Abstract
The post-transcriptional modification 2’-O-Methyl (2’OMe) could be present on the ribose of all four ribonucleosides, and is highly prevalent in a wide variety of RNA species, including the 5’ RNA cap of viruses and higher eukaryotes, as well as internally in transfer RNA and ribosomal RNA. Recent studies have suggested that 2’OMe is also located internally in low-abundance RNA species such as viral RNA and mRNA. To profile 2’OMe on different RNA species, we have developed Nm-seq, which could identify 2’OMe sites at single base resolution. Nm-seq is particularly useful for identifying 2’OMe sites located at the 3’ terminal ends of small RNAs. Here, we present an optimized protocol for Nm-seq and a protocol for applying Nm-seq to identify 2’OMe sites on small RNA 3’ terminal ends.
1. Introduction
Like modifications on DNA and protein, post-transcriptional chemical modifications on RNA play important functional roles, governing RNA function and metabolism to control gene expression [1,2]. Recent research on RNA modifications has focused primarily on modifications specific to each base. Unlike these modifications, the post-transcriptional modification 2’-O-Methyl (2’OMe) could be present at the ribose 2’ OH of all four ribonucleosides. 2’OMe is a highly prevalent modification; it is present on the 5’ RNA cap of viruses and higher eukaryotes, and marks nucleotides located at internal positions in transfer RNA and ribosomal RNA (rRNA).
Techniques to map 2’OMe transcriptome-wide using high-throughput sequencing have recently been developed, and are able to confirm established 2’OMe sites in human rRNA [3–9]. The first high-throughput technique to be developed, RiboMeth-Seq, uses alkaline RNA cleavage followed by high-throughput sequencing; because 2’OMe protects nucleotides from alkaline fragmentation, modified residues become excluded from the RNA library [4,7]. Similarly, RibOxi-seq uses alkaline RNA cleavage, and instead enriches for methylated fragments by preventing ligation of unmethylated fragments to adaptors [8]. Both techniques accurately mapped known 2’OMe sites in rRNA, demonstrating the utility of high-throughput methods over previous labor-intensive methods to map individual 2’OMe sites. Another high-throughput technique, 2’OMe-seq, sequences a cDNA library prepared using reverse transcription (RT) with limiting amounts of dNTPs, which causes RT pauses at 2’OMe sites [3]. These techniques have provided a useful window toward greater understanding of the 2’OMe modification.
Along with the development of technologies to map 2’OMe using high throughput sequencing, improvements in mass spectrometry have facilitated progress in research on 2’OMe. Studies on viruses have shown that 2’OMe is not limited to the 5’ RNA cap, but also labels non-terminal nucleotides located internally in viral RNA [10,11], showing that 2’OMe can be located internally in an RNA species that codes for protein. Other evidence has begun to suggest that 2’OMe may be located internally in mammalian mRNAs [6,9,12,13].
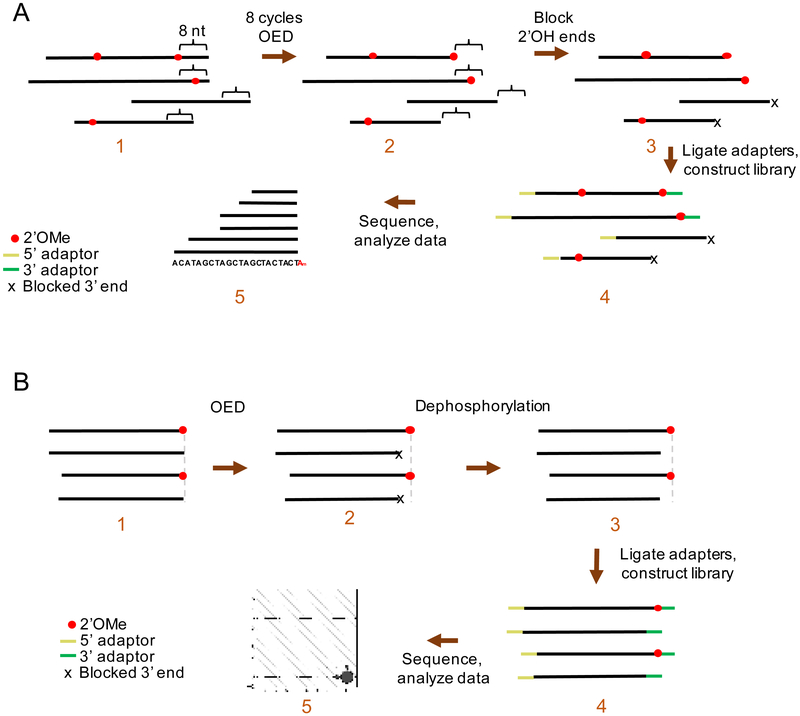
We thus aimed to develop a technique to map 2’OMe at single-nucleotide precision in different RNA species. Our lab has developed Nm-seq, which mapped hundreds of 2’OMe sites in human RNA at single-nucleotide precision [14]. Nm-seq uses multiple rounds of oxidation-elimination-dephosphorylation (OED) to iteratively eliminate 2’-unmodified nucleotides from the 3’ end of fragmented RNA (Fig. 1A). 