Abstract
Pipecolic acid or L-PA is a cyclic amino acid derived from L-lysine which has gained interest in the recent years within the pharmaceutical and chemical industries. L-PA can be produced efficiently using recombinant Corynebacterium glutamicum strains by expanding the natural L-lysine biosynthetic pathway. L-PA is a six-membered ring homolog of the five-membered ring amino acid L-proline, which serves as compatible solute in C. glutamicum.
Here, we show that de novo synthesized or externally added L-PA partially is beneficial for growth under hyper-osmotic stress conditions. C. glutamicum cells accumulated L-PA under elevated osmotic pressure and released it after an osmotic down shock. In the absence of the mechanosensitive channel YggB intracellular L-PA concentrations increased and its release after osmotic down shock was slower. The proline permease ProP was identified as a candidate L-PA uptake system since RNAseq analysis revealed increased proP RNA levels upon L-PA production. Under hyper-osmotic conditions, a ΔproP strain showed similar growth behavior than the parent strain when L-proline was added externally. By contrast, the growth impairment of the ΔproP strain under hyper-osmotic conditions could not be alleviated by addition of L-PA unless proP was expressed from a plasmid. This is commensurate with the view that L-proline can be imported into the C. glutamicum cell by ProP and other transporters such as EctP and PutP, while ProP appears of major importance for L-PA uptake under hyper-osmotic stress conditions.
Keywords: pipecolic acid, osmo regulation, compatible solute, proline, Corynebacterium glutamicum, RNAseq analysis, solute export, solute uptake
Introduction
In nature, all living organisms must respond to environmental fluctuations to survive. For example, bacteria have developed defense mechanisms for hyper- and hypo-osmotic external conditions to maintain cell viability including the de novo synthesis or uptake of osmo compatible solutes such as betaines. Since plants also contain these osmo compatible solutes, they are commonly present in production media and, thus, relevant for biotechnological fermentations (Farwick et al., 1995). The industrial workhorse Corynebacterium glutamicum disposes of mechanosensitive channels (MSCs) which perform as emergency release valves (Ruffert et al., 1999). After an osmotic downshift and to avoid cell lysis, MSCs are immediately activated by membrane turgor pressure to release solutes and to decrease in the internal osmolality (Ruffert et al., 1997; Morbach and Krämer, 2003). C. glutamicum can synthesize proline, glutamine, and trehalose and use them as compatible solutes, whereas it cannot synthesize ectoine and betaine, which therefore only function as compatible solutes when present in the environment (Frings et al., 1993; Farwick et al., 1995; Guillouet and Engasser, 1995; Wolf et al., 2003). Proline is the major de novo synthetized compatible solute in C. glutamicum (Skjerdal et al., 1996; Wolf et al., 2003). De novo biosynthesis of proline is induced under osmostress-conditions (Rönsch et al., 2003) unless nitrogen is scarce, a condition when trehalose is synthesized instead of proline (Wolf et al., 2003). Externally added proline can be taken up into the C. glutamicum cell by the import systems EctP, ProP, and PutP (Peter et al., 1998). The import of proline by the carriers EctP and ProP is osmoregulated (Peter et al., 1998), while the import of proline by PutP is not (Peter et al., 1997).
The cyclic amino acid L-pipecolic (L-PA), also known as homoproline, is a non-proteogenic amino acid and an intermediate of the catabolism of D,L-lysine (Neshich et al., 2013). L-PA is similar in chemical structure to L-proline since they only differ in ring size by one carbon (Pérez-García et al., 2016). L-PA plays many roles in microorganisms, plants, and animals; including the interactions between organisms and as precursor of natural bioactive molecules (Vranova et al., 2013). Notably, L-PA was reported as compatible solute for the microorganisms Silicibacter pomeroyi, Sinorhizobium meliloti, and Escherichia coli (Gouesbet et al., 1994; Gouffi et al., 2000; Neshich et al., 2013). E. coli does not degrade lysine to L-PA, but to cadaverine by the lysine decarboxylases LdcC or CadA (Mimitsuka et al., 2007; Kind et al., 2010). However, externally added L-PA protected E. coli cells under high osmolarity conditions (Gouesbet et al., 1994). C. glutamicum lacks lysine catabolic pathways, although the production of L-lysine-derived compounds has been established in C. glutamicum by metabolic engineering (Kind et al., 2010; Pérez-García et al., 2016; Jorge et al., 2017). A lysine producing C. glutamicum strain was engineered to overproduce L-PA by heterologous expression of the lysine dehydrogenase gene (lysDH) from S. pomeroyi and overexpression of the native pyrroline-5-carboxylate reductase gene (proC) from C. glutamicum (Pérez-García et al., 2016). However, the physiological role of L-PA for C. glutamicum has not yet been described. Here, we characterized the effect of L-PA on C. glutamicum (either added to the culture medium or synthesized de novo) under different conditions of osmotic pressure by physiological and RNAseq experiments. We provide evidence that YggB may be involved in export of L-PA and ProP in its import into the C. glutamicum cell.
Materials and Methods
Strain, Plasmids, and Culture Conditions
The strains and plasmids used in this work are listed in Table 1. E. coli strains were routinely cultivated in LB medium (10 g tryptone, 5 g yeast extract and 10 g NaCl per liter) or on LB agar plates at 37°C. C. glutamicum strains were routinely precultivated in brain heart infusion (BHI, ROTH®) plates or liquid medium overnight at 30°C. For C. glutamicum main cultures in flask, CGXII medium (Eggeling and Bott, 2005) was inoculated to an initial OD600 of 1 using 4% (w/v) glucose as sole-carbon source. For C. glutamicum main cultures in BioLector (m2p-labs, Baesweiler, Germany), growth experiments were performed in Flowerplates at 1,000 rpm, 95% humidity, 30°C and backscatter gain 15, inoculated to an initial OD600 of 1 and using 4% (w/v) glucose as sole-carbon source. When necessary, the growth medium was supplemented with kanamycin (25 μg mL-1), spectinomycin (100 μg mL-1) and isopropyl β-D-1-thiogalactopyranoside (IPTG) (1 mM). For growth in hyperosmolar conditions 100, 200, or 400 mM of NaCl were added to the medium.
Table 1.
Strains and plasmids used in this work.
| Strains and plasmids | Description | Source |
|---|---|---|
| Strains | ||
| GSL | C. glutamicum ATCC13032 with the following modifications: Δpck, ΔsugR, ΔldhA, pycP458S, homV 59A, two copies of lysCT311I, two copies of asd, two copies of dapA, two copies of dapB, two copies of ddh, two copies of lysA, two copies of lysE, in-frame deletion of prophages CGP1 (cg1507-cg1524), CGP2 (cg1746-cg1752), and CGP3 (cg1890-cg2071) | Pérez-García et al., 2016 |
| GSLΔyggB | In-frame deletion of yggB (cg1434) in GSL | This work |
| GSLΔproP | In-frame deletion of proP (cg3395) in GSL | This work |
| JJ001 | C. glutamicum ATCC13032 with the following modifications:ΔargF, ΔargR (auxotrophic for L-arginine); carrying the vector pVWEx1 | Jensen and Wendisch, 2013 |
| JJ004 | JJ001 strain carrying the vector pVWEx1-ocdPp(TAA) | Jensen and Wendisch, 2013 |
| E. coli DH5α | F-thi-1 endA1 hsdr17 (r-, m-) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 relA1 | Hanahan, 1983 |
| E. coli S17-1 | recA, thi, pro, hsd R-M+ (RP4: 2-Tc:Mu-:Km, integrated into the chromosome) | Simon et al., 1983 |
| Plasmids | ||
| pVWEx1 | KmR, C. glutamicum/E. coli shuttle vector (Ptac, lacI, pHM1519 oriVCg) | Peters-Wendisch et al., 2001 |
| pEKEx3 | SpecR, C. glutamicum/E. coli shuttle vector (Ptac, lacI, pBL1 oriVCg) | Stansen et al., 2005 |
| pVWEx1-lysDH-proC | KmR, pVWEx1 overexpressing lysDH from S. pomeroyi DSS-3 and proC from C. glutamicum ATCC 13032 | Pérez-García et al., 2016 |
| pEKEx3-yggB | SpeR, pEKEx3 overexpressing yggB from C. glutamicum ATCC 13032 | This work |
| pEKEx3-proP | SpeR, pEKEx3 overexpressing proP from C. glutamicum ATCC 13032 | This work |
| pK19mobsacB | KmR; E. coli/C. glutamicum shuttle vector for construction of insertion and deletion mutants in C. glutamicum (pK18 oriVEc sacB lacZα) | Schaffer et al., 2001 |
| pK19mobsacB-ΔyggB | pK19mobsacB with a yggB (cg1434) deletion construct | Lubitz and Wendisch, 2016 |
| pK19mobsacB-ΔproP | pK19mobsacB with a proP (cg3395) deletion construct | This work |
Molecular Biology Methods
As host for gene cloning E. coli DH5α was used (Hanahan, 1983). E. coli was transformed by heat shock following the method described elsewhere (Hanahan, 1983), while C. glutamicum was transformed by electroporation following the method described elsewhere (Eggeling and Bott, 2005). The pair of primers YgFw/YgRv (Table 2) were used to amplified yggB from genomic DNA of C. glutamicum ATCC 13032. The pair of primers PrFw/PrRv (Table 2) were used to amplified proP from genomic DNA of C. glutamicum ATCC 13032. The amplified genes were cloned by Gibson assembly (Gibson, 2011) into the vector pEKEx3 (Stansen et al., 2005) digested with BamHI, yielding the vectors pEKEx3-yggB and pEKEx3-proP. Positive clones were verified by colony PCR using the pair of primers X1Fw/X1Rv (Table 2). The up- and downstream regions of proP gene were amplified by PCR from genomic DNA of C. glutamicum ATCC 13032 using the pair of primers PrDA/PrDB and PrDC/PrDD. The up and down amplified fragments were fused by cross-over PCR with primer pair PrDA/PrDD and cloned by ligation (Eggeling and Bott, 2005) into the vector pK19mobsacB (Schäfer et al., 1994) restricted with BamHI. Positive clones were verified by colony PCR using the pair of primers 196F/197R (Table 2). The resulting vector pK19mobsacB-gdh was transferred to E. coli S17-1. In-frame deletion of the yggB and proP genes from C. glutamicum was performed via a two-step homologous recombination method (Eggeling and Bott, 2005). The pK19mobsacB vectors were transferred to the GSL strain via conjugation using E. coli S17-1 (Simon et al., 1983). The deletions of yggB and proP were verified by colony PCR using the pair of primers DE31/DE32 and PrDE/PrDF, respectively.
