Significance
Here, we describe the generation of Aedes aegypti mosquitoes that are engineered to be resistant to Zika virus (ZIKV) transmission. Our results demonstrate that engineered mosquitoes express a polycistronic cluster of synthetic small RNAs designed to target the ZIKV genome. As a result, homozygous mosquitoes were refractory to ZIKV infection, and therefore could not transmit the virus. Additionally, mosquitoes heterozygous for the transgene showed significantly lower levels of viral infection, dissemination, and transmission compared with wild-type mosquitoes; importantly, these levels were low enough to make such mosquitoes unlikely to transmit ZIKV to a susceptible host. Finally, we discuss how such an engineering approach can be used to combat the major health burden of ZIKV, and potentially other arboviruses, in the future.
Keywords: ZIKA, resistance, Aedes, aegypti, mosquito
Abstract
Recent Zika virus (ZIKV) outbreaks have highlighted the necessity for development of novel vector control strategies to combat arboviral transmission, including genetic versions of the sterile insect technique, artificial infection with Wolbachia to reduce population size and/or vectoring competency, and gene drive-based methods. Here, we describe the development of mosquitoes synthetically engineered to impede vector competence to ZIKV. We demonstrate that a polycistronic cluster of engineered synthetic small RNAs targeting ZIKV is expressed and fully processed in Aedes aegypti, ensuring the formation of mature synthetic small RNAs in the midgut where ZIKV resides in the early stages of infection. Critically, we demonstrate that engineered Ae. aegypti mosquitoes harboring the anti-ZIKV transgene have significantly reduced viral infection, dissemination, and transmission rates of ZIKV. Taken together, these compelling results provide a promising path forward for development of effective genetic-based ZIKV control strategies, which could potentially be extended to curtail other arboviruses.
Since being introduced into the Americas, Zika virus (ZIKV), a mosquito-borne flavivirus, has spread rapidly, causing hundreds of thousands of cases of ZIKV infection (1). Although most cases remain asymptomatic, infection during pregnancy has been associated with severe congenital abnormalities and pregnancy loss, presenting an unprecedented health threat with long-term consequences (2). This prompted the WHO to declare ZIKV a public health emergency of international concern in 2016 (1, 2). Currently, there are no clinically approved vaccines to prevent ZIKV and no effective treatment options for infected individuals; thus, vector control remains essential in curtailing the ZIKV epidemic. Like dengue virus (DENV) and chikungunya virus (CHIKV), ZIKV is transmitted primarily by Aedes mosquitoes, which are expanding their habitable range due to urbanization, climate change, and global trade (3). Current methods of vector control, including removal of standing water and use of insecticides, have not been entirely effective in the fight against the spread of Aedes mosquitoes (3). Therefore, novel innovative vector control strategies, including those utilizing genetically engineered mosquitoes (4), are urgently needed to combat the spread of ZIKV and other Aedes-vectored diseases worldwide.
Employment of genetically modified (or otherwise altered) insects to manipulate disease-vectoring populations was first proposed decades ago (5), and due, in part, to enabling technological advances, it has garnered increased interest in recent years (4, 6). In fact, several strategies for genetic-based vector control are currently being utilized in the field. For example, the release of insects carrying a dominant lethal system (7), a genetic-based sterile insect technique-like system, has recently been shown to be effective in reducing wild insect populations (8). Open field release trials of these genetically modified mosquitoes have been conducted in several countries, including the Cayman Islands, Malaysia, and Brazil, and are currently being considered for use in India and the United States (9–11). In addition to genetic-based vector control strategies, mosquitoes harboring artificially acquired strains of Wolbachia can be used either to reduce total insect populations (12) or to render insect populations less competent vectors of certain viruses, including ZIKV (13), and this technique has also been trialed in multiple countries to reduce the impact of mosquito-borne diseases (14–16) [although the accumulating evidence that Wolbachia can enhance certain flavivirus infections (17–19) may lead to reevaluation of this technique]. Nevertheless, while current approaches can be effective, they require inundative releases of large numbers of insects, which can be laborious and expensive, and can impede scalability and worldwide adoption.
Another category of genetic-based vector control involves an engineered gene drive system that can force inheritance in a super-Mendelian fashion, enabling it to increase itself, and any linked “cargo” genes, in frequency with each generation even without conferring fitness advantages to its host (4, 20). Such a method could be used to disseminate desirable cargo genes, such as pathogen resistance, rapidly through wild disease-transmitting populations, modifying vector populations to be disease-refractory (21). While significant efforts are currently underway to develop engineered drive systems (22–25), others are focused on creation of cargo genes that may be spread by the drive systems, and several studies have reported on the successful development of pathogen resistance cargo genes in Aedes aegypti (26–28).
