Abstract
To understand prognostic factors for outcome between differentially sequenced nivolumab and ipilimumab in a randomized phase II trial, we measured T-cell infiltration and PD-L1 by immunohistochemistry, T-cell repertoire metrics, and mutational load within the tumor. We used next-generation sequencing (NGS) and assessed the association of those parameters with response and overall survival. Immunosequencing of the T-cell receptor β-chain locus (TCRβ) from DNA of 91 pretreatment tumor samples and an additional 22 pairs of matched pre- and post-treatment samples from patients who received nivolumab followed by ipilimumab (nivo/ipi), or the reverse (ipi/nivo), was performed to measure T-cell clonality and fraction. Mutational and neoantigen load were also assessed by NGS in 82 of the 91 patients. Tumors were stained using immunohistochemistry for PD-L1+ and CD8+ T cells. Pretreatment tumor TCR clonality and neoantigen load were marginally associated with best response with nivo/ipi (P = 0.04 and 0.05, respectively), but not with ipi/nivo. Amalgamated pretreatment mutational load and tumor T-cell fraction were significantly associated with best response with nivo/ipi (P = .002). Pretreatment PD-L1 staining intensity and CD8+ T-cell counts were correlated with T-cell fraction and clonality, but not mutational or neoantigen load. Patients with increased T-cell fraction post-treatment at week 13 had a 30-fold increased likelihood of survival (P = .002). Mutational and neoantigen load, and T-cell infiltrate within the tumor, were associated with outcome of sequential checkpoint inhibition using nivolumab then ipilimumab, but not when ipilimumab was administered before nivolumab.
Keywords: PD-1, CTLA-4, tumor infiltrating lymphocytes, TCRβ sequencing, antigen
INTRODUCTION
Immunotherapy with checkpoint inhibitors as single agents or in combination has altered the landscape of cancer treatment, resulting in high response rates and impressive long-term survival for many patients with melanoma as well as other tumors [1–6]. In contrast, rates of progression-free survival remain modest at less than 12 months, and the majority of patients treated with these drugs will eventually die of their disease [1–6]. Defining predictive markers associated with outcome with these agents will be instrumental in choosing patients for the optimal therapy, as well as determining the mechanisms of action and development of resistance to treatment. Expression of programmed death-ligand 1 (PD-L1) on the surface of tumor cells and/or tumor-infiltrating immune cells has been associated with overall survival and response to treatment with PD-1 antibodies in melanoma but has not been consistently observed with other tumor types [7–9]. However, PD-L1 expression in melanoma is not an optimal predictive marker since it is inducible, varies in expression between lesions, and its detection is prone to error. Patients whose tumors are PD-L1− may still benefit from PD-1 blockade [10]. Previous work using small numbers of samples from patients receiving either the CTLA-4 blocking antibody ipilimumab or the PD-1 blocking antibody pembrolizumab demonstrated that mutational and neoantigen load were associated with response and overall survival [11–14]. The data for ipilimumab have been somewhat less certain, whereas data for pembrolizumab in melanoma and lung cancer have shown a consistent association with outcome [11–14]. Published work with pembrolizumab has examined the pretreatment tumor microenvironment T-cell infiltrate using immunohistochemistry to establish an association between CD8+/PD-1+ T-cell infiltrate and outcome [15]. Data from TCR repertoire immunosequencing have demonstrated an association between pretreatment tumor T-cell clonality and T-cell fraction and response to pembrolizumab [15]. The composition of the microbiome has also been suggested to impact on the benefit of therapy with checkpoint inhibition, both in animal models and patients [16,17]. Tumor microenvironmental factors, including PTEN deletion, altered beta-catenin signaling, absence of β2-microglobulin and the presence of uncommon inactivating JAK-STAT mutations, have also been shown to be associated with an inadequate antitumor immune response and poor outcome with checkpoint inhibition [18–23].
We analyzed pretreatment tumor samples from a randomized phase II trial of nivolumab followed by ipilimumab (nivo/ipi) compared with the reverse sequence (ipi/nivo), both followed by maintenance nivolumab. In that trial, the former sequence resulted in a higher response rate and a significantly longer survival than the latter sequence [24]. We assessed if mutational and neoantigen loads and intratumoral TCR repertoire were associated with best response in either arm, and with overall survival. A small cohort of patients with pre- and post-treatment biopsies also had mutational load and intratumoral TCR repertoire assessed. Tumor PD-L1 scoring and CD8+ T-cell infiltrate were measured by immunohistochemistry for their relationship to mutational load and TCR repertoire.
MATERIALS AND METHODS
CheckMate 064 was a randomized, open-label, phase II study [24]. Eligibility criteria have been described [24]. The protocol, amendments, and patient informed consent were approved by each institution’s Institutional Review Board (IRB) and written informed consent was obtained from each patient. Patients were randomly allocated 1:1 to arm A (nivo/ipi) or arm B (ipi/nivo) as first-line therapy. No prior DNA-damaging chemotherapeutic agents were administered to any patient.
Procedures
Dosing, treatment modification, outcomes and definition of adverse events have previously been described [24]. Patients were evaluated for tumor response (per investigator assessment) using modified Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST v1.1) [24] with computed tomography and/or magnetic resonance imaging prior to dosing at weeks 13, 25, 33, and 41, and then every 12 weeks beginning at week 49.
