Significance
RNA-directed DNA methylation (RdDM) is an important mechanism in the silencing of transposable elements in plants. The framework of RdDM has been well established, in which DNA polymerase IV (Pol IV) and Pol V act in upstream siRNA biogenesis and downstream DNA methylation, respectively. The DDR complex, consisting of DMS3, DRD1, and RDM1, is responsible for recruiting Pol V, and dysfunction of any of them causes compromised RdDM activity. However, how the association among DMS3, DRD1, and RDM1 is regulated is unexplored. We show here that the anaphase-promoting complex/cyclosome (APC/C), a key cell cycle regulatory complex, regulates the assembly of the DDR complex by targeting DMS3 for degradation, and thereby provide a perspective in understanding the regulatory mechanism of RdDM activity.
Keywords: RdDM, APC/C, Pol V, DDR, TE silencing
Abstract
During RNA-directed DNA methylation (RdDM), the DDR complex, composed of DRD1, DMS3, and RDM1, is responsible for recruiting DNA polymerase V (Pol V) to silence transposable elements (TEs) in plants. However, how the DDR complex is regulated remains unexplored. Here, we show that the anaphase-promoting complex/cyclosome (APC/C) regulates the assembly of the DDR complex by targeting DMS3 for degradation. We found that a substantial set of RdDM loci was commonly de-repressed in apc/c and pol v mutants, and that the defects in RdDM activity resulted from up-regulated DMS3 protein levels, which finally caused reduced Pol V recruitment. DMS3 was ubiquitinated by APC/C for degradation in a D box-dependent manner. Competitive binding assays and gel filtration analyses showed that a proper level of DMS3 is critical for the assembly of the DDR complex. Consistent with the importance of the level of DMS3, overaccumulation of DMS3 caused defective RdDM activity, phenocopying the apc/c and dms3 mutants. Moreover, DMS3 is expressed in a cell cycle-dependent manner. Collectively, these findings provide direct evidence as to how the assembly of the DDR complex is regulated and uncover a safeguarding role of APC/C in the regulation of RdDM activity.
The anaphase-promoting complex or cyclosome (APC/C) is a large multi-subunit complex that promotes the metaphase-to-anaphase progression and G1 arrest by targeting different substrates for ubiquitination and proteasome-mediated destruction (1). The APC/C contains at least 13 subunits, in which APC10 is involved in recognizing and recruiting substrates (2–5). The APC/C is evolutionarily conserved, as different APC/C subunits from distinct species are able to complement the corresponding yeast mutants (6–10). The APC/C, like other E3 ubiquitin ligases of the RING family, serves as a binding platform that brings together a specific substrate and an E2 coenzyme, resulting in polyubiquitination and degradation of the substrate by the 26S proteasome (11). Because most APC/C subunits are encoded by single genes, mutants are embryo and/or gametic lethal in both plants and animals (6, 10, 12–16).
The APC/C promotes degradation of more than a 100 substrates in a specific motif-dependent manner (17). Typically, substrates of APC/C contain at least one of three motifs: the destruction box (the D box, RXXLXXXN) (18), the KEN box (19), or the ABBA motif (20). Interestingly, besides the well-known cell cycle regulation-related proteins targeted by APC/C, several epigenetic regulators are substrates of APC/C in animals. For example, Dnmt1 (DNA methyltransferase) (21) and G9a (H3K9 methyltransferase) (22) were targeted by APC/C in response to DNA damage, while MIWI (the mouse homolog of Argonaute) (23) and HIWI (the human homolog of Argonaute) (24) were targeted by APC/C during male germline development. These studies provided novel insights into the function of APC/C, and connect two important regulatory activities: protein degradation and epigenetic regulation. However, although APC/C degrades DRB4, a double-stranded RNA-binding protein acting in small RNA-mediated gene silencing in plants (25), the biological importance of APC/C-involved epigenetic regulation in plants was unexplored.
In plants, gene silencing of transposable elements (TEs) is controlled by RNA-directed DNA methylation (RdDM), which depends on specialized transcriptional machineries that are performed by two plant-specific DNA-dependent RNA polymerases, polymerase IV (Pol IV) and Pol V (26). In brief, transcripts from Pol IV/RDR2 are processed by Dicer-like 3 (DCL3) into siRNAs, which are mainly loaded onto Argonaute 4 (AGO4). Nascent scaffold transcripts from regions flanking RdDM loci are produced by Pol V, which possibly facilitates the recruitment of the siRNA–AGO4 complex, DNA methyltransferases, and/or the histone modification machinery to silence TEs by DNA methylation and histone modifications. Notably, a complex termed DDR (DRD1, DMS3, and RDM1) recruits Pol V to the chromatin (27–31). Among the three DDR components, only DMS3, a protein with homology to the hinge region of structural maintenance of chromosome (SMC) proteins, was identified in a ubiquitinated proteome study (32). However, in vitro evidence for DMS3 ubiquitination, the identity of the E3 ligase, and the biological significance of DMS3 ubiquitination remain unexplored. Moreover, it remains unknown how the DDR complex is regulated.
Here, we show that APC/C regulates the association of the DDR complex by controlling DMS3 abundance as an E3 ligase. We found that a substantial subset of RdDM loci were de-repressed in apc/c mutants, without significantly disturbing Pol IV-dependent siRNA biogenesis but compromising the function of Pol V. Mechanistically, we show that APC/C targets DMS3 for ubiquitination and degradation in a D box-dependent manner, and that the level of DMS3 determines the proper association of the DDR complex. We thus provide direct evidence that APC/C-mediated DMS3 degradation is indispensable for regulation of the DDR complex.
Results
APC/C Is Required for TE Silencing in Plants.
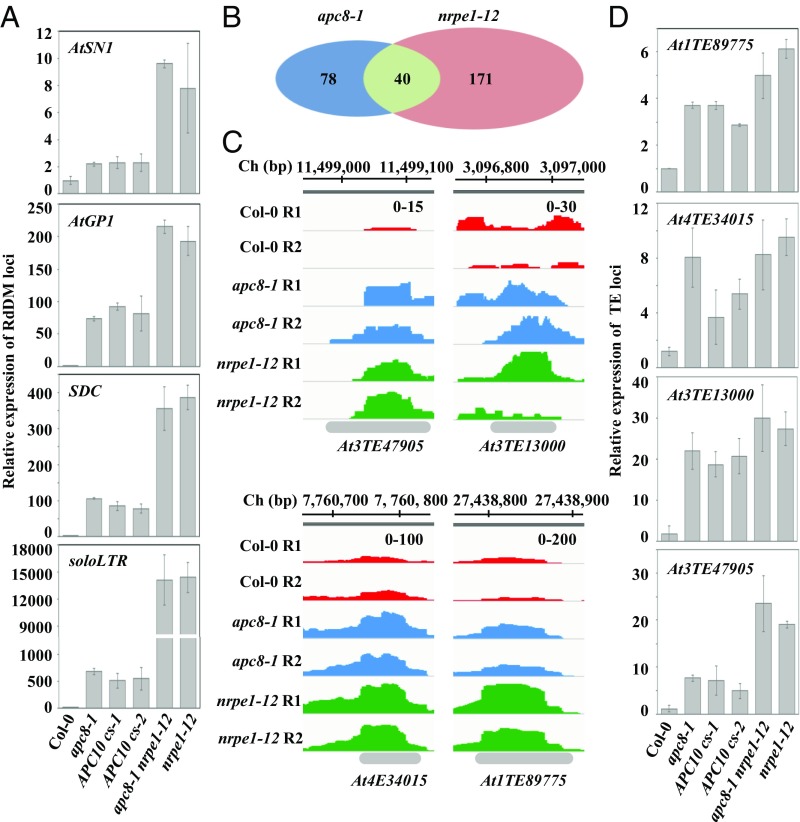
To investigate whether APC/C is important for epigenetic regulation in plants, we first examined expression of common RdDM loci using a weak allele of APC8 (8). Quantitative RT-PCR (qRT-PCR) analyses showed that AtSN1, AtGP1, SDC, and soloLTR were significantly de-repressed in apc8-1 relative to their expression levels in Col-0 (the WT control), a de-repression trend similar to that seen for nrpe1-12 (NRPE1, the largest subunit of Pol V) (Fig. 1A). Then, we determined the effects of apc8-1 on TE expression on a genome-wide scale by RNA sequencing, using the nrpe1-12 mutant as the control. We quantified transcripts derived from TEs by normalizing against total reads using featureCounts (33). Correlation analysis showed that expression data for Col-0, nrpe1-12, and apc8-1 were highly reproducible between biological replicates (SI Appendix, Fig. S1A). Overall, we identified that 211 and 118 TEs were significantly de-repressed [log2 (mutant/WT) > 1; P < 0.05] using edgeR (34) in nrpe1-12 and apc8-1, respectively (Fig. 1B and Dataset S1). Notably, as apc8-1 is a weak allele, the number of APC8-dependent TEs might be underestimated. Of the 211 TEs de-repressed in nrpe1-12, 18.9% (40 of 211) were also de-repressed in apc8-1 (Fig. 1B and Dataset S1), and of the 118 TEs de-repressed in apc8-1, 33.9% (40 of 118) were de-repressed in nrpe1-12 (Fig. 1B and Dataset S1), indicating that APC8 plays a role in the RdDM pathway, possibly by associating with NRPE1. We also confirm that de-repressed RdDM loci affected expression of their flanking regions in apc8-1 and nrpe1-12 (SI Appendix, Fig. S1B). The number of apc8-1–dependent TEs overlapping with nrpe1-12–dependent TEs was significantly larger than expected (P < 1e-10, χ2 test). Boxplot analysis further showed that the genome-wide effects of apc8-1 on TE silencing were generally similar to those of nrpe1-12 (SI Appendix, Fig. S1C). Notably, both APC8 and NRPE1 play a dominant role in a subset of TEs, respectively (SI Appendix, Fig. S1C). To validate expression of TEs identified from RNA sequencing, we used qRT-PCR to examine expression of At3TE47905, At3TE13000, At4TE34015, and At1TE89775 (Fig. 1C). We showed that these four tested TEs were indeed de-repressed in both apc8-1 and nrpe1-12 (Fig. 1D). Furthermore, these TEs that were up-regulated by APC8 and NRPE1 mostly belong to the MuDR, Helitron, Copia, and Gypsy transposon families with similar trends (SI Appendix, Fig. S1D and Dataset S2), suggesting that APC8 and NRPE1 regulate the same classes of TEs. These data indicate that APC8 is involved in TE silencing potentially by overlapping with Pol V.
