Significance
Paneth cells provide intestinal host defense against pathogens and control the healthy microbiota by secreting antimicrobial peptides. We show that the most abundant secreted Paneth cell products, human defensin HD-5 and HD-6, show a distinct susceptibility to proteolytic digestion by human duodenal fluid. While HD-5 is digested in many fragments, HD-6 is stable and still able to form nanonets. The occurring fragments of HD-5 were antimicrobially active against microorganisms. We provide proof of concept about microbiome modulating capacities in vivo, which includes an increase of Akkermansia sp. Our results indicate that fragmentation of defensins increases antimicrobial diversity and further adds to the complexity of host microbial interaction at interfaces. Fragmentation could lead to new antimicrobial peptides with possible therapeutic usage.
Keywords: antimicrobial peptides, α-defensins, intestinal barrier, proteolytic digestion, host–microbiota interaction
Abstract
Antimicrobial peptides, in particular α-defensins expressed by Paneth cells, control microbiota composition and play a key role in intestinal barrier function and homeostasis. Dynamic conditions in the local microenvironment, such as pH and redox potential, significantly affect the antimicrobial spectrum. In contrast to oxidized peptides, some reduced defensins exhibit increased vulnerability to proteolytic degradation. In this report, we investigated the susceptibility of Paneth-cell–specific human α-defensin 5 (HD-5) and -6 (HD-6) to intestinal proteases using natural human duodenal fluid. We systematically assessed proteolytic degradation using liquid chromatography–mass spectrometry and identified several active defensin fragments capable of impacting bacterial growth of both commensal and pathogenic origins. Of note, incubation of mucus with HD-5 resulted in 255–8,000 new antimicrobial combinations. In contrast, HD-6 remained stable with consistent preserved nanonet formation. In vivo studies demonstrated proof of concept that a HD-5 fragment shifted microbiota composition (e.g., increases of Akkermansia sp.) without decreasing diversity. Our data support the concept that secretion of host peptides results in an environmentally dependent increase of antimicrobial defense by clustering in active peptide fragments. This complex clustering mechanism dramatically increases the host’s ability to control pathogens and commensals. These findings broaden our understanding of host modulation of the microbiome as well as the complexity of human mucosal defense mechanisms, thus providing promising avenues to explore for drug development.
Human body surfaces, especially the intestine, are host to trillions of microbial organisms, mostly bacteria (1), and collectively termed the “microbiota.” It is widely recognized that the gut microbiota have multiple functions and that a disturbed microbial consortium has been associated with multiple diseases including multiple sclerosis, autism, obesity, and chronic inflammatory bowel diseases (2–6). Most recently, the microbiome composition has also been demonstrated to dramatically influence human response rates and survival of anticancer treatments with checkpoint inhibitors (7, 8).
It is indispensable for the host to preserve a functional intestinal barrier and to create a stable homeostasis with the microbiota (9–12). One of the most important components of the intestinal barrier are antimicrobial peptides (AMPs), which are secreted from epithelial cells throughout the gut. These peptides ensure a mostly sterile inner mucus layer and protect against different microbes, fungi, and archaea viruses (13–15). Defensins are small cationic molecules, characterized by three conserved disulphide bonds (16), and represent a main group of AMPs. In the small intestine, Paneth cells play a key role in balancing the microbiota composition and protecting the host from invading pathogens by secretion of a variety of AMPs but most abundantly α-defensin 5 (HD-5) and -6 (HD-6) (2, 17, 18). These secretory Paneth cells are located at the bottom of the crypts of Lieberkühn and are highly controlled by Wnt signaling due to their differentiation and their defensin regulation and expression (19–21). Transgenic mice expressing HD-5 or HD-6 under control of their endogenous promoters were protected against Salmonella typhimurium infections, and both peptides strongly influenced the microbiota composition (10, 22–24). A normal Paneth cell function in humans is needed to allow a balanced host–microbe relationship and a functional intestinal barrier, while an abnormal Paneth cell with reduced expression or secretion of HD-5 or HD-6 is associated with inflammatory bowel disease (2, 9, 25, 26).
Of note, HD-5 and HD-6 are functionally different. While the strong antimicrobial activity of HD-5 has been known for years (27, 28), little is known about the antimicrobial activity of HD-6. It has, however, recently been demonstrated that HD-6 forms self-assembled nanonets that trap bacteria and prevent infections (23). Moreover, under reducing conditions, which is the natural state in the gut (29), a direct antimicrobial activity of HD-6 could be observed, while the bacteria-trapping nanonet formation was still present (30). The reduction—which is naturally present owing to the low redox potential in the gut or enzymatically generated by the thioredoxin system (30–33)—leads to an opening of the three disulphide bridges, followed by a change in the tertiary structure of these peptides (31, 34). This structural change leads to a posttranslational modification of the antimicrobial spectrum (30, 31, 35). Importantly, these open, linear forms have been detected in the small intestine (31, 36). In contrast to the oxidized original peptide, we previously reported that the reduced linear peptide structures of hBD-1 were susceptible to proteases (37). It is commonly believed that the degradation process facilitates antimicrobial inactivation and/or loss of function (37). Here, we hypothesized that protein/defensin degradation is rather a novel activation mechanism mediated by the formation of new antimicrobial components. We thus investigated the susceptibility of HD-5 and HD-6 to proteolytic degradation from human duodenal fluid under reducing conditions to mimic the in vivo intestinal situation. We assessed the amino acid signature based on the exact masses and synthesized the newly identified fragments for further studies. We then systematically tested the antimicrobial spectrum of the resulting peptide fragments against a subset of commensal and pathogenic bacteria. Surprisingly, Paneth cell defensins were shown to be differentially affected by human mucus, but their overall antimicrobial spectrum was dramatically increased by the generation of new active antimicrobial fragments, thus uncovering a key mucosal defense mechanism for fine-tuning of host microbial interaction in the gut.