Table 2.
Oligonucleotide sequences used in this work for molecular cloning and in-frame deletion.
| Primer | Sequence (5→3) |
|---|---|
| PrFw | GCATGCCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGGTGAGCCCGATTCGCTC |
| PrRv | AATTCGAGCTCGGTACCCGGGGATCTTATGCGTTTTGCTTTTCAG |
| YgFw | GCATGCCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGATGATTTTAGGCGTACCC |
| YgRv | AATTCGAGCTCGGTACCCGGGGATCCTAAGGGGTGGACGTCGG |
| PrDA | GCATGCCTGCAGGTCGACTCTAGAGTTCGGTGCCCTCCACGGCAC |
| PrDB | GGGTAGGTGATTTGAATTTGTGAGTAAAACCTCTCGTCATATC |
| PrDC | ACAAATTCAAATCACCTACCCCCGTAAAGCCCGCTGCAAGG |
| PrDD | AATTCGAGCTCGGTACCCGGGGATCGTAACGATGCAGACCGCCGG |
| PrDE | CGGTGCCCTCCACGGCACC |
| PrDF | AACGATGCAGACCGCCGGCG |
| DE31 | CTTTTGGCGCTCCAAGTACT |
| DE32 | TCCTCGAGCGATCGAACAAT |
| X1Fw | CATCATAACGGTTCTGGC |
| X1Rv | ATCTTCTCTCATCCGCCA |
| 196F | CGCCAGGGTTTTCCCAGTCACGAC |
| 197R | AGCGGATAACAATTTCACACAGGA |
Internal Amino Acids Extraction
For the quantification of intracellular L-PA 2 mL of liquid medium were collected. One milliliter was centrifuged at 14,000 rpm and 4°C for 10–15 min. The resulting pellets were resuspended and treated with 5% HClO4 in an ice bath for 30 min. Then, the supernatant was neutralized with K2CO3 solution and centrifuged again at 14,000 rpm and 4°C for 10–15 min. Afterward, the supernatants were directly used for L-PA quantification or stored at -20°C (Sun et al., 2016). It has to be noted that the water space of the pellet in such a centrifugation step will contain compounds presents in the extracellular volume and this will affect the determination of the intracellular concentration. On the other hand, intracellular compounds may leak out of the cell during washing steps and this will also affect the determination of the intracellular concentration. Since all samples were processed in the same way, the possible fluctuations/errors in the measurements should affect all samples similarly. Exact quantitation would require methods such as described by Klingenberg and Pfaff (1967) which combine centrifugation through silicone oil for fast separation of supernatant and pellet by centrifugation and inactivation of the pellet by perchloric acid. The second collected mL was used to determine the biomass according to the correlation CDW [g L-1] = 0.35 OD (Bolten et al., 2007).
Determination of L-PA by High Pressure Liquid Chromatography
The concentration of L-PA was quantified by using high-pressure liquid chromatography. The samples from the cell cultures were collected by centrifugation (14,000 rpm, 15 min and at room temperature), and further used for analysis. The samples were derivatized with fluorenylmethyl chloroformate (FMOC) as described (Schneider and Wendisch, 2010). Amino acid separation was performed on a system consisting of a pre-column (LiChrospher 100 RP18 EC-5μ (40 × 4 mm), CS-Chromatographie Service GmbH, Langerwehe, Germany) and a main column (LiChrospher 100 RP18 EC-5μ (125 × 4 mm), CS Chromatographie Service GmbH). The detection was carried out with a fluorescence detector with the excitation and emission wavelength of 230 nm and 310 nm, respectively (FLD G1321A, 1200 series, Agilent Technologies).
RNAseq Analysis
For extraction of C. glutamicum bacterial cell pellets grown under the experimental conditions were harvested at mid-exponential phase. Harvesting procedure was done according to Irla et al. (2015) and cell pellets were kept at -80°C for further RNA isolation. Then, the pellets were thawed in ice and RNA was isolated individually for each sample using NucleoSpin RNA isolation kit (Macherey-Nagel, Düren, Germany). RNA samples with genomic DNA contamination were treated with the RNase-free DNase set (Qiagen, Hilden, Germany) (Brito et al., 2017). The concentration of isolated RNA was determined by DropSenseTM 16 (Trinean, Ghent, Belgium; software version 2.1.0.18). To verify the quality of RNA samples, we performed capillary gel electrophoresis (Agilent Bioanalyzer 2100 system using the Agilent RNA 6000 Pico kit; Agilent Technologies, Böblingen, Germany). The extracted RNA samples were pooled in equal parts and the pool of total RNA was subsequently used for the preparation of the cDNA libraries. The preparation and sequencing of the libraries were performed as described elsewhere (Mentz et al., 2013; Irla et al., 2015). Then, the reads were trimmed to a minimal length of 20 base pairs and in paired end mode with the Trimmotatic ver. 0.33 (Bolger et al., 2014). Trimmed reads were mapped to the reference genome of C. glutamicum ATCC13032 (Kalinowski et al., 2003) using the software Bowtie (Langmead et al., 2009). In order to perform differential gene expression analysis (DEseq) (Anders and Huber, 2010), we used the software for visualization of mapped sequences ReadXplorer (Hilker et al., 2014).
Real-Time Quantitative Reverse Transcription-PCR
The real-time quantitative reverse transcription-PCR (qRT-PCR) was performed in order to validate the data obtained by DEseq analysis by using the CFX ConnectTM Real-Time PCR Detection System (Bio-Rad Laboratories, Irvine, CA, United States). Same RNA samples utilized in the RNAseq analysis were utilized as templates for qRT-PCR. All samples RNA concentration was adjusted to 50 ng μL-1. Afterward, 1 μL for each sample was pipetted into a reaction mix of the SensiFASTTM SYBR® No-ROX Kit (Bioline, Luckenwalde, Germany), following manufacturer’s instructions. Differentially expressed genes in DEseq analysis were selected as targets for qRT-PCR amplifications (primers listed in Table 3). The melting-curve data-based quantification cycle (Cq) values, from the LightCycler® output files, were used for further calculation as it is described elsewhere (Crooks et al., 2004).
Table 3.
