Abstract
Medulloblastoma and central nervous system primitive neuroectodermal tumors (CNS-PNET) are aggressive, poorly differentiated brain tumors with limited effective therapies. Using Sleeping Beauty (SB) transposon mutagenesis, we identified novel genetic drivers of medulloblastoma and CNS-PNET. Cross-species gene expression analyses classified SB-driven tumors into distinct medulloblastoma and CNS-PNET subgroups, indicating they resemble human SHH and group 3 and 4 medulloblastoma and CNS neuroblastoma with FOXR2 activation. This represents the first genetically-induced mouse model of CNS-PNET and a rare model of group 3 and 4 medulloblastoma. We identified several putative proto-oncogenes including Arhgap36, Megf10, and Foxr2. Genetic manipulation of these genes demonstrated a robust impact on tumorigenesis in vitro and in vivo. We also determined that FOXR2 interacts with N-MYC, increases C-MYC protein stability, and activates FAK/SRC signaling. Altogether, our study identified several promising therapeutic targets in medulloblastoma and CNS-PNET.
Keywords: SB, Medulloblastoma, CNS-PNET, Arhgap36, Foxr2
Introduction
Embryonal tumors, including medulloblastoma and central nervous system primitive neuroectodermal tumors (CNS-PNETs), represent the most common malignant pediatric brain tumors (1). For ease of historical comparison, CNS-PNET is used in this manuscript according to the 2007 World Health Organization CNS tumor classification and includes CNS neuroblastomas, CNS ganglioneuroblastomas, medulloepitheliomas, and ependymoblastomas, although CNS-PNET no longer exists as an umbrella term(2). Medulloblastoma and CNS-PNET have similar histology: densely-packed, small cells with hyperchromatic nuclei and little cytoplasm. Medulloblastomas are usually cerebellar, while CNS-PNETs occur predominantly in the cerebrum. Aggressive, multi-modality treatments improve survival but produce lifelong side effects, and 5-year survival rates remain 60–65% for medulloblastoma and 20–40% for CNS-PNET(3).
Medulloblastoma and CNS-PNET are molecularly heterogenous. Medulloblastoma includes four molecular subgroups: WNT, SHH, group 3, and group 4; WNT and SHH are associated with mutations activating those pathways, but groups 3 and 4 remain less defined(4). A genomic study by Picard et al. identified three distinct CNS-PNET subgroups: primitive-neural, oligo-neural, and mesenchymal(5). Using methylation and gene expression-based analyses, Sturm et al. identified 4 molecular subgroups of CNS-PNET associated with gene fusions(6). While our understanding of the tumor biology has improved, a lack of animal models and targetable oncogenic drivers impede therapeutic development, particularly in group 3/4 medulloblastoma and CNS-PNET.
We used Sleeping Beauty (SB) transposon mutagenesis to identify novel medulloblastoma and CNS-PNET drivers. Transposition initiated in neural progenitor cells using Nestin-Cre was used alone, with Trp53lsl-R270H/+, or with Pten-deficiency to generate medulloblastomas and CNS-PNETs. These tumors resembled human medulloblastoma and CNS-PNET histologically and transcriptionally. Three candidate oncogenes, Arhgap36, Foxr2, and Megf10 were validated in vitro and in vivo and their mechanisms examined.
Materials and Methods
Generation of transgenic mice
Animal studies were conducted using procedures approved and monitored by the Institutional Animal Care and Use Committee at the University of Minnesota (UofMN). Nestin-Cre mice(7) were bred to either T2/Onc (chromosome 1/15)(8) or T2/Onc2 (chromosome 4)(9) to generate Nestin-Cre:T2/Onc(2). Rosa26lsl-SB11/+(10) were bred to either Trp53lsl-R270H/+(11) or Ptenflox/flox(12) to generate Rosa26lsl-SB11/+:Ptenflox/flox or Rosa26lsl-SB11/+:Trp53lsl-R270H/+. Nestin-Cre:T2/Onc(2) mice were bred to Rosa26lsl-SB11/+:Ptenflox/flox or Rosa26lsl-SB11/+:Trp53lsl-R270H/+ to generate mice with and without CNS-restricted SB-mutagenesis on wildtype (WT), Trp53lsl-R270H/+, and Ptenflox/+ backgrounds. T2/Onc(2) excision PCR was performed as described with primers in Supplementary Table 1(8).
Immunohistochemistry (IHC), immunoprecipitation (IP), and western blotting
Unstained tissue microarray (TMA) sections of formalin-fixed, paraffin-embedded (FFPE) human tumor specimens were obtained through the UofMN Materials Procurement Network (11 samples) and Johns Hopkins University (54 samples). FFPE tissue slides were stained with Hematoxylin and Eosin (H&E) or IHC using standard methods. IPs were done (Active Motif #54001) with 500μg total protein. Western blotting was done with whole cell lysates as described(13) using antibodies in Supplementary Table 2.
DNA-Common insertion site (D-CIS) analysis
Linker-mediated PCR to identify transposon insertion sites was performed as described(13). Transposon insertion sites were annotated using TAPDANCE(14). Non-redundant insertion sites representing >0.1% of the mapped insertions from each tumor library were used to generate CIS (P-value <0.05).
Transcriptional profiling
Isolated tumor RNA (Qiagen #75114) was assessed for quality using capillary electrophoresis (RIN>6.5, Agilent 2100 BioAnalyzer). Paired-end sequencing (30–40 million reads/sample) of TruSeq-prepared libraries was performed (Illumina HiSeq 2000). Raw FASTQ files are available at the NCBI Sequence Read Archive and linked to Gene Expression Omnibus SuperSeries (GSE122050). FASTQ files were mapped to the MM10 genome (T2/Onc and Rosa26lsl-SB11/+ as additional chromosomes)(15) using STAR-Fusion (https://github.com/STAR-Fusion/STAR-Fusion/wiki). Transcript FPKM values were computed using cuffquant and cuffnorm and adjusted by +0.1(16).
T2/Onc fusion identification
To identify T2/Onc:genome fusions, we analyzed the chimeric.out.junction and chimeric.out.sam output files from STAR-Fusion to summarize the number of junction (one read contains the T2/Onc:genome junction) and bridge (one paired-end read maps to T2/Onc and the other to the genome) reads present within 1000bp regions. Fusions supported by ≥1 junction read or ≥3 bridging reads were retained for analysis. Manual detection of T2/Onc(2):Arhgap36 transcripts was done using 500ng of purified RNA (Invitrogen #15596–018), reverse-transcribed (Invitrogen #18080–051) and amplified using primers in Supplementary Table 1.
Gene cluster similarity (GC-SIM)
GC-SIM was used for unsupervised, unbiased identification of similar gene clusters across transcriptional datasets. Transcriptional profile datasets were individually log-transformed, mean-centered, filtered for highly variant genes, and hierarchically clustered using average linkage and (1 – Pearson correlation) as the distance metric. Gene clusters with node correlation and size >respective thresholds were retained. Cross-dataset cluster pairs were tested for enrichment of common gene members (Fisher’s Exact Test) to identify conserved transcriptional patterns.
