Abstract
Tumor heterogeneity is a major challenge for cancer treatment, especially due to the presence of various subpopulations with stem cell or progenitor cell properties. In mouse melanomas, both CD34+p75− (CD34+) and CD34−p75− (CD34−) tumor subpopulations were characterized as melanoma-propagating cells (MPCs) that exhibit some of those key features. However, these two subpopulations differ from each other in tumorigenic potential, ability to recapitulate heterogeneity, and chemo-resistance. In this study, we demonstrate that CD34+ and CD34− subpopulations carrying the BRAFV600E mutation confer differential sensitivity to targeted BRAF inhibition. Through elevated KDM5B expression, melanoma cells shift towards a more drug-tolerant, CD34− state upon exposure to BRAF inhibitor or combined BRAF inhibitor and MEK inhibitor treatment. KDM5B loss or inhibition shifts melanoma cells to the more BRAF inhibitor-sensitive CD34+ state. These results support that KDM5B is a critical epigenetic regulator that governs the transition of key melanoma-propagating cell subpopulations with distinct drug sensitivity. This study also emphasizes the importance of continuing to advance our understanding of intratumor heterogeneity and ultimately develop novel therapeutics by altering the heterogeneous characteristics of melanoma.
Keywords: Melanoma, intratumor heterogeneity, histone demethylase, targeted therapy, drug tolerance
Introduction
Intratumor heterogeneity serves as a significant clinical barrier to finding effective and long-lasting treatments for various types of cancer (1). Many reports suggest that melanoma does not follow the hierarchical cancer stem cell model because tumorigenic cells within a single melanoma tumor represent more than a rare subpopulation (2-5). Meanwhile, several other studies argue that there exist more subtleties to this conclusion than previously suggested (6, 7). Our laboratory demonstrated that mouse melanoma tumors driven by different genetic alterations are composed of three major subpopulations. These three subpopulations, CD34+p75− (CD34+), CD34−p75− (CD34−), and CD34−p75+ (p75+), exhibit distinguishing cellular behaviors (8). Specifically, CD34+ cells are highly tumorigenic in vivo whereas p75+ cells are rarely tumorigenic; CD34− cells exhibit an intermediate level of tumorigenic potential but are capable of recapitulating tumor heterogeneity in vitro and in vivo. Thus, both CD34+ and CD34− cells are referred to as melanoma-propagating cells (MPCs) and are speculated to play a critical role in tumor development. In addition, these three subpopulations demonstrate varying levels of resistance to traditional cytotoxic chemotherapeutics in vitro, suggesting that they may also respond differently to more targeted treatments (8). Overall, while we have made progress in understanding the cellular characteristics representing the different tumor cell subpopulations in melanoma, we have not yet identified key regulators mediating the conversion of CD34+ and CD34− MPCs. This is an essential step to provide effective therapeutic strategies for melanoma patients considering the critical cellular features unique to these subpopulations.
Several epigenetic regulators have been used to identify heterogeneous tumor subpopulations with distinct cellular phenotypes. Non-small cell lung cancer cell lines along with several other human cancer cell lines contained a reversibly drug-resistant subpopulation that required KDM5A to maintain the drug-tolerant state (9-11). A slow-cycling subpopulation of melanoma cells expressing high levels of histone demethylase KDM5B (also known as JARID1B) was shown to be important in maintaining melanoma growth and intrinsically resistant to various cancer drugs (12, 13). Moreover, while CD34+, CD34−, and p75+ cells have been identified in mouse melanoma tumors with different genetic alterations, subpopulations expressing the same distribution of markers exhibit similar progenitor cell properties (8). Taken together, these findings indicate that epigenetic variation likely exists between the two MPC and p75+ subpopulations and may also be the explanation for their diverse cellular characteristics.
In addition, cancer cells exploit intratumor heterogeneity as a defense mechanism in response to drug therapy (9, 13, 14). This highlights the clinical need to understand the regulation of intratumor heterogeneity during tumor development and in the context of therapeutic treatment. In melanoma, over 50% of patients harbor the V600 mutation in BRAF kinase. This particular genetic alteration leads to constitutive activation of the mitogen-activated protein kinase (MAPK) pathway and contributes to uncontrolled cell proliferation (15). Unfortunately, resistance to targeted BRAF inhibitors (BRAFis) such as vemurafenib presents a continuous challenge in the clinic. Multiple resistance mechanisms have been described, including reactivation of the MAPK pathway, upregulation of the compensatory PI3K signaling, and non-genetic modifications (16, 17). In this study, we examine how CD34+ and CD34− MPC subpopulations respond to targeted therapies to better understand the link between intratumor heterogeneity and drug response. We further demonstrate that histone demethylase KDM5B is a key regulator of the conversion between MPC subpopulations of mouse melanoma.
Materials and Methods
Cell culture
Yale University Mouse Melanoma cell lines (YUMM1.1, YUMM1.3, YUMM1.7, YUMM1.9, YUMM2.1, YUMM3.3, YUMM3.4, YUMM5.1) used in this study were characterized previously (18). Cells were maintained in DMEM/F12 (1:1) media containing 10% heat-inactivated FBS, 1% penicillin streptomycin, and 1% MEM non-essential amino acids (Gibco by Life Technologies) at 37°C and 5% CO2. BRAFi-resistant YUMM1.7 (YUMM1.7R) cells were derived by continuously exposing YUMM1.7 cells to 2 μM BRAFi (PLX4032, BioVision 2235-5)(19) for 8 weeks as described previously (20) and maintained in 2 μM BRAFi media unless indicated otherwise. Human melanoma cell lines 501mel and YUMAC (within 10 passages after being obtained from Dr. Ruth Halaban from Yale University (21)) were grown in similar conditions except using OPTI-MEM (Invitrogen). Cell lines were authenticated using short tandem repeat profiling performed at the DNA Analysis Facility on Science Hill at Yale University. Sigma LookOut Mycoplasma PCR detection kit (Sigma-Aldrich, MP0035) was used for mycoplasma detection in cell culture.
For drug treatment, YUMM cells were treated with either 1.5 μM BRAFi or with a combination of 1.5 μM BRAFi and MEK inhibitor (MEKi) (Trametinib, Santa Cruz Biotechnology sc-364639). Cells were harvested for western blotting, flow cytometry, and reverse transcription and quantitative PCR (RT-qPCR) at end point. For KDM5 inhibitor (KDM5i) treatment, YUMM1.7 cells were pretreated in 1 μM KDM5-C70 (Xcess Biosciences Inc M60192-2s) (22, 23), 1 μM CPI-48 (NCGC00488278, prepared according to patent WO2015/135094)(24), or 100 μM YUKA1 (ChemBridge Hit2Lead 7870547)(11) for five days before being co-treated with 1.5 μM BRAFi for an additional three days. On the 8th day, cells were harvested for similar assays and analyzed as described above.