Oligonucleotide sequences (5→3) used for amplification of gene fragments in qRT-PCR.
| Gene identity | Forward | Reverse | Gene product length (bp) |
|---|---|---|---|
| betP | GCGGGCTTGCTTGAGAATCC | TGAAGGCCCAGCCGAGATTG | 232 |
| cg0569 | AGCTTTGGCTGCTTCAGTAG | AGATTCCATGCCGGAACTTG | 241 |
| cg1665 | GCTGCCAACTCTGCAACCTC | CCATTCGGGCCTTCTTCCAC | 245 |
| cg2677 | GGCTCTGCCTCCATTCTTTG | GGTTGTGCCTTGACCTCTTC | 210 |
| cg2851 | CAACGTGAACACGGTGTATC | CACATCGTCGAATCCGTTTG | 210 |
| cg3254 | ATGCTTGCCCTAGGTTGG | CCGAGTGAAGAACTGCACG | 255 |
| cg3282 | ATGACCTGCGGACACTGC | TCAGGACAAGACGGTGTAG | 180 |
| gntV | TCCGTCGGTAAAGCCCTAGC | CGGTTCCTGGGCATTTGGTG | 238 |
| proC | CGCGGCCAACATGAATCCAC | GGCCATGCTGACCACAACAC | 232 |
| proP | TCGACTGGTGGTGAATATGC | GAATACGCCAACCGAAATCC | 202 |
| pstC | AATGCGAACTCCTCTCAGAC | AATCCGCCAATACCTTCAGC | 206 |
| pstS | TCCGCAATGGACTACTTTGG | AACTGGGCCGATAACGAATG | 222 |
Results
C. glutamicum Can Use L-PA for Osmoprotection
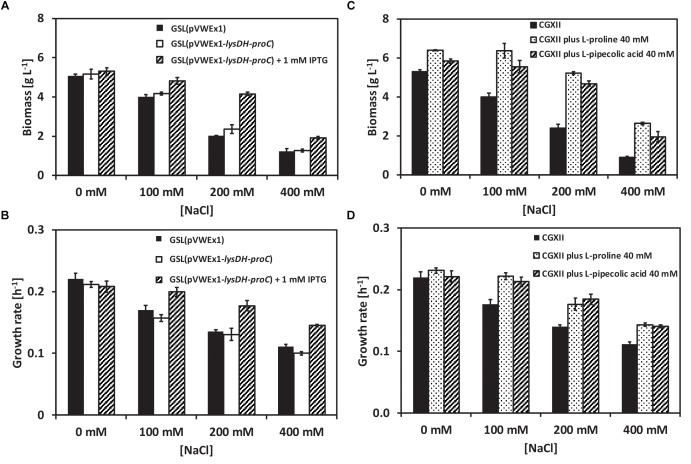
Structurally, L-PA is related to L-proline. To check whether L-PA functions as osmoprotectant in C. glutamicum the L-PA producer GSL(pVWEx1-lysDH-proC) (Pérez-García et al., 2016) was grown in glucose-minimal medium supplemented with 0, 100, 200, and 400 mM of NaCl using a BioLector system. Production of L -PA was induced by adding IPTG. When not induced for L-PA production, strains GSL(pVWEx1) and GSL(pVWEx1-lysDH-proC) showed decreasing growth rates (Figure 1A) and maximal biomass formation (Figure 1B) with increasing NaCl concentration. However, when L-PA production was induced, C. glutamicum GSL(pVWEx1-lysDH-proC) grew faster (Figure 1A) and to higher biomass concentrations (Figure 1B) in the presence of NaCl than the parent strain GSL(pVWEx1) (Figure 1). This indicated that biosynthesis of L-PA helps C. glutamicum to withstand hyperosmolar conditions.
FIGURE 1.
Growth of the C. glutamicum strains GSL(pVWEx1) and GSL(pVWEx1-lysDH-proC) under different osmotic conditions. (A) Biomass formation in g L-1 of GSL(pVWEx1) and GSL(pVWEx1-lysDH-proC) strains in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl. (B) Growth rates values in h-1 of GSL(pVWEx1) and GSL(pVWEx1-lysDH-proC) strains in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl. (C) Biomass formation in g L-1 of GSL(pVWEx1) strain in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl with 40 mM of L-proline or 40 mM of L-PA or nothing. (D) Growth rates values in h-1 of GSL(pVWEx1) strain in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl with 40 mM of L-proline or 40 mM of L-PA or nothing. Values represent means and standard deviations.
To test if also exogenously added L-PA is beneficial for C. glutamicum when grown under hyperosmolar conditions, 40 mM of either L-PA or L-proline were added to the glucose-minimal medium and growth of C. glutamicum GSL(pVWEx1) was monitored (Figure 1C,D). The exogenous addition of L-PA or L-proline improved growth of strain GSL(pVWEx1) in the presence of all NaCl concentrations tested (Figure 1C,D). Thus, under the chosen conditions L-PA functions as well as the known osmoprotectant of C. glutamicum, L-proline, in promoting growth under hyperosmolar conditions. While several transport proteins for the compatible solute L-proline are known, transport of L-PA has not yet been studied.
The Mechanosensitive Channel YggB May Play a Role as Escape Valve for L-PA
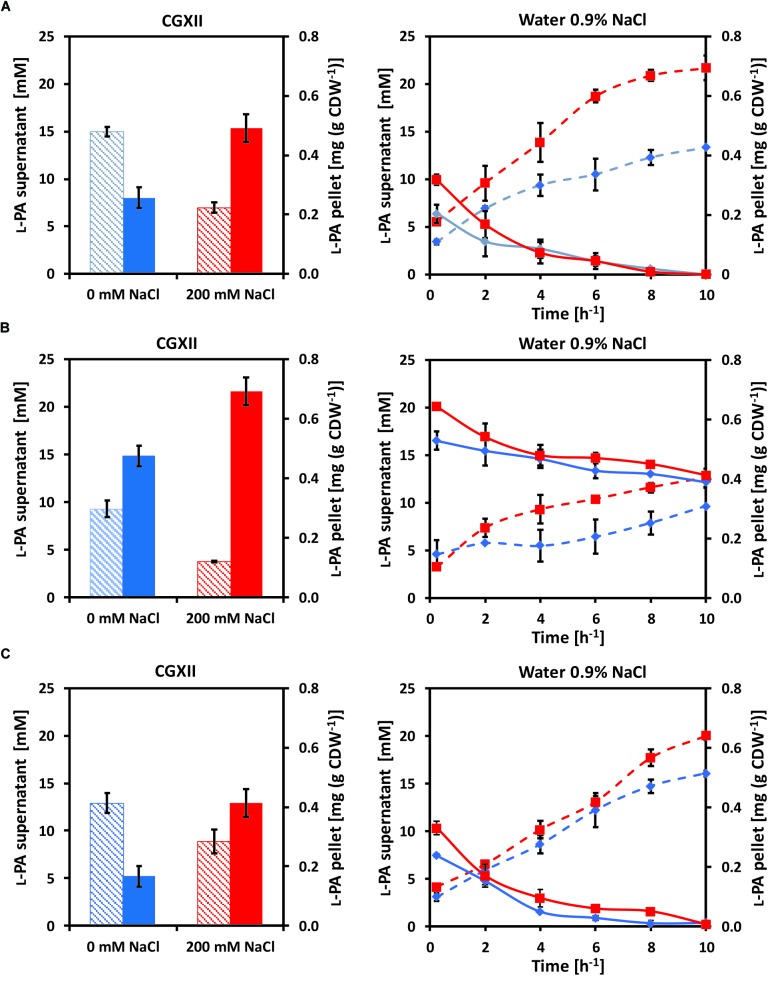
Osmo compatible solutes accumulate intracellularly under hyperosmolar conditions and are released from the cell upon osmotic downshift. Since YggB has been shown to be a key player in osmoregulation in C. glutamicum (Börngen et al., 2010), yggB deletion mutants were also analyzed. Here, the accumulation and release of L-PA by C. glutamicum cells were analyzed (Figure 2). C. glutamicum cells were cultivated in 50 mL glucose-minimal medium without (blue columns) or with (red columns) 200 mM NaCl. When glucose was depleted 1 mL supernatant and 2 mL pellet were collected to measure the extracellular (dashed columns) and intracellular (filled columns) concentrations of L-PA (Figure 2, left panels). The rest of the pellet of cells that grew in CGXII (blue lines) or CGXII + 200 mM NaCl (red lines) was transferred to 35 mL milliQ-water 0.9% NaCl to force an osmotic downshift while keeping the cells intact. The extracellular (dashed lines) and intracellular (solid lines) concentrations of L-PA were monitored over time (Figure 2, right panels). The experiment was performed with the L-PA producing strain GSL(pVWEx1-lysDH-proC) (Figure 2A); a yggB deletion mutant of this strain lacking the MSC YggB, GSLΔyggB(pVWEx1-lysDH-proC) (Figure 2B); and a derived strain expressing yggB from a plasmid for complementation of the yggB deletion, GSLΔyggB(pVWEx1-lysDH-proC)(pEKEx3-yggB) (Figure 2C). After growth in CGXII minimal medium with 200 mM NaCl, the intracellular L-PA concentrations were higher than after growth in CGXII minimal medium without added NaCl (Figure 2A), left). Upon osmotic downshift, L-PA was released from cells grown without NaCl and accumulated in the medium with a rate of 0.97 ± 0.04 mM h-1. However, when cells grown with 200 mM NaCl were subjected to osmotic downshift, L-PA was released with a 75% higher rate and to an about 1.6 higher concentration (Figure 2A, right and Table 4).
FIGURE 2.
