Abstract
We present a real-time, high-throughput, and cost-effective method of detecting apoptosis in vitro using a previously developed reagent that detects caspase activation by fluorescence. Current methods of assessing apoptosis fail to account for the dimension of time, and thus are limited in data yielded per sample. This reagent allows real-time detection of apoptosis, but until now has been restricted to a costly automated detection system. Here, we describe apoptosis detection with the Essen Bioscience IncuCyte® Caspase-3/7 Reagent using a multimode microplate reader, a common instrument in biological laboratories, which may be used prior to or in lieu of the automated system. This modified microplate reader apoptosis assay was validated against the established automated system, and was shown to detect a strong dose-response relationship (automated system r2 = 0.9968, microplate reader r2 = 0.9924). We also propose a quick and reliable method of quantifying cell density by Hoechst 33342 nuclear staining in microplates (r2 = 0.8812 between Hoechst signal and cell density). We assert that the dimension of time should not be overlooked, and that the method presented here is an accessible strategy for many researchers due to low startup cost and precise detection of apoptosis in real time.
Keywords: Apoptosis, caspase, cell death, Hoechst, live cell analysis, microplate
1. Introduction
Apoptosis is a type of regulated cell death that neutralizes and destroys cells without generating an inflammatory reaction in the surrounding tissue [1,2]. This process plays an integral role in biological functions as diverse as tissue turnover [3], adaptive immunity [4], angiogenesis [5], and normal wound healing [6,7]. Subtle alterations in apoptosis susceptibility of a particular cell population can be a necessary part of homeostasis or can contribute to disease pathogenesis [8–12]. For these reasons, apoptosis is a far-reaching and popular subject of interest in many biological research laboratories.
Current methods of assessing apoptosis in cultured cells typically require cell fixation (TUNEL or other immunohistochemistry), cell lysis (immunoblotting, PCR), or immediate disposal of assayed cells (flow cytometry), which yield a single readout per sample at a chosen timepoint. However, the process of programmed cell death involves an intricate cascade of protein activation and the execution of complex cellular events, carried out over the course of several hours [1,2]. Therefore, measuring apoptosis of a sample at a single timepoint necessarily excludes most of the time-dependent information that could be collected; an endpoint assay is a snapshot that cannot carry information about the overall timecourse of the apoptotic response. It is of course possible to collect samples from separate replicates treated at various times prior to collection; however, this is not always an ideal method of assessing apoptosis over time. First, sample availability may be limited, as is often the case with ex vivo samples, and therefore it may not be feasible to have the amount of replicates needed for the assay. Second, inter-sample variation between timepoint replicates, as well as human error, may lead to reduced precision in assessing apoptotic response over time using separate samples. Therefore, it is often preferable to use a real-time method of assessing apoptosis.
The Essen Bioscience IncuCyte® Caspase-3/7 Reagents have been developed to continuously monitor apoptosis in real time, using the proprietary IncuCyte® S3 Live-Cell Analysis System to produce and analyze fluorescent microscopy images. The IncuCyte® Caspase-3/7 Reagents are membrane-permeable and release a fluorescent DNA-intercalating dye when activated by cleaved caspases 3 and 7, which are executioner caspases cleaved as a part of the apoptotic cascade [13–16]. The IncuCyte® Caspase-3/7 Reagent also has the advantage of a persistent signal; when measuring activated caspases directly (such as by immunoblotting), the signal is ephemeral due to breakdown of activated caspases in the later stages of apoptosis. Conversely, the fluorescence of the activated IncuCyte® Caspase-3/7 Reagent is not affected by caspase degradation, providing greater assay flexibility. This signal is then captured by the automated fluorescent microscope within the incubator of the system, and images are analyzed by the accompanying IncuCyte® S3 2017A software. This method of quantifying apoptosis allows for real-time, robust, precise measurements, which can be simultaneously combined with other diverse cellular assays. However, a drawback of this live cell analysis system is cost, which may preclude many laboratories from purchasing and maintaining the system.
Here, we present a reliable method of assessing the kinetics of apoptosis in a real-time, high-throughput, microplate-based manner using the IncuCyte® Caspase-3/7 Reagent in combination with a cost-effective multimode microplate reader. Furthermore, we include a method of staining microplate samples with Hoechst 33342 to control for cell density in apoptosis assays or other experiments in which this variable is unknown.