Whole-Exome Sequencing
For each subject, tumor and normal matched DNA samples were processed for whole-exome sequencing. One normal and two tumor libraries, each with 500 ng of DNA input and constructed with individual barcoded adapters, were used. These libraries were combined into an exome-capture sequencing library using Roche NimbleGen EZ Exome v3.0 (Roche Sequencing, Pleasanton, CA), following the manufacturer’s instructions. Exome sequence data were generated as 2 × 100 base-pair reads on an Illumina HiSeq 2000 instrument (Illumina, San Diego, CA). Alignment of exome reads was performed using the Genome Modeling System (GMS) processing-profile [25], which uses BWA (v0.5.9) [26] for alignment with default parameters except for ‘-t 4 -q 5’. Sequences were aligned against human reference genome GRCh37-lite-build37, before merging and de-duplicating with Picard (v1.46). The VCF files resulting from the whole exome sequencing are deposited in the online European Variation Archive (EVA) database at: https://www.ebi.ac.uk/ena/data/view/PRJEB28604
Determination of Mutational and Neoantigen Load
Somatic mutations were identified using MuTect and Strelka algorithms by comparing tumor exome sequencing data with matched normal peripheral-blood mononuclear blood samples. Mutations called by either algorithm were pooled together, annotated using SnpEff and filtered by quality and matches to known germline polymorphisms from public databases. For mutation-load analyses, only missense mutations were counted. For neoantigen analyses, HLA-class 1 types were first inferred for each patient from exome data of the matched normal sample using OptiType software (v1.0) [27]. Mutated peptides were derived from missense mutations, and their binding affinities to corresponding HLA-class 1 types were predicted using NetMHCpan [28]. from the Immune Epitope Database. Those with predicted affinities < 500 nM were considered neoantigens.
T-Cell Receptor Variable Beta Chain Sequencing
After extracting genomic DNA, immunosequencing of the CDR3 regions of human TCRβ chains was performed using the ImmunoSEQTM Assay (Adaptive Biotechnologies, Seattle, WA). Extracted genomic DNA was amplified in a bias-controlled multiplex PCR, followed by high-throughput sequencing. Sequences were collapsed and filtered in order to identify and quantitate the absolute abundance of each unique TCRβ CDR3 region for further analysis as previously described [28,29].
Statistical Analyses of TCRβ Sequencing Results
Clonality was defined as 1- Pielou’s evenness, and was calculated on productive rearrangements as , where pi is the proportional abundance of rearrangement I and N is the total number of rearrangements. The fraction of T cells in FFPE tissue samples was calculated by normalizing TCRβ template counts to the total amount of DNA usable for TCR sequencing [29–31]. The amount of usable DNA was determined by comparison to several housekeeping genes present in all nucleated cells. Statistical analysis was performed in R v3.2. TCR sequence data are publicly available at https://clients.adaptivebiotech.com/pub/weber-2018-CIR.
Flow Cytometry Sorting
Peripheral-blood mononuclear cells (PBMCs) were collected by leukapheresis, Ficoll-Paque™ purified (GE Healthcare Bio-Sciences, AB, Uppsala Sweden), frozen in 90% human serum (Omega Scientific, Inc., Tarzana, CA), 10% DMSO (Sigma-Aldrich, St. Louis, MO) until analysis, then thawed and washed. Phenotypic T-cell markers were evaluated by flow cytometry using isotype controls with a gated lineage-negative population excluding CD19- and CD56-expressing cells using antibodies against CD19 (clone SJ25C1), CD56 (clone B159), CD3 (UCHT1) and CD8 (clone RPA-T8) from BD Biosciences (San Jose, CA). PBMCs were stained with Live/Dead violet dye (Invitrogen, Carlsbad, CA) to gate on live CD3+/CD8+ cells. Cells were sorted on an FACS Aria II flow cytometer (BD Biosciences) and analysis was performed using FlowJo version 10 software (FloJo LLC, Ashland, OR). Flow-sorted purity was verified at > 98% immediately after sorting, followed by DNA extraction. Representative gating strategy is shown in Supplementary Fig. S6.
Immunohistochemical Staining
Paraffin-embedded tumor sections were stained for PD-L1 using the DAKO 28–8 antibody as previously described [24] and scored continuously as percent staining of tumor. The CD8 (mouse clone C8/144B) antibody (Lot# 00089958, Expiration Date 11/30/2015) was purchased from Dako. The procedure for IHC analysis of CD8 (mouse clone C8/144B) with red chromogen was performed using manual detection. Staining of CD8 (mouse clone C8/144B) was evaluated by image analysis of percent positive staining within a region of interest (ROI). The ROI was circled manually to include cancer cells and intervening bands of stroma. The ROI was circled up to the margin, if present. Large bands of stroma were not included. Regions of necrosis, folds in tissue and other artifacts were excluded. The Nuclear v9 Mosaic Red-Melanin algorithm from Aperio was used and the percent positive expression was calculated.
Statistics
All statistical analyses were performed in R v3.2.4 (R Foundation for Statistical Computing, Vienna, Austria). Comparisons between responder or treatment groups were conducted with the non-parametric Mann–Whitney U test (Wilcoxon rank-sum test). To predict responses from tumor pretreatment metrics we used a multivariate logistic regression model with T-cell fraction, clonality, and mutation burden as covariates: , where θ= β0 + β1*T-cell fraction + β2*clonality + β3*mutation burden; we characterized its accuracy through receiver operator characteristics analysis and an exhaustive leave-one-out cross-validation (LOOCV). Identification of expanded clones with significantly greater frequencies in on-treatment versus baseline samples was performed as previously described [32].
