Abstract
Lipid droplets are ubiquitous dynamic organelles that contain neutral lipids surrounded by a phospholipid monolayer. They can store and supply lipids for energy metabolism and membrane synthesis. In addition, protein transport and lipid exchange often occur between LDs and various organelles to control lipid homeostasis in response to multiple stress responses and cellular signaling. In recent years, multiple membrane trafficking proteins have been identified through LD proteomics and genetic analyses. These membrane trafficking machineries are emerging as critical regulators to function in different LD-organelle interactions, e.g., for LD dynamics, biogenesis and turnover. In this review, we will summarize recent advances in regard to LD-related membrane trafficking proteins and discuss future investigations in higher plants and green algae.
Keywords: lipid droplet, membrane trafficking, small GTPase, SNARE, ESCRT, retromer, lipophagy
Introduction
Lipid droplets, which contain neutral lipids, serve as central storage organelles in eukaryotic cells to provide carbon and energy in metabolism, as well as lipids for membrane biosynthesis (Huang, 2018). While our current knowledge on the biogenesis and function of LDs in plants mainly comes from studies on oilseeds, evidence suggests that LDs also play essential roles in various physiological processes and different cell types in plants (e.g., leaves) (Pyc et al., 2017b). Green algae also accumulate considerable amounts of LDs under nitrogen deprivation or stress conditions (Tsai et al., 2015). Moreover, accumulation of LDs under nitrogen starvation is also observed in leaves in higher plant (Brocard et al., 2017). In addition, in both higher plants and green algae, a specific type of LDs called plastoglobuli are formed on the thylakoid membranes in plastids and have been suggested to function in response to oxidative stress or developmental transitions (Van Wijk and Kessler, 2017). Moreover, it has been observed that certain LDs carrying specific enzymes are recruited by the microdomains or lipid rafts in the plasmodesmata (PD) for pathogen defense or cell-to-cell signaling (Van Der Schoot et al., 2011).
The mechanism of LD biogenesis is conserved to a certain extent among yeast, mammals, higher plants and green algae. For example, the endoplasmic reticulum (ER), where enzymes for neutral lipid synthesis reside to promote LD formation and budding, also serve as a major platform for nascent LD production (Figure 1; Walther et al., 2017). Once LDs are released into the cytoplasm, they display variations in shape, size, and mobility in different plant cell types, developmental stages, and during environmental stress (Pyc et al., 2017b). In addition, to facilitate material exchange and supply, LDs build tight connections with other organelles (e.g., ER, plastids, and vacuoles) via direct membrane contact sites, or vesicle transport, which are largely depends on membrane trafficking machinery (Gao and Goodman, 2015).
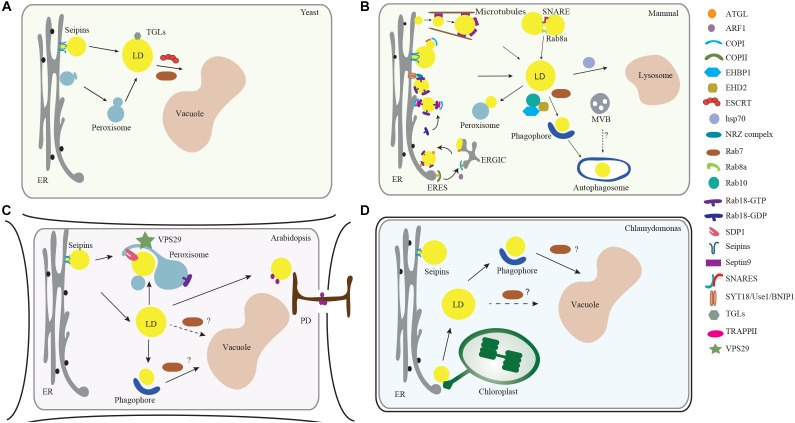
FIGURE 1.
Membrane trafficking regulators in LD dynamics, biogenesis, and turnover in model organisms. (A) In yeast, the ESCRT machinery regulates the LDs turnover via microlipophagy (Oku et al., 2017). In addition, Rab GTPase RAB7-like Ypt7 might function together with the HOPS (homotypic fusion and vacuole protein sorting) complex to mediate the fusion of LD with vacuole (Bouchez et al., 2015). (B) In mammalian cells, the ER serves as a major platform for LD biogenesis. ARF1 coordinates with COPI vesicles to deliver essential enzymes to the LD surface, while COPI subunits interact with the TRAPPII complex for its recruitment onto LD surface, which functions as a GEF (guanine nucleotide exchange factor) to activate Rab18. Rab18 also forms a complex with the ER-associated proteins NAG-RINT1-ZW10 (NRZ) and SNARE proteins (Syntaxin18, Use1, BNIP1) to control LD growth and maturation (Xu et al., 2018). Rab8 could activate Fsp27 to further promote LD fusion (Wu et al., 2014). In addition, Septin9, a filament-forming cytoskeletal GTPase, might cooperate with the microtubules to regulate LD growth (Akil et al., 2016). For the lipolysis-dependent LD turnover, both COPI and COPII vesicles mediate the delivery of the major lipase ATGL (Soni et al., 2009). During macrolipophagy-mediated LD turnover, LD is sequestered by a preautophagosome structure named phagophore, which will expand and become an autophagosome. Rab10-EHBP1-EHD2 complex as well as Rab7, are implicated to mediate the fusion between LD with MVB, autophagosomes or lysosomes (Schroeder et al., 2015). (C) In Arabidopsis plants, VPS29, a retromer subunit, has been shown to function in mediating the trafficking of SUGAR-DEPENDENT 1 (SDP1) from peroxisome to LD surface to regulate both the size and number of LDs through peroxisome tubulation (Thazar-Poulota et al., 2015). Microlipophagy-like process is also reported in plants (Poxleitner et al., 2006). LDs may also contact with the PD to deliver the specific enzymes to the cell wall (Van Der Schoot et al., 2011). Microautophagy is involved in LD turnover but the regulators in mediating the autophagosome-mediated macrolipophagy are unexplored (dashed line, question mark). (D) In the green alga Chlamydomonas, it is observed that LD formation is highly induced under stress conditions and might be initiated from the ER-chloroplast contact site (Fan et al., 2011). Similar to higher plants, it is reported that LD might be sequestered to the vacuole via microautophagy (Zhao et al., 2014). But how macrolipophagy occurs in this microalgae remains unclear (dashed line, question mark). Abbreviations: ER, endoplasmic reticulum; MVB, multivesicular bodies; LD, lipid droplet; PD, plasmodesmata.