To date, however, no anti-ZIKV refractory cargo genes in any mosquito have been developed. To fill this void, we describe here the generation of a synthetically engineered ZIKV resistance transgene comprising a polycistronic cluster of ZIKV-targeting synthetic small RNAs. We demonstrate that Ae. aegypti mosquitoes harboring this anti-ZIKV transgene express and fully process the ZIKV-targeting synthetic small RNAs in the midgut, and consequently have significantly reduced viral infection, dissemination, and transmission rates of ZIKV. Specifically, we demonstrate that mosquitoes homozygous for the anti-ZIKV transgene are fully resistant to ZIKV infection and are unable to transmit the virus. In contrast, we determine that a minority of heterozygotes for the anti-ZIKV transgene can become infected with ZIKV following exposure. However, these heterozygotes become infected at significantly lower rates than wild-type (WT) mosquitoes, and those susceptible to infection have roughly three orders of magnitude lower viral titers in their saliva, suggesting a significantly reduced possibility of viral transmission. This is supported by our finding that heterozygous anti-ZIKV mosquitoes are almost entirely incapable of in vivo ZIKV transmission in a sensitive Stat1−/− mouse model. Moreover, compared with Wolbachia wMel strain-positive mosquitoes, which have previously been shown to have reduced ZIKV vectoring competency (13), the anti-ZIKV mosquitoes perform significantly better in the ZIKV challenge assays. Taken together, these compelling results provide a promising path forward for development of effective ZIKV control, and possibly control of other clinically significant arboviruses, using genetically engineered mosquitoes.
Results
Engineering ZIKV-Resistant Mosquitoes.
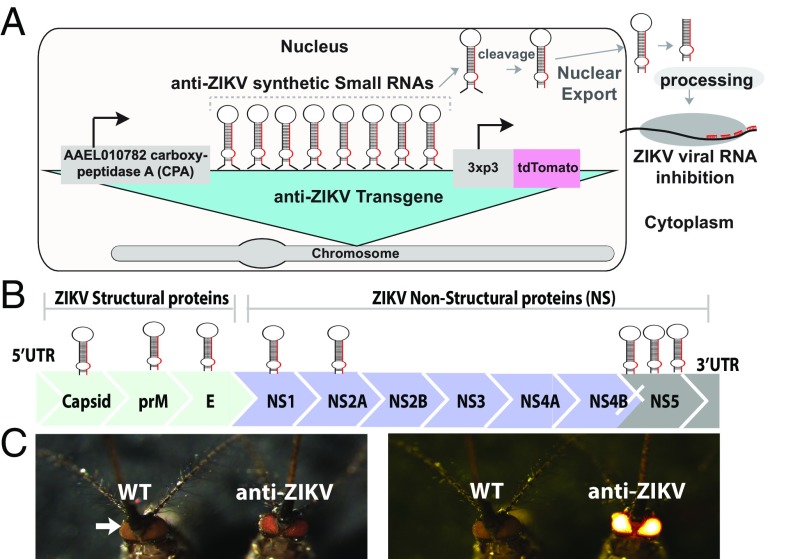
To generate a cargo gene that can confer resistance to ZIKV, we implemented a synthetic small RNA-based approach, since such an approach has previously been demonstrated to generate virus resistance phenotypes in a number of contexts (e.g., refs. 29–31), including mosquitoes (32). We engineered a piggyBac vector comprising a polycistronic cluster of eight ZIKV-targeting, miRNA-like, synthetic small RNAs (the anti-ZIKV transgene) under control of the Ae. aegypti carboxypeptidase (CPA) promoter (33) to drive expression of the synthetic small RNAs in female midguts following a blood meal (Fig. 1A). To ensure effective viral suppression and evolutionary stability, we designed each of the eight synthetic small RNAs to target six of 10 conserved protein-coding genes of French Polynesia ZIKV strain H/PF/2013 (GenBank accession no. KJ776791.2) (34), including all three structural genes (capsid, membrane precursor, and envelope) and three nonstructural genes (NS1, NS2A, and NS5). Each of these genes was targeted by a single synthetic small RNA, except for the RNA-dependent RNA polymerase NS5, which was targeted by three small RNAs due to its importance for the replication of the flaviviral RNA genome (Fig. 1B and SI Appendix, Fig. S1). The engineered anti-ZIKV transgene (termed plasmid OA959C) also contained the eye-specific 3xP3 promoter (35) driving expression of tdTomato as a transgenesis marker (Fig. 1).
Fig. 1.
Schematic of anti-ZIKV transgene, ZIKV target sites, and phenotype of transgenic mosquitoes. (A) Schematic of the anti-ZIKV transgene used in the study, consisting of a CPA (AAEL010782) promoter driving expression of a polycistronic cluster of eight synthetic small RNAs engineered to target conserved genes in the ZIKV genome. Following processing, the small RNAs and their target ZIKV viral RNA interact in the cytoplasm. (B) Schematic of the ZIKV genome, consisting of three structural proteins [capsid, membrane precursor (prM), and envelope (E)] and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), with relative synthetic small RNA targets indicated by hairpins above. (C) Higgs WT and TZIKV-C adult mosquitoes were imaged under both transmitted light and a fluorescent dsRED filter. Fluorescence is evident in mosquito eyes (indicated by white arrow).
Following embryonic microinjection, multiple transgenic lines were identified (n > 6), and four independent lines with strong expression of tdTomato fluorescence in the eyes (termed TZIKV-A, TZIKV-B, TZIKV-C, and TZIKV-D) were selected for further characterization (fluorescence in TZIKV-C is shown in Fig. 1C). To verify the transgene insertion sites, we performed an inverse PCR (iPCR) on genomic DNA extracted from transgenic mosquitoes of all four independent strains. The iPCR analysis indicated that insertion sites were on chromosome 2 (at approximate position 167,899,561) for line TZIKV-A, on chromosome 3 (at approximate position 402,525,313) for line TZIKV-B, on chromosome 3 (at approximate position 173,647,983) for line TZIKV-C, and on chromosome 1 (at approximate position 228,972,549) for line TZIKV-D when aligned to the AaegL5 assembly (GenBank assembly accession no. GCA_002204515.1) (36). To avoid any bias due to position effect variegation stemming from transgene insertion sites, all four lines were screened for midgut infection status at 4 days postinfection (dpi), and results showed that all four lines had significant reduction in midgut infection rate and viral titers compared with Higgs or Liverpool WT mosquitoes (SI Appendix, Fig. S2). Given that there was no significant difference in ZIKV reduction in midgut infection between the four lines (TZIKV-A, TZIKV-B, TZIKV-C, and TZIKV-D), the line exhibiting the strongest antiviral phenotype (TZIKV-C) was selected for further comprehensive characterization (SI Appendix, Fig. S2).