Fig. 1.
APC/C is required for a subset of TE silencing. (A) Effect of apc8-1 and cs lines of APC10 on transcriptional silencing as determined by qRT-PCR. Transcript levels of RdDM loci AtSN1, AtGP1, soloLTR, and SDC were measured. The UBQ5 gene was amplified as an internal control. Data are presented as mean ± SE (n = 3). (B) Overlap of de-repressed TEs from RNA-sequencing between apc8-1 and nrpe1-12. The Venn diagram shows the number of de-repressed TEs between apc8-1 and nrpe1-12. P = 4.217 × 10−10, calculated by the Pearson’s χ2 test with the Yates’ continuity correction. (C) Normalized expression levels of transcripts from selected TE loci in Col-0, apc8-1, and nrpe1-12. Peaks of RNA sequencing in each genotype are indicated by different colors. The x axis indicates the chromosomal location of TEs, and the y axis indicates normalized peaks. The same scales (“0–100” indicates numbers of normalized reads coverage) are identical for each genotype. Ch, chromosome; R1, replicate 1; R2, replicate 2. (D) Validation of TEs identified from RNA sequencing by qRT-PCR. UBQ5 was amplified as an internal control. Data are presented as mean ± SE (n = 3).
Because APC/C plays a role in the miRNA pathway by regulating the occupancy of Pol II at the promoter of some specific MIR genes (8), and because Pol V silences TEs mainly by the RdDM pathway (26), we then wanted to investigate whether APC/C acts in TE silencing at the step of siRNA production. Small RNA sequencing showed that 7,529 individual 24-nt siRNA clusters had lower levels in the pol iv mutant (SI Appendix, Fig. S2A and Dataset S3), while only 494 individual 24-nt siRNA clusters had lower levels in apc8-1 (SI Appendix, Fig. S2A and Dataset S3). Moreover, among the 494 siRNA clusters reduced in apc8-1, ∼30% (156 of 494) of these siRNA clusters were dependent on Pol V (35) (SI Appendix, Fig. S2B) and ∼50% (264 of 494) of the APC8-dependent siRNA clusters were coregulated by DMS3 (35) (SI Appendix, Fig. S2B). Northern blotting showed that the siRNA levels from AtRep2, AtSN1, and siR1003, three representative RdDM loci, were unaltered in apc8-1 (SI Appendix, Fig. S2C), confirming a dispensable role of APC/C in siRNA biogenesis. To further demonstrate that the role of APC8 in TE silencing is fundamental to the function of APC/C, we examined TE expression in cosuppression (cs) lines of APC10 (SI Appendix, Fig. S2D), which exhibited bonsai phenotypes similar to that of apc8-1 (36). The qRT-PCR analysis showed that four tested RdDM loci were de-repressed in cs lines of APC10 (Fig. 1A), and the extent of TE de-repression was comparable to that in apc8-1 (Fig. 1A). Considering that apc8-1 is a weak allele and cs lines of APC10 are also partially functional, there might be more severe defects on TE silencing in loss-of-function alleles of APC/C. Taken together, these results indicate that APC/C is involved in TE silencing in plants, and that APC/C functions downstream of siRNA biogenesis.
APC/C Is Involved in Pol V Recruitment and Pol V Transcription.
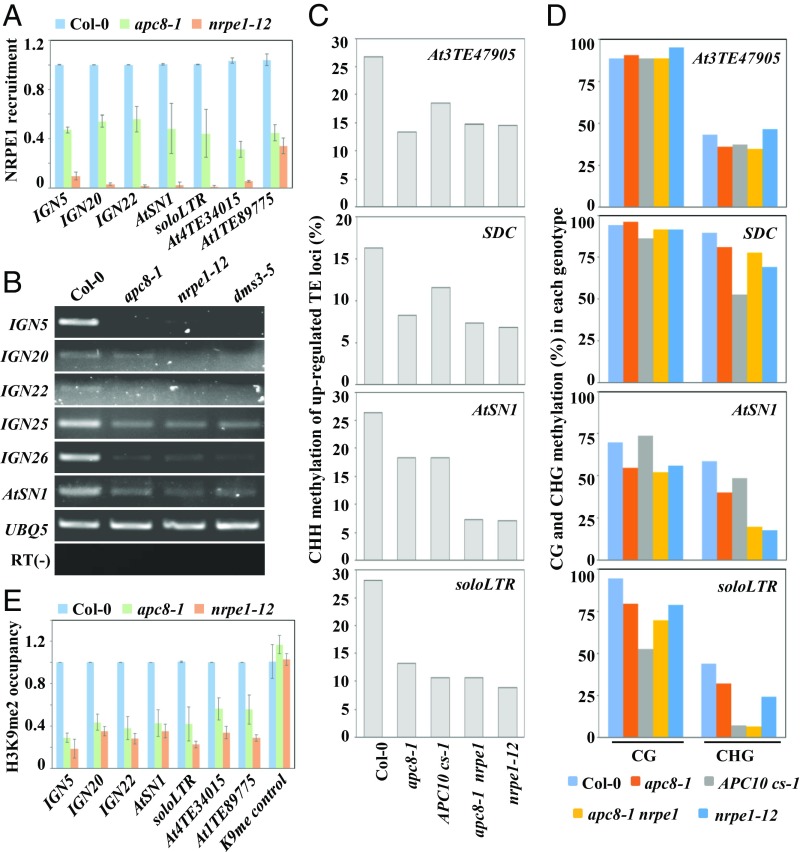
Because APC/C acts downstream of siRNA biogenesis in the RdDM pathway, and because APC/C coregulates a subset of RdDM loci with Pol V, we hypothesized that the function of Pol V might be regulated by APC/C. We therefore examined the occupancy of Pol V at TE chromatin in apc8-1. We performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) experiments using an anti-NRPE1 antibody to detect the recruitment of NRPE1 at typical Pol V-dependent intergenic noncoding (IGN) regions. Relative to occupancy in Col-0, there was an obvious reduction of Pol V occupancy at RdDM loci in apc8-1 (Fig. 2A), indicating that APC/C is involved in Pol V recruitment. Subsequently, RT-PCR analysis showed that Pol V-dependent scaffold RNAs from AtSN1, IGN, and two TE loci were significantly reduced in apc8-1, similar to reduced expression in dms3-5 and nrpe1-12 (Fig. 2B). These results indicate that APC/C facilitates TE silencing by regulating Pol V function.
