Abstract
Purpose:
Lysine-specific demethylase 1 (LSD1) regulates several biological processes via the bifunctional modulation of enhancer functions. Recently, we reported that LSD1 overexpression is a founder abnormality of T-cell leukemogenesis and is maintained in fully transformed T-cell acute lymphoblastic leukemia (T-ALL) cells. On the basis of this finding, we attempted to develop novel LSD1 inhibitors effective for T-ALL with central nervous system (CNS) involvement.
Experimental design:
We chemically modified the prototype LSD inhibitor tranylcypromine (TCP) and screened for cytotoxicity against TCP-resistant T-ALL cell lines. In vivo efficacy of novel LSD1 inhibitors was examined in immunodeficient mice transplanted with luciferase-expressing T-ALL cell lines, which faithfully reproduce human T-ALL with CNS involvement.
Results:
We found robust cytotoxicity against T-ALL cells, but not normal bone marrow progenitors, for two N-alkylated TCP derivatives, S2116 and S2157. The two compounds induced apoptosis in TCP-resistant T-ALL cells in vitro and in vivo by repressing transcription of the NOTCH3 and TAL1 genes through increased H3K9 methylation and reciprocal H3K27 deacetylation at super-enhancer regions. Both S2116 and S2157 significantly retarded the growth of T-ALL cells in xenotransplanted mice and prolonged the survival of recipients as monotherapy and in combination with dexamethasone. Notably, S2157 could almost completely eradicate CNS leukemia because of its ability to efficiently pass through the blood-brain barrier.
Conclusion:
These findings provide a molecular basis and rationale for the inclusion of a brain-permeable LSD1 inhibitor, S2157, in treatment strategies for T-ALL with CNS involvement.
Keywords: epigenetics, CNS leukemia, T-ALL, LSD1, super-enhancer
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy originated from T-cell progenitors, which accounts for 15% of pediatric and 25% of adult ALL cases (1). Despite the application of intensive multi-agent chemotherapy with stem cell support, the treatment outcome of T-ALL is worse than that of other leukemias including acute myeloid leukemia (AML) and B-cell ALL (B-ALL), mainly due to a higher prevalence of central nervous system (CNS) involvement (2, 3). Because of the low penetrance of conventional anti-leukemic agents, CNS provides a sanctuary for leukemic cells and thus is one of the most frequent sites of relapse in T-ALL patients (4, 5). Current treatments for CNS leukemias, such as intrathecal injection of anti-cancer drugs, cranial irradiation, and high-dose systemic chemotherapy, are not only curative but also associated with acute and/or long-term serious neurotoxicity (6). Given the limitation of current strategies, novel agents that can effectively cross the blood-brain barrier to target CNS lesions are in high demand.
Lysine-specific demethylase 1 (LSD1, also called KDM1A/AOF2/BHC110) is a flavin adenine dinucleotide (FAD)-dependent histone demethylase that removes the mono- and di-methyl groups from lysine-4 and lysine-9 residues of histone H3 (hereinafter H3K4 and H3K9, respectively) (7). LSD1 bifunctionally modulates the enhancer/promoter functions of target genes in a context-dependent manner: a repressive function via H3K4 demethylation as a component of the CoREST and NuRD complexes (8) and an activator function via H3K9 demethylation when associated with steroid hormone receptors (9). Hence, LSD1 plays indispensable roles in several biological processes including ontogenesis, hematopoiesis, and oncogenesis.
Using a genetic approach, we have demonstrated that LSD1 hyperactivation is a founder abnormality for the development of T-ALL (10). LSD1 overexpression primes hematopoietic stem cells for T-cell leukemogenesis and is maintained in fully transformed T-ALL cells. These findings encourage the use of small molecular compounds that inhibit LSD1 activity for the treatment of T-ALL; however, most LSD1 inhibitors, including the prototype compound tranylcypromine (trans-2-phenylcyclopropylamine; TCP) and its derivative S2101 (11), did not show robust efficacy against leukemias of T-cell origin in several preclinical studies (12-14). We therefore modified S2101 by N-alkylation to develop novel compounds S2116 and S2157. The two compounds induced apoptosis in T-ALL cell lines with significantly lower IC50 values than S2101 and other TCP derivatives. Furthermore, S2157 could eradicate CNS leukemia in mice transplanted with T-ALL cells due to its ability to efficiently pass through the blood-brain barrier. These LSD1 inhibitors may improve the treatment outcome of T-ALL, especially when complicated with CNS disease, by combining with current treatment strategies.
Materials and Methods
Cells and cell culture
We used five bona fide human T-ALL cell lines, CEM, Jurkat, MOLT4, Loucy and PEER, in this study (Health Science Research Resources Bank, Osaka, Japan). Other cell lines and their origins are HEL, MV4-11, K562, KCL22 (AML), KMS12-BM, KMS28 (multiple myeloma) and PALL2 (B-ALL). Primary T-ALL cells were isolated from the peripheral blood of patients at the time of diagnostic procedure and used when blasts were >90% of mononuclear cells. Normal human bone marrow progenitor cells were purchased from Takara Bio. (Shiga, Japan) and cultured in the presence of stem cell factor and thrombopoietin (10). We obtained written informed consent from all patients in accordance with the Declaration of Helsinki. The protocol was approved by the Institutional Review Boards of Jichi Medical University and University of Yamanashi.
Drugs
LSD1 inhibitors used in this study include RN-1 (Calbiochem, San Diego, CA), ORY-1001 (Cayman Chemical, Ann Arbor, MI), OG-L002 (Selleck Chemicals, Houston, TX), S2101 (Millipore, Temecula, CA) and two N-alkylated derivatives of S2101, S2116 and S2157 (synthesized by Tokyo Chemical Industry, Tokyo, Japan) (15). A pharmacokinetic study was conducted with a single intraperitoneal injection of either S2116 or S2157 at 50 mg/kg to nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Charles River Laboratories, Wilmington, MA), followed by serial sampling of blood at 1, 2, 4, 8 and 24 hours after administration. Brain tissues were harvested at 0.5 hour and homogenized in deionized water. Plasma and brain samples were subjected to LC-MS/MS analysis to determine the concentrations of each drug.
Semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR) and real-time quantitative RT-PCR
Total cellular RNA was isolated using an RNeasy Kit (Qiagen, Valencia, CA), reverse-transcribed into complementary DNA using ReverTra Ace and oligo(dT) primers (Toyobo, Tokyo, Japan), and subjected to subsequent RT-PCR analysis for gene expression (16). Detailed information on semi-quantitative RT-PCR primers, including sequences, corresponding nucleotide positions and PCR product sizes, is described in Supplementary Table S1. We used the Expression Assays (Hs01097987 for TAL1, Hs01128537 for NOTCH3, Hs00207691 for ZEB2, and Hs01922876 for GAPDH) and TaqMan Fast Universal PCR Master Mix (Applied Biosystems, Waltham, MA) for real-time quantitative RT-PCR (RQ-PCR).
Global analysis of gene expression
MOLT4 cells were cultured with either vehicle alone (0.1% DMSO) or 12 µM S2157 for 24 hours. RNA samples (50 ng) were T7 transcribed in the presence of Cy3- or Cy-5 CTP, and hybridized with the SurePrint G3 Human Gene Expression 8 × 60K v2 Microarray (Agilent, Santa Clara, CA). The hybridized arrays were scanned with an Agilent G2565AA scanner, and the obtained images were processed using the Agilent Feature Extraction Software (version 10.7.3.1). Microarray data have been deposited in the MIAME-compliant GEO database under accession number GSE85956.
