Abstract
Influenza A virus carries hundreds of trimeric hemagglutinins proteins (HA) on its viral envelope that interact with various sialylated glycans on a host cell. This interaction represents a multivalent binding event that is present in all the current receptor-binding assays, including those employing viruses or precomplexed HA trimers. To study the nature of such multivalent binding events, we fused a super folder GFP (sfGFP) to the C-terminus of trimeric HA to allow direct visualization of HA-receptor interactions without the need for additional fluorescent antibodies. The multivalent binding of the HA-sfGFP proteins was studied using glycan arrays and tissue staining. The HA-sfGFP with human-type receptor specificity was able to bind to a glycan array as the free trimer. In contrast, the HA-sfGFP with avian-type receptor specificity required multimerization by antibodies before binding to glycans on the glycan array could be observed. Interestingly, multimerization was not required for binding to tissues. The array data may be explained by the possible bivalent binding mode of a single human specific HA trimer to complex branched N-glycans, which is not possible for the avian specific HA due to geometrical constrains of the binding sites. The fact that this specificity pattern changes upon interaction with a cell surface probably represents the enhanced amount of glycan orientations and variable densities vs. those on the glycan array.
Keywords: Influenza, attachment protein, bidentate, multivalent, precomplexing
Introduction
Influenza A virus has the ability to infect the respiratory tract in a broad spectrum of hosts such as birds, humans, pigs, marine mammals and horses [1, 2]. A crucial determinant of the broad host tropism of the influenza A virus is the hemagglutinin (HA) protein [2]. The HA protein is a trimeric glycoprotein that contains the receptor binding domain needed for attachment to sialylated glycans located on the host cell surface [3]. Avian hosts contain glycans terminated with α2–3 linked sialic acids (or avian-type) on their surface, while human upper respiratory tissues display α2–6 linked sialic acids (or human-type). Host cell sialic acid expression therefore drives influenza A virus to be specific for either avian or human-type receptors [4–7]. This host specificity influences tissue tropism since human-type receptors are located mostly in the upper respiratory tract, whereas avian-type receptors are mostly located in the lower respiratory tract of mammals, while in birds they are present throughout the whole respiratory tract [8–11].
Sialic acids are the terminal fragments of complex glycans that contain multiple branched arms to which the HA protein can attach [12–15]. However, the abundance of complex glycans with multiple branched and extended galactose-N-acetylglucosamine (LacNAc) chain repeats terminated with human-type receptors in the upper respiratory tract of humans is low. Conversely, branched glycans with short arms terminated with avian-type receptors appear to have a high abundance in the respiratory tract of non-human species [16, 17]. This suggests that human-type HA proteins might need a higher avidity or affinity (monomeric) for human-type receptors compared to avian-type HA proteins for avian-type receptors. In this context it is relevant to note that linear glycan structures bind to a single HA site in the mM range [18–20]. In the binding of viruses, affinity is enhanced by multiple simultaneous interactions, and the virus has two possibilities for this. Firstly, branched glycans can be engaged by up to two binding sites in an HA trimer, a phenomena called bidentate binding. Secondly, the virus can simultaneously bind with multiple HA trimers, a phenomena called multivalency. Both phenomena can operate simultaneously to provide the needed avidity effects for sufficient viral binding to the cell surface.
In recent years, it has become apparent that human influenza A viruses have a preference for extended LacNAc repeats on branched glycans capped with α2–6 linked sialic acids [12, 15, 21]. We observed similar binding preferences with avian HAs that were mutated to bind human-type receptors [7, 22]. We therefore hypothesize that human-type receptor specific HAs prefer branched glycans with elongated arms. However, direct evidence for this is missing, due to the lack of branched glycan structures with a single sialic acid and the fact that all our receptor binding analyses rely on the multivalent presentation of HA. To directly visualize receptor specificities of a single trimer we fused a sfGFP open reading frame to that of an HA.
In order to gain further insight into multivalency and bidentate binding events we analyzed fluorescent HA’s in different multivalent forms, i.e. as a single fluorescent trimer and precomplexed with antibodies using different labels. Additionally, we examined the differences in binding between HA trimers that have either human- or avian-type receptor specificity. We generated a model Puerto-Rico/8/34 HA with and without a single amino acid mutation (D190E) that confers specificity for either of the two receptors [23, 24]. These HA proteins were then used in glycan array screenings and tissue staining studies. We demonstrate that the human-type HA is proficient at binding to N-glycans on a glycan array surface, while the avian-type HA is not able to engage identical avian-type receptors. The avian-type HA protein needs to be part of a multivalent representation to be able to effectively bind to the avian-type receptors on a glycan array. The human-type HA is likely capable of binding two of Sialyl-LacNAc moieties of the branched N-glycans, which is geometrically disfavored for the avian type [15]. In tissues the situation is different, and it is probably due to the complexity of glycan representation on the cell surfaces.
Results
Expression and characterization of the recombinant A/Puerto-Rico/8/34 HA sfGFP fusion proteins
With the aim to study the intrinsic receptor specificity of a single HA trimer we generated two constructs in which the HA gene was C-terminally fused to sfGFP, which differ in the presence or absence of a motif, generating PR8 D190 GCN4IL-sfGFP and PR8 D190-sfGFP (Fig. 1A). Our standard expression cassette was used as a control, encoding for PR8 D190 without sfGFP but with the trimerization domain (PR8 D190-GCN4IL) [22, 25]. The expression of GFP-fused recombinant proteins PR8 D190 GCN4IL-sfGFP and PR8 D190-sfGFP (Fig. 1B) was confirmed by fluorescence microscopy 48 hours post transfection of HEK293S GNT1(−) cells (Fig 1B). The correct nature of all three HA proteins was confirmed by the electrophoretic mobility of soluble proteins isolated from the cell culture supernatant by SDS-PAGE (Fig. 1C). Both the PR8 D190-sfGFP and PR8 D190 GCN4IL-sfGFP migrated as proteins of approximately 100kDa, which is the expected mobility of a monomer HA-sfGFP fusion protein (+ band at ca. 30 kDa). The control PR8 D190 GCN4IL had, as expected, a higher mobility of about 70kDa since it lacks sfGFP. Finally, size exclusion HPLC on a superdex200 column was performed to determine whether the GCN4IL motif resulted in formation of HA trimers (Fig. 1D). For the PR8 D190-sfGFP, monomeric HA-sfGFP proteins were formed as well as aggregates of 667kDa. As expected, the PR8 D190 GCN4IL-sfGFP and PR8 D190-GCN4IL ran at their respective expected molecular weights due to trimerization caused by the GCN4IL motif [25, 26], with the PR8 D190 GCN4IL-sfGFP having a larger molecular weight than the control due to the presence of probably three sfGFP proteins. Interestingly we observe that for HA subtypes with lower expression yields, the addition of sfGFP can increase efficiency by 5-fold (data not shown). The results confirm trimerization of a sfGFP-coupled HA does occur in the presence of GCN4IL.
Figure 1. Creation, expression, and analyses of recombinant HA proteins.
