Abstract
Lignocellulosic biomass, encompassing cellulose, lignin and hemicellulose in plant secondary cell walls (SCWs), is the most abundant source of renewable materials on earth. Currently, fast-growing woody dicots such as Eucalyptus and Populus trees are major lignocellulosic (wood fiber) feedstocks for bioproducts such as pulp, paper, cellulose, textiles, bioplastics and other biomaterials. Processing wood for these products entails separating the biomass into its three main components as efficiently as possible without compromising yield. Glucuronoxylan (xylan), the main hemicellulose present in the SCWs of hardwood trees carries chemical modifications that are associated with SCW composition and ultrastructure, and affect the recalcitrance of woody biomass to industrial processing. In this review we highlight the importance of xylan properties for industrial wood fiber processing and how gaining a greater understanding of xylan biosynthesis, specifically xylan modification, could yield novel biotechnology approaches to reduce recalcitrance or introduce novel processing traits. Altering xylan modification patterns has recently become a focus of plant SCW studies due to early findings that altered modification patterns can yield beneficial biomass processing traits. Additionally, it has been noted that plants with altered xylan composition display metabolic differences linked to changes in precursor usage. We explore the possibility of using systems biology and systems genetics approaches to gain insight into the coordination of SCW formation with other interdependent biological processes. Acetyl-CoA, s-adenosylmethionine and nucleotide sugars are precursors needed for xylan modification, however, the pathways which produce metabolic pools during different stages of fiber cell wall formation still have to be identified and their co-regulation during SCW formation elucidated. The crucial dependence on precursor metabolism provides an opportunity to alter xylan modification patterns through metabolic engineering of one or more of these interdependent pathways. The complexity of xylan biosynthesis and modification is currently a stumbling point, but it may provide new avenues for woody biomass engineering that are not possible for other biopolymers.
Keywords: xylan, cellulose, lignin, wood fiber, bioproducts, biorefinery, industrial processing, metabolism
Introduction
Lignocellulosic biomass from softwood and hardwood trees is most commonly used for construction, pulp and paper products and for biorefinery applications that entail separating the biomass into its individual components to produce various bioproducts (Xavier et al., 2010; Pei et al., 2016; Zhu et al., 2016). For over a century, the pulping industry has been mechanically or chemically deconstructing wood from hardwoods such as Eucalyptus and softwoods such as pine and spruce to produce paper and packaging products (Goswami et al., 1996; Sixta, 2006). Similar chemical processing (alkaline Kraft pulping with acidic pretreatment or acidic sulphite pulping) can be used to obtain high quality and purity cellulose for use in textiles, industrial fiber, films, food casings, plastic and various pharmaceutical related products (Klemm et al., 2005; Sixta, 2006; Sixta et al., 2013; Nasatto et al., 2015; Zhu et al., 2016). The spent chemical waste known as black (Kraft pulping) or brown (sulphite pulping) liquor can also be processed to extract valuable bioproducts such as monosaccharides, lignosulphonates and bioethanol rather than burning it to generate the heat needed for pulping liquor recovery (Hocking, 1997; Restolho et al., 2009; Xavier et al., 2010). Alternatively, after chemical or enzymatic pretreatment, the cellulosic and hemicellulosic component of lignocellulosic biomass can be subjected to saccharification and fermentation; a process which employs chemicals, enzymes and microbes to convert the polysaccharide components into ethanol for second generation biofuels and various bioproducts (Ragauskas et al., 2014).
Product value in these industries is driven by high product quality and purity, but the physical properties of the SCW biopolymers themselves impede the efficiency of deconstructing the biomass (Gübitz et al., 1998; Himmel et al., 2007; DeMartini et al., 2013; McCann and Carpita, 2015). However, several improvements have been made to woody fiber biomass processing techniques themselves which have resulted in more efficient biomass separation and higher yields (Bibi et al., 2014; Nordwald et al., 2014; Roselli et al., 2014; Chen J. et al., 2017; Shahid et al., 2017). If biomass crops which have been bred or genetically engineered for favorable processing traits were used as well, even higher yields coupled with reductions in processing costs could be achieved (Marriott et al., 2016; Zhou et al., 2017). These improvements are largely due to research that has identified genes involved in the biosynthesis and deposition of SCW biopolymers as well as the transcriptional regulation governing these processes (Persson et al., 2005; Mutwil et al., 2009; Ruprecht et al., 2011; Taylor-Teeples et al., 2015). Such research has largely been made possible by an increase in resources available for functional genomics (Oikawa et al., 2010; Gille et al., 2011a; Jensen et al., 2014), reverse genetics (Fan et al., 2015; Zhou et al., 2015; Park et al., 2017) and multi-omics approaches such as systems biology (Hillmer, 2015) analysis (Vanholme et al., 2012; Li Z. et al., 2016; Ohtani et al., 2016). The latter approach has shed valuable insight on how SCW formation is coordinated with other biological processes, what aspects of central metabolism are being drawn on and which pathways could potentially be manipulated to alter SCW polymer abundance or composition (Mizrachi et al., 2017). Systems biology approaches have been applied successfully to cellulose and lignin biosynthesis and has yielded valuable insight into their biosynthesis, regulation and metabolic dependencies. Despite these studies highlighting certain aspects of xylan biosynthesis, a xylan-centric systems biology analysis still needs to be performed to gain a holistic understanding of the process.