Chromatin immunoprecipitation assays
We used the ChIP-IT Express Enzymatic Kit (Active Motif, Carlsbad, CA) to perform chromatin immunoprecipitation (ChIP) assays (17). In brief, we fixed cells in 1% formaldehyde at room temperature for 10 minutes and isolated chromatin fractions using enzymatic shearing. After centrifugation, supernatants were incubated with antibodies of interest or isotype-matched control and protein G magnetic beads at 4 °C overnight. We purified DNA fragments from the mixture according to the manufacturer’s instructions and carried out PCR using Mighty Amp (Takara Bio.) or Power SYBR Green Master Mix (Life Technologies, Carlsbad, CA) and the primers listed in Supplementary Table S2. We outsourced ChIP-seq analyses to Active Motif and deposited the data along with a protocol in the MINSEQE-compliant GEO database under accession number GSE120314.
Establishment of T-ALL sublines
Lentiviral vectors encoding full-length NOTCH3 and TAL1 cDNAs were purchased from Addgene (Cambridge, MA) and delivered into 293FT cells with packaging plasmids for viral production. T-ALL cells were transduced with infective lentiviruses for 24 hours as previously described for luciferase-expressing sublines (18). We established stable transformants by isolating single-cell clones using limiting dilution.
Reproduction of T-ALL in mice with xenotransplantation
Luciferase-expressing T-ALL cell lines (5 × 106 cells for MOLT4 and 1 × 107 cells for Jurkat in 200 µL of Iscove’s Modified Dulbecco’s Medium) were injected via a tail vein into NOD/SCID mice. Tumor progression was monitored by measuring luciferase activity ex vivo using the noninvasive bioimaging system (Xenogen, Alameda, CA) (15-18). All animal studies were approved by the Institutional Animal Ethics Committee and performed in accordance with the Guide for the Care and Use of Laboratory Animals formulated by the National Academy of Sciences.
Other conventional techniques are described in Supplementary Materials and Methods.
Results
N-alkylated LSD1 inhibitors induce apoptotic cell death in T-ALL cells at clinically relevant concentrations in vitro
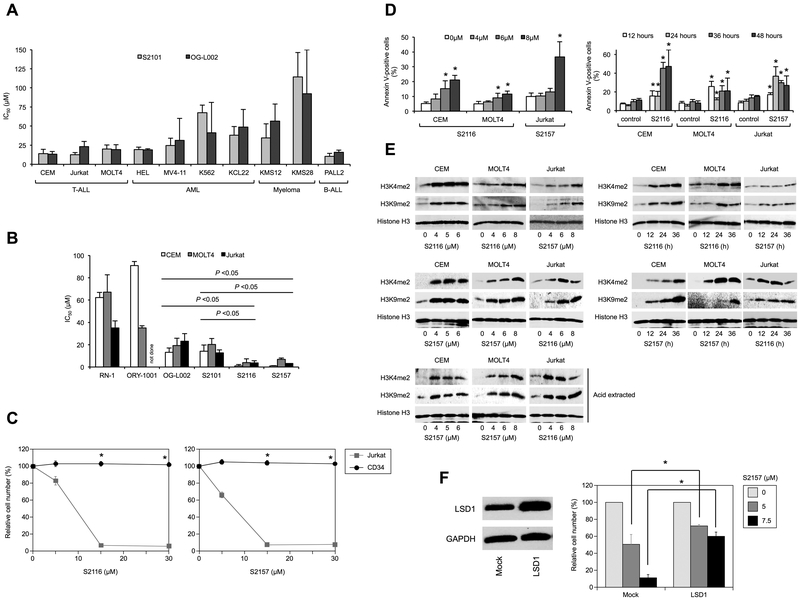
Given the fundamental role of LSD1 hyperactivity in T-cell leukemogenesis and maintenance of tumor phenotypes (10), we investigated whether T-ALL cells are vulnerable to LSD1 inhibition using conventional LSD1 inhibitors S2101 and OG-L002. As anticipated, T-ALL cell lines were more sensitive to both drugs than other malignant hematopoietic cells except for the B-ALL cell line PALL2, which was equally sensitive to LSD1 inhibitors (Figure 1A and Supplementary Figure S1). Although S2101 and OG-L002 appeared to be cytotoxic against T-ALL, the IC50 values were not clinically relevant (≥20 µM); therefore, we tested several N-alkylated derivatives of S2101 with lower Ki values and higher specificity for LSD1 than other FAD-dependent amine oxidases such as LSD2, monoamine oxidase A (MAO-A), and MAO-B (Niwa et al., manuscript in preparation and ref. 15). Among them, S2116 and S2157 were particularly effective for T-ALL cell lines with the IC50 values between 1.1 ± 0.2 µM for CEM and 6.8 ± 1.3 µM for MOLT4 (Figure 1B). The two compounds did not affect the viability of CD34-positive normal hematopoietic progenitor cells at all (Figure 1C) and modestly inhibited that of mitogen-activated normal T-lymphocytes (Supplementary Figure S2). The N-alkylated LSD1 inhibitors induced apoptosis, as evidenced by increased annexin-V reactivity on flow cytometry (Figure 1D and Supplementary Figure S3), in T-ALL cells in a dose- and time-dependent manner without affecting cell cycle distribution (Supplementary Figure S4). These effects were seemingly mediated through LSD1 inhibition because the methylation levels of H3K4 and H3K9 increased with kinetics similar to the induction of apoptosis by S2116 and S2157 (Figure 1E). Furthermore, LSD1 overexpression significantly mitigated the cytotoxicity of the inhibitor in T-ALL cells, providing an additional proof of the on-target effect (Figure 1F). However, the expression level of LSD1 was not a determinant of the sensitivity to LSD1 inhibitors (Supplementary Figure S5).
Figure 1. N-alkylated LSD1 inhibitors induce apoptotic cell death in T-ALL cells at clinically relevant concentrations in vitro.