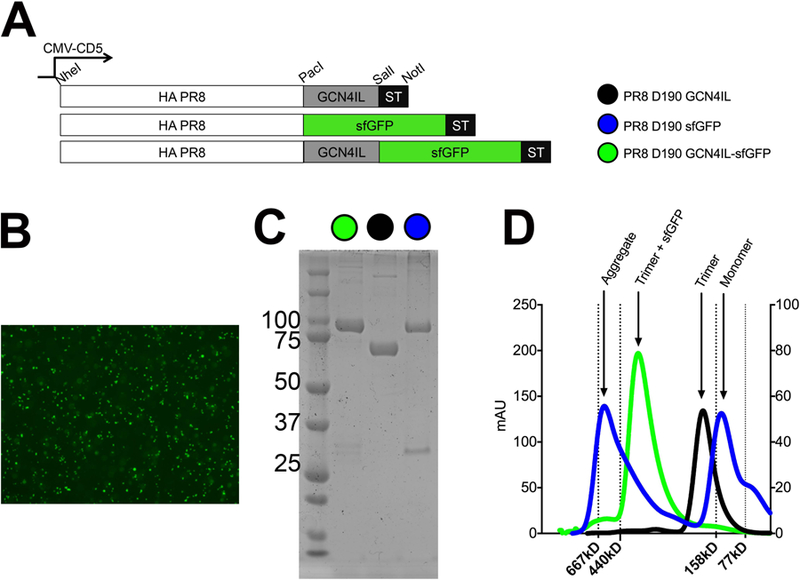
(A) Schematic view of the mammalian cell expression plasmid in which the ORF is under the control of a CMV-promoter. HA is cloned in frame with a CD5 signal peptide for efficient excretion into the cell culture supernatant. C-terminal extensions we created by cloning using PacI, SalI and NotI sites. (B) Forty-eight hours post transfection the majority of the cells were fluorescent when transfected with the sfGFP constructs. (C) Coomassie staining of the purified HA proteins, (D) Size exclusion analysis using a superdex200 column.
Molecular analysis of the PR8 fusion protein
To ensure that our trimeric HA proteins behave in a similar manner we determined the biological activity, immunogenic properties and protein stability of the HA-sfGFP fusion proteins. A hemagglutination assay was performed using human RBCs to determine HA activity (Fig. 2A). In this assay HAs were precomplexed with α-strep and α-mouse antibodies as previously described [25], the activity of PR8 D190 GCN4IL-sfGFP was not hampered by sfGFP when compared to PR8 D190 GCN4IL. However, trimerization was required to achieve HA activity as the PR8 D190-sfGFP monomer lacked hemagglutination ability. The next step was to determine if biologically and immunological relevant conformations were achieved by HA-sfGFPs with an ELISA assay (Fig. 2B). We used three previously reported conformational dependent antibodies to the stalk region [27, 28]. These were applied to HA coated plates and binding of these Abs was detected using an α-mouse secondary. Both PR8 D190 GCN4IL and PR8 D190 GCN4IL-sfGFP proteins were bound by all three conformation-dependent antibodies. Conversely, the monomeric PR8 D190 sfGFP was significantly less bound, indicating a lack of conformation. As a control, PR8 mouse serum was used to confirm that all three HA proteins were still recognized by non-conformational dependent PR8 HA antibodies. As an antigenic control we used a trimeric HA of A/Shanghai/2/13 H7N9 (Sh2) [29] that was not detected by either Ab or serum, which confirms specificity. Finally, the thermal stability of the HA-sfGFPs was determined by performing differential scanning calorimetry (Fig 2C), no significant difference in thermal stability could be observed between the PR8 D190 GCN4IL and PR8 D190 GCN4IL-sfGFP, indicating that the thermal stability was not affected by the presence of the sfGFP.
Figure 2. Biochemical characterization of PR8 D190 GCN4IL-sfGFP.
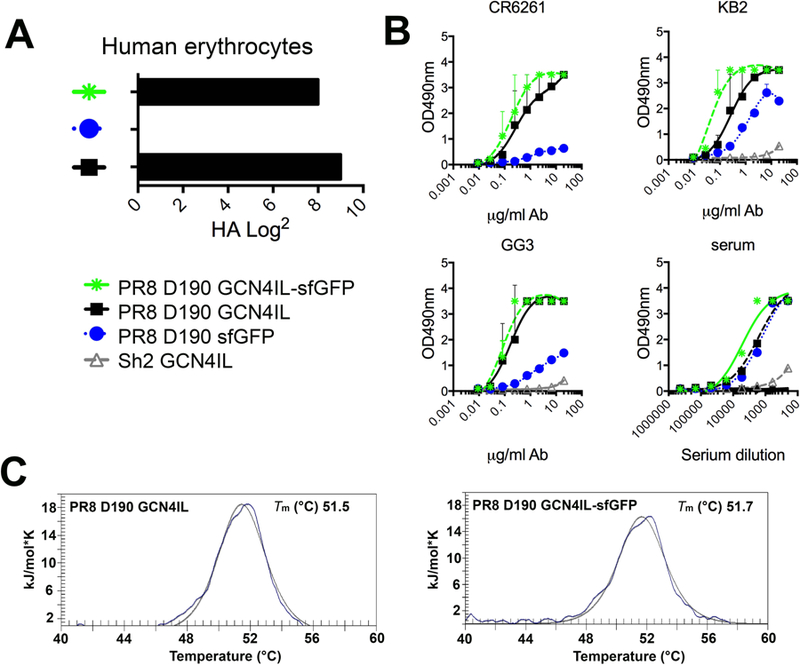
(A) Biological activity as measured by hemagglutination using human red blood cells. (B) Purified HA-sfGFP fusion proteins analyzed with conformational dependent stalk antibodies and polyclonal mouse serum. (C) Melting curves of recombinant HA proteins with or without a sfGFP fusion by Differential Scanning Calorimetry. Individual melting curves are shown with the raw data depicted in solid lines, while the fitted curves, from which the Tm values were derived, are depicted with a dotted line.
Structural analysis of the trimeric HA-sfGFP fusion protein
With the activity and thermal stability of the fusion protein confirmed, we wanted to determine if the protein folded into the correct native 3D conformation. Although a massive amount of reports are available on HA crystal structures, EM analyses of single trimers is hampered by their relatively small size. And although a plethora of studies use recombinant multimeric viral envelope proteins with a foldon, structural analyses of these proteins with a foldon is minimal. We started with a theoretical ribbon representation of the PR8 D190 GCN4IL-sfGFP (Fig. 3A), which was used as a reference for how the native conformation of the fusion protein would be under correct folding conditions. The GCN4IL motif acts as the driver for trimerization of the HA protein as well as an anchor for the sfGFP. The electron density map (Fig. 3B) confirms correct folding and 3D orientation of the HA trimer in presence of the sfGFP protein. Follow up with EM (Fig. 3C) shows that the native protein structure matched with the theoretical model (Fig. 3A). A multi-panel figure showing particle picking results can be found in the supplementary data (Fig S1). Initially, 33,397 individual particles were picked, placed into a stack, and submitted to reference free 2D classification. From the initial 2D classes, particles that did not resemble HA were removed, resulting in a final particle stack of 25,757 particles, which was then subjection to Relion 2D classification. All resultant classes demonstrated evidence of C-terminal GFP. The distinct HA-trimer, GCN4IL motif and three sfGFP protein structures can be identified in the EM image. This confirms that the PR8 D190 GCN4IL-sfGFP fusion protein attains a correctly folded native protein structure in the presence of the GCN4IL motif and sfGFP.