2’OMe-modified nucleotides resist OED, and thus become enriched at the 3’ end of fragments. After multiple rounds of OED, a final oxidation-elimination reaction performed without dephosphorylation creates unligatable 3’ monophosphate ends on fragments ending in unmodified nucleotides. In contrast, Illumina-based 3’ adaptors are ligated using the NEB 3’ Ligation Enzyme Mix onto fragments containing 2’OMe-modified ends retaining a ligatable 3’ OH so that they can be further enriched by subsequent PCR. Due to the low abundance of fragments with 2’OMe-modified ends compared with other fragments without 2’OMe-modified ends, mispriming may take place and create artifact reads [15]. We therefore designed new 3’ adaptors containing a unique barcode so that the reads from mispriming can be filtered out. PCR is then used to amplify 2’OMe-modified fractions, allowing library construction for high-throughput sequencing (Fig. 2). The 3’ end of each RNA fragment is thus the 2’OMe-modified nucleotide, which is mapped by high-throughput sequencing.
Fig. 1.
Schematic of Nm-seq. A) Schematic for mRNA library OED treatment and library preparation. RNA fragments undergo eight cycles of OED. Each cycle removes one nucleotide from unmethylated 3’ RNA ends, thus exposing 2’OMe methylated nucleotides at the 3’ end of fragments. 3’ ends are blocked by oxidation-elimination without dephosphorylation, and adaptors are ligated onto all 5’ RNA ends and onto 3’ RNA ends containing 2’OMe. High-throughput sequencing produces an asymmetric read pileup signature, where the uniform 3’ end marks the exact 2’OMe position (the sequence in panel 5 represents a reference transcriptome). B) Schematic for small RNA OED treatment and library preparation. Small RNAs undergo 1 cycle of OED. 2’OMe methylated small RNAs remain the original size, while unmethylated ones are one nucleotide smaller with 3’ ends blocked by a phosphate group. Dephosphorylation is carried out using T4 PNK to remove the 3’ blockage. Adaptors are ligated onto both RNA ends. High-throughput sequencing produces small RNA reads in different lengths for small RNAs without 2’OMe.
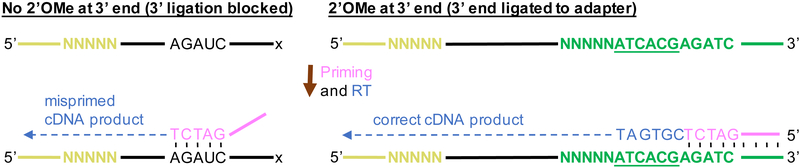
Fig. 2.
Use of a 3’ adaptor with an inline barcode prevents mispriming. Mispriming may occur on fragments containing an internal sequence complementary to the 3’ terminus of the RT primer sequence (left). However, only cDNA products produced from correctly primed products will contain the in-line barcode (5’ CGTGAT 3’), which is underlined in the figure. Reads without the in-line barcode are filtered out, thus preventing mispriming.
Nm-seq is particularly applicable to map 2’OMe residues on the 3’ terminal ends of small RNAs (Fig. 1B). It has been demonstrated that plant small RNAs and animal piwi-interacting RNAs (piRNAs) contain 3’ terminal 2’OMe, which is installed by the conserved methyltransferase HEN1 [16–19]. The terminal 2’OMe modification enhances the stability of small RNAs by inhibiting nucleotidyltransferase activities, such as uridylation, and 3’ to 5’ exoribonuclease activities, which mediate small RNA decay [20]. Methods using β-elimination followed by RNA gel blotting have been applied for 2’OMe detection, because unmodified RNA loses one base after sodium periodate treatment, resulting in a 1-2 base band shift on gel [21]. This method usually requires radioactive labeling and is not suitable for large scale detection. Based on the same principle, the Nm-seq method can be adapted for small RNA 2’OMe detection by omitting RNA fragmentation and performing just one round of OED.
We present here a detailed protocol and optimization of Nm-seq. This protocol allows mapping of 2’OMe sites in low abundance RNA species such as mRNAs and small RNAs. The bioinformatics pipeline has been described by the author of the original protocol [14].
