Significance
Outbreaks of Ebola virus disease (EVD) continue to emerge with severe and often deadly outcomes and global consequences. Novel strategies for examining host response differences that distinguish EVD survivors and fatalities can only deepen our understanding of the disease and expand diagnostic and treatment options. Lipids are major molecular constituents in human plasma with important structural, transport, energy, and signaling functions. Here we provide a comprehensive examination of the EVD lipidome. Using liquid chromatography tandem mass spectrometry, we profiled the human plasma lipidome of patients that survived and succumbed to EVD during the 2014 outbreak in Sierra Leone. The results highlight the profound impact of EVD on the host lipidome, leading to possible therapies for improving EVD survival.
Keywords: lipidomics, Ebola, critical illness, therapies, mass spectrometry
Abstract
Ebola virus disease (EVD) often leads to severe and fatal outcomes in humans with early supportive care increasing the chances of survival. Profiling the human plasma lipidome provides insight into critical illness as well as diseased states, as lipids have essential roles as membrane structural components, signaling molecules, and energy sources. Here we show that the plasma lipidomes of EVD survivors and fatalities from Sierra Leone, infected during the 2014–2016 Ebola virus outbreak, were profoundly altered. Focusing on how lipids are associated in human plasma, while factoring in the state of critical illness, we found that lipidome changes were related to EVD outcome and could identify states of disease and recovery. Specific changes in the lipidome suggested contributions from extracellular vesicles, viremia, liver dysfunction, apoptosis, autophagy, and general critical illness, and we identified possible targets for therapies enhancing EVD survival.
The West African Ebola virus (EBOV) outbreak of 2014–2016 was the most devastating human EBOV epidemic to date, with over 28,000 cases of Ebola virus disease (EVD) and greater than 11,000 deaths (1). Less than 2 years after the West African EBOV outbreak was resolved in April 2018, the virus has reemerged in the Democratic Republic of Congo, presenting a fresh reminder that the world remains ill equipped to confront the possibility of another widespread EBOV outbreak. One potential strategy for improving EVD patient outcomes is the use of supplemental supportive therapeutics (2), which may be accessible in the absence of approved antiviral or immunomodulatory therapies, or in resource-limited settings. Nutritional care, in particular, is important for recovery.
A principal target organ for EBOV infection is the liver, which is also the main site for fatty acid synthesis and lipoprotein release into the bloodstream and therefore central to lipid metabolism at the whole body level. Lipids in human blood plasma are associated with lipoprotein membranes [e.g., diacylglycerophosphocholines (PCs)], lipoprotein cargo [e.g., triacylglycerols (TGs) and cholesterol esters], and proteins [e.g., monoacylglycerophosphocholines (LPCs) by albumin], and are generated by the intestines, liver, and peripheral tissues (e.g., adipose, muscle). In addition, all human bodily fluids contain extracellular vesicles, which are thought to contribute to physiology and pathology (3), increasing or decreasing in abundance in disease states, such as sepsis (4). As such, lipid profiling of blood plasma enables the discovery of metabolic derangements (5, 6), including states of critical illness (7). During critical illness, such as sepsis, lipid abnormalities are common and can include hypertriglyceridemia, increased plasma free fatty acids, and decreased cholesterol containing lipoproteins [i.e., high-density lipoprotein (HDL) and low-density lipoprotein (LDL)] (8–10), with the levels of circulating lipids determined by the interactions between gut, liver, and adipose tissue.
Lipid involvement in EBOV infection was first elucidated in vitro, revealing the importance of lipids for both EBOV entry and egress (11–15). Recently, we performed multiplatform omics analysis of peripheral blood mononuclear cells (PBMCs) and plasma from human EVD survivors and fatalities recruited in Freetown, Sierra Leone in 2015 (16). This study demonstrated the strength of integrating omics data for biomarker discovery and also revealed immune and metabolic responses to EVD (16). In addition, we noted changes in plasma lipid subclasses that differentiated EVD survivors and fatalities and discovered lipid species that were predictive of EVD outcome; however, a comprehensive interpretation of the lipidome was not conducted. Previous studies on EVD identified blood markers of poor outcomes, including high viremia (17), lymphopenia due to lymphoid cell apoptosis (18, 19), neutrophilia, thrombocytopenia, virus-mediated tissue and organ damage, and disseminated intravascular coagulopathy (20, 21). Here, we report an in-depth analysis of the plasma lipidome from patients with EVD and show that many of the lipid alterations with EVD align with a host stress response during critical illness and infection. We also outline potential supportive therapeutics based on the detailed lipid profile of those who survived or succumbed to EVD.
Results
The Plasma Lipidome Is Dramatically Altered by EVD.
Blood was collected in 2015 from Sierra Leone healthy volunteers and EVD patients (survivors and fatalities) after initial diagnosis, with serial samples collected from survivors over the course of EVD and recovery (16). Plasma was separated from the whole blood and lipids were extracted from plasma (22) and analyzed using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (23, 24). In total, 423 lipids were identified and quantified (SI Appendix, Table S1) over 379 LC-MS lipid peaks (SI Appendix, Table S2) covering four lipid categories and 19 subclasses (16).
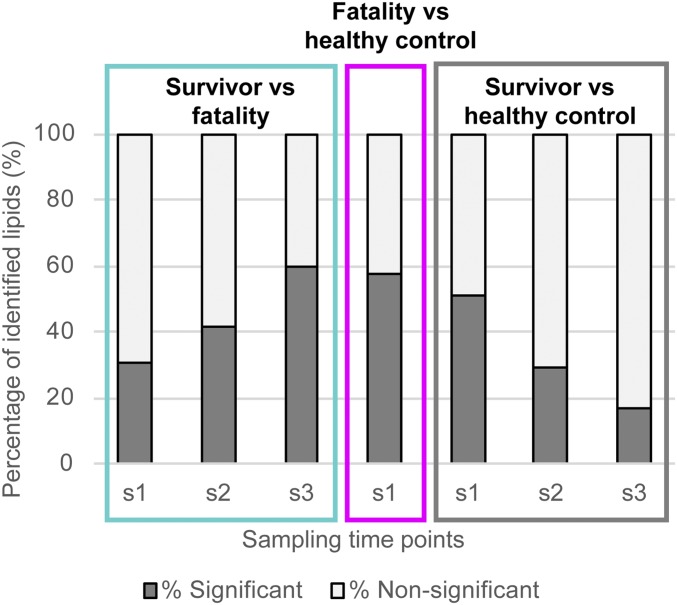
Overall, lipid metabolism was strongly altered in EVD. Samples collected from survivors, including after initial diagnosis (sample s1), and during EVD and recovery (samples s2 and s3), were compared with those from fatalities or healthy volunteers (Fig. 1). In survivors vs. fatalities, 30.6% of the identified lipids were significantly different (P < 0.05) at s1, which increased to 59.4% at s3 in survivors (Fig. 1). In contrast, the lipid profiles of survivors vs. healthy controls exhibited the opposite trend, with 50.7% of identified lipids significantly altered in survivors at s1 and only 16.6% significantly altered in survivors at s3 (Fig. 1). We hypothesized that these opposing trends most likely indicate recovery in the survivors group. Further supporting this hypothesis, 57.3% of the lipidome of fatalities was significantly different from that of controls.
Fig. 1.
Percent of the number of identified statistically significant different lipids (P < 0.05) between outcomes (survivors vs. fatalities) and compared with healthy controls. Serial samples from survivors were collected over time and annotated as s1, s2, and s3. The number of statistically significant lipids increases in EVD survivors compared with fatalities over time and becomes more similar with fewer differences between survivors and the healthy controls with EVD recovery.