Results
Natural Human Duodenal Fluid Digests HD-5, While Nanonet-Forming HD-6 Is Protease-Resistant.
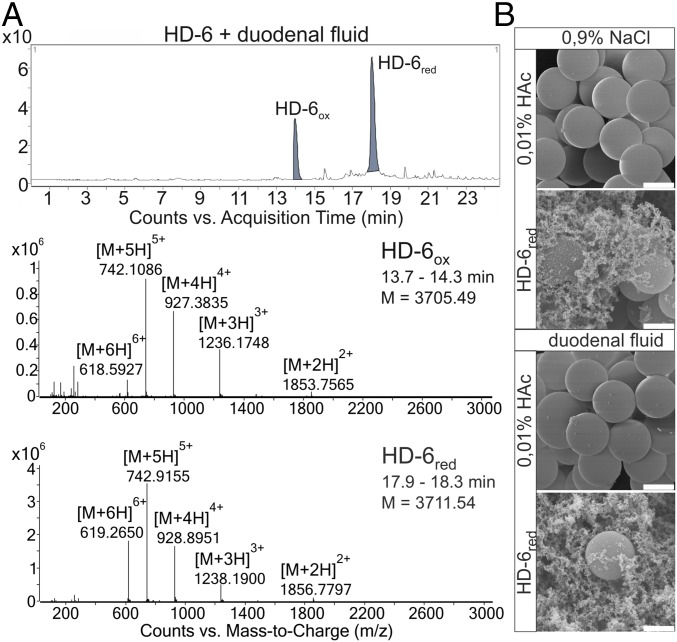
Since Paneth cell defensins can be reduced by the naturally occurring thioredoxin system (30, 33), we investigated their susceptibility to a proteolytic digest. We used the PeptideMass module of ExPASy (SIB Bioinformatics Resource Portal) for in silico digests of HD-5 and HD-6 with trypsin, chymotrypsin, or a combination of both and allowed up to five missed cleavages. Both Paneth cell defensins seemed to be susceptible to proteases although HD-6 showed a tendency of more fragmentation compared with HD-5 (SI Appendix, Table S1). We did not detect any HD-5– or HD-6–related fragments in the duodenal fluid alone (SI Appendix, Fig. S1). Therefore, we next used Tris(2-carboxyethyl)phosphine (TCEP) to reduce HD-5 or HD-6 and challenged the peptides to naturally occurring duodenal fluid, which is known to be proteolytically active (38). We detected a partial reduction with 2 mM TCEP after adding HD-6, consistent with previous our work (30). We identify only the two full-length forms, HD-6ox (expected: 3,705.49 Da) and HD-6red (3,711.54 Da), by mass-to-charge ratio (m/z) signals indicating two-, three-, four-, five-, and sixfold protonated ions (Fig. 1A). This observation demonstrates that HD-6red is protected against proteolytic digestion although the reason remains elusive, since proteolytic cleaving sites were bioinformatically predicted. It is known that both HD-6ox and HD-6red are able to form nanonets independently of their redox state (23, 30). We hypothesized that net formation protects against protease degradation and thus might provide a mechanistic explanation for the observed peptide protection. To clarify if nanonet formation leads to a more stable structure, we performed scanning electron microscopy from reduced HD-6 incubated with duodenal fluid or NaCl. Consistent with this hypothesis, we observed equal nanonets independent of duodenal fluid incubation (Fig. 1B). There is therefore a clear correlation between nanonet formation and protection against proteolytic digestion.
Fig. 1.
HD-6 nanonet formation is not affected by duodenal fluid. (A) Here we show the chromatograph of only exogenously added HD-6 incubated with duodenal fluid after reduction with 2 mM TCEP. We detected only the oxidized and reduced full-length peptides due to their retention time with their m/z graphs and their two-, three-, four-, five- and sixfold protonated ions and neutral masses. (B) We incubated beads with 200 µg/mL reduced HD-6 or 0.01% HAc (control) and duodenal fluid or 0.9% NaCl (control). The nanonet formation of HD-6 seemed to be unaffected by the duodenal fluid, because it still forms nanonets comparable to those in the 0.9% NaCl control. (Scale bar: 0.2 µm.)