2. Materials and Methods
2.1. Materials
Cell culture medium (DMEM/F12, MEM, and DMEM; cat. no. 11330, 11095, and 11965), fetal bovine serum (FBS; cat. no. 25140), L-glutamine (cat. no 25030), penicillin/streptomycin (P/S; cat. no. 15070063), gentamicin (cat. no. 15750), non-essential amino acids (NEAA; cat. no. 11140), sodium pyruvate (cat. no. 11360), trypsin-EDTA, dispase (cat. no. 17105), phosphate buffered saline pH 7.2 (PBS; cat. no. 20012), and Hank’s Balanced Salt Solution (HBSS; cat. no. 14175) were obtained from Gibco, Inc. (Gaithersburg, MD). IncuCyte® Caspase-3/7 Green Reagent (cat. no. 4440) was obtained from Essen Bioscience (Ann Arbor, MI). Staurosporine (STS; cat. no. S6942) was obtained from Sigma-Aldrich (St. Louis, MO). Hoechst 33342 (cat. no. 4082) was obtained from Cell Signaling Technology (Danvers, MA).
2.2. Animals
Female C57BL/6J mice, 6 - 8 wk of age, were obtained from Jackson Laboratory (Bar Harbor, ME) and housed five animals per cage under pathogen-free conditions and acclimated at least 2 wk prior to experimentation. Animals were fed standard laboratory diet and water ad libitum. The University Committee on Animal Resources approved all animal protocols.
2.3. Cell culture
Primary mouse lung fibroblasts (MLFs) were isolated as described by Seluanov et al. [17], with minor changes. Briefly, mice were euthanized and lungs were perfused with sterile PBS and extracted. Lungs were minced and digested in DMEM/F12 containing 1.8 U/mL dispase + 0.02% gentamicin at 37°C for 90 min, then rinsed and plated with expansion media (DMEM/F12 containing 15% FBS + 0.02% gentamicin) to facilitate the growth of primary lung fibroblasts from lung fragments. For 2 wk, lung fragment cells were allowed to expand in this media, with media changes every 2 - 3 d. Then cells were trypsinized and media was switched to selection media (MEM containing 15% FBS + 1X P/S + 1X NEAA + 1X sodium pyruvate + 0.02% gentamicin) to select for only fibroblast proliferation. At this point, the population was considered p0. For experiments described here, MLFs were used at p1 and p2. MLFs were grown in a humidified incubator at 37°C in 5 - 7% O2 and 5% CO2.
HT1080 human fibrosarcoma cells were obtained from American Type Culture Collection (ATCC, Rockville, MD) and cultured in MEM containing 10% FBS + 0.02% gentamicin in a humidified incubator at 37°C at 20 - 21% O2 and 5% CO2.
2.4. Apoptosis induction
MLFs or HT1080 cells were seeded into tissue culture treated black-sided, clear-bottom 96-well plates (cat. no. 3603, Corning Inc., Kennebunk, ME) at approximately 35,000 cells/mL (or 7,000 cells/well) and allowed to adhere and establish for 2 d. Cells were treated with 1X Essen Bioscience IncuCyte® Caspase-3/7 Green Reagent (final concentration 5 μM) and a dose of STS ranging from 0.00 μM – 1.50 μM. All treatment groups were carried out in triplicate. Data from treated plates were collected either with IncuCyte® S3 Live-Cell Analysis System or SpectraMax M5 ROM v3.0.22 multimode microplate reader.
2.4.1. Apoptosis data collection with IncuCyte® S3 Live-Cell Analysis System
Image scheduling, collection, and analysis were conducted with the IncuCyte® S3 Live-Cell Analysis System and IncuCyte S3 v2017A software. Treated plates were imaged hourly for 24 hr. At each timepoint, 2 images were taken per well in both brightfield and FITC channels. Images were analyzed for number of green objects per well. Settings for this data collection were as follows: Segmentation: “Top-Hat”, Radius (μm) “30.000”, Threshold (GCU) “3.0000”, “Edge Split On”, Edge Sensitivity “−20”; Cleanup: Hole Fill (μm2) “0.0000”, Adjust Size (pixels) “0”; Filters: Area (μm2) min “30.000”.