Figure 3. Structural analyses of HA-GCN4IL-sfGFP.
(A) Ribbon representation of the HA-sfGFP trimer. A single protomer in the HA section of the fusion protein is colored by domain. (B) Negative stain 3D reconstruction of the trimeric HA-sfGFP fusion protein. The HA trimer is well ordered while the C-terminally fused sGFP appears as irregular densities at the base of the trimer likely due to flexibility. (C) Negative stain 2D class average showing the 3D structure of the trimeric HA-sfGFP. The distinct HA, GCN4IL and sfGFP structures can be seen resembling the layout as shown in the ribbon structure.
Multivalent receptor specificity of PR8 D190 and D190E by glycan array analysis
With a biologically relevant fluorescent HA protein in hand, we wanted to analyse the intrinsic receptor specificity of the free HA-trimer only. We also wanted to determine if any multivalency enhancements could be observed using different pre-complexing antibodies to generate higher valency structures containing 4 HA trimers. We used the PR8 D190 HA with inherent α2,6-linked sialic acid specificity and the PR8 D190E protein, specific for α2,3-linked sialic acid, to analyse if the intrinsic receptor binding or multivalency effects were affected due to sialic acid linkage specificity [23, 24]. We started by applying the HA proteins as lectins on the glycan array without any detecting antibodies (Fig. 4A & B). Interestingly, only the human-type specific PR8 D190 protein was able to interact with the array whereas the PR8 D190E failed to do so. When investigating the glycans bound by PR8 D190 it was clear that the protein specifically bound branched complex glycans similarly to other human-type specific HA proteins [15]. As the readout of the array is by sfGFP fluorescence, we performed a glycan array with precomplexing the PR8 D190 GCN4IL-sfGFP with HRP- and Alexa-647-labelled antibodies to study whether this might result in detection of enhanced binding. PR8 D190 and PR8 D190E were now both able to bind their cognate receptors (Fig. 4C & D). Precomplexed PR8 D190 had an increase in receptor usage, now also binding several linear structures. Precomplexed PR8 D190E was able to efficiently bind to a subset of α2,3 glycans, suggesting that a higher order multivalent presentation of the HA protein for avian-type receptors is indeed necessary for binding to occur. Furthermore the PR8 D190E protein did not seem to have a preference for branched glycans, and bound linear structures as efficiently. Finally, we decided to precomplex both proteins with α-strep-tag and goat-α-mouse antibodies both labelled with Alexa-488 with the expectation to increase the fluorescent readout, to see whether glycans binding with low affinity would now be detectable. Again, both PR8 D190 and PR8 D190E interact efficiently with glycans on the array (Fig. 4E & F), but we were not able to detect any additional glycans bound by both proteins, indicating that the precomplexing determines binding properties and that addition of extra fluorescent probes did not result in detection of novel binding molecules. We did however see a higher intensity of some glycans especially for the linear structures bound by PR8 D190, whereas for PR8 D190E the increase of fluorescence was negligible. However, a tri-antenarry structure (#63), gave a very intense read out, obscuring the fact that all glycans in panels D and F are equally bound. From this we conclude that human-type specific HA can directly interact with branched complex glycans on a glycan-array and the avian-type specific HA cannot. Creating a multivalent HA display using antibodies caused binding to linear structures by the human-type specific HA, whereas for the avian-type specific HA multivalent display is required to see any binding at all.
Figure 4. Receptor specificity of PR8 D190 and D190E GCN4IL-sfGFP.
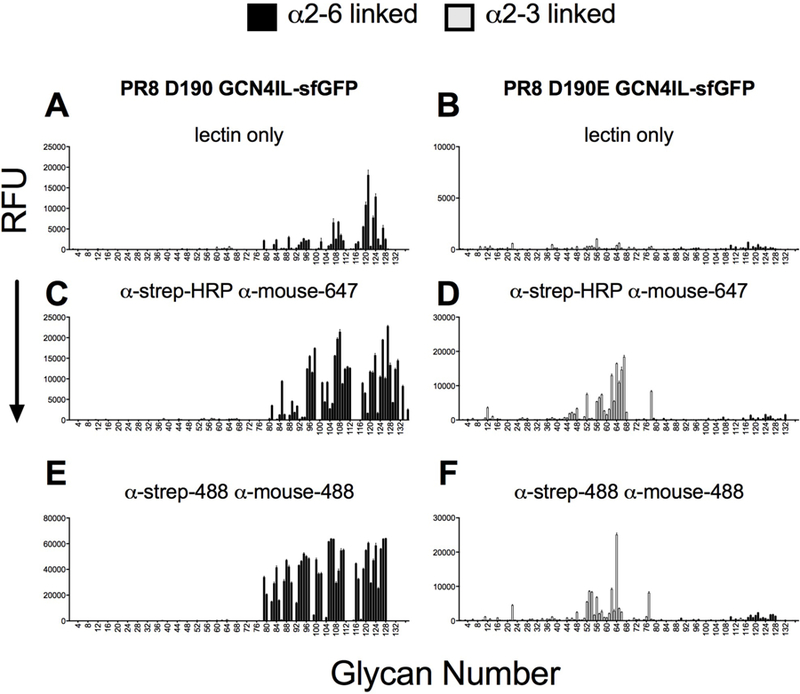
(A) PR8 D190 without precomplexing (B) PR8D190E without precomplexing (C) PR8 D190 precomplexed with α-strep-HRP and α-mouse-647 (D) PR8 D190E precomplexed with α-strep-HRP and α-mouse-647 (E) PR8 CAM precomplexed with α-Strep-488 and α-Mouse-488 (F) PR8 D190E precomplexed with α-Strep-488 and α-Mouse-488. The mean signal and standard error were calculated from six independent replicates of glycans printed on the array. The data shown is a representative of three independent assays. α2–3 linked sialosides are shown in white bars (glycans 11 to 77 on the x axis) and α2–6 linked sialosides in black (glycans 78 to 130). Glycans 1 to 10 are non-sialylated controls (see also Table S1)
Multivalent receptor specificities of different HA proteins as lectins and multivalent complexes on a directed glycan array.
In order to investigate if the avian and human-type binding specificities would hold up with other HA proteins, we next compared PR8 D190 and D190E with human H3N2 and H1N1 isolates and an avian H5N1 protein and a mutant thereof that is able to transmit between ferrets. We tested all these HA proteins fused to GCN4IL-sfGFP to our directed glycan array (Broszeit and De Vries manuscript under revision) (Fig5A), and chicken and human erythrocytes (Fig 5B). All proteins had expected binding specificities and avidities to which we will refer while describing the glycan array data.
Figure 5. Receptor specificity of different HA as lectins and multivalent complexes on a N-glycan array.
