Significance
Genomes appear similar to natural language texts, and protein domains can be treated as analogs of words. To investigate the linguistic properties of genomes further, we calculated the complexity of the “protein languages” in all major branches of life and identified a nearly universal value of information gain associated with the transition from a random domain arrangement to the current protein domain architecture. An exploration of the evolutionary relationship of the protein languages identified the domain combinations that discriminate between the major branches of cellular life. We conclude that there exists a “quasi-universal grammar” of protein domains and that the nearly constant information gain we identified corresponds to the minimal complexity required to maintain a functional cell.
Keywords: n-gram, bigram, protein domain, language, domain architecture
Abstract
From an abstract, informational perspective, protein domains appear analogous to words in natural languages in which the rules of word association are dictated by linguistic rules, or grammar. Such rules exist for protein domains as well, because only a small fraction of all possible domain combinations is viable in evolution. We employ a popular linguistic technique, n-gram analysis, to probe the “proteome grammar”—that is, the rules of association of domains that generate various domain architectures of proteins. Comparison of the complexity measures of “protein languages” in major branches of life shows that the relative entropy difference (information gain) between the observed domain architectures and random domain combinations is highly conserved in evolution and is close to being a universal constant, at ∼1.2 bits. Substantial deviations from this constant are observed in only two major groups of organisms: a subset of Archaea that appears to be cells simplified to the limit, and animals that display extreme complexity. We also identify the n-grams that represent signatures of the major branches of cellular life. The results of this analysis bolster the analogy between genomes and natural language and show that a “quasi-universal grammar” underlies the evolution of domain architectures in all divisions of cellular life. The nearly universal value of information gain by the domain architectures could reflect the minimum complexity of signal processing that is required to maintain a functioning cell.
Ever since the inception of the human genome project, the metaphorical expression “book of life,” denoting the genome sequence, has captured imaginations of both scientific and lay communities (1–3). Extending the analogy of a genome to a book (a text, or a corpus in linguistics), we can think of amino acid residues as letters, protein domains as words, and proteins as sentences consisting of ordered arrangements of protein domains (domain architectures) (4).
Genomes show remarkable similarities to natural languages. Like all cellular life forms, all natural languages are believed to have descended from a single ancestor (5) and have evolved through mechanisms comparable to biological evolution (6). In a written language, individual letters cannot carry semantic information; the smallest unit of information, therefore, is a word (7, 8). Protein domains are structural, functional, and evolutionary units of proteins (9, 10) and are thus analogous to words. This analogy is reflected in the statistical properties of the domain repertoires of diverse organisms. The frequency distribution of domains encoded in any genome follows a power law (9, 11–13). Power-law distributions have been found in numerous natural and social contexts, including a broad variety of biological systems (14–16). An important variant of power-law distributions is Zipf’s law, which describes the frequency distribution of words in natural languages (17). The slope of the curve in Zipf’s law for a natural language is approximately −1 (7), which is close to the slope of domain frequency distribution in a genome (1). Additionally, bigram (defined as two consecutive words; nonconsecutive two word combinations that co-occur in a sentence do not count as bigrams) frequency distributions in natural languages also follow power laws; in this case, with a slope of approximately −2 (18). A similar value has been reported for protein domain bigrams (19).
The function of a protein, to a large extent, is determined by the arrangement of its constituent domains—that is, its domain architecture (13, 19). All life forms possess many multidomain proteins, but both the number of unique domains and the fraction of multidomain proteins increase with the organismal complexity (defined as the number of unique cell types in an organism): Eukaryotes have more multidomain proteins than prokaryotes (4, 9, 20–25), and animals have more multidomain proteins than unicellular eukaryotes (26). This trend of increased multidomain protein formation with increasing organismal complexity is known as domain accretion (27) and apparently plays a major role in evolution, particularly in major evolutionary transitions such as the origin of multicellularity (28–33). Of the numerous possible domain combinations, only a limited subset is actually represented in genomes, suggesting that domain architectures are shaped by natural selection (10, 19, 34). It is, therefore, imperative to decipher the rules of association of protein domains.
The smallest information unit in a language is a word, and a grammar is the set of rules regulating the association of words. Given that protein domains are analogous to words, the rules of association, or the “grammar” of proteins, can be investigated using tools borrowed from linguistics. The simplest way to explore the grammar of an unknown language is to perform an n-gram analysis, a probabilistic language-modeling technique whereby consecutive words in sentences are treated as a unit to identify meaningful word associations. Depending on the number of words (n) in the unit, the analysis can be unigram (n = 1), bigram (n = 2), trigram (n = 3), and so forth. Such modeling allows determination of the conditional probabilities of a word, given the previous word(s). n-Gram language modeling has been widely employed in various text processing applications and speech recognition (7, 8).
Previously, we introduced an informal bigram analysis to explore the evolution of protein domain promiscuity—that is, the tendency of some domains to participate in many different domain architectures. A pair of domains on a protein sequence was considered a bigram, and bigram frequencies were calculated to measure promiscuity of domains in all major branches of the eukaryotic evolutionary tree (13, 19). The concept of bigrams, as applied to protein domains, has since been widely employed in studies on the evolution of protein domain architectures (35–38).
A formal n-gram modeling of domains provides a probabilistic framework for deciphering the rules of domain combination in multidomain protein architectures. Here, we analyze bigram models from all major branches of cellular life and probe the evolutionary characteristics of these models. Such modeling yields an objective measure of genome complexity from the information theory perspective and provides tools to study the evolution of complexity. Domain rearrangements and, in particular, domain accretion made major contributions to evolutionary transitions such as the origin of eukaryotes and, subsequently, the origin of multicellular organisms. Estimation of entropy (a measure of complexity) changes accompanying these events provides a quantitative framework for the analysis of these crucial aspects of evolution. We show that the loss of entropy (information gain) resulting from domain arrangements in genomes is nearly constant across the entire course of cellular life evolution and identify both similarities and dissimilarities between the “language” of proteins and natural languages.
Results and Discussion
Domain n-Gram Modeling.
Complete proteome data from the three superkingdoms of life (Bacteria, Archaea, and Eukaryota—often called domains of cellular life, a term that we do not use here to denote taxa so as to avoid confusion with protein domains), were downloaded from the UniProt (39) database (see Methods and Datasets S1 and S2), and domains were mapped onto the sequence of each protein using HMMER3 (40) and the Pfam database (41). Altogether, we identified about 23 million domains across 4,794 species. The domain maps were filtered (see Methods) to generate nonoverlapping orders of domains for each protein. Having constructed these domain maps, we proceeded to generate n-gram models for these genomes.
The n-gram models are made by calculating the conditional probability of one word (domain), given the proximal adjacent words. Depending on the number of words considered (the order), n-gram models are called unigrams (one word), bigrams (two words), trigrams (three words), and so forth. In this work, we used only bigram models, not only because these are the easiest to analyze computationally but also because the fractions of proteins with three or more domains are low in most organisms, particularly in prokaryotes (42). Thus, the higher-order n-gram models would include many missing probabilities and therefore become uninformative. The bigram models were constructed using Eq. 1, after adding the faux markers “N” and “C” to the beginning and end of each protein sequence, respectively. We also made models without the additional markers to control for the effect of these additions. Unless otherwise stated, the results are from the models with the N and C markers. In addition, we shuffled the domains in each genome and constructed bigram models from these shuffled genomes (see Methods).