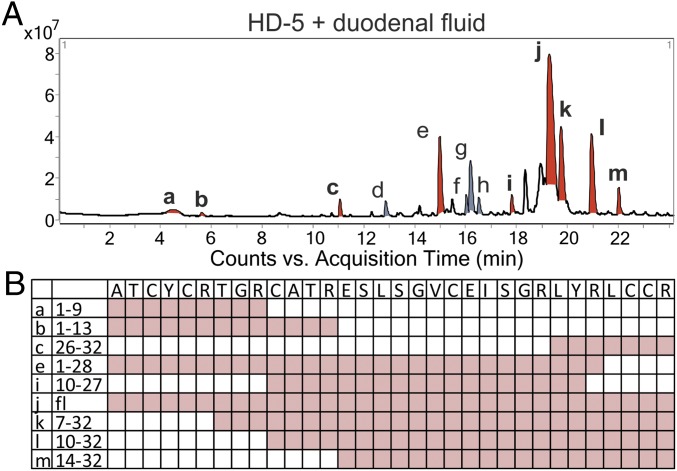
We next performed identical experiments with the second and more abundant Paneth cell defensin, HD-5. After incubation with 2 mM TCEP and duodenal fluid, HD-5ox was below the limit of detection, whereas HD-5red was highly abundant. In contrast to HD-6, duodenal fluid digestion of HD-5 induced fragmentation of the peptide, consistent with known cleaving sites (Fig. 2A and SI Appendix, Fig. S1; SI Appendix, Table S1, data highlighted in blue). The identified fragments were listed with their mass-to-charge ratios, neutral mass, and retention time, showing that fragmentation of HD-5 with duodenal fluid led to abundant fragments derived from the entire peptide sequence (SI Appendix, Fig. S2). However, we still found detectable amounts of full-length HD-5red, suggesting that the proteolytic digestion was incomplete. In summary, our data show that duodenal fluid affects HD-5 and HD-6 differently and that changes of conditions in the local microenvironment can have a major an impact on defensin fragmentation. Of note, HD-6 nanonet formation seems to protect against destruction of the reduced full-length peptide activity. Therefore, reduction changes the activity of both Paneth cell defensins, but while HD-6 gains a direct antimicrobial activity (30), HD-5 is digested by intestinal proteases to form peptide fragments with potential antimicrobial activity.
Fig. 2.
Incubation of HD-5 and duodenal fluid produced many different fragments. We incubated only HD-5 with duodenal fluid after reduction with 2 mM TCEP. (A) Here we show the overview of the chromatogram from an incubation of reduced HD-5 with duodenal fluid. All detectable fragments were marked in red or gray (a–m). All peptides marked in red were chosen for synthesis and deeper investigation of their abilities. (B) The chosen fragments with their name and amino acid sequence and their distribution over the HD-5 sequence.
HD-5 Fragments Display Antimicrobial Activity Against Commensal and Antibiotic-Resistant Bacteria.
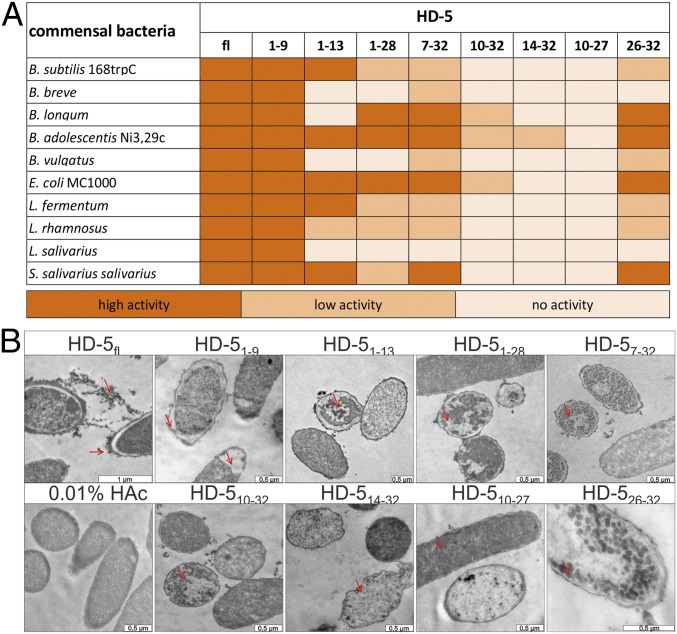
To test our hypothesis of antimicrobial activity from the HD-5 fragments, we chemically synthesized selected fragments to investigate antimicrobial functions in vitro (Fig. 2B). We performed several radial diffusion assays with of 4 µg of each fragment and 2 µg of the full-length peptide against commensal gut and pathogenic bacteria. As expected, the full-length HD-5 (HD-5fl) displayed broad antimicrobial activity. Interestingly, we observed that most of HD-5–derived fragments were antimicrobially active against these bacteria. We additionally found that the different fragments exhibited distinct activity patterns (Fig. 3A and SI Appendix, Fig. S3). Still, we identified HD-51–9 as the most bioactive peptide in terms of antimicrobial activity but also in terms of versatility since it was active against all tested bacteria. In contrast to HD-51–9, HD-510–27 did not exhibit measurable activity. Of note, HD-51–13, HD-51–28, HD-57–32, and HD-526–32 also exhibited antimicrobial properties, albeit to a lesser extent than HD-51–9. To further assess the resulting phenotype of peptide-treated bacteria, we incubated Escherichia coli MC1000 with all fragments and performed transmission electron microscopy (TEM) (Fig. 3B). We observed that HD-5fl treatment led to a detached inner membrane and small vesicular structures around the bacteria cell envelope. Similar structures were observed by Chileveru et al. (39). Of interest, exposed bacteria exhibited phenotypic discrepancies after incubation with different HD-5 fragments, indicating a fragment-specific mode of action. For HD-51–9, we identified a detached inner membrane with additional big vacuole structures at one pole of the bacteria, while HD-51–13 and HD-51–28 treatment led to bigger aggradation inside the bacteria. In contrast to HD-5fl, HD-57–32, HD-510–32, and HD-526–32 showed small vesicular structures inside the bacteria. HD-514–32 and HD-510–27 were not active in the radial diffusion assays (RDAs), as we detected only minor damage in some bacteria while the majority remained unaffected. This selectivity suggests that minor differences in fragment sequences (e.g., HD-51–9 and HD-51–13) could result in different antimicrobial activity, thus providing a fine-tuning mechanism of host–microbe interaction. Our results further indicate that the antimicrobial activity of the different peptides was strain-specific. As an example, the Bifidobacteria strains Bifidobacterium adolescentis and Bifidobacterium longum were highly susceptible to the peptides, while Bifidobacterium breve was not.