RT-PCR and 5′-rapid amplification of cDNA ends (5’-RACE)
For CNS-PNET expression analysis, cDNAs were synthesized (Applied Biosystems #4368814) and qRT-PCR was performed (Invitrogen #4369016). For Shh activation assays, purified cellular RNA (Ambion #12183025) was reverse transcribed (Invitrogen #11755050) and qRT-PCR was done in triplicate (Roche #4673492001). Shh activation was done as described(17). For 5’-RACE (Ambion #AM1700), tumor RNA was extracted from human medulloblastomas (Invitrogen #15596–018) and normal human brain RNA was purchased from BioChain (R1244039–50, R1244035–50, R1234040–10). Subsequent detection of transcripts by RT-PCR was performed with 500ng RNA (Invitrogen #18080–051). Primers and probes are listed in Supplementary Table 1.
Cell culture/assays
Cell lines were maintained, authenticated, and tested for mycoplasma as described in Supplementary Table 3. MTS (Promega G1111), soft agar assays, and transfections were done as described(13). Stable lines transfected with cDNAs (ARHGAP36 (Q6ZRI8–5), FOXR2 (Q6PJQ5–1), Megf10 (Q6DIB5–1)) were cultured as polyclonal populations in puromycin. Transient transfection in HEK293Ts was done per manufacture’s protocol (Invitrogen 11668019). CRISPR-KO clones were isolated as described(13). Briefly, Daoy cells were transfected with PiggyBAC transposase and a puromycin-selectable PiggyBAC transposon vector containing 2 FOXR2 guide RNAs (sequences in Supplementary Table 1) and Cas9. Isolated clones were sequenced to identify changes in FOXR2. Wound healing assays were performed as described(18). Primary granule neuron precursors (GNPs) were isolated from neonatal C57BL/6J and thymidine incorporation assays were performed as described(19).
in vivo assays
NRG mice (Jackson 007799) were injected as described(20). Briefly, C17.2 cells were prepared in HBSS, counted, and stored on ice prior to injection (2×105 cells/2μl injection). P0 mice were injected in the fourth ventricle (stereotactic coordinates: 1.5mm anterior to Bregma, 1.5mm deep). Successful injection was verified on P1 by luciferase imaging as described(20). Adult intracranial injections were performed as described(19). Female NU/J mice (Jackson 002019, 6–8 weeks old) were anesthetized (81 mg/kg ketamine, 13.8 mg/kg xylazine) and injected with 1×106 cells/5μl (prepared as above). For flank tumor assays, female NU/J mice (Jackson 002019, 6–8 weeks old) were injected with 1×106 C17.2 cells (prepared as above) resuspended 1:1 in HBSS and Matrigel (Corning CB-#40234C). Tumor volume = (l x w2)/2, l=length and w=width.
Results
SB Mutagenesis Promotes Medulloblastoma and CNS-PNET Formation
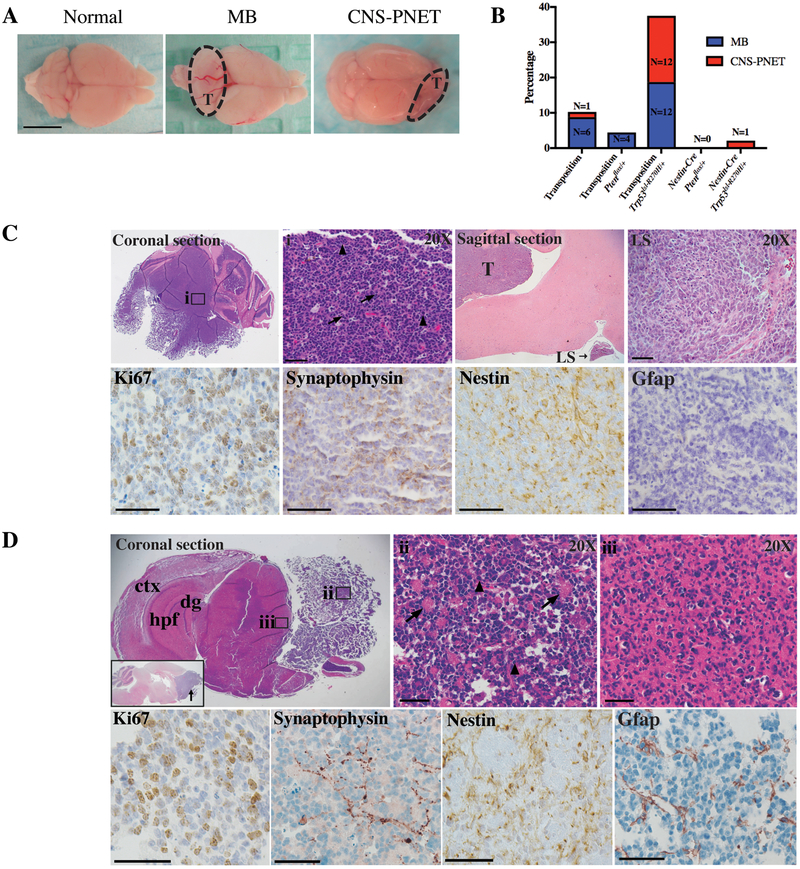
To identify genetic drivers of medulloblastoma and CNS-PNET, we targeted Nestin+ neural and glial precursor cells with SB-mutagenesis on three genetic backgrounds: wildtype (WT), Pten heterozygous (Ptenflox/+), or Trp53 mutant (Trp53lsl-R270H/+). Ptenflox/+ and Trp53lsl-R270H/+ served as sensitizing backgrounds as they are mutated in human medulloblastoma and CNS-PNET(21,22). IHC revealed SB expression throughout the developing brain, including cells within the granule layer, white matter, surrounding the fourth ventricle, subependymal midbrain, subventricular zone, and olfactory bulb (Supplementary Fig. S1A–G). Experimental cohorts harbored one of three transposon concatemers (Supplementary Fig. S1H). SB-mutagenesis significantly reduced survival in combination with Trp53lsl-R270H (Supplementary Fig. S1I–K). Upon necropsy we observed masses in the brain, testicles, bone, peripheral lymph nodes, and spleen (Supplementary Fig. S1I–K, Supplementary Table 4).
Histological analysis of brain masses revealed the presence of infratentorial medulloblastoma and supratentorial CNS-PNET (22 medulloblastomas and 14 CNS-PNETs) with highest medulloblastoma frequency in Trp53lsl-R270H/+ mice (Fig. 1A–B). The high-copy transposon (T2Onc2, chromosome 4) produced the highest proportion of medulloblastoma, while CNS-PNETs were equally derived from chromosome 4 and 15 concatemers (Supplementary Fig. S2A). Tumors expressed nuclear SB by IHC and showed transposon mobilization by PCR (Supplementary Fig. S2B–C).
Figure 1.
SB-induced medulloblastoma and CNS-PNET resemble human tumors.