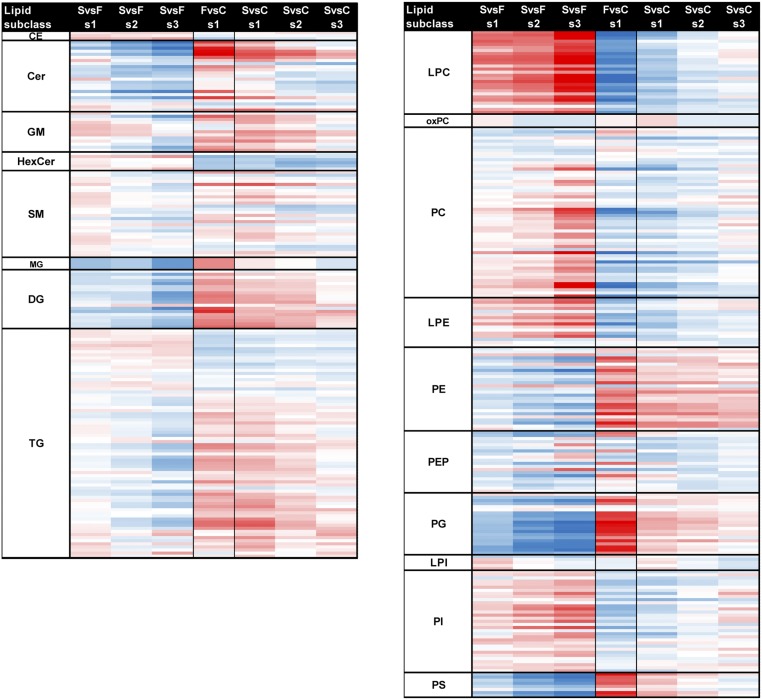
Most of the lipids were altered at the subclass level and trended with outcomes (16) (Fig. 2 and SI Appendix, Table S2). Intrasubclass variations, which were not previously described (16), also trended with outcome, both in terms of total sum of fatty acid carbons and double bonds (i.e., intact lipid species) and at the level of individual fatty acids (Fig. 2 and SI Appendix, Table S2). Statistically significant lipids in survivors of EVD contained greater amounts of longer chained polyunsaturated fatty acid (LCPUFA) PCs (average combined fatty acid length of 38 carbons and five double bonds), whereas EVD fatalities had statistically significant increases in TGs with longer unsaturated fatty acid chains (average of 55 carbons and five double bonds).
Fig. 2.
Global lipidome profile of EVD human plasma per lipid subclass. A total of 423 unique lipids across 379 LC-MS peaks were identified across four cateogories (glycerophospholipids, sterols, sphingolipids, and glycerolipids) and 19 subclasses. Average plasma lipid levels for EVD patients (F, fatalities; s1, s2, and s3, survivors’ first, second, and third samples) compared with healthy controls (C); or for s1/s2/s3 vs. F comparisons. Data in the heatmap are log2 FC values for each individual lipid. The lipids are sorted within each subclass level by the increasing total number of carbons in the hydrocarbon chains and then the total of double bonds. Values are scaled from −10 to +10 log2 FC going from blue (decrease) to white (no change) to red (increase), respectively. SvsF per s1, s2, s3 comparison values are displayed as the direction of expression in survivors. FvsC comparison values are displayed as the direction of expression in the fatalities sample. CE, cholesterol ester; GM, ganglioside; HexCer, glucosyl- or galactosylceramide; LacCer, lactosylceramide; LPE, monoacylglycerophosphoethanolamine; LPI, monoacylglycerophosphoinositol; MG, monoacylgleride; PEP, plasmalogen PE; and PG, diacylglycerophosphosphoglycerol or bis(monoacylglycerol)phosphate.
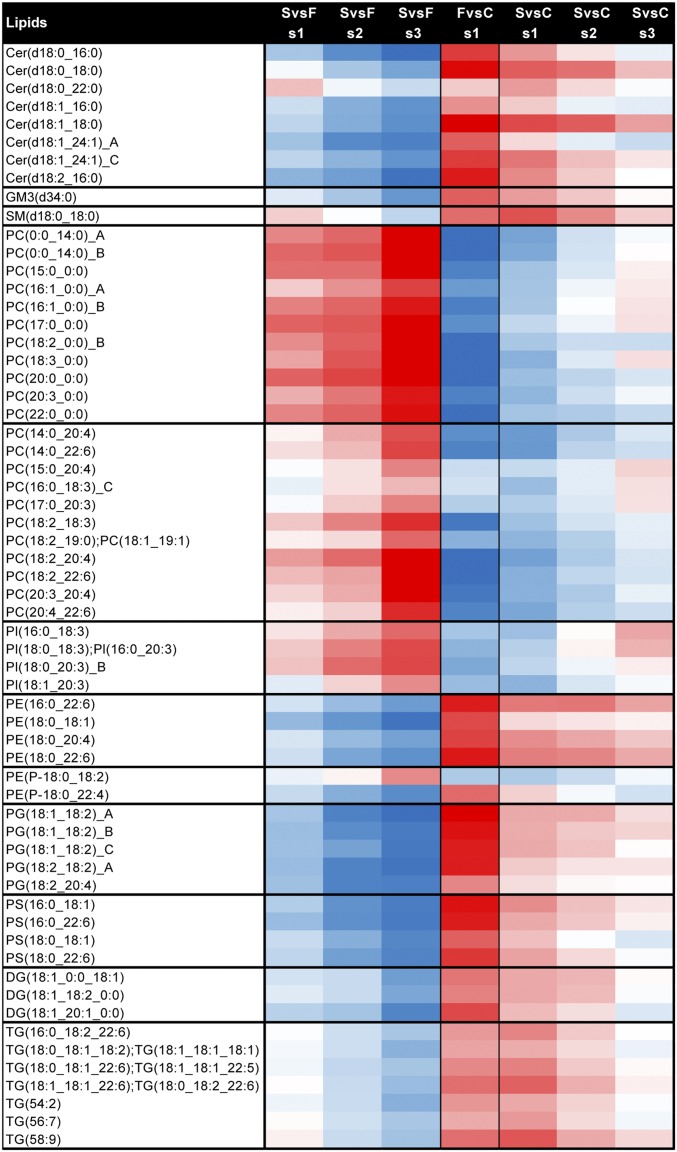
We also observed changes in the lipidome through the course of EVD. Of the lipids that underwent the greatest change [>4.5 log2 fold change (FC); typically P < 0.01] from s1 to s3 in survivors vs. fatalities or healthy controls, all ceramides (Cer) containing 16:0 or 18:0 fatty acids decreased in survivors, whereas all but one diacylglycerophosphoinositol (PI) containing 18:3 or 20:3 fatty acids increased (Fig. 3 and SI Appendix, Table S3). Additionally, 35% of all Cer, 42% of LPCs, 20% of PCs, 25% of diacylglycerophosphoglycerols (PGs), and 50% of diacylglycerophosphoserines (PSs) showed greater than 4.5 FC, indicating that these lipid classes are strongly influenced by the progress of and/or recovery from EVD.
Fig. 3.
Lipids undergoing the greatest amount of change with EVD. Lipids with a value greater than 4.5 FC or below −4.5 FC between s1 and s3 for survivors (S) vs. fatalities (F) and survivors vs. healthy controls (C). Scale of log2 FC is the same as in Fig. 1 (blue down, white no change, and red up). Values are scaled from −10 to +10 log2 FC going from blue (decrease) to white (no change) to red (increase), respectively. Direction of expression is the same as in Fig. 1. GM, ganglioside; PC with 0:0 in annotation, monoacylglycerophosphocholine; PE, diacylglycerophosphoethanolmine; PEP, plasmalogen PE; PG, diacylglycerophosphosphoglycerol OR bis(monoacylglycerol)phosphate; and PS, diacylglycerophosphoserine.
Lipids That Increased with Fatal Outcomes.
Subclasses that were increased in fatalities vs. healthy controls include PS, diacylglycerophosphoethanolamine (PE), Cer, PG, and diacylglycerol (DG). PS lipids externalized from the inner leaflet of the plasma membrane to outer leaflet is a hallmark signature of apoptosis (25). PS externalization on the surface of platelets, can also induce blood coagulation (26, 27). PS lipids also play vital roles in EBOV replication (11, 14, 28). Eight PS lipids were identified and were statistically (P value <0.05) higher in fatalities in our study, with the exception of PS(18:0_20:4) (Fig. 2 and SI Appendix, Table S2). This abundance of PS lipids in plasma is unusual because PSs are not typically found in high abundance in human blood plasma (29, 30). PSs are not associated with most plasma lipoproteins (31–33), making up <0.05 wt% of total HDL lipid in healthy individuals (34), suggesting the prevalent PS signature in EVD patients is derived from the lipoprotein-free fraction.