Domains are present in restricted contexts; only a small subset of the conditional probabilities of all possible bigrams assume nonzero values. More than 95% of all possible bigrams are not found in any genome (Dataset S3). Thus, for the analyses in which normalized data are important, such as phylogenetic tree calculation (see below), we used Good–Turing smoothing (43, 44) (see Methods) to assign nonzero probability values to all possible bigrams.
Entropy of n-Gram Models and Their Evolutionary Trends.
There are ∼7,000 languages in the world, divided into 19 linguistic families (45, 46). Notwithstanding the differences, linguistic universals have been identified in the grammar and vocabularies among all these languages (47). In a natural language, the symbols are concatenated under the constraints of syntactic rules of grammar. The resulting sequence shows a balance between order and disorder. A rigorous measure of the degree of order can be obtained by calculating the entropy of such sequences (48).
Entropic concepts are intimately linked to the concept of complexity in biology (49, 50), which has been defined in various ways. Entropic measurements of nucleotide and protein sequences have shown that entropy increases in the course of evolution (51), and this increase has been explained as a by-product of genome evolution via a largely nonadaptive, stochastic process (52). This view of evolution is buttressed by the existence of “universal laws”—that is, conserved patterns of evolutionary change that recur in all major divisions of life and do not appear to represent direct adaptations (53, 54).
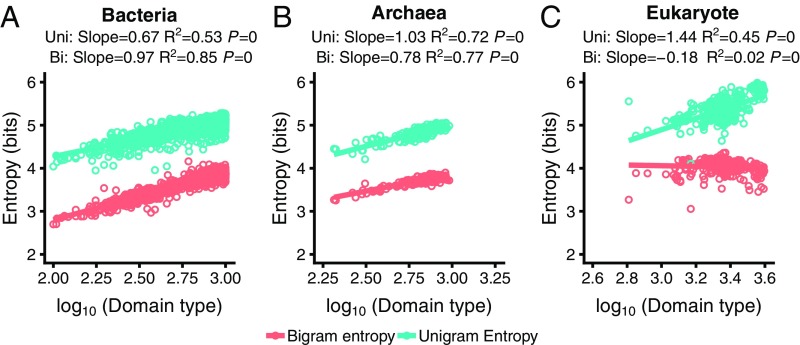
We calculated the entropy of the protein language model from each of the analyzed genomes (Dataset S3). We considered unigram models as null models, whereby the entropies are determined by the frequencies of individual domains in the genome. The entropies of the unigram and bigram models were calculated using Eqs. 3 and 4, respectively. We then investigated the relationships between the obtained entropies and various characteristics of proteomes—such as the number of unique domain families in a genome, domain count, protein count, and amino acid count—in each of the three superkingdoms of cellular life (Fig. 1 and SI Appendix, Figs. S1 and S2) and in five major kingdoms (SI Appendix, Figs. S3 and S4). The relationships were nonlinear in many cases, most prominently in Bacteria, but converting the values of the proteomic variables to the log10 scale resulted in a significant improvement of the linear regression coefficients (Fig. 1, SI Appendix, Fig. S1, and Dataset S4). As expected, for all three superkingdoms, the unigram entropy shows significant positive correlations with the number of domain families, total count of domain, number of proteins, and total amino acid count, each of which can be considered a proxy for the proteome size (and the genome size in prokaryotes) (Fig. 1 and SI Appendix, Fig. S1). Similarly, the entropy of natural languages is known to increase with the vocabulary size (8).
Fig. 1.
Log-linear regression of the unigram and bigram entropies with domain families (unique domain types in the genome) in the three superkingdoms of life: Bacteria (A), Archaea (B), and Eukaryota (C). Each point represents a genome. Some points are removed to keep all the figures in the same scale. See SI Appendix, Fig. S1 for the full data and regressions with other genomic variables. The x axis is converted to the log10 scale. See SI Appendix, Fig. S2 for the raw regression data. The slopes of the regression lines, R2, and P values are indicated on top of each plot.
For unigram entropies, the slopes of the regression lines are much greater for the number of domain families (or types) than they are for the other variables (Fig. 1 and SI Appendix, Fig. S1) because the entropy of the n-gram models, by design, is primarily determined by the diversity of domains in a genome. All regression slopes in Archaea are greater than the respective slopes in Bacteria, suggesting that, in archaeal evolution, increase in the genome size typically leads to a more pronounced innovation in the domain repertoire as compared to bacteria. This effect might be linked to the massive influx of bacterial genes that is thought to have occurred independently in several major groups of Archaea (55). The slope of the regression curve of the unigram entropy on the number of unique domain families in eukaryotes is considerably greater than in prokaryotes, indicating that the growth of the domain repertoire in eukaryotes results in more uniform domain frequency distributions than in either Bacteria or Archaea (Fig. 1). However, all of the other regression lines (SI Appendix, Fig. S1) for unigram entropies are notably flatter in eukaryotes than they are in prokaryotes, which is likely to reflect the substantially greater contribution of gene duplication to the increase of the proteome size in eukaryotes compared with prokaryotes (56–60). Analysis of individual phyla of Bacteria and Archaea shows the same trends as the bulk analysis, whereas among the eukaryotic kingdoms, unigram entropies decrease with the increase in genome size both in fungi and in plants (SI Appendix, Figs. S3 and S4). This trend is likely to stem from the major contribution of whole-genome duplications that are common in fungi and plants (61).
The bigram entropy regressions show clear differences between the two prokaryotic superkingdoms and eukaryotes (Fig. 1 and SI Appendix, Fig. S1). In prokaryotes, the bigram regression lines are roughly parallel to those for the unigrams, indicating that the diversification of domain combinations follows the growth of the domain repertoire which, at least in principle, is compatible with the notion that multidomain architectures evolve through random domain combination (11). In contrast, in eukaryotes (in bulk and in all individual kingdoms; SI Appendix, Figs. S1 and S4), the bigram entropy regression slopes are slightly negative, indicating that, with the increasing size of the proteomes, they tend to become more ordered—that is, the distributions of domain architectures become increasingly skewed. Most likely, this pattern is due to the proliferation of favorable domain combinations by gene duplication in complex multicellular eukaryotes, such as animals and plants. A striking example of such a bigram is the extensive amplification of nucleotide-binding and leucine-rich repeat proteins that combine an NTPase domain with an array of leucine-rich repeats (and, in some cases, additional domains), which are essential for both plant and animal innate immunity but, apparently, have evolved independently in plants and animals (62).
Relative Entropy of Protein Language Models.
The entropy of a language model indicates how much information is carried by the symbols of a given language in a particular text. The higher the entropy, the more uncertain we are about the information carried by the text (7, 8). The symbols can be alphabets, words, lines, or even the full corpus. A surprising and yet unexplained observation is that all known natural languages possess a nearly constant relative entropy, which is a measure of entropy loss (information gain) between a text written in the given language following the strict rules of grammar and a random sequence of words (63–66). It has been observed that for all natural languages, the information gain is about 3.6 bits, which is compatible with the existence of a universal grammar, despite some distinct, language-specific variations (46, 63).