Molecular Analysis of Synthetic Small RNA Expression and Processing.
To confirm expression and processing of the ZIKV-targeting synthetic small RNAs in TZIKV-C, we deep-sequenced small RNA populations from dissected midgut tissues isolated from blood-fed and non–blood-fed female mosquitoes using an Illumina platform. We detected expression of the nonguide (37) and mature small RNA guide strands of five of eight anti-ZIKV–targeting synthetic small RNAs (small RNAs 1, 2, 4, 6, and 8) with transcripts per million values for mature small RNA guide strands ranging from 2 to 91, 25.7 on average, indicating that these synthetic small RNAs are efficiently expressed and processed (SI Appendix, Fig. S3 and Table S1). Importantly, no anti-ZIKV–targeting small RNAs (more than one read) were identified in small RNA populations derived from Higgs WT Ae. aegypti (SI Appendix, Table S1).
We also performed RT-PCR assays on dissected midgut tissues and midgut-free carcasses from blood-fed and non–blood-fed female mosquitoes to determine whether synthetic small RNA expression was confined to the midgut. Contrary to previously published reports (33, 38), we found that the CPA promoter drove detectable expression of the synthetic small RNAs in tissues other than the midgut and that expression occurred even without a blood meal (SI Appendix, Fig. S4), suggesting that expression of the anti-ZIKV transgene may be strongly affected by its genomic insertion position. However, importantly, no anti-ZIKV–targeting small RNAs were detected in Higgs WT Ae. aegypti (SI Appendix, Fig. S4). Taken together, these results demonstrate that the anti-ZIKV transgene is stably integrated into the mosquito genome and most of the ZIKV-targeting synthetic small RNAs are expressed and processed in an appropriate context (including in the midgut) for ZIKV suppression.
Engineered Mosquitoes Are Refractory to Multiple ZIKV Strains.
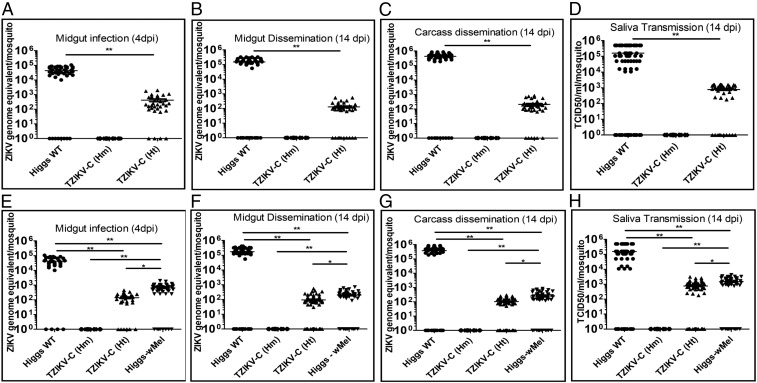
To characterize the functional significance of ZIKV-targeting synthetic small RNA expression and processing on vector competence, adult female mosquitoes (Higgs WT control and TZIKV-C) were infected with ZIKV (FSS13025, Cambodia 2010 strain; GenBank accession no. JN860885) via membrane blood feeding (39). For these experiments, we used the Cambodia ZIKV strain, which is from the Asian ZIKV lineage and in close phylogenetic proximity to the French Polynesia ZIKV strain against which the small RNAs were designed (40). Importantly, seven of eight of the ZIKV-targeting synthetic small RNA target sites are 100% conserved between the Cambodia ZIKV strain and the French Polynesia strain, allowing either strain to be used for the ZIKV challenges (SI Appendix, Fig. S1). At 4 dpi, midguts from blood-fed mosquitoes were dissected and ZIKV RNA copies were measured using real-time RT-qPCR. Results from three biological replicates revealed that none of the TZIKV-C mosquitoes homozygous for the transgene (n = 32) were positive for ZIKV infection in the midgut. ZIKV infection was detected in 87.5% (28 of 32) of the TZIKV-C mosquitoes that were heterozygous for the transgene; however, these mosquitoes had significantly (P < 0.001) lower viral RNA levels (∼2 logs) than Higgs WT (Fig. 2A and SI Appendix, Table S2).
Fig. 2.