Fig. 2.
Occupancy and transcription of Pol V are reduced in apc/c mutants. (A) Pol V occupancy in Col-0, apc8-1, and nrpe1-12 was determined by ChIP-qPCR with anti-NRPE1. ChIP signals are normalized to the input control for each genotype, and the value for apc8-1 and nrpe1-12 was then accordingly assessed relative to that of Col-0. Data are presented as mean ± SE (n = 3). (B) Pol V-dependent scaffold noncoding RNAs from IGN loci and AtSN1 were measured by semiquantitative RT-PCR. UBQ5 was used as an internal control. Representative data are presented from three biological replicates. (C and D) DNA methylation of AtSN1, SDC, soloLTR, and At3TE47905 was determined by bisulfite sequencing in Col-0, apc8-1, APC10 cs-1, apc8-1 nrpe1-12, and nrpe1-12. CHH (C) and CHG/CG (D) methylation is shown. (E) H3K9me2 occupancy in Col-0, apc8-1, and nrpe1-12 was determined by ChIP-qPCR with anti-H3K9me2. ChIP signals were normalized to the input control for each genotype, and the value for apc8-1 and nrpe1-12 was then accordingly assessed relative to that of Col-0. Data are presented as mean ± SE (n = 3).
De-repression of TEs is usually accompanied by reduced DNA methylation and H3K9 dimethylation (37). We therefore determined DNA methylation by bisulfite sequencing analysis (Dataset S4). DNA methylation of AtSN1, soloLTR, SDC, and At3TE47905 at CHH sites was substantially reduced in apc/c mutants as well as in nrpe1-12 (Fig. 2C and Dataset S4), whereas DNA methylation at CG and CHG sites was only slightly affected in apc/c mutants, except for the CHG methylation at the soloLTR locus (Fig. 2D and Dataset S4). ChIP-qPCR experiments using an anti-H3K9me2 antibody showed that H3K9me2 was significantly reduced in apc8-1 and nrpe1-12 (Fig. 2E). These results further indicate that APC/C is involved in TE silencing by the RdDM pathway.
To determine the genetic relationship of APC/C and Pol V in TE silencing, we examined TE expression in the apc8-1 nrpe1-12 double mutant. We predicted that the expression of coregulated TEs would be similar to their expression in the nrpe1-12 mutant, if APC/C and Pol V act in the same pathway to repress TEs. Indeed, the extent of de-repression of both common RdDM loci and selected TEs (identified from RNA sequencing) in apc8-1 nrpe1-12 was comparable to that in nrpe1-12 (Fig. 1). Bisulfite sequencing (Fig. 2 C and D) showed that DNA methylation of de-repressed TE loci in apc8-1 nrpe1-12 was similar to that in nrpe1-12. Taken together, these results indicate that APC/C is required for TE silencing by regulating the function of Pol V.
APC/C Interacts with DMS3 in a D Box-Dependent Manner.
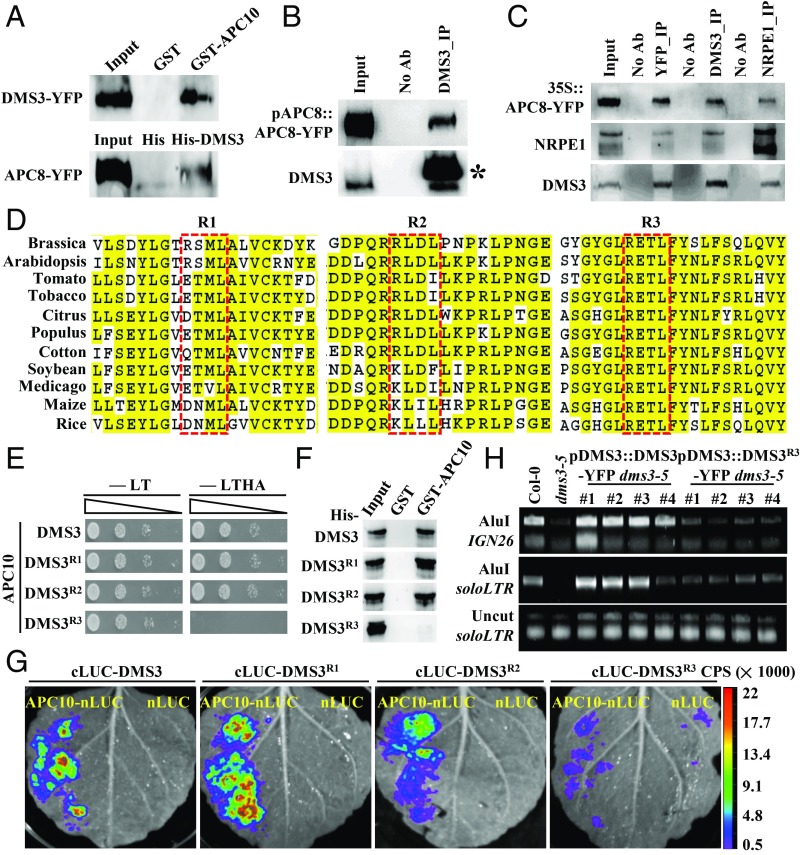
In general, APC/C functions by targeting specific substrates (i.e., APC10 recognizes and recruits substrates) (2–5). To find out the target of APC/C in regulating TE silencing in plants, we carried out yeast two hybrid (Y2H) assays to test the interaction between APC10 and components known to be essential for TE silencing, including Pol IV (38) and Pol V subunits (38, 39), SHH1 (40), AGO4 (41), KTF1 (42), SUVH proteins (35), MORC proteins (43), DRM2 (44), and DDR complex components (30, 31). DRB4 was used as the positive control (25). Under four amino acid-depleted growth conditions, only yeast cells coexpressing APC10 with DMS3, RDM1, and DRB4 exhibited normal growth (SI Appendix, Fig. S3A), suggesting a direct interaction between APC10 and these three components. To further test whether these interactions are specific to APC10, we examined the interaction between DMS3 and other APC/C subunits by Y2H assays. Indeed, DMS3 specifically interacted with APC10 (SI Appendix, Fig. S3B), which is consistent with the idea that APC10, the core subunit of the composite pocket, binds to substrates (45). To validate the interaction between APC/C and DMS3, we performed reciprocal pull-down assays using recombinant His-DMS3 and 35S::APC8-YFP transgenic plants or GST-APC10 and 35S::DMS3-YFP transgenic plants, respectively. We confirmed the interaction between APC/C and DMS3 (Fig. 3A) and the interaction between RDM1 and APC10 (SI Appendix, Fig. S3C). A coimmunoprecipitation (co-IP) assay using transgenic plants of pAPC8::APC8-YFP showed that APC8 is associated with DMS3 under the endogenously expressed APC8 condition (Fig. 3B). Of note, the association between APC8 and NRPE1 (Fig. 3C) or RDM1 (SI Appendix, Fig. S3D) was only detected when APC8 was overexpressed. Collectively, these results indicate that APC/C interacts with the DDR complex.
Fig. 3.