In recent years, increasing evidence from both yeast and mammalian cells implies that various membrane trafficking machineries play crucial roles in LD dynamics. However, the underlying molecular mechanisms for regulating LD dynamics in both higher plants and green algae are still not understood. Given the conserved roles of membrane trafficking machineries in endomembrane trafficking in eukaryotes, the working models derived from studies in yeast and mammalian cells will be important for future studies on LDs in higher plants and green algae. In this review, we summarize recent findings in the regulation of LD dynamics with a focus on the related membrane trafficking regulators, as well as in areas needing investigations in plants and green algae in the future.
LD-Associated Proteins in Higher Plants and Green Algae
The surface of LDs is decorated by the LD-associated proteins, such as oleosin (Huang, 2018). Oleosin, which is unique to higher plants and green algae, is the most abundant structural phospholipid protein found in plant seeds (Huang, 2018). With a long hairpin-like hydrophobic structure, oleosins are able to stabilize LD by inserting into the triacylglyceride (TAG) matrix, so that their hydrophilic N- and C- termini face toward the cytoplasm. Of note, mutations of oleosin can redirect the LD to the ER lumen as well as vacuoles (Huang and Huang, 2017), suggesting that oleosin might be associated with membrane trafficking regulator(s) to control LD relocalization. Recently, it has been shown that a protein family named as lipid droplet-associated proteins (LDAPs), which share high similarity to small rubber particle proteins (SRPPs) in rubber-producing plants, are abundant in non-seed cell types and might play a role in drought stress in Arabidopsis (Gidda et al., 2016; Kim et al., 2016). Through a yeast two hybrid screen, it has been shown that LDAP3 interacts with a plant-specific protein containing an amphipathic α-helix type LD targeting signal, and suppression of this protein alters LD morphology and increases neutral lipid content in seeds (Pyc et al., 2017a).
Although oleosins first evolved in green algae, it was shown that oleosins were only detected in Chlamydomonas cells at a certain developmental stage (Huang et al., 2013). By proteomic analysis, a distinct LD structural protein named Major LD Protein (MLDP), which lacks a long hydrophobic tail, has been identified. It was shown that MLDP regulates both the size and number of LDs, as well as TAG degradation. MLDP is not directly distributed on LDs but associated with ER subdomains in close proximity to LDs (Huang et al., 2013), possibly serving the recruitment of different proteins to LDs, like tubulins (Tsai et al., 2015).
In mammalian cells, PLINs (periplins), which is PAT (perilipin/ADRP/TIP47) family protein, represent the major LD-associated protein on the LD surface (Kimmel et al., 2010; Sztalryd and Kimmel, 2014). Although perilipins are less hydrophobic than oleosins, they perform a similar function by inserting into the phospholipid leaflet for binding to neutral lipids. PLINs might also function in protection of LDs from lipolysis, recruitment of lipases and interaction with mitochondria (Sztalryd and Kimmel, 2014). It has been suggested that PLINs are specific to mammalian cells, but a recent study showed that Pet10p, a protein previously identified in yeast LD proteomes, also contains a PAT domain and functions as a yeast perilipin (Gao Q. et al., 2017). Arabidopsis oleosin1 fusion proteins are also targeted to yeast LDs, but complementation assays show that only human PLIN2/3, but not plant oleosins, can restore the pet10p mutant. An interaction analysis has demonstrated that Pet10p binds to seipin, an LD assembly factor found on the ER and LD junctions. There are three isoforms of seipin in Arabidopsis, and one in Chlamydomonas (Cai et al., 2015; Lu et al., 2017). Of note, in Arabidopsis, SEIPIN1, and SEIPIN2/3 appear to regulate LD size and number with a unique tissue specificity, as SEIPIN1 is exclusively detected in the embryo, while SEIPIN2 and SEIPIN3 display a higher expression pattern in embryos and pollen (Cai et al., 2015; Lu et al., 2017; Taurino et al., 2018). Whether seipin functions in LD biogenesis in Chlamydomonas need to be explored.
