Significance
Metabolic plasticity allows cancer cells to adjust their metabolic phenotypes to adapt in hostile environments. There is an urgent need to understand the cross-talk between gene regulation and metabolic pathways underlying cancer metabolic plasticity. We establish a theoretical framework to decode the coupling of gene regulation and metabolic pathways. Our work characterizes a hybrid metabolic state where cells can use both glycolysis and oxidative phosphorylation (OXPHOS) and a possible metabolically inactive state where cells have low activity of both glycolysis and OXPHOS. We show that targeting both OXPHOS and glycolysis may be necessary to eliminate cancer aggressiveness. Our work serves as a platform to target abnormal metabolism in cancer by modulating both genes and metabolic pathways.
Keywords: TNBC, metabolic reprogramming, OXPHOS, Warburg, hybrid
Abstract
Metabolic plasticity enables cancer cells to switch their metabolism phenotypes between glycolysis and oxidative phosphorylation (OXPHOS) during tumorigenesis and metastasis. However, it is still largely unknown how cancer cells orchestrate gene regulation to balance their glycolysis and OXPHOS activities. Previously, by modeling the gene regulation of cancer metabolism we have reported that cancer cells can acquire a stable hybrid metabolic state in which both glycolysis and OXPHOS can be used. Here, to comprehensively characterize cancer metabolic activity, we establish a theoretical framework by coupling gene regulation with metabolic pathways. Our modeling results demonstrate a direct association between the activities of AMPK and HIF-1, master regulators of OXPHOS and glycolysis, respectively, with the activities of three major metabolic pathways: glucose oxidation, glycolysis, and fatty acid oxidation. Our model further characterizes the hybrid metabolic state and a metabolically inactive state where cells have low activity of both glycolysis and OXPHOS. We verify the model prediction using metabolomics and transcriptomics data from paired tumor and adjacent benign tissue samples from a cohort of breast cancer patients and RNA-sequencing data from The Cancer Genome Atlas. We further validate the model prediction by in vitro studies of aggressive triple-negative breast cancer (TNBC) cells. The experimental results confirm that TNBC cells can maintain a hybrid metabolic phenotype and targeting both glycolysis and OXPHOS is necessary to eliminate their metabolic plasticity. In summary, our work serves as a platform to symmetrically study how tuning gene activity modulates metabolic pathway activity, and vice versa.
Abnormal metabolism is an emerging hallmark of cancer (1, 2). Unlike normal cells, cancer cells largely depend on glycolysis to produce energy even in the presence of oxygen, referred to as the Warburg effect (3) or aerobic glycolysis. The up-regulation of glucose transporters (GLUTs) (4) and glycolytic enzymes, such as lactate dehydrogenase (LDH) and hexokinase 2, has been well documented in multiple types of cancer cells (2, 5). Cancer cells can use aerobic glycolysis for rapid ATP production and biomass synthesis to facilitate tumorigenesis and modulate their metastatic potential (6). Moreover, the enhanced glycolytic activity of cancer cells has been shown to be associated with increased therapy resistance (7–9). Notably, the phenomenon of aerobic glycolysis has also been observed in normal cells, such as rapidly proliferating mouse and human lymphocytes (10, 11) and embryonic stem cells (12, 13). These studies suggest that aerobic glycolysis may be a common characteristic of many proliferative animal cells.
It has been becoming clear that in addition to aerobic glycolysis, oxidative phosphorylation (OXPHOS) can also play critical roles in various types of cancer (14–17). For example, the tumor progression and metastatic propensity of several triple-negative breast cancer (TNBC) cell lines and patient-derived xenograft models are largely affected by their energy dependency on fatty acid oxidation (FAO). Pharmacologic repression of FAO activity significantly inhibits in vivo tumor growth (14, 18). The mouse basal BC cell line 4T1, which is part of an isogenic model system, is supermetastatic and commonly used to simulate the human stage IV BC (19). Compared with its isogenic nonmetastatic 67NR cells, 4T1 cells exhibit both enhanced OXPHOS and increased glycolytic activities (20). Moreover, the circulating tumor cells (CTCs) derived from 4T1 cells show significantly higher mitochondrial respiration and biogenesis activities compared with both its primary tumors and its lung metastases (15). Notably, there is no observable decrease in glycolytic activity of these 4T1 CTCs, indicating the coexistence of OXPHOS and glycolysis. The association of high OXPHOS activity with high metastatic potential has also been observed in mouse melanoma B16-M4b cells and human cervical cancer SiHa-F3 cells (16). All these indicate that cancer cells are able to utilize both glycolysis and OXPHOS, depending on its circumstances. However, it is still largely unknown how cancer cells orchestrate the metabolic pathway activity through gene regulation to facilitate malignancy.
Mathematical modeling approaches have been employed to help elucidate the aforementioned metabolic reprogramming in cancer. Constraint-based models including flux-balance analysis (FBA) based on conservation of mass (21, 22) have been the most widely used approaches to simulate cancer metabolism (23, 24). Applications include quantifying the extent of aerobic glycolysis of the NCI-60 cell lines (25), characterizing their primary uptake pathways (26), and identifying the change of their metabolic states upon gene knockdown and/or drug treatment (27). In addition to characterizing the metabolic flux change in cancer cells, modeling efforts have also been made to identify anomalous gene activity involved in cancer metabolism. Such studies include modeling the effects of reactive oxygen species (ROS) and antioxidants on hypoxia-inducible factor 1 (HIF-1), a master regulator of glycolysis (28), and our previous work modeling the interplay between HIF-1 and 5′ AMP-activated protein kinase (AMPK), a master regulator of OXPHOS and mitochondrial biogenesis (29). These computational works offer a quantitative and dynamical perspective of cancer metabolism mostly focusing on either metabolic pathway activity or gene activity. However, the alteration of the metabolic activity is coupled with the change in gene activity, and vice versa. For example, HIF-1, which is often stabilized in cancer cells due to hypoxia, promotes glycolytic activity by up-regulating the expression of GLUT genes, such as GLUT1, and glycolytic enzyme genes, such as pyruvate kinase isozyme M2 (PKM2) (30). Up-regulation of PKM2 in turn promotes HIF-1 transactivation and the products of glycolysis, such as lactate, further stabilize HIF-1, thus forming self-enforcing feedback loops of HIF-1 activity (31). Thus, to comprehensively characterize cancer metabolic reprogramming, a mathematical modeling framework integrating gene regulation with metabolic pathways is urgently needed.
Previously, to decipher the genetic interplay between glycolysis and OXPHOS, we developed a mathematical model focusing on a core regulatory circuit, composed of AMPK, HIF-1, and ROS (29). Computational modeling of this circuit showed that cancer cells can robustly acquire three stable steady states—W (HIF-1high/pAMPKlow), O (HIF-1low/pAMPKhigh), and W/O (HIF-1high/pAMPKhigh)—corresponding to a glycolysis phenotype, an OXPHOS phenotype, and a hybrid metabolic phenotype, in which cancer cells use both glycolysis and OXPHOS. Here, the AMPK activity is represented by the level of phosphorylated AMPK (pAMPK) at threonine-172 of the α subunit. The emergence of the hybrid metabolic phenotype of cancer cells from our modeling analysis has been supported by recent experimental evidence (14–17, 20). We argued that the hybrid metabolic phenotype exhibits metabolic plasticity to adapt to varying microenvironments and hence tends to be more aggressive relative to cells in a more glycolysis or OXPHOS phenotype (17).