2.4.2. Apoptosis data collection with multimode microplate reader
Data collection with SpectraMax M5 ROM v3.0.22 multimode microplate reader was conducted with SoftMax® Pro v6.4 Microplate Data Acquisition and Analysis software. Treated plates were read immediately after treatment (0 hr), and again at 8 hr, 12 hr, 16 hr, 20 hr, and 24 hr after treatment. At each timepoint, fluorescent signal was quantified with excitation and emission wavelengths of 500 nm and 530 nm, respectively, based on the excitation and emission spectra detailed by Essen Bioscience for the Caspase-3/7 Green Reagent. Raw data were reduced by subtracting mean background signal of untreated (blank) wells.
2.4.3. Apoptosis timecourse analysis
To assess robustness of the apoptotic response based on a time-dependent response, data from each treatment group were first normalized to signal from wells treated with Caspase-3/7 Green Reagent + 0 μM STS, to account for baseline apoptosis over the 24 hr period. Then, linear regression was performed on means of normalized data for each treatment group, yielding a slope value for each dose of STS. A dose-response curve was created by plotting STS dose in μM (x) against timecourse slope in A.U. (y); x values were transformed by x = log10(x), whereas y values were normalized to the highest value to facilitate comparison across experiments. Transformed data were tested for goodness of fit by sigmoidal least squares nonlinear regression. This model was chosen due to the biological nature of the apoptotic response having a both a lower and upper threshold. This regression yielded the coefficient of determination (r2), which was used to assess goodness of fit of the dose-response curve. All data were visualized and analyzed using GraphPad Prism v6.01 software.
2.5. Hoechst 33342 live cell microplate staining & fluorescence quantification
MLFs were serially diluted and seeded into a 96-well plate at cell densities ranging from 1,000 cells/mL - 40,000 cells/mL (n = 8 wells for each seeding density). After allowing cells to adhere for 1 d, media was removed and cells were rinsed 3x with HBSS. Hoechst 33342 solution (100 ug/mL in HBSS) was added to the wells and samples were incubated for 30 min at room temperature. Hoechst 33342 solution was removed and wells were rinsed 3x with HBSS, leaving the last rinse in the wells. As described above, SpectraMax M5 ROM v3.0.22 multimode microplate reader and accompanying SoftMax® Pro v6.4 Microplate Data Acquisition and Analysis software were used to quantify fluorescent signal. Fluorescent signal was quantified with excitation and emission wavelengths of 353 nm and 483 nm, respectively. Raw data were reduced by subtracting mean background signal of untreated (blank) wells.
2.5.1. Hoechst 33342 data analysis
To assess the accuracy of Hoechst 33342 live cell staining to quantify cell density in microplates, fluorescent signal from Hoechst 33342-stained cells was plotted against known seeding density, and linear regression was conducted on these data.
2.6. Fluorescent microscopy
Fluorescent signal from cells stained with Caspase-3/7 Green Reagent or Hoechst nuclear stain were imaged with a Nikon Eclipse TE2000-E Inverted Research Microscope and MetaVue v7.7.5.0 Research Imaging Software by Molecular Devices, Inc. All images shown are 40x original magnification. Images were captured with 1 ms exposure in brightfield, or 10 s exposure in FITC (for Caspase-3/7 Green Reagent imaging) or DAPI (for Hoechst 33342 imaging) channels. Images were merged using the MetaVue software.