ZIKV replication and titers in Higgs WT, TZIKV-C, and Wolbachia-infected mosquitoes challenged with either a Cambodian or Puerto Rican ZIKV strain. ZIKV genome copies and titers in Higgs WT, TZIKV-C homozygous (Hm) and heterozygous (Ht) transgenic, and Wolbachia-infected Higgs WT (Higgs-wMel) mosquitoes following a blood meal infected with a Cambodian (FSS13025, A–D) or Puerto Rican (PRVABC59, E–H) strain of ZIKV are shown. ZIKV genome-equivalents from mosquito midgut [4 dpi (A and E) and 14 dpi (B and F)] and carcass [14 dpi (C and G)] of Higgs WT and transgenic mosquitoes were determined using real-time RT-qPCR and calculated using previously published methods. (D and H) Virus titers in the saliva collected from Higgs WT and transgenic mosquitoes at 14 dpi were determined using a median tissue culture infectious dose (TCID50) on Vero cells and plotted. Higgs WT mosquitoes (●),TZIKV-C Hm transgenic mosquitoes (♦),TZIKV-C Ht mosquitoes (▲), and Higgs-wMel mosquitoes (▼) are shown. Horizontal bars represent the mean virus titer. *P < 0.05; **P < 0.001. For each experiment, data from three replicates are pooled.
To assay for viral dissemination, total RNA was collected from whole TZIKV-C mosquito carcasses and dissected midguts from both homozygous and heterozygous transgenic mosquitoes at 14 dpi. The results from three biological replicates indicated that none of the homozygous TZIKV-C mosquitoes (n = 46) were positive for viral replication (dissemination) in either the midgut or the carcass. ZIKV prevalence was detected in 74.4% (29 of 39) of heterozygous TZIKV-C mosquitoes in both the carcass and midgut; however, they had significantly (P < 0.001) lower levels of viral RNA (∼3 logs) compared with Higgs WT mosquitoes (Fig. 2 B and C and SI Appendix, Table S2). Finally, to determine viral transmission, saliva from individual mosquitoes was collected at 14 dpi and ZIKV titers were measured using a median tissue culture infectious dose assay. No ZIKV was detected in the saliva of homozygous TZIKV-C mosquitoes (n = 46). The presence of ZIKV in the saliva was detected in 74.4% (29 of 39) of heterozygous TZIKV-C mosquitoes; however, here again, the ZIKV titers were significantly (P < 0.001) lower (∼3 logs) compared with Higgs WT mosquitoes (Fig. 2D and SI Appendix, Table S2).
To determine whether the synthetic small RNAs are broadly inhibitory for ZIKV, vector competence of transgenic TZIKV-C mosquitoes was also assessed using a second contemporary ZIKV strain (PRVABC59, isolated from a US traveler to Puerto Rico in 2015; GenBank accession no. KU501215). For this strain, seven of eight ZIKV-targeting synthetic small RNA target sites (although not the same seven as for the Cambodia strain) are 100% conserved with the French Polynesia strain against which the small RNAs were designed (SI Appendix, Fig. S1). Tests for infection, dissemination, and transmission were carried out as above, and the results were comparable to those obtained with the Cambodia strain. Briefly, at 4 dpi, none of the TZIKV-C mosquitoes homozygous for the transgene (n = 32) were positive for ZIKV infection in their midgut, and while ZIKV infection was detected in 81.25% (26 of 32) of the TZIKV-C mosquitoes that were heterozygous for the transgene, these had significantly (P < 0.001) lower viral RNA levels (∼2 logs) than Higgs WT mosquitoes (Fig. 2E and SI Appendix, Table S2). TZIKV-C mosquito carcasses and dissected midguts at 14 dpi showed that none of the homozygous TZIKV-C mosquitoes (n = 70) were positive for viral replication in either the midgut or the carcass by real-time RT-qPCR, while 70% (49 of 70) of heterozygous mosquitoes had ZIKV in both the carcass and midgut, albeit with significantly (P < 0.001) lower levels of viral RNA (∼3 logs) than Higgs WT mosquitoes (Fig. 2 F and G and SI Appendix, Table S2). Finally, ZIKV titer measurements on saliva from individual mosquitoes at 14 dpi demonstrated that no ZIKV was present in homozygous TZIKV-C mosquitoes (n = 70), indicating that they would be unable to transmit the virus. Prevalence of ZIKV in saliva was detected in 70% (49 of 70) of TZIKV-C heterozygous mosquitoes; however, here again, the ZIKV titers were significantly (P < 0.001) lower (∼3 logs) compared with Higgs WT mosquitoes (Fig. 2H and SI Appendix, Table S2).
Engineered Mosquitoes Outperform Wolbachia.
We next compared the inhibitory effect of our synthetic small RNAs with ZIKV inhibition previously shown with Wolbachia (13, 41–43). Vector competence results revealed that midguts from mosquitoes (Higgs WT strain) infected with Wolbachia (wMel strain, n = 50) had significantly (P < 0.001) reduced ZIKV (Puerto Rican strain) RNA levels (∼2 logs) at 4 dpi compared with uninfected Higgs WT mosquitoes (n = 32; Fig. 2E and SI Appendix, Table S2). Similarly, viral dissemination at 14 dpi was also reduced (P < 0.001) in wMel mosquitoes (∼3 logs, n = 50; Fig. 2 F and G and SI Appendix, Table S2), and ZIKV titers in mosquito saliva at 14 dpi were significantly (P < 0.01) lower (∼2 logs) in wMel mosquitoes than in uninfected Higgs WT mosquitoes (Fig. 2H and SI Appendix, Table S2). Importantly, comparison with the TZIKV-C mosquitoes revealed that the TZIKV-C mosquitoes are significantly (P < 0.001) more effective as homozygotes, and modestly more effective as heterozygotes (P < 0.05), at blocking ZIKV infection compared with Wolbachia-infected mosquitoes.