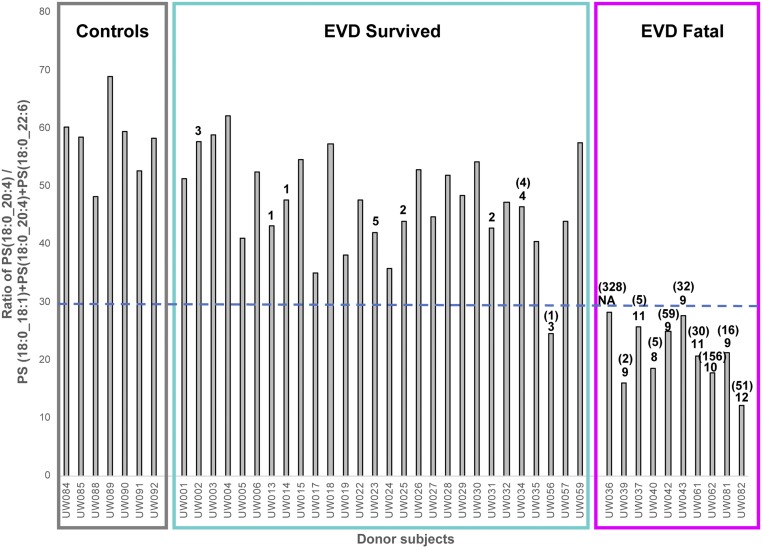
Extracellular vesicles (EVs), including exosomes and microvesicles (MVs), are part of the lipoprotein-free fraction normally present in blood, increasing in circulation in stressed systems (3). Microvesicles in particular are enriched in PS lipids (35, 36). Platelets, common sources of MVs, have been shown to externalize PS(18:0_18:1) and PS(18:0_20:4) from the inner to outer membrane when activated by thrombin (27). Lipidomics studies of exosomes generated from prostate cancer 3 (PC-3) cells specifically identified two PS lipids, PS(18:0_18:1) and PS(18:0_22:6), that were elevated and exclusively identified in exosomes vs. the parent cells, respectively (37). Raw peak intensity values of these three PS lipids from our data revealed that PS(18:0_18:1) and PS(18:0_20:4) were the most abundant PS lipids followed by PS(18:0_22:6). As previous studies have shown that lipid ratios can be predictive of certain diseases (38), we noted that when the raw intensity value of PS(18:0_20:4) was divided by the sum of the raw intensity values of these three PS lipids, all fatal outcomes had a value of less than 28.5 (average value of 21) (Fig. 4 and SI Appendix, Fig. S1). All survivors (average value 47) and healthy controls (average value 58) plotted above this value, except for donor UW056 which had a value of 24.5. Viral peptide counts and viral RNA were greatest for those with fatal outcome (Fig. 4). The PS ratio trend remained similar when PS(18:0_20:4) was divided by the sum of all PS lipids (SI Appendix, Fig. S1).
Fig. 4.
The raw LC-MS peak intensity ratio of PS(18:0_20:4) vs. 3 other PS lipids [PS(18:0_18:1), PS(18:0_22:6), PS(18:0_20:4)] identified per donor sample. The ratio values are greater for all survivors (above 28.5) vs. those EVD fatalities except UW056. This PS ratio trend also correlated with viral peptide counts (value above bars, if not listed the value is zero), as well as viral qPCR as genomic copies per nanogram of input RNA (values in parentheses, if not listed the value is below detection). Peptide and qPCR values are from Eisfeld et al. (16).
As EVs can be produced from multiple cell types (e.g., platelets, endothelium, and immune cells) (39) and increase in abundance within diseased systems, we examined transcriptomics data from the PBMCs isolated in parallel to plasma (16) for changes in transcripts related to PS and PE synthesis and translocation. The transcriptomic data did not show statistically significant gene expression involved in PS lipid biosynthesis (PTDSS1 and PTDSS2); however, the transcript data did reveal that expression of PS scramblases PLSCR1 and Xkr8 trended with the PS lipid for most outcome comparisons (SI Appendix, Table S4). Xkr8 scramblase was recently shown to be incorporated into Ebola virus-like particles (VLPs) (a surrogate for authentic virus particles) and is required for externalizing PS on the surface of the VLPs to promote Ebola virus entry (28). Interestingly, scramblase PLSCR1 exhibited the greatest log2 FC (+8.5) in fatalities among all 785 lipid-associated transcripts detected in PBMCs (SI Appendix, Table S5). Transcripts involved in PE synthesis were also examined as they are also externalized from the inner membrane to outer membrane on activated platelets. This revealed two genes, EPT1 (involved in de novo synthesis) and PISD (involved in conversion of PS to PE), that were statistically elevated in the fatalities (SI Appendix, Table S6). A previous study of platelet aminophospholipids identified seven PS and PE lipids which were externalized on activated human platelets (27), six of these lipids were identified in our data and trended similarly with EVD fatalities but not all were statistically elevated (SI Appendix, Table S7).
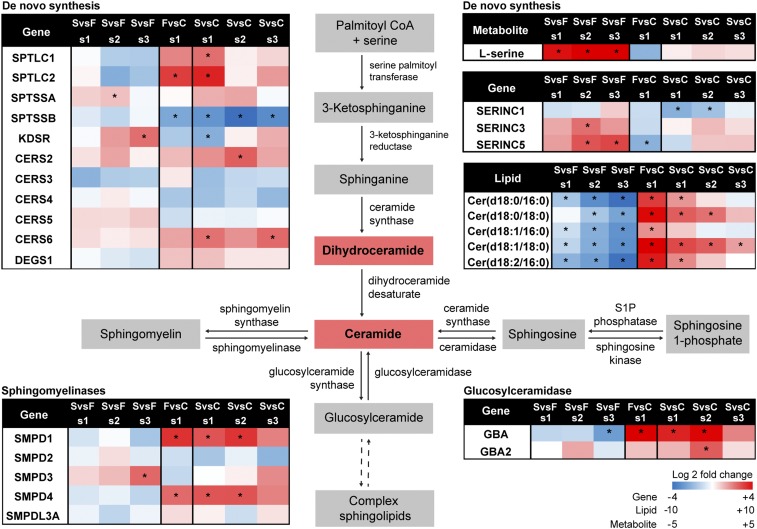
Sphingolipids have many biological functions, several of which are chain-length dependent (40). Cer containing 16:0, 18:0, and 24:1 fatty acids increased the most of all identified ceramides in EVD fatalities compared with EVD survivors (Fig. 3 and SI Appendix, Table S3). For those that survived, this ceramide signature decreased to levels close to those of healthy controls over time. In plasma, Cer, as well as sphingomyelin (SM), are enriched in LDL lipoproteins (32, 33). As SM lipids were not elevated in fatalities [with the exception of SM(d18:0_18:0)], and LDL lipoproteins are known to be lower in cases of critical illness (8, 10), the increased Cer signature is likely from additional sources. Increases in Cer with C16:0 and C18:0 have been implicated in cardiovascular health (38) and signatures of apoptosis (40). EVD-associated apoptosis is well known, especially in regards to lymphopenia (18, 19, 41). Apoptosis also occurs in multiple organs within the body (42) whose molecular signatures may leak into the bloodstream depending upon the state of vascular health, which is low in cases of severe EVD (21). Given the prevalence of lymphocyte apoptosis in those with EVD, transcripts from the PBMCs of the same patients were investigated for genes involved in sphingolipid metabolism (Fig. 5). Trends were not observed in de novo Cer synthesis; however, some similarities with gene expression were noted with two sphingomyelinases (SMPD1 and SMPD4) and a glucosylceramidase (GBA). Ceramide synthases were not significantly different except for CER6, which is specific for C14, C16, and C18, and which was significantly increased in survivors vs. healthy controls (Fig. 5).
Fig. 5.
Multiomic sphingolipid metabolic pathway of EVD. Sphingolipid related transcripts in the PBMCs, ceramide lipids in the plasma as well as l-serine from the plasma are displayed on the sphingolipid metabolic pathway. Values are log2 FC scaled going from blue (decrease) to white (no change) to red (increase), respectively. Statistically significant molecules have a *P value of <0.05.
In addition to apoptosis, lipid signatures of autophagy were also observed. The 16:0, 18:0, and 24:1 Cer described above also contain dihydroceramides (i.e., sphinganine base), which function as precursors to Cer formation (Fig. 5) but also have cellular roles in autophagy (43). In addition, PG lipids were highly elevated in those with fatal outcomes. This is unexpected as PGs are normally minor constituents of plasma (33, 44). It is possible that some or all of the PG lipids identified are actually monoacylglycerophosphomonoradylglycerols (LBPAs) [more commonly known as bis(monoacylglycerol)phosphate (BMP)], a structural isomer of PG since PG and BMP lipids have very similar tandem mass spectra. BMPs have been previously identified in plasma, with more BMPs associated with the lipoprotein-free fraction vs. lipoproteins themselves, and these lipids have been found to be elevated in plasma of those with lysosomal storage disease (45). BMP lipids are highly enriched in late endosomes and lysosomes (46) and act as lipid signatures for these organelles. LC-MS/MS analysis of reference standards BMP(14:0/14:0) and PG(14:0/14:0) revealed that BMPs eluted 1.9 min earlier than PG (SI Appendix, Fig. S2A), which is consistent with our findings (SI Appendix, Fig. S2 B–D).