3. Results and Discussion
3.1. Rate as a measure of robustness of apoptotic response
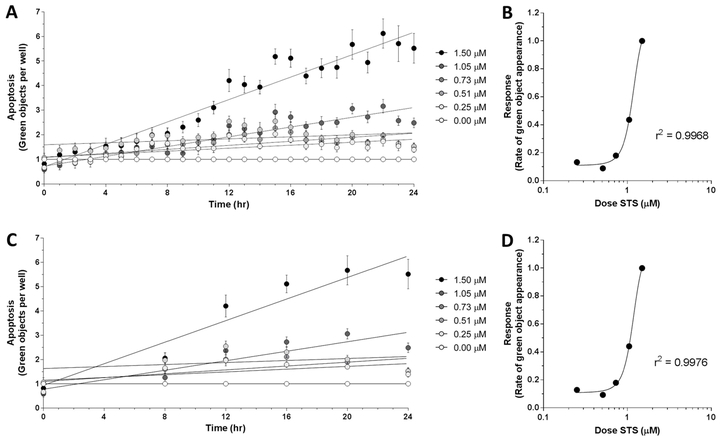
Current endpoint apoptosis assays yield a single output per sample, and therefore have relatively straightforward data analysis. In kinetic assays, the added variable of time changes statistical analysis. To investigate quantifiable differences in apoptotic response to varying strength of apoptotic stimulus over time, MLFs were treated with various doses of STS (0 μM – 1.5 μM) to induce apoptosis in addition to 1X IncuCyte® Caspase-3/7 Green Reagent. Number of green objects (i.e., fluorescing cells) per well was quantified by the IncuCyte® S3 Live-Cell Analysis System hourly for 24 hr (Fig 1A). The apoptotic response of these cells over the course of 24 hr appeared to be dose-dependent. The slope of the linear regression line for each dose of STS was plotted against the respective dose and the data were fitted with a sigmoidal nonlinear least squares regression, yielding the coefficient of determination of these two variables with respect to each other (Fig 1B, r2 = 0.9968). This statistic suggests that 99.68% of the variation in apoptotic response (i.e. slope of timecourse) is attributable to the dose of STS and is explained by the regression model. These results suggest that the slope of the linear regression of the 24 hr apoptotic timecourse – which represents rate of apoptosis – is a viable measurement of robustness of apoptotic response. Previous work using this reagent with the IncuCyte® S3 Live-Cell Analysis System reported “apoptotic index” as an output, which was defined as number of apoptotic cells per unit area at 48 hr after STS exposure [15]. However, using the slope of the timecourse has the advantage of incorporating the kinetics of the apoptotic response over time, rather than just an endpoint, and yields a robust sigmoidal dose-response curve.
Figure 1.
Apoptotic response in C57BL/6J MLFs, as quantified by IncuCyte S3 Live-Cell Analysis System. Cells were seeded at 35,000 cells/mL in 96-well plates and treated with doses of staurosporine (STS) ranging from 0.00 μM to 1.50 μM, as well as 1X IncuCyte® Caspase-3/7 Green Reagent. Using the IncuCyte® S3 Live-Cell Analysis System and accompanying software (v2017A), cellular apoptosis was assessed hourly over 24 hr by quantifying number of green objects per well (A). Rates of apoptosis in response to each dose of STS were determined by linear regression and plotted against corresponding dose of STS (B). Timecourse of apoptotic response showing fewer timepoints (0 hr, 8 hr, 12 hr, 16 hr, 20 hr, and 24 hr) is shown in C, with corresponding dose-response relationship shown in D. Goodness of fit for dose-response curves in B and D (r2 = 0.9968 and 0.9976, respectively) were determined by sigmoidal least squares nonlinear regression. Values for each timepoint in A and C represent mean and SEM of 3 replicates and are normalized to control wells (0.00 μM STS) to control for baseline apoptosis over 24 hr.
3.2. Microplate reader measurement of apoptotic response
Due to feasibility constraints of manually loading microplates into a microplate reader at each timepoint, the modified assay could not be read hourly over the 24 hr timecourse. Therefore, to address whether reading hourly is necessary to obtain a high coefficient of determination for the dose-response curve (as shown in Fig 1B), the data from Figure 1A were modified to exclude all readings except 0 hr, 8 hr, 12 hr, 16 hr, 20 hr, and 24 hr after treatment (Fig 1C). The timecourses were analyzed as described above, yielding a dose-response curve and accompanying coefficient of determination (Fig 1D, r2 = 0.9976). Since this statistic for the less densely sampled timecourse remained considerably close to 1.0000, our results indicate that that hourly readings throughout the 24 hr period are not necessary to determine robustness of apoptotic response.