Fig. 3.
HD-5 fragments are antimicrobially active peptides against commensal bacteria. (A) We tested different commensal bacteria due to their susceptibility to HD-5 fragments. In this heat map, we listed all bacteria and the activity of the fragments in RDA against them. In the RDA, we used 2 µg of the full-length peptide and 4 µg of each fragment. An inhibition zone greater than 5 mm was determined as high activity, between 2.5 and 5 mm as low activity, and 2.5 mm (the diameter of the punched well) as no activity. (B) To investigate the mode of action of the different peptides, we incubated E. coli MC1000 with all the different fragments, performed transmission electron microscopy, and analyzed the resulting phenotypes. [Scale bars of all pictures are 0.5 µm except the full-length peptide (1 µm).]
We next investigated the antimicrobial activity against pathogenic Gram-positive and Gram-negative bacteria (SI Appendix, Fig. S4). The fragments were also here found to be antimicrobially active. We observed that HD-51–9, HD-51–13, HD-57–32, and HD-5fl had a strong effect on the growth of every tested bacterium, while other fragments like HD-51–28, HD-510–32, and HD-526–32 were only minimally active or more selective in terms of different strains. In contrast, HD-514–32 and HD-510–27 were inactive against the tested bacteria under identical conditions. Of interest, the activity of HD-510–32 was limited to the two Gram-positive bacteria. Consistent with the findings described in the first experiment (Fig. 3), and in terms of their efficacy to kill commensals, HD-51–9 was active against all tested strains independent of Gram status, suggesting that this specific fragment is highly protective against bacterial barrier breach. In contrast, HD-51–13 potently regulated tested pathogenic bacteria, with the exception of Klebsiella pneumoniae 3-MRGN. Altogether, our results indicate that a proteolytic digestion of Paneth-cell HD-5 but not HD-6 leads to the generation of novel antimicrobial fragments. The significance of this finding may prove important because these fragments broaden the antimicrobial selectivity.
To gain a more detailed understanding of the antimicrobial abilities of the different HD-5 fragments and their potential therapeutic use, we next performed turbidity broth assays to determine the minimal inhibitory concentration (MIC) of these peptides, defined as the concentration (in micromoles) at which no bacterial growth was detectable after 12 h. We found that the antimicrobial activity of HD-51–9, HD-51–13, HD-51–28, HD-57–32, and HD-510–32 was in the similar range to the full-length peptide (SI Appendix, Table S2). Of note, our observed MICs were relatively high, except for HD-51–9 for Acinetobacter baumannii 4-MRGN and Enterococcus faecium 475747, suggesting that the role of antimicrobial substances in the intestinal barrier is restricted to areas exposed to high peptide concentrations, such as the crypts. This is particularly important as it points toward protection of bacterial encroachment with less severe impact on the luminal gut microbiota composition, thereby preserving host physiology.
HD-51–9 Treatment Affects Certain Bacterial Genera and Transiently Increases Microbial Diversity in the Small Intestine.
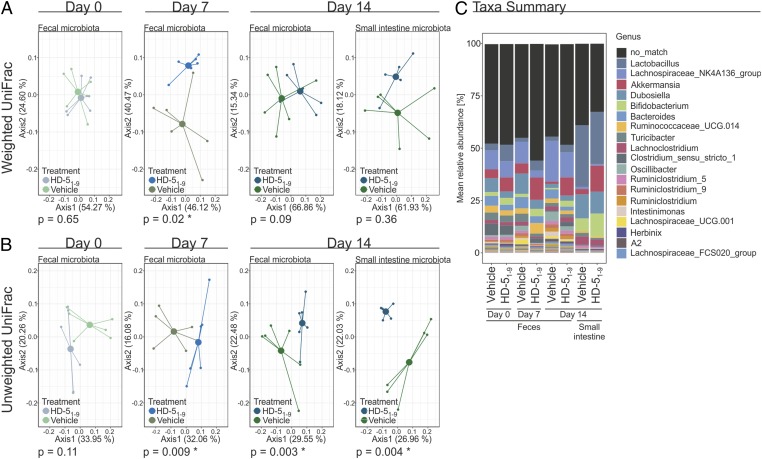
To investigate the ability of identified HD-5 fragments to modulate the microbiota, we treated mice with HD-51–9 or PBS orally for 7 d (7.19 µg/mouse), followed by a 7-d washout. The microbiota composition of the two groups of mice were indistinguishable at baseline [Adonis permutational multivariate analysis of variance (PERMANOVA) using Weighted and Unweighted Unifrac Distances, P = 0.65 and 0.11, respectively] (Fig. 4 A and B). After 7 d of HD-51–9 treatment, there was a significant divergence in the overall microbiota composition in fecal samples compared with the control (Adonis PERMANOVA using Weighted and Unweighted Unifrac Distances, P = 0.02 and 0.009, respectively, Fig. 4 A and B) but not compared with the group’s baseline microbiota (SI Appendix, Fig. S5A). As HD-5 is secreted by Paneth cells of the small intestine, we therefore investigated the microbial community of this compartment at necropsy. The microbial community structure in the small intestine of previously HD-51–9–treated mice was not affected in the overall abundances of bacteria (Weighted Unifrac Distances) after 7 d of washout (Fig. 4A, P = 0.36), but was remarkably different at the Unweighted Unifrac Distances (Fig. 4B, P = 0.004), suggesting that HD-51–9–mediated alterations were restricted to low abundant genera. The data were reproduced in two independent experiments, with and without a washout period, exhibiting consistent results.
