxisomes (33). Optimal detection of APOL1 at LDs required selection of cells with moderate APOL1 expression level. Cells highly overexpressing APOL1 localized to the ER but had increased risk of artifactual mislocalization. For example, Granado et al. (19) concluded that APOL1 highly expressed in podocytes localized predominantly to the ER, despite the presence of APOL1 in round droplet-like structures in some micrographs.
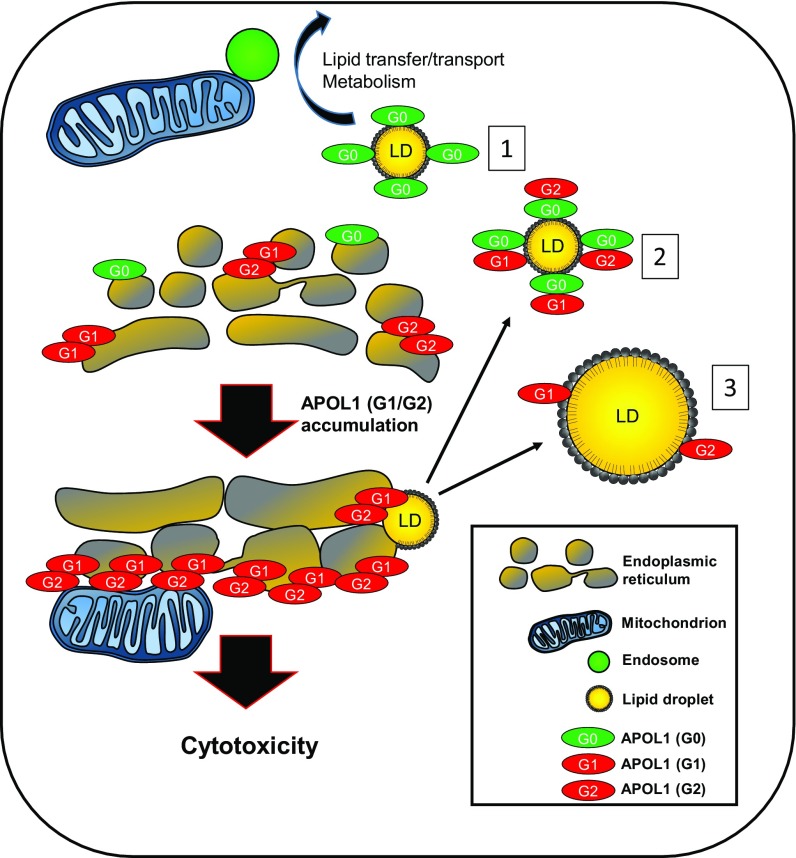
Fig. 6.
Model for APOL1-mediated cytotoxicity and its reduction by maneuvers promoting LD formation and enlargement. Under normal physiologic conditions, podocytes express low levels of APOL1 and do not cause cytotoxicity. Innate immune activation by environmental stress or viral infection are thought to up-regulate APOL1. Accumulating levels of G1 and G2 may reach a threshold level causing burden on organelles such as the ER and mitochondria, possibly via communication via mitochondrial-associated membranes. Excess APOL1 will activate cell stress pathways including stress-activated protein kinases, ER stress pathways, and mitochondrial oxidation pathways. LD formation might (partially or totally) relieve stress pathways caused by excessive APOL1 risk variant accumulation in the ER. 1: For G0, APOL1 can be efficiently diverted away from the ER to the LD (a relatively inert reservoir for lipophilic proteins) and the podocyte can process APOL1 to levels below a threshold causing toxicity. APOL1 directed to lipid droplets can interact with other organelles, including endosomes and mitochondria, which may aid the processing, transport, and metabolism of APOL1. 2: Cooverexpression of wild-type APOL1 can recruit APOL1 risk variants to lipid droplets. 3: Treatment with OA can increase LD size and number, acting as shuttles to sequester toxic APOL1 risk variant polypeptides away from the ER.
We (and others) have observed that anti-APOL1 antibodies readily detect overexpressed APOL1 at the ER, which impairs discrimination of LD-associated APOL1 from that in the ER. Despite various fixation methods and testing multiple APOL1 antibodies in podocytes stimulated with IFN gamma to increase APOL1 expression, we were unable to convincingly detect endogenous APOL1 by immunofluorescence. This may be due to very low expression of endogenous APOL1 or poor epitope unmasking. Future studies will require validation of our results for endogenous APOL1. This could be addressed, for example, by using the CRISPR/Cas9 system to create the point mutations in endogenous APOL1 with a knockin of an epitope tag.