2. Methods
2.1. Preparation of polyA-tailed RNA samples
2.1a. RNA fragmentation and 3’ end repair
Set up an RNA fragmentation reaction for each sample, using 10 μg of polyA-selected RNA for each sample. This amount of RNA is necessary, as each cycle of OED causes a loss of a portion of the RNA. Add the 10 μg of RNA, diluted in 72 μl of nuclease-free water, to 8 μl of 10x fragmentation buffer (1 M Na2CO3 buffer, pH = 9.2). Mix well by vortex and briefly centrifuge the tube. Incubate the RNA at 95°C for 18 minutes, then place the samples onto ice. This should fragment the RNA into 30-40-nucleotide long fragments. The fragmentation efficiency can be checked by bioanalyzer (i.e. Agilent 2100 Bioanalyzer, using an RNA Nano chip), and should be confirmed before moving onto the next step, as different tubes and incubators may require different fragmentation times.
Set up a 3’ end repair reaction by adding 9 μl of NEB T4 PNK buffer (10x) and 2 μl of NEB T4 PNK (M0201L, 10 units/μl) to the 80 μl of fragmented RNA. Incubate the reaction at 37°C for 30 minutes.
After the incubation, set aside 10 μl to use as input for library preparation. Perform 5’ phosphorylation on this 10 μl by adding 29 μl nuclease-free water, 5 μl NEB T4 PNK buffer (10x), 5 μl NEB ATP (P0756S, 10 mM), and 1 μl NEB T4 PNK. Incubate the 5’ phosphorylation mixture at 37°C for 30 minutes. Purify the RNA using Oligo Clean and Concentrator (Zymo Research D4061), eluting with 7 μl nuclease-free water. Elute a second time with another 7 μl water, combining the eluents. Measure the concentration, and set aside this sample, which will be used for construction of the input library.
Purify the remaining 81 μl of the 3’ end repair reaction using Oligo Clean and Concentrator, eluting with 35 μl of nuclease-free water.
2.1b. OED reagent setup
The OED cycles require that two of the reagents, lysine-HCl buffer and sodium periodate solution, be made the day of the experiment. To make 2 M lysine-HCl buffer, pH 8.5, dissolve 3.653 g of L-lysine monohydrochloride in 10 ml of nuclease free water. Titrate to pH 8.5 with sodium hydroxide, and filter through a 0.22 μm filter to help avoid the growth of bacteria. To make 200 mM sodium periodate solution, dissolve 42.778 mg of sodium periodate in 1 ml of nuclease-free water, and protect the solution from light by using a light-safe opaque microcentrifuge tube.
2.1c. OED reactions
OED is done in two successive steps: oxidation-elimination and dephosphorylation. Set up the oxidation-elimination reaction in a light-safe opaque microcentrifuge tube by adding 4.3 μl of 2 M lysine-HCl buffer, pH 8.5, and 4.3 μl of 200 mM sodium periodate solution to the 35 μl of purified fragmented and end-repaired RNA. Incubate the reaction at 37°C in a shaking incubator for 30 minutes.
Perform dephosphorylation by adding 5 μl of NEB CutSmart Buffer (10x) and 2 μl of NEB rSAP enzyme (0371S, 1 unit/μl) to the reaction. Incubate the reaction at 37°C in a shaking incubator for 30 minutes. Purify the ~50 μl reaction using Oligo Clean and Concentrator, eluting with 35 μl of nuclease-free water. Repeat the OED steps in section 2.1c 7 more times, for a total of 8 cycles of OED. Elute the last OED reaction with 38 μl of nuclease-free water.
8 cycles of OED are used to allow high sensitivity of Nm-seq. In theory, more cycles would provide higher sensitivity. However, about 10% of the starting material is lost in each iterative cycle. We found that, when starting with RNA, 8 cycles is the highest number of cycles possible before the concentration of the sample becomes too low to reliably construct OED libraries for sequencing.
2.1d. 5’ phosphorylation and final oxidation-elimination
To the 38 μl of OED-treated sample, add 5 μl of NEB T4 PNK buffer (10x), 5 μl of NEB ATP (10 mM), and 2 μl of NEB T4 PNK. Incubate the reaction at 37°C in a shaking incubator for 30 minutes. Purify the reaction using Oligo Clean and Concentrator, eluting with 40 μl of nuclease-free water.