Flow cytometry analysis and cell sorting
At the end of each in vitro experiment, cells were washed with PBS and trypsinized with 0.25% Trypsin-EDTA (Gibco by Life Technologies). Cells were stained with rat anti-CD34 (ThermoFisher 14-0341-82) and rabbit anti-Nerve Growth Factor Receptor/p75 (Millipore Sigma AB1554) in PBS supplemented with 1% bovine serum albumin (BSA, Sigma-Aldrich A9647) at 4°C for 30 minutes. Cells were washed 3 times with fluorescence-activated cell sorting (FACS) buffer and stained with Brilliant Violet 421 Donkey anti-rabbit IgG (BioLegend 406410), Alexa Fluor 647 Goat anti-rat IgG (ThermoFisher A-21247), and propidium iodide (BioLegend 421301) at 4°C for 15 minutes. Cells were washed 3 times, re-suspended in PBS, and analyzed by BD LSRII Flow Cytometer. Plots were generated using FlowJo 9.9.6. To sort CD34+ cells, YUMM1.7 cells were stained using the same protocol and subjected to single-cell sorting using BD FACSAria into a 96-well plate. Individual cells were expanded and tested by flow cytometry to ensure CD34+ purity. CD34+ and CD34− cells used for the RT-qPCR experiment were sorted using the same staining protocol but collected using 15 ml conical tubes using the BD FACSAria or Sony SH800 Cell Sorter. To analyze subpopulation distribution in grafted tumors by flow cytometry, tumors were dissociated by finely mincing and incubating in DMEM/F12 (1:1) containing 1mg/ml Collagenase/Dispase (Sigma-Aldrich 10269638001) and 1X DNase I (Qiagen 79254) at 37°C for 30 minutes. Dissociated tumors were titrated and filtered through a 70 μm filter to yield a single-cell suspension before FACS staining. Cells were stained using a similar protocol described above with an additional anti-CD45 APC-Cy7 antibody (BioLegend 103115).
Western blot analysis
Mouse cells were collected following digestion with 0.25% Trypsin-EDTA and lysed on ice with high salt lysis buffer [ 50 mM Tris-HCl pH 7.9, 0.1 mM EDTA pH 8.0, 320 mM NaCl, 0.5% NP40, 10% Glycerol, 1x Protease Inhibitor cocktail (Roche 11873580001)] to obtain whole cell lysates. The remaining pellets were re-suspended in Laemmli buffer and sonicated to extract histones. Protein concentration of whole cell lysates was measured by Bradford assay (Bio-Rad) with known BSA standards. Samples in Laemmli buffer were heated for 10 minutes at 95°C and loaded onto 7% (whole cell lysates) or 15% (histones) SDS-PAGE gels. Membranes were blocked in 4% non-fat milk in Tris-buffered saline (50 mM Tris-HCl, 138 mM NaCl, 2.7 mM KCl, pH 7.4) with 0.05% Tween (TBS-T) and incubated with primary antibodies in the same buffer or 5% BSA in TBS-T overnight at 4°C. Membranes were incubated with secondary anti-rabbit or anti-mouse antibodies for 1 hour at room temperature. Blots were visualized by ECL Western Blotting Substrate (Thermo Scientific Pierce 32106) or Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore WBKLS0100) on film or with KwikQuant Imager. Primary antibodies used included rabbit anti-KDM5B (Sigma HPA027179), mouse anti-tubulin (Sigma T5168), rabbit anti-histone H3 (Abcam ab1791), rabbit anti-H3K4me3 (C42D8, Cell Signaling Technology 9751), mouse anti-vinculin (Sigma V9131), and rabbit anti-GAPDH (14C10, Cell Signaling Technology 2118).
Proliferation assay and IC50 assays
To measure cell proliferation, cells were seeded in triplicate in 96-well tissue culture plates and grown at various experimental conditions: DMEM/F12 media, dimethyl sulfoxide (DMSO, Sigma-Aldrich 67-68-5), BRAFi, and BRAFi/MEKi. At the respective time points, cell proliferation was measured using CyQUANT NF Cell Proliferation Assay Kit (Invitrogen C35006). Values within the same group at each time point were averaged and compiled with t-test statistical analysis using Prism 7. IC50 values were determined by incubating cells in different concentrations of BRAFi at 37°C for three days. The concentrations were 0.039, 0.078, 0.156, 0.313, 0.652, 1.25, 2.5, 5 and 10 μM, in addition to DMSO controls. All samples were analyzed in triplicate and all experiments were repeated three times.
Tumor graft
To study tumor growth and response to targeted inhibitors, 100,000 YUMM1.7 cells were injected subcutaneously into each C57BL/6J mouse (Jackson Laboratory). Mice were maintained in the animal facility of Yale School of Medicine. All animal studies were approved by the Institutional Animal Care and Use Committee of Yale University. Tumor volume was monitored and determined using the following equation: width x length x height x 0.5233 (mm). Once tumor volume reached 300 mm3, mice in treatment groups were switched to a special diet with BRAFi (Plexxikon PLX4720-200ppm)(25) or combination BRAFi/MEKi (Plexxikon PLX4720/PLX2695-200/7ppm)(26) chow while mice in control groups continued with the regular chow diet. At end point, mice were euthanized and tumors were harvested for flow cytometry analysis and RT-qPCR analysis.
Generation of KDM5B knockdown cell lines
Stable knockdowns of KDM5B in mouse melanoma cell lines YUMM1.7 and YUMM3.4, and human melanoma cell lines YUMAC and 501mel were performed as described previously (27, 28) using lentiviral shRNA hairpins: pLKO.1- mouse-KDM5B sh1 (targeting GCCTACATCATGTGAAAGAAT), pLKO.1- mouse-KDM5B sh2 (targeting CCTGAAATTCAGGAGCTTTAT), pLKO.1- human-KDM5B sh1 (targeting CGAGATGGAATTAACAGTCTT), pLKO.1- human-KDM5B sh2 (targeting AGGGAGATGCACTTCGATATA), and pLKO.1-shGFP (control sh) (a gift from William Hahn, Dana-Farber Cancer Institute, Boston, MA). To generate stable cell lines, cells were infected with lentivirus for 24 hours and re-fed with fresh media with 2 µg/ml puromycin for mouse lines and 1 µg/ml puromycin for human lines until uninfected control cells were completely eliminated.
RNA purification and RT-qPCR
Total RNA was extracted from cultured mouse or human cells, or mouse tumor tissues with the RNeasy Plus Mini Kit (Qiagen 74134). cDNA was obtained using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems 4368814) according to the manufacturer’s instruction. RT-qPCR was performed with CFX96 (BioRad) using Fast SYBR Green Master Mix (Applied Biosystems 43856414). All values were normalized to the level of mouse Gapdh or human 18s abundance. Data were the average of triplicate experiments ± SEM. All primers used are described in Supplementary Table S1.
Results
CD34− cells were enriched in BRAFi-resistant melanoma cells.
