Significance
Microbial adaptation to the host environment is crucial for commensalism and pathogenesis. One adaptive strategy includes the production of lipolytic enzymes that allow acquisition of free fatty acids and disruption of lipid barriers. We demonstrate that a secreted lipase of Staphylococcus aureus also facilitates immune evasion by inactivating bacterial-derived lipoproteins, a pathogen-associated molecular pattern required for innate immune activation. Lipase activity changes the local inflammatory environment and promotes bacterial survival during infection. Thus, this work provides evidence that bacterial lipases can have unappreciated roles in immunomodulation. While many S. aureus-secreted virulence factors exert their effects directly on host cells, our study provides an interesting example of a bacterial enzyme that acts on endogenous substrates to mask immune recognition.
Keywords: lipase, MRSA, Toll-like receptor 2, immune cell activation, lipoprotein
Abstract
Commensal and pathogenic bacteria hydrolyze host lipid substrates with secreted lipases and phospholipases for nutrient acquisition, colonization, and infection. Bacterial lipase activity on mammalian lipids and phospholipids can promote release of free fatty acids from lipid stores, detoxify antimicrobial lipids, and facilitate membrane dissolution. The gram-positive bacterium Staphylococcus aureus secretes at least two lipases, Sal1 and glycerol ester hydrolase (Geh), with specificities for short- and long-chain fatty acids, respectively, each with roles in the hydrolysis of environmental lipids. In a recent study from our group, we made the unexpected observation that Geh released by S. aureus inhibits activation of innate immune cells. Herein, we investigated the possibility that S. aureus lipases interface with the host immune system to blunt innate immune recognition of the microbe. We found that the Geh lipase, but not other S. aureus lipases, prevents activation of innate cells in culture. Mutation of geh leads to enhancement of proinflammatory cytokine production during infection, increased innate immune activity, and improved clearance of the bacterium in infected tissue. These in vitro and in vivo effects on innate immunity were not due to direct functions of the lipase on mammalian cells, but rather a result of inactivation of S. aureus lipoproteins, a major pathogen-associated molecular pattern (PAMP) of extracellular gram-positive bacteria, via ester hydrolysis. Altogether, these studies provide insight into an adaptive trait that masks microbial recognition by innate immune cells through targeted inactivation of a broadly conserved PAMP.
Pathogenic and commensal microbes regularly interface with their host to promote survival. They do so through the production of myriad surface and secreted factors that facilitate nutrient acquisition, adherence, and evasion of host antimicrobial defenses (1, 2). Secreted lipases constitute a class of bacterial enzymes that play a significant role in both microbial infection and commensalism (3, 4). In lipid-rich environments, many microbes express lipases to break down host-derived lipids into free fatty acids for nutrient acquisition, which promotes bacterial colonization and can lead to disease (3, 4). Lipase activity is also critical in environments where esterified fatty acid derivatives constitute a formidable host barrier to infection. In an infectious niche, such as the cystic fibrosis lung, lipases secreted by Pseudomonas aeruginosa accelerate lung destruction by hydrolyzing esterified fatty acids within pulmonary surfactant, leading to enhancement of inflammatory responses (5–7). In skin, a host site rich in sebum triacylglycerides containing esterified fatty acids, infections caused by Propionibacterium acnes progress, in part, due to a secreted lipase that cleaves sebum triacylglycerides into glycerol and free fatty acids that ultimately cause inflammation in the sebaceous follicle (8, 9). Thus, the utility of microbial secretion of lipolytic enzymes in the host environment is seen at both the levels of microbial nutrient acquisition and immune activation.
Opportunistic pathogens, including those from the genus Staphylococcus, use lipases for both colonization and infection (10–15). The opportunistic bacterial pathogen Staphylococcus aureus is a major threat to public health and causes a range of infections from mild superficial lesions to potentially fatal deep-seated and disseminated infections (16, 17). Recent clinical studies indicate that secreted lipases produced by S. aureus are likely to contribute to the pathobiology of disease in humans (18–22). More than 80% of clinical isolates of S. aureus from patients with infections like impetigo, furunculosis, bacteremia, peritonitis, and osteomyelitis have lipolytic activities (18–21), and isolates from disseminated or deep infections have more lipolytic activity than those from localized or superficial infection sites (20). These clinical data suggest that lipases may contribute to S. aureus infection and dissemination. This is supported by experimental work, which indicates S. aureus lipases circumvent innate immunity by inactivating bactericidal lipids and possibly interfering with phagocytosis and chemotaxis of granulocytes (23, 24). Further, S. aureus harboring a mutation in the gene encoding one lipase, glycerol ester hydrolase (Geh), is attenuated in a murine peritonitis infection model (25), although a recent study from Nguyen et al. (26) did not report attenuation for this same mutant in skin and soft tissue or pneumonia models of infection. S. aureus harbors at least two lipases, Sal1 and Geh (Sal2), as well as a putative esterase, SAUSA300_0641. The enzymatic activities of Geh and Sal1 have been well characterized. Geh acts on substrates with long-chain fatty acids and has a basic pH optimum of 8.0 (27, 28), whereas Sal1 favors substrates containing short-chain fatty acids and has a more acidic pH optimum of 6.0 (28, 29). Furthermore, recent studies have demonstrated that Geh is capable of hydrolyzing host lipids to liberate free fatty acids in an adaptive strategy that allows for bacterial membrane phospholipid synthesis from host-derived free fatty acids (30). Notably, some isolates of S. aureus harbor a prophage that results in inactivation of Geh (31). Our bioinformatic analyses of S. aureus genomes deposited in the National Center for Biotechnology Information indicate 15% of strains with complete genome data (441 strains in total) harbor a prophage within the geh gene, while 85% of deposited strains contain intact Geh. Little is known about SAUSA300_0641 except its similarity to acetyl-esterases.
The first line of defense against S. aureus infection is the innate immune system, which includes such effectors as antimicrobial peptides, the complement system, and bactericidal phagocytes (32). Together, these effectors elicit an immediate and aggressive response to S. aureus that is usually sufficient to prevent disease spread and promote infection clearance (33, 34). Appropriate sensing of pathogen-associated molecular patterns (PAMPs) is critical to innate immune recognition of S. aureus (33, 34). Notably, Toll-like receptor (TLR)–mediated recognition of S. aureus PAMPs provides an essential cue for innate immune activation (35–42). Mice deficient in TLR2, which recognizes acyl chains of bacterial lipoproteins, exhibit increased sensitivity to S. aureus infection and are unable to eradicate the microbe (38, 43). Thus, TLR2-based signaling in response to S. aureus lipoproteins is central to initiation of an effective innate response, and it is possible that S. aureus has adapted in unrecognized ways to evade this recognition.
In a prior study, we conducted a transposon mutant library screen to identify S. aureus-secreted factors that disrupt innate immune activation (44). We identified the lipase Geh, but not Sal1 or SAUSA300_0641, as a broad immune-inhibitory factor that blunts primary murine macrophage activation. In this work, we sought to test the hypothesis that Geh inhibits innate immune activation to promote survival during infection. We determined that Geh is the primary immune-inhibitory lipase secreted by S. aureus. Geh has ester hydrolase activity on the esterified acyl chains of S. aureus-derived lipoproteins, the major agonists for TLR2; through this activity, it blunts lipoprotein-based immune activation. Indeed, a murine systemic infection model showed that Geh limits host proinflammatory responses, leading to delayed clearance during infection. Altogether, our findings suggest that, in addition to their activities related to nutritional adaptation, secreted lipases have the potential to contribute to infection by inactivating PAMPs, thereby blunting immune-mediated clearance.
Results
Geh Prevents Innate Immune Cell Activation.
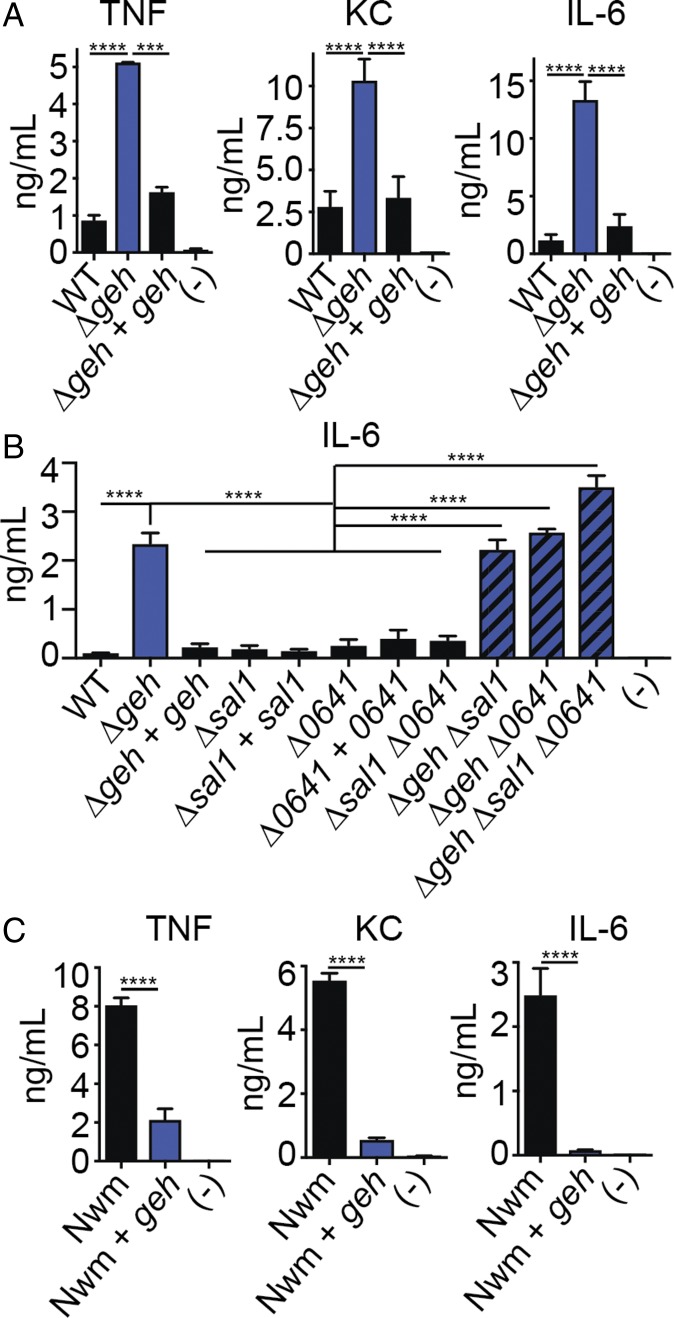
In a recent study, we screened a transposon mutant library of S. aureus to identify gene products that compromise macrophage activation (44). In total, we identified 21 transposon insertion mutants that either enhanced or prevented innate immune cell activation by S. aureus-secreted products (44, 45). One mutant identified in this screen carried a transposon insertion in SAUSA300_0320, the gene encoding Geh. Cell-free supernatant derived from the geh::Tn mutant elicited enhanced proinflammatory cytokine and chemokine production by murine macrophages (44). Because Geh is an abundant secreted protein produced by a significant proportion of S. aureus clinical isolates (21, 46), we sought to explore its role in S. aureus–immune system interactions in greater detail. To this end, we generated a Δgeh mutant (47) and Δgeh + geh single-copy chromosomal complementation strain with geh driven by its predicted native promoter in the USA300 strain LAC (47, 48). The mutant and complementation strain exhibited no notable growth deficiencies in RPMI medium devoid of exogenous fatty acids (SI Appendix, Fig. S1A). However, when 9-h bacterial culture supernatant from a Δgeh mutant was applied to primary murine bone marrow-derived macrophages (BMMs), we noted significant increases in the production of proinflammatory cytokines and chemokines, including TNF, keratinocyte chemoattractant (KC), IL-6, CCL3, and CCL4 (Fig. 1A and SI Appendix, Fig. S1B).