Intracellular (filled columns, straight lines) and extracellular (dashed columns, dashed lines) L-PA concentration profiles of the strains (A) GSL(pVWEx1-lysDH-proC), (B) GSLΔyggB(pVWEx1-lysDH-proC) and (C) GSLΔyggB(pVWEx1-lysDH-proC)(pEKEx3-yggB) after growth in glucose-minimal medium (left panels) without added NaCl (blue) or with 200 mM NaCl (red) and after osmotic downshift to 0.9% NaCl (right panels). Values represent means and standard deviations.
Table 4.
Rates of decrease of intracellular L-PA concentrations and of L-PA accumulation in the cultivation medium upon osmotic downshock of different C. glutamicum strains after growth in glucose minimal medium with or without 200 mM NaCl.
| Rates of decrease of intracellular L-PA concentrations (μg g CDW-1 h-1) | Rates of L-PA accumulation in the supernatant (mM h-1) | |||
|---|---|---|---|---|
| Strain | After growth with 0 mM NaCl | After growth with 200 mM NaCl | After growth with 0 mM NaCl | After growth with 200 mM NaCl |
| GSL(pVWEx1-lysDH-proC) | 19.3 ± 0.2 | 30.2 ± 0.4 | 0.97 ± 0.04 | 1.73 ± 0.02 |
| GSLΔyggB(pVWEx1-lysDH-proC) | 14.0 ± 0.2 | 20.8 ± 0.3 | 0.47 ± 0.13 | 0.88 ± 0.17 |
| GSLΔyggB(pVWEx1-lysDH-proC)(pEKEx3-yggB) | 22.9 ± 0.3 | 28.9 ± 0.4 | 1.37 ± 0.16 | 1.68 ± 0.14 |
Values represent means and standard deviations. Important strain features are depicted in bold.
When growing the strain GSLΔyggB(pVWEx1-lysDH-proC) on CGXII with or without 200 mM NaCl the internal accumulation of L-PA increased 80 and 40%, respectively, as compared with the strain GSL(pVWEx1-lysDH-proC) (Figure 2B, left). On 0.9% NaCl, the L-PA external accumulation rates decreased to the half (Figure 2B, right and Table 4). Therefore, the deletion of yggB slowed down L-PA export but was not enough to fully avoid it. The strain GSLΔyggB(pVWEx1-lysDH-proC)(pEKEx3-yggB) showed similar L-PA internal/external profiles in both CGXII and water as compared with the strain GSL(pVWEx1-lysDH-proC) (Figure 2C, right and Table 4).
Comparative RNAseq Analysis of a L-PA Producing C. glutamicum Strain, a L-Proline Producing Strain and the Respective Control Strains
Under the assumption that genes relevant for production of either L-PA or L-proline are differentially expressed when comparing L-PA producing C. glutamicum strain GSL(pVWEx1-lysDH-proC) with its parent strain GSL(pVWEx1) and of L-proline producing C. glutamicum strain JJ004 with its parent strain JJ001, comparative RNAseq analysis was performed. C. glutamicum strains GSL(pVWEx1-lysDH-proC), GSL(pVWEx1), JJ004 and JJ001 were grown in glucose minimal medium with 1 mM IPTG after inoculation with an initial OD600 of 1. Samples for RNA preparation were harvested after 8 h of inoculation at an OD600 of 7.6 ± 0.4 and 7.7 ± 0.4, respectively, for strains GSL(pVWEx1-lysDH-proC) and GSL(pVWEx1). Sample for strains JJ004 and JJ001 were harvested 6 h after inoculation at an OD600 of 7.6 ± 0.1 and 8.0 ± 0.2, respectively. RNA and library preparation, sequencing, read mapping and differential gene expression analysis using the statistical method DEseq (Naville et al., 2011) was performed as described in Section “Materials and Methods.” Full data is available as Gene Expression Omnibus GSE122249 data set at http://www.ncbi.nlm.nih.gov/geo/. As compared to the respective control strains, 5 genes showed increased RNA levels and 17 genes decreased RNA levels in the L-PA producing strain, while 26 genes showed increased RNA levels and 33 genes decreased RNA levels in the L-proline producing strain (Table 5).
Table 5.
Comparative RNAseq analysis of L-proline producing strain JJ004, its isogenic non-producing control strain JJ001, L-PA producing C. glutamicum strain GSL(pVWEx1-lysDH-proC) and its isogenic non-producing control strain GSL(pVWEx1).
| Log2 fold change of RNA level (P-value < 0.01) | |||
|---|---|---|---|
| Locus or gene | Product | JJ004/ control JJ001 | GSL(pVWEx1-lysDH-proC)/ control GSL(pVWEx1) |
| cg0018 | Conserved hypothetical membrane protein | 0.8 | -1.0 |
| cg0107 | Putative secreted protein | 0.3 | -1.0 |
| cg0175 | Putative secreted protein | -0.8 | -1.2 |
| cg0282 | CsbD family protein involved in stress response | -0.8 | -0.6 |
| ctpA | Putative Cu2+ transporting P-type ATPase | 1.1 | 0.2 |
| proC | Pyrroline-5-carboxylate reductase | 0.0 | 4.7 |
| glyR | Transcriptional activator of glyA, ArsR-family | 1.0 | -0.5 |
| cg0569 | Putative Cd2+ transporting P-type ATPase | 1.0 | 1.7 |
| whiB3 | Transcriptional regulator protein, WhiB-family | 2.6 | -0.5 |
| prpC2 | 2-Methylcitrate synthase | 0.9 | 0.7 |
| cg0770 | ABC-type putative iron-siderophore transporter, permease subunit | -1.9 | 0.3 |
| irp1 | ABC-type putative iron-siderophore transporter, substrate-binding lipoprotein | -2.3 | -0.3 |
| whcE | Transcriptional regulator, WhiB-family | 0.8 | -0.4 |
| pdxS | Pyridoxal 5’-phosphate (PLP) synthase subunit S | -0.8 | 0.0 |
| pdxT | Pyridoxal 5’-phosphate (PLP) synthase subunit T | -1.0 | 0.5 |
| cg0924 | ABC-type putative iron-siderophore transporter, substrate-binding lipoprotein | -1.7 | -0.5 |
| cg0926 | ABC-type putative iron-siderophore transporter, permease subunit | -1.3 | -0.2 |
| cg0935 | Conserved hypothetical protein | -0.4 | -1.3 |
| rpf1 | RPF-protein precursor | 0.9 | -0.4 |
| cg0952 | Putative integral membrane protein | -0.9 | -0.3 |
| betP | Na+/glutamate symporter | -1.2 | -0.7 |
| cg1091 | Hypothetical protein | -1.6 | -1.0 |
| cg1091 | Hypothetical protein | -1.6 | -1.0 |
| cg1109 | Hypothetical protein | -0.3 | -1.1 |
| cg1279 | Putative secreted protein | 0.8 | 0.6 |
| cg1291 | Putative membrane protein | 1.2 | -0.1 |
| cg1293 | Putative secreted protein | -0.8 | -0.4 |
| putP | Na+/proline symporter | -0.8 | -0.1 |
| cg1419 | Putative secondary Na+/bile acid symporter, bile acid:Na+ symporter (BASS) family | -1.7 | -0.2 |
| lysE | L-Lysine efflux permease | -5.4 | -0.5 |
| leuC | 3-Isopropylmalate dehydratase, large subunit | -0.8 | 0.0 |
| ptsG | Phosphotransferase system (PTS), glucose-specific enzyme IIBCA component | -0.9 | 0.1 |
| cg1604 | Secreted protein, putative channel protein | -0.9 | 0.1 |
| cg1665 | Putative secreted protein | -1.7 | -0.9 |
| cg1746 | Putative membrane protein | 0.8 | n.d. |
| cg1897 | Putative secreted protein | -3.1 | n.d. |
| cg1930 | Putative secreted hydrolase | -1.1 | n.d. |
| cg2068 | Hypothetical protein | -1.0 | n.d. |
| psp1 | Putative secreted protein | -1.6 | n.d. |
| int2’ | Putative phage Integrase (N-terminal fragment) | -1.6 | n.d. |
| cg2181 | ABC-type putative dipeptide/oligopeptide transporter, substrate-binding lipoprotein | -0.8 | 0.1 |
| xerC | Putative site-specific recombinase | -0.2 | -1.2 |
| cg2402 | Secreted protein NLP/P60 family | 0.8 | -0.1 |
| cg2425 | Putative permease | 0.7 | 0.3 |
| cg2477 | Conserved hypothetical protein | 0.0 | -0.8 |
| cg2564 | Conserved hypothetical protein | -1.7 | -0.5 |
| catA | Catechol 1,2-dioxygenase | 0.3 | 0.7 |
| cg2651 | Conserved hypothetical protein, pseudogene | -0.6 | -1.4 |
| cg2677 | ABC-type putative dipeptide/oligopeptide transporter, permease subunit | 0.0 | 0.9 |
| gntV | Gluconokinase | -0.5 | 1.8 |
| rpmJ | 50S ribosomal protein L36 | -0.3 | -1.0 |
| pstB | ABC-type phosphate transporter, ATPase subunit | 1.3 | 0.1 |
| pstA | ABC-type phosphate transporter, permease subunit | 1.2 | 0.5 |
| pstC | ABC-type phosphate transporter, permease subunit | 1.5 | 0.3 |
| pstS | ABC-type phosphate transporter, substrate-binding lipoprotein | 1.3 | 0.1 |
| cg2851 | Branched-chain amino acid aminotransferase, AT class III/4-amino-4-deoxychorismate lyase | 0.0 | -1.6 |
| cg2875 | Hypothetical protein | 0.8 | -0.3 |
| cg2908 | Putative membrane protein | -1.0 | 0.1 |
| pck | Phosphoenolpyruvate carboxykinase (GTP) | 0.8 | 0.0 |
| gntP | Gluconate:H+ symporter | -1.0 | 0.1 |
| cg3218 | Pyruvate kinase-like protein | 1.2 | 0.4 |
| ldh | L-Lactate dehydrogenase, NAD-dependent | 0.9 | 0.0 |
| cg3254 | Putative membrane protein | -0.8 | -0.9 |
| cg3271 | SAM-dependent methyltransferase | 0.3 | -1.1 |
| cg3281 | Putative Cu2+ transporting P-type ATPase | 1.2 | -0.4 |
| cg3282 | Putative Cu2+ transporting P-type ATPase | 1.4 | 0.0 |
| cg3326 | Hypothetical protein | 1.1 | 0.8 |
| mez | Malic enzyme | 0.9 | 0.3 |
| proP | Proline/betaine permease | -1.2 | -1.2 |
| cg3402 | Putative Hg2+ permease, MerTP-family | 1.2 | 0.0 |
| cg3404 | ABC-type putative iron(III) dicitrate transporter, substrate-binding lipoprotein | -2.1 | -0.4 |
| cg4014 | Conserved hypothetical protein, possibly involved in stress response | 0.3 | -1.0 |
| cg4019 | 0.8 | 0.1 | |
| cg4021 | -0.6 | -1.1 | |
Names of genes used for subsequent qRT-PCR analysis are given in bold. n.d., not detected.