Xylan, the dominant hemicellulose in hardwood biomass has been identified as a major determinant of recalcitrance, yet comparably little is known about the genetic regulation and metabolic processes governing its biosynthesis, especially in woody plants (Vanholme et al., 2012; Mizrachi et al., 2013; Yen et al., 2013). The xylan polymer is composed of a repeating β1,4 xylose residue backbone, a reducing end sequence (RES) of xylose, rhamnose and galacturonic acid and is highly modified with acetyl and (methyl)glucuronic acid side groups (Scheller and Ulvskov, 2010; Pauly et al., 2013; Rennie and Scheller, 2014). Considerable progress has been made in identifying the genes involved in xylan biosynthesis (backbone elongation, RES synthesis and modification) as well as the metabolic pathways producing precursors which are used during these processes (Pauly and Scheller, 2000; Brown et al., 2007; Urbanowicz et al., 2012; Jensen et al., 2014; Marriott et al., 2016; Zhong et al., 2017). However, the biosynthetic process itself is still poorly understood as xylan knock-out mutants often have severely stunted growth (Wu et al., 2010). Physical interactions between xylan biosynthetic proteins are weak (Lund et al., 2015; Zeng et al., 2016), the proteins which interact with each other differ between species (Jiang et al., 2016; Zeng et al., 2016), and the membrane bound nature of these proteins makes in vitro studies difficult (Urbanowicz et al., 2014; Zhong et al., 2017). Additionally, the effects of metabolic changes on xylan biosynthesis and modification are poorly understood and have only been explored in terms of carbon supply in the form of nucleotide sugar abundance (Ishihara et al., 2015; Fan et al., 2017). The modification patterns present on the xylan backbone, which are required for tight interaction with cellulose, differ from the patterns required for an interaction with lignin (Rennie and Scheller, 2014; Busse-Wicher et al., 2016a; Giummarella and Lawoko, 2016; Grantham et al., 2017; Pereira et al., 2017; Martínez-Abad et al., 2018), which begs the question whether specific combinations of modification genes are needed to set up the patterns required for each interaction. Due to the large number of genes involved in or affecting xylan biosynthesis, discovering which of these genes are co-regulated with either cellulose or lignin biosynthesis during the different stages of xylem development could provide valuable insight into which genes are responsible for the spatial and temporal changes in xylan biosynthesis and modification (Hertzberg et al., 2001; Peralta et al., 2017).
Here we focus on xylan biosynthesis and its importance for industrial processing of lignocellulosic biomass derived from dicot wood fiber. Xylan properties play important roles in biomass recalcitrance and are therefore valuable to understand from an industrial point of view. We discuss value-added products that can be derived from lignocellulosic biomass, as well as the industrial processes pertinent to these products. We discuss xylan biotechnology from two perspectives, firstly through altering the biosynthetic process by targeting the genes directly involved, and secondly through engineering of interdependent metabolic pathways. We expand on systems biology approaches that can be used to identify pathways producing nucleotide sugars, SAM and acetyl-CoA precursors required for xylan biosynthesis and modification. Finally, we propose xylan biotechnology approaches, including altering metabolic precursor supply, which can be used to obtain novel, industrially beneficial wood fiber properties.