Fig. 4.
HD-51–9 treatment did not affect the microbiota diversity but has an influence on certain bacteria strains. We treated chow-fed wild-type mice for 7 d orally with 7.19 µg/mouse HD-51–9 or PBS (six mice per group). After this period, we stopped the treatment. Fecal samples were collected for days 0, 7, and 14. Additionally, small intestine samples were taken after 14 d. (A) Here we show a principal coordinate analysis (PCoA including group mean) of fecal or small intestine microbiota composition using Weighted UniFrac Distances at days 0, 7, and 14, respectively. There was no difference in baseline microbiota between the groups at day 0 (Adonis PERMANOVA P = 0.65). After 7 d of treatment, the fecal microbial abundances were significantly different between the groups (Adonis PERMANOVA P = 0.02), but not when comparing each group to its baseline (Adonis PERMANOVA P > 0.05). At day 14, 7 d after washout, neither the fecal nor the small intestine microbial abundance was significant between the groups (Adonis PERMANOVA P = 0.09 and P = 0.36, respectively). (B) PCoA including group mean of fecal or small intestine microbiota composition using Unweighted UniFrac Distances at days 0, 7, and 14, respectively. The baseline microbiota did not differ between the groups regarding absence and presence of bacteria (Adonis PERMANOVA P = 0.11). After 7 d of treatment, the groups differed significantly in fecal microbiota calculated by Unweighted UniFrac Distances (Adonis PERMANOVA P = 0.009), which persisted after the 1-wk washout in fecal samples (Adonis PERMANOVA P = 0.003) and small intestine samples (Adonis PERMANOVA P = 0.004). Despite this, the groups did not at any time point differ significantly from their baseline composition by Unweighted UniFrac Distance (Adonis PERMANOVA P > 0.05). (C) HD-51–9 treatment affects the abundance of bacterial genera. Using a linear mixed-effect model, we identified a significant increase of Akkermansia sp. in the feces samples in mice treated with HD-51–9 (P = 0.024), while this increase was not observed in PBS-treated mice. For our small intestine samples, we observed a significantly higher abundance of Akkermansia sp. in the HD-51–9–treated than in the PBS-treated group (P = 0.008).
The Shannon diversity of the fecal microbiota composition was equal between the groups at baseline (Wilcoxon test, P > 0.99) and remained similar after 7 d of HD-51–9 treatment (Wilcoxon test, P = 0.7687) and after the washout at day 14 (Wilcoxon test, P = 0.3389; SI Appendix, Fig. S5B, Left). In contrast to these findings, we observed an increased bacterial diversity in HD-51–9–treated mice compared with PBS gavaged control mice in the small intestine (Wilcoxon test, P = 0.004; SI Appendix, Fig. S6A, Right). This change was not identified in small intestine samples after a 7-d washout period (Wilcoxon test, P = 0.45; SI Appendix, Fig. S5B, Right), suggesting that HD-51–9 induces transient, and nonpersistent, changes in the small intestinal microbiome.
The experimental design further allowed us to perform paired analyses of the fecal samples using a linear mixed model with repeated sampling from the same mice. We identified a significant effect of HD-51–9 on a few microbial genera in the fecal samples and a single biological relevant symbiont. More specifically, we observed a decrease in relative abundance of the genera Bacteroides and Lactobacillus, while Parasutterella was increased by HD-51–9 treatment. More importantly, we observed an increase of Akkermansia sp. in fecal samples of HD-51–9–treated mice (linear mixed model, P = 0.024, Fig. 4C; SI Appendix, Fig. S7A). This observation confirmed the findings of a similar experiment, without a washout period, where Akkermansia abundances were significantly affected by HD-51–9 treatment (linear mixed model, P < 0.001; SI Appendix, Fig. S6B, Left). The small intestine microbiota was broadly similar between the two groups at day 14, although the relative abundance of Akkermansia was increased by HD-51–9 (linear mixed model, P = 0.008, Fig. 4C), matching our result from the previous experiment (linear mixed model, P = 0.002, See SI Appendix, Fig. S6B, Right). In addition, the genus Ruminococcus 1 of the Ruminococcaceae family was increased in relative abundance while Intestinimonas ASF356 of the Clostridiaceae family, and Ruminococcaceae UCG-013 of the Ruminococcaceae family, were decreased in relative abundances (Fig. 4C).