DMS3 interacts with APC10 in a D box-dependent manner. (A) Reciprocal semi-in vivo pull-down assays to examine the DMS3–APC/C interaction. (Top) GST-APC10 pulled down DMS3 from 35S::DMS3-YFP transgenic plants, and GST was used as the negative control. (Bottom) His-DMS3 pulled down APC8 from 35S::APC8-YFP transgenic plants, and His was used as the negative control. Precipitates were analyzed by Western blotting using anti-GFP antibodies. (B) Co-IP between APC8 and DMS3. Anti-DMS3 antibody coupled to the protein A beads was used to immunoprecipitate (IP) APC8 from inflorescence of pAPC8::APC8-YFP transgenic plants. Anti-GFP and anti-DMS3 antibodies were used to detect APC8 (Top) and DMS3 (Bottom) by Western blotting, respectively. The asterisk indicates the band of the heavy chain resulting from the excess anti-DMS3 antibody. (C) Reciprocal co-IP between APC8, NRPE1, and DMS3. GFP-Trap beads were used to immunoprecipitate DMS3 and NRPE1 from inflorescences of 35S::APC8-YFP transgenic plants. Alternatively, anti-DMS3– or anti-NRPE1–conjugated beads were used to immunoprecipitate other proteins from the same transgenic plants. Anti-NRPE1 (Middle), anti-DMS3 (Bottom), and anti-GFP (Top) antibodies were used to detect the corresponding proteins by Western blotting. (D) Alignment of the region flanking three D boxes of DMS3 from various species. DMS3 proteins from Brassica napus (XP_013693242.1), Gossypium hirsutum (cotton, XP_016737311.1), Arabidopsis thaliana (NP_566916.1), Medicago truncatula (XP_013461420.1), Glycine max (soybean, XP_006578904.1), Nicotina tobacum (tobacco, XP_016478774.1), Populus trichocarpa (XP_002321401.2), Citrus clementina (XP_006429697), Oryza sativa (Os01g13404.1), Solanum lycopersicum (tomato, Solyc06g051990.2.1), and Zea mays (maize, GRMZM2G309152_T01) were aligned using ClustalW. The red rectangles indicate the three D boxes. Identical residues are highlighted in yellow. (E) Y2H assays to test the interaction between APC10 and mutated DMS3. Yeast cells cotransformed by the indicated combinations of plasmids were diluted and spotted onto nonselective (-LT) and selective (-LTHA) media. (F) In vitro pull-down assays to examine the APC10–mDMS3 interaction. Protein precipitates were analyzed by Western blotting using an anti-His antibody. (G) Split luciferase complementation imaging assays to examine the interaction between APC10 and mutated DMS3. nLUC was used as a negative control. (H) Complementation assay to show the function of mutated DMS3 by Chop-PCR. DMS3 or DMS3R3 was introduced into dms3-5, and genomic DNA was digested by the DNA methylation-sensitive restriction enzyme AluI and subjected to semiquantitative PCR. The soloLTR locus was amplified using the undigested genomic DNA as the template for a control.
To understand how APC/C recognizes DMS3 and RDM1, we searched these two protein sequences for the D box, KEN box, and ABBA motifs. Three putative D boxes with the core amino acids RXXL were found at amino acids 179–182 (R1), 248–251 (R2), and 291–294 (R3) of DMS3, respectively (Fig. 3D and SI Appendix, Fig. S3E). Among these putative D boxes, the R1 and R2 motifs are not conserved among plant species, and are located within the conserved SMC-hinge domain (Fig. 3D), while R3, located outside of the SMC-hinge domain (Fig. 3D), is highly conserved in flowering plants (Fig. 3D). Only one putative D box with the RXXL motif was found in RDM1 (SI Appendix, Fig. S3E). To determine whether these putative D boxes of DMS3 and RDM1 are recognition sites for APC/C, we generated corresponding variants in which the putative D boxes were mutated into AXXL (DMS3R or RDM1R) and tested their capacity to interact with APC10. The putative D box of RDM1 mutation had no effect on the interaction between APC10 and RDM1 (SI Appendix, Fig. S3 F and G). The DMS3R3 variant, but not the DMS3R1 and DMS3R2 variants, abolished interaction with APC10 (Fig. 3E), indicating that the R3 D box is essential for the interaction. Notably, the DMS3R3 variant showed a normal interaction with RDM1 (SI Appendix, Fig. S3H) and DRD1 (SI Appendix, Fig. S3I). Pull-down assays further confirmed that the R3 D box of DMS3 was essential for interaction with APC10 (Fig. 3F). To further examine the role of D boxes in the interaction with APC/C, we used a split luciferase complementation imaging assay (46) to determine the interaction between APC10 and DMS3 or RDM1 variants. We fused APC10 and DMS3 to the N-terminal fragment of luciferase (nLUC) or the C-terminal fragment of luciferase (cLUC), respectively, and introduced paired proteins into Nicotiana benthamiana leaves by infiltration. Indeed, the DMS3–APC10, DMS3R1–APC10, and DMS3R2–APC10 interactions were robust (Fig. 3G), but only a few faint signals were observed from the DMS3R3–APC10 interaction (Fig. 3G). Collectively, these results indicate that the conserved D box of DMS3 mediates the specific recognition by APC/C.
To further understand the significance of the R3 D box for DMS3 function, we introduced YFP-tagged fusions of DMS3 and DMS3R3, driven by the endogenous promoter, into the dms3-5 mutant. Since DNA methylation of RdDM loci is significantly reduced in dms3-5 (27, 28, 30), we performed Chop-PCR experiments to examine DNA methylation of soloLTR and IGN26 in these transgenic lines. As expected, the WT version of DMS3 was able to completely complement defects in DNA methylation at both soloLTR and IGN26 (Fig. 3H), demonstrating that the fusion protein of DMS3-YFP is functional in vivo. In contrast, four independent transgenic lines of DMS3R3-YFP only showed partial complementation (Fig. 3H), indicating that the R3 D box is essential for DMS3 function. To exclude the possibility that the difference was due to the different levels of DMS3R3-YFP in transgenic plants, we examined the level of DMS3 by Western blotting and found that the accumulation of DMS3R3 in transgenic plants was comparable to those in DMS3 (SI Appendix, Fig. S3I). Taken together, we concluded that APC10 interacts with DMS3 in a D box-dependent manner and that this interaction is essential for the function of DMS3.
DMS3 Is Ubiquitinated and Degraded by APC/C.
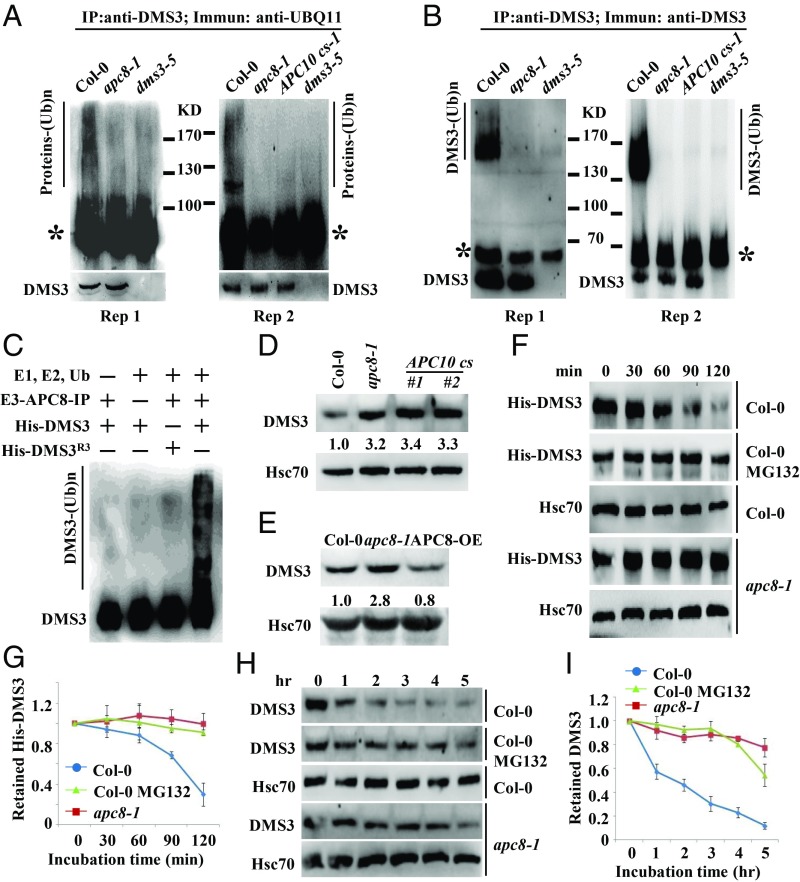
Since DMS3 interacts with APC/C and DMS3 was ubiquitinated (32), we hypothesized that DMS3 might be ubiquitinated by APC/C. Therefore, an in vivo ubiquitination assay was performed using seedlings of Col-0, apc8-1, and dms3-5 plants. Lysates were treated with MG132 to inhibit potential degradation after ubiquitination. The anti-Ubiquitin 11 (UBQ11) detected high-molecular-mass smears of protein bands above 100 kDa in the anti–DMS3-immunoprecipitated samples in Col-0 but not in apc8-1, APC10cs-1, or dms3-5 plants (Fig. 4A), indicating that proteins associated with DMS3 are ubiquitinated in vivo. To further confirm the smear shown in Fig. 4A harbors ubiquitinated DMS3, anti-DMS3 antibodies were used for Western blotting. As shown in Fig. 4B, we observed a major band together with smeared bands above 130 kDa in Col-0. Importantly, the smeared larger bands of DMS3 were eliminated in apc8-1 and dms3-5 (Fig. 4B). These results demonstrate that APC/C mediates DMS3 ubiquitination in vivo.