A, Macroscopic images of normal brain and brains with SB-induced cerebellar medulloblastoma and CNS-PNET in the cerebral cortex and olfactory bulbs. T=tumor. B, Medulloblastoma and CNS-PNET frequency across genetic backgrounds. C, Upper panels: medulloblastoma H&E. i, Rosettes (arrows), mitotic nuclei (arrowheads). Primary medulloblastoma (T) with leptomeningeal spread (LS). Lower panels: medulloblastoma IHC. D, Upper panels: CNS-PNET H&E. Cerebral cortex (ctx), hippocampal formation (hpf), dentate gyrus (dg). Inset: CNS-PNET sagittal section, olfactory bulb (arrow). ii, Bulk tumor with rosette formations (arrows) and mitotic nuclei (arrowheads). iii, Tumor cell parenchyma infiltration. Lower panels: CNS-PNET IHC. Scale bars = 50 μm.
SB-Induced Medulloblastoma and CNS-PNET Resemble Human Tumors Histologically
SB-induced medulloblastomas originated in the cerebellum whereas CNS-PNETs occurred in the rostral portion of the brain, overwhelming the olfactory bulbs, lateral ventricle, and cortex (Fig. 1A, C–D). Both tumor types resembled their human counterparts histologically (small round cells with high nuclear:cytoplasmic ratios, Homer-Wright rosettes, vascularization, and mitotic figures), expressed diagnostic markers for human medulloblastoma and CNS-PNET (Synaptophysin, Ki67, and Nestin), and stained negatively for the astrocytic marker Gfap (Fig. 1C–D). 50% of medulloblastomas and 100% of CNS-PNETs exhibited metastatic characteristics, including infiltration into the leptomeninges, parenchyma, brainstem, and cortex (Supplementary Table 4). Leptomeningeal spread is of interest due to its associated poor prognosis, prevalence (one-third of patients), and difficulty modeling(5,23).
SB-Induced Medulloblastomas Resemble Non-WNT Human Medulloblastoma
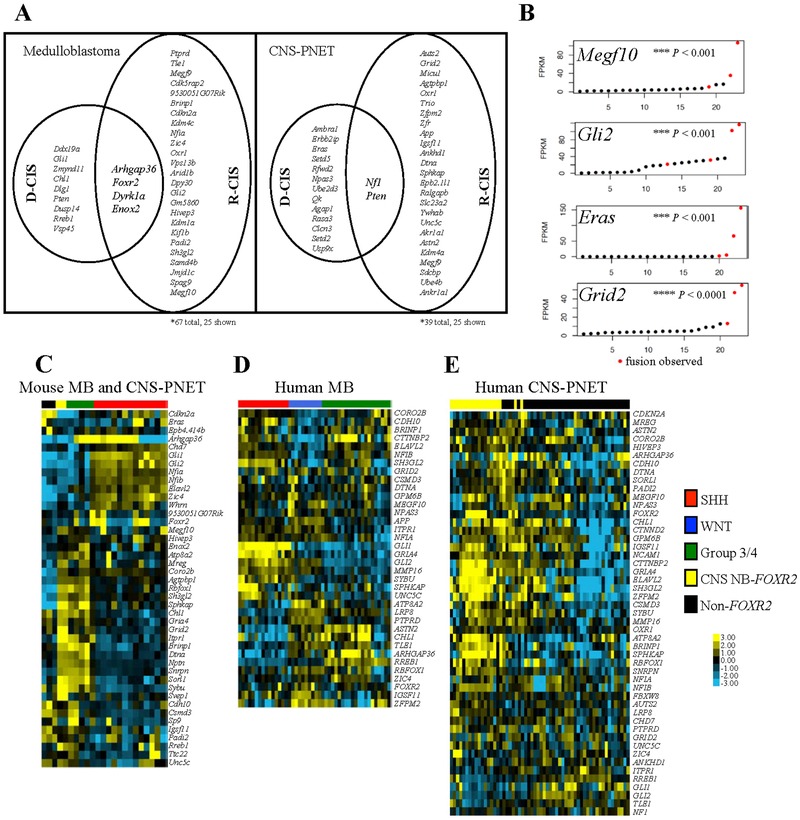
We transcriptionally profiled 18 SB-induced medulloblastomas, revealing two clear subgroups that each matched human medulloblastoma subtypes (Fig. 2A)(6). The first subgroup (N=13) showed increased Gli1, canonically associated with human SHH medulloblastoma(24). The second subgroup (N=5) showed increased Npr3 and Kcna1, markers for human group 3 and 4, respectively(24). More globally, the Gli1-overexpressing medulloblastoma gene set overlapped with highly-expressed genes in human SHH medulloblastoma (N=48, Fig. 2A, Supplementary Table 5)(6). Similarly, the Npr3 and Kcna1-overexpressing mouse gene set overlapped with highly-expressed genes in human group 3 and 4 medulloblastoma (N=120). Mouse tumors did not exhibit Wnt signatures.
Figure 2.
SB-induced tumors resemble human medulloblastoma and CNS-PNET transcriptionally.
A, Hierarchical clustering of medulloblastoma transcription profiles(6). Red and green boxes denote transcripts in SHH and group 3/4, respectively (P-value <0.002, Fisher’s Exact Test (FET)). SHH, WNT, and group 3/4 designation indicated with red, black, and green toebars, respectively. B, Hierarchical clustering of CNS-PNET transcription profiles(6). Blue boxes denote transcripts in CNS NB-FOXR2 (P-value <1.0e-8, FET). CNS NB-FOXR2 and non-FOXR2 CNS-PNET designation are shown with blue and black toebars, respectively. Log-transformed and mean-centered data with variance >0.5 for murine RNA-Seq datasets and >2.0 for human array datasets were clustered using average linkage clustering. Clusters systematically identified with node correlation >0.2.
SB-Induced CNS-PNETs Resemble Human CNS Neuroblastoma with FOXR2 Activation (CNS NB-FOXR2)
Transcriptional profiling comparing mouse CNS-PNETs (N=5) to published human CNS-PNETs (N=58) revealed two subgroups (Fig. 2B)(6). A single cluster of co-varying genes (N=298) was significantly enriched in both human and mouse CNS-PNET and contained high levels of CNS NB-FOXR2 associated genes, including MMP24, KCJN9, and CHGB (Fig. 2B, Supplementary Table 6). Consistent with CNS NB-FOXR2 classification, SB-induced CNS-PNETs showed significantly increased Olig1, Olig2, and Sox10 by qPCR and were Olig2+ by IHC (Supplementary Fig. 2D–E). The three remaining mouse CNS-PNETs had elevated expression of CNS EFT-CIC marker genes (Shc4, Argdib, and Pole) but no clearly corresponding human subgroup.
CIS reveal candidate cancer genes
We performed linker-mediated PCR on 22 medulloblastomas and 13 CNS-PNETs and identified 390,000 and 155,000 non-redundant insertions, respectively. TAPDANCE analysis(14) identified 13 medulloblastoma and 15 CNS-PNET DNA-CIS (D-CIS)(Fig. 3A, Supplementary Table 7). We also identified RNA-CIS (R-CIS) in both tumor types, defined as transposon fusion transcripts present in both ≥10% of cases and ≥1 tumor (Fig. 3A, Supplementary Table 7). For several putative oncogenes, the presence of a T2/Onc(2) fusion transcript significantly increased expression (Fig. 3B). We identified genes previously implicated in medulloblastoma, including Gli1 and Pten; upregulation of GLI1 expression and PI3K pathway activation through PTEN loss are observed in human medulloblastoma(21). Predicted transposon-mediated driving effects on Gli1 expression were confirmed by IHC (Supplementary Fig. S3A). We also confirmed Pten reduction and a corresponding increase in pAkt with Pten insertions (Supplementary Fig. 3B). Arhgap36 was most frequently modified, with insertions identified in 14 (D-CIS) and 13 (R-CIS) medulloblastomas and 2 CNS-PNETs (R-CIS). Enox2, a tumor-associated NADH oxidase involved in the growth of several cancer cell lines(25), was also a D and R-CIS in medulloblastoma (predicted SB oncogene).