Here, to capture the coupling of gene activity and metabolic pathway activity, we extend our AMPK:HIF-1:ROS model by coupling it to three distinct metabolic pathways, glycolysis, glucose oxidation, and FAO. Unlike traditional FBA, our model captures gene regulation and pathway activity and their interactions via chemical rate equations. The extended model elucidates the detailed association of AMPK and HIF-1 activities with metabolic pathway activities for each stable state. High AMPK activity associates with high OXPHOS pathway activity (glucose oxidation and/or FAO) and high HIF-1 activity associates with high glycolysis activity. Particularly, the hybrid metabolic state W/O is characterized by HIF-1high/pAMPKhigh and high activity of both glycolysis and OXPHOS (glucose oxidation and/or FAO). Our predicted association of AMPK/HIF-1 activity with metabolic pathway activity is confirmed by analyzing well-annotated metabolomic and transcriptomic data from a BC patients’ cohort. This utilizes metabolic pathway activity signatures developed in the present work as well as our published AMPK and HIF-1 signatures (29). This result was further validated in human invasive breast carcinoma and hepatocellular carcinoma (HCC) using RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA). For direct experimental validation, we use two TNBC cell lines, SUM159-PT and MDA-MB-231, and confirm that metastatic TNBC cells can acquire a stable hybrid metabolic phenotype. We further show that repressing the glycolytic activity activates AMPK and enhances OXPHOS activity, and conversely repressing mitochondrial function up-regulates multiple glycolysis genes and increases glycolytic activity. Finally, a combination of glycolytic and OXPHOS inhibitors that potentially eliminates the existence of the hybrid metabolic phenotype exhibits maximum reduction of proliferation and clonogenicity of these TNBC cells. In summary, through integrating mathematical modeling, bioinformatics, and experiments we demonstrate a direct association of the AMPK/HIF-1 activity with metabolic pathway activity and investigate the existence of the aggressive hybrid metabolic phenotype.
Results
Coupling the AMPK:HIF-1:ROS Circuit with Glycolysis/OXPHOS Pathways.
We constructed the metabolic regulatory network from an extensive literature survey featuring important gene regulation and energy pathways and their cross-talk (Fig. 1). The details of the network construction can be found in SI Appendix, section 1. To create a computational model which can capture the basic principles of the coupling between gene regulation and metabolic pathways, we coarse-grain the extensive regulatory system (Fig. 1) into a minimum network consisting of the AMPK:HIF-1:ROS core regulatory circuit and the three metabolic pathways, glycolysis, glucose oxidation, and FAO (Fig. 2). AMPK and HIF-1 form mutually inhibitory feedback loops and AMPK reduces while HIF-1 increases NOX-derived ROS (noxROS), which in turn activates both AMPK and HIF-1 (29). We now extend this AMPK:HIF-1:ROS model by explicitly including the metabolic pathways. The effective self-activation of HIF-1 is through its interaction with the glycolytic pathway and the effective self-inhibition of AMPK is due to its interaction with OXPHOS processes. One critical component of our modeling is the direct consideration of glucose levels and acetyl-CoA levels. Glucose is the competing resources for glycolysis and glucose oxidation and acetyl-CoA, as the only fuel entering the TCA cycle, is the common intermediate of both glucose oxidation and FAO. The uptake rate of glucose is regulated by AMPK and HIF-1 and the utilization rate of acetyl-CoA for the TCA cycle is restricted by the mitochondrial capacity, which is regulated by AMPK. The detailed formulation of the mathematical model representing the dynamics of pAMPK, HIF-1, mitochondrial ROS (mtROS), noxROS, ATP, glucose, and acetyl-CoA together with the rates of glycolysis, glucose oxidation, and FAO is provided in Materials and Methods. To reconcile the different time scales of gene regulation and metabolic flux, we always assume that the metabolite concentration and the pathway activities are in a steady state at certain levels of pAMPK and HIF-1 since metabolic processes are much faster than gene regulation. We propose that our coarse-grained network captures the key features of the more comprehensive set of interactions indicated in Fig. 1 and in particular is sufficient to explain important experimental observations on the coupling of gene activity and metabolic pathway activity.
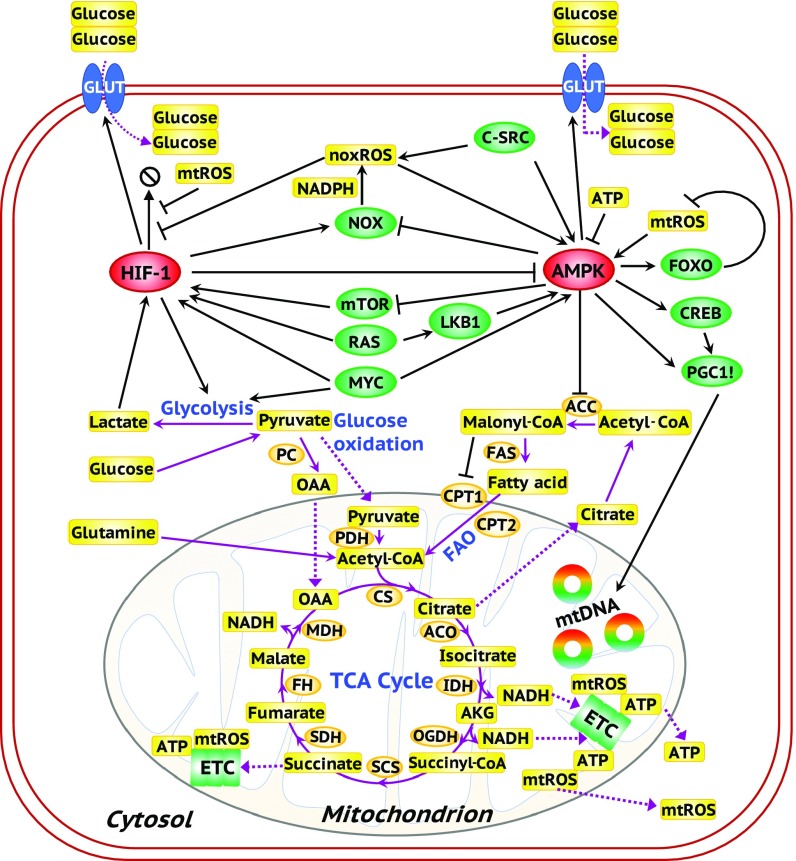
Fig. 1.
A comprehensive regulatory network of glycolysis and OXPHOS. The ovals represent genes. Red ovals highlight the master regulators AMPK and HIF-1. Green ovals represent downstream target genes of the master regulators and oncogenes. Orange ovals represent the enzyme genes. Yellow rectangles represent metabolites. Black arrows represent excitatory regulation and black bar-headed arrows represent inhibitory regulation. Purple solid lines represent the chemical reactions in metabolic pathways and purple dotted lines represent the transportation of metabolites.
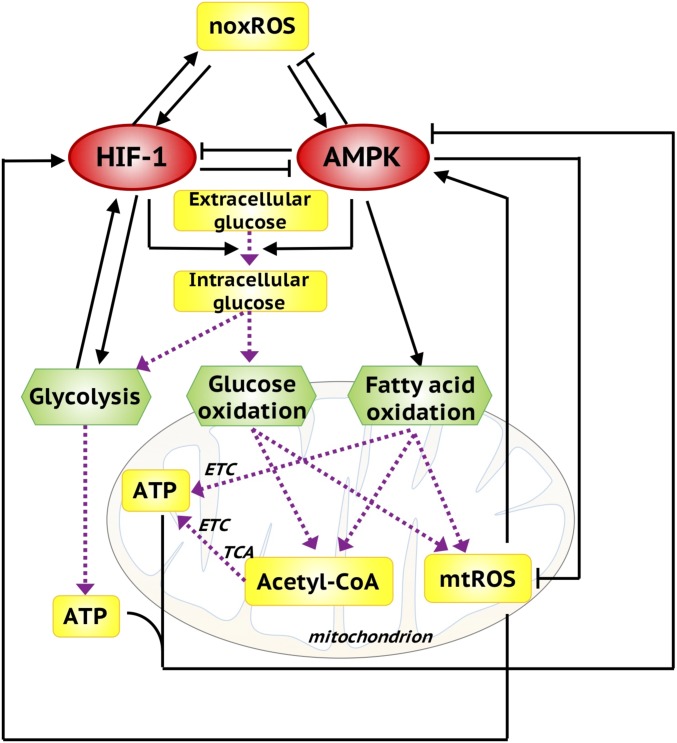
Fig. 2.
The metabolic regulatory network, coupling the core AMPK:HIF-1:ROS circuit with three metabolic pathways. AMPK and HIF-1 are the master regulators of OXPHOS and glycolysis. Both AMPK and HIF-1 can promote glucose uptake. The intracellular glucose can be used by glycolysis and glucose oxidation. Both glucose oxidation and FAO can generate acetyl-CoA to fuel the TCA cycle and consequently the production of ATP via ETC. mtROS and ATP can in turn regulate the activity of AMPK and HIF-1. Here, the black solid arrows/bar-headed arrows represent regulatory links. The purple dotted arrows represent the metabolic pathways.
Genetic and Metabolic Characterization of Each Metabolism Phenotype.