Fig. 4.
DMS3 is ubiquitinated and degraded by APC/C in vivo and in vitro. (A) Detection of ubiquitination levels of DMS3 immunoprecipitates (IP) in Col-0, dms3-5, APC10 cs-1, and apc8-1. DMS3 immunoprecipitates were immunoblotted with anti-UBQ11 to detect Ub-protein conjugates. The protein standards were indicated. The asterisk indicates the heavy chain of anti-DMS3. (Bottom) Anti-DMS3 blot to indicate the DMS3 levels in each genotype. Two biological replicates are shown. (B) Detection of DMS3 ubiquitination in Col-0, dms3-5, and apc8-1. Anti-DMS3 immunoprecipitates were immunoblotted with anti-DMS3 to detect Ub-DMS3 conjugates. The protein standards were indicated. The asterisk indicates the heavy chain of anti-DMS3. The bottom band indicates the DMS3 level in each genotype. Two biological replicates are shown. (C) APC/C-mediated ubiquitination of DMS3 in vitro. Reactions were carried out with the different combinations of E1, E2, Ub, E3 (anti-GFP immunoprecipitates from APC8-YFP transgenic plants) and the substrates His-DMS3 or His-DMS3R3. The reaction was stopped, and proteins were detected by Western blotting using an anti-DMS3 antibody. The smeared bands indicate ubiquitinated DMS3, while the bottom band indicates nonubiquitinated DMS3. Western blotting was used to determine DMS3 levels in seedlings of Col-0, apc8-1, and cs lines of APC10 (D) or APC8-OE (E). Hsc70 was the loading control. The numbers indicate the average abundance of DMS3 from three biological replicates. (F) Decay rate of His-DMS3 in a cell-free protein decay assay. His-DMS3 was incubated with equal amounts of cell lysate of Col-0 or apc8-1 with or without 50 μM MG132 for the indicated time. The levels of His-DMS3 were determined with anti-His antibody. Hsc70 was used as the loading control. (G) Statistical analysis of the decay rate of His-DMS3 in F. The levels of His-DMS3 were plotted versus the level at 0 min. Data are presented as mean ± SE (n = 3). (H) Decay rate of DMS3 in Col-0 or apc8-1. Total proteins from Col-0 and apc8-1 were extracted and incubated with or without 50 μM MG132 for indicated time. Levels of DMS3 were determined with anti-DMS3. Hsc70 was the loading control. (I) Statistical analysis of decay rate of DMS3 in H. Data are presented as mean ± SE (n = 3).
To show that Ub-DMS3 conjugates were detectable in the presence of E1, E2, and E3, we used in vitro APC/C-ubiquitination assays, in which the APC/C complex (E3) immunoprecipitated from APC8-YFP transgenic plants was incubated with Ub, ATP, E1, E2, and recombinant His-DMS3 (Fig. 4C). In contrast, when His-DMS3 was replaced by His-DMS3R3, no Ub-DMS3R3 conjugates were observed (Fig. 4C). Consistent with the finding of no visible effect on RDM1 regulated by APC/C, there were not any smeared larger bands in immunoprecipitates (SI Appendix, Fig. S4A), and RDM1 levels were not affected in apc/c mutants (SI Appendix, Fig. S4B), further supporting that RDM1 is not ubiquitinated and that RDM1 is unlikely to be a substrate of APC/C in vivo. We thus conclude that DMS3 is indeed ubiquitinated, and that APC/C is the E3 ligase responsible for DMS3 ubiquitination.
Because DMS3 is targeted by APC/C for ubiquitination, we examined the abundance of DMS3 in apc/c mutants. Western blotting showed that DMS3 consistently overaccumulated in both apc8-1 and cs lines of APC10 (Fig. 4D), while it was slightly reduced in overexpression lines of APC8 (APC8-OE) (Fig. 4E). To further confirm that the regulation of DMS3 by APC/C occurs posttranscriptionally, we performed qRT-PCR assays to examine the mRNA level of DMS3 in apc8-1 and in cs lines of APC10. As expected, the transcripts of DMS3 were largely unchanged in both (SI Appendix, Fig. S4C), indicating that APC/C degrades DMS3.
To conclusively demonstrate that APC/C targets DMS3 for degradation, we monitored the degradation of recombinant DMS3 using a cell-free degradation assay. His-DMS3 was expressed and purified from Escherichia coli, and then incubated with lysates from Col-0 and apc8-1. The degradation rate of DMS3 in apc8-1 was slower than in Col-0 (Fig. 4 F and G). To investigate whether the degradation of DMS3 was proteasome-dependent, we compared the degradation rate in the presence or absence of the protease inhibitor MG132. As shown in Fig. 4 F and G, the addition of MG132 significantly suppressed the destabilization of His-DMS3. Consistently, when His-DMS3R3 was incubated with Col-0 lysates, the degradation of His-DMS3R3 was attenuated (SI Appendix, Fig. S4 D and E). To test the proteolytic regulation of endogenous DMS3, we conducted protein decay assays in plants. Ten-day-old Col-0 and apc8-1 seedlings were treated with cycloheximide to block de novo synthesis of DMS3. In Col-0, DMS3 began to be degraded after 1 h and was at a very low level after 2 h (Fig. 4 H and I), further indicating that DMS3 is a short-lived protein. In contrast, in apc8-1, similar levels of DMS3 persisted until 4 h (Fig. 4 H and I). Moreover, the addition of MG132 significantly stabilized DMS3 (Fig. 4 H and I). Notably, both RDM1 and DRD1, two other components of the DDR complex, were much more stable proteins than DMS3 (SI Appendix, Fig. S4F). Collectively, these results demonstrate that DMS3, but not RDM1, is directly targeted by APC/C for its degradation.
APC/C Optimizes the Assembly of the DDR Complex by Maintaining a Proper Abundance of DMS3.