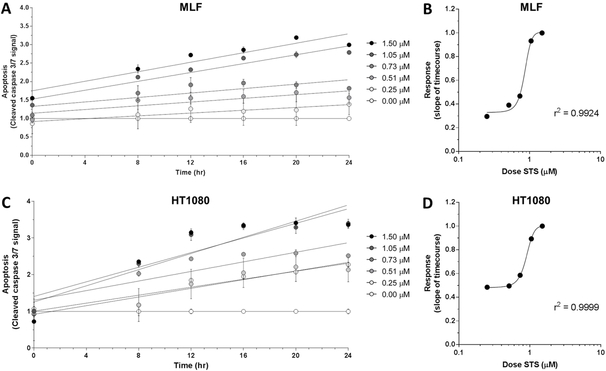
Next, to test the viability of using a microplate reader to assess apoptosis with IncuCyte® Caspase-3/7 Green Reagent, the treatment paradigm described above was repeated with MLFs and HT1080 fibrosarcoma cells, which were then analyzed using a microplate reader at the six timepoints indicated (Fig 2A, 2C). HT1080 cells were previously shown to have a dose-dependent apoptotic response using the IncuCyte® Caspase-3/7 Green Reagent and IncuCyte® S3 Live-Cell Analysis System [13]. Microplate reader data were analyzed as described above and slopes of linear regressions for each dose in both cell types were also fitted with sigmoidal nonlinear least squares regression (Fig 2B, 2D; MLF r2 = 0.9924; HT1080 r2 = 0.9999). Taken together, these data indicate that the strong dose-dependent relationship between the dose of apoptotic stimulus and the fluorescent apoptotic response can be detected by this microplate reader-based assay.
Figure 2.
Apoptotic response in C57BL/6J MLFs and HT1080 fibrosarcoma cells, as quantified by microplate reader. Cells were seeded at 35,000 cells/mL in 96-well plates and treated with doses of staurosporine (STS) ranging from 0.00 μM to 1.50 μM, as well as 1X IncuCyte® Caspase-3/7 Green Reagent. Using a microplate reader, cellular apoptosis was assessed at 6 timepoints over 24 hr by quantifying fluorescence signal from activated IncuCyte® Caspase-3/7 Green Reagent (A and C). Rates of apoptosis in response to each dose of STS were determined by linear regression and plotted against corresponding dose of STS (B and D). Goodness of fit for dose-response curves in B and D (r2 = 0.9924 and 0.9999, respectively) were determined by sigmoidal least squares nonlinear regression. Values for each timepoint in A and C represent mean and SEM of 3 replicates and are normalized to control wells (0.00 μM STS) to control for baseline apoptosis over 24 hr.
It is worth noting that, depending on the microplate reader available to a particular laboratory, plates may need to be loaded manually, which for the reading paradigm detailed here (0 hr, 8 hr, 12 hr, 16 hr, 20 hr, 24 hr), would necessitate odd hours for one or two lab personnel. However, this timecourse may be executed once or twice to obtain preliminary data for a specific cell line or type of exposure, and these findings utilized to determine a single ideal assay timepoint for future experiments, rather than completing the full timecourse.
Although two distinct cell types (MLFs and HT1080) are shown here to have dose-dependent apoptotic responses using the modified microplate reader apoptosis assay, this assay would be appropriate for virtually any adherent cell type. Furthermore, it may also have an advantage over the automated system in the analysis of nonadherent cells. The IncuCyte® S3 Live-Cell Analysis System acquires images at each scheduled timepoint, which requires a pre-determined stage height to focus clearly on the layer of cells attached to the bottom of the well. The system does not identify fluorescing cells that are dispersed throughout the vertical volume of the media. Therefore, to assess apoptosis in naturally non-adherent cells, it is recommended that the microplate wells be coated with either poly-L-ornithine or fibronectin to drive cell adherence [18]. However, poly-L-ornithine and fibronectin are extracellular matrix components that are not biologically inert [19–22] and may influence cellular signaling pathways in ways that are undesirable, thus adding unwanted variables to experimental design. Fibronectin signaling in particular has been shown to activate the PTEN/PI3K/Akt axis, which is anti-apoptotic and may directly interfere with the apoptosis assay [23]. Measurement of fluorescence with a microplate reader is based on the total fluorescent signal from a three-dimensional well, rather than analysis of a two-dimensional image, and therefore does not necessitate adherence of cells. In this sense, the modified microplate reader apoptosis assay may be preferable over the IncuCyte® S3 Live-Cell Analysis System for measuring apoptosis in non-adherent cell lines.