First, we analyze the mathematical equations that were derived to simulate the coarse-grained network using two sets of parameters corresponding to normal and cancer cells, respectively. Cancer cells often have higher mtROS production rate due to reprogrammed mitochondria (16) and more stabilized HIF-1 due to the typically hypoxic conditions. Therefore, the value of the parameters representing the production rates of mtROS during OXPHOS (glucose oxidation and FAO) is taken to be larger and the value of the parameter representing the degradation rate of HIF-1 smaller for cancer relative to normal cells. The values of the other parameters are unchanged. We will consider in more detail the effects of mtROS production rate and HIF-1 degradation rate on cancer metabolic plasticity in a later section.
To identify the robust stable metabolic states enabled by the regulatory network (Fig. 2), we utilize a parameter randomization approach. The overall strategy involves randomizing the modeling parameters for each simulation and collecting all stable steady solutions for statistical analysis, by which the most significant solution patterns can be identified (32, 33). As expected, the solution patterns are conserved even in the presence of large parameter perturbations due to restraints from the network topology (i.e., extensive cross-talk of regulatory proteins and energy pathways). We consider 1,000 sets of model parameters and for each set the value of each parameter, except for the fixed values of mtROS production rate and HIF-1 degradation rate that distinguish cancer cells from normal cells, is randomly sampled from (75%, 125%), where is the baseline value. We collect all of the stable-state solutions and use unsupervised hierarchical clustering analysis (HCA) to identify the patterns present in the solution set. HCA shows that the stable-state solutions form three large clusters; one is characterized by high pAMPK/mtROS/G1/F and low HIF-1/noxROS/G2 (G1 represents the glucose oxidation rate, F represents the FAO rate, and G2 represents the glycolysis rate), corresponding to an OXPHOS state; one is characterized by high HIF-1/noxROS/G2 and low pAMPK/mtROS/G1/F, corresponding to a glycolytic state; and one is characterized by high pAMPK/mtROS/G1/F and high HIF-1/noxROS/G2, corresponding to a hybrid metabolic state (Fig. 3A and SI Appendix, Fig. S1).
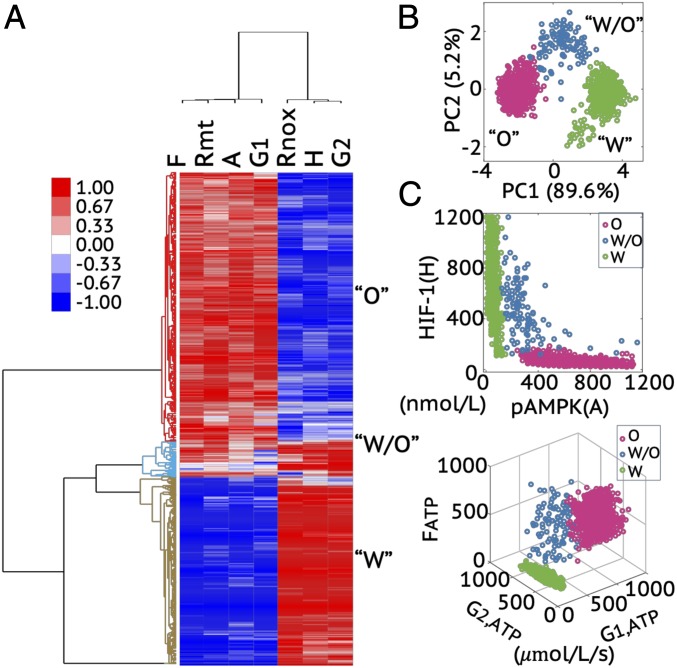
Fig. 3.
Modeling prediction of the association between AMPK/HIF-1 activity and metabolic pathway activity. (A) HCA of the stable-state solutions from 1,000 sets of parameters. Each row represents one stable-state solution, referred to as one sample here, and each column represents the levels of a regulatory protein, metabolite, or the rates of one metabolic pathway. In A, F represents FAO rate, Rmt represents mtROS level, A represents pAMPK level, G1 represents glucose oxidation rate, Rnox represents noxROS level, H represents HIF-1 level, and G2 represents glycolysis rate. The solutions can be clustered into three main groups, referred to as clusters O, W/O, and W, as marked by different colors in the dendrogram. (B) PCA of the clustered samples in A. (C, Top) The pAMPK and HIF-1 levels of the clustered samples in A. (C, Bottom) The metabolic pathway activities of the clustered samples in A. The colors representing different clusters, O, W/O, and W, are consistent with those used in B.
We also visualized the three clusters by projecting the stable-state solutions onto the first and second principal components (PC1 and PC2) through principal component analysis (PCA) of all solutions (Fig. 3B), onto the pAMPK and HIF-1 axes, and onto the G1, G2, and F axes (Fig. 3C). These results indicate that cells in the W state mostly use glycolysis for ATP production, cells in the O state mainly use OXPHOS (including glucose oxidation and FAO) for ATP production, and cells in the hybrid W/O state can utilize all three metabolic pathways to generate ATP (Fig. 3C). Thus, the modeling analysis demonstrates an association of high AMPK activity with high OXPHOS activity, and high HIF-1 activity with high glycolytic activity. This was conjectured to be the case in our previous work which, however, did not include any explicit analysis of the metabolic processes. Similarly, we performed an analogous analysis for the normal cells. However, the hybrid metabolic state is rarely observed among the stable-state solutions from 1,000 sets of randomized parameters (SI Appendix, Fig. S2).
The Effects of HIF-1 Degradation Rate and mtROS Production Rate in Modulating Cancer Metabolic Phenotypes.
The modeling framework can be utilized to analyze the effects of various kinds of perturbations on cancer metabolic phenotypes. In this section, using the HIF-1 degradation rate and the mtROS production rate as two examples, we will show how changing these two variables would modify cancer metabolism phenotype. When studying the effect of HIF-1 degradation, we keep the mtROS production rate fixed, and vice versa.
To analyze the effect of the HIF-1 degradation rate, three values of , representing relatively high, moderate, and low degradation rate of HIF-1, are selected. For each value of , we randomly sampled all other parameters using the aforementioned randomization procedure. Again, we collect all stable-state solutions from 1,000 sets of parameters and use HCA and PCA to classify the solutions. We find that increasingly stabilized HIF-1, represented by a lower degradation rate, enables a higher percentage of the hybrid metabolic and glycolytic states and lower percentage of the OXPHOS states (Fig. 4A). Similarly, we performed an analogous analysis to analyze the effect of mtROS production rate. We find that a higher mtROS production rate enables a higher percentage of the hybrid metabolic and OXPHOS states and a lower percentage of the glycolytic states (Fig. 4B). Interestingly, although both stabilization of HIF-1 and elevated production of mtROS can promote the hybrid metabolic state, their effects on the glycolysis and OXPHOS state are opposite. The results here are indicative of the fact that a hybrid metabolic state is rarely observed in normal cells and this appears to be due to both the unstable HIF-1 and relatively low production rate of mtROS in normal cells relative to cancer cells. Another interesting observation is the emergence of a cluster characterized by low activities of AMPK and HIF-1 and low rates of glycolysis and OXPHOS, referred to as the “low-low” cluster. This state appears especially when the degradation rate of HIF-1 is high and/or the production rate of mtROS is low (Fig. 4). In the section Evaluating Metabolic Pathway Activity by Enzyme Gene Expression, a subpopulation of single BC cells with the low-low characterization is identified and the significance of such a quiescent metabolic phenotype is discussed.
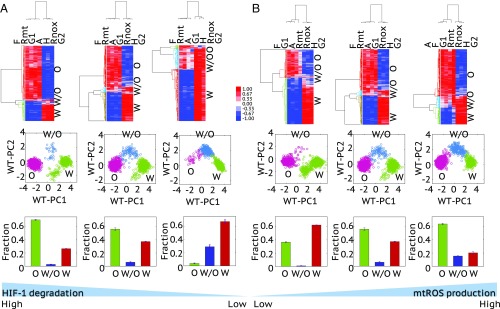
Fig. 4.