Membrane Trafficking Regulators in Mediating Ld Dynamics
Two types of degradation pathways: lipolysis and lipophagy, have been reported to regulate LD turnover (Figure 1). Lipolysis requires a set of lipases, but in the latter, LDs are delivered into the vacuole for degradation by either sequestration into a double membrane structure named an autophagosome (macrolipohagy), or by direct engulfment into the lysosome/vacuole lumen (microlipophagy) (Zechner et al., 2017). To perform these two processes, LDs require vesicle transport of the lipases onto the LD surface, and fusion with different compartments (e.g., autophagosomes and vacuole/lysosomes). Several studies have identified multiple membrane trafficking regulators, like small GTPases, as being associated with LDs in LD proteomic analyses from mammalian cells (Brasaemle et al., 2004; Liu et al., 2004). In Arabidopsis and Chlamydomonas, several proteins involved in vesicle trafficking and transport were also found by LD proteomic analysis (Nguyen et al., 2011; Zhi et al., 2017). These membrane trafficking machineries are highly conserved in most eukaryotic cells, and their possible roles in LD biogenesis are now being unraveled as discussed in more detail below (Figure 1 and Table 1).
Table 1.
Known regulators in LD dynamics and their homologs in Arabidopsis/Chlamydomonas.∗
| Regulator | Specie | Function | Plant (At) | Microalgae (Cr) |
|---|---|---|---|---|
| ESCRT | ||||
| VPS27 (ESCRT-0) | Yeast | Regulates the delivery of LDs into the vacuole | N.I. | N.I. |
| VPS4 (ESCRT-III) | Yeast | Regulates the delivery of LDs into the vacuole | SKD1/VPS4 (At2g27600) | Cre02.g079300 |
| RAB | ||||
| Rab1 | Mammal | Functions in LDs metabolism | RABD2a (At1g02130) RABD2b (At5g47200) RABD2c (At4g17530) | Cre12.g560150 |
| Rab5 | Mammal | Functions in LDs metabolism | RABF2a (At5g45130) RABF2b (At4g19640) RABF1 (At3g54840) | Cre12.g517400 |
| Rab7 | Mammal | Promotes the direct interactions between MVBs/lysosomes and LDs | RABG3a (At4g09720) RABG3b (At1g22740) RABG3c (At3g16100) RABG3d (At1g52280) RABG3e (At1g49300) RABG3f (At3g18820) | Cre06.g311900 |
| Rab8a | Mammal | Mediates the LDs fusion | RABE1a (At3g53610) RABE1b (At4g20360) RABE1c (At3g46060) RABE1d (At5g03520) RABE1e (At3g09900) | Cre15.g641800 |
| Rab10 | Mammal | Mediates the autophagic engulfment of lipid droplets | N.I. | N.I. |
| Rab18 | Mammal | Localizes on the LDs to promote LD formation and turnover | RABC1 (At1g43890) RABC2a (At5g03530) RABC2b (At3g09910) | Cre09.g386900 |
| SNARE | ||||
| SNAP23 | Mammal | Mediates LD fusion | N.I. | N.I. |
| VAMP4 | Mammal | Mediates LD fusion | N.I. | N.I. |
| Syntaxin-5 | Mammal | Mediates LD fusion | SYP31 (At5g05760) SYP32 (At3g24350) | Cre16.g692050 |
| Syntaxin-17 | Mammal | Regulates LD biogenesis | N.I. | N.I. |
| Syntaxin-18 | Mammal | Mediates the LD growth and maturation | SYP81 (At1g51740) | Cre17.g711450 |
| ARFs | ||||
| ARF1 | Mammal | Mediates the deliver of ATGL to LDs surface for lipolysis. | ARFA1a (At1g23490) ARFA1b (At5g14670) ARFA1c (At2g47170) ARFA1d (At1g70490) ARFA1e (At3g62290) ARFA1f (At1g10630) ARFB1a (At2g15310) ARFB1b (At5g17060) ARFB1c (A3g03120) ARFC1 (At3g22950) ARFD1a (At1g02440) ARFD1b (At1g02430) | Cre10.g448200 |
| Retromer | ||||
| VPS29 | Arabidopsis | Mediates peroxisome tubulation to deliver the lipase SDP1 to LDs surface | VPS29 (At3g47810) | Cre12.g499900 |
| Others | ||||
| Septin9 | Mammal | Mediate LD accumulation | N.I. | Cre12.g556250 |
∗Not all the regulators are listed. Abbreviations: At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; N.I., not identified. The gene IDs were searched manually at the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and the Joint Genome Institute (JGI, http://www.jgi.doe.gov/).
Small Gtpases in Membrane Tethering Between Ld and Other Organelles
Small GTPases are proteins that regulate membrane trafficking events by recruiting downstream effectors to promote membrane tethering and fusion. Stable connections between LDs and other organelles (e.g., ER, peroxisome, and mitochondria) have been observed in different systems (Gao and Goodman, 2015), and it is reasonable that small GTPases participate in the membrane fusion steps for LDs with other organelles, although the functional roles of these LD contacts are still obscure.