Figure 3.
CIS gene identification and expression analysis in mouse and human tumors.
A, Medulloblastoma (MB) and CNS-PNET CIS genes. B, RNA-Seq expression levels in SB-induced tumors (Student t test, two-tailed). C-E, Expression of CIS genes with highest variability in mouse tumors(C), human medulloblastoma(5)(D), and human CNS-PNET(5)(E). Log-transformed and mean-centered data with variance >1.0 were clustered using average linkage clustering. Multiple human probes corresponding to CIS were averaged to obtain a single value.
We identified known and novel molecular changes in SB-induced CNS-PNETs. Pten was a predicted tumor suppressor gene (TSG) and loss of PTEN through 10q loss or mutation is observed in human CNS-PNET(22). Novel to this study, NF1 was the most targeted CNS-PNET TSG. We identified several other predicted Ras effector gene alterations, including Eras overexpression and Erbb2ip and Rasa3 disruption (Supplementary Table 4). Tumors harboring these insertions exhibited increased pErk (Supplementary Fig. S3C, Supplementary Table 4) supporting Ras pathway activation. We also observed NF1 locus deletion in a subset of human CNS-PNETs (Supplementary Fig. S3D).
We next analyzed our CIS gene expression in published human medulloblastomas and CNS-PNETs (Fig. 3C–E)(6). GRIA4 showed high expression in both SHH medulloblastoma and CNS NB-FOXR2. FOXR2 is elevated in a subset of WNT medulloblastoma and CNS NB-FOXR2. Interestingly, ARHGAP36 is highly expressed in group 3 and 4 medulloblastoma, with low expression in SHH (Supplementary Fig. S4A), while in the mouse Arhgap36 insertions occurred in Shh and group 3/4 tumors (7/12 Shh and 4/5 group 3/4 tumors, Supplementary Table 4).
ARHGAP36 Expression is Associated with Poor Prognosis in Human Medulloblastoma
Transposon location and orientation implicate Arhgap36 as an oncogene, with insertions upstream of the locus or within intron 1 and significantly increasing gene expression (Fig. 4A–B). T2/Onc:Arhgap36 transcripts displayed precise fusion of the T2/Onc splice donor to the Arhgap36 exon 2 splice acceptor (Supplementary Fig. S4B), generating a 15 amino acid N-terminal truncation with translation from an in-frame ATG. Tumors with Arhgap36 insertions showed high levels of cytoplasmic Arhgap36 by IHC compared to tumors without Arhgap36 insertions, which displayed sparse nuclear expression similar to normal granule cells (Fig. 4C). Spatial and temporal analysis of ARHGAP36 expression in normal human and mouse cells within developing and mature cerebella showed nuclear localization throughout the molecular layer (ML), Purkinje cell layer (PC) and internal granule cell layer (IGL)(Supplementary Fig. S4C). In two combined TMAs of human medulloblastoma, 37/65 (56%) and 8/65 (12%) expressed cytoplasmic and nuclear ARHGAP36 expression, respectively, by IHC (Fig. 4D). ARHGAP36 protein was expressed across all human subgroups, although increased ARHGAP36 transcript was only expressed in group 3/4 human medulloblastoma (Fig. 4E, Supplementary Fig. S4A)(24). Overall and cytoplasmic ARHGAP36 expression correlated with accelerated mortality (Fig. 4F, Supplementary Fig. S4D).
Figure 4.
Increased Arhgap36 expression is associated with medulloblastoma.
A, Arhgap36 locus with transposon insertions (green arrowheads). B, Arhgap36 expression by RNA-Seq in SB-induced medulloblastomas (Student t test, two-tailed). C, Arhgap36 IHC in SB-induced medulloblastoma. Primary tumor (*), leptomeningeal spread (arrowhead). Nuclear expression in control tumor (arrow) compared to normal granule neural cells (inset). D, Combined TMAs analyzed for ARHGAP36 by IHC. E, ARHGAP36 positivity by IHC across subgrouped Johns Hopkins TMA. F, Kaplan-Meier analysis of patients from Johns Hopkins TMA (Log rank Mantel-Cox test). Scale bars = 50mm.
We further investigated ARHGAP36 transcript profiles using 5′-RACE on human group 3/4 medulloblastoma samples, cell lines, and normal cerebellar cells. Several ARHGAP36 amplification products were identified (Supplementary Fig. 4E–F) predicting expression of canonical isoform 1(*), 5(**), and 3(***; ****). All 3 isoforms contain intact ARHGAP36 predicted functional domains, including an arginine-rich domain (ARR), nuclear localization sequence (NLS), and GTPase-activating protein (GAP) domain. Interestingly, the 5’ ends of *** and **** begin with intron 2 sequences splicing to exon 3 and an in-frame ATG located in exon 4. Target-specific RT-PCR revealed these ARHGAP36 sequences in additional tumor samples, while normal fetal cerebellum only expressed isoform 1 (Supplementary Fig. S4G).