Anti-ZIKV Transgene Inhibits ZIKV Transmission in a Mouse Model.
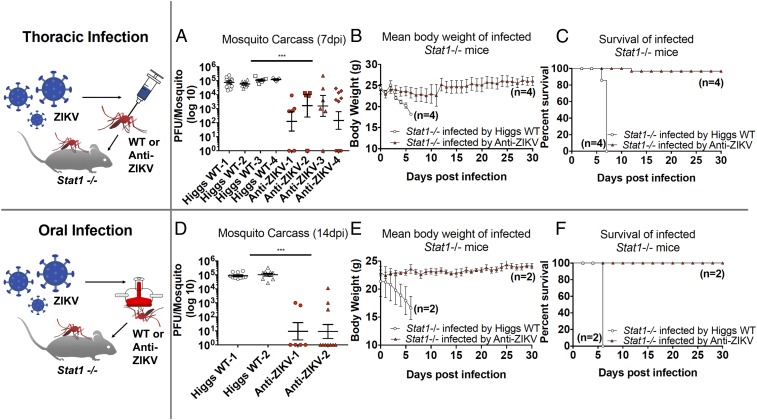
To further characterize ZIKV inhibition by the anti-ZIKV transgene, we also conducted limited tests of in vivo transmission capacity on heterozygous TZIKV-C mosquitoes. Specifically, we utilized a very sensitive STAT knockout (Stat1−/−) mouse model in which challenge with ZIKV (either intraperitoneally or via feeding by an infected mosquito) rapidly causes systemic infections presenting high viremia, resulting in significant weight loss, brain infections, and mortality (44). First, we infected adult female mosquitoes (Higgs WT and TZIKV-C) with Puerto Rican ZIKV strain (PRVABC59) via thoracic injection, which bypasses the midgut barrier, resulting in a significant viral titer in mosquitoes (44). At 7 dpi, TZIKV-C (n = 28) and Higgs WT (n = 29) mosquitoes were separately pooled into four groups (with six to 12 individuals per group; Fig. 3A), and each group was allowed to blood-feed on a Stat1−/− mouse, after which mouse weight and survival were measured daily. All mice fed on by infected Higgs WT mosquitoes (n = 4) became viremic and died before 8 dpi with significant weight loss before death (P < 0.05; Fig. 3 B and C). Conversely, of the four mice fed on by TZIKV-C mosquitoes, only one showed mortality (albeit at a later date, 12 dpi), and no significant weight loss was observed compared with the control group (P < 0.0001; Fig. 3 B and C). Measurement of ZIKV titers in the individual mosquitoes utilized for this assay demonstrated that nearly all TZIKV-C mosquitoes had significantly reduced viral titers (∼2 logs) at 7 dpi compared with Higgs WT mosquitoes (P < 0.0001; Fig. 3A).
Fig. 3.
Effect of anti-ZIKV transgene on ZIKV transmission in a mouse model. Higgs WT and heterozygous TZIKV-C mosquitoes were infected with Puerto Rican ZIKV strain PRVABC59 thoracically (A–C) or orally (D–F), and assayed for their ability to transmit ZIKV to immunocompromised Stat−/− mice. Viral titers in carcasses of mosquitoes infected thoracically (A, measured at 7 dpi) and orally (D, measured at 14 dpi) were determined by plaque assay in Vero cells and plotted. Mean body weight (B and E) and survival (C and F) of Stat−/− mice following ZIKV infection by thoracically (B and C) or orally (E and F) infected Higgs WT and TZIKV-C mosquitoes were measured and plotted. For all plots, white shapes represent results from Higgs WT mosquitoes and red shapes represent TZIKV-C mosquitoes. For viral titer plots, horizontal bars represent the mean virus titer and vertical bars represent SEM. For mean body weight plots, vertical bars represent SEM. ***P < 0.0001.
To better simulate how mosquitoes naturally obtain pathogens (i.e., from blood feeding), we also performed the above assay with mosquitoes that were infected with ZIKV (strain PRVABC59) via oral membrane blood feeding, and obtained similar results. Specifically, at 14 dpi via oral membrane blood feeding, TZIKV-C (n = 16) and Higgs WT (n = 20) mosquitoes were pooled into groups of six to 10 and allowed to feed on, and transmit the virus to, Stat1−/− mice (n = 2 for each transgenic and Higgs WT mosquito; Fig. 3D). The mice fed on by infected Higgs WT mosquitoes experienced significant weight loss and mortality before 8 dpi (P < 0.0001; Fig. 3 E and F). Conversely, mice fed on by TZIKV-C mosquitoes showed no significant change in weight and no infection-associated mortality (P < 0.0001; Fig. 3 E and F). Viral titer assays on these mosquitoes (at 14 dpi) indicated that the ZIKV infection rate was dramatically reduced in TZIKV-C mosquitoes (39% of mosquitoes infected compared with 93% of Higgs WT mosquitoes; Fig. 3D) and that viral titers of the TZIKV-C mosquitoes that were infected were significantly lower (∼2 logs) than those of Higgs WT mosquitoes (P < 0.0001; Fig. 3D). Altogether, these results demonstrate that the anti-ZIKV transgene confers robust refractoriness to ZIKV infection and transmission, and that even mosquitoes heterozygous for the transgene are unlikely to be able to transmit the virus.