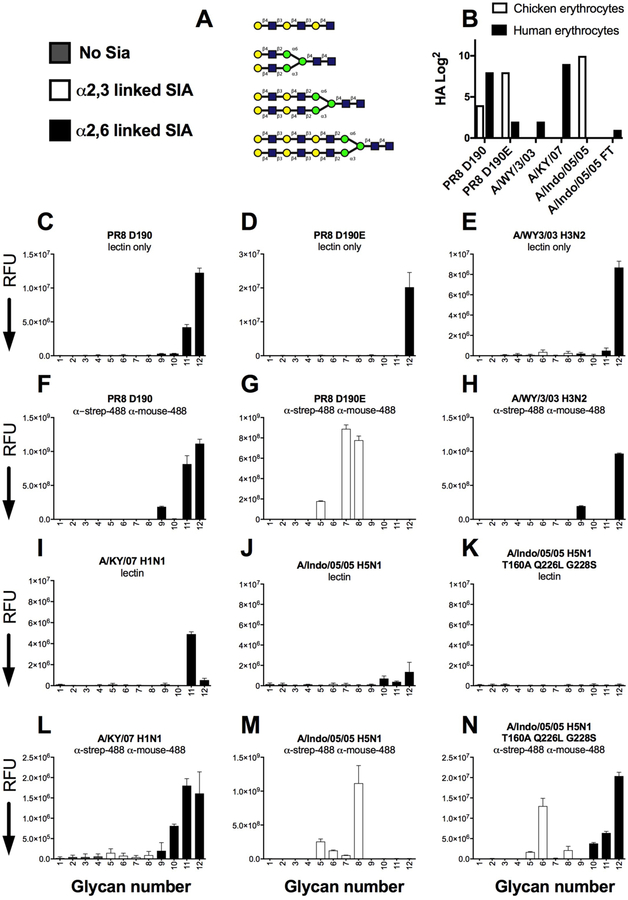
(A) Glycans imprinted on this array, depicted are the unsialylated controls that are all sialylated with either 2,3 linked and 2,6 linked sialic acids. (B) Hemaglutination assay with precomplex HA proteins toward chicken and human erythrocytes. (C–E and I–K) indicated HA proteins as lectins only. (F–H and L–N) indicated HA proteins precomplexed with α-strep and α-mouse abs both labeled with Alexa-488. The mean signal and standard error were calculated from six independent replicates of glycans printed on the array. The data shown is a representative of two independent assays. α2–3 linked sialosides are shown in white bars (glycans 5 to 8 on the x axis) and α2–6 linked sialosides in black (glycans 9 to 12). Glycans 1 to 4 are non-sialylated controls.
In the glycan array we observed that the PR8 D190 protein was able to directly interact with a N-glycan containing three LacNAc repeats and a human type receptor (Fig 5B), the same was observed for the avian-type specific PR8 D190E protein (Fig 5C), which can be explained by the residual human-type specificity to human erythrocytes, the 3 fold increase in concentration, or the sensitivity of the scanner. The human H3 protein derived from A/WY/3/03 is also able to directly interact with the same structure compared to PR8 D190 (Fig 5D) (#12). We then analysed the same proteins pre-complexed with α-strep and α-mouse antibodies with 488 labels, PR8 D190 maintained human-type receptor specificity and additionally also binds the linear structure (#9) (Fig 5E). Au contraire, the PR8 D190E now specifically binds avian-type receptors when precomplexed with antibodies (Fig 5F). The human H3 proteins, maintained human-type receptor specificity as a multivalent complex, similarly compared to PR8 D190 (Fig 5G). Interestingly the fluorescent intensities were 2 logs higher compared to the lectins only, indicating a large difference in the amount of labels that could be read by the different fluorescent slide scanner. Next we analysed a human H1 protein, derived from a seasonal strain in 2007 (A/KY/07), which we previously characterized as a strong binder specific to human-type receptors [29, 30]. This H1 protein was able to interact as a lectin albeit slightly less efficient compared to PR8 D190 and A/WY/3/03 (Fig 5I). We then also analysed an H5 protein derived from a human infection in which the virus maintained avian-type specificity and also does not transmit between humans or ferrets. In addition we used a mutant of this H5 protein that was shown to have human-type receptor specificity, albeit with very low avidity, but transmits between ferrets [20, 31, 32]. These H5 proteins were not able to directly interact with glycans on the array (Fig 5 J & K), whereas all the precomplexed HAs, displayed human-, avian- and mixed receptor specificity for A/KY07, A/Indo/05/05 and A/Indo/05/05 T160A Q226L G228S, respectively and as previously published [20, 22, 32, 33] (Fig 5L–N). From this experiment we conclude that HA proteins, with human-type receptor binding can engage these on branched N-glycans on an array, whereas low avidity binding and HAs with strict avian-type receptor specificity cannot.
Fluorescent staining of tissues demonstrates another layer of complexity for glycan binding
Binding to receptors on tissues is essential for infection and tissues may present glycans differently compared to glycans immobilized on a microarray surface. We analyzed binding of our PR8 D190 and D190E sfGFP proteins to chicken trachea and pig primary bronchus tissue sections as these tissues express mainly avian-type or human-type receptors, respectively. PR8 D190E demonstrated that binding to α2,3 linked sialic acid on host tissues was now possible without complexing with antibodies similarly, albeit with different fluorescent intensities which is due to differential expression of sialic acids, to PR8 D190 binding to pig bronchus (Fig. 6A). However, complexing the HA’s with antibodies did increase the fluorescent output of the PR8 D190 proteins confirming the action of multivalency with both human- and avian-type specific HA proteins (Fig 6B). As with the glycan array we also complexed the PR8 GCN4IL-sfGFP HA proteins with α-strep-tag and goat-α-mouse antibodies both labelled with Alexa-488 (Fig 6C). It is clear that in binding to tissues a higher number of fluorescent labels increases intensity, as one might expect. Several control stainings were preformed (Fig. 6D), first lectin histochemistry using 3-amino-9-ethylcarbazole (AEC) substrate to visualize HRPO-labelled antibody complexes was used to confirm specificity to their respective receptors on chicken trachea and pig primary bronchus. Secondly, treatment of both tissues with VCNA, which removes sialic acid residues, before application of the HA-sfGFP proteins, confirmed that both proteins bound to tissues in a sialic acid dependent manner. Reciprocal binding of PR8 to chicken and PR8 D190E to pig was not observed, confirming that both proteins were only able to bind to tissues containing their respective receptors.
Figure 6. Tissue stainings of PR8 D190 and PR8 D190E .