ARHGAP36 Promotes Tumor Formation in Neural Progenitor Cells
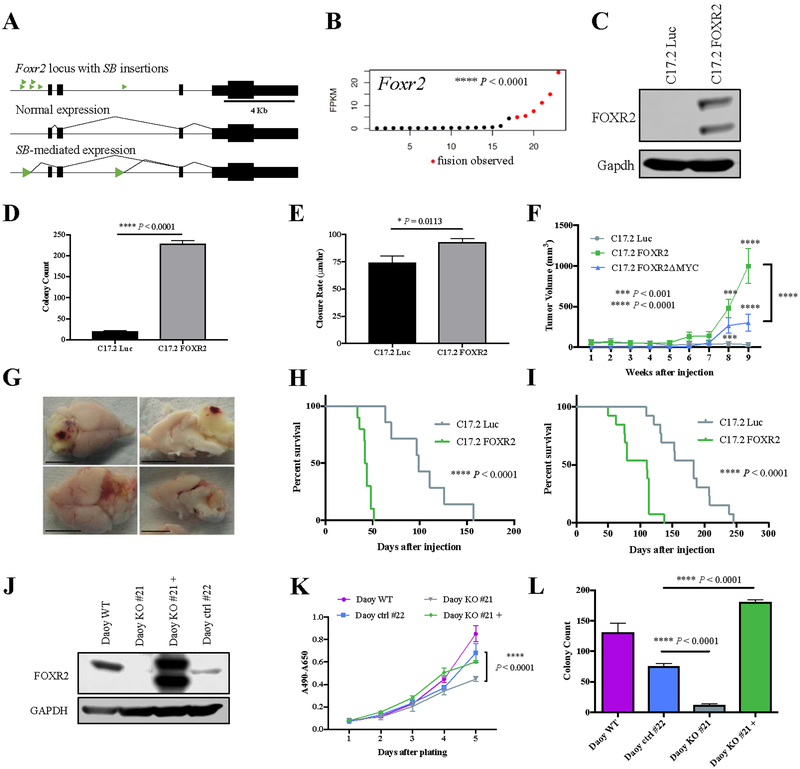
To further characterize the role of ARHGAP36 in medulloblastoma, we overexpressed truncated ARHGAP36 (isoform 5) in the mouse neural progenitor cell line C17.2 (Supplementary Fig. 5A)(26). Increased ARHGAP36 significantly enhanced soft agar colony formation but did not affect proliferation or collective cell migration rate (Fig. 5A, Supplementary Fig. S5B–C). C17.2 cells expressing ARHGAP36 formed tumors significantly faster in the flanks of NU/J mice than luciferase control cells (Fig. 5B). When injected orthotopically into adult NU/J mice, C17.2 cells localized to the granule layer of the cerebellum where ARHGAP36 expression drove leptomeningeal spread into the cerebrum and cerebellar tumor formation, reducing median survival from 99 to 71 days (Fig. 5C–D). Additionally, increased ARHGAP36 expression in primary GNPs significantly increased their proliferation (Fig. 5E). As previously reported, ARHGAP36 strongly activated Shh signaling in C17.2 cells in a ligand-independent manner, providing a potential mechanism for ARHGAP36-driven tumorigenesis (Fig. 5F)(17,27). Additionally, SB transposon insertions in the Arhgap36 locus were mutually exclusive with insertions in Gli1 and Gli2, Shh pathway activators (Fig. 5F).
Figure 5.
ARHGAP36 and Megf10 promote tumorigenesis.
A, Soft agar assay comparing C17.2 Luc and C17.2 ARHGAP36 (Student t test, two-tailed). B, Flank tumor volume of NU/J mice injected with C17.2 Luc or C17.2 ARHGAP36 (N=5, Sidak’s multiple comparisons test). C, Survival of NU/J mice injected intracranially with C17.2 Luc or C17.2 ARHGAP36 (N=7, Log rank Mantel-Cox test). D, IHC showing cerebellar and cerebral location of GFP+ C17.2 Luc or C17.2 ARHGAP36 injected into NU/J mice. E, Tritiated thymidine (3H-Td) incorporation assay in transduced GNPs (N=3, Benjamini, Krieger, and Yekutieli multiple comparisons test). F, Upper panel: qRT-PCR for Gli1 in C17.2 Luc and C17.2 ARHGAP36 (Sidak’s multiple comparison’s test). Gli1 expression is normalized to Gapdh. Lower panel: RNA-Seq of SB-induced medulloblastomas showing expression of indicated genes. G, MTS assay of C17.2 Luc and C17.2 Megf10 (Sidak’s multiple comparisons test). H, Soft agar assay of C17.2 Luc and C17.2 Megf10 (Student t test, two-tailed). I, Flank tumor volume of NU/J mice injected with C17.2 Luc (N=7) or C17.2 Megf10 (N=8)(Sidak’s multiple comparisons test). Error bars, SEM. Scale bars = 100mm.
Megf10 Promotes Transformation in vitro and in vivo
Megf10 (predicted SB oncogene) was identified as an R-CIS, with fusion transcripts in 3 medulloblastomas significantly increasing expression (Fig. 3A–B). Megf10 is expressed throughout the developing CNS and is a positive regulator of Notch signaling(28). MEGF10 is upregulated in a subset of human CNS-PNET and medulloblastoma (15/26 medulloblastomas, 14/58 CNS-PNETs, Supplementary Fig. S5D). Megf10 expression in C17.2 cells significantly enhanced colony formation in soft agar, proliferation by MTS, and flank tumor formation (Fig. 5G–I). Additionally, increased Megf10 expression in GNPs increased their proliferation by 1.7-fold, though not significantly possibly due to low sample size (Fig. 5E). Megf10 had no effect on C17.2 cell Notch signaling, however, by western blot (Supplementary Fig. S5E).
FOXR2 Promotes Transformation in Human and Mouse Cells
All Foxr2 transposon insertions were located upstream of the translation start site and drove increased Foxr2 expression, predicting an oncogenic role (Fig. 6A–B). The presence of a Foxr2 insertion significantly reduced median survival from 166.5 to 116.5 days (Supplementary Fig. S6A). FOXR2 overexpression in C17.2 cells significantly increased soft agar colony formation and collective cell migration without increasing proliferation (Fig. 6C–E, Supplementary Fig. S6B). C17.2 FOXR2 cells formed flank tumors significantly faster than C17.2 Luc controls (Fig. 6F). When injected orthotopically into adult NU/J mice, C17.2 FOXR2 cells migrated to the granule layer of the cerebellum and formed large, vascular tumors, reducing median survival from 99 to 43 days (Supplementary Fig. 6C, Fig. 6G–H). C17.2 FOXR2 cells injected orthotopically into neonatal NRG mice also significantly reduced median survival compared to C17.2 Luc and resulted in tumor formation (110 vs 183 days, Fig. 6I). Importantly, FOXR2 overexpression drove increased proliferation in primary GNPs (Fig. 5E). Using the CRISPR/Cas9 system, we knocked out (KO) FOXR2 in Daoy, a human medulloblastoma cell line. Daoy clone #21 had a nonsense mutation in exon 1 resulting in FOXR2 protein loss, decreased proliferation, and decreased soft agar colony formation, all rescued by FOXR2 cDNA expression (Fig. 6J–L).
Figure 6.
FOXR2 promotes transformation in human and mouse cells.
A, Transposon insertions (green arrowheads) in the Foxr2 locus. B, Foxr2 expression by RNA-Seq in SB-induced medulloblastoma (Student t test, two-tailed). C, Western blot showing FOXR2 expression in C17.2 Luc and C17.2 FOXR2. D, Soft agar assay comparing C17.2 Luc and C17.2 FOXR2 (Student t test, two-tailed). E, Wound closure rate of C17.2 Luc (N=14) and C17.2 FOXR2 (N=15)(Student t test, two-tailed). F, Flank tumor volume of NU/J mice injected with C17.2 Luc (N=7), C17.2 FOXR2ΔMYC (N=6) or C17.2 FOXR2 (N=8)(Sidak’s multiple comparison’s test). G, Whole and halved brains from NU/J mice injected intracranially with C17.2 FOXR2. Scale bars = 1 cm. H, Survival of NU/J mice injected intracranially with C17.2 Luc (N=7) or C17.2 FOXR2 (N=10)(Log rank Mantel-Cox test). I, Survival of NRG mice injected intracranially with C17.2 Luc or C17.2 FOXR2 (N=13)(Log rank Mantel-Cox test). J, Western blot of Daoy WT, Daoy #21 (FOXR2 KO), Daoy #21+ (FOXR2 KO with rescue FOXR2 cDNA), and Daoy #22 (has integrated CRISPR/Cas9 vector but no FOXR2 mutation). K, MTS assay of Daoy WT, Daoy #21, Daoy #21+ and Daoy #22 (Dunnett’s multiple comparison’s test). L, Soft agar assay of Daoy WT, Daoy #21, Daoy #21+ and Daoy #22 (Dunnett’s multiple comparison’s test). Error bars, SEM.