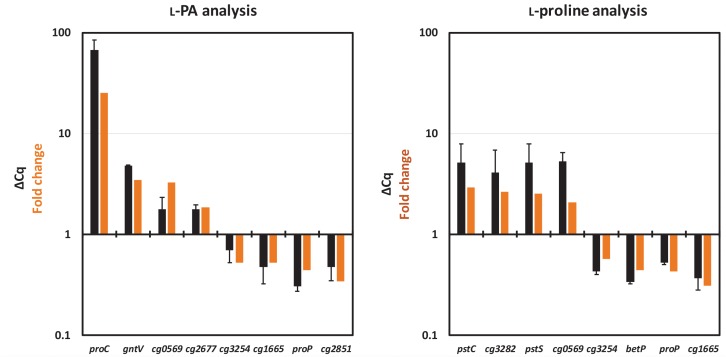
The results obtained in the RNAseq analysis were validated by the analysis of gene expression patterns by qRT-PCR. For each analysis eight genes were selected, four upregulated and four downregulated genes. As shown in Figure 3, the relative gene expression levels obtained in qRT-PCR confirmed the pattern of their differential gene expression (fold change value) obtained in the RNAseq analysis.
FIGURE 3.
Comparison of relative gene expression values obtained by qRT-PCR analysis (black bars) with those obtained by RNAseq analysis (orange bars). RNAseq data from Table 5 and qRT-PCR data (ΔCq) collected for the L-PA analysis (left) and for the L-proline analysis (right) are listed. The values from the qRT-PCR are given as means and standard deviations.
In the L-proline producing strain JJ004, pstSCAB encoding phosphate ABC uptake system, genes for divalent metal transporter proteins (ctpA, cg0569, cg3281, cg3282, and cg3402), for transcriptional regulators (glyR, whiB3, and whcE) and for enzymes of central carbon metabolism (pck, ldh and mez) showed higher RNA levels than in the control strain JJ001 (Table 5). As compared to JJ001, RNA levels were lower in JJ004 for genes encoding iron-siderophore ABC uptake systems (cg0770, irp1, cg0924 and cg0926), the lysine/arginine permease gene lysE and genes for uptake of L-proline and other compatible solutes (betP, putP and proP) (Table 5). Thus, it appears that upon overproduction of L-proline, genes for its uptake from the culture medium are downregulated.
As expected, RNAseq analysis of the L-PA producing strain revealed increased expression of pyrroline-5-carboxylate reductase gene proC since it is expressed from plasmid pVWEx1-lysDH-proC (Table 5). Other genes showing increased RNA levels upon L-PA production were the divalent metal transporter protein gene cg0569, the gluconokinase gene gntV and the catechol 1,2-dioxygenase gene catA (Table 5). Genes showing decreased RNA levels upon L-PA production were the putative site-specific recombinase gene xerC and the compatible solute transport gene proP (Table 5). Notably, the genes betP and putP coding for uptake systems of L-proline and other compatible solutes did not show increased RNA levels. Thus, ProP was chosen as potential candidate for import of L-PA into the C. glutamicum cell.
Role of the Carrier ProP During Growth With L-PA as Osmo Compatible Solute
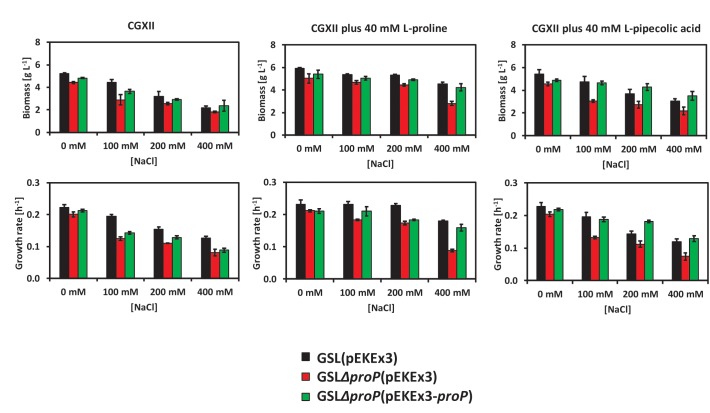
Deduced from the RNAseq data, we speculated that the carrier ProP may play a role with regard to L-PA as osmo compatible solute of C. glutamicum. ProP is used by C. glutamicum as the main osmoregulated uptake system for L-proline (Peter et al., 1998). To test if the absence of ProP affects the use of C. glutamicum of L-PA under hyperosmolar conditions, strain GSLΔproP was constructed. The strains GSL(pEKEx3) (Figure 4, black columns), GSLΔproP(pEKEx3) (Figure 4, red columns) and GSLΔproP(pEKEx3-proP) (Figure 4, green columns) were grown in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl using a BioLector system. CGXII contains 200 mM MOPS buffer, thus, has a relatively high osmolarity: about 1,1 osmol/kg without added NaCl as compared to about 1,3 osmol/kg CGXII medium with 400 mM NaCl (Börngen et al., 2010). In addition, the effect of externally added L-proline or L-PA was tested. Data for final biomass formation and growth rate was collected for all conditions (Figure 4). It was observed that the strains carrying the deletion of proP suffered more from the hyperosmotic conditions as compared to the control C. glutamicum strain GSL(pEKEx3) or the complementation strain GSLΔproP(pEKEx3-proP) (Figure 4). When 40 mM of L-proline was supplemented as osmo compatible solute to the minimal medium the growth rates and final biomass concentrations were reduced to a lesser extent than when 40 mM L-PA was added (Figure 4). Thus, ProP plays an important role when L-proline and L-PA are used as osmo compatible solutes in C. glutamicum. These findings are commensurate with the view that ProP does not only import L-proline into the C. glutamicum cell, but also L-PA. However, future in depth biochemical analysis of L-PA uptake are needed to determine the respective kinetic parameters of L-PA uptake.
FIGURE 4.
Growth behavior of the C. glutamicum strains GSL(pEKEx3) (black bars), GSLΔproP(pEKEx3) (red bars) and GSLΔproP(pEKEx3-proP) (green bars) under different osmotic conditions. Biomass formation in g L-1 (top-left) and growth rate in h-1 (down-left) when growing the strains in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl. Biomass formation in g L-1 (top-middle) and growth rate in h-1 (down-middle) when growing the strains in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl an in presence of 40 mM of L-proline. Biomass formation in g L-1 (top-right) and growth rate in h-1 (down-right) when growing the strains in glucose minimal medium supplemented with 0, 100, 200, or 400 mM of NaCl an in presence of 40 mM of L-PA. Values represent means and standard deviations.