Fig. 1.
Geh prevents innate immune activation. (A) TNF, KC, and IL-6 (ng/mL) production by BMMs after addition of cell-free supernatant from WT, Δgeh, and Δgeh + geh S. aureus. (-), medium alone. (B) IL-6 (ng/mL) production by BMMs after addition of cell-free supernatant from WT, Δgeh, Δgeh + geh, Δsal1, Δsal1 + sal1, Δ0641, Δ0641 + 0641, Δsal1 Δ0641, Δgeh Δsal1, Δgeh Δ0641, and Δgeh Δsal1 Δ0641 S. aureus. (C) TNF, KC, and IL-6 (ng/mL) production by BMMs after addition of cell-free supernatant from WT Newman (Nwm) and Nwm + geh (Nwm + pJC1112-geh). Data shown are mean ± SD from at least three representative independent experiments performed in triplicate, Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test. ***P < 0.001; ****P < 0.0001.
S. aureus is known to produce at least one additional major secreted lipase (Sal1); therefore, we tested whether or not Sal1 or an uncharacterized esterase, SAUSA300_0641, also prevents innate immune cell activation. Addition of bacterial culture supernatant from Δsal1, Δ0641, and Δsal1 Δ0641 mutants to BMMs had no effect on cytokine production (Fig. 1B and SI Appendix, Fig. S1 C–E). On the other hand, deletion of geh in all three mutant backgrounds and subsequent addition of cell-free supernatant from these strains to BMMs resulted in increased cytokine secretion at levels identical to a Δgeh mutant (Fig. 1B and SI Appendix, Fig. S1 C–E). S. aureus strain Newman, a clinical isolate of clonal complex 8, is lysogenized by a phage that disrupts the geh ORF, resulting in a lipase-null phenotype (31). Expression of geh in a single copy upon integration into the unoccupied S. aureus pathogenicity island 1 (SaPI-1) locus of strain Newman resulted in significantly reduced cytokine production compared with the parental strain after addition of supernatant to BMMs (Fig. 1C). Together, these data indicate Geh, but not other S. aureus lipases, limits activation of BMMs by S. aureus-secreted products.
Geh Reduces Inflammation and Slows Microbial Clearance During Systemic Infection.
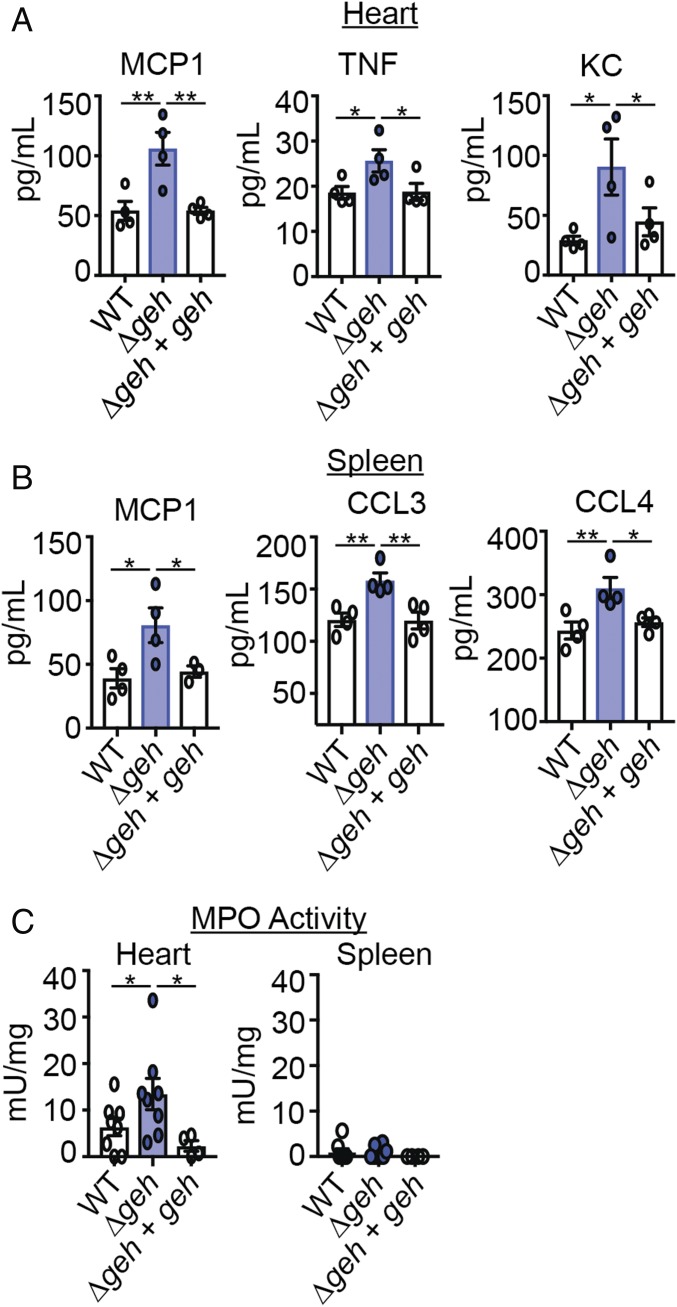
To test if the observed increase in cytokine production by BMMs after treatment with Δgeh cell-free supernatant manifests in a change in infection outcome, we employed a murine systemic infection model. C57BL/6 mice were infected in the bloodstream with WT, Δgeh, and Δgeh + geh strains. Twenty-four hours after infection, cytokine and chemokine levels were measured in the homogenates of infected tissues. We found increased levels of MCP1, TNF, and KC in the heart of infected animals (Fig. 2A and SI Appendix, Fig. S2A). Consistent with the increased levels of the major neutrophil chemotactic factors TNF and KC, we observed increased myeloperoxidase (MPO) activity, a phenotype that is suggestive of increased neutrophil activity (Fig. 2C). Although KC levels were not different and MPO activity was not detectable in the spleen, macrophage chemotactic factors MCP1, CCL3, and CCL4 were elevated (Fig. 2 B and C and SI Appendix, Fig. S2B).
Fig. 2.
Geh blunts the S. aureus-induced proinflammatory response in vivo. (A) MCP1, TNF, and KC (pg/mL) levels in hearts 24 h postinfection with WT, Δgeh, and Δgeh + geh strains (n = 4). (B) MCP1, CCL3, and CCL4 (pg/mL) levels in spleens 24 h postinfection with the indicated strains (n = 4). (C) MPO activities (mU/mg) in hearts and spleens 24 h postinfection with the indicated strains [n = 8 (WT and Δgeh), n = 4 (Δgeh + geh)]. mU, milliunits. Mean ± SD is shown. Statistical significance was determined using one-way ANOVA with Bonferroni’s post hoc test. *P < 0.05; **P < 0.01.
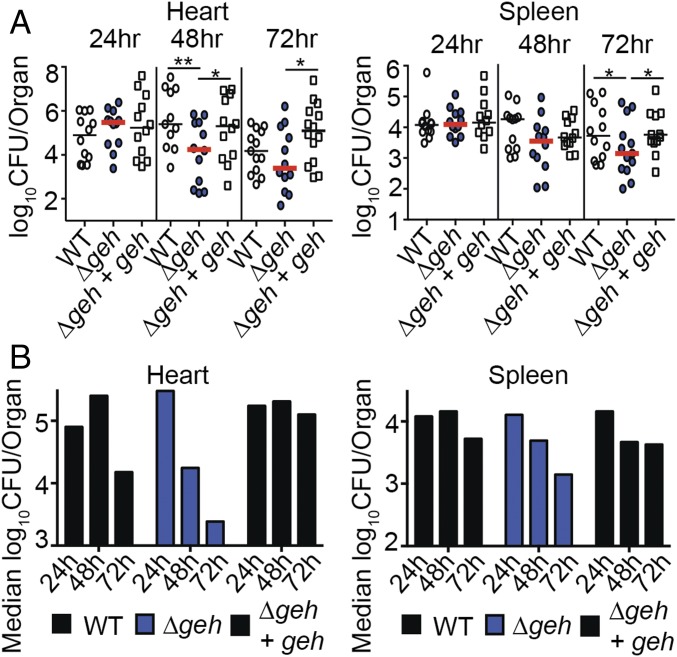
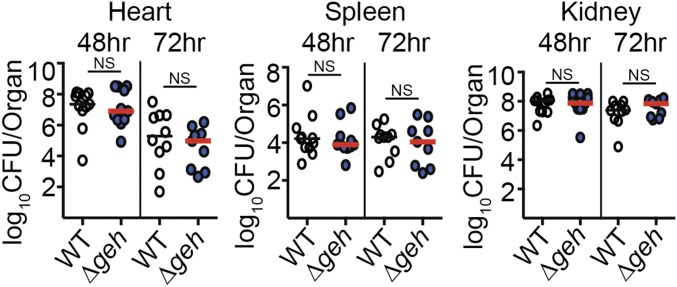
Commensurate with the increased production of macrophage- and neutrophil-responsive cytokines and increased MPO activity in heart homogenates, as well as increased macrophage-responsive chemokines in the spleen, we observed attenuation of infection in both organs (Fig. 3). This was most pronounced in the heart, where a Δgeh mutant exhibited 1- to 2-log fewer colony-forming units at 48 h and 72 h postinfection (Fig. 3A). Similar outcomes were observed in the spleen, where statistically significant reductions in bacterial colony-forming units were recorded for a Δgeh mutant at 72 h postinfection (Fig. 3A). Plotting the median colony-forming units over time demonstrated an accelerated decline in bacterial colony-forming units (approximately 2 logs over the course of 72 h) in the hearts of mice infected with a Δgeh mutant compared with 1 log for WT and no reduction in colony-forming units for the Δgeh + geh complementation strain (Fig. 3B). In the spleen, a 1-log reduction in colony-forming units was observed for a Δgeh mutant over the course of 72 h with less than a 0.5-log reduction for WT and the Δgeh + geh complementation strain (Fig. 3B). Attenuation and clearance of a Δgeh mutant were not observed in the kidney (SI Appendix, Fig. S3). These results suggest that Geh prevents beneficial inflammatory processes in vivo that would otherwise promote bacterial clearance in some, but not all, tissues during systemic infection.