We found that cells treated with OA show enhancement of both LD size and number as well as increased recruitment of APOL1 to LDs. The excellent subcellular morphology detectable in primary human podocytes (Celprogen) allowed clear discrimination of an APOL1 subpopulation localizing to LDs, but the degree of APOL1 LD association varied by genotype. APOL1 LD localization was not cell type-specific, as we observed this in Huh7, HeLa, and HEK-293 cells (SI Appendix, Fig. S2). APOL1 association with LDs may be proportional to LD size or LD surface monolayer lipid composition. The effect of APOL1 G0 accumulation on lipid droplet formation remains unclear. We observed that LDs in the presence of APOL1 G0 were generally larger than those LDs lacking surrounding G0 (Fig. 1G). APOL1 G0 occupancy on the LD phospholipid monolayer surface might prevent binding of other LD-associated proteins. Alternatively, G0 might itself promote LD formation and/or growth, or regulate steady-state LD size. Moreover, APOL1 may catalyze lipid exchange between and/or among organelles, similar to the functions proposed for VPS13A and VPS13C, proteins that bind to and tether the ER to mitochondria, endosomes, and lipid droplets (34).
LDs regulate the cytotoxicity of risk variant APOL1 polypeptides. Our results demonstrate that recruitment of APOL1 risk variants to LDs is associated with reduced cytotoxicity in the setting of independent manipulations we used to alter APOL1–LD association: (i) coexpression of G0-RFP, which recruits G1 and G2 to LDs, and (ii) oleic acid treatment, which recruits APOL1 to lipid droplets. Our results suggest that structural differences distinguishing G1 and G2 from G0 may hinder association with (binding to or incorporation into) LDs. Alternatively, risk variants may be retained at the ER by self-aggregation or interactions with resident ER proteins. If APOL1 risk variants fold improperly under certain conditions, ER retention of APOL1 G1 or G2 might activate ER stress to cause podocyte injury (12). As has been described in other settings, LDs can act as a protective reservoir for unfolded proteins and toxic aggregates by preventing interactions with other cellular compartments (35). This function appears to be relevant to control of APOL1-mediated toxicity as well.
The localization of APOL1 to lipid droplets opens the possibility of APOL1 protein transfer to various organelles including the mitochondria, ER, lysosomes, peroxisomes, and Golgi complex (33). APOL1 localized to lipid droplets may explain the promiscuous intracellular localization reported in the literature to various organelles. We anticipate that future live-cell imaging experiments will be able to better characterize APOL1 intracellular trafficking and possibly secretion via the secretory pathway or an alternative lipid droplet-dependent pathway. Recent studies have suggested that APOL1 risk variant polypeptides modulate mitochondrial function (10, 19). Altered association of APOL1 with LDs may alter APOL1 delivery to mitochondria, possibly controlling cytotoxicity. Disruption of podocyte autophagy has been shown to lead to FSGS (36). Altered autophagic flux, shown here in G1- and G2-expressing cells and perhaps modulated by altered LD-mediated trafficking of APOL1 to mitochondria, may contribute to disease pathogenesis. Stimuli that alter the size and/or number of intracellular LDs and targeting of APOL1 to and from LDs may therefore be important modulators of APOL1-associated disease, with potential therapeutic benefit. Regulation of APOL1 trafficking, turnover, and lipid binding at LDs will also be an important avenue for future research. Determining whether APOL1 structure, protein–protein interaction, or lipid droplet membrane composition mediates APOL1 binding to lipid droplets will be the next step in characterizing the function of APOL1 at lipid droplets.
How to reconcile the recessive mode of inheritance of APOL1-associated disease risk with the apparent gain-of-function effects of G1 and G2 APOL1 on cells has been an enigma (3, 37). Our results may help to explain the recessive mode of inheritance of APOL1-associated kidney disease. The ability of G0 APOL1 to recruit G1 and G2 APOL1 to lipid droplets can explain the lack of significantly increased disease risk in G0/G1 or G0/G2 humans. Coexpression of APOL1 G0 with risk variant APOL1 (G1 or G2) both increased localization of risk variant APOL1 to LDs and reduced cytotoxicity.