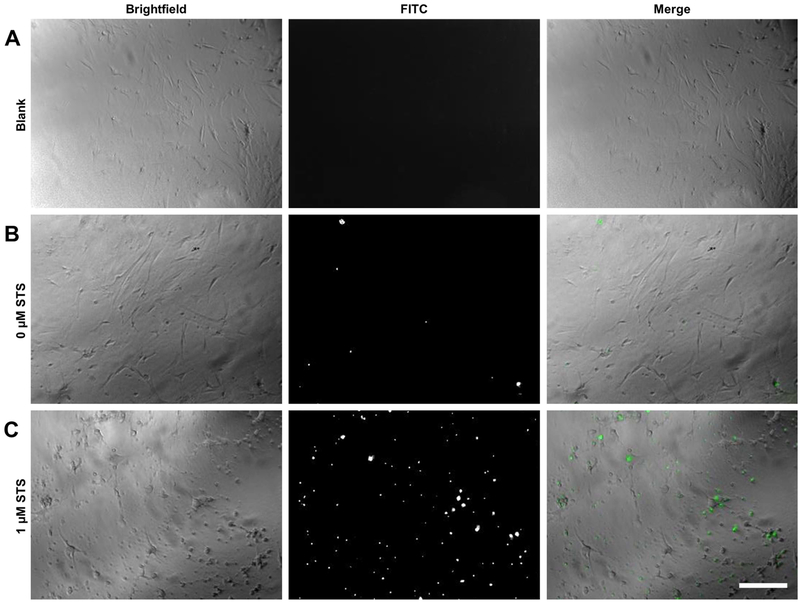
One drawback of the modified microplate reader apoptotic assay in comparison to the IncuCyte® S3 Live-Cell Analysis System is that image acquisition is not built into the system. However, images of wells may be obtained by traditional methods of fluorescent microscopy to supplement quantitative microplate reader results. To visualize the fluorescent signal produced by apoptotic cells, MLFs treated with IncuCyte® Caspase-3/7 Green Reagent and either 1.0 μM STS or vehicle were imaged 12 h after treatment. Fluorescent images obtained confirm microplate reader data; there were few fluorescing cells in control wells, but many in wells exposed to the apoptotic stimulus (Fig 3). Additionally, the difference in cell morphology was apparent in brightfield images. Control cells exhibited a flat and elongated spindle or stellate shape, typical of this cell type, whereas cells treated with STS showed advanced apoptotic morphology, including cellular shrinkage, membrane blebbing, and even detachment.
Figure 3.
Fluorescent microscopy of cells treated with IncuCyte® Caspase-3/7 Green Reagent. C57BL/6J MLFs were seeded into microplates at 35,000 cells/mL and treated with either vehicle only (A), IncuCyte® Caspase-3/7 Green Reagent only (B), or 1 μM STS plus 1X IncuCyte® Caspase-3/7 Green Reagent (C). Cells were allowed to incubate at 37°C for 12 hr, then imaged by fluorescent microscopy. Brightfield images show morphological changes in apoptotic cells treated with STS (C, as compared to A and B), and images taken in FITC channel show fluorescing IncuCyte® Caspase-3/7 Green Reagent, which localizes to nuclei and is indicative of caspase cleavage. Original magnification 40x; scale bar indicates 100 μm.
3.3. Cell density quantification in microplates with Hoechst 33342
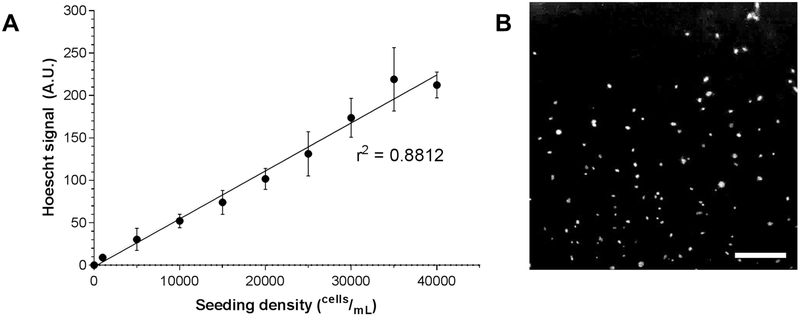
To determine whether the fluorescence readout of cells stained with Hoechst 33342 is a reliable estimation of cell density, MLFs were seeded at multiple known seeding densities into a microplate, stained with Hoechst 33342, and assayed by multimode microplate reader. Fluorescence was plotted against known seeding density, and linear regression was conducted on these data (Fig 4A; r2 = 0.8812). The high coefficient of determination suggests that this protocol yields an accurate metric of relative cell density among treatment groups. Stained nuclei were also visualized by fluorescent microscopy (Fig 4B).
Figure 4.