In plants, Rab GTPases have been extensively investigated and reported to function in multiple trafficking pathways (Nielsen et al., 2008). Rab7 proteins are well-known to function in endosome to vacuole trafficking. Thus, overexpression of a Rab7 dominant negative mutant leads to the formation of enlarged PVCs (prevacuolar compartments), fragmented vacuoles, as well as suppression of vacuolar trafficking (Cui et al., 2014). Moreover, malfunctioning of Rab7 also disturbs autophagy by blocking the fusion between autophagosomes and the vacuole membrane (Kwon et al., 2013). In mammalian cells, Rab7 knockdown, or overexpression of the Rab7 mutant form disrupts LD turnover (Liu et al., 2007). Moreover, under starvation conditions, it has been observed that Rab7 is recruited onto the LD surface, which subsequently mediates the docking of endosomes, autophagosomes and lysosomes to the LD surface to promote its degradation via lipophagy (Schroeder et al., 2015). In yeast, the Rab7-like Ypt7p also regulates LD dynamics together with the HOPS complex and V1-ATPase (Bouchez et al., 2015). In Arabidopsis, it has been shown that both oleosin and caleosin are surrounded by a tonoplast marker α-TIP during seed germination, suggesting that LDs could be engulfed by the vacuole (Poxleitner et al., 2006). In green algae, LDs are highly induced under nitrogen starvation and might be degraded by autophagy. For instance, LDs are engulfed by the vacuole through a microlipophagy-like process that is observed in heterotrophic cells in Auxenochlorella protothecoides (Zhao et al., 2014). In Micrasterias denticulata, plastid lipid bodies are sequestered by autophagosome-like structures and delivered to the vacuole for degradation (Schwarz et al., 2017). In Chlamydomonas, a set of Rab GTPases has been detected in the secreted ectosomes, including a single copy Rab7 (Long et al., 2016), implying a fundamental role of Rab7 in vesicle transport.
Apart from Rab7, other Rab proteins have been shown to be tightly associated with LDs in mammalian cells, like Rab10, and Rab18. Rab10 recruits adaptor proteins, EHBP1, and EHD2, to form a Rab10-EHBP1-EHD2 complex to promote LD sequestration into the autophagosome, while knockdown of Rab10 or overexpression of its negative mutant form causes a significant accumulation of LDs (Li et al., 2016; Xu et al., 2018). Particularly, Rab18 is the only GTPase that exclusively localizes on the LD surface, and can form distinct complexes either with the ER-associated NAG-RINT1-ZW10 (NRZ) complex or SNARE proteins (Syntaxin18, Use1, BNIP1) in mammalian cells (Martin et al., 2005; Xu et al., 2018). Moreover, activation and recruitment of Rab18 onto the LD surface are mediated by a TRAPPII complex, a Rab GEF (guanine nucleotide exchange factor) that binds to the COPI subunit (Li et al., 2017). On the other hand, Rab8 might promote LD fusion by activation of Fsp27 (Fat-specific protein), which is enriched at the LD-LD contact site for lipid transfer from smaller to larger LDs (Wu et al., 2014). Interestingly, it has been suggested that Rab8, Rab10, and Rab18 are also associated with peroxisomes (Gronemeyer et al., 2013). Except for Rab10, homologs of Rab8 and Rab18 can be found in the Arabidopsis and Chlamydomonas genomes (Table 1). Similarly, homologs of Rab8 (RABE1d) and Rab18 (RabC2a) in Arabidopsis are also associated with peroxisome (Hashimoto et al., 2008; Cui et al., 2013). Moreover, RABE1d functions in the pathogen defense response (Camacho et al., 2009), whereas RabC2a interacts with myosin XI (MYA2), which is required for rapid trafficking of peroxisomes (Hashimoto et al., 2005, 2008). Delivery of lipases from peroxisomal extensions to LDs has been observed in Arabidopsis (Thazar-Poulota et al., 2015). Therefore, there is a possibility that Rab8 and Rab18 might function in LD-peroxisome contact in Arabidopsis.
In mammalian cells, other small GTPases like ARF1 and SAR1 have also been implicated in regulating LD homeostasis, by collaboration with COPI and COPII vesicles, respectively, for delivery of lipases (e.g., ATGL) onto LD surfaces for LD turnover (Soni et al., 2009). In mammalian cells, it has been shown that Arf1, together with COPI, are recruited from the cytosol onto the LD surface for the induction of nano-LDs (about 60–80 nm diameter), which may bridge with ER for the recruitment of lipid synthesis enzymes (Wilfling et al., 2014; Li et al., 2017). It is also observed that COPI-like vesicles have interaction with LD in plants (Brocard et al., 2017). In plants, SAR1 and ARF1, as well as motor proteins like myosin, have also been detected in lipidomic and proteomic analyses of isolated LDs from mesocarp and seed kernels (Zhi et al., 2017). In Chlamydomonas, it has been suggested that lipid accumulation is also tightly correlated with COPII proteins (Valledor et al., 2014). Recently, it was reported that Septin9, a filament forming related GTPase, might cooperate together with microtubules to regulate LD growth in mammalian cells (Akil et al., 2016). However, SEPTIN can only be found in green algae but not in higher plants. Future efforts are required to validate the possible roles of these different small GTPases in LD dynamics.
Snare Proteins in Mediating Membrane Tethering During Ld Growth and Maturation
Small GTPases are often linked by distinct effectors to the membrane fusion machinery, indicating that the canonical fusion machinery is also involved in LD fusion. In Arabidopsis, 65 SNAREs have been identified, which are further divided into two groups, the Q-SNAREs with a conserved glutamine residue in the SNARE domain and the R-SNAREs with a conserved arginine residue in the SNARE domain (Bassham and Blatt, 2008). Subcellular localization analysis has revealed that these SNAREs are located on specific intracellular compartments, ranging from the ER, Golgi apparatus, trans-Golgi network, endosomes, plasma membrane, PVC to vacuoles (Uemura et al., 2004).