Metabolomics analyses reported in Eisfeld et al. (16) identified markers of abnormal lipid metabolism in the liver. Two-hydroxybutyric acid and 3-hydroxybutyric acid (a ketone body), both synthesized in the liver, decreased in survivors with time and increased in fatalities compared with healthy controls. This suggests that the patients are undergoing excessive fatty acid beta-oxidation and oxidative stress. In addition to the liver, adipose tissue plays a strong role in the circulating lipid profile, with an increase in TGs and free fatty acids in particular, and is actively influenced in critical illness (47). Those EVD patients with fatal outcomes had increases (>2 log2 FC) in TGs with longer fatty acid chains (average of 55 carbons) that were more polyunsaturated (average five double bonds) compared with those that decreased (< −2 log2 FC; average of 44 carbons and 1.8 double bonds) vs. healthy controls. Many of the plasma TGs containing 22:6 fatty acids in our data were undergoing the greatest increase in fatalities (or decrease in survivors vs. fatalities) over time (Fig. 3 and SI Appendix, Table S3). Proteomics data (16) revealed two proteins with adipose-lipid related functions (SI Appendix, Table S8). Retinoic acid receptor responder protein 2 (RARR2), an adipocyte-secreted protein which is highly expressed in adipose tissue and regulates adipogenesis and adipocyte metabolism (48, 49), trended in the same direction as the 22:6 containing TGs and was significantly elevated in fatalities. Zinc-alpha-2-glycoprotein (ZA2G), a secreted protein that stimulates lipid degradation in adipocytes and is associated with excessive weight loss and cancer cachexia (50), was elevated in survivors compared with both fatalities and healthy controls.
Lipids That Increase with Survival Outcomes.
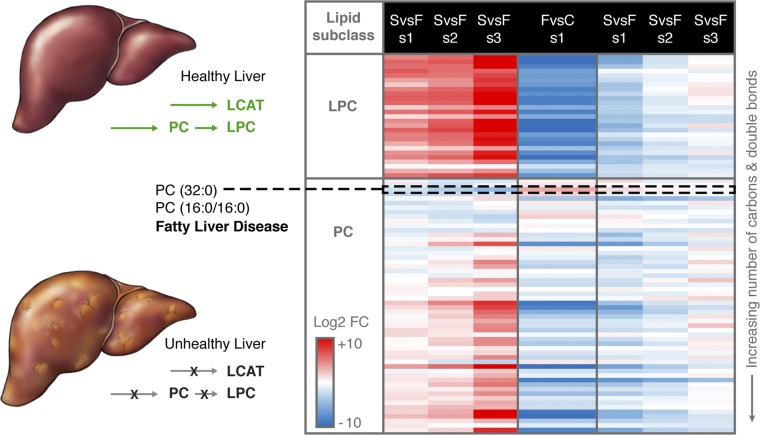
Four lipid subclasses, LPC, PC, monoacylglycerophosphoethanolamine (LPE), and PI were identified in higher abundance in the survivors vs. fatalities, with the abundances increasing with convalescence (Fig. 2). The liver is the main target for EBOV infection but also the main location of fatty acid synthesis and lipid circulation through lipoprotein synthesis and secretion. PCs are synthesized in the liver and are the only phospholipid necessary for lipoprotein assembly and excretion (51). Decreases in LPCs and PCs in blood plasma have been observed in sepsis (7, 52), cancer (53), and dengue infection (5); however, the mechanisms behind PC dysregulation in these conditions are poorly understood. In plasma, PCs are primarily associated with HDL (31–33), which has been reported as lower in critically ill patients (8–10). Of the PC lipids identified in our study, almost all decreased in fatalities, including 18 with a P value of <0.01 and 26 with a P value of <0.05. The only statistically significant PC lipid that increased in fatalities was PC(16:0/16:0) (P value 0.003) (Figs. 2 and 6). Interestingly, PC(32:0), which could correspond to PC(16:0/16:0), was recently reported as a potential signature of progressive forms of fatty liver disease (6).
Fig. 6.
LPC and PC plasma signature related to state of liver function and disease. A healthy liver produces PC lipids, which are the main phospholipids in lipoprotein membranes and are required for VLDL production from the liver. LCAT, which is also secreted by the liver, converts PCs to LPCs. In an unhealthy liver or during liver dysfunction, PC and LCAT production may be decreased. Data in the heatmap are log2 FC values for individual lipids which are sorted at the subclass level by increasing number of total hydrocarbon chain length and then the total of double bonds in hydrocarbon chains. Values are scaled from −10 to +10 log2 FC going from blue (decrease) to white (no change) to red (increase), respectively. Direction of expression is the same as in Fig. 1. The only PC lipid that is statistically significant and increased in fatal (F) vs. control (C) is PC(16:0/16:0). Gorden et al. (6) reported that PC(32:0), which could be PC(16:0/16:0), may be a lipid signature of progressive forms of fatty liver disease.
In addition to PCs, LPC circulation is also influenced by liver function. In plasma, an appreciable amount of LPC is in the lipoprotein-free fraction and is formed from lecithin:cholesterol acyltransferase (LCAT), which is primarily synthesized in the liver and bound to the surface of HDL and LDL. As outlined above, plasma HDL and LDL are decreased in critically ill patients and have been shown to be associated with fatalities due to sepsis (7, 9, 52). All but 1 of the LPCs identified in our study (27 total) were significantly altered (P value of ≤0.001; Figs. 2 and 6 and SI Appendix, Table S2) and were lower in fatalities. Proteomics data of the same patients revealed that of the 10 quantified apolipoproteins (SI Appendix, Table S8) (16), only APOD, which is mainly associated with HDL (54) and complexed with LCAT, was statistically significant (P value 0.02) in fatalities vs. controls with a log2 FC of −2.52.
Survivor vs. fatality profiles may indicate an increase in HDL with convalescence. PI lipids have a greater association with HDL (31, 33), with roles in increasing HDL-C levels during transport of cholesterol from peripheral tissues to the liver (55). Interestingly, all but 1 PI containing 18:3 or 20:3 fatty acids significantly increased (>log2 4.5 FC) in survivors vs. fatalities (Fig. 3). LCPUFA PCs (average of 38 carbons and 4.8 double bonds) also progressively increased in EVD survivors vs. fatalities with convalescence (Figs. 2 and 6 and SI Appendix, Table S2).
Discussion
Our study has shown that EVD causes profound changes in lipid metabolism and homeostasis. The ratios of certain PS lipids correlating with EVD outcome, as well as the elevated PS signature in the fatalities, could result from the presence of EVs, including exosomes and MVs. EVs are normally present in blood plasma and increase within perturbed systems (3). Within systems that trigger coagulation, such as virus infection (56), sepsis (4), and inflammation (57), cellular precursors of MVs (e.g., monocytes, platelets, and endothelial cells) become activated, moving PS lipids from the inner to the outer plasma membrane through the action of scramblases (28, 39, 58). Exosomes, which are produced from the multivesicular body of endosomes with roles related to infection response (59) also contain elevated PS lipids, but their roles in coagulation, if any, are less clear.
In addition to EVs being potential sources for the PS signatures (Figs. 2 and 4), the contribution of the signature from the virus particles themselves cannot be ruled out. Viruses, including EBOV, and EVs share many similarities (60) in both their lipid profile and route of generation. EBOV infects many of the same cell types that are known to produce MVs such as dendritic cells and endothelial cells (21, 61). MVs and EBOV bud from the plasma membrane at regions enriched in lipid rafts (13, 39, 62). EBOV is similar in size to MVs (58, 61), and EVs may be present in similar concentrations to EBOV particles during infection [∼105 to 108 EBOV RNA copies/mL (63) and >5 × 107 EVs/mL in nondiseased state (58)]. Additionally, as with activated MVs, EBOV is also believed to contain externalized PS lipids, allowing the virus to utilize PS-mediated viral apoptotic mimicry for host cell entry (12, 25, 64). Unlike in the role of coagulation activation of MVs, exposed PS lipids on EBOV particles act as a classic “eat-me” signal, attracting phagocytes, such as macrophages, that are capable of clearing apoptotic debris and enabling host entry through macropinocytosis, followed by trafficking through the endosomal system (12, 65, 66). PS lipids have additional functions in EBOV replication in that they are involved in host cell entry and egress. EBOV matrix protein VP40, which regulates budding and egress from plasma membranes (14), selectively induces vesiculation from membranes containing PSs (14). The binding of VP40 with PSs regulates VP40 localization and oligomerization of the plasma membrane inner leaflet inducing exposure of PSs to the outer leaflet at the site of egress (11). Reductions in the PS content result in decreased VP40 assembly and subsequently Ebola virus egress in some cell types (11). Recently, EBOV VP40 has also been associated with exosomes (67, 68). VP40 utilizes the endosomal pathway in formation, inducing apoptosis in recipient immune cells and is possibly responsible for bystander lymphopenia (67, 68), an established blood marker for fatal outcomes (18, 19, 41).
The exact source of the EVs in this study cannot be precisely determined. Platelet MVs are the most abundant MV in plasma (39); however, thrombocytopenia is a marker of EVD. In our study, lipids that have been previously shown to be externalized within platelets (27) (SI Appendix, Table S7) did not correlate with patient outcome, indicating a greater contribution from other cellular components. PS scramblase PLSCR1 (and Xkr8) was highly elevated in PBMCs in this study, indicating that PS lipids are being actively transported across the plasma membrane of immune cells for exposure. This translocation, however, could also be related to cellular apoptotic signaling. Damage and stress to the vascular system through the production of inflammatory molecules and nitric oxide, as well as endothelial cells being sites of EBOV replication (21), may also result in the production of EVs.