Discussion
In this study, L-PA was shown to be an osmo compatible solute for C. glutamicum. L-PA cannot be synthesized by C. glutamicum wild type, but can be imported from the environment. The synthesis and/or accumulation of compatible solutes is a widespread microbial strategy against osmolarity fluctuations (da Costa et al., 1998; Kempf and Bremer, 1998; Wood et al., 2001; Czech et al., 2018). High cytoplasmic concentrations of compatible solutes also stabilize protein folding and ribosomes and protect the DNA, increasing the resistance to other types of stress such as high and low temperatures and radiation (Li and Gänzle, 2016; Sajjad et al., 2018; Tribelli and López, 2018). The osmoprotection mechanisms for the microbial cell factories E. coli, Bacillus subtilis and C. glutamicum are described (Kempf and Bremer, 1998; Wood et al., 2001; Morbach and Krämer, 2003; Hoffmann and Bremer, 2017). These and other non-halophilic bacteria accumulate K ions, Na ions or glutamate after an osmotic upshock as first response before these ions are exchanged against compatible solutes either by synthesis or uptake (Wood, 1999). C. glutamicum either synthesizes glutamine, proline or trehalose after an osmotic upshift or imports glycine betaine, proline or ectoine (Frings et al., 1993; Guillouet and Engasser, 1995; Skjerdal et al., 1996). In C. glutamicum, biosynthesis of proline involves one set of genes (proA for g-glutamyl phosphate reductase, proB for g-glutamyl kinase and proC for pyrroline 5-carboxylate reductase), while B. subtilis possesses one set of proline biosynthesis genes for anabolic purposes and a second osmostress-induced set (Bremer, 2000). Similarly, biosynthesis and internal content of L-PA is regulated by external osmolality in Brevibacterium ammoniagenes (Gouesbet et al., 1992). It has been described that C. glutamicum prefers uptake of compatible solutes to their synthesis because uptake of compatible solutes is faster and energetically more favorable than their synthesis (Morbach and Krämer, 2003). In C. glutamicum, glycine betaine is the most effective osmoprotectant among those that can be imported from the environment, followed by ectoine and proline (Farwick et al., 1995). Here, we have shown that 40 mM L-PA are almost as effective as 40 mM proline for osmoprotection of a lysine producing C. glutamicum strain (Figure 1, 4). The improved growth in the presence of increasing salt concentrations is not due to catabolism of L-PA since C. glutamicum can use L-PA neither as carbon source nor as nitrogen source (Pérez-García et al., 2016). Rather, the external addition of L-PA is advantageous since resources for biosynthesis of L-proline for osmoprotection are not required. An osmoprotective role of L-PA has also been shown for E. coli since the addition of 1 mM of DL-PA decreased the inhibitory growth effect of 200–700 mM NaCl in 0.2% glucose minimal medium (Gouesbet et al., 1994). When growing S. pomeroyi in a mineral salts medium containing 400 mM NaCl, the supplementation of 5–20 mM of L-PA improved the growth of the microorganism (Neshich et al., 2013). In Sinorhizobium meliloti, both isomers, L-PA and D-PA have to be added for osmoprotective activity (Gouffi et al., 2000). Previously, we have developed strains for sustainable production of L-PA (Pérez-García et al., 2016, 2017a). Here, we have shown that recombinant C. glutamicum engineered for L-PA overproduction showed improved growth characteristics under hyperosmolar conditions (Figure 4). Thus, L-PA functions as osmo compatible solute not only when imported from the environment, but also when synthesized de novo. In these recombinant C. glutamicum strain synthesis is not osmostress-induced as described for certain plants. For example, in rapeseed leaf tissues, L-PA synthesis from L-lysine via the lysine-ketoglutarate reductase/saccharopine dehydrogenase pathway is osmo-dependent (Moulin et al., 2006).
Although uptake of osmo compatible solutes is important and energetically favored over their de novo synthesis, a C. glutamicum mutant devoid of the five known uptake systems for compatible solutes survived under hyperosmolar conditions (Steger et al., 2004) which has been attributed to de novo synthesis of proline, glutamine, and trehalose (Rönsch et al., 2003). The secondary carriers PutP, BetP, EctP, LcoP, and ProP differ by their substrate spectrum and substrate affinities, however, they show a degree of substrate redundancy that is typical for soil bacteria (Peter et al., 1997, 1998; Wood et al., 2001; Weinand et al., 2007). BetP is a high affinity carrier specific for betaine (Peter et al., 1997). EctP is a low affinity carrier for betaine, ectoine and proline and LcoP a low affinity carrier for betaine and ectoine, whereas the carrier ProP shows high affinity for its substrates proline and ectoine (Peter et al., 1998; Steger et al., 2004). PutP imports proline with high affinity for anabolic purposes (Peter et al., 1997). Based on differential gene expression analysis ProP was identified as possible L-PA import system in this work (Table 5). In E. coli various structural analogs of L-proline such as azetidine-2-carboxylate, L-pipecolic acid or 5-hydroxy-L-pipecolic acid enter the cell through ProP or ProU transport systems (Gouesbet et al., 1994). The E. coli proline/glycine betaine transporter ProP shares 36.8% identical amino acids with ProP from C. glutamicum. Here, growth analysis of C. glutamicum mutants lacking proP revealed perturbed growth under hyperosmolar conditions in the absence of proP (Figure 4). Since mutants lacking proP still possess functional EctP and PutP these carriers apparently do not contribute to uptake of L-PA under the chosen conditions, and, thus, ProP may act as major L-PA import system in C. glutamicum. Detailed biochemical transport assays will have to be performed in the future to characterize L-PA uptake by ProP.
It was also shown in this work that the MSC YggB performs as a major escape valve for L-PA in C. glutamicum (Figure 2). After an osmotic downshift compatible solutes are released to the medium involving MSCs (Morbach and Krämer, 2003). In particular, the MSC YggB was described as the main export system of L-glutamate in C. glutamicum (Nakamura et al., 2007). In C. glutamicum it is known that the use of biotin limitation, penicillin treatments or surfactants alter membrane tension by inhibiting lipid or peptidoglycan synthesis which triggers conformational changes in YggB allowing L-glutamate export (Duperray et al., 1992; Gutmann et al., 1992). In addition, betaine efflux induced by osmotic downshock was reduced upon deletion of yggB (Nottebrock et al., 2003). Thus, the decreased rate of L-PA accumulation in the supernatant as consequence of yggB deletion suggests that L-PA may be exported from the C. glutamicum cell by YggB. In depth biochemical analysis is required to characterize export of L-PA by YggB.
Due to their diverse applications in drug development, food industry, skin care products and cosmetics (Graf et al., 2008; Jorge C.D. et al., 2016; Li and Gänzle, 2016) the biotechnological production of compatible solutes has gained increasing momentum recently (Sauer and Galinski, 1998; Jensen and Wendisch, 2013; Tan et al., 2016; Chen et al., 2017). This included the establishment of strains that produce and secrete compatible solutes such as ectoine, L-PA or α-D-glucosylglycerol that are not synthesized by the wild-type strains (Ning et al., 2016; Pérez-García et al., 2017a,b; Ying et al., 2017; Roenneke et al., 2018). Production of L-PA by recombinant E. coli expressing the gene for lysine cyclodeaminase from Streptomyces hygroscopicus was established with a titer of 5.33 g L-1 L-PA and a yield of 0.13 g L-1 of glucose obtained in fed-batch cultivation and a titer of 0.64 g L-1 L-PA in shake flasks (Ying et al., 2017). Our previous work on establishing L-PA production in C. glutamicum led to superior values: 14.4 g L-1 L-PA and a yield of 0.20 g g-1 in fed-batch cultivation and a titer of 3.9 g L-1 L-PA in shake flasks (Pérez-García et al., 2017a). Although de novo synthesized L-PA protected C. glutamicum against high salt conditions, we have observed that in the presence of 200 mM NaCl the L-PA titer in the supernatant was reduced from about 15 mM (about 1.9 g L-1) to about half (Figure 2A, left panel). Thus, hyperosmolar conditions are not favorable for L-PA production by the C. glutamicum recombinant strains described here. However, while less L-PA was secreted under hyperosmolar conditions, more L-PA accumulated intracellularly (Figure 2A, left panel). After osmotic downshift, L-PA was released to the culture medium accumulating to about 22 mM (about 2.8 g L-1) (Figure 2A, right panel). Therefore, in principle, the described C. glutamicum strains could be used in a process called “bacterial milking” (Sauer and Galinski, 1998). The Gram-negative bacterium Halomonas elongata was grown to high-cell-density (48 g cell dry weight per liter) before being exposed to alternating hyper- and hypo-osmolar conditions. Ectoine released to the hypo-osmolar medium was harvested by crossflow filtration and by this procedure 0.16 g of ectoine per cycle per gram cell dry weight could be produced (Sauer and Galinski, 1998). As use of high-salinity media in fermentation processes is costly and poses challenges with regard to the design and durability of bioreactors, it is generally assumed that direct fermentative production is preferred over the “bacterial milking” process. To determine if this notion holds true for the L-PA producing C. glutamicum strains described here, a head-to-head comparison of these strains operated in a fed-batch fermentation process vs. a “bacterial milking” process will have to be performed after each process has been thoroughly optimized by process intensification. In addition, strain optimization by transport engineering, as described for the production of amino acids (Nakamura et al., 2007; Blombach et al., 2009), non-proteinogenic amino acids (Jorge J.M.P. et al., 2016; Pérez-García et al., 2017a), diamines (Kind et al., 2014; Nguyen et al., 2015) or organic acids (Huhn et al., 2011), may be required.