Understanding the mechanisms by which G0 binds to lipid droplets and how it recruits G1 or G2 to LDs will be an important direction for future study. APOL1 may function in a manner similar to the LD-associated protein CIDEA, which has an amphipathic helix that facilitates embedding in the phospholipid monolayer and binding to phosphatidic acid (29). Future studies of the reciprocal regulation of APOL1 targeting to LDs as a function of APOL1 genotype will be important for understanding the mechanism of kidney disease in people of recent African ancestry, and may inform the development of new approaches to disease therapy and prevention.
Materials and Methods
Chemicals and Reagents.
BODIPY 493/503 (D3922), BODIPY 665/676 (B3932), and MitoTracker Red CMXRos (M7512) were from Thermo Fisher Scientific. Oleic acid (NC9893458) and palmitic acid (NC1247921) were from Nu-Chek Prep (Thermo Fisher Scientific). Low-endotoxic, fatty acid-free BSA (Sigma-Aldrich; A8806) was used for cell culture.
Human Primary Podocytes and Cell Culture.
Human primary podocytes (Celprogen) were grown in the manufacturer’s specified medium on flasks precoated with human podocyte primary cell-culture complete extracellular matrix (Celprogen). Stable tetracycline-inducible transgenic APOL1-expressing HEK T-Rex-293 cell lines (APOL1-G0-T-Rex-293, APOL1-G1-T-Rex-293, and APOL1-G2-T-Rex-293) were grown in DMEM (Corning) supplemented with 10% tetracycline system-approved FBS (Atlanta Biologicals), 0.2 mg/mL zeocin, 2 µg/mL blasticidin, and 1% antibiotic-antimycotic (Corning) at 37 °C and 5% CO2, as previously described (8). Other HEK-293 cells were grown in the same conditions. Huh7 cells (ATCC), HEK cells (ATCC), and HeLa cells (ATCC) were cultured in DMEM with 10% FBS and penicillin-streptomycin at 37 °C and 5% CO2. Mouse podocytes of C57BL/6 strain (38) were immortalized with a temperature-sensitive T antigen (abm; LV629). Mouse podocytes and immortalized human podocytes (39) were maintained in RPMI (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific), insulin-transferrin-selenium liquid medium supplement (Sigma), and 1% penicillin-streptomycin (Thermo Fisher Scientific). For propagation, mouse and human podocytes were maintained at 33 °C and, for experimental analysis, cells were differentiated at 37 °C.
Cloning and Plasmids.
APOL1, under the control of a human CMV promoter, was cloned using the pCMV6-entry vector encoding the full-length wild-type APOL1. APOL1 cDNA was purchased from OriGene. APOL1 variants G1 and G2 were generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). APOL1-tagRFPT was cloned by insertion of tagRFPT at the C terminus of APOL1 at the Pst1/Fse1 sites (OriGene). APOL1-GFP was constructed using gBlocks Gene Fragment (Integrated DNA Technologies) for EGFP and cloned into the Pst1/Fse1 site of APOL1-tagRFPT (Integrated DNA Technologies). BFP-KDEL (Addgene; plasmid 49150), mito-BFP (Addgene; plasmid 49151), GFP-Rab7A (Addgene; plasmid 61803), and BFP-Rab5 (Addgene; plasmid 49147) were gifts from Gia Voeltz, University of Colorado at Boulder, Boulder, CO (40–42). EGFP-ADRP (Addgene; plasmid 87161) was a gift from Elina Ikonen,University of Helsinki (43).
Transfections.
Cells were transfected using Lipofectamine 3000 (Thermo Fisher Scientific) in OptiMEM (Life Technologies) using 0.5 µg plasmid cDNA and 1 µL each of P3000 and Lipofectamine reagent per well, as per the manufacturer’s instructions.
Immunoblot.