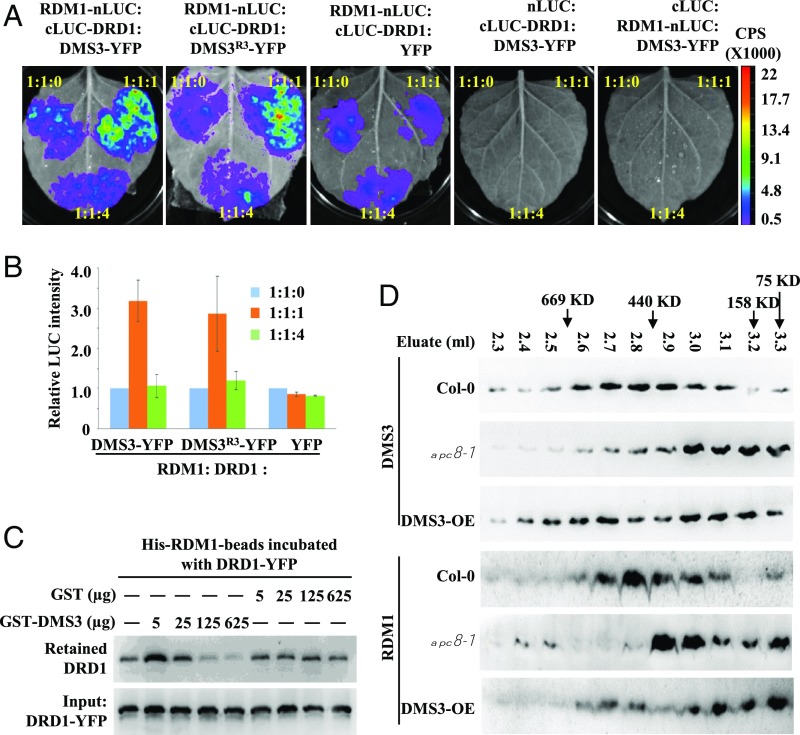
Since the protein level of DMS3 is controlled by APC/C (Fig. 4), and this regulation is important for the function of DMS3 in vivo (Fig. 3), we hypothesized that APC/C-dependent DMS3 regulation might be necessary for the assembly/association of the DDR complex. To test this hypothesis, we examined whether the amount of DMS3, RDM1, and DRD1 determines the extent of the interaction. We first evaluated the effects of DMS3 levels on the interaction between RDM1 and DRD1 by split luciferase complementation imaging assays. We fused RDM1 and DRD1 to the N-terminal fragment (nLUC) or the C-terminal fragment (cLUC), respectively, and introduced paired proteins into N. benthamiana leaves by infiltration. Consistent with previous results from co-IP and mass spectrometry analyses (30, 31), LUC activities were produced from RDM1 and DRD1 (Fig. 5 A and B), but no signal was detected when either RDM1 or DRD1 was paired with the empty vector (Fig. 5 A and B). Moreover, the LUC activity produced from RDM1 and DRD1 was significantly enhanced when an equal amount of DMS3 was supplied (Fig. 5 A and B), suggesting that the interaction between DRD1 and RDM1 was stabilized by DMS3. However, when more DMS3-YFP (fourfold more than the amount of RDM1 or DRD1) was added, the LUC activity produced from RDM1 and DRD1 was significantly attenuated (Fig. 5 A and B), although excess YFP alone showed no effect on the LUC activity produced from RDM1 and DRD1 (Fig. 5 A and B). Notably, the D box-mutated DMS3 exhibited similar effects on the DRD1–RDM1 interaction (Fig. 5 A and B), further indicating that the amount of DMS3 determines the association of the DDR components.
Fig. 5.
Proper level of DMS3 promotes the interaction between DRD1 and RDM1. (A) Split luciferase complementation imaging assays were performed to examine the RDM1–DRD1 interaction after the addition of different levels of DMS3 or DMS3R3. Three plasmids were inoculated into tobacco leaves at the indicated ratios, and YFP was used as the negative control. cLUC or nLUC was used to exclude background contamination. (B) Statistical analysis of LUC activity produced from the indicated combinations in A. The relative intensities of LUC were normalized with indiGo software (Berthhold Technologies). Data are presented as mean ± SE (n = 3). (C) Pull-down assays to analyze the interaction intensity between RDM1 and DRD1 after addition of different amounts of DMS3. Two milligrams of His-RDM1 conjugated onto nickel-nitrilotriacetic acid agarose beads was preincubated with DRD1-YFP, and different amounts of GST or GST-DMS3 were then added. After the reactions were stopped, the levels of DRD1-YFP retained on the beads (Top) and the initial amounts in each aliquot (Bottom) were quantified by Western blotting using an anti-GFP antibody. (D) Gel filtration of RDM1 and DMS3 in Col-0, apc8-1, and DMS3-OE plants. The proteins eluted from a Superose 6 column were detected by Western blotting with anti-RDM1 and anti-DMS3 antibodies. Eluate fractions and sizing standards are indicated. Specific bands are marked by arrowheads.
To further demonstrate the importance of the amount of DMS3 on assembly of the DDR complex, we performed pull-down assays to assess the role of DMS3 in the interaction intensity between RDM1 and DRD1. In this assay, the His-RDM1 beads were incubated with transiently expressed DRD1-YFP and the amounts of GST-DMS3 or GST alone gradually increased over 2 h. After stringent washing, the DRD1-YFP retained in the beads was detected by Western blotting using an anti-YFP antibody. Consistent with the results of the competitive split luciferase complementation imaging assay, the addition of excess GST-DMS3, but not of excess GST, significantly sequestered the association of DRD1 and RDM1 (Fig. 5C). These results indicate that the amount of DMS3 is important for the association of the DDR components.
Because the DMS3 level is controlled by APC/C for proper assembly of the DDR complex, we hypothesized that the association pattern of the DDR complex would be compromised in apc/c mutants. To test this hypothesis, we characterized the association between DMS3, RDM1, and Pol V by analyzing total protein extracts from Col-0 and apc8-1 by gel filtration, followed by Western blotting. The elution peak of the DDR complex is around ∼440 kDa in WT plants (30, 35). Using a Superose 6 column, we confirmed that the majority of DMS3 and RDM1 coeluted in Col-0 at ∼440 kDa (Fig. 5D) and that NRPE1 eluted as a broad high-molecular-mass peak at ∼669 kDa (SI Appendix, Fig. S5A). In contrast, the major peaks of both DMS3 and RDM1 were eluted in lower molecular-mass fractions in apc8-1 (Fig. 5D), indicating that the assembly of the DDR complex is disrupted in apc8-1. Consistent with findings that the function of the DDR complex is tightly related to Pol V function (30, 31), we showed that the Pol V-associated complex was also shifted from ∼669 kDa in Col-0 to lower molecular-mass fractions in apc8-1 (SI Appendix, Fig. S5A), further confirming the role of the DDR complex in the function of Pol V. Consistent with the requirement of APC/C in the assembly of the DDR complex, we showed that the addition of either APC10-GFP or Flag-APC10 attenuated the LUC activity produced from RDM1 and DMS3 (SI Appendix, Fig. S5 B and C), and the inhibitory effects were enhanced as the amount of APC10 increased (SI Appendix, Fig. S5 B and C). Taken together, these results indicated that APC/C optimizes the assembly of the DDR complex by maintaining a proper abundance of DMS3.
Overexpression of DMS3 Partially Phenocopies dms3 and apc/c.
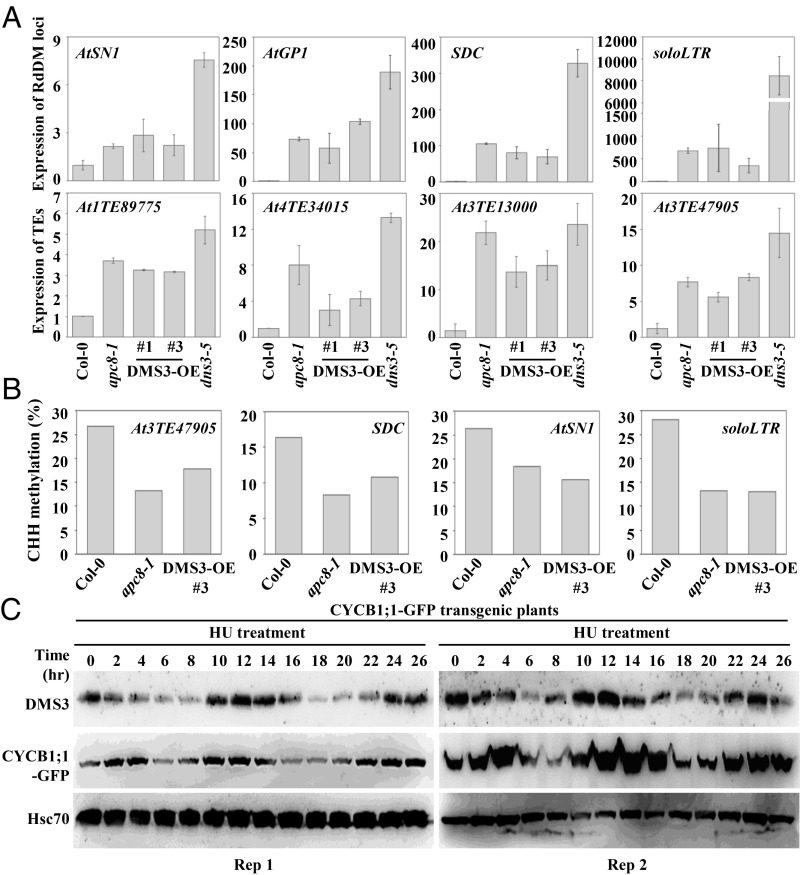
Since the level of DMS3 is important for association between two other DDR components, it was expected that overexpression of DMS3 in the WT plants would mimic or partially mimic defective RdDM activity seen in the apc/c and dms3 mutants. To test whether that was the case, we overexpressed DMS3 by introducing DMS3 driven by the 35S promoter into WT plants (Col-0) (SI Appendix, Fig. S5D). Similar to the results with the dms3 mutant, common RdDM loci, including AtSN1, AtGP1, SDC, and soloLTR, as well as TEs identified from our RNA sequencing, were de-repressed in two independent transgenic lines of DMS3-OE (overexpression of DMS3) (Fig. 6A), although the extent of de-repression was weaker than in the dms3 mutant (Fig. 6A). Moreover, bisulfite sequencing of AtSN1, soloLTR, SDC, and At3TE47905 showed that DNA methylation at CHH sites in DMS3-OE plants was decreased as in the apc8-1 mutant (Fig. 6B and Dataset S4). In contrast, DNA methylation at CHG and CG sites in DMS3-OE plants was comparable to that in Col-0 (SI Appendix, Fig. S5E and Dataset S4). Of note, an obvious reduction of CHG methylation at soloLTR was observed in DMS3-OE plants, similar to that seen in apc8-1 (SI Appendix, Fig. S5E and Dataset S4). These results show that overaccumulation of DMS3 impairs RdDM activity, further indicating the importance of APC/C-mediated surveillance of DMS3 in TE silencing.