RESULTS
Mutational and Neo-antigen load
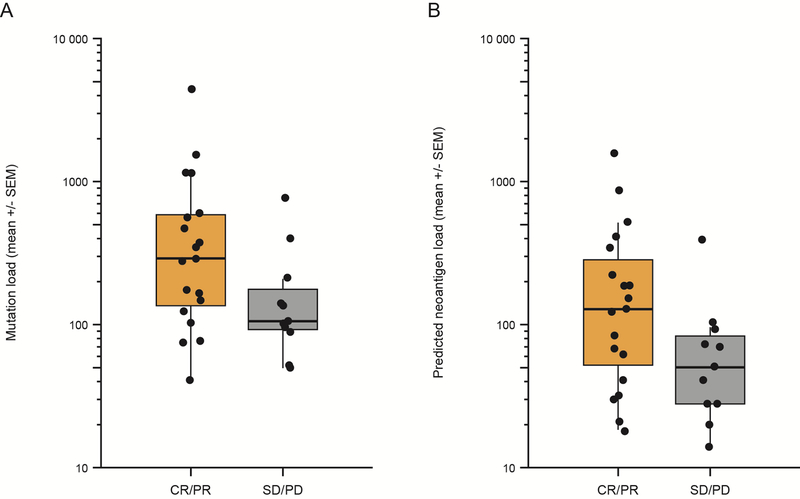
DNA extracted from pretreatment tumor and peripheral blood of 91 patients was subjected to whole-exome sequencing to assess mutational and neoantigen load. Median single nucleotide variants were 151.5 for the whole group (171 variants for arm A and 159 variants for arm B, p = 0.41, Supplementary Table S1), with a median of 70 neoantigens detected per tumor sample overall (Inter-quartile range = 29–153). Mutational and neoantigen load were significantly associated with one another (r = 0.93, P < .001, Pearson Correlation) (Supplementary Fig. S1). No significant association between pretreatment mutational load and tumor-infiltrating lymphocyte (TIL) clonality or T-cell fraction was observed. (Table 1). Within arm A (nivo/ipi), mutation burden at baseline was marginally associated with best response at week 33 defined by CR/PR versus SD/PD (N = 30, P = .06) (Fig. 1A), and neoantigen burden at baseline was associated with best response at week 33 (N = 30, P = .05) (Fig. 1B). No associations with best response for either mutational or neoantigen load were seen for arm B (ipi/nivo), for which a lower response rate and shorter survival was observed compared to arm A [24].
Table 1.
Associations Between Baseline Tumor Characteristics
| Variable | Pearson’s Rho | P value |
|---|---|---|
| CD8 versus PD-L1 | 0.440 | <.001 |
| CD8 versus clonality | 0.418 | <.001 |
| CD8 versus T-cell fraction | 0.784 | <.001 |
| CD8 versus mutation load | −0.058 | .657 |
| PD-L1 versus clonality | 0.291 | .013 |
| PD-L1 versus T-cell fraction | 0.320 | .006 |
| PD-L1 versus mutation load | 0.026 | .848 |
| Clonality versus mutation load | 0.024 | .841 |
| T-cell fraction versus mutation load | 0.036 | <.001 |
Fig 1.
Mutational burden and neoantigen load versus clinical benefit in arm A (nivo/ipi). (A) Mutational burden (P = 0.06, n = 30). (B) Neoantigen load (P = 0.05, n = 30) for all patients calculated as described in Patients and Methods is shown on a log scale on the ordinate, and benefit defined as complete and partial response as well as stable disease (CR + PR) versus stable plus progressive disease (SD+PD) shown on the abscissa. The box plots show median (horizontal middle line), the interquartile range (IQR) (shaded box region), and the minimum and maximum (lines extended above and below the box). P values were obtained from a Mann–Whitney U Test.
T-cell fraction and TIL clonality
CDR3 regions of the TCR Vβ beta chains (TCRβ) were sequenced, and clonality calculated for the productive rearrangements as described in Materials and Methods. T-cell fraction was calculated as the proportion of rearranged TCRβ relative to the total number of assayed cells. T-cell fraction and TIL clonality in pretreatment tumors were significantly correlated (R2 = 0.18, P < .001 by Pearson’s method, N = 91, Table 1). There were no significant differences in the pretreatment T-cell clonality or T-cell fraction between arms A and B (Supplementary Fig. S2 and Supplementary Table S1).
No significant association between mutational load and tumor-infiltrating lymphocyte (TIL) clonality or T-cell fraction was observed (Supplementary Fig. S3).