Fig. 3.
Geh reduces the rate of microbial clearance during systemic infection. The bacterial burden (log10 cfu per organ) (A) and median log10 cfu per organ (B) recovered from hearts and spleens 24, 48, and 72 h postinfection with WT, Δgeh, and Δgeh + geh strains (n = 12 per group) are shown. Graphs represent the combined data of three independent experiments. Statistical significance was determined using two-way ANOVA with Fisher’s least significant difference post hoc test. *P < 0.05; **P < 0.01.
Geh-Mediated Effects on BMM Activation Occur Through TLR2.
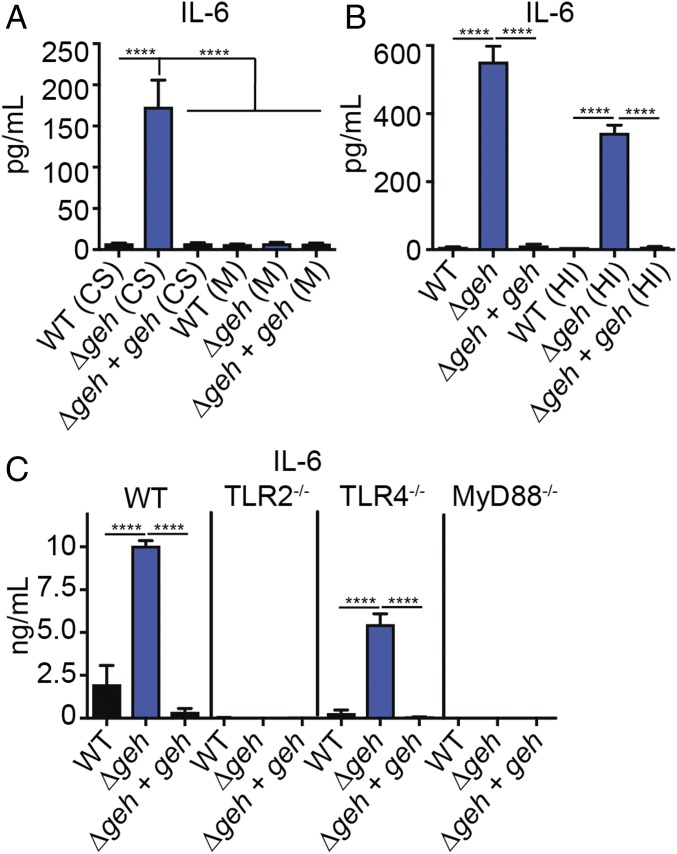
Given the clear inhibitory effects of Geh on BMM activation in vitro and analogous effects on inflammation and subsequent virulence outcomes in vivo, we sought to determine the mechanism by which Geh acts to prevent innate immune cell activation. We first determined the properties of the immune-activating signal present within Δgeh mutant supernatant. Because cell-free supernatant contains membrane components (e.g., vesicles, membranous material) that might confer an immune-stimulatory effect, we first collected cell-free supernatant, followed by ultracentrifugation, to isolate bacterial cell membrane components. Application of the membrane component fraction to BMMs had no effect on macrophage activation regardless of the strain from which it was isolated (Fig. 4A and SI Appendix, Fig. S4A). In contrast, addition of membrane-depleted supernatant from a Δgeh mutant to BMMs still resulted in enhancement in BMM cytokine production (Fig. 4A and SI Appendix, Fig. S4A). Furthermore, the stimulatory effect of Δgeh mutant supernatant was unaffected by heat treatment (Fig. 4B and SI Appendix, Fig. S4B). Our prior studies and those of others indicate the primary mode of extracellular recognition of S. aureus by innate immune cells occurs via TLR2 activation (43, 44). Indeed, macrophage cytokine production by the Δgeh mutant, as well as by WT and the Δgeh + geh complementation strain, was eliminated upon addition of supernatant to either TLR2−/− or the major adaptor protein, Myd88−/−, but not TLR4−/−, BMMs (Fig. 4C and SI Appendix, Fig. S4C). Altogether, these data indicate the immune-activating signal in Δgeh mutant supernatant is (i) not a component of extracellular membranous material, (ii) heat-resistant, and (iii) induced via the TLR2/MyD88 axis.
Fig. 4.
Geh inhibits TLR2-dependent macrophage activation. (A) IL-6 (pg/mL) production by BMMs after addition of an extracellular membrane fraction isolated from supernatant and membrane-free supernatant from WT, Δgeh, and Δgeh + geh strains. CS, cell-free supernatant lacking membrane components; M, extracellular vesicles and membrane components isolated from supernatant. (B) IL-6 (pg/mL) production by BMMs after addition of cell-free supernatant from the indicated strains with or without heat inactivation. HI, heat inactivated. (C) IL-6 (ng/mL) production by WT, TLR2−/−, TLR4−/−, and MyD88−/− BMMs after addition of cell-free supernatant from the indicated strains. Data shown are mean ± SD from three representative independent experiments performed in triplicate. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test. ****P < 0.0001.
Geh Inactivates Microbial TLR2 Ligands.
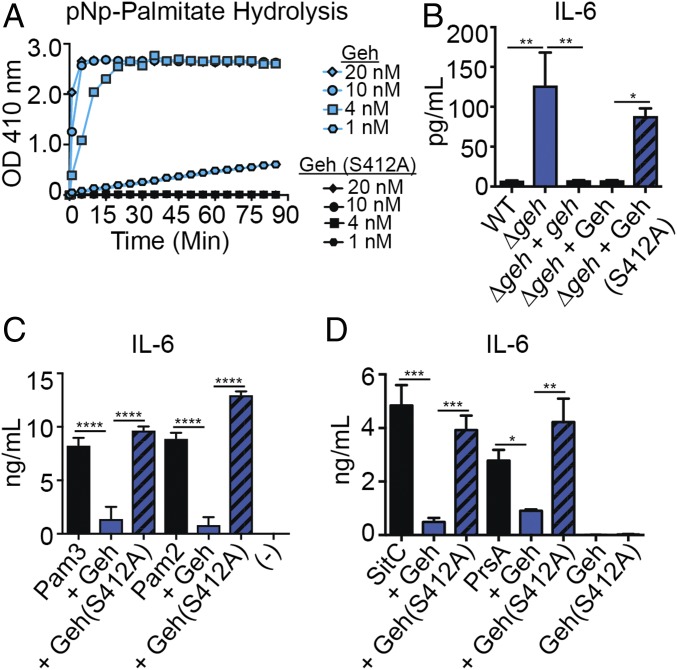
The biochemical functions of the S. aureus lipases have been interrogated since the 1960s and are known to harbor ester hydrolase activity with varying degrees of specificity for esterified lipid substrates (27–29, 49–51). Most recently, Cadieux et al. (28) demonstrated that Geh has preferential recognition of host lipids that resemble the antimicrobial fatty acid linoleic acid. However, we have observed that a Δgeh mutant appears to reduce TLR2-mediated recognition of S. aureus-secreted factors in some way, suggesting that Geh might also act on its own secreted products in an immune-inhibitory manner. To test this possibility, we first purified recombinant mature Geh from Escherichia coli and validated that it has ester hydrolase activity using the substrate para-nitrophenyl (pNp) esterified to the 16-carbon saturated fatty acid palmitate (Fig. 5A). We then determined if supplementation of Geh into Δgeh mutant supernatant was sufficient to block activation of BMMs. Supplementation of 10 nM Geh into Δgeh cultures (Fig. 5B) (corresponding to the approximate concentration of Geh found in WT S. aureus supernatant), followed by addition to BMMs, abrogated the enhanced cytokine production elicited by Δgeh mutant supernatant (Fig. 5C and SI Appendix, Fig. S5A). Addition of 10-fold higher concentrations of Geh reduced the magnitude of cytokine production even further (Fig. 5C and SI Appendix, Fig. S5A). In contrast, addition of Geh to BMMs in the absence of S. aureus supernatant had no effect on cytokine production by BMMs. These data suggest that Geh is acting on a substrate within S. aureus supernatant that ultimately prevents cytokine production and is not inducing activation itself (Fig. 5C and SI Appendix, Fig. S5A).
Fig. 5.
Geh inactivates bacterially derived PAMPs. (A) GelCode Blue-stained 4–20% gradient SDS/PAGE gel of 5 μg of purified Geh and pNp-palmitate hydrolysis (absorbance at 410 nm) in the presence of 1 nM, 4 nM, 10 nM, and 20 nM Geh. (B) GelCode Blue-stained 12% SDS/PAGE gel of TCA-precipitated exoproteins isolated from the indicated strains after 9 h of growth in RPMI (Δgeh + Geh is supernatant isolated from a Δgeh mutant that was supplemented with 10 nM Geh). The asterisk indicates the mature form of Geh. (C) IL-6 (ng/mL) production by BMMs after addition of cell-free supernatant from WT, Δgeh, and Δgeh + geh strains and from Δgeh supernatant supplemented with 1 nM, 10 nM, or 100 nM Geh or Geh alone (1 nM, 10 nM, and 100 nM). (-), medium alone. (D) IL-6 (pg/mL) production by BMMs after addition of Pam2CSK4/Pam3CSK4 (10 ng/mL) or Geh-treated Pam2CSK4/Pam3CSK4 (10 ng/mL). (E) GelCode Blue-stained 4–20% gradient SDS/PAGE gel of 1 μg of SitC/PrsA, 1 μg of SitC/PrsA + Geh, and 1 μg of FPLC-purified SitC*/PrsA* (Geh-treated). (F) IL-6 (ng/mL) production by BMMs after addition of Pam3CSK4 (10 ng/mL), SitC/PrsA (100 ng/mL), Geh-treated SitC/PrsA (SitC*/PrsA*; 100 ng/mL), and Geh (40 ng/mL). All experiments were repeated at least three times. Data shown B–D and F are mean ± SD from three representative independent experiments performed in triplicate. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Because murine BMM cytokine production in the presence of extracellular S. aureus is dependent on TLR2, we reasoned that Geh might be inactivating secreted TLR2 ligands, thereby masking detection of S. aureus by the immune system. S. aureus lipoproteins and lipopeptides are the primary inducers of TLR2 (40, 41, 43, 52). These PAMPs are either diacylated or triacylated. Both structures contain two acyl chains attached via ester bonds (53–55). Acyl chain binding induces heterodimerization of TLR2 with TLR1 or TLR6, leading to immune cell activation and production of inflammatory cytokines and chemokines (38, 54, 56). Given its ester hydrolase activity, we tested whether or not Geh was sufficient to inactivate two synthetic lipopeptide ligands of TLR2, Pam3CSK4 (TLR1/2 ligand) and Pam2CSK4 (TLR2/6 ligand). Indeed, pretreatment of Pam3CSK4 or Pam2CSK4 eliminated the ability of either ligand to activate BMMs via TLR2 (Fig. 5D and SI Appendix, Fig. S5B). We then tested whether or not Geh was sufficient to inactivate lipoprotein ligands produced endogenously by S. aureus. To this end, we purified 6xHis-tagged lipoproteins, SitC and PrsA, from the membrane of S. aureus, followed by treatment with Geh and subsequent removal of the enzyme by size exclusion chromatography (Fig. 5E). Addition of untreated SitC or PrsA, but not Geh-treated SitC* or PrsA*, to BMMs resulted in robust cytokine production (Fig. 5F and SI Appendix, Fig. S5C). These results indicate that Geh is sufficient to inactivate TLR2-inducing lipoproteins and lipopeptides produced by S. aureus, thereby preventing recognition by innate immune cells displaying TLR2 on their surface.