The effects of the HIF-1 degradation rate (A) and the mtROS production rate (B) on metabolic phenotypes. (A, Top) HCA of the stable-state solutions (referred to as samples here) of 1,000 sets of parameters with the degradation rate of HIF-1 being 0.45 , 0.25 , and 0.05 (from left to right). (A, Middle) Projection of the clustered samples at the top onto the PC1 and PC2 generated by the wild-type samples with the degradation rate of HIF-1 being 0.25 (referred to as WT-PC1 and WT-PC2). (A, Bottom) The fractions of the metabolic states, O, W/O, and W, corresponding to the top. The analysis was repeated three times and error bars were added. (B, Top) HCA of the stable-state solutions of 1,000 sets of parameters with the production rate of mtROS being 30, 50, and 80 μM/min (from left to right). (B, Middle) Projection of the clustered samples in the corresponding top onto the PC1 and PC2 generated by the wild-type samples, with the production rate of mtROS being 50 μM/min. (B, Bottom) The fractions of the metabolic states, O, W/O, and W, corresponding to the top. The analysis was repeated three times and error bars were added. The middle figures in A and B are the same with the parameters and = 50 μM/min, representing the wild type. Z-scores of the stable-state solutions were used for clustering analysis and PCA. The solutions of all scenarios here were normalized using the mean and SD of the wild type.
Evaluating Metabolic Pathway Activity by Metabolite Abundance.
To test the predicted genetic and metabolic characterization of differing cancer metabolism phenotypes, we wish to compare the AMPK/HIF-1 activity and the metabolic pathway activity using metabolomics and transcriptomics data from BC patients’ samples. Note, however, that the active form of AMPK is its phosphorylated form (pAMPK) and the most important property of HIF-1 is protein stability; neither of these features can be directly captured by the mRNA expression of AMPK and HIF-1. In the previous work, we developed AMPK and HIF-1 signatures to quantify the activity levels of AMPK and HIF-1 by evaluating the expression of their downstream target genes (a total of 33 AMPK downstream genes and 23 HIF-1 downstream genes) (29). The AMPK and HIF-1 signatures were derived by performing PCA on the gene expression data independently for AMPK- and HIF-1–downstream genes, from which the first principal components (PC1s) are used to quantify the activity of AMPK and HIF-1. The AMPK and HIF-1 signatures have been shown to capture the key metabolic features of multiple types of tumor samples from TCGA, such as invasive breast carcinoma, HCC, and lung adenocarcinoma (LUAD). Similar findings were also observed in the single-cell analysis of LUAD (29). Particularly, a significantly strong anticorrelation between the AMPK activity and the HIF-1 activity has been observed across the aforementioned tumor samples and single cells, where there is no such clear correlation observed between AMPK and HIF-1 gene expression (SI Appendix, Figs. S3 and S4).
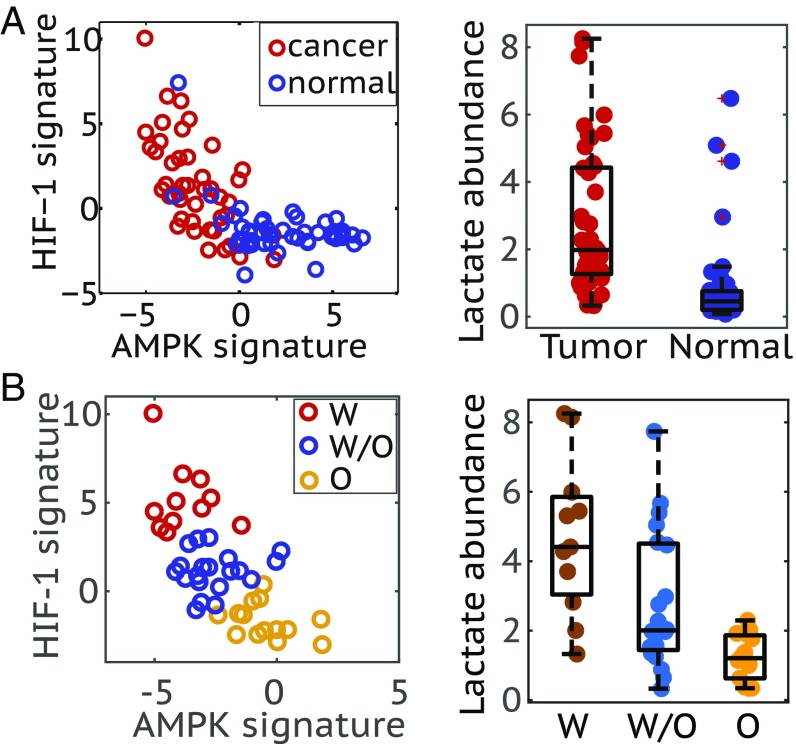
Here we apply the AMPK and HIF-1 signatures to quantify the AMPK and HIF-1 activity of 45 human BC samples and 45 corresponding adjacent benign breast tissue samples (34) (see Materials and Methods for more details). The BC samples show significantly higher HIF-1 activity and lower AMPK activity on average relative to the adjacent benign breast tissues (Fig. 5A, Left), indicating the enhanced glycolytic activity in the BC samples. Moreover, the cancer samples are more stretched in the space of AMPK and HIF-1 signatures, with some samples exhibiting very high HIF-1 activity and some samples exhibiting very low HIF-1 activity relative to benign tissues, suggesting heterogeneity in cancer metabolic activities (Fig. 5A, Left). Moreover, a strong anticorrelation between AMPK and HIF-1 activity is identified across the BC samples, which is consistent with our previous results (29).
Fig. 5.
Association of the AMPK/HIF-1 activity with lactate abundance in human breast tumor samples. (A, Left) Evaluating the AMPK and HIF-1 activities of BC patients’ samples (n = 45) and the corresponding normal samples (n = 45). (A, Right) Box plot for lactate abundance in tumor and normal samples (P < 0.0001). (B, Left) Strong anticorrelation of AMPK and HIF-1 activity of tumor samples (Pearson correlation, r = −0.67, P < 0.0001). The standard k-means clustering analysis was applied to group the tumor samples into the W, O, and W/O states. (B, Right) Box plot for lactate abundance of samples in the W, O, and W/O states (PW-W/O < 0.05, PW/O-O < 0.01).
Our model directly connects these regulatory activities to actual metabolic flux. To test our approach, we need to characterize metabolic pathway activity. We first consider using metabolite abundance. HCA were performed on BC samples and/or benign tissue samples based on the abundance of major metabolites involved in glycolysis, TCA and FAO (SI Appendix, Fig. S5). The full list of metabolites used here can be found in SI Appendix, Table S1. The clustering result shows that relative to the benign tissues BC samples have higher abundance of most metabolites (SI Appendix, Fig. S5), and among BC samples the samples exhibit either high abundance of most metabolites or low levels of almost all metabolites (SI Appendix, Fig. S6). There are no signs of specific metabolic states. These results suggest that using the static metabolite abundance may not be informative in evaluating the metabolic pathway activity. Since most of the metabolites are pathway intermediates, their abundance may not be indicative of the pathway activity since both active metabolic flux (active production) or stalled metabolic flux can cause the accumulation of an intermediate. However, if the metabolite is the end product of a metabolic pathway, its abundance should be indicative of the pathway activity. Since lactate is the end product of glycolysis, we can use its abundance to evaluate the glycolytic activity. The BC samples clearly show higher average abundance of lactate relative to the benign tissues (Fig. 5A, Right). This indicates higher glycolytic activity in cancer tissues and is consistent with the predictions by the AMPK and HIF-1 signatures.
The 45 BC tissues are further classified into three groups based on their AMPK and HIF-1 activities using k-means clustering (Fig. 5B, Left). Among the three groups, tumor samples characterized with highest HIF-1 activity, labeled as W, exhibit highest abundance of lactate, tumor samples characterized with highest AMPK activity, labeled as O, exhibit lowest abundance of lactate, and tumor samples characterized with both AMPK and HIF-1 activities, labeled as W/O, show intermediate levels of lactate (Fig. 5B, Right).
Motivated by the ability of lactate abundance to distinguish different metabolic states, we also searched for other metabolites that exhibit significantly differential levels between BC samples and benign tissue samples. Such metabolites include phosphoethanolamine, glutamate, fumarate, and cystine in addition to lactate (see the volcano plots in SI Appendix, Fig. S7). Phosphoethanolamine shows the most differential abundance between BC and benign tissues (log2 tumor/normal = 11.16) and its enrichment in BC has been frequently observed while poorly studied (35, 36). Glutamate enrichment that is often due to the overexpression of glutaminase is a characteristic of BC (37). Accumulation of fumarate, referred to as an oncometabolite, has been shown to trigger epithelial–mesenchymal transition in renal cancer (38). Last, but not least, cystine enrichment in BC samples identified here is reminiscent of the result that cystine deprivation can induce rapid programmed necrosis in the basal-like BC cells (39). We also perform an analogous analysis to identify the significantly differentially enriched metabolites among groups W, W/O, and O, and such metabolites include glutamate and lactate. Lists of the significantly differentially enriched metabolites are provided in SI Appendix, Tables S2–S4. All told, however, there is no simple way to accurately estimate metabolic pathway activity directly from metabolite abundance.
