Abstract
Purpose:
Presence of a high degree of tumor-infiltrating lymphocytes (TILs) has proven to be associated with outcome in patients with non-small cell lung cancer (NSCLC). However, recent evidence indicate that tissue architecture is also prognostic of disease specific survival and recurrence. We show a set of descriptors (SpaTIL) that capture density and spatial co-localization of TILs and tumor cells across digital images can predict likelihood of recurrence in early-stage NSCLC.
Experimental design:
The association between recurrence in early-stage NSCLC and SpaTIL features was explored on 301 patients across four different cohorts. Cohort D1 (n=70) was used to identify the most prognostic SpaTIL features and to train a classifier to predict the likelihood of recurrence. The classifier performance was evaluated in cohorts D2 (n=119), D3 (n=112), and D4 (n=112). Two pathologists graded each sample of D1 and D2; intra-observer agreement and association between manual grading and likelihood of recurrence were analyzed.
Results:
SpaTIL was associated with likelihood of recurrence in all test sets (log rank p<0.02). A multivariate Cox Proportional Hazards analysis revealed a HR of 3.08 (95% confidence interval, 2.1–4.5, p=7.3×10−5). In contrast, agreement among expert pathologists using tumor grade was moderate (Kappa=0.5), and the manual TIL grading was only prognostic for one reader in D2 (p=8.0×10−3).
Conclusion:
A set of features related to density and spatial architecture of TILs was found to be associated with a likelihood of recurrence of early-stage NSCLC. This information could potentially be used for helping in treatment planning and management of early-stage NSCLC.
Introduction
Early-stage (stages I and II) non-small cell lung cancer (NSCLC) (1,2) is typically treated with complete surgical excision. However, even after resecting the entire tumor mass, 30–55% of patients develop disease recurrence within first 5 years following surgery (3). The ability to identify patients who are at a higher risk of recurrence could help selecting those patients who may gain maximum benefit with further treatment including adjuvant chemotherapy following standard of care surgery.
Non-small cell lung cancer histopathology is characterized by a complex interplay of tumor cells, immune cells (lymphocytes, plasma cells, macrophages, and granulocytes), fibroblasts, and pericytes/endothelial cells. Recent evidence suggests that the interaction of tumor cells with immune cells has a high association with likelihood of disease progression and influences tumor development, invasion, metastasis, and patient outcome (1,2). Several independent studies (4,5,6,2) meanwhile have also shown an association between patient survival and treatment response with an increased density of Tumor-Infiltrating Lymphocytes (TILs) in diverse solid tumor types. Additionally, there is substantial evidence supporting the fact that increased TIL density is associated with better chemotherapeutic response (7,8,9,10).
Studies have also found substantial inter-reader variability in estimating TIL density using Hematoxylin & Eosin (H&E) stained slides. This has limited the routine use of TIL density as a metric in the clinic as a prognostic marker for NSCLC. Brambilla et al. (6), for instance, determined that inter-pathologist agreement was at best moderate (Kappa = 0.59) in quantifying TILs on tissue slides. While attempts have been made to establish guidelines for standardizing TIL grading in breast cancer (11), these efforts have been lagging in lung cancer.
Over the last few years there has been increasing interest in developing automated cell segmentation and detection algorithms for identifying and quantifying the extent of TILs from routine H&E pathology slide images (12,13,14). These approaches can be broadly categorized into a) involving extraction of visual features (12), b) employing morphological operations (15,16), and c) using Deep Learning based models (17). However, these approaches have primarily focused on either counting the individual TILs (13,14) or estimating the TIL grade, typically as being low, moderate, or high (12,15,16).
Interestingly, there has also been a recent surge in looking at spatial patterns of TILs and investigating their relationship with disease outcome. Multiplexed quantitative immunofluorescence (QIF) and immunohistochemistry (IHC) based methods have been employed for objectively identifying TIL subtypes and correlate the spatial arrangement and density of these TIL subtypes with overall survival (OS) in NSCLC (4,18). For instance, Schalper et al. (4) found out that increased levels of CD3 and CD8 TILs were significantly associated with improved 5-year OS. Similarly, Barua et al. (18) showed that spatial interplay between tumor and regulatory T cells was associated with OS in NSCLC. Furthermore, Liu et al. (19) demonstrated that presence of CD8+ and FOXP3+ TILs was correlated with the response of platinum-based neoadjuvant chemotherapy in advanced NSCLC.
Interestingly, computer extracted features of spatial patterns and morphologic attributes of TILs from routine H&E slides also appeared to be prognostic. Basavanhally et al. (15) explored the use of graph network algorithms to spatially characterize the arrangement of machine identified TILs in HER2+ breast cancer H&E images to predict TIL grade (i.e. high or low). Yu et al. (20) and Luo et al. (21) extracted quantitative morphological features of nuclei and the surrounding cytoplasm from H&E tissue images of early-stage NSCLC patients (e.g., area, shape, intensity, texture, density) for predicting survival. In (22), Saltz et al. used a deep learning model to identify patches of TILs in images, which were subsequently clustered using different similarity metrics. From such patch clusters, different indices were computed (Ball and Hall, Banfield and Raftery, C, determinant ratio, etc.) which was then found to be correlated with patient survival across different tumor types.
Given the recent evidence that the co-localization of immune and cancer nuclei is prognostic of disease outcome (23,20,22,21), this work aims to develop and evaluate new computer-extracted spatial TIL (SpaTIL) features relating to 1) the spatial architecture of TIL clusters, 2) co-localization of clusters of both TILs and cancer nuclei, and 3) variation in density of TIL clusters across the tissue slide image. Specifically, our goal was to evaluate the association between disease recurrence and the SpaTIL features on patients with stage I and II of non-small cell lung cancer (NSCLC). Additionally, we also sought to compare the SpaTIL features in terms of their ability to predict recurrence in these patients with NSCLC against the manually estimated degree of TILs by two thoracic pathologists.