Geh Masks Immune Cell Activation Through Hydrolysis of Immune-Activating Acyl Chains of S. aureus PAMPs.
To determine whether it is the ester hydrolase activity or some other unknown component of Geh that is responsible for inactivation of TLR2 agonists produced by S. aureus, we purified recombinant Geh harboring an S412A point mutation in a conserved serine within the ester hydrolase catalytic triad (Ser412-Asp603-His645) (28, 50, 57–59) (SI Appendix, Fig. S6A). Geh(S412A) had no activity on the synthetic substrate pNp-palmitate (Fig. 6A) and was unable to abrogate the enhancement of cytokine production induced by Δgeh mutant supernatant upon addition to BMMs (Fig. 6B and SI Appendix, Fig. S6 B and C). Furthermore, Geh(S412A) was unable to inactivate the synthetic peptides Pam3CSK4 and Pam2CSK4, as well as the S. aureus lipoproteins SitC and PrsA (Fig. 6 C and D and SI Appendix, Fig. S6 D and E). Altogether, our data suggest that the ester hydrolase activity of Geh on the esterified fatty acids of S. aureus lipoproteins prevents their recognition by TLR2 on innate immune cells.
Fig. 6.
Ester hydrolase activity of Geh is required for lipoprotein inactivation. (A) pNp-palmitate hydrolysis (absorbance at 410 nm) in the presence of 1 nM, 4 nM, 10 nM, and 20 nM Geh(S412A). (B) IL-6 (pg/mL) production by BMMs after addition of cell-free supernatant from WT, Δgeh, and Δgeh + geh strains and from Δgeh supernatant supplemented with 10 nM Geh, Δgeh supernatant supplemented with 10 nM Geh(S412A), and Geh(S412A) alone (10 nM). (C) IL-6 (ng/mL) production by BMMs after addition of Pam2CSK4/Pam3CSK4 (10 ng/mL), Geh-treated Pam2CSK4/Pam3CSK4 (10 ng/mL), and Geh(S412A)-treated Pam2CSK4/Pam3CSK4 (10 ng/mL). (-), medium alone. (D) IL-6 (ng/mL) production by BMMs after addition of Pam3CSK4 (10 ng/mL), SitC/PrsA (100 ng/mL), Geh-treated SitC/PrsA (100 ng/mL), Geh(S412A)-treated SitC/PrsA (100 ng/mL), Geh (40 ng/mL), and Geh(S412A) (40 ng/mL). All experiments were repeated at least three times. Data shown in B–D are mean ± SD from three representative independent experiments performed in triplicate. Statistical significance was determined by one-way ANOVA with Tukey’s post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Persistence of S. aureus During Infection Is Mediated by Geh-Dependent Inactivation of Microbial TLR2 Ligands.
Thus far, our in vitro data indicate that Geh is capable of hydrolyzing esterified fatty acids on bacterial lipoproteins, leading to inactivation of these important TLR2 ligands. Further, our in vivo data suggest that Geh also inactivates TLR2 ligands during infection, thereby limiting beneficial inflammatory processes and promoting persistence (Fig. 3). To determine whether or not the accelerated rate of clearance in the kidneys and spleen of Δgeh-infected mice occurs through TLR2-dependent activation of innate immunity, we infected TLR2−/− mice with WT and Δgeh S. aureus and monitored bacterial colony-forming units in the heart, spleen, and kidney at 48 h and 72 h postinfection (Fig. 7). Consistent with our hypothesis, the attenuation of the Δgeh mutant was abrogated in all tissues at 48 h and 72 h. Although there was a net reduction in colony-forming units in the heart between 48 h and 72 h, the magnitude of the reduction was equivalent between WT and Δgeh, suggesting that Geh and TLR2 are not involved in this process (Fig. 7). In summary, these data indicate that the accelerated clearance of a Δgeh mutant during systemic infection is TLR2-mediated.
Fig. 7.
Infection of TLR2−/− mice abrogates the accelerated clearance of a Δgeh mutant during systemic infection. The bacterial burden (log10 cfu per organ) was recovered from hearts, spleens, and kidneys 48 h and 72 h postinfection with WT and Δgeh strains (n = 9–11 per group). Graphs represent the combined data of two independent experiments. Statistical significance was determined using a Kruskal–Wallis test with Dunn’s posttest. NS, P > 0.05.
Discussion
Clinical reports and in vitro studies suggest that the lipases of S. aureus are likely to be instrumental virulence factors during infection (18–22). However, few studies have identified functions for the lipases that are recapitulated both in vitro and in vivo. In this study, we have determined that the major secreted lipase, Geh, facilitates S. aureus infection and persistence in the host. Further, we provide data to suggest that persistence during infection is linked to Geh-mediated lipolysis and subsequent inactivation of S. aureus-derived lipoproteins, thereby allowing the bacterium to mask itself from TLR2-mediated immune surveillance.
Our data indicate that, despite the presence of additional lipolytic enzymes (Sal1 and SAUSA300_0641), Geh is thus far the only S. aureus lipase identified to prevent immune cell activation via hydrolysis of esterified fatty acids on endogenous lipoproteins (Fig. 1 and SI Appendix, Fig. S1). We suspect Geh, but not Sal1 or SAUSA300_0641, activity on S. aureus lipoproteins is explained by the established substrate specificities of the S. aureus lipases. Sal1 preferentially cleaves substrates containing short-chain fatty acids (27, 29, 60), while Geh hydrolyzes short-chain and long-chain fatty acids, with a preference for long-chain fatty acids (28). Since the esterified fatty acids within S. aureus membranes and lipoproteins are largely made up of long-chain saturated and branched-chain fatty acids (61–64), one might anticipate preferential recognition by Geh, as seen in this study. Although our data suggest the putative esterase SAUSA300_0641 has no effect on innate immune cell activation, its substrate specificity and other enzyme properties still remain to be investigated. Further, although it is possible that alternative, or accessory, functions of the S. aureus lipases might also be important for the inhibition of innate immune recognition, we suspect this is an unlikely possibility, given the observation that addition of catalytically inactive Geh into Δgeh supernatant or purified S. aureus lipoproteins does not lead to inhibitory effects on innate immune cells (Fig. 6 and SI Appendix, Fig. S6).
Although a range of studies have implicated S. aureus lipases in pathogenesis (18–22, 25), no prior direct evidence existed to suggest that Geh interferes with the immune response during infection. Here, through monitoring bacterial colony-forming units over time during murine systemic infection, we have confirmed that Geh promotes bacterial survival during infection (Fig. 3). The innate immune response to microbial infection requires the recognition of PAMPs by TLRs, leading to early induction of proinflammatory chemokines and cytokines, which triggers recruitment of leukocytes and induction of oxidative burst (65–67). We have uncovered an early and enhanced activation of innate immunity, as evidenced by increased levels of proinflammatory cytokines in the heart and spleen and increased MPO activity in the heart of mice infected with a Δgeh mutant. The increased cytokine levels at 24 h postinfection (Fig. 2 and SI Appendix, Fig. S2) correlate well with the improved bacterial clearance within the Δgeh-infected heart and spleen at 48 h and 72 h postinfection (Fig. 3). Thus, our model suggests that Geh blunts host inflammatory responses normally induced through TLR2 activation and delays bacterial clearance in the heart and spleen during early infection. Our demonstration that infection of TLR2−/− mice abrogates the accelerated clearance of a Δgeh mutant in these tissues further supports this argument (Fig. 7). It is interesting to note that increased local inflammatory cytokines, MPO activity, and the ensuing clearance of S. aureus from infected tissue were not observed in all infection sites. In our murine bacteremia model, infected kidneys displayed increasing colony-forming units throughout the course of infection, indicating growth of the bacterium in this site (SI Appendix, Fig. S3). Thus, the effects of Geh on immune activation and bacterial clearance appear to be tissue-specific. We hypothesize this could be due to altered expression patterns of Geh in different tissues, which vary in immune cell composition, major immune cell types, and nutrient availability (68–81). This is exemplified by the transition from bacteremia to thromboembolic lesions during systemic infection, wherein S. aureus produces more adhesins, metal acquisition proteins, immune evasion factors, and toxins during the transition (82). Indeed, transcriptome studies suggest that Sal1 and Sal2 are differentially up-regulated in a range of deep tissues, but not superficial sites of infection, such as the skin (83–86). Thus, we suspect S. aureus may alter its Geh expression pattern in a manner that depends on the infection site. Clinical studies support these experimental observations and suggest that more abundant lipase activities were detected in deep tissues during disseminated infections compared with superficial localized infections (20), an observation that is also in line with recent data showing the lipase may be dispensable in models of superficial infection (26). Our study includes a time course of systemic infection in the heart, spleen, and kidney. We reason this has allowed us to uncover functions for the lipase in pathogenesis that were not observed in prior infection studies (25, 26). It is noted that a Δgeh mutant, while attenuated during murine infection, does not eliminate S. aureus colony-forming units from target organs. We surmise this is likely because Geh is one of numerous extracellular immune evasion molecules that act in concert to prevent optimal innate immune recognition (87–90).
Although our in vitro data (Fig. 1 and SI Appendix, Fig. S1) indicate Geh is thus far the only lipase that inhibits innate immune cell recognition of bacterial PAMPs and prevents their activation, we cannot yet rule out the possibility that Sal1 and SAUSA300_0641 contribute to the pathogenesis of S. aureus in unknown ways. The organs involved in lipid metabolism, such as the small intestine and liver, contain a greater diversity of lipids than organs like the heart and spleen (78, 81, 91, 92). Therefore, from a nutrient acquisition standpoint, the lipid environment of other host tissues may require a range of lipolytic activities for an invading microbe to thrive. Furthermore, S. aureus lipases have different biochemical properties, including distinct substrate specificities and pH optimums (27–29); therefore, it is possible that Geh, Sal1, SAUSA300_0641, or other uncharacterized lipases function to promote pathogenesis in a tissue environment-dependent manner.