Xylan and Its Role in the Secondary Cell Wall
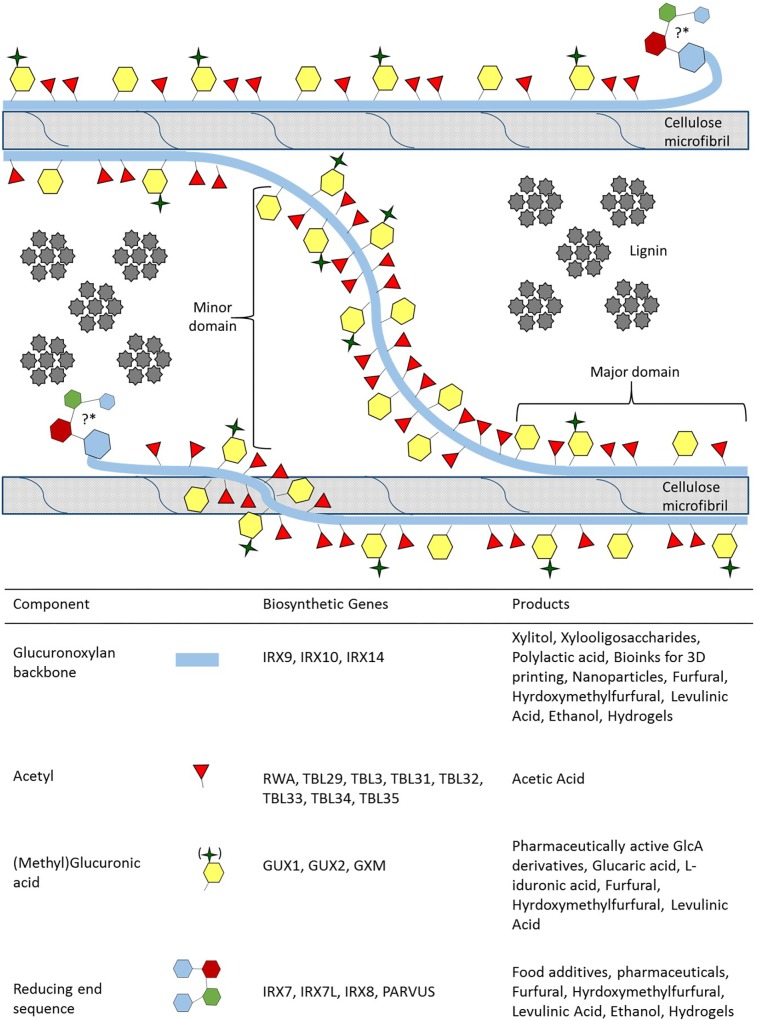
In woody dicots, the dominant hemicellulose is glucuronoxylan, however, small amounts of glucomannan and trace amount of pectins are also found in dicot SCWs (Scheller and Ulvskov, 2010). Water conducting and mechanically supportive tissues, such as the secondary xylem of woody plants, have thickened SCWs consisting of cellulose microfibrils crosslinked with hemicelluloses and fortified by a complex heterogenous matrix of lignin (Myburg et al., 2013; Plomion et al., 2001; Figure 1). The SCW is comprised of three layers: S1, S2 and S3; each of these layers has cellulose laid down at a different angle (referred to as the microfibril angle) as well as different thicknesses with the S2 layer being much thicker than S1 and S3 (Wardrop and Preston, 1947; Myburg et al., 2013). The angle of these microfibrils is determined by the orientation of the cortical microtubules as well as by hemicellulose crosslinking (Reis and Vian, 2004; Paredez et al., 2006; Watanabe et al., 2015; Schneider et al., 2017). Glucuronoxylan (xylan for short) is composed of a repeating β1,4 linked xylose backbone which is highly modified with either GlcA, which can also be methylated (MeGlcA), or an acetyl group, and a reducing end sequence (RES) composed of xylose, rhamnose and galacturonic acid (Rennie and Scheller, 2014).
FIGURE 1.
Xylan biosynthesis, its role in the secondary cell wall and xylan derived value-added products. Simplified representation of xylan, its domains and its function in the secondary cell wall, accurate modification spacing and chemical representations have been excellently review in Marriott et al. (2016) and Smith et al. (2017). Xylan can adopt two different conformations in order to interact with cellulose and lignin. The major domain of xylan is distinguished by modifications which are evenly spaced so that they face in the same direction aiding xylan in adopting a twofold helical screw conformation in order to directly interact with cellulose. The minor domain adopts the threefold helical screw conformation and acts as a linker between microfibrils, while the closely and unevenly spaced modifications play a role in establishing a hydrophobic pocket for monolignols to polymerise into lignin (Busse-Wicher et al., 2014, 2016a; Johnson et al., 2017). The position and the function of the reducing end sequence (RES) in the secondary cell wall is currently unknown, denoted by the “?∗”. We hypothesize the RES position and that it may have a role in attaching to arabinogalactan proteins, interacting with pectic polysaccharides and influences lignin polymerization (Tan et al., 2013; Hao et al., 2014; Biswal et al., 2015, 2018). For each component of xylan, the genes responsible for biosynthesis, and the value-added product that can be obtained are provided.