Association of T-cell fraction and TIL clonality with response
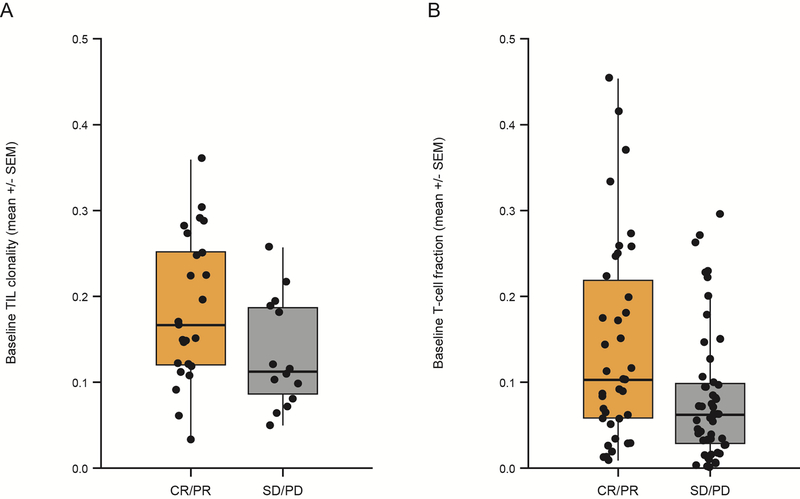
For arm A (nivo/ipi), the pretreatment TIL clonality was associated with response (P = .04) (Fig. 2A). T-cell fraction was also correlated with response in the combined dataset of both arms (P = .02, N = 89, [Fig. 2B]); however, likely due to reduced statistical power, it was not significantly correlated with response in arm A (P = .21, N = 39) or B (P = .07, N = 50) (Supplementary Fig. S4).
Fig 2.
Pretreatment TIL clonality in tumor samples of arm A, and pretreatment T-cell fraction in tumor samples from both arms A and B, correlated with clinical benefit (P = 0.04 with N = 30 and P = 0.02 with N = 89, respectively). (A) Pretreatment T-cell clonality. (B) T-cell fraction calculated as described in Patients and Methods is shown on a linear scale on the ordinate with response (CR + PR) versus non-response (SD + PD) shown on the abscissa. P values are determined from a Mann–Whitney U Test.
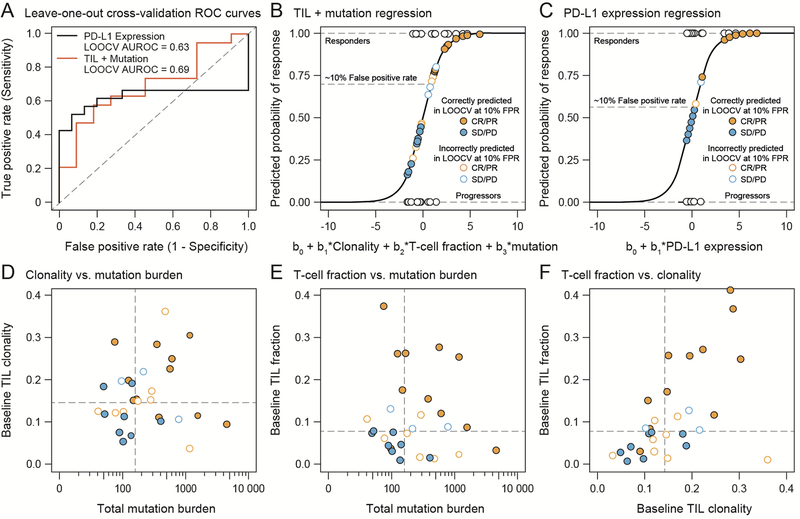
Since pretreatment T-cell fraction and clonality were independent of mutation burden, we performed an ad hoc analysis to assess whether the combination of the three genetic molecular profiling methods led to a specific and sensitive prediction of response. We performed a multivariate logistical regression analysis of samples from arm A patients (N = 30), all with baseline tumor assessments of TIL clonality, T-cell fraction, and total mutation burden. Figure 3A shows the receiver operating characteristic (ROC) curve of specificity versus sensitivity in an exhaustive, leave one out cross-validation (LOOCV) procedure to predict clinical benefit, and Fig. 3B shows the resulting fit of the regression model for the tripartite analysis. This analysis suggests that at a false positive rate of approximately 10% (falsely predicting response rather than progression 10% of the time), roughly 50% (derived from LOOCV) to 70% (derived from the full data set) of responders are correctly predicted. As seen in previous reports [5–7], PD-L1 status correlated with clinical benefit (PD-L1 ≥ 5%, P = .009, OR = 14.7 by Fisher’s exact test). Performing a univariate logistic regression on 34 patients with PD-L1 data showed similar specificity and sensitivity to the molecular profiling techniques (AUROC of 0.67 versus 0.70 in the LOOCV, Fig. 3A and C, likelihood ratio test P = .12). PD-L1 expression equal or larger to 5% corresponded to a 10% false positive rate and led to classifying approximately 55% of responders correctly. Figure 3D–F shows scatter plots of pair-wise combinations for T-cell fraction, TIL clonality, and mutation burden, as well as the values obtained from patients who were correctly (filled circles) and incorrectly (open circles) classified in the LOOCV. The performance of this classification model will need to be validated with larger numbers of patients and in a separate validation cohort. Eight patients who achieved clinical benefit but had PD-L1 values lower or equal to 5% were identified; of these, four would have been correctly classified with the trivariate biomarker model comprising mutation load, T-cell fraction, and TIL clonality. We also assessed the performance of mutation, T-cell fraction, and TIL clonality in univariate models and found the combined tripartite model to have the highest specificity and sensitivity for predicting clinical benefit (CR/PR versus SD/PD; see Fig. 3A; Fig. S5 shows each model individually with AUC values).
Fig 3.