Data Availability
The datasets generated for this study can be found in http://www.ncbi.nlm.nih.gov/geo/, GSE122249.
Author Contributions
FP-G, LB, and VW designed the study. FP-G and LB performed the experiments. FP-G, LB, and VW analyzed the data. FP-G and LB drafted the manuscript. VW finalized the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Tobias Busche and Prof. Dr. Jörn Kalinowski from the technology platform Genomics of CeBiTec for the kind assistance with the preparation and sequencing of the cDNA libraries.
Footnotes
Funding. FP-G was funded by the University of Bielefeld (Bielefeld Young Researchers Fund, Germany). LB was funded by the Science without Borders program (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil). The funding agencies were not involved in the design of the study, collection, analysis, and interpretation of data and in writing the manuscript.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B., Hans S., Bathe B., Eikmanns B. J. (2009). Acetohydroxyacid synthase, a novel target for improvement of L-lysine production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 75 419–427. 10.1128/AEM.01844-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten C. J., Kiefer P., Letisse F., Portais J.-C., Wittmann C. (2007). Sampling for metabolome analysis of microorganisms. Anal. Chem. 79 3843–3849. 10.1021/ac0623888 [DOI] [PubMed] [Google Scholar]
- Börngen K., Battle A. R., Möker N., Morbach S., Marin K., Martinac B., et al. (2010). The properties and contribution of the Corynebacterium glutamicum MscS variant to fine-tuning of osmotic adaptation. Biochim. Biophys. Acta 1798 2141–2149. 10.1016/j.bbamem.2010.06.022 [DOI] [PubMed] [Google Scholar]
- Bremer E. (2000). Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes in B. subtilis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 126:17. [Google Scholar]
- Brito L. F., Irla M., Kalinowski J., Wendisch V. F. (2017). Detailed transcriptome analysis of the plant growth promoting Paenibacillus riograndensis SBR5 by using RNA-seq technology. BMC Genomics 18:846. 10.1186/s12864-017-4235-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Zhu L., Lv L., Yao S., Li B., Qian J. (2017). Optimization of the extraction and purification of the compatible solute ectoine from Halomonas elongata in the laboratory experiment of a commercial production project. World J. Microbiol. Biotechnol. 33:116. 10.1007/s11274-017-2281-y [DOI] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech L., Hermann L., Stöveken N., Richter A. A., Höppner A., Smits S. H. J., et al. (2018). Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes 9:E177. 10.3390/genes9040177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa M. S., Santos H., Galinski E. A. (1998). An overview of the role and diversity of compatible solutes in bacteria and archaea. Adv. Biochem. Eng. Biotechnol. 61 117–153. [DOI] [PubMed] [Google Scholar]
- Duperray F., Jezequel D., Ghazi A., Letellier L., Shechter E. (1992). Excretion of glutamate from Corynebacterium glutamicum triggered by amine surfactants. Biochim. Biophys. Acta Biomembr. 1103 250–258. 10.1016/0005-2736(92)90094-3 [DOI] [PubMed] [Google Scholar]
- Eggeling L., Bott M. (2005). Handbook of Corynebacterium glutamicum. Boca Raton, FL: CRC Press. [Google Scholar]
- Farwick M., Siewe R. M., Krämer R. (1995). Glycine betaine uptake after hyperosmotic shift in Corynebacterium glutamicum. J. Bacteriol. 177 4690–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frings E., Kunte H. J., Galinski E. A. (1993). Compatible solutes in representatives of the genera Brevibacterium and Corynebacterium: occurrence of tetrahydropyrimidines and glutamine. FEMS Microbiol. Lett. 109 25–32. 10.1111/j.1574-6968.1993.tb06138.x [DOI] [Google Scholar]
- Gibson D. G. (2011). Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498 349–361. 10.1016/B978-0-12-385120-8.00015-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouesbet G., Blanco C., Hamelin J., Bernard T. (1992). Osmotic adjustment in Brevibacterium ammoniagenes: pipecolic acid accumulation at elevated osmolalities. Microbiology 138 959–965. 10.1099/00221287-138-5-959 [DOI] [Google Scholar]
- Gouesbet G., Jebbar M., Talibart R., Bernard T., Blanco C. (1994). Pipecolic acid is an osmoprotectant for Escherichia coli taken up by the general osmoporters ProU and ProP. Microbiol. Read. Engl. 140(Pt 9), 2415–2422. 10.1099/13500872-140-9-2415 [DOI] [PubMed] [Google Scholar]
- Gouffi K., Bernard T., Blanco C. (2000). Osmoprotection by pipecolic acid in Sinorhizobium meliloti: specific effects of d and l isomers. Appl. Environ. Microbiol. 66 2358–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R., Anzali S., Buenger J., Pfluecker F., Driller H. (2008). The multifunctional role of ectoine as a natural cell protectant. Clin. Dermatol. 26 326–333. 10.1016/j.clindermatol.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Guillouet S., Engasser J. M. (1995). Sodium and proline accumulation in Corynebacterium glutamicum as a response to an osmotic saline upshock. Appl. Microbiol. Biotechnol. 43 315–320. 10.1007/BF00172831 [DOI] [Google Scholar]
- Gutmann M., Hoischen C., Krämer R. (1992). Carrier-mediated glutamate secretion by Corynebacterium glutamicum under biotin limitation. Biochim. Biophys. Acta 1112 115–123. [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166 557–580. [DOI] [PubMed] [Google Scholar]
- Hilker R., Stadermann K. B., Doppmeier D., Kalinowski J., Stoye J., Straube J., et al. (2014). ReadXplorer–visualization and analysis of mapped sequences. Bioinformatics 30 2247–2254. 10.1093/bioinformatics/btu205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann T., Bremer E. (2017). Guardians in a stressful world: the Opu family of compatible solute transporters from Bacillus subtilis. Biol. Chem. 398 193–214. 10.1515/hsz-2016-0265 [DOI] [PubMed] [Google Scholar]
- Huhn S., Jolkver E., Krämer R., Marin K. (2011). Identification of the membrane protein SucE and its role in succinate transport in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 89 327–335. 10.1007/s00253-010-2855-1 [DOI] [PubMed] [Google Scholar]
- Irla M., Neshat A., Brautaset T., Rückert C., Kalinowski J., Wendisch V. F. (2015). Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genomics 16:73. 10.1186/s12864-015-1239-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. V. K., Wendisch V. F. (2013). Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb. Cell Fact. 12:63. 10.1186/1475-2859-12-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge C. D., Borges N., Bagyan I., Bilstein A., Santos H. (2016). Potential applications of stress solutes from extremophiles in protein folding diseases and healthcare. Extremophiles 20 251–259. 10.1007/s00792-016-0828-8 [DOI] [PubMed] [Google Scholar]
- Jorge J. M. P., Nguyen A. Q. D., Pérez-García F., Kind S., Wendisch V. F. (2016). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars and xylose. Biotechnol. Bioeng. 114 862–873. 10.1002/bit.26211 [DOI] [PubMed] [Google Scholar]
- Jorge J. M. P., Pérez-García F., Wendisch V. F. (2017). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245 1701–1709. 10.1016/j.biortech.2017.04.108 [DOI] [PubMed] [Google Scholar]
- Kalinowski J., Bathe B., Bartels D., Bischoff N., Bott M., Burkovski A., et al. (2003). The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104 5–25. [DOI] [PubMed] [Google Scholar]
- Kempf B., Bremer E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170 319–330. [DOI] [PubMed] [Google Scholar]
- Kind S., Jeong W. K., Schröder H., Wittmann C. (2010). Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 12 341–351. 10.1016/j.ymben.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Kind S., Neubauer S., Becker J., Yamamoto M., Völkert M., Abendroth G., et al. (2014). From zero to hero - production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab. Eng. 25 113–123. 10.1016/j.ymben.2014.05.007 [DOI] [PubMed] [Google Scholar]
- Klingenberg M., Pfaff E. (1967). “Means or terminating reactions,” in Methods of Enzymology, Vol. X, eds Estabrook R. W., Pullamnn M. R. (New York, NY: Academic press; ), 680–684. [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gänzle M. (2016). Some like it hot: heat resistance of Escherichia coli in food. Front. Microbiol. 7:1763. 10.3389/fmicb.2016.01763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz D., Wendisch V. F. (2016). Ciprofloxacin triggered glutamate production by Corynebacterium glutamicum. BMC Microbiol. 16:235. 10.1186/s12866-016-0857-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentz A., Neshat A., Pfeifer-Sancar K., Pühler A., Rückert C., Kalinowski J. (2013). Comprehensive discovery and characterization of small RNAs in Corynebacterium glutamicum ATCC 13032. BMC Genomics 14:714. 10.1186/1471-2164-14-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitsuka T., Sawai H., Hatsu M., Yamada K. (2007). Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci. Biotechnol. Biochem. 71 2130–2135. 10.1271/bbb.60699 [DOI] [PubMed] [Google Scholar]