(A) The IC50 values of S2101 (gray column) and OG-L002 (black column) were calculated from dose-response curves of the indicated cell lines obtained at 72 hours of culture. The mean ± S.D. (bars) of three independent experiments. The results of statistical analyses are shown in Supplementary Figure S1. (B) The IC50 values of the indicated LSD1 inhibitors were calculated from dose-response curves of T-ALL cell lines obtained at 72 hours of culture. The means ± S.D. (bars) of three independent experiments. P values were calculated by one-way ANOVA with Tukey’s multiple comparison test. (C) We cultured CD34-positive bone marrow progenitor cells from healthy volunteers with stem cell factor and thrombopoietin in the absence or presence of various concentrations of S2116 (left panel) or S2157 (right panel) for 72 hours. Jurkat cells were simultaneously treated to serve as a control. Cell viability was determined by the MTT reduction assay and expressed as a percentage of untreated controls. Each point represents the mean ± S.D. (bars) of three independent experiments. *P <0.05 by one-way ANOVA with the Bonferroni post-hoc test. (D) Left panel: T-ALL cells were cultured with various concentrations of S2116 or S2157 for 24 hours. Right panel: CEM and MOLT4 cells were cultured in the absence (control) or presence of 6 µM and 8 µM S2116, respectively, and Jurkat cells were cultured in the absence (control) or presence of 6 µM S2157 for the indicated periods. Cells were stained with annexin-V for the assessment of apoptosis by flow cytometry. The means ± S.D. (bars) of three independent experiments. *P <0.05 against untreated control by one-way ANOVA with Tukey’s multiple comparison test. See Supplementary Figure S3 for other combinations. (E) Whole cell lysates were prepared during the experiments described in panel D and Supplementary Figure S3, and subjected to immunoblotting for the indicated molecules. We performed the same set of experiments using acid-extracted histone proteins for comparison (Acid extracted). (F) Left panel: MOLT4 cells were transduced with either an empty lentiviral vector (Mock) or LSD1-overexpressing vector (LSD1) to establish stable transformants. The expression level of LSD1 was determined by immunoblotting. Right panel: We cultured each transformant with S2157 at the indicated concentrations for 72 hours. Cell viability was determined by the MTT reduction assay and expressed as a percentage of the values of untreated cells. The means ± S.D. (bars) of three independent experiments are shown. *P <0.05 by paired Student’s t test.
LSD1 inhibitors modify the gene expression program in favor of cell death in T-ALL cells
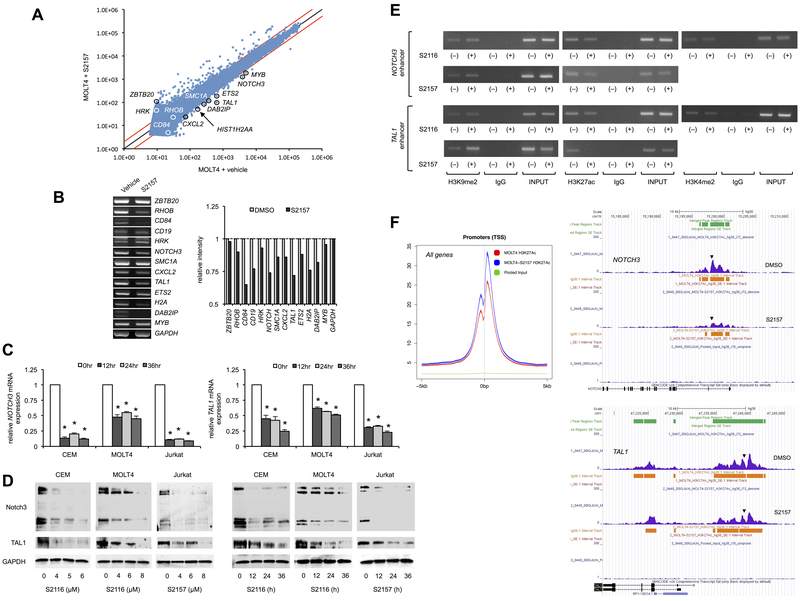
Next, we investigated the molecular mechanisms by which the N-alkylated LSD1 inhibitors cause T-ALL cell death. Toward this end, we initially performed a global screening for LSD1 inhibition-mediated changes in the gene expression program of T-ALL cells. As illustrated in Figure 2A, S2157 induced greater than 2-fold increase and decrease in the abundance of 909 and 148 genes, respectively, among 32,078 genes on the array in MOLT4 cells (P <0.05 with an FDR threshold of 0.05). No significant change was detected in the expression levels of NOTCH1, constitutive activation of which is the most frequent abnormality in T-ALL (18, 19), and its downstream effectors such as HES1, CYLD, GATA3 and RUNX3 (Supplementary Figure S6 and the data deposited in GEO #GSE85956), in line with our previous finding that LSD1 and Notch1 act redundantly with NOTCH1 mutations as a later event during T-cell leukemogenesis (10). The latter notion was also verified in human T-ALL by single-cell clonal analysis (20). Instead, the down-regulated genes include NOTCH3 and TAL1, both of which are known to be oncogenic drivers confounding with NOTCH1 in T-ALL cells (21-24). The decrease in NOTCH3 and TAL1 expression was confirmed in other combinations of LSD1 inhibitors and T-ALL cell lines by semiquantitative RT-PCR (Figure 2B), RQ-PCR (Figure 2C and Supplementary Figure S7) and immunoblotting (Figure 2D and Supplementary Figure S8). Furthermore, the expression level of Notch3 and TAL1, but not Notch1, was correlated with the cellular sensitivity to S2116 and S2157 (Supplementary Figures S5 and S9). Regarding the mechanisms of NOTCH3 and TAL1 down-regulation, we found that both S2116 and S2157 readily increased the methylation level of H3K9 and reciprocally reduced the acetylation level of H3K27 at super-enhancer regions of the NOTCH3 and TAL1 genes (GRCh38/hg38: 15,198,031-15,197,862 and GRCh38/hg38: 47,239,435-47,239,119, respectively) (25, 26) using ChIP assays (Figure 2E and Supplementary Figure S10 for data quantification) and ChIP with quantitative PCR (Supplementary Figure S11). In addition, global ChIP-seq analyses revealed that the acetylation level of H3K27 was readily decreased by LSD1 inhibition through the entire range of NOTCH3 and TAL1 enhancers (Figure 2F, right panel). However, S2157 rather increased H3K27 acetylation at the whole-genome level (Figure 2F, left panel, and Supplementary Figure S12), in agreement with the microarray data that up-regulated genes were 6-fold higher in number than down-regulated genes (Figure 2A). These results suggest that the N-alkylated LSD1 inhibitors selectively cease hyperactive oncogenic enhancers, leading to apoptosis of addicted cells.
Figure 2. LSD1 inhibitors modify the gene expression program in favor of cell death in T-ALL cells.
(A) MOLT4 cells were cultured with either vehicle alone (0.1% DMSO) or 12 µM S2157 for 24 hours. RNA samples were subjected to microarray analysis for 32,078 genes including microRNAs. Red lines correspond to 2-fold changes (P <0.05 with an FDR threshold of 0.05). Some genes related to T-ALL biology are annotated. (B) Left panel: The expression of the annotated genes in panel A was examined by semiquantitative RT-PCR with suboptimal amplification cycles (40 cycles) in S2157-treated MOLT4 cells (see Supplementary Table S1 for primer sequences). Right panel: The signal intensities of bands in the left panel were quantified and are shown as relative values to those of GAPDH. (C) CEM and MOLT4 cells were cultured with 6 µM S2116, and Jurkat cells were cultured with 8 µM S2157 for the indicated periods. The expression level of NOTCH3 (left panel) and TAL1 (right panel) was determined by RQ-PCR, normalized to that of GAPDH, and quantified by the 2−∆∆Ct method with the T-0 values set at 1.0. The means ± S.D. (bars) of three independent experiments. *P <0.05 vs. T-0 control by one-way ANOVA with Tukey’s multiple comparison test. See Supplementary Figure S7 for other combinations. (D) Left panel: T-ALL cells were cultured with various concentrations of S2116 or S2157 for 24 hours. Right panel: CEM and MOLT4 cells were cultured with 6 µM S2116, and Jurkat cells were cultured with 8 µM S2157 for the indicated periods. Whole cell lysates were subjected to immunoblotting for Notch3, TAL1 and GAPDH expression. See Supplementary Figure S8 for other combinations. (E) Using ChIP assays, we analyzed the status of chromatin modifications at super-enhancer regions of the NOTCH3 and TAL1 genes in MOLT4 cells treated with either S2116 or S2157 at 12 µM for 24 hours. Representative data of 40 cycles are shown. Input indicates that PCR was performed with genomic DNA. (F) Purified chromatin was isolated from MOLT4 cells treated with either 0.1% DMSO (vehicle) or 12 µM S2157 for 24 hours, followed by immunoprecipitation with anti-histone H3K27ac antibody. Genomic DNA fragments were purified from the precipitants and subjected to high-throughput sequencing. Left panel: The status of H3K27 acetylation surrounding transcription-start sites (TSS) in the entire genome. Red; vehicle-treated control, Blue; S2157-treated MOLT4 cells, Green; Pooled input. Right panel: ChIP seq tracks of normalized tags showing enrichment of histone H3K27 acetylation (purple) in the enhancer regions of the NOTCH3 (upper panel) and TAL1 (lower panel) genes visualized on the UCSC genome browser. Arrowheads indicate the regions examined in ChIP assays (panel E) and ChIP-qPCR (Supplementary Figure S11).