Materials and methods
Datasets
Tissue microarrays (TMAs) obtained from H&E-stained slides collated from three independent and well-characterized early-stage NSCLC cohorts in were included in this study, representing a total of n=301 patients. The three cohorts are represented by D1 (n=70), D2 (n=119), and D3 (n=112). A fourth dataset, named D4 (n=112), was also included, containing tissue cores corresponding to the same patients in D3 but extracted from different regions of the tumor. The corresponding clinico-pathological and outcome information from patients in D1-D4 was obtained from retrospective chart review from the institutions at which the datasets were collated after obtaining the respective IRB approval.
Patients from D1 and D2 comprised formalin-fixed paraffin-embedded (FFPE) tumor sections from previously reported retrospective collections of NSCLC patients (4). 0.6-mm cores from each tumor were then arrayed to make up the TMA. A further 116 patients provided two cores from the same tumor, to make up TMA datasets D3 and D4. The standard TMA preparation protocol is described in (24). D1 and D2 were scanned and digitized using an Aperio Scanscope CS whole slide imager at 20× magnification. D3 and D4 were scanned and digitized at 20× using a Ventana iScan HT Scanner (serial #: BI15N7205). Finally, a 1500×1500 pixel image at 20× magnification was extracted and used to represent a unique tumor sample derived from each patient D1 was employed for feature discovery and model training. This dataset included samples from 350 patients and was collected independently at Sotiria General Hospital and Patras University General Hospital between 1991 and 2001. D2 and D3 were used for independently validating the trained classifier. D2 comprised samples from 202 patients and was collected at Yale Pathology between 1988 and 2003. D3 and D4 (tissue cores corresponding to same patients in D3 but from a different portion of the tumor) comprised tissue images from 189 patients and was collected at the Cleveland Clinic between 2004 and 2014. D3 and D4 were used to quantitatively assess the ability of the approach to deal with intra-tumoral heterogeneity. Figure S1 illustrates the inclusion and exclusion criteria for patient selection for this study.
Automatic characterization of TILs
Identification of Lymphocytes
The first step in the process was to identify the spatial location of the TILs on digitized H&E images. A watershed-based algorithm (25) was used for automatically detecting the nuclei. This method applies a set of mathematical operations (fast radial symmetry transform and regional minima) at different scales to identify candidate locations for nuclei. This method was chosen based off its documented advantages, including simplicity, speed, and the ease with which parameters can be adjusted and fine-tuned.
Once nuclei were detected, the approach described in (26) was used to distinguish lymphocytes from non-lymphocytes. This approach takes advantage of the fact that TILs tend to be smaller compared to cancerous nuclei, they also tend to be more rounded and with a darker, more homogeneous staining. Once all the candidate nuclei were identified via the watershed approach described above, a machine classifier using 7 image derived features related to texture, shape, and color attributes of the segmented nuclei was used to classify the individual nuclei as corresponding either to TILs or non-TILs (See Figures 1-b and 1-e).
Figure 1.
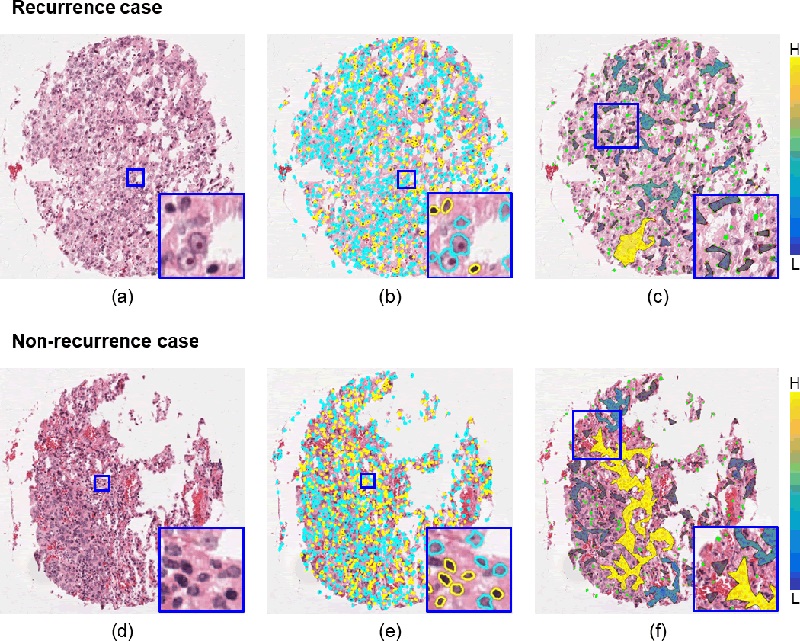
Representative TMA tissue spots of recurrent (top row) and non-recurrent (bottom row) early-stage NSCLC cases. The first column (a, d) shows the original H&E-stained images. Identification of TILs (yellow) and non-TILs (cyan) is presented in the second column (b, e). The third column (c, f) illustrates the qualitative representation of one of the SpaTIL features overlaid on the original images, specifically, the variation in the density of lymphocyte clusters. The color bars represent the density measurement (H stands for highly dense clusters while L stands for low-density or sparse clusters). Non-recurrence cases are characterized by the presence of more high-density clusters while recurrence cases were comprised of a larger number of low-density clusters.
Quantitative evaluation of spatial arrangement of TILs
Spatial TIL Graph Construction
A graph is a mathematical construct comprising of a finite sets of objects (nodes) that capture global and local relationships via pairwise connections (edges) between the nodes. Graphs have been previously used to quantitatively characterize nuclear architecture in histopathologic images by representing the nuclei as nodes and subsequently quantifying neighborhood relationships (e.g., proximity) and spatial arrangement between the nodes (15,27,28)
In order to evaluate a spatial network of TILs and to extract the corresponding SpaTIL features, we first identified sets of clusters of proximal TILs and non-TILs respectively. We represented the centroids of each of the individual TILs and non-TILs as nodes of a graph. Using the approach described in (27,28), each node was connected to others based off the weighted Euclidean norm where a weighting function favors the connectivity between proximal nodes. Utilizing this process resulted in multiple disconnected subgraphs or clusters of TILs being generated. This process was also repeated separately for all the non-TILs.