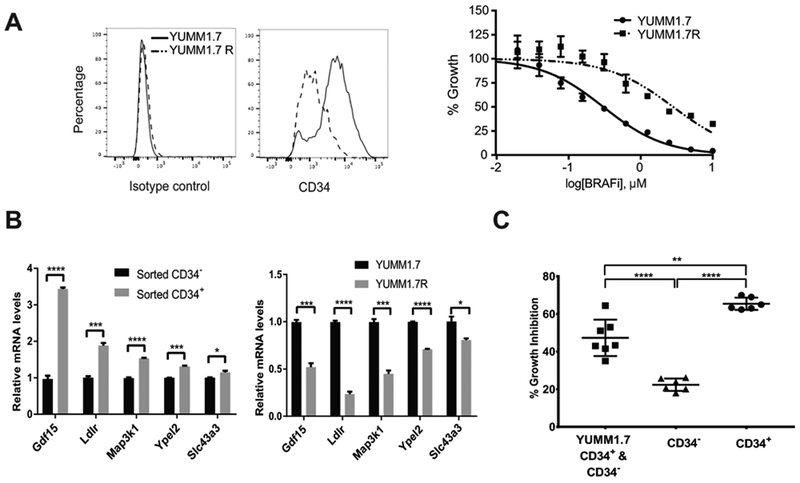
Several mouse melanoma cell lines known as YUMMs were recently derived from genetically engineered mouse models (GEMM) with relevant human driver mutations (18). By flow cytometry, we identified both CD34+ and CD34− MPC subpopulations at various percentage breakdowns in these cell lines across different genotypes (Supplementary Table S2). As a result, YUMMs served as an ideal experimental model to study potential epigenetic differences between subpopulations and to identify key factors that regulate intratumor heterogeneity. In YUMM1.7R cells, a BRAFi-resistant mouse melanoma line (BRAFi IC50=2.8 μM) derived from BRAFi-sensitive YUMM1.7 cells (BRAFi IC50=0.3 μM), we detected a major shift in CD34 positivity compared to parental YUMM1.7 cells by flow cytometry (Fig. 1A). Specifically, YUMM1.7R was composed of mostly CD34− cells whereas CD34+ cells existed at a higher percentage in parental YUMM1.7. It is worth noting that YUMM1.7 cells do not have p75+ subpopulations as in some other cell lines (Supplementary Table S2). To further compare the properties of these cells, we examined the expression of BRAFV600E-responsive genes Gdf15, Ldlr, Map3k1, Ypel2, and Slc43a3, which are highly expressed in BRAFi-sensitive human melanoma cell lines and are downregulated by BRAFi treatment (29). We found that, in comparison to sorted CD34− cells, sorted CD34+ cells exhibited significantly higher mRNA levels of these BRAFV600E-responsive genes (Fig. 1B, left panel). Consistently, these BRAF-responsive genes are expressed at higher levels in YUMM1.7 cells than in YUMM1.7R cells (Fig.1B, right panel), suggesting that the resistant cells adapted to BRAFi treatment by dampening the expression of these genes. Sorted CD34+ cells also demonstrated stronger growth inhibition by BRAFi treatment than CD34− or unsorted YUMM1.7 cells consisting of both CD34+ and CD34− cells (Fig. 1C). These data suggest that CD34+ and CD34− subpopulations are intrinsically distinct in response to targeted BRAF inhibition, in addition to their known differences in tumorigenicity, ability to recapitulate heterogeneity, and chemo-resistance (8). Furthermore, these findings suggest that mouse melanoma cells undergo changes upon exposure to drug treatment that alter intratumor heterogeneity and shift subpopulations towards a more drug tolerant state.
Figure 1.
CD34− cells were enriched in BRAFi-resistant melanoma cells. (A) Compared to BRAFi-sensitive YUMM1.7 (IC50=0.3 μM), BRAFi-resistant YUMM1.7R (IC50=2.8 μM) had increased CD34− cells. Shown are flow cytometry plot (left panel) and dose response curve of YUMM1.7 and YUMM1.7R cells to BRAFi (right panel). Isotype control, IgG2aκ. (B) Expression of BRAFV600E–responsive genes in different cell populations. Left panel, sorted CD34+ YUMM1.7 cells demonstrated higher expression of BRAFV600E–responsive genes than sorted CD34− YUMM1.7 cells. Right panel, YUMM1.7 cells expressed higher levels of BRAFV600E-responsiveness genes than YUMM1.7R cells (*, p≤0.05; ***, p≤0.001; ****, p≤0.0001). (C) Treatment with 1.5 μM BRAFi for three days led to more growth inhibition of sorted CD34+ cells than CD34− cells, as well as unsorted YUMM1.7 cells consisting of both subpopulations (**, p≤0.01; ****, p≤0.0001).
BRAFi induced reversible subpopulation shifting from CD34+ to CD34− cells.
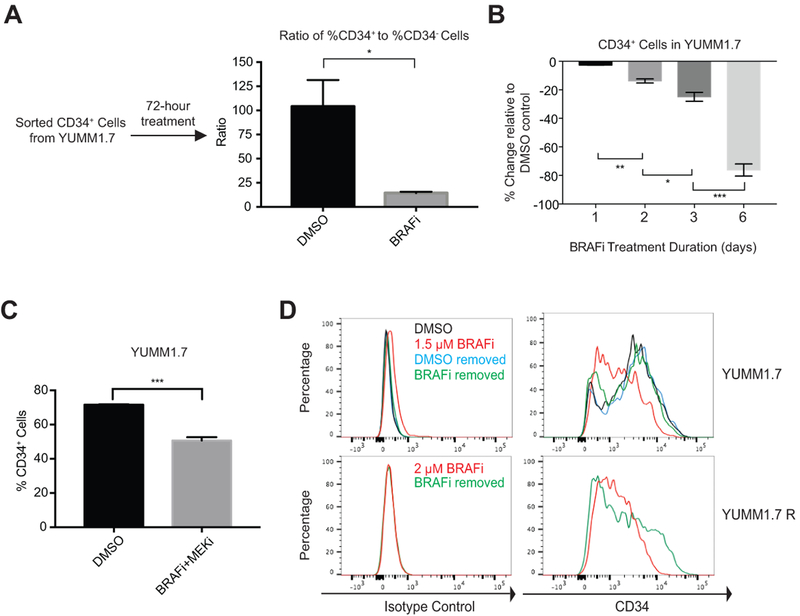
To evaluate the effect of BRAFi on subpopulation composition in YUMMs, we treated sorted CD34+ cells with either DMSO or 1.5 μM BRAFi for three days and subjected the cells to flow cytometry analysis (Fig. 2A). As described previously (8), sorted CD34+ cells treated with DMSO vehicle mainly remained CD34 positive (Supplementary Fig. S1A). In contrast, BRAFi induced emergence of a small subpopulation of CD34− cells from sorted CD34+ cells, as shown by the reduced ratio of %CD34+ to %CD34− cells (Fig. 2A and Supplementary Fig. S1A). Consistently, in parental YUMM1.7 cells, in a treatment time course from one to six days, the degree of subpopulation shifting from CD34+ towards CD34− increased (Fig. 2B). To examine whether this was not just a genotype-specific phenomenon, we conducted similar experiment using YUMM3.3 cells. Unlike YUMM1.7 cells (BrafV600EPten−/−Cdkn2a−/−), YUMM3.3 cells (BrafV600ECdkn2a−/−) carry wild type Pten. Despite the difference in their genotypes, YUMM3.3. cells showed a similar shifting from CD34+ to CD34− cells after three days of 1.5 μM BRAFi treatment (Supplementary Fig. S1B). Furthermore, we treated YUMM1.7 cells with a combination of 1.5 μM BRAFi and MEKi for three days and observed a comparable degree of CD34+ to CD34− cell shifting compared to BRAFi treatment alone (Fig. 2B and C). These data suggest that BRAFi alone is sufficient to induce changes in MPC subpopulations. To investigate whether subpopulation shifting is stable, we withdrew BRAFi from treated YUMM1.7 cells after three days of treatment. The percentage of CD34+ cells was restored in both YUMM1.7 and YUMM1.7R cells after withdrawal of BRAFi for six days (Fig. 2D). These data strongly indicate that BRAFi-induced subpopulation shifting between CD34+ and CD34− cells is reversible within a short period of time and therefore likely caused by epigenetic changes. In addition, propidium iodide staining revealed that BRAFi treatment or long-term exposure to BRAFi did not induce significant cell death in both YUMM1.7 and YUMM1.7R cells, respectively (Supplementary Fig. S1C-D), suggesting that the rise of CD34− subpopulation was not driven by selective pressure.
Figure 2.
BRAFi induced reversible subpopulation shifting from CD34+ to CD34− cells. (A) The ratio of %CD34+ to %CD34− cells in sorted CD34+ cells decreased significantly after three days of 1.5 μM BRAFi treatment (*, p ≤0.05). (B) The degree of subpopulation shifting from CD34+ to CD34− cells with 1.5 μM BRAFi increased over time (*, p≤0.05; **, p≤0.01; ***, p≤0.001). (C) Combined treatment of BRAFi and MEKi (1.5 μM each) led to subpopulation shifting from CD34+ to CD34− cells after three days (***, p≤0.001). (D) Withdrawal of 1.5 μM BRAFi from YUMM1.7 cells after three days of treatment reversed the CD34+ to CD34− subpopulation shifting phenotype at day 6. Withdrawal of 2 μM BRAFi media from YUMM1.7R cells for six days partially restored the CD34+ subpopulation (bottom). Isotype control, IgG2aκ.