We compared the relative entropy values (Hg in Eq. 5) of protein languages across the major prokaryotic and eukaryotic taxa (Dataset S3). Because the unigram entropy is derived from frequencies of individual domains in a genome, it can be considered the entropy of a random disorganized genome. Thus, we calculated the relative entropy (information gain) by subtracting the bigram entropy from the unigram entropy for each genome (Hg in Eq. 5). The difference between the unigram and bigram entropy measures the amount of information that is gained upon transition from a random collection of domains in the genome (unigrams) to the observed domain architectures (bigrams). This difference in entropy is a measure of the order imposed on the domain architectures by the rules of domain association forced by the biological functions that are relevant for the particular organism—that is, the grammar of the protein language. Clearly, the relative entropy calculated using only bigrams is but an approximation that ignores the information gain from more complex domain architectures (trigrams, tetragrams, etc.). However, given the relatively low fraction of proteins with more than two domains in proteomes (9), these relative entropy values can be expected to accurately reflect proteomic complexity.
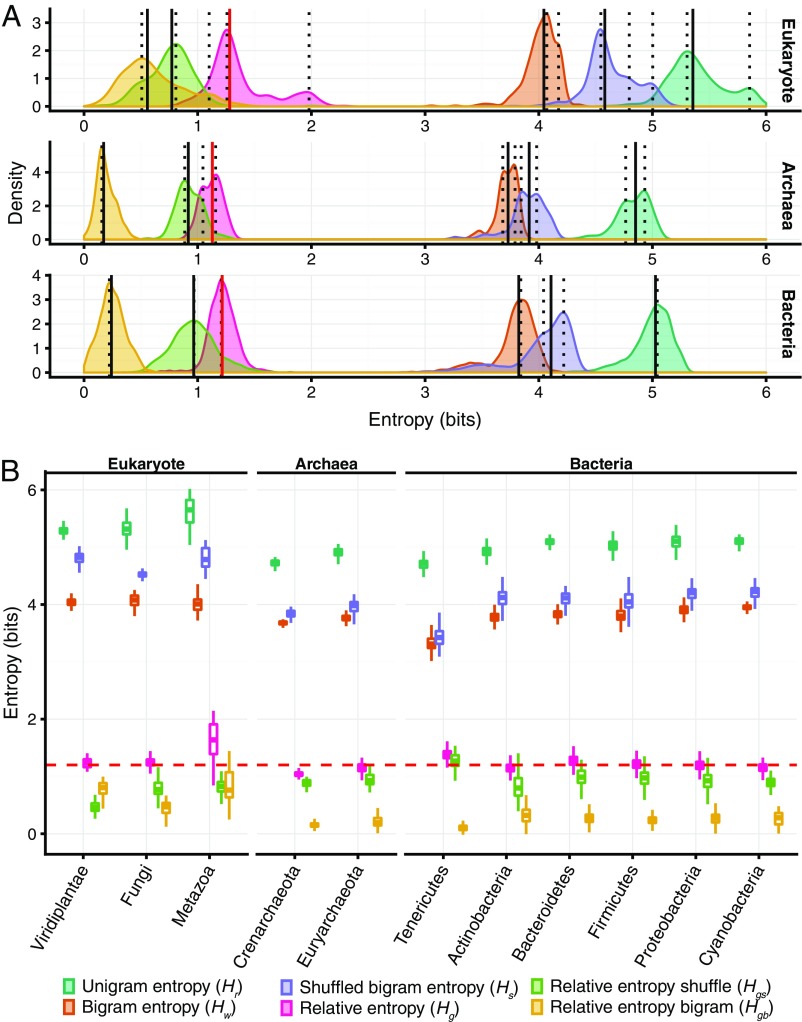
In both the unigram and the bigram entropy distributions, the median values increase in the following order: Archaea < Bacteria < Eukaryota (Fig. 2A and Dataset S5). This trend is not surprising because archaeal genomes are typically smaller in size and encode fewer domain families than bacterial, let alone eukaryotic genomes (67). The median values of the relative entropy (Hg) follow the same order: Archaea (1.13 bits) < Bacteria (1.21 bits) < Eukaryota (1.28 bits) (Fig. 2A and Dataset S5). The differences between these median values of the relative entropy for the three superkingdoms are statistically significant according to the permutation test (Dataset S6). Nevertheless, the three distributions strongly overlap as shown by counting discordant points and calculating Bhattacharyya coefficients (68) for pairs of distributions (Fig. 2A and Dataset S6).
Fig. 2.
Distributions of the unigram, bigram, and the three relative entropies. (A) Density plot of entropy values for the three superkingdoms. Each panel represents one superkingdom (from top to bottom: Eukaryota, Archaea, and Bacteria). Peaks in distributions are marked with dotted lines. The median values are indicated with solid lines. The median values of relative entropies (Hg) are marked with solid red lines. The x axis represents entropy in bits. (B) Box plots of the unigram, bigram, and the three relative entropies in three eukaryotic kingdoms (green plants, fungi, animals); two archaeal phyla (Crenarchaeota and Euryarchaeota); and six bacterial phyla (Tenericutes, Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Cyanobacteria). For the full list of median entropy values in all of the groups, see Dataset S5. The dashed horizontal red line represents the near-universal relative entropy of 1.2 bits.
For both Archaea and Eukaryota, the distributions of relative entropies are bimodal. The bimodality of the distribution in Archaea is mainly due to the difference between two groups, one of which consists of Euryarchaeota and Nanoarchaeota, and the other consisting of Crenarchaeota and other archaeal taxa. Euryarchaeota and Nanoarchaeota show an information gain value close to that in Bacteria, ∼1.2 bits (Fig. 2B and Dataset S5), whereas the rest of the archaea have a lower value of ∼1.04 bits. Thus, these archaea are characterized by anomalously low proteomic complexity. In eukaryotes, the two peaks correspond to plants and fungi (∼1.2 bits) and animals (>1.6 bits) (Fig. 2B and Dataset S5). Thus, animals show the highest information gain among the analyzed groups, in accord with the notion that domain architectures in animals are more elaborate and evolve under stronger constraints than those in other organisms (27). In contrast to archaea and eukaryotes, bacterial phyla exhibit remarkable conservation of relative entropy: Except Tenericutes, all analyzed bacteria have similar relative entropy close to ∼1.2 bits.
The above calculations of entropies are based on n-gram models with added N and C markers that potentially could bias the entropy calculations, especially for smaller genomes. To control for any such effect, we calculated the relative entropies without using the end markers (Dataset S3). As expected, this approach resulted in a substantial increase in unigram, bigram, and relative entropy. Nevertheless, the relative entropy values across all taxa were closely similar, ∼7 bits; moreover, all of the clade-specific trends noticed with the first approach remained valid (SI Appendix, Fig. S5).
Thus, apart from the two notable deviations, namely, the low information gain (low complexity) in a subset of Archaea and the high information gain (high complexity) in animals, the median relative entropies lie within a narrow range between ∼1.1 and ∼1.3 bits in many groups of highly diverse organisms. Together, these findings suggest the existence of a “quasi-universal” grammar of protein domain architecture.