SpaTIL features
We extracted two separate sets of SpaTIL features. The first set comprised of 20 features related to spatial arrangement of TILs, extracted from the TIL cluster graphs. These features included first-order statistics (e.g. mean, mode, median) of the following representative descriptors: number of lymphocytes within the clusters, ratio between the area of the TIL clusters and area of the TMA spot, ratio between the numbers of TILs within the cluster and the cluster area. The second set included 65 features describing the relationship between TIL and non-TIL clusters extracted for each image. These included the ratio between the density (ratio between the number of nuclei within the cluster and the cluster area) of a non-lymphocyte cluster and the density of its closest lymphocyte cluster, the intersecting areas of the lymphocyte and non-lymphocyte clusters, a value indicating if the nearest neighbor of a lymphocyte cluster is either a lymphocyte or a non-lymphocyte cluster. The 85 features are listed in the supplementary material (See Table S1).
Feature selection
The Minimum Redundancy Maximum Relevance (mRMR) feature selection method (29) was employed to identify the SpaTIL features that most correlated with recurrence in the discovery set D1 while also eliminating features which were grossly similar to each other to prevent redundancy.
Comparative strategies
Inter-reader variability in TIL estimation by human readers
Two expert thoracic pathologists experienced in grading TILs were asked to determine the infiltration grade for each of the images in D1 and D2 via visual evaluation of digitized H&E stained images. An in-house custom web application was used by the readers to assign an infiltration score to each image. Infiltration options were defined based on findings reported in (4,6) as 0) no-infiltration (virtual absence of TILs), 1) low (1%−33%), 2) moderate (34%−66%), and 3) high (67%−100%).
The agreement among pathologists during the TIL-grading task was measured via two quantitative indices: Spearman’s correlation coefficient (30) and Cohen’s Kappa coefficient (31). The Kappa index is a widely used measure to determine the agreement among a set of experts making categorical judgments, considering agreement may occur by chance.
Computer based estimation of TIL-density
We also extracted TIL-density-based (DenTIL) features and compared the prognostic performance of these DenTIL features against the SpaTIL features. The DenTIL features included ratio between the number of TILs and the TMA spot area, ratio between the total regions of the TMA spot covered by TILS to the total area of the corresponding TMA spot, ratio between the number of TILs and the number of non-TILs within a TMA spot, and a grouping value indicating how close the TILs are to each other (computed as the sum of the inverse distances between TILs).
Statistical analysis
Classification
A Quadratic Discriminant Analysis (QDA) classifier was trained using the top SpaTIL features (QS) identified from D1 to separate the patients into two classes: recurrence and non-recurrence. We chose this classifier owing to the fact that it has no hyper parameters to tune and is able to learn quadratic boundaries. Therefore, it has been shown to be more flexible compared to linear classifiers. Additionally, another QDA classifier was trained using DenTIL features (QD) on the training set D1. Following parameter optimization, the classifier was locked down using D1.
The performance of the locked down QDA classifiers QS and QD in distinguishing between early-stage NSCLC patients who did and did not have recurrence was evaluated on the independent validation sets D2, D3, and D4. Classifiers QS and QD assigned a probability of recurrence to each image in the test sets. Classifier performance was evaluated via the concordance statistic or C-index. The recurrence and non-recurrence labels predicted by QS and QD were compared with the ground truth labels (true patient outcomes) to determine classifier accuracy and C-index. The C-index obtained for QS on D3 and D4 were quantitatively compared to evaluate the effect of spatial tissue sampling on the classifier performance.
Statistical and Survival Analysis
Recurrence-free survival (RFS) was defined as the time interval between the date of diagnosis and the date of death or recurrence (whichever happened first). Patients who were still alive without recurrence at the last reported date were labeled as censored. Kaplan-Meier survival analysis was used to examine the difference of RFS between different patient groups categorized by the classifier output and the difference of RFS in each group was assessed by the log-rank test. Multivariate Cox regression analysis was employed to examine the predictive ability of the QS and QD classifiers when controlling for the effects of clinical and pathological parameters including gender, age, T-stage, and N-stage. P-values were two-sided and all values under 0.05 were considered to be statistically significant.
A Kaplan-Meier analysis was also carried out for the DenTIL approach and for the TIL density estimation by the human readers. In the case of manual estimation of TIL density, patients were split into two groups: High-TIL and Low-TIL. Since pathologists graded each case from 0 to 3, case patients with TIL categories of 0 to 2 were considered as being part of the low-TIL group and those with scores of 3 were grouped in as part of the high-TIL tumor category. This strategy was based on the approaches described in (4,6).
Experimental Results
Clinico-pathologic Features of the Patient Cohorts
The median follow-up for patients was 40.91 months, 45.33 months, and 70.92 months for D1, D2, and D3, respectively. By the end of the study/follow-up, 34 out of 70 patients (48.6%) in D1, 54 of 119 (45.4%) in D2, and 34 out of 112 (30.4%) in D3 had developed recurrence. Table 1 presents a summary of clinical and pathological features for the whole dataset (D1, D2, D3, and D4). Supplemental Table S3 presents the clinical and pathological features of cohorts D1, D2, D3, and D4 individually.