In this work, we show that Geh prevents TLR2-dependent immune cell activation by inactivating S. aureus lipoproteins (Figs. 4–6 and SI Appendix, Figs. S4–S6). Our study used murine macrophages as a representative TLR2-expressing immune cell; however, given the conserved detection of lipoproteins by TLR2, it is likely that recognition of lipoproteins will be masked on all TLR2-expressing cells (42, 43, 56). Although previous studies suggested purified preparations of Geh affect chemotaxis and phagocytosis of immune cells (23, 24), our data indicate it is unlikely that Geh directly causes immune cell activation, since inactivation only occurred when Geh, but not its catalytically inactive mutant (S412A), was used to treat supernatant, synthetic lipopeptides, or purified S. aureus lipoproteins, followed by addition to macrophages (Figs. 5 and 6 and SI Appendix, Figs. S5 and S6). TLRs are important sensors of PAMPs, initiating immediate responses that can promote clearance and restrict infection dissemination (35–37). It is well understood that lipoproteins are the major S. aureus PAMPs that activate TLR2 to trigger an innate immune response. S. aureus is capable of releasing diacylated and triacylated lipoproteins (53–55). Diacylated lipoproteins contain two esterified lipid chains, whereas triacylated lipoproteins contain the same ester-bound lipids along with an additional amino-linked lipid chain (53–55). In either example, the esterified lipids are essential for triggering strong inflammatory signals through TLR2 (43, 54, 56). From a structure/function viewpoint, our observation that the ester hydrolase activity of Geh is required for inactivation of lipoprotein (Fig. 6 and SI Appendix, Fig. S6) indicates Geh must hydrolyze at least one, but likely both, ester bond(s) linking the fatty acid chains to the N terminus of the lipoprotein. Further biochemical studies are in process to determine the extent of lipoprotein targeting by Geh.
Diverse bacterial species produce lipases that bear similarities to Geh. Pseudomonas spp., Burkholderia spp., and streptococcal species produce lipoprotein lipases (EC 3.1.1.34) that act on mammalian lipoproteins (93–95) but, incidentally, can also remove esterified fatty acids from a diverse repertoire of bacterial lipoproteins, including those produced by S. aureus (53, 96–101). Due to their promiscuous esterase activity, the lipoprotein lipases of Burkholderia cepacia and Pseudomonas fluorescens have been used in commercial settings to remove lipoprotein contamination or harnessed to characterize lipoprotein structure and function (53, 96, 100, 101). This observation opens up the possibility that the immune evasion mechanism described in this work for S. aureus could be broadly applicable to other gram-positive and gram-negative species that produce similar lipases and may have important implications for mammalian infection or colonization.
S. aureus Geh was recently reported to hydrolyze mammalian low-density lipoproteins to obtain free fatty acids and incorporate them into bacterial membrane phospholipids (30). Thus, in addition to this important role in nutrient acquisition, we have found that S. aureus Geh is an immunomodulatory factor. It facilitates infection by inactivating bacterial lipoproteins to mask the bacterium from recognition by TLR2. Therefore, Geh represents a multifaceted virulence factor with important roles in nutrient acquisition and immune evasion.
Materials and Methods
Bacterial Strains and Growth Conditions.
S. aureus strains used in this study were derived from methicillin-resistant S. aureus USA300 LAC (AH-1264, WT) (102), RN4220 (103), and Newman (104) strains (SI Appendix, Table S1). Strains were grown in either tryptic soy broth (TSB; Criterion) or RPMI 1640 medium (Corning), supplemented with 1% casamino acids (Amresco) and 0.2% sodium bicarbonate (Amresco), at 37 °C with shaking at 200 rpm in an Innova 42 shaker incubator (New Brunswick) unless otherwise noted. E. coli lysY/Iq was used for Geh and Geh(S412A) purification. E. coli DH5α and DC10B were used for passaging pIMAY, pJC1111, pJC1112, pQE60, and pOS1 plasmids. All E. coli strains were cultivated in lysogeny broth, Miller formulation (LB; BD Biosciences) at 37 °C with shaking at 200 rpm in an Innova 42 shaker incubator (New Brunswick). Antibiotics were supplemented when necessary. For E. coli strains, 100 μg/mL ampicillin (Gold Biotechnology), 25 μg/mL kanamycin (Amresco), and 25 μg/mL neomycin (Amresco) were used. For S. aureus strains, 1 μg/mL anhydrous tetracycline (Amresco), 10 μg/mL chloramphenicol (Amresco), 3.5 μg/mL erythromycin (Amresco), 50 μg/mL kanamycin (Amresco), 50 μg/mL neomycin (Amresco), and 0.15 mM CdCl2 (Amresco) were used.
Construction of In-Frame Deletion Mutants.
Five hundred-base pair regions of homology upstream and downstream of each gene of interest were amplified from WT S. aureus genomic DNA (a list of primers is provided in SI Appendix, Table S2). The amplicons were used in a splicing by overlap extension (SOE) PCR to obtain the final amplicon, which was subcloned into the pIMAY plasmid (47). Mutagenesis was carried out as previously described (44).
Construction of Marked Deletion Mutants Δgeh::kan.
A kanamycin resistance cassette (aphA3) was amplified from plasmid pBTK (105) with primer set KanF KasI and KanR KasI (SI Appendix, Table S2), followed by subcloning into pIMAY harboring the 500-bp upstream and downstream regions of homology that flank the geh gene using a unique KasI restriction site engineered between the two flanking regions. The pIMAY-geh-kanr was transformed into WT AH-1264 LAC, and the procedure described above for generation of in-frame deletion mutants was performed. The gene replacement was selected for by plating on kanamycin + neomycin and confirmed by PCR.
Construction of Double- or Triple-Lipase Deletion Mutants.
To generate double- or triple-deletion mutants (Δsal1 Δgeh::kan; Δ0641 Δgeh::kan; and Δsal1 Δ0641 Δgeh::kan), bacteriophage Φ11 was used to package the Δgeh::kan mutation from the S. aureus AH-1264 LAC Δgeh::kan strain and transduced into the Δsal1, Δ0641, and Δsal1 Δ0641 mutant backgrounds following previously described protocols (106, 107). The potential double- or triple-deletion mutants were selected for by plating on kanamycin + neomycin and confirmed by PCR.
Generation of Complementation Strains.
Complementation strains were generated using pJC1111 or pJC1112 plasmid (48). The genes of interest were amplified using their corresponding forward and reverse primers (SI Appendix, Table S2) and cloned into pJC1111 at the KpnI and SacI restriction sites or into pJC1112 at the BamHI and EcoRI restriction sites. Plasmids were propagated in E. coli DH5α and transformed into SaPI-1 integrase-expressing S. aureus (RN9011) to allow single-copy integration at the SaPI-1 site in the chromosome (48). Bacteriophage Φ11 was used to package the integrated complementation plasmid from RN9011, and was subsequently transduced into the desired in-frame deletion mutant strains. Complementation strains were selected for based on CdCl2 resistance (pJC1111) or erythromycin resistance (pJC1112) and confirmed by PCR.
Generation of Geh-6xHis Expressing E. coli lysY/Iq.
C-terminally 6xHis-tagged S. aureus Geh was expressed in E. coli lysY/Iq. The geh ORF was amplified by PCR from WT S. aureus genomic DNA with primer pair pQE60-Geh F NcoI and pQE60-Geh R BamHI (SI Appendix, Table S2). The amplicon was digested with BamHI and NcoI restriction endonucleases and ligated into plasmid pQE60, which encodes a 6xHis at the 3′ end of the gene. The resulting pQE60-geh-6xHis plasmid was passaged in E. coli DH5α and then transformed into E. coli lysY/Iq.
Generation of Geh(S412A)-6xHis Expressing E. coli lysY/Iq.
The S412A Geh mutant was constructed by site-directed mutagenesis. The entire pQE60-Geh-6xHis plasmid was amplified using the 5′-phosphorylated mutagenic primers pQE60-GehS412A forward and reverse (SI Appendix, Table S1), which contained the S412A (AGT→GCT) mutation, followed by DpnI treatment to remove the methylated template plasmid. The linear plasmid amplicons were circularized by a ligation reaction with T4 ligase (New England Biolabs) at room temperature for 2 h, followed by transformation into E. coli DC10B and E. coli lysY/Iq. The mutation was confirmed through sequencing analysis.
Generation of SitC-6xHis and PrsA-6xHis Expressing S. aureus RN4220.
C-terminally 6xHis-tagged SitC or PrsA was generated by amplification of sitC or prsA from WT genomic DNA using primer pairs SitC SOE3/SitC SOE4 Sal1 and PrsA SOE3/PrsA SOE4 EcoRI. The nucleotide sequence corresponding to 6xHis was incorporated into primers SitC SOE4 Sal1 and PrsA SOE4 EcoRI. The resulting sitC-6xHis and prsA-6xHis amplicons were fused to the constitutive S. aureus sarA promoter followed by the sod ribosome binding site by amplifying PsarA-sodRBS from plasmid pOS1-PsarA-sodRBS-sGFP (108) using primer pairs SitC/PrsA SOE1 PstI and SitC SOE2 or PrsA SOE2. The resulting amplicon was fused to sitC-6xHis and prsA-6xHis by SOE-PCR to generate PsarA-sodRBS-sitC-6xhis and PsarA-sodRBS-prsA-6xhis. These amplicons were digested using PstI and Sal1 or PstI and EcoRI restriction endonucleases and ligated into plasmid pOS1 (109, 110). The resulting plasmids were propagated in E. coli DH5α and subsequently transformed into S. aureus RN4220.
Generation of a Geh Expressing S. aureus Newman Strain.
To generate a Geh expressing Newman strain, bacteriophage Φ80α was used to package the integrated pJC1112-geh from S. aureus RN9011 (as discussed above) and transduced into the SaPI-1 attachment site within the chromosome of strain Newman.
Growth Curves.
Strains were grown in triplicate in 200 μL of fresh RPMI or TSB medium in a 96-well, round-bottom plate (CELLTREAT) overnight at 37 °C. The bacteria were then subcultured into 198 μL of fresh media (1:100) using a 96-well, flat-bottom plate at 37 °C. The optical density at 550 nm (OD550) was measured every hour over a 10-h period until reaching stationary phase.
Trichloroacetic Acid Precipitation.
Strains were grown overnight in 3 mL of RPMI at 37 °C. The bacteria were then subcultured (1:100) in 5 mL of fresh RPMI for 9 h. A total of 1.3 mL of cell-free supernatant was collected, and the exoproteins were precipitated overnight at 4 °C by mixing the supernatant with ice-cold 100% trichloroacetic acid (TCA; 10:1 vol/vol; Alfa Aesar). The precipitated supernatant proteins were washed with 100% acetone, air-dried, and dissolved in TCA SDS reducing buffer [0.5 M Tris⋅HCl (Amresco) at pH 8.0 + 2XSDS + β-mercaptoethanol (Sigma)] mixed 1:1 with 4% SDS (Amresco) in water. Samples were normalized to the highest OD600 to ensure equivalent loading in relation to biomass and separated on 12% SDS polyacrylamide gels followed by staining with GelCode Blue (Thermo).