The difference in entropy between the unigram and the bigram distributions comes from two constraints on the bigram distributions: first, the genome-specific distribution of proteins by the number of domains (relative frequencies of single- and multidomain proteins); and second, the biologically permissible and preferred domain combinations in the multidomain proteins. To differentiate between these two effects, we shuffled the domains in each genome, keeping constant the number of proteins and the number of domains in each protein (see Methods). This shuffling procedure does not change the unigram entropies of the genomes because the frequencies of domains do not change, but the shuffling changes the bigram entropies because the domain combinations are randomized. We then subtracted the shuffled bigram entropy (Hs) from the unigram entropy (Hr) to estimate the relative shuffled entropy (Hgs in Eq. 6) and subtracted the bigram entropy before shuffling (Hw) from the shuffled bigram entropy (Hs) to derive the relative bigram entropy (Hgb in Eq. 7) (Fig. 2 and Dataset S5). The bigram entropies calculated from these shuffled genomes (Hs) are, as expected, lower than the corresponding unigram entropies (Hr) but higher than the empirical bigram entropies (Hw) (Fig. 2). The only exception is Nanoarchaeota, where Hs is equal to or even slightly less than Hw. The bigram entropy gain due to randomization (relative bigram entropy; Hgb in Eq. 7) should be less in smaller genomes with fewer multidomain proteins. This is indeed the case, with Eukaryota having higher Hgb (0.56 bits), compared with Archaea (0.17 bits) and Bacteria (0.24 bits) (Fig. 2 and Dataset S5). This difference measures the information gain due to nonrandom, biologically meaningful domain combinations that are maintained by selection. In contrast, the difference between the unigram and shuffled bigram entropies (relative shuffled entropy; Hgs in Eq. 6) reflects the contribution of the global domain architecture—that is, the distribution of domains among the existing number of proteins. We found these values to be lower in Eukaryota (0.77 bits) than in Archaea (0.92 bits) and Bacteria (0.96 bits). Thus, in complex organisms, the effect of the global domain architecture, although greater than the contribution from specific domain combinations, plays a relatively less important role.
Using Cross-Entropy of Bigram Models to Build an Evolutionary Tree.
Several studies, including our own earlier work, have shown that domain frequency as well as domain architectures carry phylogenetic information (19, 69, 70). Therefore, it could be expected that this signal strengthens with the refinement of models of domain architecture. A domain-based phylogeny might be helpful to address certain long-standing questions in evolution, such as finding the root of the eukaryotic tree. Such problems are difficult to solve using traditional phylogenetic methods, and there is considerable interest in harnessing rare genomic changes (RGCs) for this purpose (71), given that they are less prone to various phylogeny-construction artifacts (71–73). Such RGCs could include diagnostic domain architectures—that is, taxon-specific domain combinations that are features for specific taxa.
A probabilistic language model can be used to determine the probability of a given genome by computing the conditional probabilities of all domain combinations it encodes. Given a set of n-gram models, we can calculate which model predicts the highest probability for a given genome. This value can be calculated directly by measuring perplexity, or the cross-entropy (7, 8), of a given genome under all the models. Perplexity is a measure of how well a given n-gram model describes a language. The model that has the maximum prediction power (lowest cross-entropy or lowest perplexity) is considered optimal.
Given two n-gram models generated from two separate genomes and a target genome, the cross-entropy calculation allows one to select the model that has a higher probability, or lower perplexity, for the target genome (see Eq. 8 and Methods). Domain models created from phylogenetically closer taxa are expected to possess higher predictive power (lower perplexity) compared with distant taxa. Thus, the pairwise cross-entropies can be represented as distances and, accordingly, can be used to create a whole-genome phylogenetic tree.
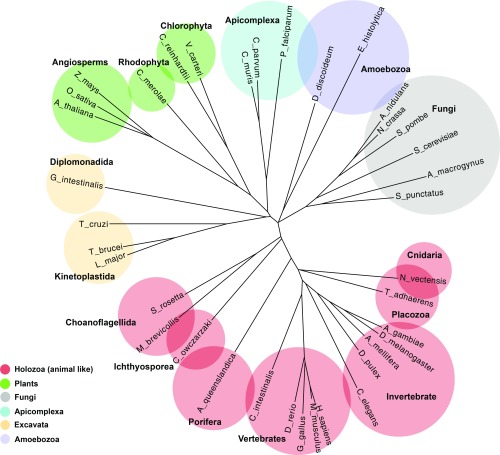
We calculated pairwise cross-entropies for 37 selected eukaryotic genomes (Dataset S2; see Methods for the selection procedure), and the resulting values were used to build a distance tree (Eq. 9 and Fig. 3). We focused on eukaryotes because of the well-defined topology of the main clades as opposed to the case of prokaryotes. The tree (Fig. 3) exhibits a near-perfect separation of the established major groups of eukaryotes. Depending on the placement of the root, the tree can be interpreted as being compatible with the unikont–bikont topology (74, 75). Under this root placement, the eukaryotic tree consists of three major clades: (i) bikonts, which include Archaeplastida (Viridiplantae and Rhodophyta) and Apicomplexa; (ii) unikonts (Amoebozoa, fungi, and animals); and (iii) Excavata (Diplomonadida and Kinetoplastida) (74, 76). The internal branching in the tree is also compatible with modern phylogenies. Examples include monophyly of mammals (human and mouse), insects (Apis mellifera, Drosophila melanogaster, and Anopheles gambei), fungi (Aspergillus nidulans, Neurospora crassa, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Allomyces macrogynus, and Spizellomyces punctatus), angiosperms (Oryza sativa, Arabidopsis thaliana, and Zea mays), choanoflagellates (Monosiga brevicollis and Salpingoeca rosetta), and excavates (diplomonads and kinetoplastids). Furthermore, the tree correctly positions Choanozoa and Ichthyosporea at the base of the animal clade.
Fig. 3.
Phylogenetic tree built from cross-entropy values. Domain bigram models were generated from 37 selected eukaryotic clades (Dataset S2) from the main branches of Eukaryota. The cross-entropies of bigram models were calculated in an all-vs.-all comparison. The entropy values were then normalized to create a distance matrix (see Methods for details), and the tree was constructed using the neighbor-joining method. The major groups are colored as shown in the legend.
The congruence of the resulting tree with traditional, sequence-based phylogenies indicates that the language models of domain architectures indeed carry robust phylogenetic information and that the models generally coevolve with the core genes that are used for phylogeny construction. However, the tree also shows some notable deviations from the accepted phylogeny; in particular, the apparent monophyly of Rhodophyta and angiosperms (Fig. 3). Such deviations from the species tree could reflect anomalous changes in domain architectures and, in this particular case, probably in Chlorophyta.
Clade-Specific Signatures of Domain Architecture.
Formation of new domain combinations, particularly domain accretion that leads to increased functional complexity, is an important avenue of protein evolution (27). In our previous analyses, we investigated the evolution of eukaryotes based on domain architecture evolution by measuring promiscuity of protein domains (13, 19). The results revealed that the repertoires of promiscuous domains (and especially domain architectures) are specific to major clades of eukaryotes and apparently reflect distinct biological functionalities. To a large extent, the evolution of domain architectures appears to be governed by natural selection (19, 34).
We identified signature bigrams for different major branches of organisms and examined their potential functional implications. To this end, we analyzed bigram language models of major subdivisions of life (bacteria, archaea, green plants, fungi, and animals) by using sparse partial least squares discriminant analysis (sPLS-DA) (77) to identify the bigrams that contributed maximally to the differentiation of each clade from the rest. The bigram entropy values (Eq. 10) were weighted as features for species of Proteobacteria (n = 1,345), Crenarchaeota (n = 36), Euryarchaeota (n = 111), Viridiplantae (n = 57), Fungi (n = 187), and Metazoa (n = 154), with each proteome containing at least 1,000 proteins.