Table 1:
Summary of clinical and pathological features for the patients in the whole dataset.
| Variable | Sub variables | N (%) |
|---|---|---|
| Number of patients | 301 | |
| Average age | 64.3 +/− 10.5 | |
| Sex | Male Female |
176 (58.5) 125 (41.5) |
| N-Pathological | 0 1 |
205 (68.1) 96 (31.9) |
| T-Pathological | 1 2 |
135 (44.9) 166 (55.1) |
| Stage | I/IA/IB II/IIA/IIB |
221 (73.4) 80 (26.6) |
| Recurrence | Non-recurrence Recurrence |
179 (59.5) 122 (40.5) |
| Tumor types | Adenocarcinoma Squamous Cell Carcinoma Others |
135 (44.9) 89 (29.6) 77 (25.6) |
The images are all pre-treatment tissue slides. The analysis is performed during the diagnosis phase (no treatment at all). Obviously, the treatment may influence the survival variable because of the inherent biological variability, i.e., each individual response is different, but once the patient is categorized, the treatment is the same for that particular category. The kind of treatment is shown in Supplementary Table S5.
Experiment 1: Prognostic ability of SpaTIL in early-stage NSCLC
Supplemental Table S2 and Figure S3 illustrate the top SpaTIL features identified via feature selection and their corresponding boxplots, respectively.
Figures 2-c, 3-c, 4-a, and 4-d illustrates the ROC curves and corresponding C-indices for the SpaTIL classifier (Qs) for predicting recurrence in NSCLC on D1, D2, D3, and D4. C-indices of the binary classifier for the 4 datasets were 0.70, 0.73, 0.70, and 0.71, respectively.
Figure 2.
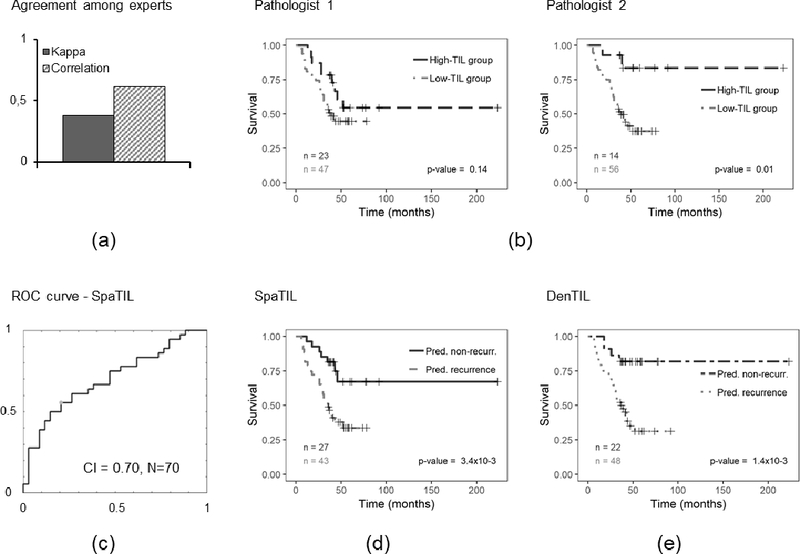
Prognostic prediction results for human readers, QD, and QS for D1. a) Bar plot illustrating the Kappa index and correlation coefficient computed between readers 1 and 2, b) Kaplan-Meier curves for readers 1 and 2, c) ROC curve and corresponding CI for QS, d) Kaplan-Meier plot for QS classifier using recurrence free survival as endpoint, e) Kaplan-Meier plot for QD classifier. The number of cases in each category is indicated in the charts.
Figure 3.
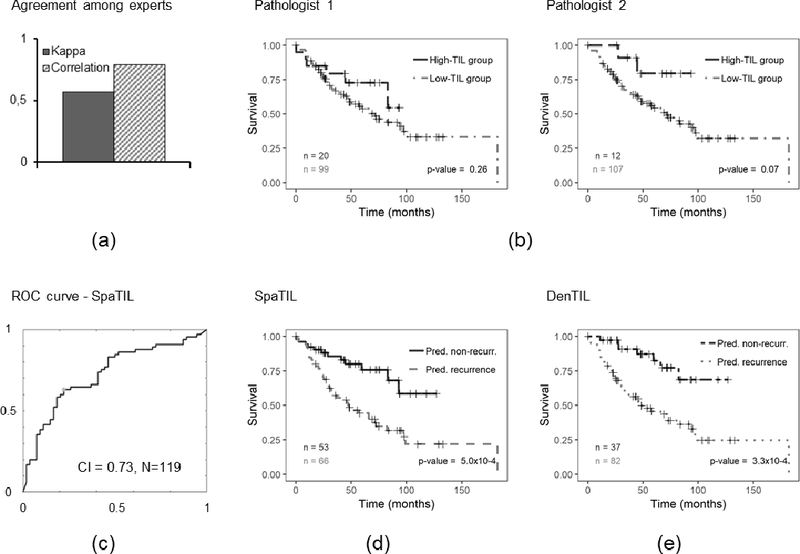
Prognostic prediction results for human readers, QD, and QS for D2. a) Bar plot illustrating the Kappa index and correlation coefficient computed between readers 1 and 2, b) Kaplan-Meier curves for readers 1 and 2, c) ROC curve and corresponding CI for QS, d) Kaplan-Meier plot for QS classifier using recurrence free survival as endpoint, e) Kaplan-Meier plot for QD classifier. The number of cases in each category is indicated in the charts.
Figure 4.