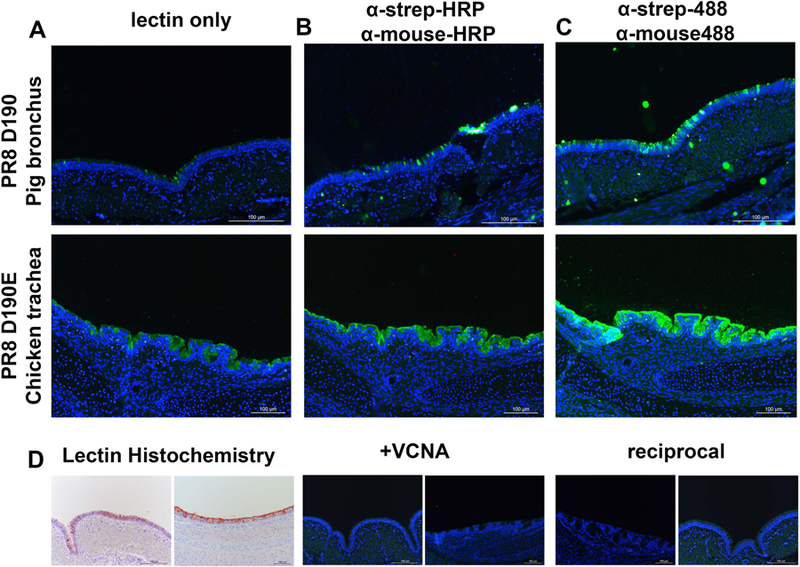
(A) Staining of chicken trachea with PR8 D190 and pig primary bronchus with PR8 D190E (B) Staining of tissues with PR8 D190 and PR8 D190E precomplexed with α-strep-tag mouse antibody and goat-α-mouse-HRP in a 4:2:1 molar ratio (C) Staining of tissue with PR8 D190 and PR8 D190E precomplexed with α-strep-tag-488 conjugated mouse antibody and goat-α-mouse-488 conjugated antibody in a 4:2:1 molar ratio (D) Left: lectin histochemistry using the PR8 proteins precomplexed with HRP labelled antibodies using AEC as a readout. Middle, neuraminidase treatment of the tissues and stained with PR8 D190 for pig bronchus (left) and PR8 D190E on chicken trachea (right) to deterimine sialic acid dependency. Right, reciprocal stainings using PR8 D190 on chicken trachea and PR8 D190E on pig bronchus to confirm sialic acid linkage specificity of both proteins, respectively.
Bidentate versus multivalent receptor binding
One major observation we made is that human-type specific HA is able to bind N-glycans on a glycan array as a single trimer, while the avian-type specific HA was unable to do so. However this avian-type specific HA is able to visibly bind as a single trimer to glycans on a tissue. This observation can be explained by looking at the interaction of a complex multi-arm glycan as present on the glycan array. Based on our previous modeling a human-type HA should bind far stronger due to its ability to bind two sites simultaneously (Fig. 7A) [7, 15]. Such binding can lead to orders of magnitude affinity enhancements. Geometric constraints preclude this binding mode for the avian specific HA, for which only the weak millimolar binding option remains, which is undetectable by microarray experiments (Fig. 7B,C). Nevertheless on a cell surface the glycan presentation is more dense and simultaneous binding to multiple HA binding sites becomes possible for the free avian-type specific HA trimer (Fig. 7D). Two features in the glycan are required for biantennary binding to a single HA. Firstly, the branches of the glycan must be long enough to span the distance between two RBSs without steric blocking by HA surface residues or by HA glycosylation. Secondly, the shapes of the termini of individual glycan branches (i.e. the shape generated by the α2–6 versus α2–3 linkage), must be such that they allow the sialic acid residues to fit correctly into each binding site as discussed by Ji et al. (see Supplementary Figure 2). Only α2–6 receptors in a curled anti-ψ conformation satisfy the terminal shape requirement for bidentate binding. Although the α2–3 oligosaccharides in the –gauche φ-conformation reach upward from the RBS rather than away, their spatial divergence from each other coupled with the orientation of the α2–3 linkage precludes biantennary binding (see Supplementary Figure 2).
Figure 7: Presentation of N-glycans capped with α2,3 linked sialic acids on a glass slide lacks complexity for multivalent interaction with an HA trimer.
The HA binding sites are colored cyan (to indicate multivalent binding) or red (no multivalent binding). (A) Bidentate binding of a HA to an α2–6 capped, branched N-glycan with three LacNAc repeats on each branch. (B) Bidentate binding is not possible between the equivalent glycan capped with α2–3 linked sialic acid and a HA, due to the different orientation of the sialic acid. (C) On a slide surface glycans capped with α2–3 linked sialic acid must be spaced correctly to form multivalent interactions with a HA, and no recruitment of the glycans is possible. (D) Multivalent binding of avian HA to glycans capped with α2–3 linked sialic acid may be possible on a cell surface, due to the higher density of glycans on cell surface glycoproteins (example shown is ICAM-1, with glycans attached to N-glycosylation sites). HA forms a multivalent interaction with glycans present on one ICAM-1 molecule, or between two different ICAM-1 molecules.
Discussion
In this study we generated fluorescent trimeric influenza hemagglutinins to study the intrinsic binding specificity of a single HA trimer. It was previously shown that sequential detection by antibodies fails to visualize binding to glycans [34], which was a motivator for us to create fluorescent hemagglutinins. Furthermore, it was previously shown that monomeric or trimeric HAs have low millimolar affinity for linear structures as e.g. measured by NMR [18, 19]. It was therefore interesting to observe that fluorescent HAs are able to interact with the more elaborate multi-arm N-glycans on a glycan-array. Since clear binding of a free HA trimer of human-type specificity was observed, strong support was obtained for a bidentate binding mode.
Protein-carbohydrate interactions are involved in a plethora of biological events, these interactions are in general rather weak and therefore employ multivalency. Multivalent protein-carbohydrate interactions are intensively studied especially in the field of glycan binding pathogens and glycan binding antibodies [35, 36]. Yet multivalent receptor-binding specificity studies using influenza viruses or HA proteins are scarce [37, 38]. Despite this, multivalency is heavily used by medicinal chemists looking for high affinity inhibitors against influenza, which include polymers, dendrimers and nanoparticles [39–41], and our data on bidentate binding will aid in the design of such anti-infectives. Recently, a similar bidentate binding was invoked for glycan functionalized DNA-PNA hybrids [37].
In conclusion, we demonstrate that a single HA trimer with human-type receptor specificity can engage a branched N-glycan in a bidentate manner, as we have previously modeled [7, 15]. This binding mode could explain why human and human-adapted viruses gain specificity to complex glycans that are specific for the human upper respiratory tract [16] but are displayed at low abundance. Although avian type specific HA is not able to interact with avian-type receptors as a single trimer on an array, a multivalent presentation of these receptors as present on the respiratory epithelium (this study) or HA on viral particles [42, 43], can overcome this.
Material and Methods
Genes and expression vectors
pCD5 plasmids were used which contained a HA sequence originating from A/Puerto Rico/8/1934/H1N1 (PR8 Cambridge strain (CAM)) (GenBank: NP_040980) and were previously described in [24]. pCD5-PR8D GCN4IL-TEV-sfGFP plasmid encodes for a GCN4IL leucine zipper trimerization motif.(IL:RMKQIEDKIEEIESKQKKIENEIARIKK) [44], followed by a seven amino acid cleavage recognition sequence (ENLYFQG) of tobacco etch virus (TEV) and a sfGFP fused to a Strep-tag II (WSHPQFEKGGGSGGGSWSHPQFEK; IBA, Germany) C-terminally [25, 45]. Furthermore, variants without a GCN4IL leucine zipper, pCD5-PR8D-TEV-sfGFP-Strep-tag (PR8 D190 sfGFP), and a vector without the sfGFP, pCD5-PR8D GCN4IL-TEV-Strep-tag (PR8 D190 GCN4IL), were generated.