A final oxidation-elimination step is performed without dephosphorylation to create unligatable 3’ monophosphate ends on unmodified nucleotides. To the 40 μl of 5’ phosphorylated OED-treated RNA, add 5 μl of 2 M lysine-HCl buffer, pH 8.5, and 5 μl of 200 mM sodium periodate solution. Incubate the reaction at 37°C in a shaking incubator for 30 minutes. Purify the RNA using Oligo Clean and Concentrator, eluting with 7 μl nuclease-free water. Elute a second time with another 7 μl water, combining the eluents. Measure the concentration. The sample should contain around 10 ng of RNA per μl, for a total of approximately 140 ng of RNA. This sample can be used to construct the OED libraries.
2.2. Preparation of small RNA samples
2.2a. Reagent setup
Prepare reagents as described in section 2.1b.
2.2b. Oxidation-elimination and 3’ end repair
Prepare 10-20 μg of total RNA. Set aside one third of the total RNA, which will be used for construction of the input library. Adjust the volume of the remaining two thirds of total RNA to 40 μl.
Set up a 50 μl oxidation-elimination reaction in a light-safe opaque microcentrifuge tube by adding 5 μl of 2 M lysine-HCl buffer, pH 8.5, and 5 μl of 200 mM sodium periodate solution to the 40 μl of total RNA. Incubate the reaction at 37°C in a shaking incubator for 30 minutes. Purify the RNA from the oxidation-elimination reaction using Oligo Clean and Concentrator, eluting with 42 μl nuclease-free water.
Remove 3’ phosphates resulting from the oxidation-elimination reaction by adding 5 μl of NEB T4 PNK buffer, 2 μl of T4 PNK, and 1 μl of 5% Triton-X100 (to maximize the 3’ phosphatase activity of T4 PNK). Incubate the reaction at 37°C in a shaking incubator for 30 minutes. Purify the RNA from the oxidation-elimination reaction using Oligo Clean and Concentrator, eluting with 10 μl nuclease-free water.
2.2c. Gel electrophoresis and RNA recovery
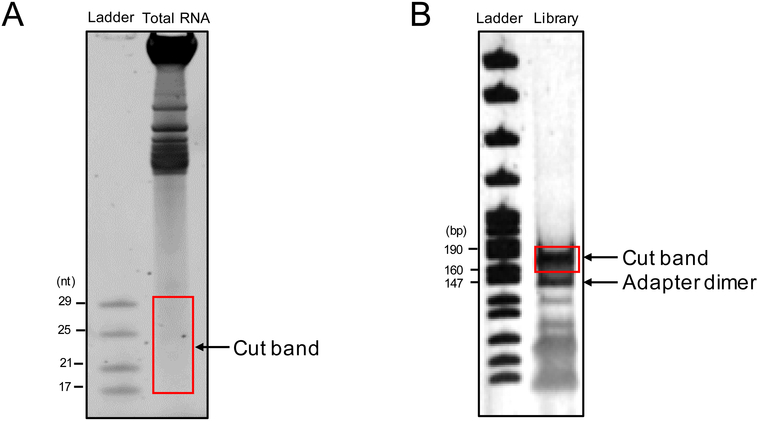
Denature the half of the total RNA set aside at the beginning of step 2.2b, the 10 μl of oxidation-elimination treated RNA, and 10 μl of ZR small-RNA ladder (Zymo Research R1090) in RNA Loading Dye (NEB B0363S) at 70°C for 5 minutes. Run the samples and ladder on a 15% TBE urea PAGE gel until the dye front comes close to the end of the gel (i.e. 180V, 75 minutes). Stain the gel with SYBR Gold Nucleic Acid Gel Stain (Thermo S11494) for 5 min at room temperature according to the manufacturer’s instructions, and cut out the small RNA (17-29 nt) portion of the gel, using the ladder as a reference (Fig. 3A). To recover piRNAs, cut the gel band between 17 to just over 30 nt. Recover the small RNAs using ZR small-RNA PAGE Recovery Kit (Zymo Research R1070), eluting with 6 μl of nuclease-free water.
Fig. 3.
Expected band sizes for gel cutting. A) An example image of a 15% TBE urea PAGE gel of total RNA from Arabidopsis seedlings. The red box indicates the area to be cut from the gel for small RNA recovery. B) An example image of a 6% PAGE gel after small RNA library construction. The red box indicates the area to be cut from the gel for library recovery, and the band of adapter dimer, which is 20-30 bp smaller, is also indicated.