BRAFi induced reversible upregulation of KDM5B
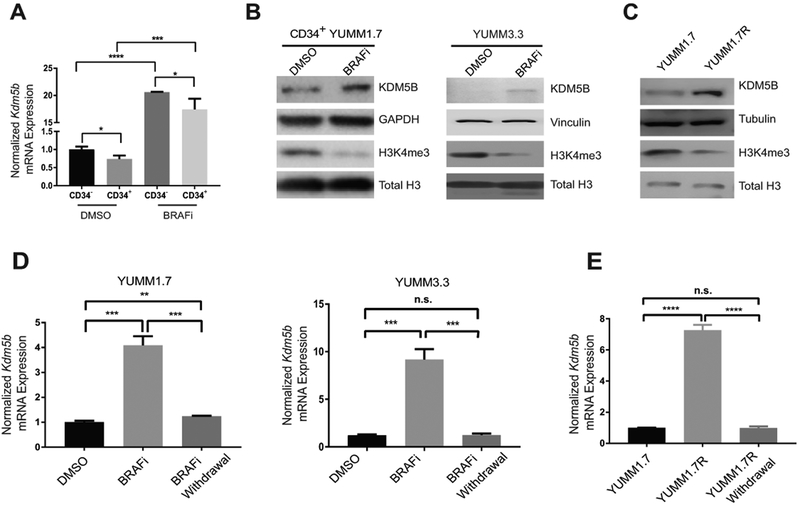
A small subpopulation of slow-cycling, KDM5B (JARID1B)high human melanoma cells was shown to confer intrinsic multi-drug resistance (12). To determine whether KDM5B level is associated with drug resistance and varies between CD34+ and CD34− cells, we treated YUMM1.7 cells for three days with 1.5 μM BRAFi or DMSO control, sorted CD34+ and CD34− subpopulations from YUMM1.7 cells and evaluated Kdm5b expression in these cells. After BRAFi treatment, Kdm5b mRNA level significantly increased in both subpopulations (Fig. 3A). Consistently, KDM5B protein level increased significantly under BRAFi treatment in CD34+ YUMM1.7 cells (Fig. 3B, left panel) and in YUMM3.3 cells (Fig. 3B, right panel). Moreover, KDM5B protein level is significantly higher in YUMM1.7R cells than that in YUMM1.7 cells (Fig. 3C). Although we also noticed that Kdm5b was expressed at a slightly higher level in CD34− cells than in CD34+ cells (Fig. 3A), this small difference is unlikely to be biologically important. These results suggested that BRAFi induced KDM5B expression is not simply due to subpopulation switching. To investigate the effects of upregulated KDM5B, we compared the levels of KDM5B substrate H3K4me3 in DMSO control and 1.5 μM BRAFi-treated cells. We found that BRAFi-treated cells had a lower level of H3K4me3 than DMSO control cells, consistent with the increased expression of KDM5B after BRAFi treatment (Fig. 3B). Similarly, the increased KDM5B level in YUMM1.7R cells compared to YUMM1.7 cells correlated with decreased H3K4me3 level (Fig. 3C). We further demonstrated that withdrawing BRAFi for six days from treated YUMM1.7 and YUMM3.3 (Fig. 3D) and BRAFi resistant YUMM1.7R (Fig. 3E) significantly lowered their Kdm5b mRNA levels by RT-qPCR, suggesting that induction of KDM5B expression by BRAFi is reversible. Taken together, these results suggest that KDM5B plays an important role in regulating BRAFi-induced subpopulation switching.
Figure 3.
BRAFi induced reversible upregulation of KDM5B. (A) BRAFi induced upregulation of Kdm5b mRNA level in both CD34+ and CD34− cells sorted after three days of BRAFi treatment. (*, p≤0.05; ***, p≤0.001; ****, p≤0.0001). (B) KDM5B protein level was upregulated in established clone of CD34+ YUMM1.7 cells (left panel) and unsorted YUMM3.3 cells (right panel) by treatment with 1.5 μM BRAFi for three days. H3K4me3 levels decreased correspondingly in these BRAFi-treated cells. (C) YUMM1.7R cells cultured in 2 μM BRAFi media expressed high levels of KDM5B and had lower level of H3K4me3 than YUMM1.7 cells. (D) Withdrawing 1.5 μM BRAFi from YUMM1.7 and YUMM3.3 after three days of treatment and culturing them in regular DMEM/F12 media with vehicle control DMSO for additional six days restored the Kdm5b level to the baseline level (**, p≤0.01; ***, p≤0.001; n.s., not significant). (E) Replacement of 2 μM BRAFi media with regular DMEM/F12 media with vehicle control DMSO in YUMM1.7R for six days reduced Kdm5b level (****, p≤0.0001; n.s., not significant).
KDM5B regulated the transition between CD34+ and CD34− subpopulations.
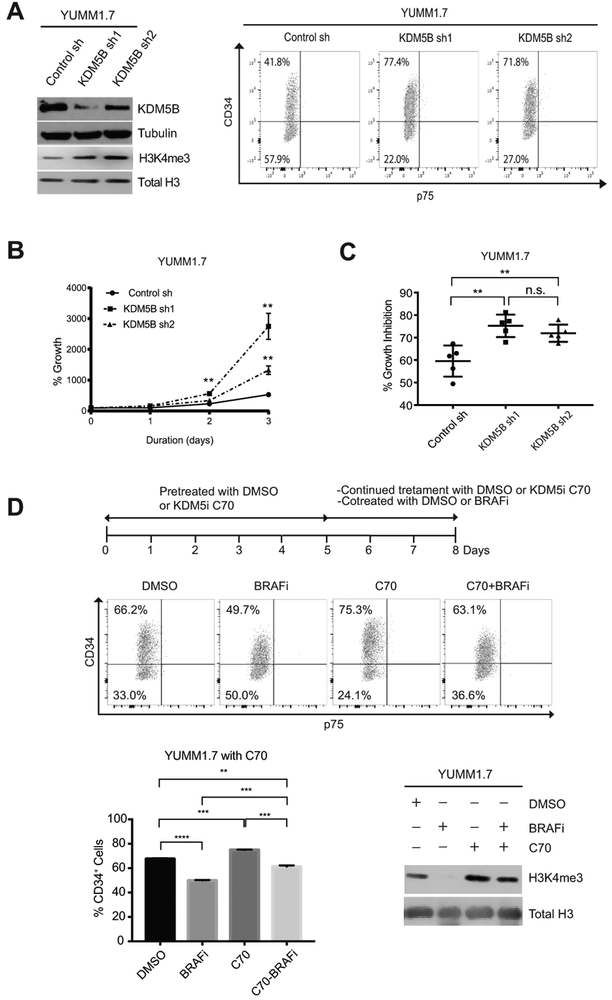
To further investigate whether KDM5B is important in changing MPC subpopulation switching in melanoma, we generated KDM5B knockdown derivatives in different YUMM cell lines. By flow cytometry, we observed an approximately 1.8-fold increase in the percentage of CD34+ subpopulation in YUMM1.7 cells with KDM5B shRNA-mediated knockdown (77.4% or 71.8% vs 41.8%) (Fig. 4A). The percentage of CD34− cells in the KDM5B knockdown cell lines decreased correspondingly, with no emergence of p75+ cells (Fig. 4A). Similarly, knocking down KDM5B in YUMM3.4 cells also led to an increase in the percentage of CD34+ cells (Supplementary Fig. S2A). Since CD34+ and CD34− cells exhibit different cellular features in terms of proliferation and sensitivity to BRAFi, we examined whether these characteristics changed in the KDM5B knockdown cells as a result of subpopulation shifting. Indeed, compared to control shRNA cells, YUMM1.7 cells with KDM5B knockdown exhibited significantly faster cell proliferation (Fig. 4B) and increased sensitivity to BRAFi, shown by with higher growth inhibition after three days of 1.5 μM BRAFi treatment (Fig. 4C, and Supplementary Fig. S2B).