Notch3 and TAL1 are bonafide targets of the N-alkylated LSD1 inhibitors in T-ALL
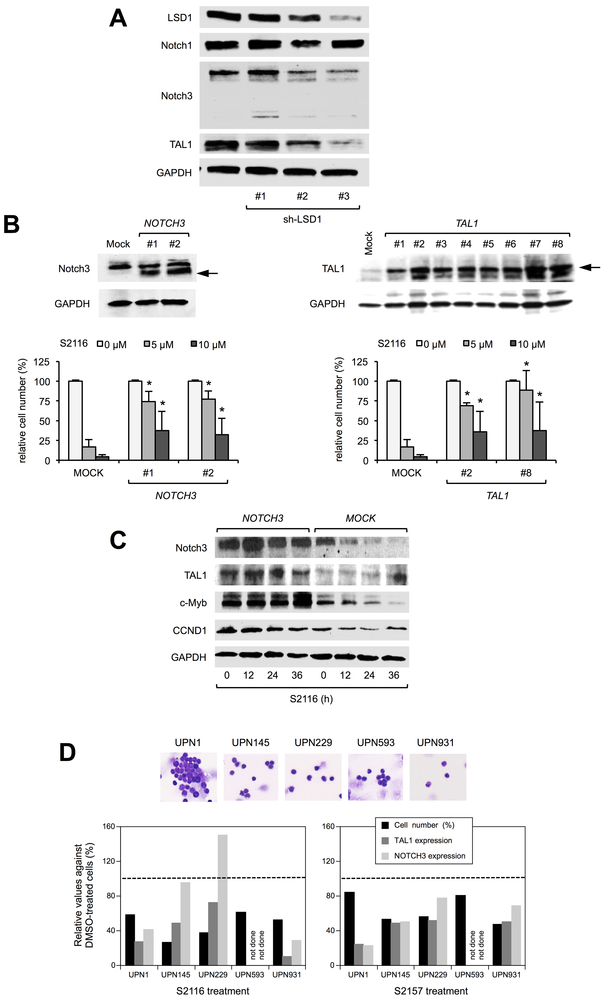
To verify that Notch3 and TAL1 are targets of LSD1 inhibitors, we performed shRNA-mediated knockdown and found that the expression levels of Notch3 and TAL1, but not Notch1, declined in correlation with the decreased abundance of LSD1 in T-ALL cells (Figure 3A). In addition, we established Notch3- and TAL1-overexpressing MOLT4 sublines (Figure 3B, upper panels). As anticipated, forced expression of either Notch3 or TAL1 imparted the resistance to S2116 and S2157 in T-ALL cells (Figure 3B, lower panels). However, dual expression of Notch3 and TAL1 could not completely rescue T-ALL cells from the effect of LSD1 inhibitors (Supplementary Figure S13), because TAL1 expression was not further up-regulated in double-transformants, probably due to redundancy of the two oncogenes (21). In fact, Notch3 overexpression increased the abundance of several interrelated and/or downstream molecules such as TAL1, c-Myb and Cyclin D1 (Figure 3C). Upon overexpression, c-Myb and Cyclin D1 deregulate cellular homeostasis and may act as proapoptotic factors (27, 28), providing an explanation of the failure of complete rescue from the drug effects in Notch3/TAL1 double-transfected cells. In patient-derived primary T-ALL cells, the reduction in viable cell numbers was mostly associated with the decreased expression of TAL1 and NOTCH3 under treatment with S2116 and S2157 (Figure 3D). These results imply that the cytotoxic effect of N-alkylated LSD1 inhibitors is at least partly mediated through the down-regulation of Notch3 and TAL1 in T-ALL cells, but the involvement of other targets cannot be ruled out.
Figure 3. Notch3 and TAL1 are bona fide targets of N-alkylated LSD1 inhibitors.
(A) MOLT4 cells were transduced with lentiviral vectors encoding shRNA against LSD1 (sh-LSD1 #1, #2 and #3) or an ineffective control (sh-Control). Whole cell lysates were prepared after 72 hours to examine the expression of the indicated molecules with immunoblotting. (B) Upper panel: Immunoblot analyses of Notch3 and TAL1 in lentivirus-transduced stable MOLT4 transformants. Lower panel: We cultured selected clones with S2116 at the indicated concentrations for 72 hours. Cell viability was determined by the MTT reduction assay and expressed as a percentage of the values for corresponding untreated cells. The means ± S.D. (bars) of three independent experiments are shown. *P <0.05 against Mock-transfected cells by paired Student’s t test. (C) We cultured a Mock-transformant and the NOTCH3-stable transformant #2 with 8 µM S2116 for the indicated periods. Whole cell lysates were subjected to immunoblot analyses for the expression of some T-cell oncogene products. (D) Upper panel: Wright-Giemsa staining of primary leukemic cells (> 90% purity) from T-ALL patients. Lower panel: Cells were cultured with vehicle (0.1% DMSO), 1 µM S2116 (left panel) or 1 µM S2157 (right panel) for 48 hours, and subjected to the MTT reduction assay for relative cell numbers (%Control) and RQ-PCR for TAL1 and NOTCH3 mRNA expression. Broken lines indicate the values for corresponding DMSO-treated cells.