Purification of Geh-6xHis and Geh(S412A)-6xHis.
To purify Geh-6xHis and Geh(S412A)-6xHis, E. coli lysY/Iq containing pQE60-geh-6xHis and pQE60-geh(S412A)-6xHis was grown in 20 mL of LB overnight at 37 °C. The bacteria were subcultured (1:100) in 500 mL of fresh LB and allowed to grow at 37 °C with shaking at 220 rpm in an Innova 42 shaker incubator (New Brunswick) until reaching an OD600 of 0.6. Isopropyl β-d-1-thiogalactopyranoside (Gold Biotechnology) was added to the culture at a final concentration of 0.5 mM to induce Geh-6xHis or Geh(S412A)-6xHis expression and was allowed to incubate at 16 °C for 16 h. Bacteria were pelleted by centrifugation at 12,210 × g for 10 min at 4 °C, supernatant was decanted, and pellets were stored at −80 °C overnight. Bacterial pellets were thawed on ice and resuspended in lysis buffer [10 mM Tris⋅HCl, 300 mM NaCl, 10 mM imidazole (Alfa Aesar) at pH 8.0] supplemented with 2 mM phenylmethane sulfonyl fluoride (PMSF; Acròs Organics). The cells were lysed on ice via sonication (Branson) at 20-s intervals with a rate of 0.8 s per pulse and an output of 340 W for a total of 30 min. The bacterial debris was pelleted by centrifugation at 16,000 × g for 1 h. Lysates were collected, filtered through a 0.22-μm polyethersulfone (PES) membrane, and incubated for 1 h at 4 °C with nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) that was preequilibrated with lysis buffer. The protein-bound resin was washed three times with wash buffer (10 mM Tris⋅HCl, 300 mM NaCl, 50 mM imidazole at pH 8.0) and subsequently eluted with elution buffer (10 mM Tris⋅HCl, 300 mM NaCl, 500 mM imidazole at pH 8.0) in 6 × 1-mL fractions. The protein fractions were combined and dialyzed three times against 1 L of dialysis buffer (10 mM Tris⋅HCl, 300 mM NaCl at pH 8.0) containing a decreasing concentration of imidazole (100 mM, 25 mM, and 0 mM) for 12 h with buffer changed every 4 h. Endotoxin was removed using endotoxin removal resin following the manufacturer’s instructions (Pierce High Capacity Endotoxin Removal Resin; Thermo Fisher). Protein purity was confirmed by SDS/PAGE [12% or 4–20% gradient SDS polyacrylamide gels (Bio-Rad)], and concentrations were estimated using a BCA Colorimetric Assay Kit (Thermo Fisher Scientific) following the manufacturer’s protocol. Proteins were stored at −80 °C.
Purification of SitC-6xHis and PrsA-6xHis.
SitC-6xHis and PrsA-6xHis were purified as previously described (44).
pNp-Palmitate Ester Hydrolase Activity Assay.
The lipase activity of Geh was assayed spectrophotometrically using pNp-palmitate as a substrate. The reaction buffer was composed of 50 mM Tris⋅HCl at pH 8.0, arabic gum (1 mg/mL), 0.005% Triton X-100, and 795 μM pNp-palmitate (Sigma). Varied concentrations of purified Geh were added to the reaction buffer in a total reaction volume of 1 mL. Samples were incubated at 25 °C in the dark, and the absorbance at 410 nm was measured every 10 min using a Genesys 10s UV-Vis Spectrophotometer (Thermo Fisher).
Geh-6xHis/Geh(S412A)-6xHis Lipopeptide or Lipoprotein Processing.
For Geh-6xHis or Geh(S412A)-6xHis lipopeptide reactions, 300 ng of LPS-free Geh-6xHis or Geh(S412A)-6xHis was incubated with 10 ng of Pam3CSK4 (Invivogen) or Pam2CSK4 (Invivogen) at 37 °C for 2 h. Geh- or GehS412A-treated Pam3CSK4 and Pam2CSK4 or untreated controls were incubated with Ni-NTA resin for 1 h at room temperature, followed by centrifugation at 21,130 × g for 5 min at 4 °C to remove Geh-6xHis– and Geh(S412A)-6xHis–containing resin. The Geh-6xHis/Geh(S412A)-6xHis–treated Pam3CSK4 and Pam2CSK4 or untreated controls were applied to macrophages (10 ng/mL), followed by collection of macrophage supernatant after overnight incubation. For Geh-6xHis/Geh(S412A)-6xHis lipoprotein processing experiments, reactions were conducted in 50 μL of reaction buffer (20 mM Tris⋅HCl, 50 mM NaCl at pH 8.0) with 20 μM substrate (PrsA or SitC) and 5 μM LPS-free Geh-6xHis or GehS412A-6xHis. The reactions were incubated at 37 °C for 3 h, followed by removal of Geh-6xHis from the reactions by size exclusion chromatography using a Superose 6 Increase 10/300 GL column (GE Healthcare), and purity was confirmed by SDS/PAGE [12% or 4–20% gradient SDS polyacrylamide gels (Bio-Rad)]. Assessment of Geh-mediated lipoprotein modifications were made by determining whether or not the Geh- or Geh(S412A)-treated lipoproteins retained the ability to activate macrophages (discussed below).
Cell-Free Supernatant Collection.
Bacterial strains were grown in triplicate in 150 μL of fresh RPMI medium in 96-well, round-bottom plates overnight at 37 °C. The bacteria were subcultured into 150 μL of fresh RPMI medium (1:100) in a 96-well, round-bottom plate at 37 °C for 9 h. The OD550 was measured, followed by centrifugation at 2,510 × g for 15 min to pellet bacteria. Cell-free supernatants were collected after filtration (0.22-μm pore size PES membrane filter; Corning) and stored at −80 °C. For heat treatment, the collected cell-free supernatants were heated to 100 °C for 15 min and cooled on ice before being applied to macrophages.
Removal of Bacterial Extracellular Vesicles and Membrane Components.
Extracellular vesicles and other extracellular membrane components were isolated from S. aureus culture supernatant by ultracentrifugation. Overnight cultures of S. aureus were subcultured into TSB (1:100). After 9 h of growth, culture density was determined by measuring the OD600 and cell-free supernatants were collected by centrifugation at 10,000 × g for 30 min at 4 °C, followed by filtration (0.22-μm pore size PES membrane filter). The resulting filtrate was subjected to ultracentrifugation at 150,000 × g for 3 h at 4 °C. The resulting extracellular vesicles and membrane components (diluted in 1× PBS) or the membrane component-free supernatants were collected and applied onto murine macrophages.
Supplementation of Geh-6xHis or Geh(S412A)-6xHis into Bacterial Cultures.
Strains were grown overnight in 150 μL of RPMI at 37 °C in a 96-well, round-bottom plate. The bacteria were subcultured in 150 μL of fresh RPMI (1:100) for 6 h, followed by addition of purified LPS-free Geh-6xHis (1 nM, 10 nM, and 100 nM) or Geh(S412A)-6xHis (10 nM) and incubation for an additional 3 h. After 9 h, bacterial density was measured at OD550, followed by centrifugation at 2,510 × g for 15 min to collect cell-free supernatants. The desired concentration of cell-free supernatant applied to macrophages was adjusted based on the strain with highest OD550 values.
Isolation of BMMs.
Murine BMMs were derived from progenitor cells isolated from tibias and femurs of C57BL/6 mice (JAX stock no. 000664) (WT, tlr2−/−, tlr4−/−, and myd88−/−) as previously described (44).
In Vitro Macrophage Experiments.
Murine BMMs were seeded onto 96-well tissue culture-treated plates (Corning) at 65,000 cells per well in 90 μL of BMM medium supplemented with 100 μg/mL penicillin/streptomycin and 10 μg/mL gentamicin cultured at 37 °C in 5% CO2 for 24 h. To measure the effects of bacterial supernatant on macrophage responses, 10 μL of S. aureus cell-free supernatants, membrane components, or membrane component-free supernatant was applied to the macrophages, after first normalizing to bacterial density (OD550). To determine whether Geh-treated lipopeptides/lipoproteins change macrophage responses relative to native lipopeptides/lipoproteins, Pam3CSK4/Pam2CSK4 (10 ng/mL) and native S. aureus lipoproteins (100 ng/mL), with or without prior Geh treatment, were added to each well. After 24 h, macrophage supernatants were collected and the secreted cytokine and chemokine profile was analyzed using a customized cytometric bead array (CBA) flex set (BD Biosciences) (discussed below) and flow cytometry (LSR Fortessa cell analyzer).
Murine Systemic Infection Model.
Overnight cultures of WT, Δgeh, and Δgeh + geh strains were subcultured (1:100) in 15 mL of TSB medium for 3 h. After centrifugation at 3,234 × g for 15 min, bacterial pellets were washed three times in 5 mL of 1× PBS and adjusted to an OD600 value of 0.32 (∼1 × 108 cfu/mL). The infection dose was confirmed by plating serial dilutions of inoculum on TSA plates. C57BL/6J mice (6 wk old; JAX stock no. 000664) or B6.129-Tlr2 < tm1Kir>/J (Tlr2−/− ; JAX stock no. 004650) (111) were retro-orbitally infected with 107 cfu (100 μL) of OD-normalized bacteria. The infected mice were euthanized by CO2 narcosis, followed by cervical dislocation at 24, 48, or 72 h postinfection. Hearts, kidneys, livers, and spleens were collected and homogenized in 5 mL of 1× PBS, and 10 μL of organ homogenates was serially diluted and plated on a TSA plate to assess the bacterial burden in each organ. One hundred microliters of organ homogenates was also plated when serial dilution yielded no colonies on agar plates. At the 24-h time point, supernatant from the homogenized organs was collected and assessed for local inflammation by measuring cytokine or chemokine levels via a customized CBA kit following the manufacturer’s protocol.
Tissue-Specific MPO Assay.
Twenty-four hours after initiation of systemic infection, hearts and spleens were collected from C57BL/6J mice (6 wk old; JAX stock no. 000664) and homogenized in 1 mL of homogenizing solution [50 μM taurine (Sigma), 0.1% Nonidet P-40 (Sigma) in 1× PBS]. Twenty-five microliters of organ homogenates was then used in a commercially available MPO assay kit (Biovision) following the manufacturer’s protocol.
Cytometric Bead Array.
Ten microliters of bead mixture and 5 μL of macrophage supernatant or organ homogenate were incubated in a 96-well, V-bottom plate (Corning) for 90 min at 25 °C with shaking at 600 rpm in a Thermomixer C (Eppendorf). The samples were washed and resuspended in 200 μL of fluorescence-activated cell sorting buffer [0.05% sodium azide (Sigma) + 2% heat-inactivated FBS (Seradigm) in 1× PBS] and analyzed by flow cytometry using an LSR Fortessa cell analyzer.