Figure 4.
KDM5B regulated the transition between CD34+ and CD34− subpopulations. (A) Western blot (left) and FACS (right) analyses of YUMM1.7 cells with the indicated shRNAs. shRNA knockdown of KDM5B in YUMM1.7 led to an increase of the percentage of CD34+ cells (right). (B-C). Loss of KDM5B in YUMM1.7 enhanced CD34+-like cellular features, faster proliferation (B) and higher BRAFi sensitivity (C) (**, p≤0.01; n.s., not significant). (D) 1 μM pan-KDM5i C70 induced a consistent phenotype in YUMM1.7 as knockdown of KDM5B. A combined treatment of 1 μM C70 and 1.5 μM BRAFi weakened the subpopulation shifting effect observed with 1.5 μM BRAFi treatment alone for three days (**, p≤0.01; ***, p≤0.001; ****, p≤0.0001; n.s., not significant) (top and left bottom). Western blotting (right bottom) showed that in YUMM1.7 cells, 1.5 μM BRAFi treatment decreased H3K4me3 level, 1 μM KDM5i C70 increased H3K4me3 level and reversed the effects of 1.5 μM BRAFi treatment.
We then asked whether the histone demethylase activity of KDM5B contributed to the subpopulation transition. As no KDM5B specific inhibitor is current available, we examined the effects of pan-KDM5 enzymatic inhibitors (KDM5i) KDM5-C70 (now referred to as C70) (22, 23), CPI-48 (24, 30), which inhibits KDM5A-D, and a KDM5A/C inhibitor YUKA1 (11). We pretreated YUMM1.7 cells with 1 μM KDM5i C70 for five days before subjecting them to BRAFi or combined BRAFi and KDM5i treatment. On the 8th day, we harvested all cells and examined subpopulation changes by flow cytometry (Fig. 4D). Consistent with the phenotype induced by KDM5B knockdown on YUMM1.7 and YUMM3.4 cells, KDM5i C70 increased the percentage of CD34+ cells of YUMM1.7 cells. Furthermore, KDM5i C70 blocked BRAFi-induced decrease of CD34+ cells (Fig. 4D). Treatment of YUMM1.7 cells with 1 μM CPI-48, another pan-KDM5i delivered consistent results as KDM5i C70 (Supplementary Fig. S2C). In contrast, pre-treatment with 100 μM YUKA1, a KDM5A/C inhibitor, had no effect on YUMM1.7 CD34+ and CD34− subpopulations and did not affect the BRAFi-induced phenotype (Supplementary Fig. S2C). Western blotting analyses showed that C70 and CPI-48 induced dramatic induction of the H3K4me3 levels, while the effect of YUKA1 was more modest, suggesting that KDM5B is the dominant H3K4me3 demethylase in these melanoma cells (Fig. 4D and Supplementary Fig S2C). Moreover, KDM5i C70 blocked the reduction of H3K4me3 level by BRAFi (Fig. 4D). These data suggest that the enzymatic activity of KDM5B is critical for the subpopulation transition between CD34+ and CD34− MPCs.
KDM5B modulates MPC-like features of human melanoma cells.
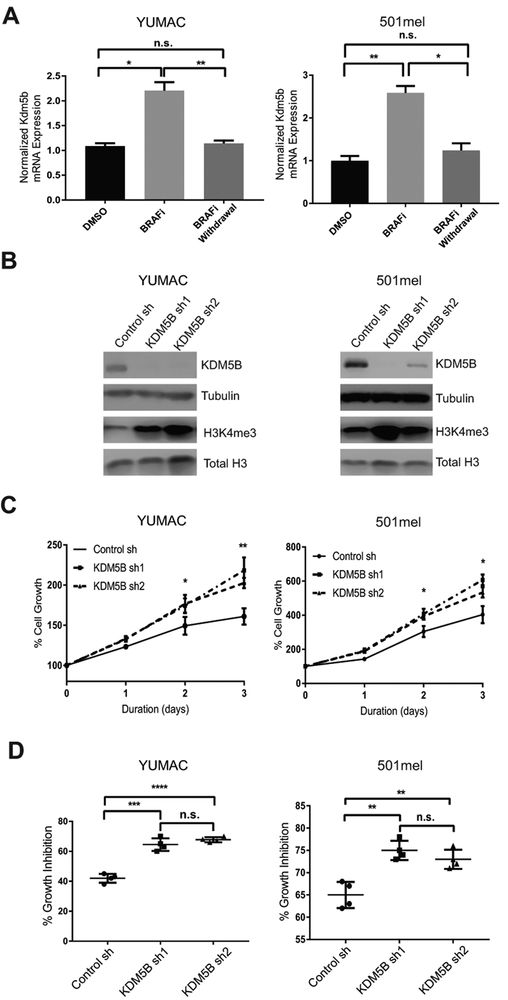
To investigate whether KDM5B plays a similar role in regulating tumor cell behaviors in human melanoma, we examined the levels of KDM5B in YUMAC and 501mel human melanoma cells treated with DMSO control, 1.5 μM BRAFi for three days, or 1.5 μM BRAFi for three days followed by six days of BRAFi withdrawal. Similar to the YUMM cells, KDM5B levels increased significantly in both YUMAC and 501mel cells after BRAFi treatment and then decreased to DMSO control level after BRAFi removal (Fig. 5A). Furthermore, knockdown of KDM5B with two independent shRNA hairpins in 501mel and YUMAC significantly increased H3K4me3 level (Fig. 5B), cell proliferation (Fig. 5C), and growth inhibition by 1.5 μM BRAFi (Fig. 5D). In mouse melanoma, enhanced cell proliferation and sensitivity to BRAFi are characteristics unique to CD34+ cells as opposed to CD34− cells. Here we demonstrate that KDM5B loss in human melanoma cell lines also leads to phenotypic changes towards a cellular state that resembles that of the CD34+ subpopulation in mouse melanoma.
Figure 5.
KDM5B modulated MPC-like features of human melanoma cells. (A) After three days of 1.5 μM BRAFi treatment, withdrawing the drug from YUMAC and 501mel cells and culturing them in regular OPTI-MEM with vehicle DMSO for additional six days restored their KDM5B mRNA levels closer to the DMSO baseline(*, p≤0.05; **, p≤0.01, n.s., not significant). (B) Western blot analysis of YUMAC and 501mel cells with the indicated shRNAs. KDM5B knockdown increased H3K4me3 levels in both YUMAC and 501mel cells. (C) Cell growth assays of YUMAC and 501mel cells with indicated shRNAs. KDM5B knockdown caused faster proliferation (*, p≤0.05; **, p≤0.01). (D) YUMAC and 501mel cells with KDM5B shRNAs showed higher growth inhibition by three days of 1.5 μM BRAFi treatment than shRNA control cells (**, p≤0.01; ****, p≤0.000, n.s., not significant ).
In vivo BRAFi therapy triggers reversible MPC subpopulation changes.