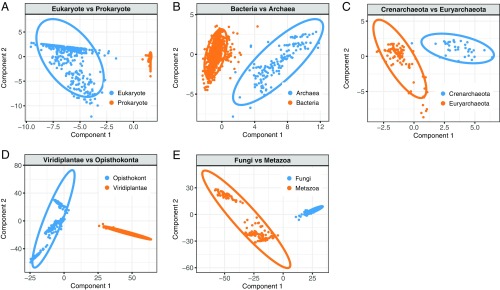
We used the splsda method from the R package mixOmics (78) and performed feature selection after variable tuning. Two methods were employed for feature selection, one using multiclass and the other using binary classes. The first method included a recursive technique by eliminating one clade at a time from the datasets and repeating sPLS-DA on the remaining clades (SI Appendix, Fig. S6). Under this method, sPLS-DA was carried out using the multiclass output. In the second method, we used binary classes for each partition and merged classes based on their taxonomic supergroup (Fig. 4). In the first method, the first round of sPLS-DA was run on the entire dataset, with six classes as output (Proteobacteria, Crenarchaeota, Euryarchaeota, Viridiplantae, Fungi, and Metazoa) (SI Appendix, Fig. S6A). Based on the component showing the maximum difference, the second round of sPLS-DA was run on two datasets, one of which included the three prokaryotic groups (SI Appendix, Fig. S6B) and the second including the three eukaryotic groups (SI Appendix, Fig. S6D). The process was iterated until all of the groups selected for the analysis were classified (SI Appendix, Fig. S6 C and E). In the second method, the output classes were always kept binary. In the first round, the sPLS-DA merged the prokaryotes (Bacteria and Archaea) as one combined output class, and eukaryotes (green plants, fungi, and metazoans) as the other (Fig. 4A). In subsequent rounds of sPLS-DA, prokaryotes and eukaryotes were split into the corresponding subdivisions: prokaryotes were split into Proteobacteria and Archaea (Fig. 4B), and eukaryotes were split into Viridiplantae and Opisthokonta (Fig. 4D). An additional round of sPLS-DA was carried out with Archaea (Fig. 4C) and Opisthokonta (Fig. 4E) to classify all of the groups. These guided, nested sPLS-DA analyses allowed us to identify the domain bigrams that are characteristic in each clade.
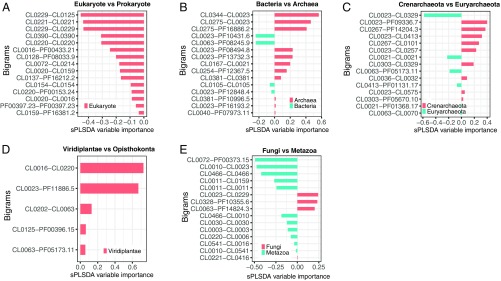
Fig. 4.
Bigram feature selection using sPLS-DA. (A–E) Weighted bigram probabilities were calculated from each species, and sPLS-DA was carried out using binary classes as outcome vectors. For multiclass classification, see SI Appendix, Fig. S6. The analyses were carried out in a nested hierarchical manner, beginning with the eukaryotes and prokaryotes at the top level, where all the species were binned into these two divisions (A). In the subsequent rounds, each division was split into the respective subdivisions: prokaryotes were split into Bacteria and Archaea (B); Archaea was split into Crenarchaeota and Euryarchaeota (C); eukaryotes were first split into Viridiplantae (green plants) and Opisthokonta (fungi and animals) (D); and finally, Opisthokonta was split into Fungi and Metazoa (animals) (E). For each analysis, a biplot with component 1 on the x axis and component 2 on the y axis is shown. The ellipses represent the 95% confidence area of each cluster.
In both methods, we visually selected the components that showed maximum separation between the classes and extracted the features that contributed to these components using the selectVar function of mixOmics package (Fig. 5 and SI Appendix, Fig. S7). In most cases, features selected using multiclass sPLS-DA were identical to or comprised a subset of the features selected using binary classes. In the resulting clustering, prokaryotes are perfectly separated from eukaryotes along component 1 (Fig. 4A, SI Appendix, Fig. S6A, and Dataset S7). We identified 15 bigrams (Fig. 5A, SI Appendix, Fig. S7A, and Dataset S7) that differentiate the two clusters. All these bigrams are overrepresented in eukaryotes, indicating that eukaryotes and prokaryotes are separated mostly through gain of new domain architectures (and, by inference, new functions) in eukaryotes. The proteins that contain these bigrams are involved in signature eukaryotic functions such as ubiquitin signaling pathways and splicing (Dataset S8). In particular, the three top bigrams are CL0229 (RING)-CL0125 (Peptidase_CA), CL0221 (RRM)-CL0221(RRM), and CL0229 (RING)-CL0229 (RING). The first and the third of these are involved in the ubiquitin network, whereas the second is represented in spliceosome subunits.
Fig. 5.
Discriminating bigrams selected using sPLS-DA. (A–E) For each analysis in Fig. 4, the bigram variables were selected for the component that showed maximum separation between the binary classes. For multiclass selection, see SI Appendix, Fig. S7. The bar plots show loading values on the x axis for each selected bigram on the y axis. The color of the bar indicates the group shown in the legend. The bar plots show the selected bigrams and their loading weights for the following binary classes: eukaryotes vs. prokaryotes (A), Bacteria vs. Archaea (B), Crenarchaeota vs. Euryarchaeota (C), Viridiplantae (green plants) vs. Opisthokonta (fungi plus animals) (D), and Fungi vs. Metazoa (animals) (E).
Within the prokaryotic cluster (Fig. 4B and SI Appendix, Fig. S6B), Proteobacteria and Archaea are also clearly separated along component 1, with 15 bigrams differentiating the archaeal and bacterial clusters, of which 13 are shared by the binary and multiclass sPLS-DA (Fig. 5, SI Appendix, Fig. S7B, and Dataset S7). Many of these domain combinations are involved in cell-wall biogenesis (Dataset S8), which is biologically plausible, given the distinct molecular structures of the cell walls of archaea and bacteria (79).
Crenarchaeota and Euryarchaeota also form nonoverlapping clusters separated on component 1 (Fig. 4C and SI Appendix, Fig. S6C), with 15 discriminating bigrams that are common in both the binary and multiclass sPLS-DA (Fig. 5C, SI Appendix, Fig. S7C, and Dataset S7). These domain combinations are mostly involved in ATP hydrolysis, DNA replication, and proteolysis, apparently related to heat-shock response (Datasets S7 and S8).
Comparing green plants (Viridiplantae) with fungi/metazoans (Opisthokonta), we found that the discriminating bigrams are all plant specific, indicating gain of function in plants (Figs. 4D and 5D and SI Appendix, Figs. S6D and S7D). Altogether, there are 10 such bigrams (Dataset S7), five of which are common to both multiclass and binary sPLS-DA. The discriminating domain combinations mostly included NTPases and protein kinases (Dataset S8).
Finally, Fungi and Metazoa are distinguished by 15 bigrams (Fig. 4E, SI Appendix, Fig. S6E, and Dataset S7), most of which are dominant in Metazoa (Fig. 5E and SI Appendix, Fig. S7E). The Metazoa-specific bigrams are related to cell–cell adhesion, including the hedgehog family proteins and various membrane proteins, channels, and kinases—that is, functions mostly associated with metazoan multicellularity (Dataset S8).
Together, these findings indicate that domain bigram models readily and cleanly distinguish between the major divisions of cellular life. Following the linguistic metaphor, the protein languages in different divisions of cellular life are clearly distinct, the quasi-universal grammar notwithstanding.