2.3. Construction of OED libraries
Before library preparation, prepare custom 3’ and 5’ SR adapters (stock concentrations of 11.25 μM). The custom adapters are used due to the possibility of mispriming of the RT transcription primer. In Nm-seq, only a small proportion of RNA fragments contain 2’OMe at the 3’ end, and thus become ligated to the 3’ RNA adaptor. The reverse transcription primer, which is intended to hybridize with the 3’ RNA adaptor, may then hybridize to RNA fragments with internal sequences complementary to 3’ terminal ends of the RT primer (Fig. 2, top). To prevent mispriming from occurring, use the custom 3’ adaptor with a six-letter in-line barcode (ATCACG) at its 5’ end. Correct hybridization of the RT primer with the 3’ adaptor creates cDNAs containing the complementary sequence of the in-line barcode (Fig. 2, bottom). On the other hand, cDNAs created by mispriming do not contain the complementary sequence of the in-line barcode, as only the sequence of the RNA transcript is transcribed. By using only reads generated by cDNAs containing the in-line barcode, Nm-seq largely prevents false positives from mispriming. Moreover, both the custom 3’ adaptor and custom 5’ adaptor contain a sequence of 5 random nucleotides at the ligation junctions to reduce ligation-associated bias and to function as unique molecular identifiers to detect and exclude PCR duplicates.
3’ adaptor: 5’-AppNN NNN ATC ACG AGA TCG GAA GAG CAC ACG TCT-3’
5’ adaptor: 5’-GUU CAG AGU UCU ACA GUC CGA CGA UC NNN NN-3’
For library preparation of polyA-tailed RNAs, use 6 μl of the OED library and 1 μl of the input library. For library preparation of small RNAs, use all of the OED and input samples.
Library construction is performed using the NEBnext Small RNA Library Prep Set for Illumina (E7330S) with minor changes to the manufacturer protocol. Particularly:
In step 1.1 of the NEB protocol, replace the NEB provided 3’ SR Adaptor for Illumina (green cap) with 1 μl of a 1:4 dilution of the custom Nm-seq 3’ adaptor (5’-AppNN NNN ATC ACG AGA TCG GAA GAG CAC ACG TCT-3’, stock concentration: 11.25 μM). Dilution is necessary, as only a small fraction of the OED library will be ligatable by the 3’ adaptor. Dilution helps prevent the formation of 143nt adaptor dimers.
In step 1.4 of the NEB protocol, incubate at 16°C overnight (instead of at 25°C for one hour).
In step 2.1 of the NEB protocol, dilute the (pink) SR RT Primer for Illumina 1:4 in nuclease-free water before use.
Skip steps 3.1-3.2 of the NEB protocol, as a custom 5’ adaptor is used.
In steps 3.3-3.4, replace the NEB provided 5’ SR Adaptor for Illumina (yellow cap) with a 1:4 dilution of the custom Nm-seq 5’ adaptor (5’-GUU CAG AGU UCU ACA GUC CGA CGA UC NNN NN-3’, stock concentration: 11.25 μM).
The size of polyA-tailed RNA libraries should be around 165-170 nt, while small RNA libraries should be around 160 nt. Use gel electrophoresis to isolate the RNA libraries while removing adapter dimers, which are present at around 147 nt (Fig. 3B). PCR-amplified libraries may be stored at −20°C until sequencing. Deep sequence the libraries using an Illumina NextSeq platform (or similar).
2.4. Data processing
2.4a. Pre-filtering
Pre-process sequencing reads from the Illumina NextSeq-500 by removing standard adapters “AGATCGGAAGAGCACACGTCT” using Cutadapt [22]. Next, filter out misprimed reads by removing reads that do not carry the correct in-line barcode sequence at the 3’ end of the read (Fig. 2). Filter out PCR duplicates by using the 5 nucleotides at the 5’ and 3’ ends of reads as barcodes. Use only reads that have passed all of these filtering steps for further steps.
2.4b. Alignment and 2’OMe site identification
Map the filtered reads to the appropriate reference genome and transcriptome with TopHat [23]. To find significantly enriched sites carrying 2’OMe, use the 3’ end of sequencing reads to calculate the depth of each position of all RNA molecules. Signals on a potential 2’OMe site should present significant enrichment over the other regions on the same transcript, as well as over the same position of the matched input. Thus, on each nucleotide, calculate the sequencing depth in both the OED library and input library. Specifically, calculate the following numbers and perform a Chi-squared test: the depth of the current site (Dbase) in both OED and input samples, the total depth of the transcript (DSum) in nptj OED and input samples, and the average depth of the transcript in the OED sample (DAve). Positive sites are sites that meet the following criteria: (1) Chi(Dbase_Nm, DAve_Nm, Dbase_input, DAve_input): log2(ratio)>=1 & Dbase_Nm >= 10 (2) Chi(Dbase_Nm, DSum_Nm, DAve_Nm, DSum_Nm): p-value < 0.01 & log2(Dbase_Nm / DAve_Nm)>=2.