Fig. 6.
Overexpression of DMS3 mimics impaired RdDM activity in dms3 and apc/c. (A) qRT-PCR to determine transcript levels of the RdDM loci AtSN1, AtGP1, soloLTR, SDC, At1TE89775, At4TE34015, At3TE13000, and At3TE47905 in Col-0, apc8-1, and DMS3-OE plants. Total RNA from inflorescences was analyzed. The UBQ5 gene was amplified as an internal control. Data are presented as mean ± SE (n = 3). (B) CHH methylation of AtSN1, SDC, soloLTR, and At3TE47905 was determined by bisulfite sequencing in Col-0, apc8-1, and DMS3-OE plants. (C) Western blotting analysis detected the endogenous DMS3 levels of root tips at indicated time points after HU treatments. DMS3 levels using anti-DMS3 antibody (Top), the CYCB1;1 level using anti-GFP antibody (a positive control regulated by APC/C) (Middle), and the loading control using anti-Hsc70 antibody (Bottom) are indicated. R1, biological replicate 1; R2, biological replicate 2.
To further determine whether impaired assembly of the DDR complex occurred in the DMS3-OE plants, we performed gel filtration followed by Western blotting to detect associations between DMS3 and RDM1 in Col-0 or DMS3-OE. As in the dms3 mutant (Fig. 5D), we found that DMS3 was distributed evenly in all fractions in the DMS3-OE plants (Fig. 5D), possibly due to significantly higher DMS3 levels. Interestingly, although the availability of DMS3 is enhanced, the major RDM1 peak was shifted to smaller molecular-mass fractions in DMS3-OE plants (Fig. 5D), notably different from the major RDM1 peak at ∼440 kDa in Col-0 (Fig. 5D), indicating that the assembly of the DDR complex is altered in DMS3-OE plants, similar to that in the dms3 and apc/c mutants. Collectively, these data indicate that a proper level of DMS3 is critical for the assembly of a functional DDR complex.
The Level of DMS3 Is Expressed in a Cell Cycle-Dependent Manner.
Because APC/C is a core regulator during cell cycle regulation, and because the substrates of APC/C are generally expressed in a cell cycle-dependent manner (18, 22, 47), the degradation of DMS3 by APC/C prompted us to investigate whether the protein level of DMS3 is expressed in a cell cycle-dependent manner. To this end, root tips from the pCYCB1;1::CYCB1-GFP reporter line were synchronized on medium containing hydroxyurea (HU). About 1-wk-old indicated plants were transferred to medium containing 2 mM HU; harvested; and ground for total protein extraction at 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, and 26 h after transfer. We showed that CYCB1;1-GFP, a well-known substrate of APC/C, stabilized after 10–16 h of synchronization and decreased after ∼18 h of synchronization, indicating that the HU treatment is effective. Interestingly, the endogenous DMS3 exhibited a pattern very similar to that of CYCB1;1 (Fig. 6C). By contrast, Hsc70, the loading control, remained unchanged after synchronization. The similarity of DMS3 and CYCB1;1 indicates that DMS3 is indeed regulated in a cell cycle-dependent manner.
To further conclusively demonstrate that DMS3 is regulated at the posttranscriptional level during cell cycle progression, root tips from the 35S::DMS3-YFP transgenic plants were also synchronized by HU. Unexpectedly, the level of DMS3-YFP showed a cell cycle-dependent trend similar to the endogenous DMS3 (SI Appendix, Fig. S6). Notably, the observation that the cell cycle-dependent amplitude of the endogenous DMS3 is much more obvious than that of the transgenic DMS3-YFP could be caused by the constitutive expression of DMS3-YFP in root cells. Taken together, these results indicate that DMS3 is a cell cycle-dependent component.
Discussion
An important question in the RdDM pathway is how the DDR complex is regulated to recruit Pol V. We provide evidence that APC/C, an E3 complex traditionally functioning in control of cell cycle, targets DMS3 for its degradation, and thus regulates the assembly of the DDR complex by maintaining a proper level of DMS3. Our findings provide a direct molecular link between selective protein degradation and TE silencing in plants, shedding light on how assembly of the DDR complex is exquisitely regulated. Moreover, together with the findings that APC/C targets DnMT1 (21), G9a (22), DRB4 (25), and MIWI (23), the role of APC/C in gene silencing is widely conserved in eukaryotes. In addition, since the R3 D box is highly conserved from monocots to dicots, it is plausible that the regulation of APC/C for DMS3 was positively selected for effective TE silencing during plant evolution.
Because DMS3 overaccumulated in apc/c mutants (Fig. 4), it was unexpected that TEs were also de-repressed in apc/c mutants, as shown in dms3 mutants (27–31). Mechanistically, we provide evidence that a precise stoichiometry among the three DDR components is critical for the formation of a functional DDR complex. Stoichiometric imbalances can sequester complex partners and disrupt the DDR multiprotein complex into nonfunctional subassemblies, such as DD (DRD1 and DMS3), DR (DMS3 and RDM1), or RD (RDM1 and DRD1). Among three DDR components, DMS3 has been found only to assemble with the DDR complex, whereas both RDM1 and DRD1 are also associated with other complexes. For example, RDM1 associates with Pol II (29), and DRD1 associates with Pol V (30). Therefore, it is reasonable why the amount of DMS3 has to be tightly controlled, because sorting of RDM1 and DRD1 among different protein complexes potentially attenuates the disadvantages of the imbalanced amounts.
In fission yeast, RNA interference (RNAi)-mediated histone methylation is regulated by the cell cycle. Centromeric repeats are transiently transcribed in the S phase of the cell cycle, and RNAi occurs specifically in the S phase to reconstitute histone modifications after DNA replication (48, 49). In plants, how heterochromatin silencing is inherited during cell divisions, and whether RdDM activity is regulated during the cell cycle, remains unexplored. Our finding that APC/C is involved in the regulation of RdDM activity provides a potential molecular link that RdDM activity might be dynamically regulated during the cell cycle. APC/C is activated from the late G2 phase, remains active throughout mitosis, and is specifically inactivated at the G1-to-S phase transition (1). If RdDM activity, similar to RNAi in yeast, is selectively activated during the S phase, it is plausible that the level of DMS3 is tightly controlled by APC/C before and after the S phase, thus delimiting a window for effective RdDM activity during cell cycle progression. Indeed, we show that DMS3 fluctuates during cell cycle progression (Fig. 6C and SI Appendix, Fig. S6). In conclusion, this study offers a perspective for the regulation of RdDM and uncovers the importance of a proper level of DMS3 controlled by APC/C.