Evaluating Metabolic Pathway Activity by Enzyme Gene Expression.
To more directly evaluate the metabolic pathway activities, we develop a metabolic pathway scoring metric by evaluating the gene expression of key enzymes involved in specific metabolic pathways. The assumption here is that higher metabolic pathway activity would require higher levels of enzymes functioning in that pathway. With this assumption, a total of 14 enzyme genes of FAO, 10 enzyme genes of TCA, and 8 enzyme genes of glycolysis were selected as the signature genes to evaluate the activities of OXPHOS and glycolysis. The full list of all enzyme genes used here can be found in Materials and Methods. To unbiasedly test for the association of the AMPK and HIF-1 activities with the metabolic pathway activities, we ensured that the genes used in constructing the AMPK and HIF-1 signatures do not overlap with the genes comprising the metabolic pathway scoring metric. The metabolic pathway scoring metric is defined as the average gene expression of relevant enzymes for each pathway. A detailed formulation of this metric can be found in Materials and Methods.
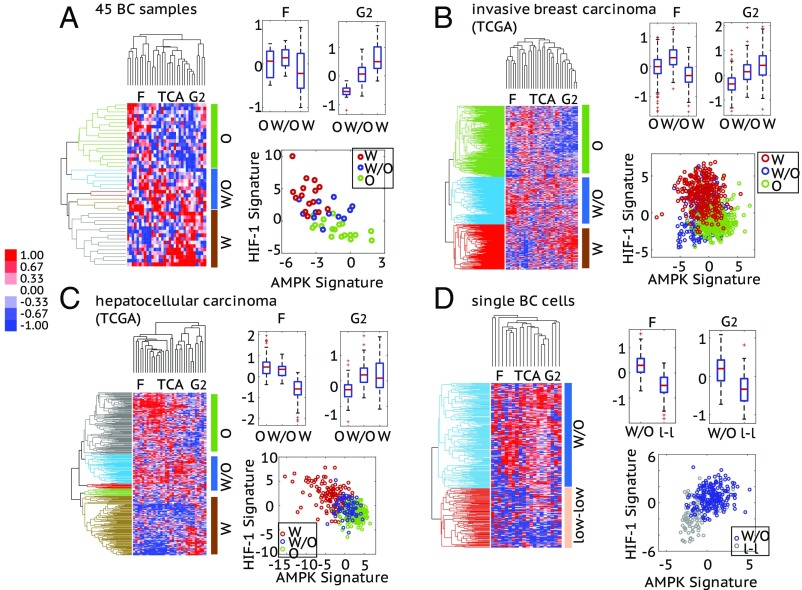
First, we use the metabolic pathway scoring metric to quantify the pathway activities of the 45 BC patients’ samples. We perform HCA on the microarray data for the enzyme genes and classify the samples into three groups with the most significant gene expression patterns; group W contains samples that have high expression of glycolytic enzyme genes and low expression of OXPHOS enzyme genes, group O contains samples that have high expression of OXPHOS enzyme genes and low expression of glycolytic enzyme genes, and group W/O contains samples that have high expression of both OXPHOS and glycolytic enzyme genes, indicating the use of both metabolism phenotypes by the tumor (Fig. 6A, Left). Then, we calculate the pathway scores for each group. The glycolysis score shows a significantly decrease from group W, to group W/O to group O, and the FAO score of groups O and W/O is significantly higher than that of group W (Fig. 6A, Right). We then evaluate the activity of AMPK and HIF-1 of each sample by the AMPK and HIF-1 signatures. Strikingly, members of group W characterized by high expression of glycolytic enzymes show high HIF-1 activity and members of group O characterized by high expression of TCA and FAO enzymes show high AMPK activity and, finally, members of group W/O characterized by high expression of both glycolysis and OXPHOS enzyme genes show both AMPK and HIF-1 activities (Fig. 6A, Right). Importantly, although there are only 45 samples analyzed, a distinct gene expression pattern of enzymes is clearly observed and strongly associated with the AMPK/HIF-1 activities, as we predicted using our model.
Fig. 6.
Association of the AMPK/HIF-1 activity with the metabolic pathway activity in 45 BC samples (A), 1,100 invasive breast carcinoma samples from TCGA (B), 373 HCC samples from TCGA (C), and 317 single BC cells (D). (A–C, Left) Average linkage HCA of enzyme gene expression (microarray data for A and RNA-seq data for B and C) of patient samples. The similarity metric used here is based on Pearson correlation. Each row represents a patient sample and each column represents the expression of one enzyme gene. Three major enzyme gene states, O, W/O, and W, are identified and highlighted by different colors in the dendrogram. The cutoff values to get these clusters are 0.04 for A, 0.1 for B, and 0.06 for C. (Right, Top) Box plots for the FAO (F) and glycolysis (G2) scores of the clustered patient samples. For 45 BC samples: FAO score, , , ; glycolysis score, , , . For invasive breast carcinoma: FAO score, , , ; glycolysis score, , , . For HCC: FAO score, , , ; glycolysis score, , , . (Right, Bottom) The AMPK and HIF-1 signatures of the clustered patient samples. Here, each hollow dot represents one patient sample. (D, Left) Average linkage HCA of the RNA-seq data of enzyme genes of single BC cells. The enzyme genes whose expression was detected in more than 150 single cells (approximately half of the total) were used to do clustering. The similarity metric used here is based on Pearson correlation. Each row represents a single cell and each column represents the expression of one enzyme gene. Two major enzyme gene states—W/O and low-low—are identified and highlighted by different colors in the dendrogram. The cutoff value to get these two clusters is 0.009. (Right, Top) Box plots for the F and G2 scores of cells in the W/O and low-low clusters. FAO score, ; glycolysis score, . (Right, Bottom) The AMPK and HIF-1 signatures of the single cells in the W/O cluster and the low-low cluster. For the heat maps in A–D, the names of the columns are listed as the names of the metabolism pathways and the full list of enzyme genes can be found in SI Appendix, Table S8. The TCA score of each case is shown in SI Appendix, Fig. S8. For all box plots here, P values for a balanced one-way ANOVA are calculated.
We further extended the analysis of pathway activity and AMPK/HIF-1 activities using patient data from TCGA. We used HCA to classify 1,100 invasive breast carcinoma samples (Fig. 6B, Left) and 373 HCC samples (Fig. 6C, Left) into three groups respectively based on the aforementioned enzyme gene expression patterns and calculated their pathway scores. Consistent with what we found in the 45 BC samples, TCGA analysis also allows for the identification of three groups (W, O, and W/O). Since we have data from a relatively large number of samples, the three metabolic groups of invasive breast carcinoma and HCC samples exhibit more significant differences, especially among the FAO and glycolysis scores (Fig. 6 B and C, Right). As predicted, invasive breast carcinoma and HCC samples with high glycolysis score showed high HIF-1 activity, samples with high FAO score showed high AMPK activity, and samples with high/intermediate scores of FAO and glycolysis exhibited both AMPK and HIF-1 activities (Fig. 6 B and C, Right). All these confirm the modeling-predicted association of AMPK/HIF-1 activity with pathway activities.
We further validated the association of the AMPK/HIF-1 activity with pathway activity at the single-cell level. We looked into the single-cell RNA-seq data of 317 BC cells (40), which are classified into a W/O cluster and a low-low cluster (Fig. 6D, Left), containing cells having high and low expression of both glycolytic and OXPHOS enzyme genes, respectively. Cells in the W/O cluster exhibit significantly higher FAO, TCA, and glycolysis scores compared with cells in the low-low cluster (Fig. 6D, Right and SI Appendix, Fig. S8D). As predicted, cells in the W/O cluster exhibit both high AMPK and high HIF-1 activity and cells in the low-low cluster exhibit low activity of both (Fig. 6D, Right). To compare the metabolic state of these BC single cells with benign breast tissues, we projected these BC cells to the AMPK/HIF-1 axes generated by the 45 BC samples and 45 adjacent benign tissue samples (SI Appendix, Fig. S9). A consistent AMPK/HIF-1 activity characterization is observed for cells in the W/O cluster and low-low cluster. Intriguingly, some cells in the low-low cluster exhibit even lower HIF-1 activity relative to benign tissues (SI Appendix, Fig. S9). These results confirm the association of the AMPK/HIF-1 activity with pathway activity at the single-cell level and also demonstrate the presence of single-cell hybrids.