A detailed command structure can be found at https://github.com/malijia/2Ome
3. Results and discussion
Nm-seq allows high-throughput identification of sites of 2’OMe. Our method could detect 2’OMe in abundant RNA species such as rRNA, but also 2’OMe sites in lower abundance RNA species such as mRNA. The method can be adapted to detect 2’OMe at the 3’ terminal ends of small RNAs as we have shown here. In addition, further optimization of our protocol [14] includes custom 3’ and 5’ adaptors allows filtering out PCR duplicates and RT mispriming products.
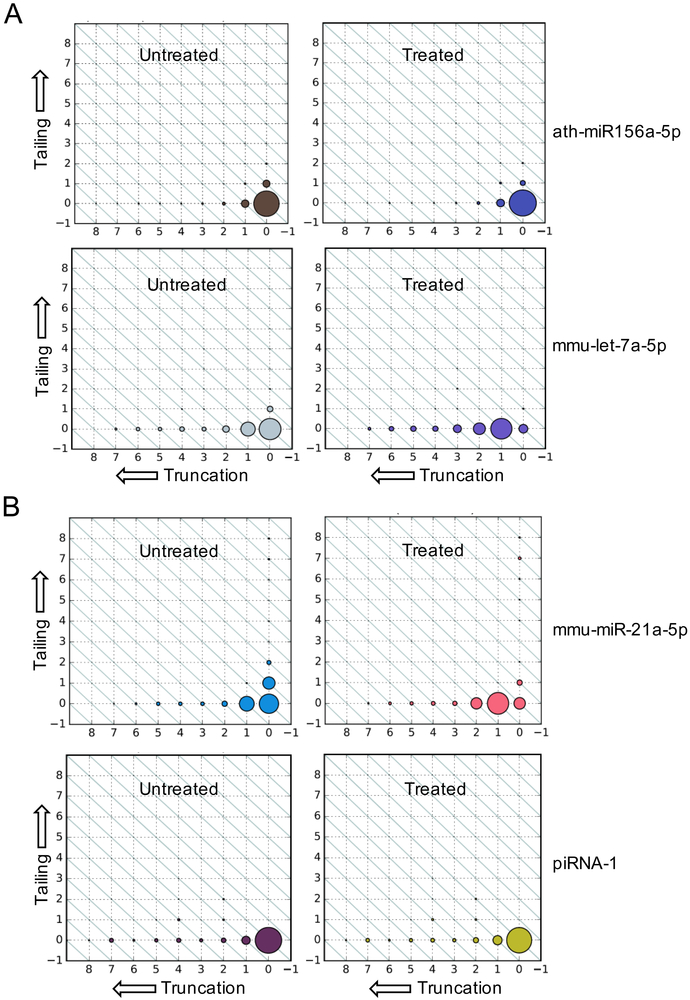
The optimized Nm-seq protocol could identify 2’OMe sites in various RNA species, including mRNA and viral RNAs. Detection of 2’OMe at the 3’ terminal ends of small RNAs using Nm-seq is also possible by comparing length distributions of miRNAs using a previously established pipeline [24]. We used Nm-seq to sequence small RNAs from mouse brain samples, which do not contain a 3’ terminal 2’OMe [25,26], and from Arabidopsis seedlings, which contain miRNAs with 3’ terminal 2’OMe [19]. In mouse brain samples, we observed a uniform size shift of the miRNA let-7a-5p after OED treatment (Fig. 4A, lower panel). However, in Arabidopsis seedlings, the sizes of miRNAs such as miR156a-5p, which has a 3’ terminal 2’OMe, are the same in OED treated and untreated samples (Fig. 4A, upper panel). Similar results were found in mouse testis samples (Fig. 4B). These results suggest that Nm-seq can be applied for small RNA terminal 2’OMe detection in a large scale.
Fig. 4.
Nm-seq detects 2’OMe at the 3’ terminal ends of small RNAs. A) Size distributions of (top) miRNAs ath-miR156a-5p from Arabidopsis, which contains 2’OMe and is not truncated, and (bottom) mmu-let-7a-5p from mouse brain, which does not contain 2’OMe and is truncated by one nucleotide. Dot sizes indicate the relative abundances of different lengths of a miRNA. X-axes represent the extent of 3’ truncation from the canonical, nontruncated miRNA length at the 0 position (right). Y-axes represent 3’ tailing, with the canonical miRNA length at the 0 position (bottom). Dots on diagonals represent reads of the same length with different statuses of truncation/tailing. B) Size distributions of a miRNA (miR-21a-5p) and a piRNA (named piRNA-1 here, with the sequence “TCCTGTTCTGATTTCCTTTGACTGTGATGT”) in a mouse testis sample.