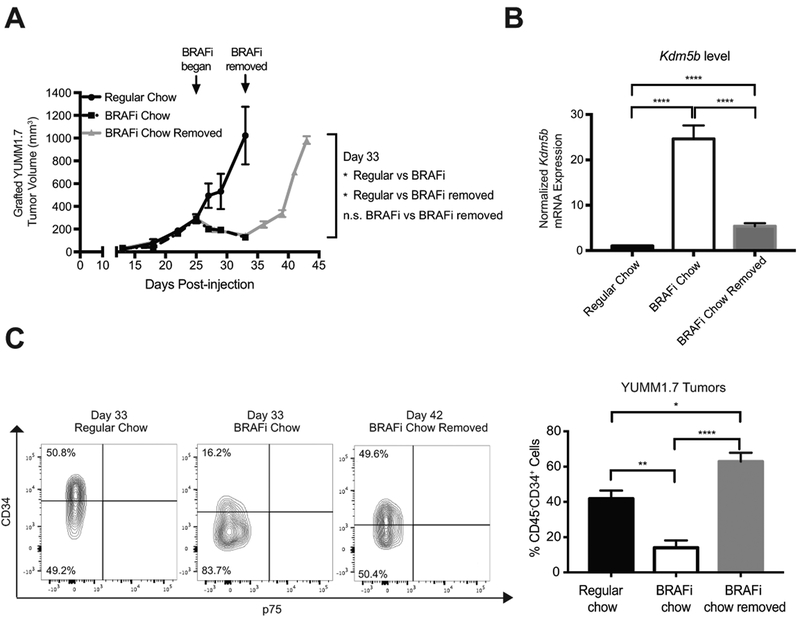
To evaluate how heterogeneous subpopulations of melanoma cells respond to BRAFi in vivo, we engrafted 100,000 YUMM1.7 cells subcutaneously in C57BL/6J mice. When tumors reached a volume of 300 mm3 (Day 25), these mice were provided with BRAFi or regular chow diet. Tumor growth curves revealed that grafted YUMM1.7 tumors responded to BRAFi chow (Fig. 6A). At day 33, replaced BRAFi chow with regular chow for some BRAFi treated mice, and we call this group of mice “BRAFi chow removed” group. Tumors re-grew in these mice and reached 1,000 mm3 at day 42, which allowed us to harvest tumors of similar size to those in the “regular chow” group on day 33. We then evaluated the tumors for Kdm5b level and percentage of CD34+ cells in tumors for “regular chow” group on day 33, “BRAFi chow” group on day 33, and “BRAFi chow removed” group on day 42. Using RT-qPCR, we observed an increased Kdm5b level in tumors collected on day 33 from BRAFi-treated mice compared to that in tumors from mock treated mice, while this increase was partially reversed in tumors harvested on day 42 from mice with BRAFi removed (Fig. 6B). This pattern of fluctuating Kdm5b expression in grafted tumors was consistent with our observation in vitro. We analyzed these tumors by flow cytometry for CD45− cells to exclude hematopoietic cells and enrich for tumor cells. We demonstrated a reduced percentage of CD45−CD34+ cells in BRAFi-treated tumors compared with tumors that received regular chow (Fig. 6C). Tumors that grew out after BRAFi chow removal exhibited a higher percentage of CD45−CD34+ cells than BRAFi-treated tumors on day 33 (Fig. 6C). In addition, we examined the subpopulation composition of several small “baseline” tumors from day 25 before providing any BRAFi chow to exclude the possibility that the difference observed between “regular chow” and “BRAFi chow” was due to changes in tumor volume. Indeed, there was no significant difference in the percentage of CD45−CD34+ cells between “baseline” and “regular chow” tumors, suggesting that tumor size does not affect the percentage of CD45−CD34+ cells (Supplementary Fig. S3A). Furthermore, combined treatment of BRAFi and MEKi chow in grafted YUMM1.7 tumors led to an increase in Kdm5b expression (Supplementary Fig. S4A-B) and a reduced percentage of CD45−CD34+ cells, when compared with tumors from the control group (Supplementary Fig. S4C). These results indicate that in vivo tumor MPC subpopulations respond to BRAFi or BRAFi/MEKi therapies similarly to in vitro cultured cells by altering KDM5B expression in a reversible manner.
Figure 6.
In vivo BRAFi therapy triggered reversible MPC subpopulation changes. (A) Subcutaneously grafted YUMM1.7 tumors in C57BL/6J mice exhibited a sensitive drug response to BRAFi chow (n=4 to 6 per treatment condition). Arrows indicated that BRAFi chow was provided to mice in the treatment group on day 25 and removed on day 33. BRAFi-treated tumors were harvested on day 33 and BRAFi-removed tumors were harvested on day 42. (B) Kdm5b mRNA level increased in BRAFi treated-tumors and was reduced closer to the untreated tumors nine days after treatment removal (****, p≤0.0001). (C) The percentage of CD34− tumor cells increased after eight days of BRAFi chow treatment (*, p≤0.05; **, p≤0.01; ****, p≤0.0001). Shown are flow cytometry plots (left) and quantification of flow cytometry analysis (right). Flow cytometry analysis gated only live, non-immune cells with negative propidium iodide and CD45 staining.
Discussion
In mouse melanoma, while both CD34+ and CD34− cells are referred to as MPCs, the two subpopulations differ significantly in their tumorigenic potential, in addition to several other critical cellular features, such as cell proliferation, resistance to chemotherapy, and ability to recapitulate heterogeneity (8). Specifically, CD34− cells are capable of re-establishing cellular heterogeneity both in vitro and in vivo whereas CD34+ cells mainly produce CD34+ progeny. In this study, we identify a key regulator that modulates the CD34+ and CD34− MPC transition and shed light on novel therapeutic strategies that exploit the heterogeneous nature of melanoma.
YUMM1.7 cells consist of approximately 75% CD34+, 25% CD34− cells, and no p75+ cells (Supplementary Table S2). Despite long-term in vitro culturing and passaging, the subpopulation composition of YUMM1.7 remains relatively consistent. While YUMM3.3 and several other genotypically different YUMM cell lines are composed of varying percentages of CD34+, CD34− and p75+ cells, the size of each subpopulation does not change significantly over time in vitro. Nevertheless, in YUMM1.7R cells, which are derived from parental YUMM1.7 cells, we demonstrate a noticeable increase in the percentage of CD34− cells. These observations suggest that a well-controlled mechanism regulates the conversion of CD34+ and CD34− MPCs which is altered upon exposure to targeted therapies, shifting the subpopulation equilibrium in favor of a more drug tolerant state. Indeed, compared to CD34+ cells, CD34− cells demonstrate lower expression of several BRAFV600E-responsive genes (29) (Fig. 1B), indicating that CD34− cells are likely more resistant to BRAFi treatment. Consistent with these observations, sorted CD34+cells are indeed more sensitive to BRAFi treatment than sorted CD34− and parental YUMM1.7 (Fig. 1C).
Of the MPC subpopulations of CD34+ and CD34− cells, it is previously suggested that only the CD34− subpopulation is capable of generating all subpopulations (8). Here, we provide evidence that under certain circumstances, CD34+ cells can re-establish intratumor heterogeneity under drug treatment (Fig. 2A and B). Our findings are also consistent with several previous studies that have shown stressful exposures like drug treatment could trigger phenotype switching in tumor cells as a transient defense mechanism (20, 31, 32). We observed that subpopulation shifting from CD34+ to CD34− of YUMM1.7 cells occurs as early as one day after the initial BRAFi treatment (Fig. 2B). Moreover, the more drug-tolerant CD34− subpopulation expands under BRAFi exposure and the degree of subpopulation shifting intensifies as treatment prolongs (Fig. 2B). Subpopulation shifting from CD34+ to CD34− MPCs also occurs in BRAFi/MEKi co-treated YUMM1.7 cells (Fig. 2C), suggesting that the subpopulation shifting phenotype is not limited to BRAFi monotherapy and has broader clinical implications, as combined BRAFi and MEKi treatment is becoming the standard of care (33, 34). Similar to the findings in recent studies (35, 36), we observed reversibility in subpopulation shifting upon BRAFi withdrawal. The reversible phenotype applies to both BRAFi-sensitive YUMM1.7 and YUMM3.3 cells, and BRAFi-resistant YUMM1.7R cells, suggesting that BRAFi-resistance of YUMM1.7R cells is reversibly regulated.