Cell lysates prepared from cells washed with ice-cold PBS were lysed in 1% Nonidet P-40 lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40; Boston BioProducts) supplemented with cOmplete, Mini, EDTA-free Protease Inhibitor Mixture (Sigma-Aldrich) and PhosSTOP (Sigma-Aldrich). Lysates cleared by 10-min centrifugation at 16,000 × g and 4 °C were boiled 5 min in SDS sample buffer with β-mercaptoethanol and separated by SDS/PAGE (Bio-Rad). Proteins transferred to PVDF membranes were blocked in 5% (wt/vol) skim milk in Tris-buffered saline, 0.05% Tween 20 (TBST) for 1 h, and then incubated overnight at 4 °C with primary antibodies (1:1,000 unless otherwise specified). Immunoblots were washed with TBST and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (1:2,500; Santa Cruz Biotechnologies), visualized by ECL chemiluminescence (SuperSignal West Dura or Femto Kit; Life Technologies), and imaged (ProteinSimple FluorChem E or R; Bio-Techne). Band intensity was quantitated by densitometric analysis using ImageJ (version 1.47; NIH). Antibodies were from the following sources: APOL1 (1:1,000; Sigma-Aldrich; HPA018885), LC3A/B [1:1,000; Cell Signaling Technologies (CST); 12741], LC3B (1:1,000; CST; 2275), Beclin-1 (1:1,000; CST; 1395), ATG5 (1:1,000; CST; 12994), vinculin (1:2,500; Sigma-Aldrich; V9131); WT1 (1:100; Santa Cruz; sc-192), podocin (1:1,000; Sigma; P0732), and nephrin (1:1,000; Abcam; 80299).
Immunoprecipitation and Elution of APOL1-FLAG.
Human primary podocytes transiently transfected with APOL1-FLAG (G0 or G2) or empty vector for 16 to 20 h were rinsed with cold PBS, pelleted, and lysed in 1% lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40). Lysates were incubated with anti–FLAG-agarose affinity gel (Sigma-Aldrich; F2426) at 4 °C overnight. The agarose beads were then washed three times with ice-cold TBS (50 mM Tris⋅HCl, 150 mM NaCl, pH 7.4) and incubated with 150 ng/µL 3× FLAG peptide (Sigma-Aldrich; F4799) in TBS at 4 °C overnight to elute APOL1-FLAG.
APOL1-FLAG Solid-Phase Lipid Binding Assay.
Affinity-purified APOL1-FLAG (G0, G2) or 3× FLAG peptide (negative control) was used in a lipid overlay assay as per the manufacturer’s instructions. Prespotted membrane lipid strips (P-6002; Echelon Biosciences) were blocked in 3% fatty acid-free BSA (A7030; Sigma-Aldrich) for 1 h at room temperature and incubated with the purified APOL1-FLAG protein (∼500 ng/mL) in 3% fatty acid-free BSA in TBST for 1 h at room temperature with gentle agitation. Membranes were probed with primary anti-APOL1 antibody (1:1,000; Sigma-Aldrich; HPA018885), followed by secondary goat anti-rabbit antibody conjugated to horseradish peroxidase, and ECL chemiluminescence detection and imaging as described for the immunoblots.
Bright-Field Microscopy.
Bright-field microscopy was performed using a CKX31 inverted microscope (Olympus) with a CAch N 10×/0.25 PhP (infinity)/1/FN22 or LCAch N 20×/0.40 PhP (infinity)/1/FN22 objective equipped with an Exmor RS IMX315 12 MP camera (Sony) for image acquisition.
Immunofluorescence and Confocal Microscopy.
Cells grown on fibronectin-coated glass coverslips were transiently transfected with untagged or C-terminally tagged RFPT, GFP, or FLAG-tagged APOL1 (G0, G1, G2). Cells were fixed for 20 min with 4% paraformaldehyde, quenched with 50 mM ammonium chloride, and permeabilized with 0.3% Triton X-100. Fixed cells were blocked with 0.2% gelatin in PBS followed by incubation with primary antibodies for 1 h or overnight. For perilipin-2 staining, an additional postfixation permeabilization step using 1% SDS in PBS for 1 min improved fluorescence signal at LDs. Primary antibodies included APOL1 (1:100; Sigma-Aldrich; HPA018885), ADRP/perilipin-2 (1:100; Proteintech; 15294-1-AP), calnexin (1:100; CST; 2679), EEA1 (1:100; CST; 3288), LC3A/B (1:100; CST; 12741), LAMP2 (1:50; Santa Cruz; sc-18822), TOM20 (1:100; Millipore; MABT166), and Rab7 (1:100; Sigma; R8779). Washed cells were then incubated 1 h with Alexa Fluor 488-, 555-, or 647-labeled secondary antibodies (Thermo Fisher Scientific). Fixed, stained cells were mounted with ProLong Gold Antifade Reagent with or without DAPI (Thermo Fisher Scientific). For lipid droplet staining, cells were incubated 5 to 15 min with BODIPY 493/503 (1 µg/mL) or BODIPY 665/676 (1 µg/mL). Confocal images were acquired by LSM 880 laser scanning microscope (Zeiss) with a 63× oil lens, N.A. 1.4.