- Morbach S., Krämer R. (2003). Impact of transport processes in the osmotic response of Corynebacterium glutamicum. J. Biotechnol. 104 69–75. [DOI] [PubMed] [Google Scholar]
- Moulin M., Deleu C., Larher F., Bouchereau A. (2006). The lysine-ketoglutarate reductase–saccharopine dehydrogenase is involved in the osmo-induced synthesis of pipecolic acid in rapeseed leaf tissues. Plant Physiol. Biochem. 44 474–482. 10.1016/j.plaphy.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Nakamura J., Hirano S., Ito H., Wachi M. (2007). Mutations of the Corynebacterium glutamicum NCgl1221 gene, encoding a mechanosensitive channel homolog, induce L-glutamic acid production. Appl. Environ. Microbiol. 73 4491–4498. 10.1128/AEM.02446-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M., Ghuillot-Gaudeffroy A., Marchais A., Gautheret D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8 11–13. [DOI] [PubMed] [Google Scholar]
- Neshich I. A. P., Kiyota E., Arruda P. (2013). Genome-wide analysis of lysine catabolism in bacteria reveals new connections with osmotic stress resistance. ISME J. 7 2400–2410. 10.1038/ismej.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. Q. D., Schneider J., Reddy G. K., Wendisch V. F. (2015). Fermentative production of the diamine putrescine: system metabolic engineering of Corynebacterium glutamicum. Metabolites 5 211–231. 10.3390/metabo5020211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y., Wu X., Zhang C., Xu Q., Chen N., Xie X. (2016). Pathway construction and metabolic engineering for fermentative production of ectoine in Escherichia coli. Metab. Eng. 36 10–18. 10.1016/j.ymben.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Nottebrock D., Meyer U., Krämer R., Morbach S. (2003). Molecular and biochemical characterization of mechanosensitive channels in Corynebacterium glutamicum. FEMS Microbiol. Lett. 218 305–309. [DOI] [PubMed] [Google Scholar]
- Pérez-García F., Max Risse J., Friehs K., Wendisch V. F. (2017a). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12:1600646. 10.1002/biot.201600646 [DOI] [PubMed] [Google Scholar]
- Pérez-García F., Ziert C., Risse J. M., Wendisch V. F. (2017b). Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J. Biotechnol. 258 59–68. 10.1016/j.jbiotec.2017.04.039 [DOI] [PubMed] [Google Scholar]
- Pérez-García F., Peters-Wendisch P., Wendisch V. F. (2016). Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 100 8075–8090. 10.1007/s00253-016-7682-6 [DOI] [PubMed] [Google Scholar]
- Peter H., Bader A., Burkovski A., Lambert C., Krämer R. (1997). Isolation of the putP gene of Corynebacterium glutamicum and characterization of a low-affinity uptake system for compatible solutes. Arch. Microbiol. 168 143–151. [DOI] [PubMed] [Google Scholar]
- Peter H., Weil B., Burkovski A., Krämer R., Morbach S. (1998). Corynebacterium glutamicum is equipped with four secondary carriers for compatible solutes: identification, sequencing, and characterization of the proline/ectoine uptake system, ProP, and the ectoine/proline/glycine betaine carrier, EctP. J. Bacteriol. 180 6005–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters-Wendisch P. G., Schiel B., Wendisch V. F., Katsoulidis E., Möckel B., Sahm H., et al. (2001). Pyruvate carboxylase is a major bottleneck for glutamate and lysine production by Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 3 295–300. [PubMed] [Google Scholar]
- Roenneke B., Rosenfeldt N., Derya S. M., Novak J. F., Marin K., Krämer R., et al. (2018). Production of the compatible solute α-D-glucosylglycerol by metabolically engineered Corynebacterium glutamicum. Microb. Cell Fact. 17:94. 10.1186/s12934-018-0939-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönsch H., Krämer R., Morbach S. (2003). Impact of osmotic stress on volume regulation, cytoplasmic solute composition and lysine production in Corynebacterium glutamicum MH20-22B. J. Biotechnol. 104 87–97. [DOI] [PubMed] [Google Scholar]
- Ruffert S., Berrier C., Krämer R., Ghazi A. (1999). Identification of mechanosensitive ion channels in the cytoplasmic membrane of Corynebacterium glutamicum. J. Bacteriol. 181 1673–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffert S., Lambert C., Peter H., Wendisch V. F., Krämer R. (1997). Efflux of compatible solutes in Corynebacterium glutamicum mediated by osmoregulated channel activity. Eur. J. Biochem. FEBS 247 572–580. [DOI] [PubMed] [Google Scholar]
- Sajjad W., Qadir S., Ahmad M., Rafiq M., Hasan F., Tehan R., et al. (2018). Ectoine: a compatible solute in radio-halophilic Stenotrophomonas sp. WMA-LM19 strain to prevent ultraviolet-induced protein damage. J. Appl. Microbiol. 125 457–467. 10.1111/jam.13903 [DOI] [PubMed] [Google Scholar]
- Sauer T., Galinski E. A. (1998). Bacterial milking: a novel bioprocess for production of compatible solutes. Biotechnol. Bioeng. 57 306–313. [PubMed] [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145 69–73. [DOI] [PubMed] [Google Scholar]
- Schaffer S., Weil B., Nguyen V. D., Dongmann G., Günther K., Nickolaus M., et al. (2001). A high-resolution reference map for cytoplasmic and membrane-associated proteins of Corynebacterium glutamicum. Electrophoresis 22 4404–4422. [DOI] [PubMed] [Google Scholar]
- Schneider J., Wendisch V. F. (2010). Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88 859–868. 10.1007/s00253-010-2778-x [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Skjerdal O. T., Sletta H., Flenstad S. G., Josefsen K. D., Levine D. W., Ellingsen T. E. (1996). Changes in intracellular composition in response to hyperosmotic stress of NaCl, sucrose or glutamic acid in Brevibacterium lactofermentum and Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 44 635–642. 10.1007/BF00172497 [DOI] [Google Scholar]
- Stansen C., Uy D., Delaunay S., Eggeling L., Goergen J. L., Wendisch V. F. (2005). Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl. Environ. Microbiol. 71 5920–5928. 10.1128/AEM.71.10.5920-5928.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger R., Weinand M., Krämer R., Morbach S. (2004). LcoP, an osmoregulated betaine/ectoine uptake system from Corynebacterium glutamicum. FEBS Lett. 573 155–160. 10.1016/j.febslet.2004.07.067 [DOI] [PubMed] [Google Scholar]
- Sun Y., Guo W., Wang F., Peng F., Yang Y., Dai X., et al. (2016). Transcriptome and multivariable data analysis of Corynebacterium glutamicum under different dissolved oxygen conditions in bioreactors. PLoS One 11:e0167156. 10.1371/journal.pone.0167156 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tan X., Luo Q., Lu X. (2016). Biosynthesis, biotechnological production, and applications of glucosylglycerols. Appl. Microbiol. Biotechnol. 100 6131–6139. 10.1007/s00253-016-7608-3 [DOI] [PubMed] [Google Scholar]
- Tribelli P. M., López N. I. (2018). Reporting key features in cold-adapted bacteria. Life 8:E8. 10.3390/life8010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranova V., Lojkova L., Rejsek K., Formanek P. (2013). Significance of the natural occurrence of L- versus D-pipecolic acid: a review. Chirality 25 823–831. 10.1002/chir.22237 [DOI] [PubMed] [Google Scholar]
- Weinand M., Krämer R., Morbach S. (2007). Characterization of compatible solute transporter multiplicity in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 76 701–708. 10.1007/s00253-007-0938-4 [DOI] [PubMed] [Google Scholar]
- Wolf A., Krämer R., Morbach S. (2003). Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Mol. Microbiol. 49 1119–1134. [DOI] [PubMed] [Google Scholar]
- Wood J. M. (1999). Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63 230–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M., Bremer E., Csonka L. N., Kraemer R., Poolman B., van der Heide T., et al. (2001). Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 130 437–460. [DOI] [PubMed] [Google Scholar]
- Ying H., Tao S., Wang J., Ma W., Chen K., Wang X., et al. (2017). Expanding metabolic pathway for de novo biosynthesis of the chiral pharmaceutical intermediate L-pipecolic acid in Escherichia coli. Microb. Cell Fact. 16:52. 10.1186/s12934-017-0666-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in http://www.ncbi.nlm.nih.gov/geo/, GSE122249.