In addition to PS lipids, the PG lipids identified in this study were unexpected, given the low level of PGs in human plasma (29). It is possible that some of the PGs observed in this study are the structural isomers BMPs, as some of the PGs and putative BMP isomers were separated by the same retention time difference observed between the BMP and PG analytical standards. Two of the PG lipids undergoing the greatest decrease in survivors vs. fatalities were the earlier eluting isomers, suggesting these lipids are indeed BMPs (SI Appendix, Fig. S2 and Table S3). Elevated BMPs in EVD fatalities may be associated with the presence of exosomes as they are generated from late endosomes. Recently, BMP was suggested to be a candidate biomarker of endolysosomal dysfunction associated with neurodegenerative disorders (69). Elevated BMPs may also be a signature of enhanced autophagy, which is also supported by the dihydroceramide trend observed in our data (Fig. 3). EBOV utilizes the autophagy endosomal pathway for cellular infection (66), and it is thought that autophagy assists the host’s immune response against EBOV infection (70). In addition, autophagy is elevated in individuals with fatty liver disease as well as nutrient deprivation (71). For the elevated PG lipids in fatalities, we hypothesize that since these lipids are required for BMP synthesis, which takes place in late endosomes (35), they are elevated during enhanced endosomal pathway activity.
In addition to the activation of the coagulation cascade, PS lipids are also a hallmark signature of apoptosis when externalized to the outer plasma membrane leaflet. Pathology reports from over 89 autopsies of individuals infected with EBOV identified apoptosis in the liver, spleen, lymph nodes, and kidneys (42). Given vascular integrity of EVD, organ and tissue apoptosis contributing to blood plasma lipid signatures is likely. EVD-associated apoptosis is well known, and our data showed two signatures consistent with this: PS and Cer. Specifically Cer containing C16:0 and C18:0 fatty acids (72), were elevated in fatalities and at onset in EVD survivors compared with healthy controls. Plasma Cer are largely associated with LDL particles; however, SMs are also largely associated with LDLs (32) but were not significantly elevated in EVD, with the exception of SM(d18:0_18:0), indicating the Cer signature is not solely due to the presence of LDLs. EVD-related lymphopenia results as circulating T lymphocytes undergo bystander apoptosis (18, 19, 41), possibly due to EVs (67, 68). Apoptotic blebs, another form of EVs (61), could also be the source of this chain-specific Cer signature. Examination of transcripts from the PBMCs (16) (SI Appendix, Table S5) in our data did not reveal a clear alteration in Cer synthesis pathways, indicating that the Cer apoptotic signature in the plasma is likely from multiple sources. Plasma Cer with C16:0 and C18:0 have been shown to be predictors of cardiovascular death (38), associated with metabolic disorders (73), and activated by proinflammatory molecules (e.g., NF-κB and TNFα) (74). The apoptotic signature may also be related to nutritional factors. Choline, an essential nutrient, can also induce apoptosis when deficient in the diet (75, 76). Many of the patients in EVD treatment centers (and critically ill patients in general) have difficulty eating, with many of the disease symptoms (e.g., vomiting, loss of appetite, diarrhea, nausea,) having a direct or indirect impact on nutrition (77). Nutritional support during critical illness is complex, with requirements (e.g., energy, protein, micronutrient) differing with physiological and pathological conditions (78). Those admitted to Ebola treatment centers underwent nutritional management similar to patients with severe sepsis or shock, although care priorities varied depending on the stage of illness, underlying nutritional status, and the workload of health-care staff (77). Fasting has resulted in a choline-deficient diet that can lead to liver and muscle damage with greater rates of associated lymphocyte apoptosis and DNA damage, but without affecting the mean total lymphocyte count (76). Apoptosis is reversible when a normal choline diet is restored along with restored liver and muscle function (75, 79).
Choline has additional importance in that most is converted into PC in the liver (79). Steatosis has been identified in those who succumbed to EVD (42), and lesser forms of fatty liver disease are also associated with choline deficiency as the choline-based lipids (e.g., PCs and LPCs) are not being synthesized or are synthesized at a reduced rate (79). PC is synthesized in the liver using dietary choline and also through the phosphatidylethanolamine N-methyltransferase (PEMT) pathway, the latter of which is critical for maintaining PC supply in the liver as PCs are the only phospholipid necessary for lipoprotein (VLDL and HDL) assembly and excretion (51). If PC synthesis is decreased or a choline deficiency develops, then lipoprotein synthesis and secretion (e.g., VLDL) and corresponding blood plasma PC and LPC abundance would decrease and lipids, in particular TGs, would not be transported out of the liver, thus leading to the accumulation of TGs within hepatocytes with subsequent fatty liver disease. These signatures were identified in those with fatal outcomes (Fig. 6). Only one PC in our study significantly increased in fatalities, PC(16:0/16:0). The exclusive increase of PC(16:0/16:0) supports fatty liver disease with EVD, as PC(32:0) was recently reported as a potential signature of progressive forms of fatty liver disease (6). Proteomics data (16) support a decrease in lipoprotein LPC transport, since APOD, which is complexed with LCAT, was decreased in fatalities (SI Appendix, Table S8). The increase in PC and LPCs in survivors with time suggests that liver function can be restored while recovering from EVD, possibly partly due to restoring dietary choline intake.
In addition, adipose tissue is important in adaptations of critical illness (47), responding to hormonal and energetic signals (80). During a starvation response due to critical illness, lipolysis of adipose tissue increases, converting TG to free fatty acids, glycerol, and DG but also resulting in enhanced recycling of the fatty acids back into TGs. Our study supports this critical illness response. Proteomics data (16) (SI Appendix, Table S8) supports that lipolysis increased in EVD regardless of outcome. Lipidomics data indicate that excessive fatty acid breakdown is occurring in fatalties by the increase in DG along with long chained PUFA TGs. The specific increase in TG containing C22:6 in fatalities (or decrease in survivors with EVD progression), is unclear given the lack of nutrition while ill. During a stress starvation response, as occurs during severe illness, excessive amounts of fatty acids can be released to a level that exceeds energy needs, resulting in some of the fatty acids being reesterfied into TGs. Those that succumbed to Ebola were typically within the treatment center for a mean of 4 d and were likely malnourished upon arrival, with little feeding at the center given their state of illness (77). As adipose tissue is the main fatty acid storage site, with a TG half-life of 6–9 mo (81), and acting as a reservoir of dietary history, it is possible that these fatty acids are coming from adipose (or intramyocellular) tissue. In normal human adults from developed populations, 22:6 fatty acids comprise ∼0.4 mol% of plasma TG fatty acids and 0.1 mol% in adipose tissue (82). These values would be expected to be greater in Sierra Leoneans as their daily intake of 22:6 in the form of docosahexaenoic acid (DHA) is greater than most other countries, summarized by Forsyth et al. (83). As free fatty acids are toxic, they are reesterified back into TGs. Intramyocellular TGs could also be a source, as during critical illness lean muscle wasting is common. Preliminary analysis of survivors with post-Ebola syndrome have shown that 53% have musculoskeletal complications, including muscle weakness (84).
Lastly, survivors are marked with an increase in PI lipids containing 18:3 and 20:3 fatty acids (Fig. 3). The increase in PI lipids with EVD survivors (SI Appendix, Table S5) was also reported in transcriptomics data from PBMCs (16). In the metabolomics data (16), myoinositol, a metabolite used in de novo PI synthesis inversely correlated with the lipid data presented here. The increase in PIs with 18:3 and 20:3 is unclear but may be associated with lipid signaling and/or dietary influences. The 18:3 gamma-linolenic acid (GLA) is a direct precursor to 20:3 dihomo-gamma-linolenic acid (DGLA) (SI Appendix, Fig. S3). PI lipids are common sources for fatty acids converted into signaling molecules. Metabolites of DGLA have predominantly antiinflammatory properties (85) but also with reported roles inhibitory platelet aggregation (86). DGLA also has been shown to compete with arachidonic acid (AA) synthesis through the expression (or lack thereof) of FADS1, which converts DGLA to AA (85).
The particular increase in 18:3 and 20:3 with EVD progression may also reflect food intake at the Ebola treatment center as patients recover. Patients are provided local foods along with ready-to-use therapeutic food (RUTF) biscuits, which are 45–60% fat in the form of canola and soybean oil (77), and containing 26% and 51% 18:2 n-6 (a precursor to GLA) total unsaturated lipid, respectively. Why these specific fatty acid conversion products would be predominant in PI lipids is unclear, but previous studies have shown that supplemental intake of GLA resulted in an increase of DGLA in neutrophil PE lipids (87).