Ethics Statement.
All experiments were performed following the ethical standards of the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee (IACUC) at Loyola University Chicago, Health Sciences Division. The institution is approved by the Public Health Service (PHS; A3117-01 through 02/028/2022), is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (000180, certification dated 11/17/2016), and is registered/licensed by the US Department of Agriculture (USDA; 33-R-0024 through 08/24/2020). All animal experiments were performed in Animal Biosafety Level 2 facilities using IACUC-approved protocols (IACUC 2017028) under the guidance of the Office of Laboratory Animal Welfare following the USDA and PHS Policy on Humane Care and Use of Laboratory Animals guidelines.
Quantification and Statistical Analyses.
All experiments were repeated at least three independent times. For in vitro macrophage data, statistical significance was analyzed from representative experiments conducted in triplicate and repeated a minimum of three independent times. All statistical significance was analyzed using GraphPad Prism version 7.0 with the statistical tests specified in the figure legends. Before conducting statistical analyses on data derived from animal studies, a D’Agostino and Pearson or Shapiro–Wilk normality test was conducted. Based on normality testing, either ANOVA or nonparametric one-way ANOVA was performed. Post hoc statistical significance was calculated for the following comparisons (WT vs. Δgeh and Δgeh vs. Δgeh + geh) using Bonferroni or Fisher’s least significant difference post hoc test where described. The number of animals per treatment group is indicated as “n” in the figure legends. For all other data, statistical significance (P < 0.05) was determined by one-way ANOVA with Tukey’s post hoc test.
Supplementary Material
Acknowledgments
We thank members of the F.A. laboratory for helpful discussions and for critically reviewing this manuscript. This work was supported by NIH Grant R01 AI120994 (to F.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817248116/-/DCSupplemental.
References
- 1.Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hornef MW, Wick MJ, Rhen M, Normark S. Bacterial strategies for overcoming host innate and adaptive immune responses. Nat Immunol. 2002;3:1033–1040. doi: 10.1038/ni1102-1033. [DOI] [PubMed] [Google Scholar]
- 3.Jaeger KE, et al. Bacterial lipases. FEMS Microbiol Rev. 1994;15:29–63. doi: 10.1111/j.1574-6976.1994.tb00121.x. [DOI] [PubMed] [Google Scholar]
- 4.Bender J, Flieger A. Handbook of Hydrocarbon and Lipid Microbiology. Springer; Berlin: 2010. pp. 3241–3258. [Google Scholar]
- 5.Griese M, et al. BEAT Study Group Pulmonary surfactant, lung function, and endobronchial inflammation in cystic fibrosis. Am J Respir Crit Care Med. 2004;170:1000–1005. doi: 10.1164/rccm.200405-575OC. [DOI] [PubMed] [Google Scholar]
- 6.Son MS, Matthews WJ, Jr, Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty AL, Malloy JL, Wright JR. Pseudomonas aeruginosa degrades pulmonary surfactant and increases conversion in vitro. Am J Respir Cell Mol Biol. 2005;32:128–134. doi: 10.1165/rcmb.2004-0276OC. [DOI] [PubMed] [Google Scholar]
- 8.Miskin JE, Farrell AM, Cunliffe WJ, Holland KT. Propionibacterium acnes, a resident of lipid-rich human skin, produces a 33 kDa extracellular lipase encoded by gehA. Microbiology. 1997;143:1745–1755. doi: 10.1099/00221287-143-5-1745. [DOI] [PubMed] [Google Scholar]
- 9.Tan HH. Topical antibacterial treatments for acne vulgaris: Comparative review and guide to selection. Am J Clin Dermatol. 2004;5:79–84. doi: 10.2165/00128071-200405020-00002. [DOI] [PubMed] [Google Scholar]
- 10.Asada Y. Lipolytic activity of resident flora of the skin: Some observations on lipase activity of Corynebacterium acnes and Staphylococcus epidermidis compared with Staphylococcus aureus. Skin Res. 1968;10:585–593. [Google Scholar]
- 11.Kellum RE, Strangfeld K. Acne vulgaris. Studies in pathogenesis: Fatty acids of corynebacterium acnes. Arch Dermatol. 1970;101:337–339. doi: 10.1001/archderm.101.3.337. [DOI] [PubMed] [Google Scholar]
- 12.Kligman AM. An overview of acne. J Invest Dermatol. 1974;62:268–287. doi: 10.1111/1523-1747.ep12676801. [DOI] [PubMed] [Google Scholar]
- 13.Longshaw CM, Farrell AM, Wright JD, Holland KT. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: Comparison of sequence with those of other staphylococcal lipases. Microbiology. 2000;146:1419–1427. doi: 10.1099/00221287-146-6-1419. [DOI] [PubMed] [Google Scholar]
- 14.Pospiil L. Quntitative lipase best immung der aus verschiedenen dermatosen isolierten staphylokokken. Dermatol Wochenschr. 1966;48:1335–1339. German. [PubMed] [Google Scholar]
- 15.Bowden MG, et al. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J Biol Chem. 2002;277:43017–43023. doi: 10.1074/jbc.M207921200. [DOI] [PubMed] [Google Scholar]
- 16.Archer GL. Staphylococcus aureus: A well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 17.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG., Jr Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedström SÅ. Lipolytic activity of Staphylococcus aureus strains from cases of human chronic osteomyelitis and other infections. Acta Pathol Microbiol Scand B. 1975;83:285–292. doi: 10.1111/j.1699-0463.1975.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 19.Hedström SA, Nilsson-Ehle P. Trioleoylglycerol lipolysis by Staphylococcus aureus strains from recurrent furunculosis, pyomyositis, impetigo and osteomyelitis. Acta Pathol Microbiol Immunol Scand B. 1983;91:169–173. doi: 10.1111/j.1699-0463.1983.tb00028.x. [DOI] [PubMed] [Google Scholar]
- 20.Rollof J, Hedström SA, Nilsson-Ehle P. Lipolytic activity of Staphylococcus aureus strains from disseminated and localized infections. Acta Pathol Microbiol Immunol Scand B. 1987;95:109–113. doi: 10.1111/j.1699-0463.1987.tb03096.x. [DOI] [PubMed] [Google Scholar]
- 21.Al-Wali WI, Elvin SJ, Mason CM, Clark A, Tranter HS. Comparative phenotypic characteristics of Staphylococcus aureus isolates from line and non-line associated septicaemia, CAPD peritonitis, bone/joint infections and healthy nasal carriers. J Med Microbiol. 1998;47:265–274. doi: 10.1099/00222615-47-3-265. [DOI] [PubMed] [Google Scholar]
- 22.Hoff GE, Hølby N. Staphylococcus aureus in cystic fibrosis: Antibiotic sensitivity and phage types during the latest decade. Investigation of the occurrence of protein A and some other properties of recently isolated strains in relation to the occurrence of precipitating antibodies. Acta Pathol Microbiol Scand B. 1975;83:219–225. [PubMed] [Google Scholar]
- 23.Rollof J, Braconier JH, Söderström C, Nilsson-Ehle P. Interference of Staphylococcus aureus lipase with human granulocyte function. Eur J Clin Microbiol Infect Dis. 1988;7:505–510. doi: 10.1007/BF01962601. [DOI] [PubMed] [Google Scholar]
- 24.Tyski S, Tylewska S, Hryniewicz W, Jeljaszewicz J. Induction of human neutrophils chemotaxis by staphylococcal lipase. Zentralbl Bakteriol Mikrobiol Hyg A. 1987;265:360–368. doi: 10.1016/s0176-6724(87)80254-7. [DOI] [PubMed] [Google Scholar]
- 25.Hu C, Xiong N, Zhang Y, Rayner S, Chen S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem Biophys Res Commun. 2012;419:617–620. doi: 10.1016/j.bbrc.2012.02.057. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen MT, et al. Staphylococcal (phospho)lipases promote biofilm formation and host cell invasion. Int J Med Microbiol. 2018;308:653–663. doi: 10.1016/j.ijmm.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Rollof J, Hedström SA, Nilsson-Ehle P. Purification and characterization of a lipase from Staphylococcus aureus. Biochim Biophys Acta. 1987;921:364–369. doi: 10.1016/0005-2760(87)90038-5. [DOI] [PubMed] [Google Scholar]
- 28.Cadieux B, Vijayakumaran V, Bernards MA, McGavin MJ, Heinrichs DE. Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J Bacteriol. 2014;196:4044–4056. doi: 10.1128/JB.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simons JW, et al. The lipase from Staphylococcus aureus. Expression in Escherichia coli, large-scale purification and comparison of substrate specificity to Staphylococcus hyicus lipase. Eur J Biochem. 1996;242:760–769. doi: 10.1111/j.1432-1033.1996.0760r.x. [DOI] [PubMed] [Google Scholar]
- 30.Delekta PC, Shook JC, Lydic TA, Mulks MH, Hammer ND. Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of exogenous fatty acids. J Bacteriol. 2018;200:e00728-17. doi: 10.1128/JB.00728-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae T, Baba T, Hiramatsu K, Schneewind O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol Microbiol. 2006;62:1035–1047. doi: 10.1111/j.1365-2958.2006.05441.x. [DOI] [PubMed] [Google Scholar]
- 32.Medzhitov R, Janeway C., Jr Innate immunity. N Engl J Med. 2000;343:338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 33.Fournier B, Philpott DJ. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev. 2005;18:521–540. doi: 10.1128/CMR.18.3.521-540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rooijakkers SH, van Kessel KP, van Strijp JA. Staphylococcal innate immune evasion. Trends Microbiol. 2005;13:596–601. doi: 10.1016/j.tim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Aderem A. Role of Toll-like receptors in inflammatory response in macrophages. Crit Care Med. 2001;29(Suppl):S16–S18. doi: 10.1097/00003246-200107001-00008. [DOI] [PubMed] [Google Scholar]
- 36.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 37.Akira S. Mammalian toll-like receptors. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 38.Yimin, et al. Contribution of toll-like receptor 2 to the innate response against Staphylococcus aureus infection in mice. PLoS One. 2013;8:e74287. doi: 10.1371/journal.pone.0074287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aliprantis AO, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–739. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen MT, Götz F. Lipoproteins of gram-positive bacteria: Key players in the immune response and virulence. Microbiol Mol Biol Rev. 2016;80:891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci USA. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fournier B. The function of TLR2 during staphylococcal diseases. Front Cell Infect Microbiol. 2013;2:167. doi: 10.3389/fcimb.2012.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi O, Hoshino K, Akira S. Cutting edge: TLR2-deficient and MyD88-deficient mice are highly susceptible to Staphylococcus aureus infection. J Immunol. 2000;165:5392–5396. doi: 10.4049/jimmunol.165.10.5392. [DOI] [PubMed] [Google Scholar]
- 44.Grayczyk JP, Harvey CJ, Laczkovich I, Alonzo F., 3rd A lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host Microbe. 2017;22:678–687.e9. doi: 10.1016/j.chom.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensson B, Fehrenbach FJ, Hedström SA. A new serological assay for Staphylococcus aureus infections: Detection of IgG antibodies to S. aureus lipase with an enzyme-linked immunosorbent assay. J Infect Dis. 1985;152:286–292. doi: 10.1093/infdis/152.2.286. [DOI] [PubMed] [Google Scholar]