Although little evidence is available that points to SNAREs function in LD dynamics in higher plants and green algae, a growing body of data demonstrates the crucial roles of SNAREs in endomembrane trafficking events. Some plant SNAREs appear to be distributed on more than a single organelle, suggesting an ability to transfer between distinct compartments by paring with other SNAREs (Fujiwara et al., 2014). In support of this, a recent study has uncovered that two distinct sets of SNAREs and Rabs complexes are involved in membrane fusion for endosome-vacuolar and vacuole-vacuole in Arabidopsis, respectively (Takemoto et al., 2018). In a recent proteomics study, two SNARE proteins, AT4G04910, and AT3G56190, have been identified in the isolated LD proteome analysis (Brocard et al., 2017). In Marchantia polymorpha, it has been observed that two Q-SNAREs, MpSYP13A, and MpSYP12B, localize to the oil body membrane (Kanazawa et al., 2016). It was also reported that two SNARE proteins (Vmpl2 and Vamp74) in Chlamydomonas are highly induced during autophagy (Ramundo and Rochaix, 2014). Particularly, Chlamydomonas cells lacking CrVMP1, which contains a predicted SNARE domain, exhibit defects in macroautophagy and TAG metabolism, indicating a possible role of CrVMP1 in autophagy-mediated LD degradation (Tenenboim et al., 2014). Indeed, a VMP1 homolog in mammalian cells functions in ER-LD contacts to contribute to autophagosome formation (Zhao et al., 2017). Future studies should provide novel insights by investigating how these SNAREs are targeted to LDs and regulate LD-organelle contacts.
Interestingly, previous studies in mammalian cells have shown that LD might sequester a novel v-/t-SNARE complex containing SNAP23, syntaxin-5, and VAMP4, to regulate the exocytosis of glucose transporters (Bostrom et al., 2007). Knockdown of SNAP23, syntaxin-5, or VAMP4, all suppress the fusion and impair the size of LDs. In addition, it has been shown that knockdown of SNAP23 interrupts the interaction between mitochondria and LD (Jagerstrom et al., 2009). LD-mitochondrial contacts have been implicated to mediate the direct flow of fatty acids from LD to mitochondria for further oxidation to release energy (Gao and Goodman, 2015). Moreover, SNAP23 also collaborates with Syntaxin17 to regulate the distribution of the ACSL3 (acyl-CoA synthetase 3), an important enzyme localized on the ER for LD biogenesis (Kimura et al., 2018). A recent study showed that another SNARE protein, Syntaxin18, works together with the Rab18 and NRZ (NAG-RINT1-ZW10) to establish a direct contact between LD and ER allowing LD growth and maturation (Xu et al., 2018). These studies all support the notion that LDs employ different combinations of SNAREs to mediate LD tethering with distinct compartments.
The Escrt Machinery in Mediating the Ld Degradation
The ESCRT machinery, which is conserved in eukaryotic cells, is essential for the scission of narrow membrane necks during the budding of intraluminal vesicles (ILVs) into the multivesicular body (MVB) (Gao C. et al., 2017). Recently, yeast ESCRT components have also been demonstrated to function in LD turnover during microautophagy, although independent of the core Autophagy-related (ATG) genes (Oku et al., 2017). It has been shown that yeast Vps27 may interact with clathrin and translocate into the vacuole after diauxic shift, which leads to the incorporation of LDs into the vacuole for LD degradation. In addition, mutation in Vps4, which functions at the final step for ESCRT disassembly, leads to a failure in LD degradation (Vevea et al., 2015). In a recent study by genetic screening, several ESCRTIII deletion mutants have been identified, displaying defects in LD metabolism in a lipophagy independent manner (Mast et al., 2018). With its membrane scission ability, it is very likely that the ESCRT machinery may directly induce the invagination of LD by the vacuolar membrane.
In plants, most ESCRT homologs play conserved roles in ILV formation and MVB biogenesis (Gao C. et al., 2017). Plant unique ESCRT components such as FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1) and POSITIVE REGULATOR OF SKD1 (PROS) have also been recently identified (Gao C. et al., 2014; Reyes et al., 2014). Recent studies have implied that FREE1 might play a dual role in regulating autophagosome-vacuole fusion and vacuole biogenesis in Arabidopsis (Gao et al., 2015; Kalinowska et al., 2015). In addition, CHMP1 has also been involved in the autophagic turnover of plastid materials (Spitzer et al., 2015). However, it remains unexplored as to whether the ESCRT components are involved in LD turnover in plants. Of note, several ESCRT proteins have been identified by analyzing the ectosomes released from the flagella lipids in Chlamydomonas, and ectosome secretion is disturbed in ESCRT mutants (Long et al., 2016). More investigation is needed to figure out if ESCRT can play a directly role in LD degradation in plants and algae.
Retromer and Transport Between Ld and Peroxisomes
The retromer complex mediates retrograde transport from the endosome to other compartments (Heucken and Ivanov, 2018). In Arabidopsis, the retromer complex consists of SNX1, SNX2a/b, VPS35a/b/c, VPS26a/b, and VPS29 (Heucken and Ivanov, 2018). Intriguingly, a recent study has uncovered a novel function of retromer in LD homeostasis. At the early stage of seed germination, SDP1 (SUGAR-DEPENDENT 1), the major lipase for LD degradation, is localized on the peroxisome, and then transported to the LD surface via peroxisome tubulation (Thazar-Poulota et al., 2015). However, in vps29 mutant, the formation of the SDP1 peroxisomal tubulations is inhibited, resulting in a suppression in both SDP1 migration and lipid mobilization. The molecular mechanism of retromer-mediated peroxisomal tubulation in LD homeostasis remains to be elucidated, and it is possible that retromer functions in recycling of other regulators from peroxisomes. Particularly, the role of retromer in LD turnover has so far only been reported in plants, and whether it also operates in other eukaryotes is unclear.