In summary, although the AMPK and HIF-1 activity and the metabolic pathway activity were evaluated independently using different sets of genes, the strong correlation observed at both the tumor level and the single-cell level supports the prediction of the model regarding the genetic and metabolic characterization of each metabolic phenotype.
Experimental Validation of the Coupling of AMPK/HIF-1 Activity with Metabolic Pathway Activity in Metastatic Cancer Cells.
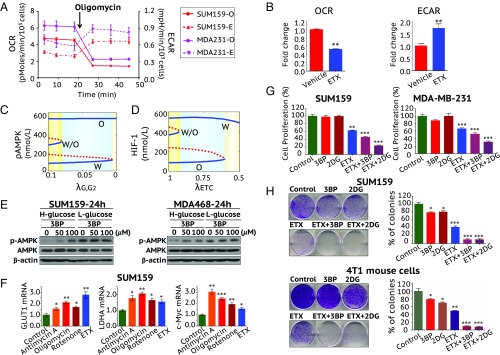
To further validate the predictions regarding the coupling of gene activity and metabolic pathway activity in cancer cells, we used various metabolic inhibitors to perturb the glycolysis and/or FAO activity and analyzed the change in AMPK and HIF-1 activity in BC cells. First, we show that the metastatic TNBC cells MDA-MB-231 and SUM-159-PT, that have a significant dependency on mitochondrial FAO (14), exhibit a hybrid metabolic phenotype with both high OXPHOS and high glycolysis, as measured by Seahorse respiration analysis (Fig. 7 A and B). When these TNBC cells were treated with the mitochondrial complex V inhibitor oligomycin or the FAO inhibitor etomoxir (ETX), which prevents the entry of fatty acid into mitochondria by inhibiting the FAO rate-limiting enzyme CPT1, oxygen consumption rate (OCR) was sharply decreased in both MDA-MB-231 and SUM-159-PT cells (Fig. 7 A and B). Interestingly, oligomycin- or ETX-mediated reduction in respiration resulted in a simultaneous increase in glycolysis represented by extracellular acidification rate (ECAR). This confirmed the metabolic plasticity of these cells.
Fig. 7.
TNBC cells exhibiting a hybrid metabolic phenotype that requires treatment with both glycolytic and OXPHOS inhibitors. (A) Seahorse XF analysis suggesting that the metastatic TNBC cells MDA-MB-231 and SUM159-PT exhibit a hybrid metabolic phenotype. While addition of the respiratory inhibitor oligomycin decreased the mitochondrial respiration (OCR, solid lines), it immediately increased the glycolysis (ECAR, dotted lines). (B) Fold change in OCR and ECAR after FAO inhibitor ETX treatment in SUM159-PT cells. (C) Bifurcation diagram of pAMPK levels in response to inhibition of the glycolytic pathway (G2). represents the strength of inhibition of G2. The smaller the , the stronger the inhibition. (D) Bifurcation diagram of HIF-1 levels in response to inhibition of ETC. represents the strength of inhibition of ETC. The smaller the , the stronger the inhibition. More details to calculate D can be found in SI Appendix, section 5. (E) Western blotting analysis of AMPK and pAMPK after the cells were treated with 3BP for 24 h in high-glucose (4.5 g/L) or low-glucose (1 g/L) medium. (F) Cells were treated with the mitochondrial ETC complex I inhibitor rotenone (10 nM), the complex III inhibitor antimycin-A (10 μM), the complex V inhibitor oligomycin (5 μg/mL), or ETX (100 μM) for 24 h. Expression of glycolytic genes (GLUT1, LDHA, and c-Myc) was determined by qPCR. Glycolytic and OXPHOS inhibitors inhibited the cell proliferation (G) and the colony formation (H) in SUM159-PT and mouse metastatic basal BC 4T1 cells. Use of both glycolytic and OXPHOS inhibitors effectively eliminates the colony-formation potential of SUM159-PT and 4T1 cells. All data are presented as the means ± SE. *P < 0.05, **P < 0.01, ***P < 0.001 (two-tailed t test).
To analyze how altering metabolic pathway activity affects the gene activity, we first use the model (Fig. 2) to simulate how repressing the glycolytic pathway affects AMPK activity. Increasing inhibition of glycolysis first excludes the existence of the hybrid metabolic state, and then the glycolytic state, and finally only the OXPHOS state remains (Fig. 7C). In other words, the cell populations are expected to automatically switch to increased OXPHOS via increased average pAMPK levels upon glycolytic inhibition. We also analyze how repressing the electron transport chain (ETC) affects HIF-1 activity. Increasing inhibition of ETC first excludes the existence of the hybrid state, and then the OXPHOS state, and finally only the glycolysis state remains (Fig. 7D). To test the model prediction, we used TNBC metastatic cell models SUM-159-PT, MDA-MB-468, and MDA-MB-231. Cells were treated with a glycolytic inhibitor, 3-bromopyruvate (3BP), in low- and high-glucose medium. We found that AMPK was activated, that is, the levels of pAMPK were up-regulated by 3BP treatment, especially in the high-glucose condition (Fig. 7E and SI Appendix, Fig. S10). Combination therapy with the FAO inhibitor ETX partially abolished the 3BP-induced up-regulation of pAMPK (SI Appendix, Fig. S10). When the mitochondrial function was inhibited using ETC or FAO inhibitors, the expression of the glycolysis-relevant genes, including GLUT1, LDHA, and c-Myc, was significantly up-regulated, indicating the increased glycolytic activity at the population level (Fig. 7F). These experimental results support the model prediction and indicate that cancer cells have the metabolic plasticity to switch between glycolysis and OXPHOS as a compensatory strategy in response to metabolic drugs.
To further understand the functional significance of various metabolism phenotypes in the tumor properties, we performed proliferation assays and clonogenic assays using SUM-159-PT and mouse metastatic basal BC 4T1 cells. While the glycolytic inhibitors 3BP and 2-deoxyglucose (2-DG) showed minor inhibition in their clonogenic potential but not in proliferation, their combination with ETX caused a major reduction in both the proliferation and clonogenic potential of both of the cell lines (Fig. 7 G and H). This suggests that disrupting the survival potential of either tumors or individual cells with hybrid metabolic status requires targeting both metabolic pathways.
Discussion
For a long time, aerobic glycolysis has been regarded as the dominant metabolic phenotype in cancer. However, there has recently been increasing experimental work demonstrating a critical role of OXPHOS and mitochondrial biogenesis in tumorigenesis and metastasis. Apparently, cancer cells are able to adjust their metabolism phenotypes to adapt to the microenvironment. In this regard, we showed that cancer cells can acquire a stable hybrid metabolic state by a demand-sensitive cross-talk of regulatory proteins and energy pathways (29).
In this study, we established a theoretical framework (Fig. 2) that couples the gene regulatory circuit with metabolic pathways to explore the genetic and metabolic interplay between glycolysis and OXPHOS. The model predicts a direct association of high AMPK activity with high OXPHOS activity and high HIF-1 activity with high glycolysis activity. To validate this prediction, we developed signatures to quantify the activity of metabolic pathways and could therefore show the association of the pathway activity with the AMPK and HIF-1 activities, evaluated by our previously defined AMPK and HIF-1 signatures. By applying the pathway signatures and the AMPK/HIF-1 signatures, we confirmed their association and the existence of the hybrid metabolic phenotype at both the tumor level and the single-cell level. Furthermore, we performed experiments in TNBC using MDA-MB-231 and SUM-159-PT cells, which stably maintain a hybrid metabolic phenotype. Inhibiting the glycolytic activity in these TNBC cells activates AMPK. With the use of selective inhibitors, we show that cells in the hybrid metabolic phenotype exhibit maximum proliferation and clonogenicity relative to cells in a more glycolytic or more OXPHOS phenotype. Our work couples gene regulation with metabolic regulation to elucidate cancer metabolic plasticity, through integrating modeling, data analysis, and experiments.