Live-Cell, Time-Lapse Imaging by Confocal Superresolution Microscopy with Airyscan.
For live-cell imaging, cells were cultured in glass-bottomed microwell dishes (MatTek) and imaged in the incubation module of the LSM 880 with Airyscan (Zeiss) at 37 °C and 5% CO2. ZEN black edition software version 2 (Zeiss) was used for acquisition and analysis.
All images and movies were acquired in superresolution, Fast Airyscan mode using an oil immersion objective Plan-Apochromat 63×/1.4 oil DIC M27. Detector gain and pixel dwell times were adjusted to the lowest operationally possible values to minimize saturation and bleaching effects. ImageJ (version 1.51n) was used to correct for photobleaching using the histogram matching bleach correction method. Colocalization analysis of APOL1 with organelles (MitoTracker Red and calnexin) was performed manually using Volocity 6.3 (PerkinElmer).
Superresolution Structured Illumination.
For SR-SIM, cells were imaged using the ELYRA PS.1 illumination system (Zeiss) with a 63× oil objective lens, N.A. 1.4. Four lasers were used in image acquisition (exciting at 642, 561, 488, and 405 nm). Three orientation angles of the excitation grid were acquired in each Z plane. Raw images were SIM-processed in ZEN Black (Zeiss) and exported in TIFF format. The selected cell images were cropped using Adobe Photoshop CC 2017.
Cytotoxicity Assay.
HEK T-Rex-293 cells were plated at 1 × 105 per well in 96-well plates (Corning). Cells induced with 10 ng/mL tetracycline for 16 to 22 h were subjected to testing by the MultiTox-Fluor Multiplex cytotoxicity/viability assay (Promega) per the manufacturer’s instructions using a SpectraMax M5 microplate reader (Molecular Devices) as previously described (8).
Statistical Analysis.
Analysis among three groups, each including at least three biological replicates, was by ANOVA followed by Tukey’s multiple comparison test. Data are presented as means ± SD with P values as indicated (*P < 0.05, **P < 0.01, ***P < 0.001; ns, nonsignificant). GraphPad Prism 5 was used to calculate statistical significance.
Supplementary Material
Acknowledgments
Dr. Doug Richardson and Sven Terclavers from the Harvard Center for Biological Imaging provided technical assistance on the Zeiss Elyra and LSM 880 Airyscan. Dr. Lay Hong Ang and Mr. Aniket Gad (Beth Israel Deaconess Medical Center) provided technical assistance for confocal microscopy and image analysis. This work was supported by grants from the NIH (MD007898), DoD (W81XWH-14-1-0333), NephCure Foundation, Vertex Pharmaceuticals, and Ellison Foundation. J.C. was supported by an Alberta Innovates Health Solutions Clinician Fellowship and is a KRESCENT Postdoctoral Fellow.
Footnotes
Conflict of interest statement: M.R.P. and D.J.F. have filed patents related to APOL1-associated kidney disease, and D.J.F. and M.R.P. own equity in ApoLo1 Bio, LLC.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820414116/-/DCSupplemental.