Hoechst 33342 live cell nuclear staining and fluorescence quantification. C57BL/6J MLFs were serially diluted and plated at various known seeding densities ranging from 1,000 cells/mL to 40,000 cells/mL (n = 8 wells per seeding density) and stained with Hoechst 33342. Fluorescent signal was quantified by microplate reader and shown to be highly correlated to known seeding density (A; linear regression r2 = 0.8812). In B, cells seeded at 40,000 cells/mL and stained with Hoechst 33342 and imaged by fluorescence microscopy. Original magnification 40x; scale bar indicates 100 μm.
This microplate-based nuclear stain protocol has a broad range of potential applications. It can be used alongside the modified microplate reader apoptosis assay to control for cell density, particularly if an endpoint is being measured, rather than the full timecourse. Hoechst 33342 microplate staining may also be used in studies assessing proliferation, or in essentially any experiment in which it is useful to determine cell density in microplates.
4. Conclusions
The modified microplate reader apoptosis assay detailed here used in combination with the Essen Bioscience IncuCyte® Caspase-3/7 Reagent provides the advantage of being effective, real-time, high-throughput, and cost-efficient. As described above, a real-time kinetic assay has the benefit of conveying more information about the same samples. This confers increased statistical power onto the data, as the need to account for inter-sample variation is obviated. Continuously monitoring apoptosis in this way also allows for the possibility of exploring apoptotic molecular mechanisms by intervening with one or more reagents while apoptosis is ongoing. Furthermore, the high throughput capability of the assay allows for more treatment groups or increased sample size per treatment group. Additionally, when total cell numbers are limited, the microplate format allows for relatively few cells per sample (7,000 cells/well and possibly fewer). The main advantage of this microplate reader-based assay in comparison to the IncuCyte® S3 Live-Cell Analysis System specifically is that it lacks a substantial start-up cost, as a basic microplate reader is a common piece of equipment in biological laboratories.
In summary, continuously monitoring apoptosis in a high-throughput manner is a useful and generalizable tool with applications in many fields of biological research. The modified microplate reader apoptosis assay provides an inexpensive way of doing so with minimal startup cost; consequently, this robust method of assessing apoptosis is accessible to a much wider range of researchers.
Highlights.
Popular methods of apoptosis detection yield a single reading per sample
Accounting for kinetics of the apoptotic response provides more robust and precise data outputs
Current real time methods of apoptosis detection are either expensive or have low sample capacity
A high throughput microplate reader based assay is shown here to accurately detect apoptosis in real time
Acknowledgements
We thank Paul Carman and Dr. Anu Konduru for assistance with the IncuCyte® S3 Live-Cell Analysis System, Devin Chandler-Militello for providing HT1080 cells, and Dr. Tanzy Love for statistics advice. We also thank all members of our lab for their insights and helpful discussions.
Funding
This work was supported by a National Institutes of Health (NIH) National Institute of Environmental Health Sciences (NIEHS) Institutional Training Grant (T32-ES007026-40), a NIH NIEHS Environmental Health Sciences Core (EHSC) Center Grant (P30-ES01247), a NIH National Institute of Allergy and Infectious Diseases (NIAID) Centers for Medical Countermeasures against Radiation Program (CMCR; U19-AI067773), and a University of Rochester Medical Center (URMC) Lung Biology Program pilot grant.