FOXR2 Has a Multi-Faceted Mechanism Including Effects on MYC and FAK
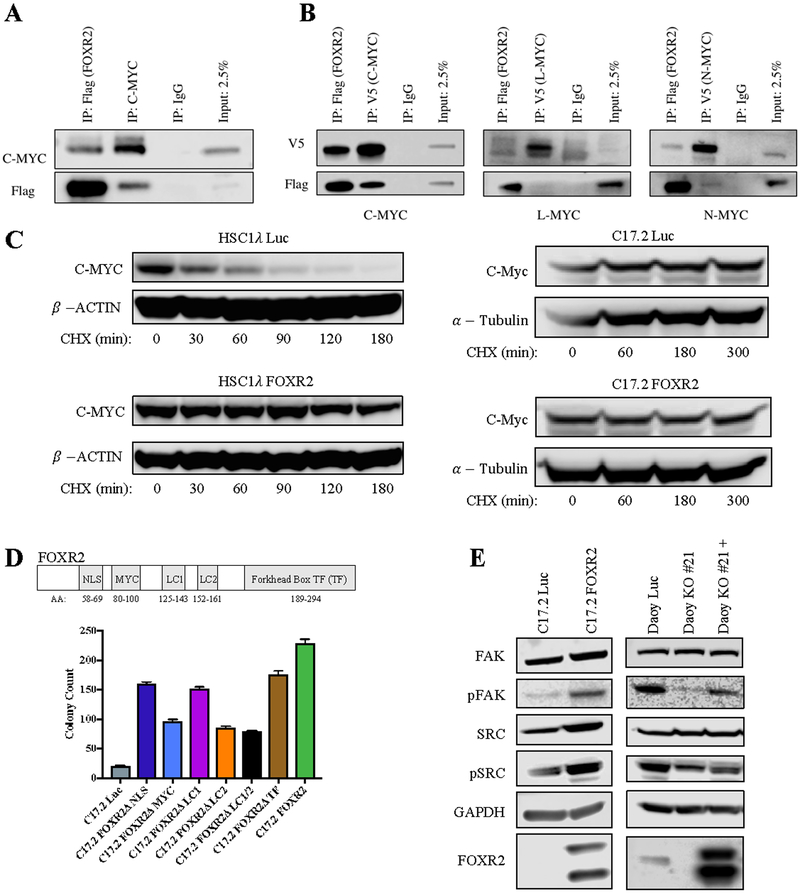
FOXR2 has many suggested oncogenic mechanisms, including interaction with C-MYC(29). We confirmed this interaction by CoIP in a human Schwann cell line, HSC1λ, and C17.2 cells stably expressing FOXR2 (Fig.7A, Supplementary Fig. 6D). To determine if FOXR2 interacts with other forms of MYC, we transiently transfected HEK293T cells with V5-tagged C-MYC, L-MYC and N-MYC. We observed reduced interaction of FOXR2 with N-MYC and minimal interaction with L-MYC (Fig. 7B). To determine if FOXR2 stabilizes C-MYC, we treated cells with cycloheximide (CHX) to inhibit translation. Almost all C-MYC protein was degraded in control HSC1λ cells, but with FOXR2, C-MYC levels were only reduced by half after 3 hours, indicating FOXR2 is stabilizing (Fig. 7C). Interestingly, C-MYC was highly stable in C17.2 cells regardless of FOXR2 expression, likely due to their immortalization by V-Myc(26), implying that FOXR2 transforms C17.2 cells through alternative mechanisms.
Figure 7.
FOXR2 interacts with C-MYC and N-MYC and activates FAK/SRC signaling.
A, CoIP of endogenous C-MYC with flag-tagged FOXR2 in HSC1λ. B, CoIP of V5-tagged C-MYC, L-MYC and N-MYC with flag-tagged FOXR2 in HEK293T. C, Western blot showing cycloheximide (CHX)-treated HSC1λ and C17.2 with and without FOXR2. CHX treatment (100 ug/ml in DMSO) was done for time indicated. D, Upper panel: putative FOXR2 protein domains. Lower panel: soft agar assay of C17.2 Luc or indicated FOXR2 deletion mutants. Error bars, SEM. E, Western blot showing effects of FOXR2 expression changes on FAK/SRC signaling.
To further characterize the oncogenic mechanism of FOXR2, we synthesized FOXR2 cDNA constructs missing the following predicted domains: NLS(ΔNLS)(30), MYC interaction (ΔMYC)(29), low complexity regions (ΔLC1, ΔLC2 and ΔLC1/2)(31), and forkhead box transcription factor (ΔTF)(31)(Fig. 7D). We stably expressed each mutant and performed soft agar assays in C17.2 and HSC1λ cells. Surprisingly, no single deletion mutant completely ablated the colony formation promoting capacity of FOXR2 in either line, but loss of the Myc interaction and LC2 domains significantly reduced colony formation (Fig. 7D, Supplementary Fig. 6E). C17.2 FOXR2ΔMYC also had an intermediate phenotype in the flank (Fig. 6F). We verified that the FOXR2ΔMYC mutant did not bind C-Myc (Supplementary Fig. 6F). Given no single domain loss completely ablated colony formation but some did reduce it, we conclude FOXR2 has a multi-faceted mechanism. We observed a slight change in the actin cytoskeleton of FOXR2-expressing cells, prompting assessment of focal adhesion kinase (FAK) activation. C17.2 cells expressing FOXR2 displayed increased Fak phosphorylation (Y397), resulting in increased (activating) phosphorylation at Src Y416 (Fig. 7E). This effect is Myc-independent (Supplementary Fig. 6G). Correspondingly, FOXR2 loss in Daoy cells resulted in decreased pFAK and pSRC rescuable by FOXR2 cDNA expression (Fig. 7E).
Discussion
We used SB transposon mutagenesis to identify novel drivers of medulloblastoma and CNS-PNET. Over half of our D-CIS and several of our R-CIS were reported in previous SB medulloblastoma screens(23,32–34), including Pten, Wac, Arid1b, Arhgap36, Foxr2, and Megf10, making them especially compelling candidates (Supplementary Table 8). Notably, several R-CIS are located on chromosomes 4 and 15, the locations of the T2/Onc2 and T2/Onc concatemers, respectively. Although local hopping may account for bias toward genes on these chromosomes, several are implicated in cancer, including Tle1 and Ptprd (35,36). Additionally, these concatemers have been previously used to identify R-CIS in osteosarcoma, and only one R-CIS gene was common to both studies (Cdkn2a, Supplementary Table 9)(15). Our medulloblastoma R-CIS include several highly compelling targets, including Megf10. We identified a novel, oncogenic role for Megf10 in neural progenitor cells, the mechanism for which warrants further study.