The N-alkylated LSD1 inhibitors induce apoptotic cell death in T-ALL cells in vivo
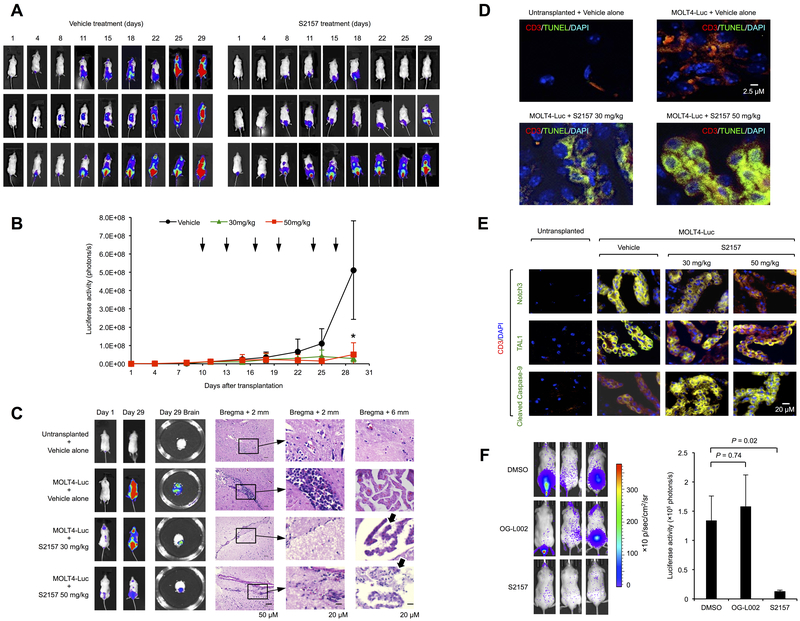
Having demonstrated the robust cytotoxicity against T-ALL cells in vitro, we sought to investigate therapeutic effects of the N-alkylated LSD1 inhibitors in vivo using xenograft systems. First, we carried out a pilot experiment, in which anti-leukemic activity and safety were tested in NOD/SCID mice xenografted with T-ALL cells (18). The maximum tolerated dose (MTD) was not determined for S2116 and S2157 because no obvious changes were observed in complete blood count, liver and kidney functions, or a histopathological examination of some organs in NOD/SCID mice treated with up to 150 mg/kg of the drugs (Supplementary Figure S14), whereas MTD was defined to be 12.5 mg/kg for OG-L002 as recipients died immediately after intraperitoneal injection of the drug ≥25 mg/kg for unknown causes. The size of subcutaneous tumors reduced to less than 20% of that in the untreated control by intraperitoneal injection of either S2116 or S2157 at 50 mg/kg for 3 times a week (Supplementary Figure S15A) with negligible side effects, such as moderate leukocytosis and thrombocytopenia without bleeding tendency (Supplementary Figure S16). In addition, S2116 significantly prolonged the overall survival of MOLT4-inoculated mice (Supplementary Figure S15B). Encouraged by these results, we examined therapeutic effects of LSD1 inhibitors on systemic T-ALL disease generated by tail-vein injection of luciferase-expressing MOLT4 cells into NOD/SCID mice. Injected MOLT4 cells were widely distributed within sacral and vertebral bone marrow (7 days after transplantation), followed by CNS involvement (10 days after transplantation) that faithfully reproduces human leukemic meningitis with subsequent parenchymal invasion (Figure 4A and Supplementary Figure S17). Because of a high incidence of CNS involvement, one of the most important requirements of novel therapeutic agents for T-ALL is the ability to pass through the blood-brain barrier. A pharmacokinetic study revealed that S2157 had approximately 50-fold higher maximal brain concentrations and a >75-fold higher brain/plasma concentration ratio than S2116 after a single intraperitoneal injection at 50mg/kg in NOD/SCID (Table 1) and ICR mice (Supplementary Table S3). Based on the pharmacokinetic data, S2157 was chosen for the treatment of mice harboring systemic T-ALL disease. As shown in Figure 4A/B, S2157 almost completely suppressed the growth of MOLT4 cells in most but not all mice by twice-per-week injection at either 30 mg/kg or 50 mg/kg. Histopathological examination revealed that CNS leukemia nearly disappeared with some residual cells undergoing apoptosis as demonstrated by the presence of apoptotic bodies (Figure 4C), DNA fragmentation (Figure 4D), and caspase-9 cleavage (Figure 4E) in human CD3-expressing cells in the brain of recipient mice. It is noted that Notch3 and TAL1 were readily down-regulated in MOLT4 cells in S2157-treated mice, suggesting that the two molecules are targets of LSD1 inhibition also in vivo (Figure 4E). In addition, we carried out the same experiment using a luciferase-expressing Jurkat subline and verified a therapeutic benefit of S2157 including the eradication of CNS disease (Figure 4F). In contrast, another LSD1 inhibitor, OG-L002, failed to show anti-leukemic activity at its MTD in this system.
Figure 4. The N-alkylated LSD1 inhibitor S2157 eradicates CNS leukemia in murine xenotransplanted models.
(A) We injected 5 × 106 luciferase-expressing MOLT4 cells into NOD/SCID mice via a tail vein and started treatments at day 10 after transplantation with either vehicle alone (0.1% DMSO) or 50 mg/kg S2157 twice a week for 3 weeks (vertical arrows in panel B). Tumor-derived luciferase activity was measured ex vivo by the IVIS Imaging System after D-luciferin injection on the indicated days. (B) Quantitative data of in vivo bioluminescence imaging as photon units (photons/s). *P <0.05 vs. the vehicle-treated group determined by one-way ANOVA with Tukey’s multiple comparison test (n=3). (C) Brains were resected from each mice on day 29 and subjected to a whole-mount luciferase assay (Day 29 Brain) and histopathological examination. Representative hematoxylin-eosin staining of bregma + 2 mm and + 6 mm sections is shown. Arrows indicate apoptotic bodies in the bregma + 6 mm panel. (D) Day 29 brain was subjected to an in situ TUNEL assay to detect apoptoic cells (green). Transplanted MOLT4 cells were identified by human CD3 expression (red) and DAPI staining (blue). (E) Immunohistochemical staining of brain tissues with specific antibodies recognizing Notch3, TAL1 and cleaved caspase-9 (green). Transplanted MOLT4 cells were identified by human CD3 expression (red) and DAPI staining (blue). Only merged images are shown. Data shown are representative of multiple independent samples collected on day 29. (F) We injected 1 × 107 luciferase-expressing Jurkat cells into NOD/SCID mice via a tail vein and started treatments at day 20 after transplantation with vehicle alone (0.1% DMSO), 12.5 mg/kg OG-L002, or 50 mg/kg S2157 twice a week for 3 weeks. Left panel: Representative photographs of mice on day 56. Right panel: Quantitative data of tumor-derived luciferase activity on day 56 (photons/s). The means ± S.D. (bars) of three independent experiments are shown. P values were calculated by a paired Student’s t test.
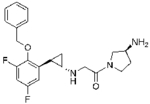
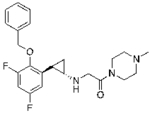
Table 1.
Pharmacokinetic parameters of the developed LSD1 inhibitors*
| S2116 | S2157 | |
|---|---|---|
 |
 |
|
| (±)-trans | (±)-trans | |
| Cmax (μM) | 3.77 ± 0.56 | 4.33 ± 3.07 |
| T1/2 (h) | 10.88 ± 2.97 | 0.88 ± 0.04 |
| AUC (μM•h) | 41.78 ± 8.66 | 5.75 ± 3.67 |
| nAUC (μM•h)** | 0.84 ± 0.17 | 0.11 ± 0.07 |
| Plasma concentration at 0.5 h (μM) | 8.17 ± 2.80 | 5.71 ± 2.47 |
| Brain concentration at 0.5 h (μmol/kg) | 1.35 ± 0.39 | 64.0 ± 3.46 |
| Brain/Plasma ratio | 0.17 ± 0.03 | 13.0 ± 6.26 |
Pharmacokinetic parameters were determined in NOD.CB17-PrkdcSCID/J mice treated with a single intraperitoneal injection of either S2116 or S2157 in 5 mL of 6.25% DMSO-saline at 50 mg/kg (n = 3).