Future Perspectives
Emerging findings have greatly expanded our current knowledge about the crucial roles of LDs in eukaryotic cells. While most of the work has focussed on the characterization of physiological roles of LDs in higher plants and green algae, the underlying mechanism of LD homeostasis is still unclear. In regard to the complex cross-talk between LDs and other compartments, more attention should be given to a molecular understanding of LD dynamics mediated by membrane trafficking systems. Future studies with genetic screens, proteomics, as well as mechanistic studies of the candidate proteins will shed new light on this field. We anticipate that these basic studies underlying lipid droplet dynamics would provide useful tools to be applied in plant stress response, seed oil production and biofuels.
Author Contributions
XZ designed the concept and the organization of the manuscript. All authors wrote the manuscript. XZ and LJ edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to researchers whose work has not been included in this manuscript owing to space limit.
Footnotes
Funding. This work was supported by grants from the National Natural Science Foundation of China (31470294, 31670179, and 91854201), the Research Grants Council of Hong Kong (G-CUHK403/17, G-CUHK404/18, CUHK14130716, CUHK14 102417, 14100818, C4011-14R, C4012-16E, C4002-17G, R4005-18F, and AoE/M-05/12), and CUHK Research Committee.
References
- Akil A., Peng J., Omrane M., Gondeau C., Desterke C., Marin M., et al. (2016). Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat. Commun. 7:12203. 10.1038/ncomms12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham D. C., Blatt M. R. (2008). SNAREs: cogs and coordinators in signaling and development. Plant Physiol. 147 1504–1515. 10.1104/pp.108.121129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., et al. (2007). SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 9 1286–1293. 10.1038/ncb1648 [DOI] [PubMed] [Google Scholar]
- Bouchez I., Pouteaux M., Canonge M., Genet M., Chardot T., Guillot A., et al. (2015). Regulation of lipid droplet dynamics in Saccharomyces cerevisiae depends on the Rab7-like Ypt7p, HOPS complex and V1-ATPase. Biol. Open 4 764–775. 10.1242/bio.20148615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004). Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279 46835–46842. 10.1074/jbc.M409340200 [DOI] [PubMed] [Google Scholar]
- Brocard L., Immel F., Coulon D., Esnay N., Tuphile K., Pascal S., et al. (2017). Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 8:894. 10.3389/fpls.2017.00894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Goodman J. M., Pyc M., Mullen R. T., Dyer J. M., Chapman K. D. (2015). Arabidopsis SEIPIN Proteins Modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27 2616–2636. 10.1105/tpc.15.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L., Smertenko A. P., Perez-Gomez J., Hussey P. J., Moore I. (2009). Arabidopsis Rab-E GTPases exhibit a novel interaction with a plasma-membrane phosphatidylinositol-4-phosphate 5-kinase. J. Cell Sci. 122 4383–4392. 10.1242/jcs.053488 [DOI] [PubMed] [Google Scholar]
- Cui S., Fukao Y., Mano S., Yamada K., Hayashi M., Nishimura M. (2013). Proteomic analysis reveals that the Rab GTPase RabE1c is involved in the degradation of the peroxisomal protein receptor PEX7 (peroxin 7). J. Biol. Chem. 288 6014–6023. 10.1074/jbc.M112.438143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Zhao Q., Gao C., Ding Y., Zeng Y., Ueda T., et al. (2014). Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for pvc-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26 2080–2097. 10.1105/tpc.114.123141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Andre C., Xu C. (2011). A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585 1985–1991. 10.1016/j.febslet.2011.05.018 [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Uemura T., Ebine K., Nishimori Y., Ueda T., Nakano A., et al. (2014). Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 55 781–789. 10.1093/pcp/pcu038 [DOI] [PubMed] [Google Scholar]
- Gao C., Luo M., Zhao Q., Yang R., Cui Y., Zeng Y., et al. (2014). A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24 2556–2563. 10.1016/j.cub.2014.09.014 [DOI] [PubMed] [Google Scholar]
- Gao C., Zhuang X., Shen J., Jiang L. (2017). Plant ESCRT complexes: moving beyond endosomal sorting. Trends Plant Sci. 22 986–998. 10.1016/j.tplants.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Gao Q., Binns D. D., Kinch L. N., Grishin N. V., Ortiz N., Chen X., et al. (2017). Pet10p is a yeast perilipin that stabilizes lipid droplets and promotes their assembly. J. Cell Biol. 216 3199–3217. 10.1083/jcb.201610013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C. J., Zhuang X. H., Cui Y., Fu X., He Y. L., Zhao Q., et al. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. U S A. 112 1886–1891. 10.1073/pnas.1421271112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Goodman J. M. (2015). The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 3:49. 10.3389/fcell.2015.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda S. K., Park S., Pyc M., Yurchenko O., Cai Y., Wu P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170 2052–2071. 10.1104/pp.15.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer T., Wiese S., Grinhagens S., Schollenberger L., Satyagraha A., Huber L. A., et al. (2013). Localization of rab proteins to peroxisomes: a proteomics and immunofluorescence study. FEBS Lett. 587 328–338. 10.1016/j.febslet.2012.12.025 [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Igarashi H., Mano S., Nishimura M., Shimmen T., Yokota E. (2005). Peroxisomal localization of a myosin XI isoform in Arabidopsis thaliana. Plant Cell Physiol. 46 782–789. 10.1093/pcp/pci085 [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Igarashi H., Mano S., Takenaka C., Shiina T., Yamaguchi M., et al. (2008). An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J. Exp. Bot. 59 3523–3531. 10.1093/jxb/ern202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heucken N., Ivanov R. (2018). The retromer, sorting nexins and the plant endomembrane protein trafficking. J. Cell Sci. 131:jcs203695. 10.1242/jcs.203695 [DOI] [PubMed] [Google Scholar]