Ours is the first transposon screen to produce CNS-PNETs. We identified several genes with known roles in neural cancer not previously implicated in CNS-PNET, including Setd2, Ambra1, and Usp9x(37–39). Several Ras-associated genes were mutated in our screen, including Nf1, Eras, Pten, and Ras3, suggesting an importance of Ras pathway activation and cooperation with p53 loss in PNETagenesis. Additionally, we identified NF1 loss in human CNS-PNETs. Activated RAS/MAPK signaling with p53 loss has been shown to drive CNS NB-FOXR2 formation in zebrafish(40), and somatic PTEN loss is associated with human CNS-PNET(22). Interestingly, we did not recover any CNS-PNETs on the Pten-deficient background, possibly indicating that p53 loss creates a permissive cell with subsequent Ras activation.
FOXR2 is a member of the forkhead-box (FOX) transcription factor family, which contribute to a wide variety of cellular processes(41). FOXR2 acts as an oncogene in several neural cancers including: malignant peripheral nerve sheath tumors, glioma, CNS-PNET, and medulloblastoma(6,13,34,42). Interestingly, although FOXR2 has been shown to be upregulated in CNS NB-FOXR2(6), we did not recover Foxr2 insertions in the SB-induced CNS-PNETs, including 2 tumors that resembled CNS NB-FOXR2 transcriptionally. Other insertions may mimic the CNS NB-FOXR2 phenotype; these 2 tumors exclusively harbored insertions in Epb4.1l1, Itpr1, Rbfox1, and Sphkap.
The mechanism of FOXR2-driven tumorigenesis has proven diverse and elusive. FOXR2 can promote WNT signaling, activate SHH signaling, promote EMT, and affect cell cycle (34,42–45). We examined each of these pathways in C17.2 cells and found no effect of FOXR2 on B-catenin localization, Axin2, Gli1, p21, or CyclinD1 mRNA expression, or N-Cadherin, E-Cadherin, or Vimentin protein levels (Supplementary Fig. 7A–D). We found that FOXR2 binds and stabilizes C-MYC. Mouse models with C-MYC-driven tumors show an addiction to C-MYC expression, suggesting C-MYC is a strong therapeutic target in cancer(46). However, directly targeting C-MYC has been difficult. Therefore, targeting C-MYC interacting proteins, such as FOXR2, may prove useful for cancer therapy. We also found that FOXR2 promotes activation of the FAK/SRC signaling pathway. FAK activation is associated with poor prognosis and drug resistance in a variety of cancers and targeting FAK produces deleterious off-target effects(47). Interestingly, co-targeting of FAK and C-MYC was recently shown to have synergistic effects in ovarian cancer(48). The ability of FOXR2 to activate both of these pathways makes it an excellent candidate for targeted therapy. Additionally, FOXR2 has minimal expression in adult tissues, making off-target toxicity risk low(13).
We identified ARHGAP36 as both a mouse and human medulloblastoma oncogene. ARHGAP36 expression in C17.2 cells promoted anchorage independent growth, tumor formation, leptomeningeal spread, and SHH activation. Current therapies targeting an upstream pathway member, Smoothened (SMO), have been met with resistance through SMO mutations(49). Since its interactions with PKA and SUFU are both downstream of SMO, ARHGAP36 poses a good target for treatment-resistant, SHH-driven medulloblastoma(17,27). Additionally, Arhgap36 was the most up-regulated gene in mouse allografts propagated in the presence of a SMO antagonist(17). Interestingly, Arhgap36 insertions occurred in mouse Shh and group 3/4 tumors, and ARHGAP36 is expressed across all subgroups of human medulloblastoma, indicating ARHGAP36 may also have non-SHH pro-tumorigenic effects.
We identified several candidate driver genes in medulloblastoma and CNS-PNET relevant to human cancer. To our knowledge, this is the first study to present a genetically-induced CNS-PNET mouse model, providing an opportunity for studying this rare and aggressive tumor. We also present tumors that resemble group 3/4 medulloblastoma with high incidence of leptomeningeal spread, again providing a needed mouse model for these tumors. Interestingly, these diverse tumor types were driven with the same, Nestin-driven Cre recombinase, indicating that the cell of origin of non-Wnt medulloblastoma and CNS-PNET is Nestin+ or a close descendent. We used RNA-Seq to identify CIS genes and subtype mouse SB-induced tumors based on human expression data. Arhgap36, our top CIS gene, was shown to transform a mouse neuroblast line. Foxr2 was identified as a proto-oncogene and shown to promote C-MYC stability and FAK pathway activation. Both of these genes offer promise as novel therapeutic targets in human medulloblastoma and warrant additional study. Further functional testing of additional CIS genes may reveal additional treatment options for embryonal tumors.
Supplementary Material
Significance.
A transposon-induced mouse model identifies several novel genetic drivers and potential therapeutic targets in medulloblastoma and CNS-PNET.
Acknowledgements
This work was supported by UofMN Genomics Center, UofMN Biology Materials Procurements Network, Research Animal Resources, and University Imaging Centers that are supported by the National Cancer Institute.
Financial support: Support for this research was provided by The American Cancer Society (Research Professor Award #123939 to DAL), the National Institutes of Health (U54CA210190 to DAL and DJO, R01CA113636 to DAL, T32 T32GM113846 to PJB, R50-CA211249 to ALS, T32 AI083196 to BRT, T32CA009138 to JDL, and R01CA172986 to DJO), the Children’s Cancer Research Fund, and the Hedberg Family Chair (to DAL).
Footnotes
Disclosures: DAL is the co-founder and co-owner of several biotechnology companies including NeoClone Biotechnologies, Inc., Discovery Genomics, Inc. (recently acquired by Immunsoft, Inc.), and B-MoGen Biotechnologies, Inc. He consults for Surrogen, Inc. Genentech, Inc. is funding some of his research. The business of all these companies is unrelated to the contents of this manuscript. Other authors have no conflicts of interest.