Combined, these results demonstrate that HD-51–9 alters the proportion of certain bacteria in the fecal microbiota without affecting the overall community structure or diversity in healthy chow-fed mice.
Encouraged by the increased Akkermansia abundances, we next assessed whether the bacterium was either susceptible to or protected from HD-51–9–killing activity using a turbidity broth assay. Despite broad antimicrobial activity against both commensal and pathogenic bacteria, HD-51–9 failed to kill this important symbiont even at high concentrations. We did, however, observe mild nonsignificant obstruction of growth rate already at the lowest concentration tested. Importantly, this modest impact on growth rate was not further promoted by increasing concentration of HD-51–9, suggesting that this specific fragment preserved Akkermansia presence in the gastrointestinal tract (SI Appendix, Fig. S7B).
Discussion
During this study we made a number of observations regarding α-defensins and their susceptibility to proteases. Based on indications from the in silico digest of HD-6, it was rather surprising that HD-6 was unaffected by the naturally occurring proteases in the duodenal fluid. Unlike HD-6, HD-5 was degraded and its fragments contained antimicrobial activity against commensal and pathogenic bacteria. In several studies, HD-6 has been described as a nanonet-forming defensin in both oxidized and reduced forms (23, 30). This net formation seems to play a key role as part of the mucosal defense in shielding the host from bacteria penetrating the mucus layer. This concept was put forward by Chu et al. (23) who showed that HD-6 transgenic mice were resistant to S. typhimurium infection because of these nanonets. Based on our experiments these nanonets seem to underlie the stability of HD-6 against proteolytic digestion. In contrast to HD-6, we identified a number of different HD-5 fragments after incubation of the reduced HD-5 with human duodenal fluid. However, the digestion of HD-5 depended on the environmental conditions. The antimicrobial spectrum was different from that of the full-length molecule, and these HD-5 fragments thus seem to add further bacterial killing and/or microbiota modulation capability. Additionally, our results suggest fragment-specific killing efficacy, where each fragment exhibits a unique antimicrobial strategy as revealed by TEM experiments. This interesting phenomenon also contributes to the understanding of how a few intestinal defensins can modulate or support very different commensal colonization statuses in different parts of the intestine (SI Appendix, Fig. S8). Although not formally assessed in this report, we hypothesize that the mentioned degradation occurs in biologically relevant amounts in the crypts of human hosts, even in the absence of duodenal fluid. In support of this hypothesis, HD-5 exists in high concentrations (0.5–2.5 mg/g) in the crypts of terminal ileum mucosa, is activated by trypsin with most of our cleavage sites predicting tryptic digestion, and is reduced by thioredoxin, which occurs in human crypts (32, 40).
Moreover, the used fragments were shown to be antimicrobially active against some pathogenic bacteria. Surprisingly, we could not identify any antimicrobial activity against K. pneumoniae 3-MRGN, while we were able to detect some active fragments in the RDA. Consistent with the data shown here and previous findings (38), it is obvious that the antimicrobial activities in the RDA and turbidity broth assays are not easy to compare. We observed significant differences between these two methods, especially for HD-51–13 and HD-526–32 fragments. As evident from previous studies, the local microenvironment controls the antimicrobial function of different defensins including hBD-1 and HD-6 (30, 31, 38). The local conditions inherent in those two assays are very different, with particular emphasis on the very high local concentration of peptides in the RDA. This could, together with the immobilization of the bacteria in the used gel, be potential explanations for different findings. However, taken together, our results indicate a generally strong antimicrobial potential in vitro. Future research studies are warranted to carefully characterize potential additive, synergistic, or antagonistic synergies between in situ-generated fragments. However, preliminary studies from the B.A.H.J. laboratory indicate that combining the most potent fragments, i.e., HD-51–9, HD-51–28, and HD-57–32, preserve the selective increase of Akkermansia sp. observed in this report and also specifically diminish Helicobacter abundance, an important pathobiont.
It has been shown that HD-5 changes the microbiome of mice (10, 24). We thus wanted to investigate if HD-51–9 possessed similar microbiota-modulating effects. In contrast to the effects of a standard antibiotics treatment, we did not detect loss of bacteria strains. Instead, we observed increased bacterial richness, likely rooted in a reduction of targeted high abundant bacteria, creating new niches for otherwise low abundant bacteria. In addition to the effects on some low abundant bacteria strains, our results showed that mice treated orally with HD-51–9 had an increased amount of Akkermansia sp. compared with nontreated mice. Moreover, in contrast to other bacteria, Akkermansia muciniphila was not susceptible to HD-51–9 in a turbidity broth assay, thus aligning our in vitro findings with our in vivo microbiome analysis in which Akkermansia sp. was increased. Such an influence on A. muciniphila has not previously been described for the full-length HD-5 peptide, thus underlining the different spectra between the full-length peptides and its fragments. Additionally, it is known that a normal HD-5 expression and secretion is instrumental for a normal, healthy microbiota, while a lower amount of HD-5 or HD-6 in the intestine is associated with inflammatory bowel diseases including Crohn’s disease of the small intestine (9, 20). This is affected by varying mechanisms such as Wnt-dependent transcription (21) and PPAR signaling via NOD2 (41). Collectively, these findings corroborate a significant role of defensins in intestinal barrier functions and homeostasis. Our results suggest that defensin-derived fragments with antimicrobial activity might be an important part of this barrier function, influencing the existing microbiota by controlling important bacteria like Akkermansia sp. Also, previous studies have shown that a decreased abundance of A. muciniphila correlated with Crohn’s disease and ulcerative colitis (42, 43). Other studies have demonstrated that an increased amount of A. muciniphila is associated with a healthy microbiome and a decreased risk of metabolic syndrome (44).