Protein Expression and purification
pCD5-HA- +/− GCN4IL-sfGFP expression vectors were transfected into HEK293S GNT1(−) cells (which are modified HEK293S cells lacking glucosaminyltransferase I activity (ATCC® CRL-3022™)) with polyethyleneimine I (PEI) in a 1:8 ratio (µg DNA:µg PEI) as previously described [25]. The transfection mix was replaced after 6 hours by 293 SFM II suspension medium (Invitrogen, 11686029, supplemented with glucose 2.0 gram/L, sodium bicarbonate 3.6 gram/L, primatone 3.0 gram/L (Kerry), 1% glutaMAX (Gibco), 1.5% DMSO and 2mM valproic acid). Culture supernatants were harvested 5 days post-transfection. The HA expression was analysed with SDS-PAGE followed by Western-blot on PVDF membrane (Biorad) using α-strep-tag mouse antibodies 1:3000 (IBA Life Sciences). Subsequently, HA proteins were purified with sepharose strep-tactin beads (IBA Life Sciences) as previously described [26].
High pressure liquid chromatography size exclusion
Oligomerization status of the HA proteins was determined by analyzing the elution profile with high-pressure liquid chromatography (HPLC) using a Superdex 200 10/300 GL column (GE Healthcare).
Hemagglutination biological activity assay
Hemagglutination assay was performed by precomplexing HA protein, α-strep-tag mouse antibody (IBA Life Sciences) and goat-α-mouse antibody (Novus Biologicals) for 30 minutes at a ratio of 4:2:1. Human RBCs were washed with PBS and diluted to 0,5%, before incubation with serial dilutions of the HA-α-strep-tag-α-mouse complex.
ELISA protein folding analysis
For the ELISA 96-wells plates were coated with 2 µg/mL HA protein in PBS for 16 hours at 4°C, followed by blocking by 3% bovine serum albumin (BSA, VWR, 421501J) in phosphate buffered saline-Tween 0,1% (PBS-T 0,1%). After the block, the wells were treated with 40 µg/ml with the indicated antbodies or serum for 60 minutes at room temperature. Next, wells were treated with rabbit-α-mouse HRP secondary antibody (IBA Life Sciences) for 1 hour at room temperature. HA binding was detected using ODP and measured in an UV reader (Polarstar Omega, BMG Labtech) at 490 nm.
Negative stain electron microscopy structural analysis
PR8 D190 GCN4IL-sfGFP in 10mM Tris, 150mM NaCl at 4°C was deposited on 400 mesh copper negative stain grids and stained with 2% uranyl formate. The grid was imaged on a 120KeV Tecnai Spirit electron microscope with a LaB6 filament and a 4k x 4k TemCam F416 camera. Micrographs were collected using Leginon [46] and then uploaded to Appion [47]. Particles were picked using DoGPicker [48], stacked, and aligned using MSA/MRA [49]. Further 2D and 3D processing was undertaken using Relion. Images showing HA with C-terminal sGFP were colored using Photoshop.
Glycan microarray binding of HA
Purified, soluble trimeric HA was directly apllied to the array; or pre-complexed with HRP or Alexa488-linked α-strep-tag mouse antibody and with HRP or Alexa488-linked α-mouse IgG (4:2:1 molar ratio) prior to incubation for 15 min on ice in 100 µl PBS-T and incubated on the array surface in a humidified chamber for 90 minutes [15]. Slides were subsequently washed by successive rinses with PBS-T, PBS, and deionized H2O. Washed arrays were dried by centrifugation and immediately scanned for FITC signal on a Perkin-Elmer ProScanArray Express confocal microarray scanner. Fluorescent signal intensity was measured using Imagene (Biodiscovery) and mean intensity minus mean background was calculated and graphed using MS Excel. For each glycan, the mean signal intensity is calculated from 6 replicates spots. The highest and lowest signals of the 6 replicates are removed and the remaining 4 replicates are used to calculate the mean signal, standard deviation (SD), and standard error measurement (SEM). Bar graphs represent the averaged mean signal minus background for each glycan sample and error bars are the SEM value. A list of glycans on the microarray is included in Table S1. A near identical experiment was done with the directed glycan array with minor modifications. Here glycans were printed by a non-contact scienion S3 printer. The array was read out with the Innopsys 1100 scanner in Carbonne France, and the images where analysed using Innoscan software and processed with an in-house developed excel macro.
Tissue staining
The binding of PR8 D190 GCN4IL-sfGFP and PR8 D190E GCN4IL-sfGFP to glycan structures on tissues was analysed by using tissue slides containing either chicken trachea or pig bronchus sections. The tissues were subsequently deparaffinised, rehydrated and then antigen retrieved using a citric acid buffer (pH 6). Blocking was done by 3% BSA in PBS overnight at 4 °C. For the staining of tissues, the HA proteins were either directly applied or precomplexed with either α-strep-tag mouse antibody and goat-α-mouse-HRP or α-strep-tag mouse-488 and rabbit-α-mouse-488 antibody in a 4:2:1 ratio in PBS for 20 minutes on ice, followed by incubating the tissues for 90 minutes with the complexed HA-antibody mix at room temperature. Counterstain was performed with DAPI 1:1000 for 5 minutes at room temperature. The stained tissues were covered with cover slides using Fluorsave (Merck Millipore).
Generation of 3D models
Structures of HA bound to biantennary N-glycans -
The structures of human and avian HAs bound by an α2–6 or α2–3 biantennary trilacNAc N-linked glycans, respectively, were generated using the crystal structures (PDB ID 3UBE and 1RVX) of HA bound to Neu5Acα2–6/3Gal and modelling the larger glycan, as described previously [15].
Models of avian HA bound to a glycosylated array surface -
Models of an array surface with α2–3 biantennary trilacNAc N-linked glycans attached via the consortium for functional glycomics linker Sp24 was generated. The array surface and linker portion beta linked to a single GlcNAc residue were generated as described previously [50]. The α2–3 biantennary trilacNAc N-linked glycan bound to an avian HA generated above was superimposed via the reducing terminal GlcNAc onto the GlcNAc-Sp24-surface, using UCSF Chimera [51].
Membrane-embedded, glycosylated ICAM -
A 3D structure of ICAM with domains 1–5 was generated by combining the crystal structures of D1-D2 (PBD ID 1ZXQ) and D3-D5 (PDB ID 1P53 [52]), following the procedure outlined in Yang et al [52]. A biantennary, pentalacNAc N-glycan capped with α2–3 sialic acid was generated using the carbohydrate builder available on Glycam-Web [53] (www.glycam.org/cb) and attached to each of the seven identified glycosylation sites in ICAM [52]. A glycoprotein builder webtool under development at (dev.glycam.org/gp, which will replace the builder now available at www.glycam.org/gp) was used to attach the glycans and resolve any glycan-glycan or glycan-protein overlaps. Briefly, the glycan is attached onto the Asn sidechain, with the sidechain χ1 and χ2 dihedral angles were set to 180°. Any overlaps between the attached glycan and protein or other glycans were resolved by adjusting the χ1 and χ2 dihedral angles using a Monte Carlo algorithm. All atomic overlaps were successfully removed with this approach.