The hybrid metabolic phenotype, characterized by high HIF-1/AMPK activities and high glycolysis/OXPHOS (glucose oxidation and FAO) activities, enables for tumors and even for individual cancer cells the metabolic plasticity to utilize various kinds of nutrients, such as glucose and fatty acid. It allows the cells to efficiently produce energy through multiple metabolism pathways and meanwhile synthesize biomass for rapid proliferation using by-products from glycolysis. In addition, the hybrid metabolic phenotype maintains the cellular ROS at a moderate level so that cancer cells can benefit from ROS signaling (41) and still avoid DNA damage due to excessive ROS (42). Moreover, the hybrid metabolic phenotype may be specifically associated with metastasis as supported by the experimental studies in highly metastatic mouse 4T1 BC cells (15, 20), B16-M4b melanoma cells (16), human SUM-159 and MDA-MB-231 TNBC cells (14), and SiHa-F3 cervix squamous cell carcinoma cells (16). Also, hybrid states may be preferentially associated with the survival and propagation of therapy-resistance cancer stem cells (43–45). As we show in Fig. 7, a combination of the glycolytic and OXPHOS inhibitors effectively eliminates the tumor survival potential of hybrid cells. Dual inhibition of glycolysis (by 2-DG) and OXPHOS (by metformin) has been shown to effectively repress tumor growth and metastasis across multiple preclinical cancer models (46). Moreover, the hybrid metabolic phenotype has recently been observed in immune cells. The regulatory T cells using both glycolysis and FAO exhibit more successful expansion than the conventional T cells using primarily glycolysis (47). Future work should develop in more detail the nature of the hybrid metabolic phenotype and its coupling with other hallmarks of cancer, such as metastasis and stem-like properties. Another intriguing result is the emergence of a low-low metabolic state with the characteristic feature of low AMPK/HIF-1 activity and low OXPHOS/glycolysis activity, especially when the HIF-1 degradation rate is high or mtROS production rate is low, as predicted by the model (Fig. 4) and shown by gene expression data analysis (Fig. 6D). This metabolically inactive state has been observed in bacterial persister cells (48) and its significance in cancer is worthy of future study.
We found that enzyme gene expression performs better than metabolite abundance in evaluating metabolic pathway activity. This may be due to the fact that in metabolic processes chemical reactions are much faster than gene regulation and consequently gene expression is more “stable” than metabolite abundance. In addition, most metabolites are reaction intermediates and their accumulation is not particularly indicative of the level of pathway activity. We note that intermediate metabolite concentrations also do not enter into other approaches such as FBA (discussed below). In this regard, the end products of metabolic pathways are expected to be more informative, and this is indeed what we have shown.
It is important to point out that there are other well-established methodologies such as metabolomics and FBA to quantitatively measure and simulate metabolism. Metabolomics utilizes cellular metabolite profiling to infer cellular metabolic activity. To interpret metabolomics data, systems biology approaches have been applied to identify causative mechanisms underlying the observed metabolite profiles (49). As we have already stated, this inference could be problematic. Alternatively, FBA is a widely used method to predict the steady-state flux distribution of metabolites that satisfies the imposed constraints and optimizes an independently chosen objective function such as maximizing biomass production. FBA is often computationally inexpensive and can be adapted to analyze the effect of specific reactions on the objective function via reaction deletion or inhibition (21, 22). Under many circumstances this is a reasonable strategy. However, the predictions of FBA depend on the imposed constraints, constraints which may not be an accurate characterization of what the coupled genetic system is attempting to impose (21). Our approach, although at the moment much more coarse-grained than a full FBA analysis, allows the cell to make its own decisions about how to direct its metabolic fluxes based on its own available information, with no extra assumptions needed.
In this work, we focused on the genetic and metabolic interplay of glycolysis and OXPHOS (FAO and glucose oxidation). It is worth noting that glutamine oxidation, which is often driven by the oncogene MYC, can also play a critical role in regulating tumor growth and metastasis (50, 51). Other HIF isoforms, such as HIF-2, which plays an important role in regulating lipid metabolism in clear cell renal cell carcinoma (52), could be integrated into the modeling framework in future to analyze the effect of various HIF isoforms in regulating cancer metabolism. Moreover, abnormal metabolism, as a hallmark of cancer, involves not only cancer cells. The surrounding glycolytic cancer-associated fibroblasts, the stromal cells which often dominate the tumor microenvironment, can provide energy-rich metabolites to promote OXPHOS activity and anabolic metabolism of cancer cells (53). These multicellular aspects, although beyond the scope of this work, are definitely worthy of further investigation combining both theoretical and experimental efforts.
Materials and Methods
Formulation of the Mathematical Model.
The generic deterministic equations representing the temporal dynamics of pAMPK, HIF-1, mtROS, and noxROS are given by
where and represent the basal production and degradation rates of component and and are two functions representing the regulation of ’s production and degradation due to the cross-talk between and other components. Since the chemical reactions in metabolism processes are much faster than the gene regulation, we assume the metabolite concentrations and the metabolic pathways are in the equilibrium state at a certain level of pAMPK and HIF-1. The uptake of glucose is regulated by AMPK and HIF-1. The intracellular glucose is shared by glucose oxidation and glycolysis. The production of acetyl-CoA is from glucose oxidation and FAO. The generation of ATP takes place from all three metabolic pathways studied here. The detailed modeling procedure can be found in SI Appendix, sections 2 and 3 and the parameter values can be found in SI Appendix, Table S5.
The Metabolic Pathway Scoring Metric.
The metabolic pathway score is defined as
where represents the expression of enzyme gene and represents the total number of genes analyzed for pathway .
The genes used to evaluate the FAO are ECHS1, HADH, ACAA1, ACAA2, CD36, SLC25A20, IVD, ACADS, GCDH, ACADVL, ACADSB, ACAD8, ACAD9, and ACAD10 (a total of 14 genes). The genes used to evaluate TCA are ACO2, IDH1, OGDH, SDHA, SDHC, FH, MDH1, CS, PC, and PDHA1 (a total of 10 genes). The genes used to evaluate glycolysis are HK1, GPI, PFKM, TPI1, GAPDH, PGAM2, ENO1, and PKM (a total of eight genes). A detailed explanation of the selection of these enzyme genes can be found in SI Appendix, section 4.
Forty-Five BC Samples and 45 Benign Tissue Samples.
Sixty-seven human BC samples and corresponding adjacent benign breast tissue samples are provided in ref. 34. The microarray data and metabolite data of 45 BC samples and their paired adjacent benign tissue samples are selected for analysis in the present study. For other paired BC and benign samples, the AMPK and HIF-1 downstream gene expression of either BC or benign sample is missing. The sample ID used in this study is listed in SI Appendix, Table S6. Analysis has also been performed on all samples with available AMPK and HIF-1 gene expression data and lactate abundance. The analysis results are shown in SI Appendix, Figs. S11 and S12, which are consistent with the results presented in Fig. 5.
Experimental Design.
Cell culture and chemicals.
Human BC cell line MDA-MB-231 and MDA-MB-468 cells were from the American Type Culture Collection; SUM-159-PT cells from Asterand Bioscience were provided by Venkata Lokesh Battula, The University of Texas MD Anderson Cancer Center, Houston, TX, and 4T1 cells were provided by Sendurai A. Mani, The University of Texas MD Anderson Cancer Center. Cells were cultured in DMEM supplemented with 100 μg/mL of streptomycin, 100 U/mL of penicillin, and 10% heat-inactivated FBS (Genedepot). ETX (Tocris), 2-DG (Sigma), and 3BP (Sigma) were dissolved according to the manufacturer’s instructions.
Cell respiratory assay.
The XF24 extracellular flux analyzer (Seahorse Biosciences) was used to measure the OCR and ECAR using the procedure described before (14, 54). Metabolic inhibitors used were oligomycin (500 nM final concentration) and ETX (100 µM final concentration). After the assay was completed, viable cells in each well were counted using a cell counter and the cell counts used to normalize the values (54).
Cell proliferation assay.
Cell proliferation was analyzed by the sulforhodamine B (SRB)-based colorimetric assay (55). Briefly, cells were fixed with 5% trichloroacetic acid to terminate reaction, and 0.4% SRB (Sigma) in 1% acetic acid was added to each well. After 30-min incubation, the plates were washed with 1% acetic acid, and dyes were dissolved by 10 mM Tris buffer. Then, the absorbance density values were read by Infinite M200 PRO reader (510 nm). Experiments were performed in triplicate.