In addition to their microbiota-modulating abilities, another important application for antimicrobial peptides is the rapidly increasing number of antibiotic-resistant bacteria (45, 46). The antimicrobial spectrum of defensins and their peptide fragments thus deserve more investigations to reveal if these peptides could be a source of new antibiotics against multi-drug–resistant bacteria. Also, the discovery of these easy-to-produce and low-cost peptide fragments could represent a therapeutic alternative to manipulating the microbiome composition and treating Paneth-cell–associated diseases such as Crohn’s disease of the small intestine.
Materials and Methods
Collection of Duodenal Fluid During Gastroscopy.
Human duodenal fluid was collected during a routine gastroscopy from three healthy individuals. The duodenum was washed with 0.9% NaCl solution, which was recollected. Patients gave their written and informed consent. The sample collection had been previously approved by the Ethical Committee of the University Hospital of Tübingen, Germany.
Screening for Fragments of HD-5 and HD-6 Using Liquid Chromatography–Mass Spectrometry.
We incubated 2.5 µg of HD-5 or HD-6 in 50 mM NH4HCO3 buffer (pH 8.0) (Fluka) with 2 mM Tris (2-carboxyethyl) phosphine for 15 min at 37 °C. Afterward, we added human duodenal fluid and incubated it for an additional 30 min at 37 °C and analyzed the fluid using a liquid chromatography–mass spectrometry system.
Scanning Electron Microscopy.
As previously described (23), Protein-A–coated beads (Spherotech) were incubated with reduced HD-6 (200 µg/mL) for 1.5 h at 37 °C to allow net formation. We subsequently incubated the whole sample with duodenal fluid for an additional 30 min at 37 °C. As a control, we used 0.01% acetic acid (HAc). The sample fixation, preparation, and analysis were performed at the Max Planck Institute for Developmental Biology (Tübingen, Germany).
Transmission Electron Microscopy.
As previously described (31), we incubated 6 × 108 cfu E. coli MC1000 with 200 µg/mL of each peptide for 2 h. Fixed bacteria were cut (30-nm sections) and analyzed with a Zeiss LIBRA 120 transmissions electron microscope.
Radial Diffusion Assay.
Antimicrobial activity of all peptides were tested with a modified radial diffusion assay from Lehrer et al. (47). In brief, log-phase bacteria were grown (anaerobic bacteria with AnaeroGen Oxoid in anaerobic jars) in liquid tryptic soy broth (Becton Dickinson). We used 4 × 106 cfu/mL in each assay. The antimicrobial activity was determined by the diameter of the inhibition zones the next day.
Turbidity Broth Assay.
To investigate the MIC of each peptide, we incubated 5 × 105 cfu/mL bacteria with a defined peptide concentration and measured the bacterial growth by the optical density of 600 nm (Spark 10M; Tecan).
In Vivo Microbiota Analysis.
We treated wild-type C57BL/6J mice by oral gavage for 7 d with 7.19 µg/mouse HD-51–9 administered in 100 µL of PBS solution, followed by a 7-d wash period. Control mice were treated with an equal volume of PBS. Fresh feces samples were collected from individual mice at days 0, 7, and 14. At day 14, mice were euthanized, and the small intestine content was collected. All animal protocols were conducted according to guidelines set out by the Laval University Animal Care and Handling Committee. C57BL/6J male mice (Jackson Laboratories) were housed in a pathogen-free, temperature-controlled environment under a 12:12-h light-dark cycle and fed ad libitum standard rodent chow diet (Harlan Teklad T-2018) for the 5 wk of accommodation in our animal facility (3 wk of acclimatization and 2 wk of experimental protocol). For the microbiota analysis, bacterial DNA was extracted from snap-frozen feces and content of the small intestine at necropsy by the NuceloSpin 96 soil kit (Macherey-Nagel) following the manufacturer’s instructions.
A more detailed description of all methods can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Marion Strauss and Jutta Bader for excellent technical assistance; Brigitte Fehrenbacher for providing the TEM pictures; and Dr. Thibault Varin (Quebec Heart and Lung Institute, Department of Medicine, Faculty of Medicine, Cardiology Axis, Laval University) for expert bioinformatics assistance. This study was supported by the European Union ERC Starting Grant DEFENSINACTIVITY (J. Wehkamp) and the Deutsche Forschungsgemeinschaft. J. Wehkamp holds a Heisenberg Professorship. B.A.H.J. is the recipient of the Lundbeck Foundation Grant R232-2016-2425 and Novo Nordisk Foundation Postdoctoral Research Fellowship (Grant NNF17OC0026698), both of which have supported this work. This work was also supported by Excellence cluster EXC2124.
Footnotes
Conflict of interest statement: J. Wehkamp and D.E. are planning to apply for a patent on the HD-5 fragments.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817376116/-/DCSupplemental.