- Huang A. H. C. (2018). Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 176 1894–1918. 10.1104/pp.17.01677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. Y., Huang A. H. C. (2017). Unique motifs and length of hairpin in oleosin target the cytosolic side of endoplasmic reticulum and budding lipid droplet. Plant Physiol. 174 2248–2260. 10.1104/pp.17.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N. L., Huang M. D., Chen T. L., Huang A. H. (2013). Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 161 1862–1874. 10.1104/pp.112.212514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagerstrom S., Polesie S., Wickstrom Y., Johansson B. R., Schroder H. D., Hojlund K., et al. (2009). Lipid droplets interact with mitochondria using SNAP23. Cell Biol. Int. 33 934–940. 10.1016/j.cellbi.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Kalinowska K., Nagel M. K., Goodman K., Cuyas L., Anzenberger F., Alkofer A., et al. (2015). Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3. Proc. Natl. Acad. Sci. U S A. 112 E5543–E5551. 10.1073/pnas.1510516112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T., Era A., Minamino N., Shikano Y., Fujimoto M., Uemura T., et al. (2016). SNARE Molecules in Marchantia polymorpha : unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57 307–324. 10.1093/pcp/pcv076 [DOI] [PubMed] [Google Scholar]
- Kim E. Y., Park K. Y., Seo Y. S., Kim W. T. (2016). Arabidopsis small rubber particle protein homolog srps play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170 2494–2510. 10.1104/pp.16.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Brasaemle D. L., Mcandrews-Hill M., Sztalryd C., Londos C. (2010). Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51 468–471. 10.1194/jlr.R000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Arasaki K., Ohsaki Y., Fujimoto T., Ohtomo T., Yamada J., et al. (2018). Syntaxin 17 promotes lipid droplet formation by regulating the distribution of acyl-CoA synthetase 3. J. Lipid Res. 59 805–819. 10.1194/jlr.M081679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. I., Cho H. J., Kim S. R., Park O. K. (2013). The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 161 1722–1736. 10.1104/pp.112.208108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Luo X., Zhao S., Siu G. K., Liang Y., Chan H. C., et al. (2017). COPI-TRAPPII activates Rab18 and regulates its lipid droplet association. EMBO J. 36 441–457. 10.15252/embj.201694866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Schulze R. J., Weller S. G., Krueger E. W., Schott M. B., Zhang X., et al. (2016). A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2:e1601470. 10.1126/sciadv.1601470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Bartz R., Zehmer J. K., Ying Y. S., Zhu M., Serrero G., et al. (2007). Rab-regulated interaction of early endosomes with lipid droplets. Biochim. Biophys. Acta 1773 784–793. 10.1016/j.bbamcr.2007.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Ying Y., Zhao Y., Mundy D. I., Zhu M., Anderson R. G. (2004). Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279 3787–3792. 10.1074/jbc.M311945200 [DOI] [PubMed] [Google Scholar]
- Long H., Zhang F., Xu N., Liu G., Diener D. R., Rosenbaum J. L., et al. (2016). Comparative analysis of ciliary membranes and ectosomes. Curr. Biol. 26 3327–3335. 10.1016/j.cub.2016.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wang X., Balamurugan S., Yang W.-D., Liu J.-S., Dong H.-P., et al. (2017). Identification of a putative seipin ortholog involved in lipid accumulation in marine microalga Phaeodactylum tricornutum. J. Appl. Phycol. 29 2821–2829. 10.1007/s10811-017-1173-8 [DOI] [Google Scholar]
- Martin S., Driessen K., Nixon S. J., Zerial M., Parton R. G. (2005). Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J. Biol. Chem. 280 42325–42335. 10.1074/jbc.M506651200 [DOI] [PubMed] [Google Scholar]
- Mast F. D., Herricks T., Strehler K. M., Miller L. R., Saleem R. A., Rachubinski R. A., et al. (2018). ESCRT-III is required for scissioning new peroxisomes from the endoplasmic reticulum. J. Cell Biol. 217 2087–2102. 10.1083/jcb.201706044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. M., Baudet M., Cuine S., Adriano J. M., Barthe D., Billon E., et al. (2011). Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11 4266–4273. 10.1002/pmic.201100114 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Cheung A. Y., Ueda T. (2008). The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 147 1516–1526. 10.1104/pp.108.121798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Maeda Y., Kagohashi Y., Kondo T., Yamada M., Fujimoto T., et al. (2017). Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216 3263–3274. 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poxleitner M., Rogers S. W., Lacey Samuels A., Browse J., Rogers J. C. (2006). A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 47 917–933. 10.1111/j.1365-313X.2006.02845.x [DOI] [PubMed] [Google Scholar]
- Pyc M., Cai Y., Gidda S. K., Yurchenko O., Park S., Kretzschmar F. K., et al. (2017a). Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92 1182–1201. 10.1111/tpj.13754 [DOI] [PubMed] [Google Scholar]