Impact of Anti-ZIKV Transgene on Mosquito Fitness.
Finally, to determine whether the anti-ZIKV transgene had any significant fitness effects on transgenic mosquitoes, we assessed several fitness parameters, including larval-to-pupal development time, male and female fecundity and fertility, male mating success, adult wing length (as a proxy for body size), and longevity (SI Appendix, Table S3). No significant differences between Higgs WT and TZIKV-C mosquitoes were observed when examining male mating success, fecundity, and fertility (all P > 0.9); female fecundity (P > 0.05); and male and female wing length (P > 0.05). Conversely, we observed a significant difference (P < 0.01) in hatching rates of eggs laid by Higgs WT versus TZIKV-C female mosquitoes (with the latter having lower hatching rates) and a significant difference between larval-to-pupal development time (P < 0.01), with TZIKV-C individuals developing faster. When assessing adult mosquito survivorship, no significant differences were observed between Higgs WT and TZIKV-C male mosquitoes (P > 0.05; SI Appendix, Fig. S5 and Table S3), while Higgs WT female mosquitoes survived slightly longer than TZIKV-C female mosquitoes (P < 0.0001; SI Appendix, Fig. S5 and Table S3). Furthermore, there was no significant difference in survival at 14 dpi between Higgs WT and TZIKV-C mosquitoes infected with the Cambodia ZIKV strain (P > 0.05; SI Appendix, Table S4) and, similarly, no significant difference in survival between Higgs WT, Higgs wMel-infected, and TZIKV-C mosquitoes infected with the Puerto Rico ZIKV strain (P > 0.05; SI Appendix, Table S4). Based on the above observations, it appears that although the anti-ZIKV transgene did negatively affect female mosquito longevity and egg-hatching rate, it did not result in significant changes to most fitness parameters measured, including fecundity and fertility, male mating success, and body size.
Discussion
Taken together, our results demonstrate that targeting conserved genes in the ZIKV genome by expressing an engineered polycistronic cluster of synthetic small RNAs confers to homozygous mosquitoes complete refractoriness to multiple strains of ZIKV infection, dissemination, and transmission. Although incomplete, heterozygous mosquitoes also display partial refractoriness to ZIKV infection, dissemination, and transmission, with significant reduction of viral titers in the saliva (>2 logs compared with WT). This significant reduction of ZIKV is greater than the viral inhibition effect of Wolbachia, and may be sufficient to ensure these heterozygous mosquitoes are unable to transmit ZIKV to a susceptible host. Indeed, this latter point is supported by our finding that heterozygotes were largely unable to transmit ZIKV to immunocompromised (Stat1−/−) mice after infection via thoracic injection, and completely unable to transmit ZIKV after infection via membrane feeding. As intrathoracic injection generates unnaturally high infection levels by bypassing the mosquito midgut and lumen barriers (44), it is perhaps unsurprising that one mouse (possibly fed on by one of the mosquitoes with relatively high viral titers; Fig. 3A) experienced ZIKV-associated mortality. However, the observation that most of the heterozygous anti-ZIKV mosquitoes infected via thoracic injection, and all of the mosquitoes infected via membrane feeding, were unable to transmit ZIKV to a susceptible mouse model strongly suggests that even heterozygotes are unlikely to be capable of ZIKV transmission in the wild.
While the robust resistance observed in thoracic injection experiments may seem unexpected because this method of infection bypasses the midgut, where the CPA promoter is canonically expected to drive expression, our RT-PCR data indicated that the anti-ZIKV small synthetic RNAs were expressed in tissues other than the midgut. This is likely due to genomic position effects, which have been previously observed with this promoter (38), and could be addressed in future work by optimization of midgut-specific expression [although RNA-sequencing data indicate that CPA is actually expressed in multiple tissues other than the midgut across various developmental time points (36, 45), and the CPA promoter may therefore also act systemically].
Previously in Ae. aegypti, resistance to DENV has been engineered by transgenic activation of antiviral pathways (26), transgene-based RNAi in either the midgut (27, 46) or salivary glands (28), and antiviral hammerhead enzymes (47), and expression of synthetic miRNAs has also been demonstrated to induce partial resistance to DENV-3 and CHIKV (32). However, similar approaches have not been successfully demonstrated for ZIKV, and the currently described system is potentially especially advantageous since targeting six of 10 conserved protein-coding genes from the ZIKV genome with eight separate synthetic small RNAs may reduce the possibility of escapee mutants, and thus ensure evolutionary stability.
That said, it remains uncertain how many synthetic small RNAs are necessary to ensure robust disease refractoriness and evolutionary stability in a wild population. In our small RNA-sequencing efforts, we only detected expression/processing of five of the eight synthetic small RNAs, suggesting that perhaps the small RNA processing machinery is overloaded, that the CPA promoter is not strong enough to ensure robust expression from all eight synthetic small RNAs, or that some synthetic small RNAs are possibly unstable and get quickly degraded after processing. The latter hypothesis is supported by the fact that expression was only detected from synthetic small RNAs 1, 2, 4, 6, and 8, while synthetic small RNAs 3, 5, and 7 were undetected; given that these are arranged numerically in a linear array (i.e., small RNAs 1–8), small RNAs 3, 5, and 7 must have been expressed/processed in order for small RNAs 4, 6, and 8 to be expressed/processed/detected. Moreover, contrary to expectations when using a blood meal-inducible promoter such as CPA, levels of multiple synthetic small RNAs were lower in post–blood meal-fed transgenic samples than in nonblood-fed ones. The lack of clear blood meal expression induction is not inconsistent with previous findings regarding use of the CPA promoter to drive transgene expression (38), and likely arises due to genomic position effects associated with the transgene integration site. In any case, even without full expression driven by the CPA promoter, it is clear that synthetic small RNA levels are sufficient enough to bring about robust ZIKV resistance in multiple genetic backgrounds. However, future efforts should be focused on addressing the above-mentioned open questions regarding small RNA processing and specificity of expression.