Dose-normalized AUC
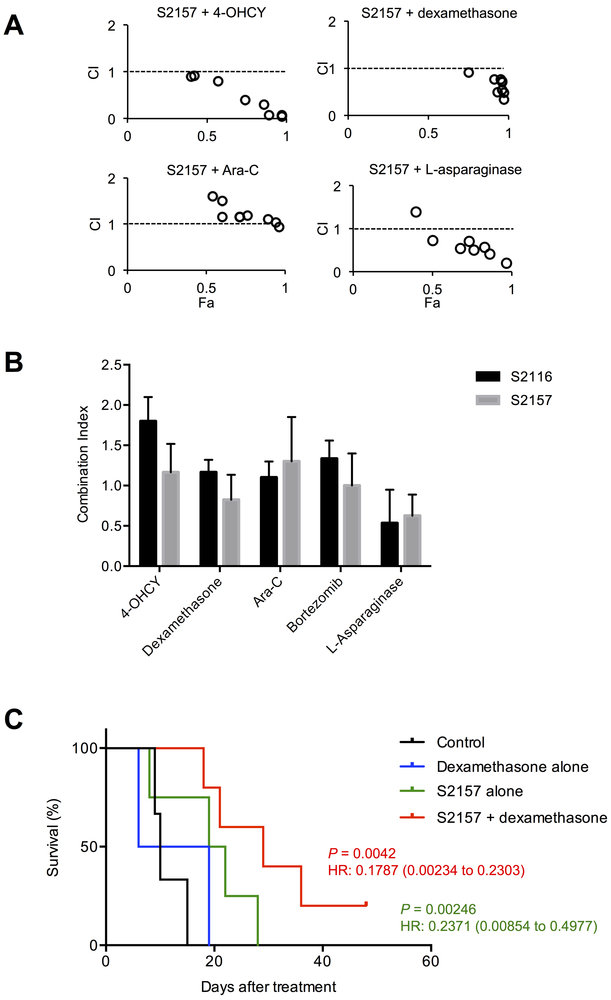
LSD1 inhibitors exhibit synergistic effects with conventional anti-leukemic agents in vitro and in murine xenografts
Recent clinical trials clearly demonstrated that the addition of molecular target agents to standard regimens conferred significant therapeutic benefits to patients with intractable malignancies such as Philadelphia chromosome-positive B-ALL (29) and AML with FLT3 mutations (30). The developed LSD1 inhibitors may be combined with conventional anti-leukemic drugs to increase the therapeutic index in T-ALL patients. To validate this assumption, we investigated whether S2116 and S2157 exerted synergistic effects with four drugs commonly used for the initial treatment of T-ALL and bortezomib, whose activity against T-ALL was proved in preclinical studies (18, 31) and in some clinical trials (32). An isobologram analysis revealed that S2157 was synergistic with dexamethasone and L-asparaginase as well as additive with 4-hydroxycyclophosphamide (4-OHCY) and bortezomib (Figure 5A/B), whereas S2116 was only synergistic with L-asparaginase and made additive combinations with dexamethasone and cytosine arabinoside (Figure 5B and Supplementary Figure S18). The synergy between S2157 and either dexamethasone or L-asparaginase was also observed in primary T-ALL cells (Supplementary Figure S19). The superb combination of LSD1 inhibitors and dexamethasone is at least partly explained by our previous finding that dexamethasone down-regulates the expression of mutant Notch1, which is unaffected by LSD1 inhibitors, at transcriptional levels in T-ALL cells (18). From these results, we decided to test the combined effects of S2157 and dexamethasone in a murine xenograft model, because L-asparaginase has a narrow therapeutic window and is too toxic for NOD/SCID mice (33). As shown in Figure 5C, single-agent chemotherapy with S2157 but not dexamethasone significantly prolonged the survival of T-ALL transplanted mice (P = 0.0246 with a hazard ratio of 0.2371 against untreated mice by the log-rank test), but the effect of combination was more striking with a hazard ratio of 0.1787 against untreated control. These observations confirm the results of in vitro experiments and further suggest that combination therapies containing a brain-permeable LSD1 inhibitor may be effective for T-ALL in a clinical setting, especially when complicated with CNS disease.
Figure 5. The LSD1 inhibitors exhibit synergistic effects with conventional anti-leukemic agents in vitro and in murine xenotransplanted models.
(A) MOLT4 cells were cultured with four anti-leukemia drugs in the absence or presence of S2157 for 72 hours to obtain dose-response curves for each combination. The combination index plots were generated by the CompuSyn software according to the method of Chou and Talalay. A combination index (CI) <1.0 indicates synergism of the two drugs. (B) The means ± S.D. (bars) of CI between the indicated drugs and either S2116 or S2157 from three independent experiments are shown. (C) We injected 5 × 106 luciferase-expressing MOLT4 cells into NOD/SCID mice via a tail vein and randomized them into four treatment groups at day 10 after transplantation: the vehicle alone (0.1% DMSO, intraperitoneally, 5 times a week; n=3), dexamethasone alone (10 mg/kg, intraperitoneally, 5 times a week; n=3), S2157 alone (50 mg/kg, intraperitoneally, twice a week; n=4) and the combination of S2157 and dexamethasone (n=5). Kaplan-Meier survival curves were generated and statistically analyzed by the log-rank test using the Prism software (GraphPad Software, La Jolla, CA). P-values and hazard ratios (95% confidential interval) between the vehicle-treated group (Control) and the S2157-alone (green) or combined-treatment group (red) are shown.
Discussion
Several lines of evidence indicate that LSD1 is overexpressed and hyperactivated in various malignancies, including T-ALL in which LSD1 plays an initiating role and is indispensable for the maintenance of tumor phenotypes (10). Hence, specific inhibitors for LSD1 hold a promise as a novel type of anti-cancer agents (34, 35). In fact, several LSD1 inhibitors were developed and their anti-cancer efficacy has been investigated in preclinical studies. For example, LSD1 binds to leukemic fusion proteins, such as MLL-AF9 and PML-PARα, and acts as a direct effector to sustain the leukemic stem cell potential and prevent normal differentiation (36, 37). A prototype LSD1 inhibitor, tranylcypromine (TCP), could remodel the oncogenic program to induce differentiation and cell death in AML carrying leukemic fusion proteins (37, 38). However, TCP may not be used for cancer treatment, because it inhibits other FAD-dependent enzymes, such as MAO-A and MAO-B, more potently due to covalent binding to FAD, raising the concern of neuronal toxicity especially when applied at high doses in cancer patients. To circumvent this issue, a series of TCP derivatives with increased specificity to LSD1 have been developed, including S2101 (11), OG-L002 (39), RN-1 (12), ORY-1001 (40), GSK2879552 (14, 41), IMG-7289 (34), NCD38 (42), and T-3775440 (13, 43, 44). Among them, phase I/II clinical trials of ORY-1001, GSK2879552 and IMG-7289 are ongoing for patients with AML and small cell lung cancer (34). It is of note that S2101 could specifically target glioblastoma stem cells, albeit at preclinical levels (45), consistent with the initiating role of LSD1 in oncogenesis.