Conclusions
The similarities between natural languages and genomes are apparent when domains are treated as functional analogs of words in natural languages. Here, we investigated these similarities using modifications of methods employed by computational linguistics. Using domain bigram models, we show that for most groups of organisms (both prokaryotes and eukaryotes), the information gain in the observed domain architectures lies within the narrow interval between 1.1 and 1.3 bits. As shown by our analysis of shuffled-genome entropy, these nearly universal values can be decomposed into distinct contributions from the global domain architecture and the specific domain combinations. This finding implies the existence of a quasi-universal grammar of domain architectures. The nature of the rules that underlie this universal grammar remains to be further investigated. Generally, multidomain architectures are most common among proteins that are involved in signal transduction, regulatory processes, and immune functions (19, 80). Conceivably, the near-universal value of information gain by the genome-wide domain architectures represents the minimum complexity that is required to maintain a functioning cell capable of adequately processing internal and external signals. The significant deviations from the universal value of the information gain in a subset of archaeal phyla seem to represent streamlining under extreme conditions to the lowest limit of complexity sustainable for autonomous cells. Conversely, in animals, the high value of the information gain reflects the exceptional complexity that is incumbent in multicellular organisms with differentiated tissues.
The near-universal information gain relates the protein languages of biology to human natural languages. However, the characteristic values of the information gain are substantially different, namely, ∼1.2 bits in biology and ∼3.6 bits in linguistics. Thus, both the protein and the human (and formal) languages seem to be based on quasi-universal grammars, but the complexity (orderliness) of the latter is substantially greater than that of the former. This difference could be expected because, unlike human languages, all proteomes are rich in single-domain proteins (one-word sentences) (4, 25) and because the role of the stochastic component in the evolution of protein languages appears to be much greater than it is in the evolution of natural languages (9, 11).
We also show that domain bigram models can be used to generate evolutionary trees by measuring the probability of a genome, given the language models from other genomes. This cross-entropy largely accurately reflects the topology of the currently accepted sequence-based trees, showing that domain architectures (languages) mostly coevolve with the core components of genomes. Along similar lines, using feature selection, we identified the sets of bigrams that discriminate between the major clades and indeed seem to correspond to clade-specific functions such as ubiquitin signaling and splicing in eukaryotes. Under the linguistic metaphor, these bigrams are signature phrases of the respective protein languages, and the tree generated using cross-entropy values as distances reflects the evolution of these languages.
Methods
Genomes Used in the Study.
Reference proteome FASTA files were downloaded from UniProt (www.uniprot.org/proteomes/) consisting of 4,159 bacterial, 187 archaeal, and 448 eukaryotic genomes. The eukaryotic genomes were a subset selected based on whether the genome has an isoform file. Only genomes with isoforms were used in the analysis (n = 448; Dataset S1). However, isoform data were not included in this analysis. Only the canonical proteins were used for all further calculations. A subset of the eukaryotic genomes (n = 37) was manually selected for phylogenetic analysis to maximize coverage of the main branches of the eukaryotic tree topology (Dataset S2).
Domain Structure Determination.
We used HMMER (v. 3.1b) (40) and Pfam-A database (release 30) (41) to identify domains in each genome. The details are provided in SI Appendix, Supplementary Methods.
The n-Gram Model of a Protein Language.
Modeling domains under a first-order Markov process, in which the probability of a domain depends only on the one preceding domain , is called a bigram model, and the domain pair is called a bigram. We estimated the conditional probability of the domain , given the preceding domain , using maximum likelihood estimation (MLE) (see SI Appendix for details):
| [1] |
where is the count of the bigram in the genome, and is the count of all bigrams in which the first domain is , which is equivalent to the count of domain in the genome.
In bigram models of natural languages, the beginnings and the ends of sentences are marked with faux word markers (7, 8). Similarly, the start and the end of protein sequences were marked with the two artificial domain markers N and C, respectively. The addition of the end markers allows us to include all domains in the analysis, including those that solely occur in the single-domain proteins; otherwise, bigram models can only be constructed for multidomain proteins. The addition of these markers is a common practice in n-gram modeling in linguistics that makes these models probabilistic and generative (7, 8). However, the addition of these markers has large effects on small genomes; therefore, we also analyzed models without these additional markers. In this case, although models are not truly probabilistic, we could compare them against models created without additional markers to measure the effect of these markers on small genomes.
Good–Turing Smoothing.
The biggest problem with n-gram models is the sparsity of the data. Most of the domains are present in strictly constrained contexts and participate only in a restricted number of domain pairs. Therefore, a large number of the conditional probabilities are 0 in a genome. We used a smoothing technique called Simple Good–Turing (SGT) (43) to assign counts to missing bigrams in the genomes (SI Appendix). Once the SGT counts were estimated, these counts were used to calculate the conditional probabilities as shown in Eq. 1.
Entropy and Relative Entropy of n-Gram Language Models.
In general, entropy of a probabilistic system, as defined by Shannon (81) is
| [2] |
where is the probability of the event . In this equation, entropy is the sum of weighted log probabilities converted to a positive value.
Entropy of a unigram model (a random genome) is calculated by simply replacing in Eq. 2 with the frequency of the domains in a genome, which can be simplified as follows:
| [3] |
where is a domain, is its count, is its frequency, and is the total number of domains in the genome. This entropy calculation effectively gives the entropy of the genome after a random shuffling of all of the domains in the genome, disregarding the protein structures. In other words, it is a “bag of words” model in which the probability of a domain in the genome is proportional to its frequency.
According to Eq. 2, the entropy of a system is the sum of weighted probabilities of the events. The entropy of a bigram model is defined as a weighted average branching factor of a language (7, 8), where each conditional probability is weighted by the frequency of the particular bigram in the genome. This can be simplified as follows:
| [4] |
where is the count of the bigram, is the count of the first domain in the bigram domain pair, and Nb is the total count of bigrams in the genome.
The loss of entropy resulting from the transition from the completely random version of the genome to the observed domain architectures can, therefore, be calculated as
| [5] |
where is the entropy loss (information gain), is the entropy of the unigram (random) model, and is the entropy of the bigram model.
Entropy of a Shuffled Genome.
We also estimated the entropy of a genome after shuffling all its constituent domains. The domains encoded in a genome were shuffled in such a way that the total numbers of proteins, domains, and domain families, as well as the number of domains in each protein and the positions of the N and C markers were kept unchanged, but all of the n-grams were randomized. We then estimated the bigram entropy from this shuffled genome following the procedure described above. The shuffling was repeated 100 times and the average bigram entropy from these 100 runs was taken as the shuffled bigram entropy . This entropy value for each genome was then subtracted from the corresponding unigram entropy to derive the relative shuffled entropy :
| [6] |
The bigram entropy before shuffling for each genome were then subtracted from to derive relative bigram entropy :
| [7] |
Cross-Entropy (Perplexity).
Given that n-gram language models are generative, cross-entropy or perplexity is a measure of how well a given language model describes a language (8). Perplexity can be utilized to evaluate the probability of a genome given a bigram model, where the conditional probabilities of the bigrams come from the model, but the weight for each bigram is derived from the genome being evaluated. The better the model describes the genome, the higher the probability and the lower the perplexity (see SI Appendix for details). Given a language model from a source genome , the cross-entropy of a target genome can be calculated as
| [8] |
where the function is the count of the bigram in the target genome , the function is the conditional probability of the same bigram in the source genome provided by the model from the source genome, is the count of the bigram in the source genome, and is the number of bigrams in the target genome .