Nonetheless, there is much room for improvement in the Nm-seq technique. For mRNAs, the need for eight cycles of OED makes the procedure laborious; they alone take over 10 hours to perform, which does not include the fragmentation, 3’ end repair, final phosphorylation, final oxidation-elimination, or library preparation steps. A method to automate Nm-seq would be highly useful. It could be possible to modify Nm-seq to involve fixing RNA fragments to magnetic beads, thus allowing the use of existing automated platforms for Nm-seq. Another limitation of Nm-seq is the large amount of RNAs needed for the procedure, as each cycle of OED causes a loss of around 10% of the RNA sample. A method that does not cause such a large loss of sample would allow Nm-seq to be performed on more limited samples, such as specific cell types or human patient samples.
Recently, a method to map 2’OMe sites at single-nucleotide precision on mRNA in yeast was developed [9]. This method, MeTH-seq, identified hundreds of novel 2’OMe sites on mRNA genome-wide, and showed that most 2’OMe sites are located on Um. The technique maps 2’OMe by using limited concentrations of magnesium to promote the pausing of reverse transcriptase one nucleotide 3’ to 2’OMe methylated sites. To detect false positives, MeTH-seq validates all 2’OMe sites by depletion of 2’OMe methyltransferases. Thus, MeTH-seq is a powerful tool for 2’OMe detection when combined with the manipulation of methyltransferases. Future work to discover 2’OMe methyltransferases in mRNA and further enhance techniques such as Nm-seq and MeTH-seq would greatly accelerate the characterization of functional roles of 2’OMe.
Other studies using β-elimination for small RNA library construction have also proved useful for identifying 2’OMe sites on small RNAs. In these studies, 3’ phosphate groups at the ends of unmethylated small RNAs are removed, preventing unmethylated small RNAs from ligating to a 3’ adapter [18,27]. Thus, unmethylated small RNAs are excluded from the final library preparation. These studies provide a useful method to sequence small RNAs with 2’OMe. However, this method without the dephosphorylation step does not retain information about small RNAs that are partially methylated. Increasing evidence shows that some small RNAs are partially methylated in diverse species, and varied methylation status may endow distinct biological functions [28,29]. In contrast, because our method includes a dephosphorylation step, the library preparation includes both methylated and unmethylated small RNAs, with unmethylated small RNAs appearing 1 nt shorter. Thus, our method could capture partially methylated small RNAs based on the 3’ truncation analysis. With further improvements, our method can potentially quantify 2’OMe levels in small RNAs.
Highlights for “Single base resolution mapping of 2’-O-methylation sites in human mRNA and in 3’ terminal ends of small RNAs”
Nm-seq can be used to identify 2’OMe sites at single base resolution on various RNAs
Nm-seq is particularly useful for detecting 2’OMe sites located at the 3’ terminal ends of small RNAs
Our optimization to include a redesigned custom 3’ adapter improves the quality of the data by filtering out misprimed reads
Further enhancements to Nm-seq and similar techniques will prove useful in the characterization of functional roles of 2’OMe
Acknowledgments
PH is supported by NIH/NIAID Ruth L Kirchstein National Research Service Award F30 AI136318 and NIH Medical Scientist National Research Service Award T32 GM007281. CH is an investigator of the Howard Hughes Medical Institute and is supported by NIH/NHGRI HG008935.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of potential conflicts of interest
C.H. is a scientific founder of Accent Therapeutic, Inc.