Mutation, T-cell fraction, and TIL clonality in pretreatment tumors predict clinical benefit in arm A. (A) ROC curves generated from an exhaustive leave-one-out cross-validation (LOOCV) procedure with a trivariate model (red line) with mutation, T-cell fraction, and TIL clonality compared to a univariate model (black line) employing PD-L1 expression. (B) Outcome of the multivariate logistic regression model, which included data from 34 patients. (C) Outcome of the univariate logistic regression model, including data from 30 patients. Two-dimensional projections of the TIL clonality and mutation burden in (D), T-cell fraction and mutational burden in (E), and T-cell fraction and mutation burden in (F). The color of the point or circle indicates clinical benefit, with closed circles indicating patients correctly classified and open circles indicating patients incorrectly classified in the LOOCV procedure.
Association of changes in T-cell fraction and TIL clonality with response
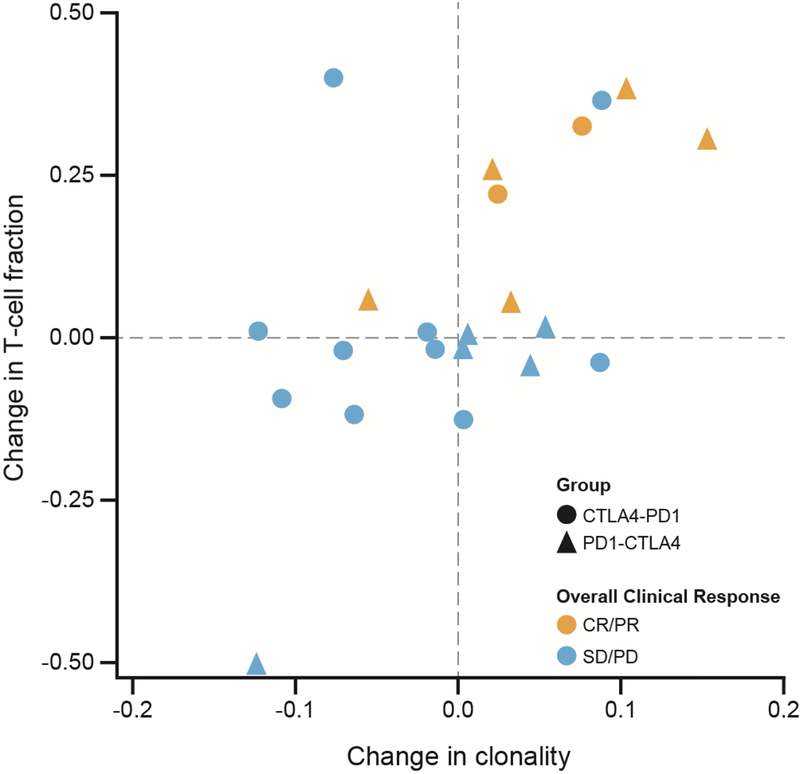
Twenty-two matched pairs of tumors from pretreatment and 13-week post-treatment biopsies after either nivolumab alone (arm A) or ipilimumab alone (arm B), respectively, were obtained, of which 80% were from the same lesion. At week 13, individuals in either arm that showed increased T-cell fraction and TIL clonality were 30 times more likely to achieve a best clinical response at week 33 than those without evidence of change (P = .002; OR = 30, Fisher’s exact test) (Fig. 4).
Fig 4.
Increased on-treatment TIL clonality and T-cell fraction in tumor samples correlate with clinical benefit (P = 0.002, OR = 30). Changes in TIL clonality and T-cell fraction were determined by subtracting values determined at baseline from those determined in week 13 biopsies. P value was obtained from a Fisher’s exact test of CR/PR versus SD/PD comparing the top right quadrant to the remaining three quadrants.
Tumor staining for T cells and PD-L1
We also assessed whether the parameters of pretreatment mutation load, T-cell fraction, and TIL clonality were correlated with immunohistochemical staining for CD3+ T-cells, CD8+ T-cells, and PD-L1 staining. The data in Table 1 suggest that there was an interdependence of CD8+ T-cell counts or PD-L1 immunohistochemical staining with the measures of both T-cell clonality and T-cell fraction. In contrast, there was no clear association of quantitation of CD8+ T cells or PD-L1 by immunohistochemical staining, with mutational load (Pearson’s coefficients = −0.058 and 0.026, respectively). These results are consistent with the independence of TIL clonality and T-cell fraction from mutational load described above.
Associations with survival:
An assessment of the association of the above tumor microenvironment parameters with overall survival was also performed. Data from 89 patients were available for survival analysis. We fit overall survival data with a Cox proportional hazards model conditional on treatment arm and showed the resulting hazard ratios (+/− 95% Confidence Interval) and P values from a Wald test for analysis with mutation burden, PD-L1 expression, TIL clonality, and TIL infiltration in pretreatment tumor as single-covariates in the model (Table 2). The results in Table 2 show that increasing mutation burden, PD-L1 expression, TIL clonality, and TIL infiltrate are associated with a reduced hazard ratio and increased overall survival in patients treated sequentially with PD-1 blockade before CTLA-4 blockade (Arm A).
Table 2.