Phylogeny Construction Using Cross-Entropy Data.
A phylogenetic tree was built from the cross-entropy data from all-vs.-all comparison of 37 selected species representing all major divisions of the eukaryotes. Because cross-entropy requires assigning probability to a genome, given the bigram model from another genome, missing bigrams in the target genome have to be taken into consideration. We used SGT-smoothed bigrams for this analysis. We first normalized the pairwise cross-entropy values (Eq. 8) using the self-entropy (entropy calculated using the models created from the same genome). Given genomes with models and target genomes , the distance between the two genomes is calculated as
| [9] |
where is the cross-entropy of genome (under the bigram model ), and is the self-entropy (the model is derived from the same genome for which the entropy is being calculated). Because self-entropy is the lowest for any genome, the self-distance , where . The cross-entropy value is unidirectional; therefore, we took the average of and to derive the final distance between the two genomes. The pairwise distance table created in this manner was used to build a neighbor-joining tree using the R package APE (82).
sPLS-DA.
After creating the cross-entropy tree, we sought to identify the sets of bigrams defining the major branches in the tree. To this end, supervised feature selection classifying each clade was performed using sPLS-DA (77) as implemented in the R package mixOmics (78). For this analysis, we only used bigrams that were actually present in the genome (N and C markers were removed from the analysis). We also used only species having more than 1,000 proteins in the genomes taken from the following major subdivisions of each superkingdom: Proteobacteria (n = 1,345), Crenarchaeota (n = 36), Euryarchaeota (n = 111), Fungi (n = 187), Viridiplantae (n = 57), and Metazoa (n = 154).
For each genome, all bigram conditional probabilities were weighted by their counts in the genome to calculate entropy values for individual bigrams using the following formula:
| [10] |
where is calculated as in Eq. 1, and is the count of bigram in the genome. This was divided by the total number of domains (N) in the genome to normalize this value across all species. The formula calculates individual entropy of a given bigram measuring the uncertainty that a particular domain chooses the next domain with the probability .
sPLS-DA uses a feature matrix () and a group or outcome vector (). The feature matrix in this case was the bigram entropy values (Eq. 10) of each bigram on rows and of each species on columns. Absent bigrams in a genome were all given 0 values. We performed two types of analyses using two types of vectors. In the first category, each species in the study was assigned its known clade, and sPLS-DA was carried out using this multiclass outcome vector. In the second analysis, we performed a binary classification using sPLS-DA, whereby each species was preclassified into two supergroup clades.
We tuned (tune.splsda function in the mixOmics package) sPLS-DA runs using fivefold cross-validation repeated 10 times to determine the best parameters of the classification (optimum number of components). After each run, we determined the classification accuracy using the area under the receiver operating characteristic curve (AUROC). We visually selected the components that maximally separated each clade from the rest of the clades and selected features (bigrams) using the selectVar function from the mixOmics package.
Ontology Analysis of Domains.
After the feature selection, we performed ontology analysis of the bigrams to find the functional significance of each bigram. First, we identified all the proteins in the clades that were used in the analysis that contained the identified bigrams. We then searched the UniProt database using the REST application programming interface (https://www.uniprot.org/help/programmatic_access) to identify Gene Ontology (GO) terms corresponding to those proteins. We then calculated the frequency of each GO term in the result and kept only the most frequent GO terms.
Supplementary Material
Acknowledgments
The computational analyses were carried out on the Genifx computational facility (genifx.ifx.uab.edu) at the University of Alabama at Birmingham. This work was partially supported by grants from the University of Alabama Health Services Foundation (M.K.B.). Y.I.W. and E.V.K. are supported by intramural funds of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All data and code are available from the GitHub repository, https://github.com/malaybasu/2019-domain_pnas.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814684116/-/DCSupplemental.
References
- 1.Searls DB. The language of genes. Nature. 2002;420:211–217. doi: 10.1038/nature01255. [DOI] [PubMed] [Google Scholar]
- 2.Scaiewicz A, Levitt M. The language of the protein universe. Curr Opin Genet Dev. 2015;35:50–56. doi: 10.1016/j.gde.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.List J-M, Pathmanathan JS, Lopez P, Bapteste E. Unity and disunity in evolutionary sciences: Process-based analogies open common research avenues for biology and linguistics. Biol Direct. 2016;11:39. doi: 10.1186/s13062-016-0145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scaiewicz A, Levitt M. Unique function words characterize genomic proteins. Proc Natl Acad Sci USA. 2018;115:6703–6708. doi: 10.1073/pnas.1801182115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruhlen M. The Origin of Language : Tracing the Evolution of the Mother Tongue. Wiley; New York: 1994. [Google Scholar]
- 6.Atkinson QD, Meade A, Venditti C, Greenhill SJ, Pagel M. Languages evolve in punctuational bursts. Science. 2008;319:588. doi: 10.1126/science.1149683. [DOI] [PubMed] [Google Scholar]
- 7.Manning C, Schütze H. Foundations of Statistical Natural Language Processing. MIT Press; Cambridge, MA: 1999. [Google Scholar]
- 8.Jurafsky D, Martin JH. Speech and Language Processing. 2nd Ed Prentice Hall, Upper Saddle River, NJ; 2008. [Google Scholar]
- 9.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- 11.Karev GP, Wolf YI, Rzhetsky AY, Berezovskaya FS, Koonin EV. Birth and death of protein domains: A simple model of evolution explains power law behavior. BMC Evol Biol. 2002;2:18. doi: 10.1186/1471-2148-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuznetsov VA. Computational and Statistical Approaches to Genomics. Kluwer; Boston: 2002. [Google Scholar]
- 13.Basu MK, Poliakov E, Rogozin IB. Domain mobility in proteins: Functional and evolutionary implications. Brief Bioinform. 2009;10:205–216. doi: 10.1093/bib/bbn057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luscombe NM, Qian J, Zhang Z, Johnson T, Gerstein M. The dominance of the population by a selected few: Power-law behaviour applies to a wide variety of genomic properties. Genome Biol. 2002;3:RESEARCH0040. doi: 10.1186/gb-2002-3-8-research0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barabási A-L. Linked : The New Science of Networks. Perseus Books Group; New York: 2002. [Google Scholar]
- 16.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 17.Zipf GK. Human Behaviour and the Principle of Least Effort. Addison-Wesley; Boston: 1949. [Google Scholar]
- 18.Krishna M, Hassan A, Liu Y, Radev D. 2011 The effect of linguistic constraints on the large scale organization of language. Available at https://arxiv.org/abs/1102.2831. Accessed August 15, 2011.