Despite the fundamental role of LSD-1 in T-cell leukemogenesis, the first-generation TCP derivatives are not effective for T-ALL in contrast with their robust activity against AML. It has been reported that the IC50 values of RN-1, T-3775440 and NCD38 were >10 µM, whereas all three agents induced myeloid differentiation of AML and preleukemic MDS cells by disrupting the leukemogenic LSD1-GFI1B complex at <1 µM (12-14, 42). A recent elegant study by Goossens et al. provides a mechanistic insight into the resistance of T-ALL cells to LSD1 inhibition (14). One of the TCP derivatives, GSK2879552, failed to show pro-apoptotic activity in murine genetically engineered T-ALL and human T-ALL cell lines such as MOLT4, Jurkat, HPB-ALL and KARPAS45. They identified that ZEB2, a homeobox transcription factor (46), forms an oncogenic complex with LSD1 in early T-cell progenitor ALL (ETP-ALL) and determines the sensitivity to LSD1 inhibitors. GSK2879552-resistant T-ALL cells lack the expression of ZEB2 and restore the drug response upon the reinstitution of ZEB2. In striking contrast, S2116 and S2157 were effective for T-ALL cells lacking ZEB2, but relatively ineffective for ZEB2-positive ETP-ALL cell lines such as Loucy and PEER (Supplementary Figure S20). Moreover, the absence of Notch3 and TAL1 overexpression may account for the insensitivity of ETP-ALL cells to S2116 and S2157 (24-26) (Supplementary Figure S21). Nonetheless, further investigations, especially crystallographic studies, are required to elucidate the mechanisms by which S2116 and S2157 are only effective for ZEB2-deficient T-ALL cells.
Super-enhancers were originally defined as the regulatory region highly enriched for the binding of the Mediator complex and transcriptional activators through unique histone modifications, providing a platform for cell-specific transcriptional outputs that drive identity in particular cell types, especially pluripotent stem cells and cancer cells (25, 26, 47, 48). In a subset of T-ALL including Jurkat and MOLT4 cell lines, heterozygous somatic mutations create the super-enhancers upstream of T-ALL oncogenes such as TAL1 and NOTCH 2/3 (25, 26). Our data suggest that LSD1 inhibition induces specific, but not global, enhancer decommissioning in Notch3/TAL1-dependent T-ALL cells. It is possible that S2116 and S2157 remodel the enhancer function by switching LSD1 complexes from transcriptional activators to repressors at specific regions. This scenario is partly supported by the preliminary finding that these compounds did not affect the binding of HDACs to LSD1 (Supplementary Figure S22).
The CNS involvement is a critical determinant of the prognosis of T-ALL patients (1, 6). Most of anti-cancer drugs used for the treatment of T-ALL cannot pass through the blood-brain barrier unless administered at high doses or via direct intrathecal injection. The high penetrance of LSD1 inhibitors into the brain is an obvious advantage over other drugs in T-ALL treatment (49). In particular, S2157 showed a very high brain/blood concentration ratio at PK/PD studies and was indeed effective for the eradication of CNS disease in murine xenografts without detectable neurotoxicity because of its negligible effects on brain monoamine oxidases. It is possible that LSD1 inhibitors prevent the invasion of T-ALL cells into the brain. To test this hypothesis, we compared the effects of S2157, a brain-permeable LSD1 inhibitor, and S2116, a brain-impermeable LSD1 inhibitor (Table 1 and Supplementary Table S3), on CNS involvement in a murine xenotransplant model. As shown in Supplementary Figure S23, S2116 failed to eradicate CNS disease although the efficacy on bone marrow lesions was comparable to that of S2157. Moreover, S2157 did not significantly alter the expression of molecules facilitating CNS invasion of T-ALL cells such as CCR7 (4) and ITGA6 (5) (Supplementary Table S4). These results suggest that therapeutic effects of S2157 on CNS leukemia are mainly direct and local, although the suppression of CNS invasion could be an additional mechanism of action. In summary, our findings provide a molecular basis and rationale for the inclusion of N-alkylated LSD1 inhibitors in treatment strategies for T-ALL, especially when complicated with or at high-risk of CNS involvement.
Supplementary Material
Translational Relevance.
Treatment outcome of T-cell acute lymphoblastic leukemia (T-ALL) is still worse than that of other hematologic malignancies. A critical determinant of a poor prognosis is arguably a high incidence of central nervous system (CNS) involvement because of the low penetrance of commonly used anti-ALL drugs into the brain. Despite the fundamental role of lysine-specific demethylase 1 (LSD1) in T-cell leukemogenesis, conventional LSD1 inhibitors, such as tranylcypromine (TCP), were shown to be ineffective for T-ALL cells in several preclinical studies. We therefore modified TCP to develop novel LSD1 inhibitors with higher activity and specificity for T-ALL. One of the newly developed inhibitors, S2157, showed robust cytotoxicity against TCP-resistant T-ALL cells both in vitro and in xenotransplanted mice. Notably, S2157 could eradicate CNS leukemia because of its ability to efficiently pass through the blood-brain barrier. This brain-permeable LSD1 inhibitor is promising as a novel therapeutic agent for T-ALL with CNS involvement.
Acknowledgments
The authors are grateful to Ms. Akiko Yonekura, Ms. Mayuka Shiino, and Ms. Michiko Ogawa for their technical assistance, Dr. Takehiro Fukami (RIKEN) for his support in chemical and pharmacokinetic analyses, and Tokyo Chemical Industry Co., Ltd. (TCI) for chemical synthesis.
Financial support: This work was supported in part by the High-Tech Research Center Project for Private Universities: Matching Fund Subsidy from MEXT, a Grant-in-Aid for Scientific Research from JSPS, Research Grants from the Japan Leukemia Research Fund, Yasuda Memorial Cancer Foundation, Takeda Science Foundation, Novartis Foundation (to JK and YF), Mother and Child Health Foundation, Children’s Cancer Association of Japan (to JK) and the RIKEN Program for Drug Discovery and Medical Technology Platforms (to TU). YF received the Kobayashi Foundation Cancer Research Award.