Association of Baseline Tumor Characteristics and Survival by Arm
| Arm A | Arm B | ||||
|---|---|---|---|---|---|
| Unit for HR | HR (95%CI) | P value | HR (95%CI) | P value | |
| Mutation burden | Log(x) mutations | 0.44 (0.17 – 1.11) | 0.080 | 1.03 (0.73 – 1.43) | 0.887 |
| PD-L1 Expression | 1 % | 0.65 (0.34 – 1.24) | 0.194 | 0.96 (0.92 – 1.01) | 0.125 |
| TIL Clonality | 0.1 value | 0.40 (0.15 – 1.10) | 0.075 | 0.89 (0.56 – 1.42) | 0.634 |
| T-cell fraction | Log(1%) | 0.64 (0.36 – 1.13) | 0.121 | 0.73 (0.57 – 0.95) | 0.017 |
Mutation burden: Arm A (N = 30, 7 events), Arm B (n = 38, 26 events)
PD-L1: Arm A (N = 34, 8 events), Arm B (N = 37, 27 events)
TIL Clonality & T-cell fraction: Arm A (N = 39; 9 events), Arm B (N = 50, 32 events)
Association of T-cell fraction and mutational burden with survival:
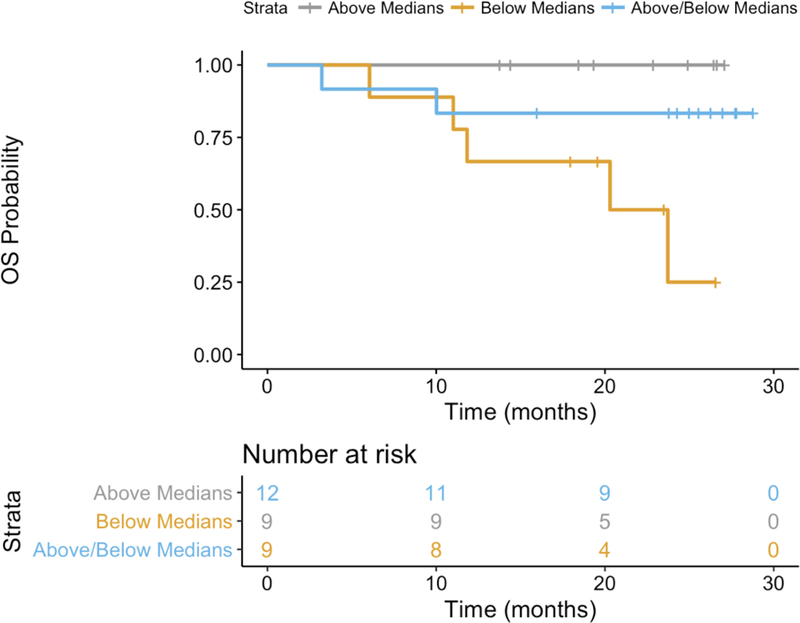
Since the NGS-based tumor microenvironment parameters (TcR clonality and mutational burden) were not correlated with one another, we hypothesized that the combination of mutation burden and TIL assessments could provide a superior prognostic model compared to any single variate. There were 68 patients with 33 events that had data for mutation burden, T-cell fraction, and TCRVß clonality. In an aggregate analysis of both arms A and B, T cell fraction was significantly associated with better survival (p = 0.003, likelihood ratio test) as seen in Supplementary Table S2. This result is biased toward outcomes in Arm B, in which 64% of patients died during the observation period compared to 23% in Arm A and is consistent with the single variate analysis shown in Table 2. Results from patients within Arm A (N = 30, 7 events) suggested that the combination of T cell fraction and mutation burden could best predict survival in patients who received PD-1 antibody but did not have prior ipilimumab exposure (p = 0.08, likelihood ratio test). In Fig. 5, survival curves are shown for those same patients stratified by whether the parameters were above or below the median: for example, both parameters above; both parameters below; above-median T-cell fraction and below-median mutational burden; or below-median T-cell fraction and above-median mutational burden. These data suggest that the best survival was associated with high T-cell fraction and high mutational burden.
Fig 5.
Overall survival curves for patients in arm A (30) with high T-cell fraction and mutational burden shown in orange, high T-cell fraction and low mutational burden in blue and low T-cell fraction and mutational burden in brown. High versus low was determined by the medians for each parameter.
DISCUSSION
The definition of useful predictive and pharmacodynamic biomarkers for the clinical efficacy of checkpoint blockade has remained elusive. Blockade of PD-1 to restore a dysfunctional antitumor immune response is thought to act primarily within the tumor microenvironment. The presence of a CD8+ T-cell infiltrate detected by immunohistochemistry within tumors and tumor T-cell clonality prior to treatment was associated with a favorable outcome with pembrolizumab [15]. A tumor signature of genes from the interferon-gamma signaling pathway was associated with outcome for pembrolizumab [33]. An emerging concept is that of the “hot” versus the “cold” tumor, in which a hot tumor is defined by an inflamed microenvironment associated with expression of genes in the interferon-gamma pathway, and decreased expression of epithelial mesenchymal transition and wound healing pathway genes [34,35]. Genetic alterations in tumors have been identified that promote a “cold” microenvironment, including PTEN deletions and mutations in the beta-catenin pathway [18,19]. Alterations or deletions in beta-2 microglobulin may lead to inadequate recognition of tumors by T cells due to deficient antigen presentation [21]. Non-synonymous tumor mutations are associated with clinical outcome using checkpoint blockade and may provide potential neoantigen targets for T-cell recognition [10–13, 36–38]. No single biomarker, however useful for stratification, has been adequate as a predictive biomarker for the efficacy of checkpoint blockade.