- 19.Basu MK, Carmel L, Rogozin IB, Koonin EV. Evolution of protein domain promiscuity in eukaryotes. Genome Res. 2008;18:449–461. doi: 10.1101/gr.6943508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf YI, Brenner SE, Bash PA, Koonin EV. Distribution of protein folds in the three superkingdoms of life. Genome Res. 1999;9:17–26. [PubMed] [Google Scholar]
- 21.Apic G, Gough J, Teichmann SA. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 22.Ekman D, Björklund AK, Elofsson A. Quantification of the elevated rate of domain rearrangements in metazoa. J Mol Biol. 2007;372:1337–1348. doi: 10.1016/j.jmb.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Rost B. CHOP proteins into structural domain-like fragments. Proteins. 2004;55:678–688. doi: 10.1002/prot.20095. [DOI] [PubMed] [Google Scholar]
- 24.Novozhilov AS, Karev GP, Koonin EV. Biological applications of the theory of birth-and-death processes. Brief Bioinform. 2006;7:70–85. doi: 10.1093/bib/bbk006. [DOI] [PubMed] [Google Scholar]
- 25.Levitt M. Nature of the protein universe. Proc Natl Acad Sci USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tordai H, Nagy A, Farkas K, Bányai L, Patthy L. Modules, multidomain proteins and organismic complexity. FEBS J. 2005;272:5064–5078. doi: 10.1111/j.1742-4658.2005.04917.x. [DOI] [PubMed] [Google Scholar]
- 27.Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 28.Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- 29.Koonin EV, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 31.Nichols SA, Dirks W, Pearse JS, King N. Early evolution of animal cell signaling and adhesion genes. Proc Natl Acad Sci USA. 2006;103:12451–12456. doi: 10.1073/pnas.0604065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusserow A, et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- 33.Marsh JA, Teichmann SA. How do proteins gain new domains? Genome Biol. 2010;11:126. doi: 10.1186/gb-2010-11-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forslund K, Henricson A, Hollich V, Sonnhammer ELL. Domain tree-based analysis of protein architecture evolution. Mol Biol Evol. 2008;25:254–264. doi: 10.1093/molbev/msm254. [DOI] [PubMed] [Google Scholar]
- 35.Dong Q, Wang K, Liu X. Identifying the missing proteins in human proteome by biological language model. BMC Syst Biol. 2016;10:113. doi: 10.1186/s12918-016-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie X, Jin J, Mao Y. Evolutionary versatility of eukaryotic protein domains revealed by their bigram networks. BMC Evol Biol. 2011;11:242. doi: 10.1186/1471-2148-11-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seidl MF, Van den Ackerveken G, Govers F, Snel B. A domain-centric analysis of oomycete plant pathogen genomes reveals unique protein organization. Plant Physiol. 2011;155:628–644. doi: 10.1104/pp.110.167841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner J, 3rd, Moore AD, Bornberg-Bauer E. Just how versatile are domains? BMC Evol Biol. 2008;8:285. doi: 10.1186/1471-2148-8-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bateman A, et al. The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ekman D, Björklund AK, Frey-Skött J, Elofsson A. Multi-domain proteins in the three kingdoms of life: Orphan domains and other unassigned regions. J Mol Biol. 2005;348:231–243. doi: 10.1016/j.jmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 43.Gale WA, Sampson G. Good‐turing frequency estimation without tears. J Quant Linguist. 1995;2:217–237. [Google Scholar]
- 44.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 45.Lewis M, editor. Ethnologue: Languages of the World. 16th Ed SIL International, Dallas; 2009. [Google Scholar]
- 46.Montemurro MA, Zanette DH. Universal entropy of word ordering across linguistic families. PLoS One. 2011;6:e19875. doi: 10.1371/journal.pone.0019875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenberg JH. Language universals: A research frontier. Science. 1969;166:473–478. doi: 10.1126/science.166.3904.473. [DOI] [PubMed] [Google Scholar]
- 48.Shannon CE. Prediction and entropy of printed English. Bell Syst Tech J. 1951;30:50–64. [Google Scholar]
- 49.Adami C, Ofria C, Collier TC. Evolution of biological complexity. Proc Natl Acad Sci USA. 2000;97:4463–4468. doi: 10.1073/pnas.97.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adami C. What is complexity? BioEssays. 2002;24:1085–1094. doi: 10.1002/bies.10192. [DOI] [PubMed] [Google Scholar]
- 51.Koonin EV. A non-adaptationist perspective on evolution of genomic complexity or the continued dethroning of man. Cell Cycle. 2004;3:280–285. [PubMed] [Google Scholar]
- 52.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 53.Koonin EV. Are there laws of genome evolution? PLoS Comput Biol. 2011;7:e1002173. doi: 10.1371/journal.pcbi.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press Science; Upper Saddle River, NJ: 2011. [Google Scholar]
- 55.Nelson-Sathi S, et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature. 2015;517:77–80. doi: 10.1038/nature13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 57.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 58.Van de Peer Y. Computational approaches to unveiling ancient genome duplications. Nat Rev Genet. 2004;5:752–763. doi: 10.1038/nrg1449. [DOI] [PubMed] [Google Scholar]
- 59.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7:e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou X, Lin Z, Ma H. Phylogenetic detection of numerous gene duplications shared by animals, fungi and plants. Genome Biol. 2010;11:R38. doi: 10.1186/gb-2010-11-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urbach JM, Ausubel FM. The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci USA. 2017;114:1063–1068. doi: 10.1073/pnas.1619730114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunn M, Greenhill SJ, Levinson SC, Gray RD. Evolved structure of language shows lineage-specific trends in word-order universals. Nature. 2011;473:79–82. doi: 10.1038/nature09923. [DOI] [PubMed] [Google Scholar]
- 64.Rao RPN, et al. A Markov model of the Indus script. Proc Natl Acad Sci USA. 2009;106:13685–13690. doi: 10.1073/pnas.0906237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rao RPN, et al. Entropic evidence for linguistic structure in the Indus script. Science. 2009;324:1165. doi: 10.1126/science.1170391. [DOI] [PubMed] [Google Scholar]
- 66.Greenberg JH. Universals of Human Language. MIT Press; Cambridge, MA: 1963. Some universals of grammar with particular reference to the order of meaningful elements. [Google Scholar]
- 67.Koonin EV, Wolf YI. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhattacharyya A. On a measure of divergence between two statistical populations defined by their probability distributions. Bull Calcutta Math Soc. 1943;35:99–109. [Google Scholar]
- 69.Yang S, Doolittle RF, Bourne PE. Phylogeny determined by protein domain content. Proc Natl Acad Sci USA. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M, Caetano-Anollés G. Global phylogeny determined by the combination of protein domains in proteomes. Mol Biol Evol. 2006;23:2444–2454. doi: 10.1093/molbev/msl117. [DOI] [PubMed] [Google Scholar]
- 71.Rogozin IB, Basu MK, Csuros M, Koonin EV. Analysis of rare genomic changes does not support the unikont-bikont phylogeny and suggests cyanobacterial symbiosis as the point of primary radiation of eukaryotes. Gen Biol Evol. 2009;1:99–113. doi: 10.1093/gbe/evp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo Y, Fu C, Zhang D-Y, Lin K. Overlapping genes as rare genomic markers: The phylogeny of gamma-Proteobacteria as a case study. Trends Genet. 2006;22:593–596. doi: 10.1016/j.tig.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 73.Rokas A, Holland PW. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 74.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 75.Keeling PJ. Genomics. Deep questions in the tree of life. Science. 2007;317:1875–1876. doi: 10.1126/science.1149593. [DOI] [PubMed] [Google Scholar]
- 76.Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 77.Lê Cao K-A, Boitard S, Besse P. Sparse PLS discriminant analysis: Biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lombard J. Early evolution of polyisoprenol biosynthesis and the origin of cell walls. PeerJ. 2016;4:e2626. doi: 10.7717/peerj.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vogel C, Bashton M, Kerrison ND, Chothia C, Teichmann SA. Structure, function and evolution of multidomain proteins. Curr Opin Struct Biol. 2004;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 81.Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423. [Google Scholar]
- 82.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.