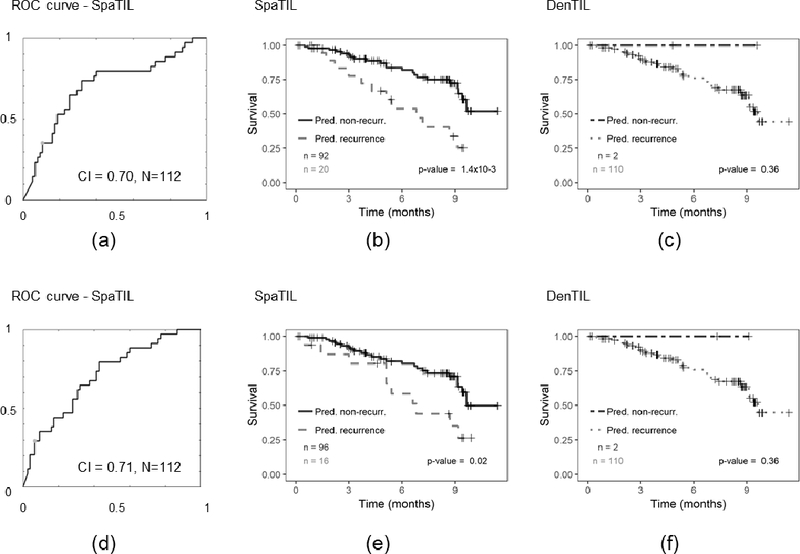
Prognostic prediction results for QD and QS for D3 (top row) and D4 (bottom row). First column (a, d) shows the ROC curve and corresponding CI for QS, second column (b, e) presents the Kaplan-Meier plots for QS classifier using recurrence free survival as endpoint, and third column (c, f) illustrates the Kaplan-Meier plot for QD classifier. The number of cases in each category is indicated in the charts.
Figures 2-d, 3-d, 4-b, and 4-e illustrate the Kaplan-Meier plots corresponding to the SpaTIL features for D1, D2, D3, and D4, respectively. Qs was found to be prognostic for D2 (p=5.0×10−4, HR: 2.80, 95% confidence interval: 1.64 – 4.80), D3 (p-value = 1.4×10−3, HR: 4.45, 95% confidence interval: 1.76 – 11.25), and D4 (p-value = 0.02, HR: 4.45, 95% confidence interval: 1.26 – 8.46).
Significance of clinical and pathologic variables with patients’ survival time in the test sets was evaluated via the log-rank test (Figure S2). A multivariate analysis, controlling the effect of major pathological and clinical variables, was performed for the three test cohorts (D2, D3, and D4). Table 2 presents the results of the analysis for the three cohorts together while Supplemental Table S4 shows the results for each cohort individually. Patients identified by the Qs classifier as having poor prognosis had statistically significantly worse disease-specific survival. The hazard ratio (HR) was 3.08 (95% confidence interval: 2.1 – 4.5, p=7.3×10−5), meaning that patients with recurrence were approximately 3 times more likely to develop disease recurrence and die from the disease.
Table 2:
Multivariable survival analysis performed on the test sets (D2, D3, and D4) including SpaTIL.
| Variable | Hazard Ratio (95% Confidence Interval) |
p-value |
|---|---|---|
| Gender Male vs. Female |
1.18 (0.85–1.65) | 0.32 |
| T-stage T1 vs. T2 |
0.98 (0.68–1.40) | 0.90 |
| N-stage N0 vs. N1 |
0.91 (0.59–1.41) | 0.68 |
| Stage Stage I vs. Stage II |
1.01 (0.65–1.58) | 0.96 |
| Tumor subtypes ADCs vs. SCC vs. others |
0.92 (0.72–1.19) | 0.53 |
| SpaTIL recurrence vs. non-recurrence |
3.08 (2.10–4.51) | 7.3×10−5 |
| Two-sided p < 0.05 (in bold) was considered as statistically significant. | ||
Experiment 2: Comparison of Human and Machine based assessment of TIL Density for Predicting Recurrence in Early stage NSCLC
Figures 2-a and 3-a show the computed Spearman’s correlation and Kappa index values for the two pathologists for D1 and D2, respectively. The overall computed Kappa (considering both analyzed datasets) was 0.50. When computed independently for each dataset, Kappa indices were 0.38 and 0.57 for D1 and D2, respectively. On the other hand, the correlation coefficients were 0.61 for D1 and 0.79 for D2.
Figures 2-b and 3-b illustrate the Kaplan-Meier plots for both pathologists on D1 and D2, respectively. For reader 1, no statistically significant correlation between TIL estimation and outcome was observed for D1 (p=0.14) nor D2 (p=0.26). Conversely, for reader 2, a significant statistical correlation was observed for set D1 (p=0.01) but not for set D2 (p=0.07).
Figures 2-e, 3-e, 4-c, and 4-f illustrate the Kaplan-Meier plots corresponding to the QD classifier on D1, D2, D3, and D4, respectively. The QD classifier was found to have a statistically significant correlation between the classifier and patient outcome for D2 (p=3.4×10−4); the predictions, however, were not statistically significant for D3(p=0.36) and D4 (p=0.36), respectively.
Discussion
Different studies reported the importance of immune response in different cancers (32). Such studies have demonstrated a high correlation between TILs density and both disease outcome and treatment response, particularly for lung, breast, ovarian, pancreatic, colorectal, and skin cancer, to name a few (12,13,11,1,33,6,2,32,22). Pathologists have aimed to quantify the number of immune cells and their relationships within a tumor. For example, Galon et al. (34) classified tumors with an Immunoscore that relates the density and location of immune cells within the tumor. They showed this score yields a prognostic value that can complement or even replace the standard TNM classification in colorectal cancer. The authors have subsequently made a dedicated effort to promote the Immunoscore in routine clinical settings (See http://www.immunoscore.org/). Likewise, Salgado et al. (11), as part of the International Immuno-oncology Biomarker Working Group on Breast Cancer (See https://www.tilsinbreastcancer.org/), have performed the largest effort to date to establish a TIL based quantification protocol. The authors constructed a set of guides that standardize the methodology for visual assessment of TILs on H&E slide sections. In lung cancer, however, there is a conspicuous lack of a standardized guidelines for TIL scoring/use. Although different works have demonstrated the prognostic value of TILs in lung cancer, in each of these studies (4,19), the degree of TILs was estimated by visual observation, a time consuming, subjective, and often error-prone task. Moreover, several studies have shown a limited reproducibility in visual estimation of TILs (6,12).