Footnotes
Conflict of interest
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Nomenclature Committee on Cell Death, Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018, Cell Death Differ. 25 (2018) 486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Green DR, Llambi F, Cell Death Signaling, Cold Spring Harb. Perspect. Biol. 7 (2015) a006080. doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Arandjelovic S, Ravichandran KS, Phagocytosis of apoptotic cells in homeostasis, Nat. Immunol. 16 (2015) 907–917. doi: 10.1038/ni.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yang S-H, Gao C-Y, Li L, Chang C, Leung PSC, Gershwin ME, Lian Z-X, The molecular basis of immune regulation in autoimmunity., Clin. Sci. 132 (2018) 43–67. doi: 10.1042/CS20171154. [DOI] [PubMed] [Google Scholar]
- [5].Watson EC, Grant ZL, Coultas L, Endothelial cell apoptosis in angiogenesis and vessel regression, Cell. Mol. Life Sci. 74 (2017) 4387–4403. doi: 10.1007/s00018-017-2577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Desmoulière a, Redard M, Darby I, Gabbiani G, Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar., Am. J. Pathol. 146 (1995) 56–66. [PMC free article] [PubMed] [Google Scholar]
- [7].Hinz B, The role of myofibroblasts in wound healing, Curr. Res. Transl. Med. 64 (2016) 171–177. doi: 10.1016/j.retram.2016.09.003. [DOI] [PubMed] [Google Scholar]
- [8].Thannickal VJ, Horowitz JC, Evolving concepts of apoptosis in idiopathic pulmonary fibrosis, Proc. Am. Thorac. Soc. 3 (2006) 350–356. doi: 10.1513/pats.200601-001TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mohammad RM, Muqbil I, Lowe L, Yedjou C, Hsu HY, Lin LT, Siegelin MD, Fimognari C, Kumar NB, Dou QP, Yang H, Samadi AK, Russo GL, Spagnuolo C, Ray SK, Chakrabarti M, Morre JD, Coley HM, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Helferich WG, Yang X, Boosani CS, Guha G, Bhakta D, Ciriolo MR, Aquilano K, Chen S, Mohammed SI, Keith WN, Bilsland A, Halicka D, Nowsheen S, Azmi AS, Broad targeting of resistance to apoptosis in cancer, Semin. Cancer Biol. 35 (2015) S78–S103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Radi E, Formichi P, Battisti C, Federico A, Apoptosis and oxidative stress in neurodegenerative diseases, in: J. Alzheimer’s Dis., 2014: pp. S125–S152. doi: 10.3233/JAD-132738. [DOI] [PubMed] [Google Scholar]
- [11].Hwang HS, Kim HA, Chondrocyte apoptosis in the pathogenesis of osteoarthritis, Int. J. Mol. Sci. 16 (2015) 26035–26054. doi: 10.3390/ijms161125943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu H, Fu S, Zhao M, Lu L, Lu Q, Dysregulation of cell death and its epigenetic mechanisms in systemic lupus erythematosus, Molecules. 22 (2017) 1–12. doi: 10.3390/molecules22010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].IncuCyte® by Essen Bioscience, IncuCyte® caspase-3/7 reagents for apoptosis product data sheet, (2017) 1–2. [Google Scholar]
- [14].Artymovich K, Appledorn DM, A multiplexed method for kinetic measurements of apoptosis and proliferation using live-content imaging, Methods Mol. Biol. 1219 (2015) 35–42. doi: 10.1007/978-1-4939-1661-0. [DOI] [PubMed] [Google Scholar]
- [15].Artymovich K, Nelson T, Dale T, Alcantara S, Endsley E, Appledorn DM, Trezise D, Groppi V, CellPlayer™ 96-well kinetic caspase-3/7 apoptosis assay, (2015) 1–8. [Google Scholar]
- [16].Ramirez MLG, Salvesen GS, A primer on caspase mechanisms, Semin. Cell Dev. Biol. (2018) 1–7. doi: 10.1016/j.semcdb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Seluanov A, Vaidya A, Gorbunova V, Establishing primary adult fibroblast cultures from rodents., J. Vis. Exp. (2010) 1–4. doi: 10.3791/2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].IncuCyte® by Sartorius, IncuCyte® apoptosis assay protocol, (2017) 1–4. [Google Scholar]
- [19].Ye Z-C, Sontheimer H, Modulation of glial glutamate transport through cell interactions with the extracellular matrix., Int. J. Dev. Neurosci. 20 (2002) 209–217. doi: 10.1016/S0736-5748(02)00048-5. [DOI] [PubMed] [Google Scholar]
- [20].Ge H, Tan L, Wu P, Yin Y, Liu X, Meng H, Cui G, Wu N, Lin J, Hu R, Feng H, Poly-L-ornithine promotes preferred differentiation of neural stem/progenitor cells via ERK signalling pathway, Sci. Rep. 5 (2015). doi: 10.1038/srep15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bentzinger CF, Wang YX, Von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA, Fibronectin regulates Wnt7a signaling and satellite cell expansion, Cell Stem Cell. 12 (2013) 75–87. doi: 10.1016/j.stem.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Labat-Robert J, Cell-Matrix interactions, the role of fibronectin and integrins. A survey, Pathol. Biol. 60 (2012) 15–19. doi: 10.1016/j.patbio.2011.10.003. [DOI] [PubMed] [Google Scholar]
- [23].Duronio V, The life of a cell: apoptosis regulation by the PI3K/PKB pathway, Biochem. J. 415 (2008) 333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]