To understand how PD-1 and CTLA-4 blockade promote immune recognition of tumors, we analyzed tumor samples from a randomized sequential trial of PD-1 and CTLA-4 blockade whose clinical results have been reported [24]. The week 13 data from that trial allowed an assessment of response to either drug alone, and the data on best overall response at week 33 demonstrated the effects of both drugs sequentially. In that trial, the response rates and overall survival were superior in arm A (NIVO/IPI) compared with arm B (IPI/NIVO) [24]. Immunosequencing of 91 pretreatment tumor TcRVβ segments was performed, followed by assessment of clonality and T-cell fraction, as well as immunohistochemical staining for CD8+ T cells and PD-L1. Whole-exome sequencing was performed on 82 tumors and matched normal tissue to assess mutational and neoantigen load.
The data herein indicate that an amalgamated biomarker of T-cell fraction, TIL clonality, and mutational load, which were independent variables, were significantly associated with response for patients receiving nivolumab followed by ipilimumab, but not ipilimumab followed by nivolumab. T-cell fraction and mutation load were also associated with overall survival for those treated with nivolumab followed by ipilimumab, but not ipilimumab followed by nivolumab. The area under the ROC curve for the amalgamated marker of T-cell fraction, TIL clonality, and mutational load for arm A were as good as or better than those for PD-L1 staining, seen in Figure 3. Clonality and T-cell fraction were weakly associated with one another in this cohort. The ability to predict a responder or long-term survivor using only the mutational or neoantigen load was inadequate, since some responders had a low mutational load, and some non-responders displayed a high load [10–13]. There was no statistical association between the mutational load and the following biomarkers: intratumoral T-cell fraction, TIL clonality, PD-L1, or CD8 staining. The lower response rate in arm B could not account for the lack of association of mutational load and clonality with outcome, since baseline clonality, mutational load, and T-cell fraction were fairly well-balanced between the arms (Supplementary Table S1), and mutational load was previously shown to be associated with ipilimumab alone with a response rate of less than 20% [11]. There was a modest imbalance, but not statistically significant difference, in PD-L1 staining between arms A and B, which may have accounted for some of the difference. Tumor PD-L1 staining was associated with metrics of T-cell infiltration, and analysis of larger data sets will determine if it adds to the predictive and prognostic value of parameters of clonality and T-cell fraction and whether TcR metrics are not dependent on mutational or neoantigen load.
For 22 patients with pretreatment and post-treatment samples at week 13, increased T-cell fraction and TIL clonality over baseline had an odds ratio of 30 for clinical benefit, suggesting that an influx of clonal T-cells within the tumor microenvironment was advantageous for the benefit of PD-1 or CTLA-4 blockade, albeit with small numbers of responders. These data confirm previously published data [14] but need to be validated with a larger dataset from an independent clinical trial. The presence of tumor PD-L1 staining and the influx of T cells as measured by immunohistochemical staining were significantly correlated with T-cell fraction and clonality determined by immunosequencing and were both independent of mutational load. This on-treatment measurement was the most significant marker measured that was associated with response.
These results reinforce the view that pretreatment tumor characteristics can determine the benefit of checkpoint blockade but also indicate that on-treatment metrics are informative. These data also suggest that mutational or neoantigen load compared to T-cell parameters including clonality, T-cell fraction, CD8+ T-cell infiltrate and PD-L1 staining might have independent predictive value, but that each parameter alone has fairly weak predictive value. Loss of neoantigens has been found to be associated with resistance to checkpoint blockade, reinforcing the importance of mutational and neo-antigen load as a prognostic and predictive marker [39]. Both pretreatment or post-treatment tumors can be infiltrated with clonal T cells, and peripheral blood may reflect the tumor-infiltrating clonal T-cell populations associated with clinical benefit. The data herein also suggest that the mutational or neoantigen load was weakly associated with benefit for ipilimumab followed by nivolumab, but more clearly so with PD-1 blockade then ipilimumab, and that the same pattern was seen for an amalgamated biomarker using mutational load and T-cell fraction or clonality. Our results also indicate that predictive biomarkers for the benefit of checkpoint inhibition should include characteristics of the target and the infiltrating effectors. The absence of β2-microglobulin, for example, might compromise immune recognition independently of mutational or neoantigen load and has been associated with adaptive resistance to PD-1 blockade [21]. The lower response rate and shorter survival observed with ipilimumab followed by nivolumab may reflect the inability of patients with an inflamed microenvironment, as defined by the amalgamated mutational load and T-cell fraction, to benefit from nivolumab, compared with those who received nivolumab first. This suggests the possibility of transient resistance to PD-1 blockade induced by CTLA-4 abrogation, the etiology of which is being actively investigated. A composite biomarker reflecting qualities of the tumor and the host, and the short-term effects of prior immune therapy, will most likely be required to provide clinically useful prediction of outcome with checkpoint inhibition.
Supplementary Material
ACKNOWLEDGMENTS
Data presented in part at the 2016 European Society for Medical Oncology meeting in Copenhagen, Denmark. Funding was provided by the National Cancer Institute (grant 5R01CA175732), the Chris Sullivan Foundation, Adaptive Bioptechnologies and Bristol-Myers Squibb. Editorial assistance was provided by Amy Agbonbhase (Spark-Medica), funded by Bristol-Myers Squibb.