Computer based approaches for automatic estimation of TILs (12,13,14,15) have helped to mitigate the subjectivity and low reproducibility associated with human TIL grading. However, different recent works have shown that, besides TIL density, the spatial location of immune cells is useful in predicting patient prognosis (32). Such studies appear to suggest that the spatial organization of lymphocytic infiltration in the context of nearby cancer cells is an important prognostic hallmark of certain types of tumors. This suggests that the study of the immune response with respect to patient outcome should take into account not only the quantity of immune cells, but also the spatial arrangement of the cancerous and surrounding immune cells (20,21,23,35). Previous related studies have found a strong association between the spatial location of nuclei and surrounding cytoplasmic features with OS (20,21) and RFS (23,4) in patients with early-stage NSCLC.
In this work, we presented a set of features based on the spatial architecture of TILs (SpaTIL), devised to capture the TIL local density and variation in the density, architecture, and co-localization of TIL and cancerous cells.
On two TMA datasets comprising of 119 and 112 patients, the SpaTIL classifier yielded CIs of 0.73, and 0.70, respectively. A Kaplan-Meier analysis along utilizing the log-rank test showed a strong association between the predictions of the SpaTIL classifier and recurrence for D2 (p=5.0×10−4), D3 (p=1.0×10−3), and D4 (p=0.01). Likewise, a multivariate Cox proportional survival analysis revealed a HR of 3.08 (95% confidence interval: 2.1 – 4.5, p=7.3×10−5).
Notably, the cohorts were obtained from different institutions with local (and presumably variable) tissue processing and preservation protocols. In addition, slides were stained in different institutions and digitalized with two different instruments. The latter appears to reflect the robustness of the SpaTIL classifier, its relative resilience to image and color variance on account of major pre-analytical variables and with samples from multiple different sites.
We also compared the prognostic performance of TIL estimation carried out by two human readers. A Kaplan-Meier analysis was conducted for each pathologist; results showed that no significant statistical correlation was found between Pathologist 1 and prognosis for any dataset (p>0.05) while there was a significant statistical correlation between TIL grade estimation of Pathologist 2 and patient outcome for D1 (p=6.0×10−3). In addition, the agreement among expert pathologists for D1 and D2 was found to be moderate (K=0.50) and comparable to the values previously reported for NSCLC (K=0.59) (6) and breast pathology (K=0.72) (36).This moderate agreement might be due to the fact that TIL grading in lung pathology lacks a standardized scoring system, hence each pathologist might preferentially focus on different areas of the tissue during examination (e.g., epithelium or stroma) or consider different cell populations within the “TIL” infiltration (e.g., mononuclear cells beyond lymphocytes such as plasmocytes and myeloid cells). Finally, different pathologists may have variable expertise evaluating immune cell infiltrates or natural individual variation in their perception of colors, shapes, and relative amounts/proportions. In contrast, the results obtained by using SpaTIL features are objectively measured and suggest that the spatial arrangement of TILs and tumor cells were strongly associated with recurrence in early stage NSCLC (p<0.05).
The two papers most closely related to the work presented here are that of Khan et al. (37) and Saltz et al. (22). Khan et al. (37) measured the degree of TIL infiltration for each core of a TMA as the ratio of lymphocytes to all cells. The infiltration score was found to be associated with poor prognosis in breast cancer, specifically in HER2-amplified/positive cases (p=0.02). In (22), Saltz et at. used a deep learning model to identify patches from whole slide images (WSIs) containing TILs. From these patch clusters, cluster indices were calculated, namely Ball and Hall (38), Banfield and Raftery (39), C (40), determinant ratio (41), among others. Five associations between these cluster indices and patient survival were found to be significant for different tumor types: Ball-Hall for breast invasive carcinoma (p=7×10−3), C-index for lung adenocarcinoma (p=3.0×10−3), Banfield-Raftery for prostate adenocarcinoma (p=0.01), Determinant Ratio for prostate adenocarcinoma (p=0.01), and Banfield-Raftery for skin cutaneous melanoma (p=1.0×10−3). While this study (22) clearly suggests the prognostic relevance of spatial arrangement of TILs, the SpaTIL features capture not just the spatial arrangement of TILs, but also the spatial interplay and co-localization of TILs and cancer nuclei. While studies involving IHC and QIF images (4,18,19) have shown the importance of looking at the spatial architecture and interplay of different TIL families, to the best of our knowledge, the SpaTIL approach represents the first attempt to capture the spatial architecture and arrangement of TILs and non-TILs from routine H&E images alone.
Additionally, the approaches of both Khan et al. (37) and Saltz et al. (22) are actually more similar to the DenTIL classifier which we employed to compare against the SpaTIL features, one that invoked the ratio of lymphocytes to all cells in a TMA core and also included other TIL based density features. While the results of the DenTIL classifier were found to be statistically significant in predicting recurrence for D2, the results were not significant for either D3 or D4 hence showing the lack of reproducibility. The corresponding HR and CI values for the DenTIL classifier were also found to be lower compared to the SpaTIL features across the validation sets D2, D3, and D4.
Our work did however have a few limitations. First, unlike the study of Saltz et al. (22), our approach was evaluated on TMAs and not using WSIs. The SpaTIL features were however evaluated and compared from tissue punches from different parts of the same tumor (D3 and D4). The SpaTIL features were found to prognostic of recurrence in both D3 and D4 (p≤0.01) and the corresponding C-index and Hazard ratios were also found to be comparable (CI=0.70, HR: 4.45 for D3; CI=0.70, HR: 3.26 D4). These findings appear to be in concordance with previous studies involving TIL based biomarkers that have suggested that results using TMAs are concordant with findings from WSIs (11,4,37). However, additional studies comparing the results in TMAs vs WSI will help establish the minimum amount of tumor tissue required for optimally calculating SpaTIL features that are prognostic of recurrence.