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 3.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 4.Uronis JM, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cryan JF, Dinan TG. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 6.Cekanaviciute E, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopalakrishnan V, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matson V, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359:104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wehkamp J, et al. Reduced Paneth cell alpha-defensins in ileal Crohn’s disease. Proc Natl Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bevins CL, Salzman NH. The potter’s wheel: The host’s role in sculpting its microbiota. Cell Mol Life Sci. 2011;68:3675–3685. doi: 10.1007/s00018-011-0830-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 14.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 15.Wilson SS, Wiens ME, Smith JG. Antiviral mechanisms of human defensins. J Mol Biol. 2013;425:4965–4980. doi: 10.1016/j.jmb.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrer RI, Ganz T, Selsted ME. Defensins: Endogenous antibiotic peptides of animal cells. Cell. 1991;64:229–230. doi: 10.1016/0092-8674(91)90632-9. [DOI] [PubMed] [Google Scholar]
- 17.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: Implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–192. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 18.Wehkamp J, Stange EF. Paneth’s disease. J Crohn’s Colitis. 2010;4:523–531. doi: 10.1016/j.crohns.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Wehkamp J, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn’s disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 20.Clevers HC, Bevins CL. Paneth cells: Maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 21.Courth LF, et al. Crohn’s disease-derived monocytes fail to induce Paneth cell defensins. Proc Natl Acad Sci USA. 2015;112:14000–14005. doi: 10.1073/pnas.1510084112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 23.Chu H, et al. Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science. 2012;337:477–481. doi: 10.1126/science.1218831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salzman NH. Paneth cell defensins and the regulation of the microbiome: Détente at mucosal surfaces. Gut Microbes. 2010;1:401–406. doi: 10.4161/gmic.1.6.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzman NH, Bevins CL. Dysbiosis: A consequence of Paneth cell dysfunction. Semin Immunol. 2013;25:334–341. doi: 10.1016/j.smim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Stappenbeck TS, McGovern DPB. Paneth cell alterations in the development and phenotype of Crohn’s disease. Gastroenterology. 2017;152:322–326. doi: 10.1053/j.gastro.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porter EM, van Dam E, Valore EV, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396–2401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ericksen B, Wu Z, Lu W, Lehrer RI. Antibacterial activity and specificity of the six human α-defensins. Antimicrob Agents Chemother. 2005;49:269–275. doi: 10.1128/AAC.49.1.269-275.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy E. Anaerobic infections: Update on treatment considerations. Drugs. 2010;70:841–858. doi: 10.2165/11534490-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 30.Schroeder BO, et al. Paneth cell α-defensin 6 (HD-6) is an antimicrobial peptide. Mucosal Immunol. 2015;8:661–671. doi: 10.1038/mi.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 32.Jaeger SU, et al. Cell-mediated reduction of human β-defensin 1: A major role for mucosal thioredoxin. Mucosal Immunol. 2013;6:1179–1190. doi: 10.1038/mi.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Cougnon FBL, Wanniarachchi YA, Hayden JA, Nolan EM. Reduction of human defensin 5 affords a high-affinity zinc-chelating peptide. ACS Chem Biol. 2013;8:1907–1911. doi: 10.1021/cb400340k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehrer RI. Immunology: Peptide gets in shape for self-defence. Nature. 2011;469:309–310. doi: 10.1038/469309a. [DOI] [PubMed] [Google Scholar]
- 35.Hein KZ, et al. Disulphide-reduced psoriasin is a human apoptosis-inducing broad-spectrum fungicide. Proc Natl Acad Sci USA. 2015;112:13039–13044. doi: 10.1073/pnas.1511197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, et al. Reduction impairs the antibacterial activity but benefits the LPS neutralization ability of human enteric defensin 5. Sci Rep. 2016;6:22875. doi: 10.1038/srep22875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schroeder BO, Stange EF, Wehkamp J. Waking the wimp: Redox-modulation activates human beta-defensin 1. Gut Microbes. 2011;2:262–266. doi: 10.4161/gmic.2.4.17692. [DOI] [PubMed] [Google Scholar]
- 38.Raschig J, et al. Ubiquitously expressed human beta defensin 1 (hBD1) forms bacteria-entrapping nets in a redox dependent mode of action. PLoS Pathog. 2017;13:e1006261. doi: 10.1371/journal.ppat.1006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chileveru HR, et al. Visualizing attack of Escherichia coli by the antimicrobial peptide human defensin 5. Biochemistry. 2015;54:1767–1777. doi: 10.1021/bi501483q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh D, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 41.Bonen DK, et al. Crohn’s disease-associated NOD2 variants share a signaling defect in response to lipopolysaccharide and peptidoglycan. Gastroenterology. 2003;124:140–146. doi: 10.1053/gast.2003.50019. [DOI] [PubMed] [Google Scholar]
- 42.Png CW, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 43.Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 44.Cani PD. Human gut microbiome: Hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.WHO 2017 Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO. Available at https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. Accessed October 26, 2017.
- 46.Brötz-Oesterhelt H, Sass P. Postgenomic strategies in antibacterial drug discovery. Future Microbiol. 2010;5:1553–1579. doi: 10.2217/fmb.10.119. [DOI] [PubMed] [Google Scholar]
- 47.Lehrer RI, Rosenman M, Harwig SS, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.