Materials and Methods
Plant Materials, Regents, and Plasmid Construction.
T-DNA insertion mutants of nrpe1-12, pol iv, and dms3-5 were obtained from the Arabidopsis Information Resource. Details are provided in SI Appendix, Supplemental Materials and Methods.
RNA Sequencing and Small RNA Sequencing.
The raw and processed data for both RNA sequencing and small RNA sequencing have been deposited into the National Center for Biotechnology Information Gene Expression Omnibus database under the series accession number GSE106972. The original data can be reached by the link https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE106972 and the secure token: afyhqmwgxnqnlep. Details are provided in SI Appendix, Supplemental Materials and Methods.
Cell-Free Decay Assay.
Recombinant proteins were incubated with cell lysates from plants and analyzed by SDS/PAGE, followed by immunoblots. Details are provided in SI Appendix, Supplemental Materials and Methods.
In Vitro Ubiquitination Assay.
In vitro APC/C-mediated ubiquitination was carried out as described (50). Details are provided in SI Appendix, Supplemental Materials and Methods.
Gel Filtration.
Gel filtration was carried out as described (30) with modifications. Details are provided in SI Appendix, Supplemental Materials and Methods.
DNA Methylation Assay.
Genomic DNA was treated using the BisulFlash DNA Modification Kit (catalog no. P-1026-050; Epigentek). Details are provided in SI Appendix, Supplemental Materials and Methods.
Synchronization by HU Treatment.
The synchronization was performed as described (47). Details are provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Xiuren Zhang and Prof. Sheila McCormick for critically reading the manuscript. We thank Dr. Xinjian He for providing seeds of dms3-5, ros1-1 rdm1-1, and 35S::DRD1-Flag. We thank Dr. Qi Xie for sharing regents and providing valuable suggestions for in vitro ubiquitination assays. We are grateful to Drs. Xinjian He, Heng Zhang, and Weiqiang Qian for valuable suggestions on bisulfite sequencing analyses. This work was supported by the National Natural Science Foundation of China (Grants 31830045, 31470281, and 31671261), the Shanghai Pujiang Program (Grant 13PJ1401200), the Higher Education of China Program (Grant 20120071120012), and the Recruitment Program of Global Expects (China).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE106972).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816652116/-/DCSupplemental.
References
- 1.Chang LF, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kominami K, Seth-Smith H, Toda T. Apc10 and Ste9/Srw1, two regulators of the APC-cyclosome, as well as the CDK inhibitor Rum1 are required for G1 cell-cycle arrest in fission yeast. EMBO J. 1998;17:5388–5399. doi: 10.1093/emboj/17.18.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grossberger R, et al. Characterization of the DOC1/APC10 subunit of the yeast and the human anaphase-promoting complex. J Biol Chem. 1999;274:14500–14507. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- 4.Wendt KS, et al. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 5.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capron A, et al. The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell. 2003;15:2370–2382. doi: 10.1105/tpc.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwickart M, et al. Swm1/Apc13 is an evolutionarily conserved subunit of the anaphase-promoting complex stabilizing the association of Cdc16 and Cdc27. Mol Cell Biol. 2004;24:3562–3576. doi: 10.1128/MCB.24.8.3562-3576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng B, Chen X, McCormick S. The anaphase-promoting complex is a dual integrator that regulates both MicroRNA-mediated transcriptional regulation of cyclin B1 and degradation of cyclin B1 during Arabidopsis male gametophyte development. Plant Cell. 2011;23:1033–1046. doi: 10.1105/tpc.111.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy O, et al. lemmingA encodes the Apc11 subunit of the APC/C in Drosophila melanogaster that forms a ternary complex with the E2-C type ubiquitin conjugating enzyme, Vihar and Morula/Apc2. Cell Div. 2012;7:9. doi: 10.1186/1747-1028-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. The Arabidopsis APC4 subunit of the anaphase-promoting complex/cyclosome (APC/C) is critical for both female gametogenesis and embryogenesis. Plant J. 2012;69:227–240. doi: 10.1111/j.1365-313X.2011.04785.x. [DOI] [PubMed] [Google Scholar]
- 11.Davey NE, Morgan DO. Building a regulatory network with short linear sequence motifs: Lessons from the degrons of the anaphase-promoting complex. Mol Cell. 2016;64:12–23. doi: 10.1016/j.molcel.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley AM, Williams BC, Goldberg ML, Andres AJ. Phenotypic characterization of Drosophila ida mutants: Defining the role of APC5 in cell cycle progression. J Cell Sci. 2002;115:949–961. doi: 10.1242/jcs.115.5.949. [DOI] [PubMed] [Google Scholar]
- 13.Deak P, Donaldson M, Glover DM. Mutations in mákos, a Drosophila gene encoding the Cdc27 subunit of the anaphase promoting complex, enhance centrosomal defects in polo and are suppressed by mutations in twins/aar, which encodes a regulatory subunit of PP2A. J Cell Sci. 2003;116:4147–4158. doi: 10.1242/jcs.00722. [DOI] [PubMed] [Google Scholar]
- 14.Kwee HS, Sundaresan V. The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J. 2003;36:853–866. doi: 10.1046/j.1365-313x.2003.01925.x. [DOI] [PubMed] [Google Scholar]
- 15.Guo L, et al. The anaphase-promoting complex initiates zygote division in Arabidopsis through degradation of cyclin B1. Plant J. 2016;86:161–174. doi: 10.1111/tpj.13158. [DOI] [PubMed] [Google Scholar]
- 16.Guo L, Jiang L, Lu XL, Liu CM. ANAPHASE PROMOTING COMPLEX/CYCLOSOME-mediated cyclin B1 degradation is critical for cell cycle synchronization in syncytial endosperms. J Integr Plant Biol. 2018;60:448–454. doi: 10.1111/jipb.12641. [DOI] [PubMed] [Google Scholar]
- 17.Min M, Lindon C. Substrate targeting by the ubiquitin-proteasome system in mitosis. Semin Cell Dev Biol. 2012;23:482–491. doi: 10.1016/j.semcdb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 19.Pfleger CM, Kirschner MW. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- 20.Di Fiore B, et al. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell. 2015;32:358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghoshal K, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Takahashi A, et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol Cell. 2012;45:123–131. doi: 10.1016/j.molcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24:13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Gou LT, et al. Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell. 2017;169:1090–1104.e13. doi: 10.1016/j.cell.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrocco K, et al. APC/C-mediated degradation of dsRNA-binding protein 4 (DRB4) involved in RNA silencing. PLoS One. 2012;7:e35173. doi: 10.1371/journal.pone.0035173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 27.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 28.Kanno T, et al. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- 29.Gao Z, et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law JA, et al. A protein complex required for polymerase V transcripts and RNA- directed DNA methylation in Arabidopsis. Curr Biol. 2010;20:951–956. doi: 10.1016/j.cub.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19:870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maor R, et al. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu ZW, et al. The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 2014;10:e1003948. doi: 10.1371/journal.pgen.1003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC. The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development. 2009;136:1475–1485. doi: 10.1242/dev.035535. [DOI] [PubMed] [Google Scholar]
- 37.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 39.Li CF, et al. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 40.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013;498:385–389. doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 42.He XJ, et al. An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moissiard G, et al. MORC family ATPases required for heterochromatin condensation and gene silencing. Science. 2012;336:1448–1451. doi: 10.1126/science.1221472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 45.Buschhorn BA, et al. Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat Struct Mol Biol. 2011;18:6–13. doi: 10.1038/nsmb.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cools T, Iantcheva A, Maes S, Van den Daele H, De Veylder L. A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J. 2010;64:705–714. doi: 10.1111/j.1365-313X.2010.04361.x. [DOI] [PubMed] [Google Scholar]
- 48.Chen ES, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 49.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kraft C, Gmachl M, Peters JM. Methods to measure ubiquitin-dependent proteolysis mediated by the anaphase-promoting complex. Methods. 2006;38:39–51. doi: 10.1016/j.ymeth.2005.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.