Future work will include additional analysis among larger patient cohorts and eventually using prospectively collected samples. Although our cohorts were not characterized using alternative prognostic molecular signatures for NSCLC (4,18,19), we anticipate the SpaTIL to be complementary with such tests. Additional studies will be required to determine the relative contributions, feasibility, and cost-effectiveness of each approach; and the possible role of such metrics to predict sensitivity/resistance to novel anti-cancer immunostimulatory therapies.
In summary, we presented a novel computational image-analysis based approach that exploits a set of density and spatial topological features related to the arrangement of clusters of TILs and non-lymphocytes within the tumor area using standard H&E histology preparations. We identified a metric that was consistently associated with recurrence/prognosis in early stage NSCLC. This work represents the first preliminary step in the development of an image-based companion diagnostic test to help identify early-stage lung cancer patients at increased risk for recurrence and hence potential candidates for adjuvant therapy (e.g., chemotherapy or immunotherapy) following standard of care surgery
Supplementary Material
Translational Relevance.
The presence of a high degree of tumor-infiltrating lymphocytes (TILs) has been found to be associated with better prognosis in non-small cell lung cancer (NSCLC). There is a moderate agreement between pathologists when assessing the degree of TIL presence from histopathologic tissue specimens. In this study, we present a new set of computer-extracted quantitative features (SpaTIL) related to the spatial architecture of TILs, the co-localization of TILs and cancer nuclei, and the density variation of TIL clusters from H&E images. We demonstrate that SpaTIL can predict the likelihood of recurrence in early-stage NSCLC. Compared to clinico-pathologic features, these features were independently prognostic of disease recurrence. Following additional, independent, multi-site validation, this SpaTIL test could be used in a manner similar to genomic-based companion diagnostic tests to identify those patients who have a higher chance of recurrence and who might thus potentially receive added benefit from adjuvant chemotherapy following surgical resection.
Acknowledgement of research support:
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers: 1U24CA199374–01, R01CA202752–01A1, R01CA208236–01A1, R01 CA216579–01A1, R01 CA220581–01A1; the National Center for Research Resources under award number 1 C06 RR12463–01; the DOD Prostate Cancer Idea Development Award; the DOD Lung Cancer Idea Development Award; the DOD Peer Reviewed Cancer Research Program W81XWH-16–1-0329; the Ohio Third Frontier Technology Validation Fund; the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure of Conflicts of Interest: A. Madabhushi: Inspirata-Stock Options/Consultant/Scientific Advisory Board Member, NIH Academic-Industry Partnership grants, Sponsored Research, Elucid Bioimaging Inc.-Stock Options, PathCore Inc-NIH Academic Industrial Partnership, Philips – Sponsored Research. D. L. Rimm: Consultant or advisor to Astra Zeneca, Agendia, Agilent, Biocept, BMS, Cell Signaling Technology, Cepheid, Merck, Perkin Elmer, PAIGE, and Ultivue; equity in PixelGear; Astra Zeneca, Cepheid, Navigate/Novartis, NextCure, Lilly, Ultivue, and Perkin Elmer fund research in his lab. K. A. Schalper: Speaker for Takeda, Merck; Consultant for Moderna, Celgene, Shattuck Labs; Research funding from: Genoptix (Novartis), Vasculox/Tioma, Tesaro, Onkaido, Takeda, Surface Oncology, Pierre Fabre.
References
- 1.Geng Y, Shao Y, He W, Hu W, Xu Y, Chen J, et al. Prognostic Role of Tumor-Infiltrating Lymphocytes in Lung Cancer: a Meta-Analysis. Cellular Physiology and Biochemistry 2015;37:1560–1571. [DOI] [PubMed] [Google Scholar]
- 2.Bremnes RM, Donnem T, Busund LT. Importance of tumor infiltrating lymphocytes in non-small cell lung cancer? Annals of Translational Medicine 2016;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uramoto H, Tanaka F. Recurrence after surgery in patients with NSCLC. Translational Lung Cancer Research 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, et al. Objective Measurement and Clinical Significance of TILs in Non–Small Cell Lung Cancer. Journal of the National Cancer Institute 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnem T, Hald SM, Paulsen EE, Richardsen E, Al-Saad S, Kilvaer TK, et al. Stromal CD8+ T-cell Density - A Promising Supplement to TNM Staging in Non-Small Cell Lung Cancer. Clinical Cancer Research 2015;21:2635–2643. [DOI] [PubMed] [Google Scholar]
- 6.Brambilla E, Teuff GL, Marguet S, Lantuejoul S, Dunant A, Graziano S, et al. Prognostic Effect of Tumor Lymphocytic Infiltration in Resectable Non–Small-Cell Lung Cancer. Journal of Clinical Oncology 2016;34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed quantitative analysis of CD3, CD8, and CD20 predicts response to neoadjuvant chemotherapy in breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2014;20(23):5995–6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Xu J, Zhang T, Xue D. Tumor-infiltrating lymphocytes in breast cancer predict the response to chemotherapy and survival outcome: A meta-analysis. Oncotarget 2016;7(28):44288–44298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kochi M, Iwamoto T, Niikura N, Bianchini G, Masuda S, Mizoo T, et al. Tumour-infiltrating lymphocytes (TILs)-related genomic signature predicts chemotherapy response in breast cancer. Breast cancer research and treatment 2017. [DOI] [PubMed] [Google Scholar]