Contributor Information
Erik Yusko, Adaptive Biotechnologies, Seattle, WA.
Marissa Vignali, Adaptive Biotechnologies, Seattle, WA.
Richard K. Wilson, McDonnell Genome Institute, Washington University in St. Louis, St. Louis, MO
Elaine R. Mardis, McDonnell Genome Institute, Washington University in St. Louis, St. Louis, MO
F. Stephen Hodi, Dana-Farber Cancer Center, Boston, MA.
Christine Horak, Bristol-Myers Squibb, Princeton, NJ.
Han Chang, Bristol-Myers Squibb, Princeton, NJ.
David M. Woods, Laura and Isaac Perlmutter Cancer Center at NYU Langone Medical Center, New York, NY
Harlan Robins, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, and Adaptive Biotechnologies, Seattle, WA.
Jeffrey Weber, Laura and Isaac Perlmutter Cancer Center at NYU Langone Medical Center, New York, NY.
REFERENCES
- 1.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L et al. : Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015. 372:320–330 [DOI] [PubMed] [Google Scholar]
- 2.Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B et al. : Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. The Lancet Oncol 2015. 16:375–384 [DOI] [PubMed] [Google Scholar]
- 3.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L et al. : Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 2015. 372:2521–2532 [DOI] [PubMed] [Google Scholar]
- 4.Ribas A, Puzanov I, Dummer R, Schadendorf D, Hamid O, Robert C et al. : Pembrolizumab versus investigator-choice chemotherapy for ipilimumab-refractory melanoma (KEYNOTE-002): a randomised, controlled, phase 2 trial. The Lancet Oncol 2015. 16:908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, Ribas A, Hamid O, Daud A, Wolchok JD, Joshua AM et al. Durable Complete Response After Discontinuation of Pembrolizumab in Patients With Metastatic Melanoma. J Clin Oncol. 2018. 36(17):1668–1674 [DOI] [PubMed] [Google Scholar]
- 6.Callahan MK, Kluger H, Postow MA, Segal NH, Lesokhin A, Atkins MB et al. Nivolumab Plus Ipilimumab in Patients With Advanced Melanoma: Updated Survival, Response, and Safety Data in a Phase I Dose-Escalation Study. J Clin Oncol. 2017. 36(4):391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS et al. : Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014. 515:563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL et al. : Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med 2012. 4:127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. : Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 2013. 5:200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel SP and Kurzrock R: PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther 2015. 14:847–856 [DOI] [PubMed] [Google Scholar]
- 11.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A et al. : Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014. 371:2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ et al. : Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015. 348:124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L et al. : Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015. 350:207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK et al. : Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016. 351:1463–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L et al. : PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014. 515:568–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM et al. : Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015. 350:1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B et al. : Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity 2016. 44:1255–1269 [DOI] [PubMed] [Google Scholar]
- 18.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT et al. : Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016. 6:202–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spranger S, Bao R and Gajewski TF: Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015. 523:231–235 [DOI] [PubMed] [Google Scholar]
- 20.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE et al. : Direct Activation of STING in the Tumor Microenvironment Leads to Potent and Systemic Tumor Regression and Immunity. Cell Rep 2015. 11:1018–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S et al. : Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016. 375:819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D et al. : Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med 2015. 372:2006–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD et al. : Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med 2015. 373:23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber JS, Gibney G, Sullivan RJ, Sosman JA, Slingluff CL Jr, Lawrence DP et al. : Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial. The Lancet Oncol 2016. 17:943–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffith M, Griffith OL, Smith SM, Ramu A, Callaway MB, Brummett AM et al. : Genome Modeling System: A Knowledge Management Platform for Genomics. PLoS Comput Biol 2015. 11:e1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H and Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009. 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, and Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014. 30:3310–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O et al. : NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 2009. 61:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O et al. : Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009. 114:4099–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM et al. : Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013. 4:2680. [DOI] [PubMed] [Google Scholar]
- 31.Robins H, Desmarais C, Matthis J, Livingston R, Andriesen J, Reijonen H et al. : Ultra-sensitive detection of rare T cell clones. J Immunol Methods 2012. 375:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeWitt WS, Emerson RO, Lindau P, Vignali M1, Snyder TM1, Desmarais C et al. : Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol 2015. 89:4517–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S et al. : Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016. 165:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gajewski TF: The Next Hurdle in Cancer Immunotherapy: Overcoming the Non-T-Cell-Inflamed Tumor Microenvironment. Semin Oncol 2015. 42:663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teng MW, Ngiow SF, Ribas A, and Smuth MJ: Classifying Cancers Based on T-cell Infiltration and PD-L1. Cancer Res 2015. 75:2139–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacher TN and Schreiber RD: Neoantigens in cancer immunotherapy. Science 2015. 348:69–74 [DOI] [PubMed] [Google Scholar]
- 37.Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov Aet al A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017. 551:517–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stronen E, Toebes M, Kelderman S, Buuren MM2, Yang W1, van Rooij N et al. : Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 2016. 352:1337–1341 [DOI] [PubMed] [Google Scholar]
- 39.Anagnostou V, Smith KN, Forde PM Niknafs N, Bhattacharya R, White J et al. Evolution of Neoantigen Landscape during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Discov. 2017. 3:264–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.