References
- 1.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman DJ, Pollak MR. Apolipoprotein L1 and kidney disease in African Americans. Trends Endocrinol Metab. 2016;27:204–215. doi: 10.1016/j.tem.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman DJ, Kozlitina J, Genovese G, Jog P, Pollak MR. Population-based risk assessment of APOL1 on renal disease. J Am Soc Nephrol. 2011;22:2098–2105. doi: 10.1681/ASN.2011050519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akilesh S, et al. Arhgap24 inactivates Rac1 in mouse podocytes, and a mutant form is associated with familial focal segmental glomerulosclerosis. J Clin Invest. 2011;121:4127–4137. doi: 10.1172/JCI46458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasembeli AN, et al. APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol. 2015;26:2882–2890. doi: 10.1681/ASN.2014050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olabisi OA, et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci USA. 2016;113:830–837. doi: 10.1073/pnas.1522913113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan X, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol. 2014;307:F326–F336. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckerman P, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat Med. 2017;23:429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhaorigetu S, Wan G, Kaini R, Jiang Z, Hu CA. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy. 2008;4:1079–1082. doi: 10.4161/auto.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen H, et al. APOL1 risk variants cause podocytes injury through enhancing endoplasmic reticulum stress. Biosci Rep. 2018;38:BSR20171713. doi: 10.1042/BSR20171713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heymann J, Winkler CA, Hoek M, Susztak K, Kopp JB. Therapeutics for APOL1 nephropathies: Putting out the fire in the podocyte. Nephrol Dial Transplant. 2017;32(Suppl 1):i65–i70. doi: 10.1093/ndt/gfw402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duchateau PN, et al. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J Biol Chem. 1997;272:25576–25582. doi: 10.1074/jbc.272.41.25576. [DOI] [PubMed] [Google Scholar]
- 15.Lugli EB, Pouliot M, Portela MdPM, Loomis MR, Raper J. Characterization of primate trypanosome lytic factors. Mol Biochem Parasitol. 2004;138:9–20. doi: 10.1016/j.molbiopara.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Thomson R, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA. 2014;111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruggeman LA, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol. 2014;25:634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman BI, et al. APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation. 2016;100:194–202. doi: 10.1097/TP.0000000000000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granado D, et al. Intracellular APOL1 risk variants cause cytotoxicity accompanied by energy depletion. J Am Soc Nephrol. 2017;28:3227–3238. doi: 10.1681/ASN.2016111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhavan SM, et al. APOL1 variants change C-terminal conformational dynamics and binding to SNARE protein VAMP8. JCI Insight. 2017;2:92581. doi: 10.1172/jci.insight.92581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanhollebeke B, Pays E. The function of apolipoproteins L. Cell Mol Life Sci. 2006;63:1937–1944. doi: 10.1007/s00018-006-6091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith EE, Malik HS. The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res. 2009;19:850–858. doi: 10.1101/gr.085647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dummer PD, et al. APOL1 kidney disease risk variants: An evolving landscape. Semin Nephrol. 2015;35:222–236. doi: 10.1016/j.semnephrol.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shu X, Ryan RO, Forte TM. Intracellular lipid droplet targeting by apolipoprotein A-V requires the carboxyl-terminal segment. J Lipid Res. 2008;49:1670–1676. doi: 10.1194/jlr.M800111-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- 26.Shu X, Chan J, Ryan RO, Forte TM. Apolipoprotein A-V association with intracellular lipid droplets. J Lipid Res. 2007;48:1445–1450. doi: 10.1194/jlr.C700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Wan G, et al. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fei W, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barneda D, et al. The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix. eLife. 2015;4:e07485. doi: 10.7554/eLife.07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto Y, et al. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Gluchowski NL, et al. Identification and characterization of a novel DGAT1 missense mutation associated with congenital diarrhea. J Lipid Res. 2017;58:1230–1237. doi: 10.1194/jlr.P075119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fornoni A, Merscher S, Kopp JB. Lipid biology of the podocyte—New perspectives offer new opportunities. Nat Rev Nephrol. 2014;10:379–388. doi: 10.1038/nrneph.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valm AM, et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar N, et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol. 2018;217:3625–3639. doi: 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartleben B, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010;120:1084–1096. doi: 10.1172/JCI39492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruggeman LA, O’Toole JF, Sedor JR. APOL1 polymorphisms and kidney disease: Loss-of-function or gain-of-function? Am J Physiol Renal Physiol. 2019;316:F1–F8. doi: 10.1152/ajprenal.00426.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian B, et al. Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int. 2016;90:363–372. doi: 10.1016/j.kint.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saleem MA, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 40.Friedman JR, et al. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK. ER contact sites define the position and timing of endosome fission. Cell. 2014;159:1027–1041. doi: 10.1016/j.cell.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salo VT, et al. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 2016;35:2699–2716. doi: 10.15252/embj.201695170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.