Altogether, this strategy may provide a suitable cargo gene for practical use with a gene drive system to reduce/eliminate vector competence of mosquito populations. For example, previous reports have shown that Cas9-mediated, homing-based gene drive can be used for population modification of the malaria vector mosquito, Anopheles stephensi (22), and it should be possible to develop similar systems in Ae. aegypti. Given that homing-based drive systems quickly convert heterozygotes into homozygotes (4), linking the anti-ZIKV transgene described here to such a system could quickly convert an entire mosquito population into anti-ZIKV homozygotes that would be 100% resistant to ZIKV transmission. Recent ZIKV outbreaks have shown that vector control remains an essential part of reducing the health burden of emerging arboviruses. Although the aim of this study was to illustrate the feasibility of producing ZIKV-refractory mosquitoes, similar genetic engineering strategies could be used to develop [or improve on (32)] single transgenes that render mosquitoes completely resistant to multiple arboviruses like DENV and CHIKV. Given the increasing incidence of these viral infections worldwide, such transgenes (coupled with gene drive systems) can provide an effective, sustainable, and comprehensive strategy for reducing the impact of arboviral mosquito-borne diseases.
Methods
Mouse Experiments.
All mouse-related experiments were conducted in compliance with the guidelines of the Laboratory Animal Center of National Health Research Institutes (NHRI) in Taiwan. The animal protocol (NHRI-IACUC-105111) was approved by the Institutional Animal Care and Use Committee of the NHRI, according to the Guide for the Care and Use of Laboratory Animals (48). Management of animal experiments and animal care and use practices of the NHRI have been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Data Availability Statement.
All sequencing data associated with this study are available from the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP150144, BioProject accession no. PRJNA475410). The complete annotated plasmid sequence and DNA are publicly available at Addgene (plasmid no. 104968). Transgenic mosquitoes will be made available by the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Prof. Robert Tesh and Dr. Nikos Vasilakis (University of Texas Medical Branch) for providing the ZIKV FSS13025 and PRVABC59 strains and Lee Trinidad for preparing viral stocks. We thank Christian Bowman for helping with the inverse PCR assay. We also thank Prof. Scott O’Neill (Institute of Vector-Borne Diseases, Monash University) and the World Mosquito Program for providing Wolbachia-infected mosquito eggs. Finally, we thank Dr. Guann-Yi Yu (National Tsing Hua University) for providing the Stat−/− mice used in this study. This work was supported, in part, by an NIH-K22 Career Transition Award (5K22AI113060), an NIH Exploratory/Developmental Research Grant Award (1R21AI123937 to O.S.A.), and CSIRO internal funding (to P.N.P.).
Footnotes
Conflict of interest statement: A.B. and O.S.A have submitted a provisional patent application on this technology. All other authors declare no competing financial interests.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data associated with this study are available from the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP150144, BioProject accession no. PRJNA475410). The complete annotated plasmid sequence and DNA are publicly available at Addgene (plasmid no. 104968).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810771116/-/DCSupplemental.
References
- 1.Chitti SV, Prasad AK, Saxena SK. Emerging Zika virus disease: A public health emergency of global concern. Virusdisease. 2016;27:211–214. doi: 10.1007/s13337-016-0317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization 2017 Zika virus and complications: Questions and answers. Available at https://www.who.int/features/qa/zika/en/. Accessed July 27, 2017.
- 3.Huang YS, Higgs S, Vanlandingham DL. Biological control strategies for mosquito vectors of arboviruses. Insects. 2017;8:E21. doi: 10.3390/insects8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet. 2016;17:146–159. doi: 10.1038/nrg.2015.34. [DOI] [PubMed] [Google Scholar]
- 5.Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature. 1968;218:368–369. doi: 10.1038/218368a0. [DOI] [PubMed] [Google Scholar]
- 6.Kandul NP, et al. 2018. Transforming insect population control with precision guided sterile males. bioRXiv:10.1101/377721.
- 7.Alphey L, et al. Genetic control of Aedes mosquitoes. Pathog Glob Health. 2013;107:170–179. doi: 10.1179/2047773213Y.0000000095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris AF, et al. Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat Biotechnol. 2012;30:828–830. doi: 10.1038/nbt.2350. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho DO, et al. Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLoS Negl Trop Dis. 2015;9:e0003864. doi: 10.1371/journal.pntd.0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollack A. (January 31, 2016) New weapon to fight Zika: The mosquito. NY Times, Health Section, p A1.
- 11.Doyle M. 2016. The need for self-dispersing mosquito control technologies in urban areas: Update on releases of genetically modified male mosquitoes for suppression of Ae. aegypti in the Florida Keys. 2016 International Congress of Entomology (Entomological Society of America, Annapolis, MD), 10.1603/ICE.2016.108064.