The glycosylated ICAM 3D structure was embedded into the 3D structure of a membrane taken from PDB ID 2MLR [54] using UCSF Chimera [51]. Periodic images of the membrane were generated within VMD [55] to create a larger surface.
Supplementary Material
Supplementary Figure 1.
All of the reference free 2D class averages contain the C-terminal GFP at the base of the HA trimer.
Supplementary Figure 2.
In previous work, a 3D structure of an α2–6 biantennary N-glycan with three lacNAc repeats was produced in a bidentate co-complex with HA (Upper Panel). This was modeled by first superimposing the terminal disaccharide onto a disaccharide in a crystal structure co-complex. Then the glycosidic linkages were adjusted within their low energy ranges and dihedral angles were selected that brought the second, unbound motif towards a second binding site. The binding motif was pulled into position during an all-atom molecular dynamics simulation and finally, the restraints were released to determine if the structure was stable. This method produced a bidentate co-complex for α2–6 biantennary N-glycan with three, four or five lacNAc repeats. In contrast, this method did not find bidentate N-glycans with α2–3 linkages. Although N-glycans containing α2–3 linkages and five lacNAc repeats could span both sites (Lower panel), the α2–3 linkage places the motif in the opposite orientation required by the binding site.
Acknowledgements
R.P.dV is a recipient of a VENI grant from the Netherlands Organization for Scientific Research (NWO). M.H.V is a recipient of a MEERVOUD grant from the NWO. This work was funded in part by National Institutes of Health grant R01 AI114730 and the Kwang Hua Educational Foundation to J.C.P. Several glycans used for HA binding assays were provided by the Consortium for Functional Glycomics (http://www.functionalglycomics.org/) funded by NIGMS grant GM62116 (J.C.P.). M.H.A. was funded by the Saudi Food and Drug Authority (Riyadh, Saudi Arabia) via a PhD scholarship. R.J.W. thanks the National Institutes of Health for support (U01 CA207824, P41 GM103390). The authors would like to thank Rogier Sanders, Ronald Derking and Marielle van Breemen at the Department of Medical Microbiology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands, for technical assistance. The authors would like to acknowledge the Center for Cell Imaging at the Faculty of Veterinary Medicine, Utrecht University.
Abbreviations:
- CMV
Cytomegalovirus
- DSC
Differential scanning calorimetry
- FITC
Fluorescein isothiocyanate
- HA
hemagglutinin
- HRP
Horse Radish Peroxidase
- IHC
immunohistochemistry
- sfGFP
superfolder Green Fluorescent Protein
- ODP
o-phenylenediamine dihydrochloride
- RBC
Red Blood Cell
- PR8 CAM
Puerto-Rico/8/34 Cambridge strain
- VCNA
Vibrio cholorae neuraminidase
Footnotes
Accession numbers
The 3D reconstruction of HA-GCN4IL-sfGFP has been deposited into the Electron Microscopy Databank under the accession code EMD-9061.
References
- [1].Horimoto T, Kawaoka Y, Influenza: lessons from past pandemics, warnings from current incidents, Nature Rev Micro 3 (2005) 591–600. [DOI] [PubMed] [Google Scholar]
- [2].De Graaf M, R.a.M. Fouchier, Role of receptor binding specificity in influenza A virus transmission and pathogenesis, EMBO J 33 (2014) 1614–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Imai M, Kawaoka Y, The role of receptor binding specificity in interspecies transmission of influenza viruses, Curr Opp Virol 2 (2012) 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, et al. , Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities, J Mol Bio 355 (2006) 1143–1155. [DOI] [PubMed] [Google Scholar]
- [5].Stevens J, Blixt O, Paulson JC, Wilson IA, Glycan microarray technologies: tools to survey host specificity of influenza viruses, Nature Rev Micro 4 (2006) 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, et al. , Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site, Virology 233 (1997) 224–234. [DOI] [PubMed] [Google Scholar]
- [7].De Vries RP, Peng W, Grant OC, Thompson AJ, Zhu X, Bouwman KM, et al. , Three mutations switch H7N9 influenza to human-type receptor specificity, PLoS Pathog 13 (2017) e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lipsitch M, Barclay W, Raman R, Russell CJ, Belser JA, Cobey S, et al. , Viral factors in influenza pandemic risk assessment, eLife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Van Riel D, Munster VJ, De Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, et al. , Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals, Am J Pathol 171 (2007) 1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y, Avian flu: influenza virus receptors in the human airway, Nature 440 (2006) 435–436. [DOI] [PubMed] [Google Scholar]
- [11].Costa T, Chaves AJ, Valle R, Darji A, Van Riel D, Kuiken T, et al. , Distribution patterns of influenza virus receptors and viral attachment patterns in the respiratory and intestinal tracts of seven avian species, Vet Res 43 (2012) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bateman AC, Karamanska R, Busch MG, Dell A, Olsen CW, Haslam SM, Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAcα2–6 glycans, J Biol Chem 285 (2010) 34016–34026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, et al. , Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses, Proc Natl Acad Sci U S A 111 (2014) E2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nycholat CM, Mcbride R, Ekiert DC, Xu R, Rangarajan J, Peng W, et al. , Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins, Angew Chem Int Ed Engl 51 (2012) 4860–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Peng W, De Vries RP, Grant OC, Thompson AJ, Mcbride R, Tsogtbaatar B, et al. , Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity, Cell Host Microbe 21 (2017) 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Walther T, Karamanska R, Chan RW, Chan MC, Jia N, Air G, et al. , Glycomic analysis of human respiratory tract tissues and correlation with influenza virus infection, PLoS Pathog 9 (2013) e1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jia N, Barclay WS, Roberts K, Yen HL, Chan RW, Lam AK, et al. , Glycomic characterization of respiratory tract tissues of ferrets: implications for its use in influenza virus infection studies, J Biol Chem 289 (2014) 28489–28504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sauter NK, Bednarski MD, Wurzburg BA, Hanson JE, Whitesides GM, Skehel JJ, et al. , Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study, Biochemistry 28 (1989) 8388–8396. [DOI] [PubMed] [Google Scholar]
- [19].Xu H, Shaw DE, A simple model of multivalent adhesion and its application to influenza infection, Biophys J 110 (2016) 218–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, Mccauley JW, et al. , Receptor binding by a ferret-transmissible H5 avian influenza virus, Nature 497 (2013) 392–396. [DOI] [PubMed] [Google Scholar]
- [21].Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, et al. , Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin, Nat Biotechnol 26 (2008) 107–113. [DOI] [PubMed] [Google Scholar]
- [22].Peng W, Bouwman KM, Mcbride R, Grant OC, Woods RJ, Verheije MH, et al. , Enhanced human-type receptor binding by ferret-transmissible H5N1 with a K193T mutation, J Virol 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, et al. , The structure and receptor binding properties of the 1918 influenza hemagglutinin, Science 303 (2004) 1838–1842. [DOI] [PubMed] [Google Scholar]