Footnotes
Conflict of interest: The authors have no potential conflicts of interest
References
- 1.Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukaemia. Lancet 2013; 381: 1943–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lazarus HM, Richards SM, Chopra R, et al. Central nervous system involvement in adult acute lymphoblastic leukemia at diagnosis: results from the international ALL trial MRC UKALL XII/ECOG E2993. Blood 2006; 108: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fielding AK, Richards SM, Chopra R, et al. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood 2007; 109: 944–950. [DOI] [PubMed] [Google Scholar]
- 4.Bounamici S, Trimarchi T, Ruocco MG, Reavie L, Cathelin S, Mar BG, et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 2009; 459: 1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao H, Price TT, Cantelli G, Ngo B, Warner MJ, Oliviere L, et al. Leukaemia hijacks a neuronal mechanism to invade the central nervous system. Nature 2018; 560: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frishman-Levy L, Izraeli S. Advances in understanding the pathogenesis of CNS acute lymphoblastic leukaemia and potential for therapy. Br J Haematol. 2017; 176: 157–167. [DOI] [PubMed] [Google Scholar]
- 7.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13: 297–311. [DOI] [PubMed] [Google Scholar]
- 8.Hino S, Sakamoto A, Nagaoka K, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun 2012; 3: 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai C, He HH, Chen S, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 2011; 20: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wada T, Koyama D, Kikuchi J, Honda H, Furukawa Y. Overexpression of the shortest isoform of histone demethylase LSD1 primes hematopoietic stem cells for malignant transformation. Blood 2015; 125: 3731–3746. [DOI] [PubMed] [Google Scholar]
- 11.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Structurally designed trans-2-phenylcyclopropylamine derivatives potently inhibit histone demethylase LSD1/KDM1. Biochemistry 2010; 49: 6494–6503. [DOI] [PubMed] [Google Scholar]
- 12.McGrath JP, Williamson KE, Balasubramanian S, et al. Pharmacological inhibition of the histone lysine demethylase KDM1A suppresses the growth of multiple acute myeloid leukemia subtypes. Cancer Res 2016; 76: 1975–88. [DOI] [PubMed] [Google Scholar]
- 13.Ishikawa Y, Gamo K, Yabuki M, et al. A novel LSD1 inhibitor T-3775440 disrupts GFI1B-containing complex leading to transdifferentiation and impaired growth of AML cells. Mol Cancer Ther 2016; 16: 273–284. [DOI] [PubMed] [Google Scholar]
- 14.Goossens S, Peirs S, Van Loocke W, et al. Oncogenic ZEB2 activation drives sensitivity toward KDM1A inhibition in T-cell acute lymphoblastic leukemia. Blood 2017; 129: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osada N, Kikuchi J, Umehara T, et al. Lysine-specific demethylase 1 inhibitors prevent teratoma development from human induced pluripotent stem cells. Oncotarget 2018; 9: 6450–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi J, Koyama D, Wada T, Izumi T, Hofgaard PO, Bogen B, Furukawa, Y. Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J Clin Invest 2015; 125: 4375–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi J, Kuroda Y, Koyama D, et al. Myeloma cells are activated in bone marrow microenvironment by the CD180/MD-1 complex, which senses lipopolysaccharide. Cancer Res 2018; 78: 1766–78. [DOI] [PubMed] [Google Scholar]
- 18.Koyama D, Kikuchi J, Hiraoka N, Wada T, Kurosawa H, Chiba S, Furukawa, Y. Proteasome inhibitors exert cytotoxicity and increase chemosensitivity via transcriptional repression of Notch1 in T-cell acute lymphoblastic leukemia. Leukemia 2014; 28: 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271. [DOI] [PubMed] [Google Scholar]
- 20.De Bie J, Demeyer S, Alberti-Servera L, Geerdens E, Segers H, Broux M, et al. Single-cell sequencing reveals the origin and the order of mutation acquisition in T-cell acute lymphoblastic leukemia. Leukemia 2018; 32: 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talora C, Cialfi S, Oliviero C, et al. Cross talk among Notch3, pre-TCR, and Tal1 in T-cell development and leukemogenesis. Blood 2006; 107: 3313–3320. [DOI] [PubMed] [Google Scholar]
- 22.Bernasconi-Elias P, Hu T, Jenkins D, et al. Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies. Oncogene 2016; 35: 6077–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SH, Severson E, Pear WS, Liu XS, Aster JC, Blacklow SC. The common oncogenic program of NOTCH1 and NOTCH3 signaling in T-cell acute lymphoblastic leukemia. PLoS One 2017; 12: e0185762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet 2017; 49: 1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansour MR, Abraham BJ, Anders L, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014; 346: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med 2013; 210: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albero R, Enjuanes A, Demajo S, et al. Cyclin D1 overexpression induces global transcriptional downregulation in lymphoid neoplasms. J Clin Invest 2018; 128: 4132–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanada M, Takeuchi J, Sugiura I, et al. High complete remission rate and promising outcome by combination of imatinib and chemotherapy for newly diagnosed BCR-ABL-positive acute lymphoblastic leukemia: a phase II study by the Japan Adult Leukemia Study Group. J Clin Oncol 2006; 24: 460–466. [DOI] [PubMed] [Google Scholar]
- 30.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 2017; 377: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi K, Inukai T, Imamura T, et al. Anti-leukemic activity of bortezomib and carfilzomib on B-cell precursor ALL cell lines. PLoS One 2017; 12: e0188680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messinger YH, Gaynon PS, Sposto R, et al. Bortezomib with chemotherapy is highly active in advanced B-precursor acute lymphoblastic leukemia: Therapeutic Advances in Childhood Leukemia & Lymphoma (TACL) Study. Blood 2012; 120: 285–290. [DOI] [PubMed] [Google Scholar]
- 33.Mar BG, Chu SH, Kahn JD. et al. SETD2 alterations impair DNA damage recognition and lead to resistance to chemotherapy in leukemia. Blood 2017; 130: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa H, Umehara T. Structural insight into inhibitors of flavin adenine dinucleotide-dependent lysine demethylases. Epigenetics 2017; 12: 340–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shortt J, Ott CJ, Johnstone RW, Bradner JE. A chemical probe toolbox for dissecting the cancer epigenome. Nat Rev Cancer 2017; 17: 160–183. [DOI] [PubMed] [Google Scholar]
- 36.Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012; 21: 473–487. [DOI] [PubMed] [Google Scholar]
- 37.Schenk T, Chen WC, Göllner S, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nat Med 2012; 18: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakamoto K, Imamura T, Yano M, et al. Sensitivity of MLL-rearranged AML cells to all-trans retinoic acid is associated with the level of H3K4me2 in the RARα promoter region. Blood Cancer J. 2014; 4: e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Quenelle D, Vogel J, Mascaro C, Ortega A, Kristie TM. A novel selective LSD1/KDM1A inhibitor epigenetically blocks herpes simplex virus lytic replication and reactivation from latency. mBio 2013; 4: e00559–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maes T, Mascaró C, Tirapu I, Estiarte A, Ciceri F, Lunardi S, et al. ORY-1001, a potent and selective covalent KDM1A inhibitor, for the treatment of acute leukemia. Cancer Cell 2018; 33: 495–511. [DOI] [PubMed] [Google Scholar]
- 41.Mohammad HP, Smitheman KN, Kamat CD, et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 2015; 28: 57–69. [DOI] [PubMed] [Google Scholar]
- 42.Sugino N, Kawahara M, Tatsumi G, et al. A novel LSD1 inhibitor NCD38 ameliorates MDS-related leukemia with complex karyotype by attenuating leukemia programs via activating super-enhancers. Leukemia 2017; 31: 2303–2314. [DOI] [PubMed] [Google Scholar]
- 43.Takagi S, Ishikawa Y, Mizutani A, et al. LSD1 Inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res 2017; 77(17): 4652–4662. [DOI] [PubMed] [Google Scholar]
- 44.Ishikawa Y, Nakayama K, Morimoto M, et al. Synergistic anti-AML effects of the LSD1 inhibitor T-3775440 and the NEDD8-activating enzyme inhibitor pevonedistat via transdifferentiation and DNA rereplication. Oncogenesis 2017; 6: e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suvà ML, Rheinbay E, Gillespie SM, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 2014; 157: 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun 2015; 6: 5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian J, Wang Q, Dose M, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell 2014; 159: 1524–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelish HE, Liau BB, Nitulescu II, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 2015; 526: 273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan MNA, Suzuki T, Miyata N. An overview of phenylcyclopropylamine derivatives: biochemical and biological significance and recent developments. Med Res Rev 2013; 33(4): 873–910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.