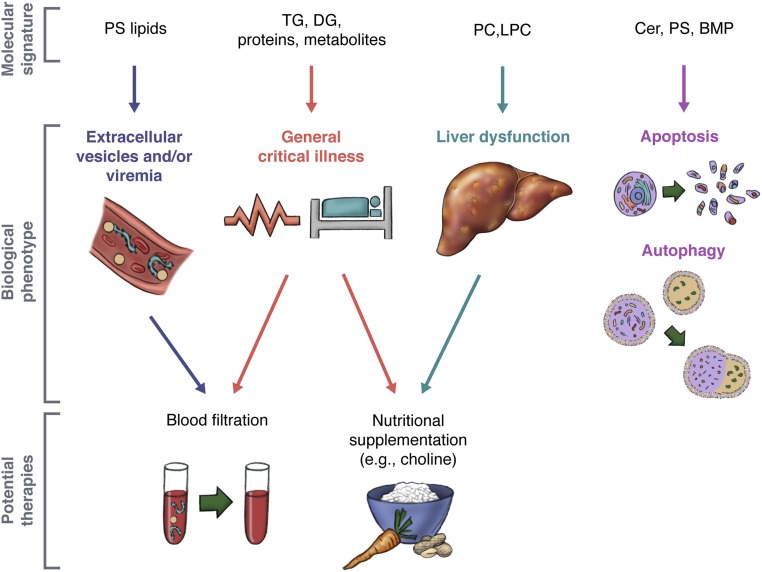
EVD promotes profound changes in the human blood plasma lipidome. Differences in the lipidome of fatalities and survivors highlights the state of disease, but also recovery. As it is difficult to decipher EVD-specific alterations, targeting treatment toward host response instead of virus may be a beneficial step forward. Based on the lipidome, we present the following suggestions for therapeutic consideration (Fig. 7). Nutritional choline supplementation may improve liver function and lipoprotein circulation, including minimizing instances of fatty liver disease (42). Blood plasma filtration to remove MVs may also assist apoptosis, coagulation, and EBOV load. MVs, and platelet microparticles in particular, found within platelet concentrate transfusions have led to venous thrombosis and embolisms (88). Chou et al. (89), found that 75 nm filtrated leukoreduced plasma may help normalize hemostatic activity while maintaining the lipoprotein profile, coagulation factor content, and global coagulation activity. Lastly, given EBOV reliance on PS lipids for egress and its entry, therapeutics toward PS lipids creates the opportunity of targets for antiviral therapy (12, 90). These suggested therapies may be applicable with other infectious diseases with similar pathologies and viruses that utilize PS for replication.
Fig. 7.
Summary of EVD molecular signatures to potential therapies. The molecular signatures of EVD (i.e., lipid, transcript, and protein) highlighted instances of critical illness, disease, and recovery in humans infected with EVD. Given the biological interpretation of signatures of the human blood plasma lipidome, potential supportive therapies are suggested that may alleviate symptoms and improve outcome.
Materials and Methods
Methods used to generate the proteomics and transcriptomics data presented here are provided in Eisfeld et al. (16)’s supplementary data file where experimental design, sample collection, analyte extraction, statistical analysis, and processing methods are reported in detail. Below is a brief description of the sample collection, lipid extraction and lipid mass spectrometry analysis as the focus of this paper is the lipidome.
Ethics and Human Subjects.
As detailed in Eisfeld et al. (16), all work conducted in this study was approved by the Sierra Leone Ethics and Scientific Review Committee, the Research Ethics Review Committee of the Institute of Medical Science at the University of Tokyo, and the University of Wisconsin (UW)–Madison Health Sciences Institutional Review Board (IRB). Before sample shipment, IRB approval was obtained at Pacific Northwest National Laboratory (PNNL). Consent was gathered from all subjects before enrollment, and for subjects under the age of 18, consent was provided by the children’s parent or guardian.
Sample Collection.
Peripheral blood samples were collected from 11 EVD survivors (total of 29 samples), 9 EVD fatalities (total of 9 samples), and 7 healthy volunteers (total of 7 samples) as controls for a total of 45 samples. EBOV-positive subjects were recruited from three Ebola treatment centers (ETCs) in Freetown, Sierra Leone [located at the Joint Military Unit (JMU) 34th Regimental Military Hospital at Wilberforce; the Hastings Police Training School 1; or the Police Training School 2] from February through May of 2015. Healthy subjects were recruited from healthcare workers and laboratory technicians at JMU during the same time period, none of whom had previously experienced EBOV disease. For the survivors, the number of days between sample collection s1 to s2 was 5–7 d and s2 to s3 was 5–11 d as shown in figure 1a in ref. 16.
Analyte Extraction.
Within 3 h of blood collection, samples were processed to separate PBMCs and plasma using Ficoll-Paque Plus. Briefly, lipids, proteins, and metabolites were extracted from 150 µL of plasma that underwent a modified Folch extraction (22) whereupon the total lipid and metabolite extracts were collected, evaporated to dryness using a speedvac, frozen at −80 °C, and shipped to UW–Madison, and then to the Pacific Northwest National Laboratory. For proteomics extraction, although the 14 most abundant plasma proteins [albumin, α1-antitrypsin, transferrin, haptoglobin, α2-macroglobulin, α1-acid glycoprotein (orosomucoid), fibrinogen, complement C3, IgG, IgA, IgM, HDL (apolipoproteins A-I and A-II), and LDL (mainly apolipoprotein B)] were simultaneously immunodepleted by using Seppro IgY14 spin columns (Sigma-Aldrich) (16), apolipoproteins were still identified and quantified (SI Appendix, Table S8) as complete depletion is difficult to achieve.
Mass Spectrometry Analysis.
The total lipid extracts were analyzed by LC-MS/MS using a Waters NanoAcquity UPLC system interfaced with a Velos Orbitrap mass spectrometer (Thermo Fisher Scientific). Lipid extracts were reconstituted in 200 µL of methanol. Seven microliters was injected and separated over a 90-min gradient elution [mobile phase A: ACN/H2O (40:60) containing 10 mM ammonium acetate; mobile phase B: ACN/IPA (10:90) containing 10 mM ammonium acetate] at a flow rate of 30 µL/min. Samples were analyzed in both positive and negative ionization (full scan range of 200–2,000 m/z) using higher-energy collision dissociation (HCD) and collision-induced dissociation (CID) to obtain high coverage of the lipidome, a normalized collision energy of 30 and 35 arbitrary units for HCD and CID, respectively. Both CID and HCD were set with a maximum charge state of 2 and an isolation width of 2 m/z units. Confident lipid identifications were made using LIQUID (24) in which the tandem mass spectra and corresponding fragment ions, the isotopic profile, precursor extracted ion chromatogram, mass parts per million error and retention time were examined.
Lipid standards BMP(14:0/14:0) and PG(14:0/14:0) were purchased from Avanti Polar Lipids, Inc., and analyzed using the same LC-MS parameters as the plasma samples.
Statistical Analysis.
Statistical analysis including normalization was conducted as outlined in ref. 16 except the FC values presented here are standardized FC which explicitly display FC and significance. Briefly, normalization of positive and negative mode lipidomics data were performed separately. Peak apex intensities for each identified lipid was log2 transformed and normalized using median scaling (91).
Data Availability.
All mass spectrometry datasets generated during this study have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) at the University of California at San Diego, (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp), under the ID code MSV000080129 (92).
Supplementary Material
Acknowledgments
We thank Zachary Najacht and Daniel Beechler (University of Wisconsin–Madison) and Thomas Korfeh, Francis Khoryama, Alex Bockarie, and Mohamed Nyallay (34th Regimental Military Hospital at Wilberforce, Freetown, Sierra Leone) for technical assistance; Amy E. S. Kuehn and Alexander Karasin (Univesity of Wisconsin–Madison) and Ishamil Barrie (Project 1808; Freetown, Sierra Leone) for excellent administrative support; and Rose Perry for graphic art assistance. This study was funded by a Health and Labor Sciences Research grant (Japan); grants for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grants 16H06429, 16K21723, and 16H06434); the Emerging/Re-Emerging Infectious Diseases Project of Japan; and an administrative supplement to Grant U19AI106772, provided by the National Institute of Allergy and Infectious Diseases (NIAID), NIH. Lipidomics, proteomics, and metabolomics analyses were performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy (DOE) Office of Biological and Environmental Research, and located at PNNL. Additional support was provided by National Institute of General Medical Sciences Grant P41 GM103493. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All mass spectrometry datasets generated during this study have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) at the University of California at San Diego, (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp), under the ID code MSV000080129.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815356116/-/DCSupplemental.