- 12.Bourtzis K, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132:S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Dutra HLC, et al. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe. 2016;19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waltz E. US government approves “killer” mosquitoes to fight disease. Nature. November 6, 2017 doi: 10.1038/nature.2017.22959. [DOI] [Google Scholar]
- 15.Marris E. Bacteria could be key to freeing South Pacific of mosquitoes. Nature. 2017;548:17–18. doi: 10.1038/548017a. [DOI] [PubMed] [Google Scholar]
- 16.Callaway E. Rio fights Zika with biggest release yet of bacteria-infected mosquitoes. Nature. 2016;539:17–18. doi: 10.1038/nature.2016.20878. [DOI] [PubMed] [Google Scholar]
- 17.Dodson BL, et al. Wolbachia enhances West Nile virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 2014;8:e2965. doi: 10.1371/journal.pntd.0002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amuzu HE, et al. Wolbachia enhances insect-specific flavivirus infection in Aedes aegypti mosquitoes. Ecol Evol. 2018;8:5441–5454. doi: 10.1002/ece3.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King JG, Souto-Maior C, Sartori LM, Maciel-de-Freitas R, Gomes MGM. Variation in Wolbachia effects on Aedes mosquitoes as a determinant of invasiveness and vectorial capacity. Nat Commun. 2018;9:1483. doi: 10.1038/s41467-018-03981-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 22.Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA. 2015;112:E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akbari OS, et al. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth Biol. 2014;3:915–928. doi: 10.1021/sb300079h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C-H, et al. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- 26.Jupatanakul N, et al. Engineered Aedes aegypti JAK/STAT pathway-mediated immunity to dengue virus. PLoS Negl Trop Dis. 2017;11:e0005187. doi: 10.1371/journal.pntd.0005187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franz AW, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci USA. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathur G, et al. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2010;19:753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niu Q-W, et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- 30.Saha A, Bhagyawant SS, Parida M, Dash PK. Vector-delivered artificial miRNA effectively inhibited replication of Chikungunya virus. Antiviral Res. 2016;134:42–49. doi: 10.1016/j.antiviral.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie P-W, et al. Inhibition of dengue virus 2 replication by artificial micrornas targeting the conserved regions. Nucleic Acid Ther. 2013;23:244–252. doi: 10.1089/nat.2012.0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yen P-S, James A, Li J-C, Chen C-H, Failloux A-B. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Commun Biol. 2018;1:11. doi: 10.1038/s42003-017-0011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira LA, et al. Robust gut-specific gene expression in transgenic Aedes aegypti mosquitoes. Proc Natl Acad Sci USA. 2000;97:10895–10898. doi: 10.1073/pnas.97.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baronti C, et al. Complete coding sequence of Zika virus from a French polynesia outbreak in 2013. Genome Announc. 2014;2:e00500-14. doi: 10.1128/genomeA.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 36.Matthews BJ. 2017. Improved Aedes aegypti mosquito reference genome assembly enables biological discovery and vector control. bioRXiv:10.1101/240747.
- 37.Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: Follow the leader? Biochem Soc Trans. 2014;42:1135–1140. doi: 10.1042/BST20140142. [DOI] [PubMed] [Google Scholar]
- 38.Franz AWE, et al. Comparison of transgene expression in Aedes aegypti generated by mariner Mos1 transposition and ΦC31 site-directed recombination. Insect Mol Biol. 2011;20:587–598. doi: 10.1111/j.1365-2583.2011.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladner JT, et al. Complete genome sequences of five Zika virus isolates. Genome Announc. 2016;4:e00377-16. doi: 10.1128/genomeA.00377-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gubler DJ, Vasilakis N, Musso D. History and emergence of Zika virus. J Infect Dis. 2017;216(Suppl 10):S860–S867. doi: 10.1093/infdis/jix451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caragata EP, Dutra HLC, Moreira LA. Inhibition of Zika virus by Wolbachia in Aedes aegypti. Microb Cell. 2016;3:293–295. doi: 10.15698/mic2016.07.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultz MJ, et al. Variable inhibition of Zika virus replication by different Wolbachia strains in mosquito cell cultures. J Virol. 2017;91:e00339-17. doi: 10.1128/JVI.00339-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aliota MT, Peinado SA, Velez ID, Osorio JE. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci Rep. 2016;6:28792. doi: 10.1038/srep28792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo Y-P, et al. Establishment of a mouse model for the complete mosquito-mediated transmission cycle of Zika virus. PLoS Negl Trop Dis. 2018;12:e0006417. doi: 10.1371/journal.pntd.0006417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akbari OS, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3 (Bethesda) 2013;3:1493–1509. doi: 10.1534/g3.113.006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franz AWE, et al. Fitness impact and stability of a transgene conferring resistance to dengue-2 virus following introgression into a genetically diverse Aedes aegypti strain. PLoS Negl Trop Dis. 2014;8:e2833. doi: 10.1371/journal.pntd.0002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra P, Furey C, Balaraman V, Fraser MJ. Antiviral hammerhead ribozymes are effective for developing transgenic suppression of Chikungunya virus in Aedes aegypti mosquitoes. Viruses. 2016;8:E163. doi: 10.3390/v8060163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data associated with this study are available from the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP150144, BioProject accession no. PRJNA475410). The complete annotated plasmid sequence and DNA are publicly available at Addgene (plasmid no. 104968). Transgenic mosquitoes will be made available by the corresponding author upon request.