References
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol 2013;15 Suppl 2:ii1–56 doi 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114(2):97–109 doi 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan TSY, Wang X, Spence T, Taylor MD, Huang A Embryonal brain tumors. New York: Springer; 2015. [Google Scholar]
- 4.Northcott PA, Buchhalter I, Morrissy AS, Hovestadt V, Weischenfeldt J, Ehrenberger T, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547(7663):311–7 doi 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard D, Miller S, Hawkins CE, Bouffet E, Rogers HA, Chan TS, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol 2012;13(8):838–48 doi 10.1016/S1470-2045(12)70257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, et al. New Brain Tumor Entities Emerge from Molecular Classification of CNS-PNETs. Cell 2016;164(5):1060–72 doi 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet 1999;23(1):99–103 doi 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 8.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 2005;436(7048):272–6 doi 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 9.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 2005;436(7048):221–6 doi 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 10.Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res 2009;69(20):8150–6 doi 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119(6):847–60 doi 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Xiao A, Yin C, Yang C, Di Cristofano A, Pandolfi PP, Van Dyke T. Somatic induction of Pten loss in a preclinical astrocytoma model reveals major roles in disease progression and avenues for target discovery and validation. Cancer Res 2005;65(12):5172–80 doi 10.1158/0008-5472.CAN-04-3902. [DOI] [PubMed] [Google Scholar]
- 13.Rahrmann EP, Watson AL, Keng VW, Choi K, Moriarity BS, Beckmann DA, et al. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 2013;45(7):756–66 doi 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarver AL, Erdman J, Starr T, Largaespada DA, Silverstein KA. TAPDANCE: an automated tool to identify and annotate transposon insertion CISs and associations between CISs from next generation sequence data. BMC Bioinformatics 2012;13:154 doi 10.1186/1471-2105-13-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Temiz NA, Moriarity BS, Wolf NK, Riordan JD, Dupuy AJ, Largaespada DA, et al. RNA sequencing of Sleeping Beauty transposon-induced tumors detects transposon-RNA fusions in forward genetic cancer screens. Genome Res 2016;26(1):119–29 doi 10.1101/gr.188649.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott MC, Temiz NA, Sarver AE, LaRue RS, Rathe SK, Varshney J, et al. Comparative Transcriptome Analysis Quantifies Immune Cell Transcript Levels, Metastatic Progression, and Survival in Osteosarcoma. Cancer Res 2018;78(2):326–37 doi 10.1158/0008-5472.CAN-17-0576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rack PG, Ni J, Payumo AY, Nguyen V, Crapster JA, Hovestadt V, et al. Arhgap36-dependent activation of Gli transcription factors. Proc Natl Acad Sci U S A 2014;111(30):11061–6 doi 10.1073/pnas.1322362111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marko TA, Shamsan GA, Edwards EN, Hazelton PE, Rathe SK, Cornax I, et al. Slit-Robo GTPase-Activating Protein 2 as a metastasis suppressor in osteosarcoma. Sci Rep 2016;6:39059 doi 10.1038/srep39059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008;14(2):135–45 doi 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015;6:7391 doi 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012;488(7409):49–56 doi 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraus JA, Felsberg J, Tonn JC, Reifenberger G, Pietsch T. Molecular genetic analysis of the TP53, PTEN, CDKN2A, EGFR, CDK4 and MDM2 tumour-associated genes in supratentorial primitive neuroectodermal tumours and glioblastomas of childhood. Neuropathol Appl Neurobiol 2002;28(4):325–33. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature 2012;482(7386):529–33 doi 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 2011;29(11):1408–14 doi 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin MH, Lee YH, Cheng HL, Chen HY, Jhuang FH, Chueh PJ. Capsaicin Inhibits Multiple Bladder Cancer Cell Phenotypes by Inhibiting Tumor-Associated NADH Oxidase (tNOX) and Sirtuin1 (SIRT1). Molecules 2016;21(7) doi 10.3390/molecules21070849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartweieg EA, Cepko CL. Multipotent Neural Cell Lines Can Engraft and Participate in Development of Mouse Cerebellum. Cell 1992;68:33–51. [DOI] [PubMed] [Google Scholar]
- 27.Eccles RL, Czajkowski MT, Barth C, Muller PM, McShane E, Grunwald S, et al. Bimodal antagonism of PKA signalling by ARHGAP36. Nat Commun 2016;7:12963 doi 10.1038/ncomms12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holterman CE, Le Grand F, Kuang S, Seale P, Rudnicki MA. Megf10 regulates the progression of the satellite cell myogenic program. J Cell Biol 2007;179(5):911–22 doi 10.1083/jcb.200709083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Wang W, Xi Y, Gao M, Tran M, Aziz KE, et al. FOXR2 Interacts with MYC to Promote Its Transcriptional Activities and Tumorigenesis. Cell Rep 2016;16(2):487–97 doi 10.1016/j.celrep.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin JR, Mondal AM, Liu R, Hu J. Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinformatics 2012;13:157 doi 10.1186/1471-2105-13-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res 2018;46(D1):D754–D61 doi 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genovesi LA, Ng CG, Davis MJ, Remke M, Taylor MD, Adams DJ, et al. Sleeping Beauty mutagenesis in a mouse medulloblastoma model defines networks that discriminate between human molecular subgroups. Proc Natl Acad Sci U S A 2013;110(46):E4325–34 doi 10.1073/pnas.1318639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lastowska M, Al-Afghani H, Al-Balool HH, Sheth H, Mercer E, Coxhead JM, et al. Identification of a neuronal transcription factor network involved in medulloblastoma development. Acta Neuropathol Commun 2013;1:35 doi 10.1186/2051-5960-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koso H, Tsuhako A, Lyons E, Ward JM, Rust AG, Adams DJ, et al. Identification of FoxR2 as an oncogene in medulloblastoma. Cancer Res 2014;74(8):2351–61 doi 10.1158/0008-5472.CAN-13-1523. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz B, Fabius AW, Wu WH, Pedraza A, Brennan CW, Schultz N, et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proc Natl Acad Sci U S A 2014;111(22):8149–54 doi 10.1073/pnas.1401952111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dali R, Verginelli F, Pramatarova A, Sladek R, Stifani S. Characterization of a FOXG1:TLE1 transcriptional network in glioblastoma initiating cells. Mol Oncol 2018. doi 10.1002/1878-0261.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 2013;125(5):659–69 doi 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci 2015;72(11):2075–89 doi 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol 2015;17(1):20–30 doi 10.1038/ncb3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Modzelewska K, Boer EF, Mosbruger TL, Picard D, Anderson D, Miles RR, et al. MEK Inhibitors Reverse Growth of Embryonal Brain Tumors Derived from Oligoneural Precursor Cells. Cell Rep 2016;17(5):1255–64 doi 10.1016/j.celrep.2016.09.081. [DOI] [PubMed] [Google Scholar]
- 41.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett 2013;328(2):198–206 doi 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Liu N, Yue C, Wang D, Qi Z, Tu Y, et al. FoxR2 promotes glioma proliferation by suppression of the p27 pathway. Oncotarget 2017;8(34):56255–66 doi 10.18632/oncotarget.17447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X, He B, Gao Y, Li Y. FOXR2 contributes to cell proliferation and malignancy in human hepatocellular carcinoma. Tumour Biol 2016;37(8):10459–67 doi 10.1007/s13277-016-4923-3. [DOI] [PubMed] [Google Scholar]
- 44.Lu SQ, Qiu Y, Dai WJ, Zhang XY. FOXR2 Promotes the Proliferation, Invasion, and Epithelial-Mesenchymal Transition in Human Colorectal Cancer Cells. Oncol Res 2017;25(5):681–9 doi 10.3727/096504016X14771034190471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu W, Chang J, Liu G, Du X, Li X. Knockdown of FOXR2 suppresses the tumorigenesis, growth and metastasis of prostate cancer. Biomed Pharmacother 2017;87:471–5 doi 10.1016/j.biopha.2016.12.120. [DOI] [PubMed] [Google Scholar]
- 46.D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med 2001;7(2):235–9 doi 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 47.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer 2014;14(9):598–610 doi 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu B, Lefringhouse J, Liu Z, West D, Baldwin LA, Ou C, et al. Inhibition of the integrin/FAK signaling axis and c-Myc synergistically disrupts ovarian cancer malignancy. Oncogenesis 2017;6(1):e295 doi 10.1038/oncsis.2016.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong X, Wang C, Chen Z, Zhao W. Overcoming the resistance mechanisms of Smoothened inhibitors. Drug Discov Today 2018. doi 10.1016/j.drudis.2018.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.