- 10.Kashiwagi S, Asano Y, Goto W, Takada K, Takahashi K, Noda S, et al. Use of Tumor-infiltrating lymphocytes (TILs) to predict the treatment response to eribulin chemotherapy in breast cancer. PLoS ONE 2017;12:e0170634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Annals of Oncology 2015;26:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allard MA, Bachet JB, Beauchet A, Julie C, Malafosse R, Penna C, et al. Linear quantification of lymphoid infiltration of the tumor margin: a reproducible method, developed with colorectal cancer tissues, for assessing a highly variable prognostic factor. Diagnostic Pathology 2012;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robins HS, Ericson NG, Guenthoer J, O’Briant KC, Tewari M,W.Drescher C, et al. Digital Quantification of Tumor Infiltrating Lymphocytes. Science Translational Medicine 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksen AC, Andersen JB, Kristensson M, dePont Christensen R, Hansen TF, Kjær-Frifeldt S, et al. Computer-assisted stereology and automated image analysis for quantification of tumor infiltrating lymphocytes in colon cancer. Diagnostic Pathology 2017;12:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, et al. Computerized Image-Based Detection and Grading of Lymphocytic Infiltration in HER2+ Breast Cancer Histopathology. IEEE Transactions on Biomedical Engineering 2010;57:642–653. [DOI] [PubMed] [Google Scholar]
- 16.Fatakdawala H, Xu J, Basavanhally A, Bhanot G, Ganesan S, Feldman M, et al. Expectation-Maximization-Driven Geodesic Active Contour With Overlap Resolution (EMaGACOR): Application to Lymphocyte Segmentation on Breast Cancer Histopathology. IEEE Transactions on Biomedical Engineering 2010;57(7):1676–1689. [DOI] [PubMed] [Google Scholar]
- 17.Janowczyk A, Madabhushi A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. Journal of Pathology Informatics 2016;7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barua S, Fang P, Sharma A, Fujimoto J, Wistuba I, Rao AUK, et al. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer 2018;117:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu H, Zhang T, Ye J, Li H, Huang J, Li X, et al. Tumor-infiltrating lymphocytes predict response to chemotherapy in patients with advance non-small cell lung cancer. Cancer Immunology, Immunotherapy 2012;61(10):1849–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nature Communications 2016;7:12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Zang X, Yang L, Huang J, Liang F, Rodriguez-Canales J, et al. Comprehensive computational pathological image analysis predicts lung cancer prognosis. Journal of Thoracic Oncology 2017;12(3):501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Reports;23(1):181–193.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Janowczyk A, Zhou Y, Thawani R, Fu P, Schalper K, et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Scientific Reports 2017;7:13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nature Medicine 2002;8:1323– [DOI] [PubMed] [Google Scholar]
- 25.Veta M, van Diest P, Kornegoor R, Huisman A, Viergever MA, Pluim JPW. Automatic Nuclei Segmentation in H&E Stained Breast Cancer Histopathology Images. PLoS ONE 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corredor G, Wang X, Lu C, Velcheti V, Romero E, Madabhushi A. A watershed and feature-based approach for automated detection of lymphocytes on lung cancer images. In SPIE Medical Imaging; 2018. [Google Scholar]
- 27.Ali S, Lewis J, Madabhushi A. Spatially aware cell cluster(spACC1) graphs: predicting outcome in oropharyngeal pl6+ tumors. In International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2013. p. 412–419. [DOI] [PubMed] [Google Scholar]
- 28.Ali S, Veltri R, Epstein JA, Christudass C, Madabhushi A. Cell cluster graph for prediction of biochemical recurrence in prostate cancer patients from tissue microarrays. In SPIE Medical Imaging; 2013. p. 86760H–86760H. [Google Scholar]
- 29.Peng H, Long F, Ding C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Transactions on Pattern Analysis and Machine Intelligence 2005;27:1226–1238. [DOI] [PubMed] [Google Scholar]
- 30.Myers L, Sirois MJ. Spearman Correlation Coefficients, Differences between In Encyclopedia of Statistical Sciences.: American Cancer Society; 2006. [Google Scholar]
- 31.Carletta J Assessing Agreement on Classification Tasks: The Kappa Statistic. Comput. Linguist. 1996;22:249–254. [Google Scholar]
- 32.Yuan Y Modelling the spatial heterogeneity and molecular correlates of lymphocytic infiltration in triple-negative breast cancer. Journal of The Royal Society Interface 2015;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savas P, Salgado R, Denkert C, Sotiriou C, Darcy PK, Smyth MJ, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol 2016;13:228–241. [DOI] [PubMed] [Google Scholar]
- 34.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pag, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science 2006;313:1960–1964. [DOI] [PubMed] [Google Scholar]
- 35.Nawaz S, Yuan Y. Computational pathology: Exploring the spatial dimension of tumor ecology. Cancer Letters 2016;380:296–303. [DOI] [PubMed] [Google Scholar]
- 36.Buisseret L, Desmedt C, Garaud S, Fornili M, Wang X, Van den Eyden G, et al. Reliability of tumor-infiltrating lymphocyte and tertiary lymphoid structure assessment in human breast cancer. Modern Pathology 2017;30:1204. [DOI] [PubMed] [Google Scholar]
- 37.Khan AM, Yuan Y. Biopsy variability of lymphocytic infiltration in breast cancer subtypes and the ImmunoSkew score. Scientific Reports 2016;6:36231– [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ball GH, Hall DJ. Isodata, a Novel Method of Data Analysis and Pattern Classification: Stanford Research Institute; 1965. [Google Scholar]
- 39.Banfield JD, Raftery AE. Model-Based Gaussian and Non-Gaussian Clustering. Biometrics 1993;49:803–821. [Google Scholar]
- 40.Hubert L, Schultz J. Quadratic assignment as a general data analysis strategy. British Journal of Mathematical and Statistical Psychology 1976;29:190–241. [Google Scholar]
- 41.Scott AJ, Symons MJ. Clustering Methods Based on Likelihood Ratio Criteria. Biometrics 1971;27:387–397. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