- [24].Meng B, Marriott AC, Dimmock NJ, The receptor preference of influenza viruses, Influenza Other Respir Viruses 4 (2010) 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].De Vries RP, De Vries E, Bosch BJ, De Groot RJ, Rottier PJM, De Haan C.a.M., The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity, Virology 403 (2010) 17–25. [DOI] [PubMed] [Google Scholar]
- [26].Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P, A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates, PLoS One 7 (2012) e43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Throsby M, Van Den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, et al. , Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells, PLoS One 3 (2008) e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Heaton NS, Leyva-Grado VH, Tan GS, Eggink D, Hai R, Palese P, In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies, J Virol 87 (2013) 8272–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xu R, De Vries RP, Zhu X, Nycholat CM, Mcbride R, Yu W, et al. , Preferential recognition of avian-like receptors in human influenza A H7N9 viruses, Science 342 (2013) 1230–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].De Vries RP, De Vries E, Moore KS, Rigter A, Rottier PJ, De Haan CA, Only two residues are responsible for the dramatic difference in receptor binding between swine and new pandemic H1 hemagglutinin, J Biol Chem 286 (2011) 5868–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, De Wit E, Munster VJ, et al. , Airborne transmission of influenza A/H5N1 virus between ferrets, Science 336 (2012) 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang W, Shi Y, Lu X, Shu Y, Qi J, Gao GF, An airborne transmissible avian influenza H5 hemagglutinin seen at the atomic level, Science 340 (2013) 1463–1467. [DOI] [PubMed] [Google Scholar]
- [33].De Vries RP, Zhu X, Mcbride R, Rigter A, Hanson A, Zhong G, et al. , Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses, J Virol 88 (2014) 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Srinivasan A, Viswanathan K, Raman R, Chandrasekaran A, Raguram S, Tumpey TM, et al. , Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses, Proc Natl Acad Sci U S A 105 (2008) 2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wittmann V, Pieters RJ, Bridging lectin binding sites by multivalent carbohydrates, Chem Soc Rev 42 (2013) 4492–4503. [DOI] [PubMed] [Google Scholar]
- [36].Garces F, Sok D, Kong L, Mcbride R, Kim HJ, Saye-Francisco KF, et al. , Structural evolution of glycan recognition by a family of potent HIV antibodies, Cell 159 (2014) 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bandlow V, Liese S, Lauster D, Ludwig K, Netz RR, Herrmann A, et al. , Spatial screening of hemagglutinin on influenza A virus particles: sialyl-LacNAc displays on DNA and PEG scaffolds reveal the requirements for bivalency enhanced interactions with weak monovalent binders, J Am Chem Soc 139 (2017) 16389–16397. [DOI] [PubMed] [Google Scholar]
- [38].Liese S, Netz RR, Quantitative prediction of multivalent ligand-receptor binding affinities for influenza, cholera, and anthrax inhibition, ACS Nano 12 (2018) 4140–4147. [DOI] [PubMed] [Google Scholar]
- [39].Gambaryan AS, Tuzikov AB, Chinarev AA, Juneja LR, Bovin NV, Matrosovich MN, Polymeric inhibitor of influenza virus attachment protects mice from experimental influenza infection, Antiviral Res 55 (2002) 201–205. [DOI] [PubMed] [Google Scholar]
- [40].Landers JJ, Cao Z, Lee I, Piehler LT, Myc PP, Myc A, et al. , Prevention of influenza pneumonitis by sialic Acid-conjugated dendritic polymers, J Infect Dis 186 (2002) 1222–1230. [DOI] [PubMed] [Google Scholar]
- [41].Waldmann M, Jirmann R, Hoelscher K, Wienke M, Niemeyer FC, Rehders D, et al. , A nanomolar multivalent ligand as entry inhibitor of the hemagglutinin of avian influenza, J Am Chem Soc 136 (2014) 783–788. [DOI] [PubMed] [Google Scholar]
- [42].Shriver Z, Raman R, Viswanathan K, Sasisekharan R, Context-specific target definition in influenza a virus hemagglutinin-glycan receptor interactions, Chem Biol 16 (2009) 803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, et al. , Influenza virus pleiomorphy characterized by cryoelectron tomography, Proc Natl Acad Sci U S A 103 (2006) 19123–19127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Harbury PB, Zhang T, Kim PS, Alber T, A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants, Science 262 (1993) 1401–1407. [DOI] [PubMed] [Google Scholar]
- [45].Sliepen K, Van Montfort T, Ozorowski G, Pritchard LK, Crispin M, Ward AB, et al. , Engineering and characterization of a fluorescent native-like HIV-1 envelope glycoprotein trimer, Biomolecules 5 (2015) 2919–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, et al. , Leginon: a system for fully automated acquisition of 1000 electron micrographs a day, Ultramicroscopy 77 (1999) 153–161. [DOI] [PubMed] [Google Scholar]
- [47].Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, et al. , Appion: An integrated, database-driven pipeline to facilitate EM image processing, Journal of Structural Biology 166 (2009) 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B, DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy, Journal of Structural Biology 166 (2009) 205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ogura T, Iwasaki K, Sato C, Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking, J Struct Bio 143 (2003) 185–200. [DOI] [PubMed] [Google Scholar]
- [50].Grant OC, Smith HM, Firsova D, Fadda E, Woods RJ, Presentation, presentation, presentation! Molecular-level insight into linker effects on glycan array screening data, Glycobiology 24 (2014) 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. , UCSF Chimera--a visualization system for exploratory research and analysis, J Comput Chem 25 (2004) 1605–1612. [DOI] [PubMed] [Google Scholar]
- [52].Yang Y, Jun CD, Liu JH, Zhang R, Joachimiak A, Springer TA, et al. , Structural basis for dimerization of ICAM-1 on the cell surface, Mol Cell 14 (2004) 269–276. [DOI] [PubMed] [Google Scholar]
- [53].Woods-Group. GLYCAM Web In: Woods RJ, editor. Athens, GA: Complex Carbohydrate Research Center, The University of Georgia; 2005 – 2018. [Google Scholar]
- [54].Koppisetti RK, Fulcher YG, Jurkevich A, Prior SH, Xu J, Lenoir M, et al. , Ambidextrous binding of cell and membrane bilayers by soluble matrix metalloproteinase-12, Nat Commun 5 (2014) 5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Humphrey W, Dalke A, Schulten K, VMD: visual molecular dynamics, J Mol Graph 14 (1996) 33–38, 27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1.
All of the reference free 2D class averages contain the C-terminal GFP at the base of the HA trimer.
Supplementary Figure 2.
In previous work, a 3D structure of an α2–6 biantennary N-glycan with three lacNAc repeats was produced in a bidentate co-complex with HA (Upper Panel). This was modeled by first superimposing the terminal disaccharide onto a disaccharide in a crystal structure co-complex. Then the glycosidic linkages were adjusted within their low energy ranges and dihedral angles were selected that brought the second, unbound motif towards a second binding site. The binding motif was pulled into position during an all-atom molecular dynamics simulation and finally, the restraints were released to determine if the structure was stable. This method produced a bidentate co-complex for α2–6 biantennary N-glycan with three, four or five lacNAc repeats. In contrast, this method did not find bidentate N-glycans with α2–3 linkages. Although N-glycans containing α2–3 linkages and five lacNAc repeats could span both sites (Lower panel), the α2–3 linkage places the motif in the opposite orientation required by the binding site.