References
- [1].Roundtree IA, Evans ME, Pan T, He Chuan, Dynamic RNA Modifications in Gene Expression Regulation, Cell. 169 (2017) 1187. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang Y, Hsu PJ, Chen YS, Yang YG, Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism, Cell Res 28 (2018) 616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Incarnato D, Anselmi F, Morandi E, Neri F, Maldotti M, Rapelli S, Parlato C, Basile G, Oliviero S, High-throughput single-base resolution mapping of RNA 2-O-methylated residues, Nucleic Acids Res 45 (2017) 1433–1441. doi: 10.1093/nar/gkw810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H, Profiling of ribose methylations in RNA by high-throughput sequencing, Angew. Chemie - Int. Ed 54 (2015) 451–455. doi: 10.1002/anie.201408362. [DOI] [PubMed] [Google Scholar]
- [5].Krogh N, Jansson MD, Häfner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, Lund AH, Nielsen H, Profiling of 2’-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity., Nucleic Acids Res. 44 (2016) gkw482. doi: 10.1093/nar/gkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gumienny R, Jedlinski DJ, Martin G, Vina A, Zavolan M, High--throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq, Nucleic Acids Res 45 (2017)2341–2353. doi: 10.1101/037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y, Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA., Nucleic Acids Res 44 (2016) gkw547. doi: 10.1093/nar/gkw547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu Y, Pirnie SP, Carmichael GG, High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq), RNA. 23 (2017) 1303–1314. doi: 10.1261/rna.061549.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bartoli KM, Schaening Cassandra, Carlile TM, Gilbert WV, Conserved Methyltransferase Spb1 Targets mRNAs for Regulated Modification with 2′-O-Methyl Ribose, BioRxiv. (2018). doi: 10.1101/271916. [DOI] [Google Scholar]
- [10].Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM, Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells, Nat. Microbiol (2016) 16011. doi: 10.1038/nmicrobiol.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Martin B, Coutard B, Guez T, Paesen GC, Canard B, Oise Debart F, Vasseur J-J, Grimes JM, Decroly E, The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure, Nucleic Acids Res (2018) 1–11. doi: 10.1093/nar/gky637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Hüttenhofer A, Erlacher MD, Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code, Nucleic Acids Res 44 (2016) 852–862. doi: 10.1093/nar/gkv1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Choi J, Indrisiunaite G, DeMirci H, leong K-W, Wang J, Petrov A, Prabhakar A, Rechavi G, Dominissini D, He C, Ehrenberg M, Puglisi JD, 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation, Nat. Struct. Mol. Biol 25 (2018). doi: 10.1038/s41594-018-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C, Nm-seq maps 2′-O-methylation sites in human mRNA with base precision, Nat. Methods (2017). doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gillen AE, Yamamoto TM, Kline E, Hesselberth JR, Kabos P, Improvements to the HITS-CLIP protocol eliminate widespread mispriming artifacts, BMC Genomics. 17 (2016) 338. doi: 10.1186/s12864-016-2675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK, The caenorhabditis elegans HEN1 Ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs, PLoS Genet 8 (2012). doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kamminga LM, Luteijn MJ, Den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF, Hen1 is required for oocyte development and piRNA stability in zebrafish, EMBO J. 29 (2010) 3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SEJ, Ruvkun G, PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1, PLoS Genet 8 (2012). doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X, Methylation as a crucial step in plant microRNA biogenesis, Science (80-. ). 307 (2005) 932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhao Y, Mo B, Chen X, Mechanisms that impact microRNA stability in plants, RNA Biol 9 (2012) 1218–1223. doi: 10.4161/rna.22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang Z, Vilkaitis G, Yu B, Klimašauskas S, Chen X, Approaches for Studying MicroRNA and Small Interfering RNA Methylation In Vitro and In Vivo, Methods Enzymol 427 (2007) 139–154. doi: 10.1016/S0076-6879(07)27008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Martin M, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads, EMBnet.Journal 17 (2011) 10–12. [Google Scholar]
- [23].Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions, Genome Biol 14 (2013) R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Patel P, Ramachandruni SD, Kakrana A, Nakano M, Meyers BC, miTRATA: A web-based tool for microRNA Truncation and Tailing Analysis, Bioinformatics. 32 (2015) 450–452. doi: 10.1093/bioinformatics/btv583. [DOI] [PubMed] [Google Scholar]
- [25].Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T, The 3’ termini of mouse Piwi-interacting RNAs are 2’-O-methylated, Nat. Struct. Mol. Biol 14 (2007) 349–350. [DOI] [PubMed] [Google Scholar]
- [26].Kirino Y, Mourelatos Z, Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini, Nat. Struct. Mol. Biol 14(2007) 347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- [27].Williams Z, Morozov P, Mihailovic A, Lin C, Puvvula PK, Juranek S, Rosenwaks Z, Tuschl T, Discovery and Characterization of piRNAs in the Human Fetal Ovary, Cell Rep 13 (2015) 854–863. doi: 10.1016/j.celrep.2015.09.030. [DOI] [PubMed] [Google Scholar]
- [28].Abe M, Naqvi A, Hendriks GJ, Feltzin V, Zhu Y, Grigoriev A, Bonini NM, Impact of age-associated increase in 2’-O-methylation of miRNAs on aging and neurodegeneration in Drosophila, Genes Dev 28 (2014) 44–57. doi: 10.1101/gad.226654.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fei Q, Yu Y, Liu L, Zhang Y, Baldrich P, Dai Q, Chen X, Meyers BC, Biogenesis of a 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation, Proc. Natl. Acad. Sci 115 (2018) 201807403. doi: 10.1073/pnas.1807403115. [DOI] [PMC free article] [PubMed] [Google Scholar]