- 47.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio. 2012;3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid. 2014;76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vadehra DV, Harmon LG. Factors affecting production of staphylococcal lipase. J Appl Bacteriol. 1969;32:147–150. doi: 10.1111/j.1365-2672.1969.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 50.Götz F, Verheij HM, Rosenstein R. Staphylococcal lipases: Molecular characterisation, secretion, and processing. Chem Phys Lipids. 1998;93:15–25. doi: 10.1016/s0009-3084(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 51.Rosenstein R, Götz F. Staphylococcal lipases: Biochemical and molecular characterization. Biochimie. 2000;82:1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- 52.Hashimoto M, et al. Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int Immunol. 2006;18:355–362. doi: 10.1093/intimm/dxh374. [DOI] [PubMed] [Google Scholar]
- 53.Asanuma M, et al. Structural evidence of α-aminoacylated lipoproteins of Staphylococcus aureus. FEBS J. 2011;278:716–728. doi: 10.1111/j.1742-4658.2010.07990.x. [DOI] [PubMed] [Google Scholar]
- 54.Tawaratsumida K, et al. Characterization of N-terminal structure of TLR2-activating lipoprotein in Staphylococcus aureus. J Biol Chem. 2009;284:9147–9152. doi: 10.1074/jbc.M900429200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurokawa K, et al. Environment-mediated accumulation of diacyl lipoproteins over their triacyl counterparts in Staphylococcus aureus. J Bacteriol. 2012;194:3299–3306. doi: 10.1128/JB.00314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang JY, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 57.Brady L, et al. A serine protease triad forms the catalytic centre of a triacylglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 58.Jäger S, Demleitner G, Götz F. Lipase of Staphylococcus hyicus: Analysis of the catalytic triad by site-directed mutagenesis. FEMS Microbiol Lett. 1992;100:249–254. doi: 10.1111/j.1574-6968.1992.tb14048.x. [DOI] [PubMed] [Google Scholar]
- 59.Ollis DL, et al. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 60.Nikoleit K, Rosenstein R, Verheij HM, Götz F. Comparative biochemical and molecular analysis of the Staphylococcus hyicus, Staphylococcus aureus and a hybrid lipase. Indication for a C-terminal phospholipase domain. Eur J Biochem. 1995;228:732–738. [PubMed] [Google Scholar]
- 61.Nguyen MT, Hanzelmann D, Härtner T, Peschel A, Götz F. Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect Immun. 2015;84:205–215. doi: 10.1128/IAI.00822-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White DC, Frerman FE. Fatty acid composition of the complex lipids of Staphylococcus aureus during the formation of the membrane-bound electron transport system. J Bacteriol. 1968;95:2198–2209. doi: 10.1128/jb.95.6.2198-2209.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sen S, et al. Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS One. 2016;11:e0165300. doi: 10.1371/journal.pone.0165300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh VK, et al. Insertional inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl Environ Microbiol. 2008;74:5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843:2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 66.Medzhitov R, Janeway CA., Jr Innate immunity: The virtues of a nonclonal system of recognition. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 67.Janeway CA, Travers P, Walport M, Shlomchik MJ. Immunobiology: The Immune System in Health and Disease. Garland Science Publishing; New York: 2005. [Google Scholar]
- 68.Li L, Okusa MD. Macrophages, dendritic cells, and kidney ischemia-reperfusion injury. Semin Nephrol. 2010;30:268–277. doi: 10.1016/j.semnephrol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Velásquez-Lopera MM, Correa LA, García LF. Human spleen contains different subsets of dendritic cells and regulatory T lymphocytes. Clin Exp Immunol. 2008;154:107–114. doi: 10.1111/j.1365-2249.2008.03734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott RP, Quaggin SE. Review series: The cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pinto AR, et al. Revisiting cardiac cellular composition. Circ Res. 2016;118:400–409. doi: 10.1161/CIRCRESAHA.115.307778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koeppen BM. Renal regulation of acid-base balance. Adv Physiol Educ. 1998;275:S132–S141. doi: 10.1152/advan.00054.2009. [DOI] [PubMed] [Google Scholar]
- 74.Knolle PA, Gerken G. Local control of the immune response in the liver. Immunol Rev. 2000;174:21–34. doi: 10.1034/j.1600-0528.2002.017408.x. [DOI] [PubMed] [Google Scholar]
- 75.Kmieć Z. Cooperation of Liver Cells in Health and Disease. Springer; Berlin: 2001. Cooperation of liver cells in the synthesis and degradation of eicosanoids; pp. 51–59. [Google Scholar]
- 76.Ding C, et al. A cell-type-resolved liver proteome. Mol Cell Proteomics. 2016;15:3190–3202. doi: 10.1074/mcp.M116.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ackerman JJ, Lowry M, Radda GK, Ross BD, Wong GG. The role of intrarenal pH in regulation of ammoniagenesis: [31P]NMR studies of the isolated perfused rat kidney. J Physiol. 1981;319:65–79. doi: 10.1113/jphysiol.1981.sp013892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ohlrogge JB, Emken EA, Gulley RM. Human tissue lipids: Occurrence of fatty acid isomers from dietary hydrogenated oils. J Lipid Res. 1981;22:955–960. [PubMed] [Google Scholar]
- 79.Elvehjem C, Peterson W. The iron content of animal tissues. J Biol Chem. 1927;74:433–441. [Google Scholar]
- 80.Kotronen A, et al. Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity (Silver Spring) 2010;18:937–944. doi: 10.1038/oby.2009.326. [DOI] [PubMed] [Google Scholar]
- 81.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jenkins A, et al. Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. MBio. 2015;6:e02272-14. doi: 10.1128/mBio.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Szafranska AK, et al. High-resolution transcriptomic analysis of the adaptive response of Staphylococcus aureus during acute and chronic phases of osteomyelitis. MBio. 2014;5:e01775-14. doi: 10.1128/mBio.01775-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu Y, et al. In vivo gene expression in a Staphylococcus aureus prosthetic joint infection characterized by RNA sequencing and metabolomics: A pilot study. BMC Microbiol. 2016;16:80. doi: 10.1186/s12866-016-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Date SV, et al. Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J Infect Dis. 2014;209:1542–1550. doi: 10.1093/infdis/jit668. [DOI] [PubMed] [Google Scholar]
- 86.Malachowa N, Kobayashi SD, Sturdevant DE, Scott DP, DeLeo FR. Insights into the Staphylococcus aureus-host interface: Global changes in host and pathogen gene expression in a rabbit skin infection model. PLoS One. 2015;10:e0117713. doi: 10.1371/journal.pone.0117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zecconi A, Scali F. Staphylococcus aureus virulence factors in evasion from innate immune defenses in human and animal diseases. Immunol Lett. 2013;150:12–22. doi: 10.1016/j.imlet.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 88.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 89.Bohach GA. Gram-Positive Pathogens. 2nd Ed. American Society of Microbiology; Washington, DC: 2006. Staphylococcus aureus endotoxins; pp. 464–477. [Google Scholar]
- 90.Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ito M, Adachi-Akahane S. Inter-organ communication in the regulation of lipid metabolism: Focusing on the network between the liver, intestine, and heart. J Pharmacol Sci. 2013;123:312–317. doi: 10.1254/jphs.13r09cp. [DOI] [PubMed] [Google Scholar]
- 92.Velagapudi VR, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: Structure, function, regulation, and role in disease. J Mol Med (Berl) 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 94.Medh JD, et al. Lipoprotein lipase binds to low density lipoprotein receptors and induces receptor-mediated catabolism of very low density lipoproteins in vitro. J Biol Chem. 1996;271:17073–17080. doi: 10.1074/jbc.271.29.17073. [DOI] [PubMed] [Google Scholar]
- 95.Beisiegel U, Weber W, Bengtsson-Olivecrona G. Lipoprotein lipase enhances the binding of chylomicrons to low density lipoprotein receptor-related protein. Proc Natl Acad Sci USA. 1991;88:8342–8346. doi: 10.1073/pnas.88.19.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jain S, Coats SR, Chang AM, Darveau RP. A novel class of lipoprotein lipase-sensitive molecules mediates Toll-like receptor 2 activation by Porphyromonas gingivalis. Infect Immun. 2013;81:1277–1286. doi: 10.1128/IAI.01036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shimada E, Kataoka H, Miyazawa Y, Yamamoto M, Igarashi T. Lipoproteins of Actinomyces viscosus induce inflammatory responses through TLR2 in human gingival epithelial cells and macrophages. Microbes Infect. 2012;14:916–921. doi: 10.1016/j.micinf.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 98.Hashimoto M, Asai Y, Ogawa T. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int Immunol. 2004;16:1431–1437. doi: 10.1093/intimm/dxh146. [DOI] [PubMed] [Google Scholar]
- 99.Stevens DL. Streptococcal Pharyngitis. Karger Publishers; Basel, Switzerland: 2004. Virulence factors of streptococcal pharyngitis; pp. 3–15. [Google Scholar]
- 100.Machata S, et al. Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J Immunol. 2008;181:2028–2035. doi: 10.4049/jimmunol.181.3.2028. [DOI] [PubMed] [Google Scholar]
- 101.Kurokawa K, et al. Novel bacterial lipoprotein structures conserved in low-GC content gram-positive bacteria are recognized by Toll-like receptor 2. J Biol Chem. 2012;287:13170–13181. doi: 10.1074/jbc.M111.292235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fairweather N, Kennedy S, Foster TJ, Kehoe M, Dougan G. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect Immun. 1983;41:1112–1117. doi: 10.1128/iai.41.3.1112-1117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Duthie ES, Lorenz LL. Staphylococcal coagulase; mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 105.Fuller JR, et al. Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front Cell Infect Microbiol. 2011;1:19. doi: 10.3389/fcimb.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zorzoli A, Grayczyk JP, Alonzo F., 3rd Staphylococcus aureus tissue infection during sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathog. 2016;12:e1005933. doi: 10.1371/journal.ppat.1005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Laczkovich I, et al. Increased flexibility in the use of exogenous lipoic acid by Staphylococcus aureus. Mol Microbiol. 2018;109:150–168. doi: 10.1111/mmi.13970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.DuMont AL, et al. Staphylococcus aureus elaborates leukocidin AB to mediate escape from within human neutrophils. Infect Immun. 2013;81:1830–1841. doi: 10.1128/IAI.00095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 111.Wooten RM, et al. Toll-like receptor 2 is required for innate, but not acquired, host defense to Borrelia burgdorferi. J Immunol. 2002;168:348–355. doi: 10.4049/jimmunol.168.1.348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.