Clonogenic assay.
Clonogenic assay was performed with minor modification to the previously published protocol (56). Briefly, cells (1 × 103 per well) were seeded into each well of a six-well plate in triplicate. After overnight culture, the media with the drug was changed every 3 d for 2 wk. Cells were rinsed with PBS, fixed in fixation solution (10% methanol, 10% acetic acid, and 80% H2O) for 10 min, and stained with 0.5% crystal violet (20% methanol and 80% H2O) for 30 min. Crystal violet was solubilized with 10% acetic acid for 15 min and quantified by absorbance at 590 nm and colonies were interpreted according to the absorbance.
RNA isolation and qPCR.
Total RNA was isolated using mRNeasy extraction kit (Qiagen) and cDNA was amplified using primers for GLUT1, LDHA, and c-Myc (SI Appendix, Table S7) in a qPCR System (MX3000P; Stratagene) with SYBR Green Supermix (Bio-Rad). The relative mRNA was quantified using β-actin mRNA expression.
Western blotting.
Cells were washed with ice-cold PBS and cell lysates were prepared in RIPA buffer containing phosphatase inhibitors and protease inhibitors (Gendepot). Cell lysates were centrifuged at 20,800 × g for 15 min at 4 °C, and supernatants were collected. Protein were separated by SDS/PAGE gel and transferred onto nitrocellulose membranes (Bio-Rad). The membrane was blocked with a 5% skim milk solution and incubated with anti–phospho-AMPK, anti-AMPK, and anti–β-actin antibodies (Cell Signaling) and detected using the ClarityTM western ECL substrate kit (Bio-Rad). Figures are presented with β-actin as the loading control for pAMPK.
Supplementary Material
Acknowledgments
We thank Dr. Donti and Ms. Vithayathil for the Seahorse experiments, and Dr. Ambs [National Cancer Institute (NCI)] for sharing data from breast cancer and benign breast tissues. This work was supported by Physics Frontiers Center NSF Grants PHY-1427654 and PHY-1605817 and John S. Dunn Foundation Collaborative Research Award (to D.J., J.N.O., and H.L.); start-up funds from The Jackson Laboratory, NCI Grant P30CA034196, and the National Institute of General Medical Sciences Grant R35GM128717 (to M.L.); and NCI Grants R21CA173150, R21CA179720, and R03CA212816, Department of Defense Grant W81XWH-18-1-0714, the Collaborative Faculty Research Investment Program, the Breast Cancer Research Foundation, the Dan L Duncan Comprehensive Cancer Center, and the Charles A. Sammons Cancer Center (J.H.P., K.H.J., and B.A.K.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 3370.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816391116/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Ward PS, Thompson CB. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Szablewski L. Expression of glucose transporters in cancers. Biochim Biophys Acta. 2013;1835:164–169. doi: 10.1016/j.bbcan.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Ganapathy-Kanniappan S, Geschwind J-FH. Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol Cancer. 2013;12:152. doi: 10.1186/1476-4598-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Payen VL, Porporato PE, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 1: Tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol Life Sci. 2016;73:1333–1348. doi: 10.1007/s00018-015-2098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya B, et al. Increased drug resistance is associated with reduced glucose levels and an enhanced glycolysis phenotype. Br J Pharmacol. 2014;171:3255–3267. doi: 10.1111/bph.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br J Pharmacol. 2016;173:970–979. doi: 10.1111/bph.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Icard P, et al. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1–11. doi: 10.1016/j.drup.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J Immunol. 2004;172:4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- 11.Menk AV, et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 2018;22:1509–1521. doi: 10.1016/j.celrep.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folmes CDL, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JH, et al. Fatty acid oxidation-driven Src links mitochondrial energy reprogramming and oncogenic properties in triple-negative breast cancer. Cell Rep. 2016;14:2154–2165. doi: 10.1016/j.celrep.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003, 1–15. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porporato PE, et al. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 17.Jia D, Park JH, Jung KH, Levine H, Kaipparettu BA. Elucidating the metabolic plasticity of cancer: Mitochondrial reprogramming and hybrid metabolic states. Cells. 2018;7:E21. doi: 10.3390/cells7030021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camarda R, et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao K, Fang M, Alroy J, Sahagian GG. Imagable 4T1 model for the study of late stage breast cancer. BMC Cancer. 2008;8:228. doi: 10.1186/1471-2407-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuy F, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22:577–589. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raman K, Chandra N. Flux balance analysis of biological systems: Applications and challenges. Brief Bioinform. 2009;10:435–449. doi: 10.1093/bib/bbp011. [DOI] [PubMed] [Google Scholar]
- 23.Nilsson A, Nielsen J. Genome scale metabolic modeling of cancer. Metab Eng. 2017;43:103–112. doi: 10.1016/j.ymben.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 24.Medina MÁ. Mathematical modeling of cancer metabolism. Crit Rev Oncol Hematol. 2018;124:37–40. doi: 10.1016/j.critrevonc.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Yizhak K, et al. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol Syst Biol. 2014;10:744. doi: 10.15252/msb.20134993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zielinski DC, et al. Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci Rep. 2017;7:41241. doi: 10.1038/srep41241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qutub AA, Popel AS. Reactive oxygen species regulate hypoxia-inducible factor 1alpha differentially in cancer and ischemia. Mol Cell Biol. 2008;28:5106–5119. doi: 10.1128/MCB.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu L, et al. Modeling the genetic regulation of cancer metabolism: Interplay between glycolysis and oxidative phosphorylation. Cancer Res. 2017;77:1564–1574. doi: 10.1158/0008-5472.CAN-16-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Saedeleer CJ, et al. Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS One. 2012;7:e46571. doi: 10.1371/journal.pone.0046571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang B, et al. Interrogating the topological robustness of gene regulatory circuits by randomization. PLoS Comput Biol. 2017;13:e1005456. doi: 10.1371/journal.pcbi.1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang B, et al. RACIPE: A computational tool for modeling gene regulatory circuits using randomization. BMC Syst Biol. 2018;12:74. doi: 10.1186/s12918-018-0594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest. 2014;124:398–412. doi: 10.1172/JCI71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kano-Sueoka T, Watanabe T, Miya T, Kasai H. Analysis of cytosolic phosphoethanolamine and ethanolamine and their correlation with prognostic factors in breast cancer. Jpn J Cancer Res. 1991;82:829–834. doi: 10.1111/j.1349-7006.1991.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah T, et al. Molecular causes of elevated phosphoethanolamine in breast and pancreatic cancer cells. NMR Biomed. 2018;31:e3936. doi: 10.1002/nbm.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budczies J, et al. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int J Cancer. 2015;136:1619–1628. doi: 10.1002/ijc.29152. [DOI] [PubMed] [Google Scholar]
- 38.Sciacovelli M, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature. 2016;537:544–547. doi: 10.1038/nature19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X, et al. Cystine addiction of triple-negative breast cancer associated with EMT augmented death signaling. Oncogene. 2017;36:4235–4242. doi: 10.1038/onc.2016.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishikawa K, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 42.Piskounova E, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527:186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sancho P, et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 44.De Luca A, et al. Mitochondrial biogenesis is required for the anchorage-independent survival and propagation of stem-like cancer cells. Oncotarget. 2015;6:14777–14795. doi: 10.18632/oncotarget.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamb R, et al. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: Treating cancer like an infectious disease. Oncotarget. 2015;6:4569–4584. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheong J-H, et al. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol Cancer Ther. 2011;10:2350–2362. doi: 10.1158/1535-7163.MCT-11-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacella I, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc Natl Acad Sci USA. 2018;115:E6546–E6555. doi: 10.1073/pnas.1720113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosato A, et al. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics. 2018;14:37. doi: 10.1007/s11306-018-1335-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu B, et al. HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 54.Donti TR, et al. Screen for abnormal mitochondrial phenotypes in mouse embryonic stem cells identifies a model for succinyl-CoA ligase deficiency and mtDNA depletion. Dis Model Mech. 2014;7:271–280. doi: 10.1242/dmm.013466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 56.Singleton KR, et al. Melanoma therapeutic strategies that select against resistance by exploiting MYC-driven evolutionary convergence. Cell Rep. 2017;21:2796–2812. doi: 10.1016/j.celrep.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.