References
- 1.World Health Organization . Interim Guideline: Clinical Care for Survivors of Ebola Virus Disease. WHO; Geneva: 2016. [Google Scholar]
- 2.West TE, von Saint André-von Arnim A. Clinical presentation and management of severe Ebola virus disease. Ann Am Thorac Soc. 2014;11:1341–1350. doi: 10.1513/AnnalsATS.201410-481PS. [DOI] [PubMed] [Google Scholar]
- 3.Yuana Y, Sturk A, Nieuwland R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013;27:31–39. doi: 10.1016/j.blre.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwland R, et al. Cellular origin and procoagulant properties of microparticles in meningococcal sepsis. Blood. 2000;95:930–935. [PubMed] [Google Scholar]
- 5.Cui L, et al. Serum metabolome and lipidome changes in adult patients with primary dengue infection. PLoS Negl Trop Dis. 2013;7:e2373. doi: 10.1371/journal.pntd.0002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorden DL, et al. Biomarkers of NAFLD progression: A lipidomics approach to an epidemic. J Lipid Res. 2015;56:722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrario M, et al. Mortality prediction in patients with severe septic shock: A pilot study using a target metabolomics approach. Sci Rep. 2016;6:20391. doi: 10.1038/srep20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpentier YA, Scruel O. Changes in the concentration and composition of plasma lipoproteins during the acute phase response. Curr Opin Clin Nutr Metab Care. 2002;5:153–158. doi: 10.1097/00075197-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Cirstea M, et al. Decreased high-density lipoprotein cholesterol level is an early prognostic marker for organ dysfunction and death in patients with suspected sepsis. J Crit Care. 2017;38:289–294. doi: 10.1016/j.jcrc.2016.11.041. [DOI] [PubMed] [Google Scholar]
- 10.van Leeuwen HJ, et al. Lipoprotein metabolism in patients with severe sepsis. Crit Care Med. 2003;31:1359–1366. doi: 10.1097/01.CCM.0000059724.08290.51. [DOI] [PubMed] [Google Scholar]
- 11.Adu-Gyamfi E, et al. Host cell plasma membrane phosphatidylserine regulates the assembly and budding of Ebola virus. J Virol. 2015;89:9440–9453. doi: 10.1128/JVI.01087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amara A, Mercer J. Viral apoptotic mimicry. Nat Rev Microbiol. 2015;13:461–469. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller ME, Adhikary S, Kolokoltsov AA, Davey RA. Ebolavirus requires acid sphingomyelinase activity and plasma membrane sphingomyelin for infection. J Virol. 2012;86:7473–7483. doi: 10.1128/JVI.00136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soni SP, Stahelin RV. The Ebola virus matrix protein VP40 selectively induces vesiculation from phosphatidylserine-enriched membranes. J Biol Chem. 2014;289:33590–33597. doi: 10.1074/jbc.M114.586396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisfeld AJ, et al. Multi-platform 'Omics analysis of human ebola virus disease pathogenesis. Cell Host Microbe. 2017;22:817–829.e8. doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanini S, et al. INMI-EMERGENCY EBOV Sierra Leone Study Group Blood kinetics of Ebola virus in survivors and nonsurvivors. J Clin Invest. 2015;125:4692–4698. doi: 10.1172/JCI83111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElroy AK, et al. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci USA. 2015;112:4719–4724. doi: 10.1073/pnas.1502619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4:e837. doi: 10.1371/journal.pntd.0000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisbert TW, et al. Mechanisms underlying coagulation abnormalities in ebola hemorrhagic fever: Overexpression of tissue factor in primate monocytes/macrophages is a key event. J Infect Dis. 2003;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- 21.Ansari AA. Clinical features and pathobiology of Ebolavirus infection. J Autoimmun. 2014;55:1–9. doi: 10.1016/j.jaut.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Nakayasu ES, et al. MPLEx: A robust and universal protocol for single-sample integrative proteomic, metabolomic, and lipidomic analyses. mSystems. 2016;1:e00043-16. doi: 10.1128/mSystems.00043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dautel SE, et al. Lipidomics reveals dramatic lipid compositional changes in the maturing postnatal lung. Sci Rep. 2017;7:40555. doi: 10.1038/srep40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kyle JE, et al. LIQUID: An-open source software for identifying lipids in LC-MS/MS-based lipidomics data. Bioinformatics. 2017;33:1744–1746. doi: 10.1093/bioinformatics/btx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birge RB, et al. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016;23:962–978. doi: 10.1038/cdd.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lentz BR. Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res. 2003;42:423–438. doi: 10.1016/s0163-7827(03)00025-0. [DOI] [PubMed] [Google Scholar]
- 27.Clark SR, et al. Characterization of platelet aminophospholipid externalization reveals fatty acids as molecular determinants that regulate coagulation. Proc Natl Acad Sci USA. 2013;110:5875–5880. doi: 10.1073/pnas.1222419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nanbo A, et al. Ebola virus requires a host scramblase for externalization of phosphatidylserine on the surface of viral particles. PLoS Pathog. 2018;14:e1006848. doi: 10.1371/journal.ppat.1006848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quehenberger O, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ulmer CZ, et al. LipidQC: Method validation tool for visual comparison to SRM 1950 using NIST interlaboratory comparison exercise lipid consensus mean estimate values. Anal Chem. 2017;89:13069–13073. doi: 10.1021/acs.analchem.7b04042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serna J, et al. Quantitative lipidomic analysis of plasma and plasma lipoproteins using MALDI-TOF mass spectrometry. Chem Phys Lipids. 2015;189:7–18. doi: 10.1016/j.chemphyslip.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Wiesner P, Leidl K, Boettcher A, Schmitz G, Liebisch G. Lipid profiling of FPLC-separated lipoprotein fractions by electrospray ionization tandem mass spectrometry. J Lipid Res. 2009;50:574–585. doi: 10.1194/jlr.D800028-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Dashti M, et al. A phospholipidomic analysis of all defined human plasma lipoproteins. Sci Rep. 2011;1:139. doi: 10.1038/srep00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontush A, Lhomme M, Chapman MJ. Unraveling the complexities of the HDL lipidome. J Lipid Res. 2013;54:2950–2963. doi: 10.1194/jlr.R036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 37.Llorente A, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Laaksonen R, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J. 2016;37:1967–1976. doi: 10.1093/eurheartj/ehw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yáñez-Mó M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grösch S, Schiffmann S, Geisslinger G. Chain length-specific properties of ceramides. Prog Lipid Res. 2012;51:50–62. doi: 10.1016/j.plipres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Ruibal P, et al. Unique human immune signature of Ebola virus disease in Guinea. Nature. 2016;533:100–104. doi: 10.1038/nature17949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol. 2015;235:153–174. doi: 10.1002/path.4456. [DOI] [PubMed] [Google Scholar]
- 43.Hernández-Tiedra S, et al. Dihydroceramide accumulation mediates cytotoxic autophagy of cancer cells via autolysosome destabilization. Autophagy. 2016;12:2213–2229. doi: 10.1080/15548627.2016.1213927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camont L, et al. Small, dense high-density lipoprotein-3 particles are enriched in negatively charged phospholipids: Relevance to cellular cholesterol efflux, antioxidative, antithrombotic, anti-inflammatory, and antiapoptotic functionalities. Arterioscler Thromb Vasc Biol. 2013;33:2715–2723. doi: 10.1161/ATVBAHA.113.301468. [DOI] [PubMed] [Google Scholar]
- 45.Meikle PJ, et al. Effect of lysosomal storage on bis(monoacylglycero)phosphate. Biochem J. 2008;411:71–78. doi: 10.1042/BJ20071043. [DOI] [PubMed] [Google Scholar]
- 46.Hullin-Matsuda F, Taguchi T, Greimel P, Kobayashi T. Lipid compartmentalization in the endosome system. Semin Cell Dev Biol. 2014;31:48–56. doi: 10.1016/j.semcdb.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 47.Ilias I, et al. Adipose tissue lipolysis and circulating lipids in acute and subacute critical illness: Effects of shock and treatment. J Crit Care. 2014;29:1130.e5–1130.e9. doi: 10.1016/j.jcrc.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Goralski KB, et al. Chemerin, a novel adipokine that regulates adipogenesis and adipocyte metabolism. J Biol Chem. 2007;282:28175–28188. doi: 10.1074/jbc.M700793200. [DOI] [PubMed] [Google Scholar]
- 49.Roh SG, et al. Chemerin–A new adipokine that modulates adipogenesis via its own receptor. Biochem Biophys Res Commun. 2007;362:1013–1018. doi: 10.1016/j.bbrc.2007.08.104. [DOI] [PubMed] [Google Scholar]