Furthermore, we provide evidence that increase of the percentage of CD34− cells is not driven by selective pressure due to BRAFi-induced cell death, as there is no significant cell death after BRAFi treatments. It is important to note that CD34+ cells actually exhibit greater survival in the presence of chemotherapeutic agents such as cisplatin and temozolomide than CD34− cells and are the main subpopulation present after treatment (8). In contrast to BRAFi, both drugs are cytotoxic and induce excessive cell death. Therefore, loss of CD34− cells after chemotherapy is likely a consequence of selection of CD34+ cells rather than subpopulation shifting. MPC subpopulations’ distinct responses to different categories of therapeutic drugs highlights a potential benefit of combining cytotoxic chemotherapies and targeted therapies (37).
Moreover, since shifting of MPC subpopulations in YUMM cells occurs relatively quickly upon initial exposure to BRAFi and is a reversible phenotype, we suspect that epigenetic regulators mediate this transition. In fact, subpopulations enriched with epigenetic regulators such as KDM5A or KDM5B confer intrinsic drug tolerability in cancers including melanoma and non-small cell lung cancer (9, 13, 38). However, it is unclear how these epigenetic regulators are modulated during the development of drug resistance. It has been reported that BRAFi treatment up-regulates MITF/PGC1α signaling in BRAF mutated melanoma cells (39). It is possible that the induced MITF/PGC1α signaling set the stage for the increased KDM5B expression. Our study shows that BRAFi treatment leads to an increase in KDM5B and decrease in H3K4me3, suggesting that induced KDM5B is functional and re-shapes the epigenetic landscape. In addition, we demonstrate that knockdown of KDM5B in YUMM1.7 and YUMM3.4 increases the percentage of CD34+ cells (Fig. 4A and Supplementary Fig. S2A). Compared to shRNA control cells, YUMM1.7 cells with KDM5B shRNAs exhibit increased cell proliferation and higher sensitivity to BRAF inhibition, both of which are the cellular features of CD34+ cells (8). KDM5B loss induced cell proliferation in YUMAC and 501mel human melanoma cells were consistent with previous studies of WM3734, WM3899 and WM35 human melanoma cells (12). Taken together, our results show that KDM5B promotes the switching from CD34+ to CD34− MPCs of YUMM1.7 and YUMM3.4 cells. Further studies with the appropriate mouse melanoma cell lines are necessary to identify and characterize the regulators of transition between MPCs and the non-tumorigenic p75+ subpopulation.
We show that pan-KDM5 inhibitors C70 or CPI-48 mitigate the degree of BRAFi-sensitive CD34+ cells shifting towards more BRAFi-tolerant CD34− cells (Fig. 4D and Supplementary Fig. S2C), while YUKA1, an enzymatic inhibitor specific to KDM5A and KDM5C (11, 40, 41) has minimal impact on subpopulation conversion (Supplementary Fig. S2C). These data suggest that the enzymatic activity of KDM5B plays major roles in subpopulation switching. However, the effects of pan-KDM5i are milder than that of KDM5B knockdown (compare Fig. 4D and Supplementary Fig. S2C with Fig. 4A). Although we cannot fully exclude the possibility that pan-KDM5 inhibitors has broader effects on multiple KDM5 family members than knockdown of only KDM5B, these differences suggest that both histone demethylase catalytic activity-dependent and -independent functions of KDM5B contribute to its regulation of subpopulation shifting. Consistent with this idea, in addition to the JmjC domain that mediates histone demethylation, KDM5 enzymes have several other domains involved in binding specific histone markers or interacting with other chromatin complex (42-44). Nonetheless, the KDM5i treatment tactic has the potential to prevent melanoma cells from transitioning to a more intrinsic drug-tolerant state, limiting the potential for developing acquired resistance. Taken together, these data emphasize the importance of understanding the precise cellular features and roles of different tumor subpopulations and identifying their key regulator(s) to develop novel and effective therapeutic paradigms.
The tumor microenvironment has been reported to influence tumor cell plasticity (45-47). In this study, by using immune-competent C57BL/6J mice grafted with YUMM1.7 cells, we provide evidence that a similar subpopulation shifting phenotype from CD34+ to CD34− tumor cells occurs with BRAFi or combined BRAFi and MEKi treatments in vivo. In addition, treated tumors are composed of a higher percentage of CD34− cells that express elevated levels of Kdm5b. Consistent with our in vitro findings, the subpopulation phenotype is also reversible upon treatment withdrawal and Kdm5b levels fluctuate accordingly. Although we have not studied the interaction between tumor subpopulations and the immune/stromal cells in the microenvironment, it is possible that the more drug-tolerant CD34− population are more immunosuppressive than CD34+ cells in immunocompetent mice. Consistently, studies have shown benefits to combining targeted therapies and immune checkpoint inhibitors for melanoma treatment (48, 49). Therefore, it will be interesting to examine whether the tumor microenvironment plays any role in shifting CD34+ and CD34− subpopulations in vivo. This could offer insight into designing more complex and effective treatment plans.
In summary, our study suggests that KDM5B is a key factor modulating the conversion of CD34+ and CD34− MPC subpopulations. It also demonstrates that by altering expression of KDM5B and therefore the level of H3K4me3, melanoma cells treated with BRAFi or BRAFi/MEKi are capable of shifting towards a more drug-tolerant CD34− state as a defense mechanism. Furthermore, this study suggests that novel combination treatment using pan-KDM5 or KDM5B-specific inhibitors with BRAFi could be used to increase drug sensitivity of tumor cells to BRAFi and thus improve overall therapeutic efficacy. It sheds light on the importance to continue advancing our understanding of intratumor heterogeneity in melanoma and designing more effective treatment paradigms by manipulating the tumor heterogeneity.
Supplementary Material
Acknowledgements
We would like to thank Dr. Ruth Halaban from the Department of Dermatology at Yale University for kindly providing the human melanoma cell lines used in this study. We thank Plexxikon, Inc for providing PLX4720-200ppm and PLX4720/PLX2695-200/7ppm chow. We thank the members of the Bosenberg and Yan laboratories for their helpful discussions and support. We thank the whole KDM5 team from Emory Specialized Center and the National Center for Advancing Translational Sciences (NCATS) in the Chemical Biology Consortium and the University of Texas MD Anderson Cancer Center, especially Dr. Haian Fu, Dr. Xiaodong Cheng, Dr. Paula M. Vertino, Dr. John R. Horton, Ajit Jadhav, Dr. David J. Maloney, Dr. Margaret A. Johns, John R. Shanks, Dr. Philip J. Webber, and Dr. Melinda S. Hanes for their essential contribution to the selection and characterization of KDM5 inhibitors used in this manuscript. We thank Drs. Anthony Welch and Barbara Mroczkowski (National Cancer Institute) and Drs. Andrew Flint and Dane Liston (Frederick National Laboratory for Cancer Research) for administrative support.