- Pyc M., Cai Y., Greer M. S., Yurchenko O., Chapman K. D., Dyer J. M., et al. (2017b). Turning over a new leaf in lipid droplet biology. Trends Plant Sci. 22 596–609. 10.1016/j.tplants.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Ramundo S., Rochaix J. D. (2014). Loss of chloroplast ClpP elicits an autophagy-like response in Chlamydomonas. Autophagy 10 1685–1686. 10.4161/auto.29960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F. C., Buono R. A., Roschzttardtz H., Di Rubbo S., Yeun L. H., Russinova E., et al. (2014). A novel endosomal sorting complex required for transport (ESCRT) component in Arabidopsis thaliana controls cell expansion and development. J. Biol. Chem. 289 4980–4988. 10.1074/jbc.M113.529685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B., Schulze R. J., Weller S. G., Sletten A. C., Casey C. A., Mcniven M. A. (2015). The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61 1896–1907. 10.1002/hep.27667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz V., Andosch A., Geretschlager A., Affenzeller M., Lutz-Meindl U. (2017). Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J. Plant Physiol. 208 115–127. 10.1016/j.jplph.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Soni K. G., Mardones G. A., Sougrat R., Smirnova E., Jackson C. L., Bonifacino J. S. (2009). Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 122 1834–1841. 10.1242/jcs.045849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Li F., Buono R., Roschzttardtz H., Chung T., Zhang M., et al. (2015). The endosomal protein charged multivesicular body protein1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27 391–402. 10.1105/tpc.114.135939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd C., Kimmel A. R. (2014). Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie 96 96–101. 10.1016/j.biochi.2013.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K., Ebine K., Askani J. C., Kruger F., Gonzalez Z. A., Ito E., et al. (2018). Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 115:E2457–E2466. 10.1073/pnas.1717839115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino M., Costantini S., De Domenico S., Stefanelli F., Ruano G., Delgadillo M. O., et al. (2018). SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 176 1531–1546. 10.1104/pp.17.01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenboim H., Smirnova J., Willmitzer L., Steup M., Brotman Y. (2014). VMP1-deficient Chlamydomonas exhibits severely aberrant cell morphology and disrupted cytokinesis. BMC Plant Biol. 14:121. 10.1186/1471-2229-14-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazar-Poulota N., Miquel M., Fobis-Loisya I., Gaudea T. (2015). Peroxisome extensions deliver the Arabidopsis SDP1 lipase to oil bodies. Proc. Natl. Acad. Sci. U S A. 112 4158–4163. 10.1073/pnas.1403322112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. H., Zienkiewicz K., Amstutz C. L., Brink B. G., Warakanont J., Roston R., et al. (2015). Dynamics of protein and polar lipid recruitment during lipid droplet assembly in Chlamydomonas reinhardtii. Plant J. 83 650–660. 10.1111/tpj.12917 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29 49–65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- Valledor L., Furuhashi T., Recuenco-Munoz L., Wienkoop S., Weckwerth W. (2014). System-level network analysis of nitrogen starvation and recovery in Chlamydomonas reinhardtii reveals potential new targets for increased lipid accumulation. Biotechnol. Biofuels 7:171. 10.1186/s13068-014-0171-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Schoot C., Paul L. K., Paul S. B., Rinne P. L. (2011). Plant lipid bodies and cell-cell signaling: a new role for an old organelle? Plant Signal Behav. 6 1732–1738. 10.4161/psb.6.11.17639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wijk K. J., Kessler F. (2017). Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 68 253–289. 10.1146/annurev-arplant-043015-111737 [DOI] [PubMed] [Google Scholar]
- Vevea J. D., Garcia E. J., Chan R. B., Zhou B., Schultz M., Di Paolo G., et al. (2015). Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35 584–599. 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., Chung J., Jr., Robert V. F. (2017). Lipid droplet biogenesis. Ann. Rev. Cell Dev. Biol. 33 491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfling F., Thiam A. R., Olarte M. J., Wang J., Beck R., Gould T. J., et al. (2014). Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3:e01607. 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Xu D., Zhou L., Xie B., Yu L., Yang H., et al. (2014). Rab8a-AS160-MSS4 regulatory circuit controls lipid droplet fusion and growth. Dev. Cell. 30 378–393. 10.1016/j.devcel.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Xu D., Li Y., Wu L., Li Y., Zhao D., Yu J., et al. (2018). Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217 975–995. 10.1083/jcb.201704184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner R., Madeo F., Kratky D. (2017). Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18:671. 10.1038/nrm.2017.76 [DOI] [PubMed] [Google Scholar]
- Zhao L., Dai J., Wu Q. (2014). Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front. Plant Sci. 5:400. 10.3389/fpls.2014.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. G., Chen Y., Miao G., Zhao H., Qu W., Li D., et al. (2017). The ER-Localized transmembrane protein EPG-3/VMP1 regulates SERCA activity to control er-isolation membrane contacts for autophagosome formation. Mol. Cell 67:e976. 10.1016/j.molcel.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Zhi Y., Taylor M. C., Campbell P. M., Warden A. C., Shrestha P., El Tahchy A., et al. (2017). Comparative lipidomics and proteomics of lipid droplets in the mesocarp and seed tissues of chinese tallow (Triadica sebifera). Front. Plant Sci. 8:1339. 10.3389/fpls.2017.01339 [DOI] [PMC free article] [PubMed] [Google Scholar]