- 50.Bing C, et al. Zinc-alpha2-glycoprotein, a lipid mobilizing factor, is expressed in adipocytes and is up-regulated in mice with cancer cachexia. Proc Natl Acad Sci USA. 2004;101:2500–2505. doi: 10.1073/pnas.0308647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole LK, Vance JE, Vance DE. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2012;1821:754–761. doi: 10.1016/j.bbalip.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Park DW, et al. Impact of serial measurements of lysophosphatidylcholine on 28-day mortality prediction in patients admitted to the intensive care unit with severe sepsis or septic shock. J Crit Care. 2014;29:882.e5–882.e11. doi: 10.1016/j.jcrc.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 53.Taylor LA, Arends J, Hodina AK, Unger C, Massing U. Plasma lyso-phosphatidylcholine concentration is decreased in cancer patients with weight loss and activated inflammatory status. Lipids Health Dis. 2007;6:17. doi: 10.1186/1476-511X-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McConathy WJ, Alaupovic P. Isolation and partial characterization of apolipoprotein D: A new protein moiety of the human plasma lipoprotein system. FEBS Lett. 1973;37:178–182. doi: 10.1016/0014-5793(73)80453-3. [DOI] [PubMed] [Google Scholar]
- 55.Burgess JW, et al. Phosphatidylinositol increases HDL-C levels in humans. J Lipid Res. 2005;46:350–355. doi: 10.1194/jlr.M400438-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Assinger A. Platelets and infection–An emerging role of platelets in viral infection. Front Immunol. 2014;5:649. doi: 10.3389/fimmu.2014.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cloutier N, et al. The exposure of autoantigens by microparticles underlies the formation of potent inflammatory components: The microparticle-associated immune complexes. EMBO Mol Med. 2013;5:235–249. doi: 10.1002/emmm.201201846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arraud N, et al. Extracellular vesicles from blood plasma: Determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12:614–627. doi: 10.1111/jth.12554. [DOI] [PubMed] [Google Scholar]
- 59.Hosseini HM, Fooladi AA, Nourani MR, Ghanezadeh F. The role of exosomes in infectious diseases. Inflamm Allergy Drug Targets. 2013;12:29–37. doi: 10.2174/1871528111312010005. [DOI] [PubMed] [Google Scholar]
- 60.Nolte-’t Hoen E, Cremer T, Gallo RC, Margolis LB. Extracellular vesicles and viruses: Are they close relatives? Proc Natl Acad Sci USA. 2016;113:9155–9161. doi: 10.1073/pnas.1605146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.György B, et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bavari S, et al. Lipid raft microdomains: A gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med. 2002;195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Towner JS, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol. 2004;78:4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morizono K, Chen IS. Role of phosphatidylserine receptors in enveloped virus infection. J Virol. 2014;88:4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nanbo A, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pleet ML, DeMarino C, Lepene B, Aman MJ, Kashanchi F. The role of exosomal VP40 in Ebola virus disease. DNA Cell Biol. 2017;36:243–248. doi: 10.1089/dna.2017.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pleet ML, et al. Ebola VP40 in exosomes can cause immune cell dysfunction. Front Microbiol. 2016;7:1765. doi: 10.3389/fmicb.2016.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miranda AM, et al. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat Commun. 2018;9:291. doi: 10.1038/s41467-017-02533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falasca L, et al. Molecular mechanisms of Ebola virus pathogenesis: Focus on cell death. Cell Death Differ. 2015;22:1250–1259. doi: 10.1038/cdd.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rautou PE, et al. Autophagy in liver diseases. J Hepatol. 2010;53:1123–1134. doi: 10.1016/j.jhep.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 72.Thomas RL, Jr, Matsko CM, Lotze MT, Amoscato AA. Mass spectrometric identification of increased C16 ceramide levels during apoptosis. J Biol Chem. 1999;274:30580–30588. doi: 10.1074/jbc.274.43.30580. [DOI] [PubMed] [Google Scholar]
- 73.Chaurasia B, Summers SA. Ceramides–Lipotoxic inducers of metabolic disorders. Trends Endocrinol Metab. 2015;26:538–550. doi: 10.1016/j.tem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 74.Osawa Y, et al. Roles for C16-ceramide and sphingosine 1-phosphate in regulating hepatocyte apoptosis in response to tumor necrosis factor-alpha. J Biol Chem. 2005;280:27879–27887. doi: 10.1074/jbc.M503002200. [DOI] [PubMed] [Google Scholar]
- 75.da Costa KA, Badea M, Fischer LM, Zeisel SH. Elevated serum creatine phosphokinase in choline-deficient humans: Mechanistic studies in C2C12 mouse myoblasts. Am J Clin Nutr. 2004;80:163–170. doi: 10.1093/ajcn/80.1.163. [DOI] [PubMed] [Google Scholar]
- 76.da Costa KA, Niculescu MD, Craciunescu CN, Fischer LM, Zeisel SH. Choline deficiency increases lymphocyte apoptosis and DNA damage in humans. Am J Clin Nutr. 2006;84:88–94. doi: 10.1093/ajcn/84.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.World Health Organization/UNICEF/WFP . Interim Guideline: Nutritional Care of Children and Adults with Ebola Virus Disease in Treatment Centres. WHO; Geneva: 2014. [Google Scholar]
- 78.Preiser JC, et al. Metabolic and nutritional support of critically ill patients: Consensus and controversies. Crit Care. 2015;19:35. doi: 10.1186/s13054-015-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Z, Vance DE. Phosphatidylcholine and choline homeostasis. J Lipid Res. 2008;49:1187–1194. doi: 10.1194/jlr.R700019-JLR200. [DOI] [PubMed] [Google Scholar]
- 80.Rutkowski JM, Stern JH, Scherer PE. The cell biology of fat expansion. J Cell Biol. 2015;208:501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strawford A, Antelo F, Christiansen M, Hellerstein MK. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am J Physiol Endocrinol Metab. 2004;286:E577–E588. doi: 10.1152/ajpendo.00093.2003. [DOI] [PubMed] [Google Scholar]
- 82.Hodson L, Skeaff CM, Fielding BA. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog Lipid Res. 2008;47:348–380. doi: 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 83.Forsyth S, Gautier S, Salem N., Jr Global estimates of dietary intake of docosahexaenoic acid and arachidonic acid in developing and developed countries. Ann Nutr Metab. 2016;68:258–267. doi: 10.1159/000446855. [DOI] [PubMed] [Google Scholar]
- 84.Burki TK. Post-Ebola syndrome. Lancet Infect Dis. 2016;16:780–781. doi: 10.1016/S1473-3099(15)00259-5. [DOI] [PubMed] [Google Scholar]
- 85.Sergeant S, Rahbar E, Chilton FH. Gamma-linolenic acid, dihommo-gamma linolenic, eicosanoids and inflammatory processes. Eur J Pharmacol. 2016;785:77–86. doi: 10.1016/j.ejphar.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Willis AL, Comai K, Kuhn DC, Paulsrud J. Dihomo-gamma-linolenate suppresses platelet aggregation when administered in vitro or in vivo. Prostaglandins. 1974;8:509–519. doi: 10.1016/0090-6980(74)90063-x. [DOI] [PubMed] [Google Scholar]
- 87.Johnson MM, et al. Dietary supplementation with gamma-linolenic acid alters fatty acid content and eicosanoid production in healthy humans. J Nutr. 1997;127:1435–1444. doi: 10.1093/jn/127.8.1435. [DOI] [PubMed] [Google Scholar]
- 88.Burnouf T, et al. An overview of the role of microparticles/microvesicles in blood components: Are they clinically beneficial or harmful? Transfus Apher Sci. 2015;53:137–145. doi: 10.1016/j.transci.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 89.Chou ML, Lin LT, Devos D, Burnouf T. Nanofiltration to remove microparticles and decrease the thrombogenicity of plasma: In vitro feasibility assessment. Transfusion. 2015;55:2433–2444. doi: 10.1111/trf.13162. [DOI] [PubMed] [Google Scholar]
- 90.Dowall SD, et al. Effective binding of a phosphatidylserine-targeting antibody to Ebola virus infected cells and purified virions. J Immunol Res. 2015;2015:347903. doi: 10.1155/2015/347903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Webb-Robertson BJ, Matzke MM, Jacobs JM, Pounds JG, Waters KM. A statistical selection strategy for normalization procedures in LC-MS proteomics experiments through dataset-dependent ranking of normalization scaling factors. Proteomics. 2011;11:4736–4741. doi: 10.1002/pmic.201100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith RD. 2016 Human Ebola virus disease pathogenesis multi-platform omics analysis. Mass Spectrometry Interactive Virtual Environment. Available at https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=c68a6c834301476fa590f1f85d6c0362. Deposited September 7, 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mass spectrometry datasets generated during this study have been deposited at the Mass Spectrometry Interactive Virtual Environment (MassIVE) at the University of California at San Diego, (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp), under the ID code MSV000080129 (92).