These studies were supported in part by grants from by the NIH Yale SPORE in Skin Cancer, NCI P50 CA121974 (to M.W. Bosenberg and Q. Yan), Department of Defense Peer-Reviewed Cancer Research Program Award W81XWH-13-10235 (to Q. Yan) and W81XWH-16-1-0306 (to S-M. Zhang), the Melanoma Research Foundation (to M.W. Bosenberg and Q. Yan), R01 CA196660 (to M.W. Bosenberg), P01 CA128814 (to M.W. Bosenberg and D.P. Kelly), the Melanoma Research Alliance (M.W. Bosenberg), the American Skin Association (M.W. Bosenberg), the Hervey Family Foundation (M.W. Bosenberg), by pilot funding from the Yale Cancer Center, NCI P30 CA16359 (M.W. Bosenberg and Q. Yan), T32CA193200 - Yale Cancer Biology Training Program (to M.K. McGeary), F30 CA228444 (to I. Krykbaeva), and by federal funds from the NCI, NIH, under Chemical Biology Consortium Contract No. HHSN261200800001E, and the intramural research program of the National Center for Advancing Translational Sciences (NCATS), NIH (to D.J. Jansen, S.C. Kales, A. Simeonov, M.D. Hall). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflicts of Interest Statement
M. Bosenberg serves as a consultant for Eli Lilly and Company. Other authors declare no conflicts of interest.
References
- 1.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25:3357–64. [DOI] [PubMed] [Google Scholar]
- 3.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–9. [DOI] [PubMed] [Google Scholar]
- 4.Fukunaga-Kalabis M, Roesch A, Herlyn M. From cancer stem cells to tumor maintenance in melanoma. J Invest Dermatol. 2011;131:1600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292–306. [DOI] [PubMed] [Google Scholar]
- 7.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Held MA, Curley DP, Dankort D, McMahon M, Muthusamy V, Bosenberg MW. Characterization of melanoma cells capable of propagating tumors from a single cell. Cancer research. 2010;70:388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, et al. An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat Chem Biol. 2016;12:531–8. [DOI] [PubMed] [Google Scholar]
- 11.Gale M, Sayegh J, Cao J, Norcia M, Gareiss P, Hoyer D, et al. Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance. Oncotarget. 2016;7:39931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer cell. 2013;23:811–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somasundaram R, Villanueva J, Herlyn M. Intratumoral heterogeneity as a therapy resistance mechanism: role of melanoma subpopulations. Adv Pharmacol. 2012;65:335–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 16.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manzano JL, Layos L, Buges C, Gil M de Los Llanos, Vila L, Martinez-Balibrea E, et al. Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med. 2016;4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meeth K, Wang JX, Micevic G, Damsky W, Bosenberg MW. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment cell & melanoma research. 2016;29:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment cell & melanoma research. 2010;23:190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton JR, Liu X, Gale M, Wu L, Shanks JR, Zhang X, et al. Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem Biol. 2016;23:769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12:539–45. [DOI] [PubMed] [Google Scholar]
- 24.Liang J, Zhang B, Labadie S, Ortwine DF, Vinogradova M, Kiefer JR, et al. Lead optimization of a pyrazolo[1,5-a]pyrimidin-7(4H)-one scaffold to identify potent, selective and orally bioavailable KDM5 inhibitors suitable for in vivo biological studies. Bioorganic & medicinal chemistry letters. 2016;26:4036–41. [DOI] [PubMed] [Google Scholar]
- 25.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorganic & medicinal chemistry letters. 2008;18:6501–4. [DOI] [PubMed] [Google Scholar]
- 27.Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, et al. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. The Journal of biological chemistry. 2013;288:9408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Minamishima YA, Yan Q, Schlisio S, Ebert BL, Zhang X, et al. pVHL acts as an adaptor to promote the inhibitory phosphorylation of the NF-kappaB agonist Card9 by CK2. Mol Cell. 2007;28:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Cao J, Cai WL, Lang SM, Horton JR, Jansen DJ, et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS biology. 2018;16:e2006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemper K, de Goeje PL, Peeper DS, van Amerongen R. Phenotype switching: tumor cell plasticity as a resistance mechanism and target for therapy. Cancer research. 2014;74:5937–41. [DOI] [PubMed] [Google Scholar]
- 32.Menon DR, Das S, Krepler C, Vultur A, Rinner B, Schauer S, et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34:4545. [DOI] [PubMed] [Google Scholar]
- 33.Welsh SJ, Rizos H, Scolyer RA, Long GV. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer. 2016;62:76–85. [DOI] [PubMed] [Google Scholar]
- 34.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371:1877–88. [DOI] [PubMed] [Google Scholar]
- 35.Fallahi-Sichani M, Becker V, Izar B, Baker GJ, Lin JR, Boswell SA, et al. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol Syst Biol. 2017;13:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richard G, Dalle S, Monet MA, Ligier M, Boespflug A, Pommier RM, et al. ZEB1-mediated melanoma cell plasticity enhances resistance to MAPK inhibitors. EMBO Mol Med. 2016;8:1143–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masui K, Gini B, Wykosky J, Zanca C, Mischel PS, Furnari FB, et al. A tale of two approaches: complementary mechanisms of cytotoxic and targeted therapy resistance may inform next-generation cancer treatments. Carcinogenesis. 2013;34:725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn A, Chatterjee A, Eccles MR. The Slow Cycling Phenotype: A Growing Problem for Treatment Resistance in Melanoma. Mol Cancer Ther. 2017;16:1002–9. [DOI] [PubMed] [Google Scholar]
- 39.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer cell. 2013;23:302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Oto A, Tian QW, Davidoff AM, Yang J. Histone demethylases and their roles in cancer epigenetics. J Med Oncol Ther. 2016;1:34–40. [PMC free article] [PubMed] [Google Scholar]
- 41.Blair LP, Cao J, Zou MR, Sayegh J, Yan Q. Epigenetic Regulation by Lysine Demethylase 5 (KDM5) Enzymes in Cancer. Cancers (Basel). 2011;3:1383–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou MR, Cao J, Liu Z, Huh SJ, Polyak K, Yan Q. Histone demethylase jumonji AT-rich interactive domain 1B (JARID1B) controls mammary gland development by regulating key developmental and lineage specification genes. The Journal of biological chemistry. 2014;289:17620–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J, Liu Z, Cheung WK, Zhao M, Chen SY, Chan SW, et al. Histone demethylase RBP2 is critical for breast cancer progression and metastasis. Cell reports. 2014;6:868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein BJ, Piao L, Xi Y, Rincon-Arano H, Rothbart SB, Peng D, et al. The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell reports. 2014;6:325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albini A, Bruno A, Gallo C, Pajardi G, Noonan DM, Dallaglio K. Cancer stem cells and the tumor microenvironment: interplay in tumor heterogeneity. Connect Tissue Res. 2015;56:414–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–34. [DOI] [PubMed] [Google Scholar]
- 47.Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X, et al. Recurrent Tumor Cell-Intrinsic and -Extrinsic Alterations during MAPKi-Induced Melanoma Regression and Early Adaptation. Cancer Discov. 2017;7:1248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu-Lieskovan S, Mok S, Homet Moreno B, Tsoi J, Robert L, Goedert L, et al. Improved antitumor activity of immunotherapy with BRAF and MEK inhibitors in BRAF(V600E) melanoma. Sci Transl Med. 2015;7:279ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, Mayes PA, Eastman S, Shi H, Yadavilli S, Zhang T, et al. The BRAF and MEK Inhibitors Dabrafenib and Trametinib: Effects on Immune Function and in Combination with Immunomodulatory Antibodies Targeting PD-1, PD-L1, and CTLA-4. Clin Cancer Res. 2015;21:1639–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.