(Me)GlcA modifications occur exclusively on the O-2 position of xylose residues, whereas acetyl groups can occur on both O-2 and O-3 positions with modifications being either evenly or unevenly spaced across xylose residues (Ebringerová and Heinze, 2000; Busse-Wicher et al., 2014). The spacing of the modifications is vital to the formation of two distinct domains proposed for xylan (Bromley et al., 2013; Busse-Wicher et al., 2014; Smith et al., 2017; Figure 1). The major domain has evenly spaced modifications (from every second to every tenth residue) that allows the xylan to adopt a twofold helical screw conformation similar to the glucan chains of cellulose (Simmons et al., 2016; Grantham et al., 2017; Smith et al., 2017; Figure 1). The modification present at the O-2 position sterically reduces fluctuations in xylan backbone conformation, thus promoting stable interaction with the hydrophilic face of the cellulose microfibril via hydrogen bonding (Grantham et al., 2017; Pereira et al., 2017). More densely and unevenly spaced modifications (every fifth, sixth and seventh residue are the most common) are characteristic of the minor domain which adopts a threefold helical screw conformation (Bromley et al., 2013; Busse-Wicher et al., 2014; Simmons et al., 2016; Smith et al., 2017). The minor domain can potentially interact with the hydrophobic face of cellulose, act as a linker between microfibrils, and may create a hydrophobic space between the microfibrils for monolignol polymerisation (Figure 1). While the exact mechanism of how these two domains are formed is currently unclear, the function of each domain is vital for SCW formation. An abolishment of the evenly spaced modifications in mutants of the Eskimo1/ TRICHOME BIREFRINGENCE-LIKE (TBL) 29 gene (esk1 in Arabidopsis)lead to thin SCWs, collapsed vessels and stunted growth that is characteristic of the irregular xylem (irx) phenotype (Xiong et al., 2013; Yuan et al., 2013; Grantham et al., 2017). Furthermore, altered acetylation in both domains leads to altered lignin composition (Pawar et al., 2017a,b; Yang et al., 2017). The occurrence and the type of interactions (covalent or hydrogen bonding) between xylan, glucomannan and pectin in the SCW remains to be fully resolved. Additionally, the types of linkages in lignin are thought to be influenced by polysaccharides in close proximity of the monolignols during polymerisation (Lairez et al., 2005; Li et al., 2015). Therefore, both the interaction of polysaccharides and the modification patterns on the xylan backbone may influence lignin composition. It is therefore likely that, in addition to xylan’s role in stabilizing and orientating cellulose microfibrils, but it also affects lignin content, solubility and composition. Consequently, xylan traits are often strongly correlated with processability traits (Hao et al., 2014; Johnson et al., 2017; Lyczakowski et al., 2017). A greater understanding of xylan biosynthesis and modification would therefore be useful to engineer woody biomass for improved processing traits.
Biorefinery Associated Industrial Processes
Industrial Processes
Due to the abundance and renewable nature of wood, it has always been a sought after natural commodity. Historical evidence shows that wood was used for structural timber as long as 2.6 million years ago (Klein and Grabner, 2015). The use of wood pulp as a paper source is a much more recent innovation with commercial-scale wood pulp and paper production starting with the invention of large, industrial mechanical pulping plants in the 1840s (Hunter and De la Mare, 1978). Soon after the wide-scale adoption of mechanical pulping, chemical pulping became common with sulphite pulping being the dominant form until the 1940’s when sulfate (kraft) pulping with the addition of pre-boilers became prevalent and today remains the preferred pulping process for paper production. During the pulping process, lignin is removed from the biomass leaving behind cellulose and xylan. The pulp is most often used for paper production and, due to its renewable nature, it is also increasingly being used for the manufacturing of paper bags and boxes for packaging as an alternative to non-renewable plastics. The lignin which is removed during pulping is, due to its high calorific value, typically burned to produce the heat required for the pulping process. Recently, rather than burning, lignin is isolated for the production of various value added products. Industries which process wood fiber derived biomass have increasingly begun adopting biorefinery approaches toward converting all of the wood biomass into a wide variety of valuable renewable bioproducts (products listed in Table 1). Processing plants aim to either obtain the three individual components of the biomass for biopolymer or biorefinery applications, or for simple sugars for fermentation into biofuels (Klemm et al., 2005; Restolho et al., 2009; Ragauskas et al., 2014; Nasatto et al., 2015). However, due to the recalcitrance of lignocellulosic biomass, industrial deconstruction has had to deal with several costly hurdles to increase quality and quantity of the desired constituents (Sixta, 2006; Himmel et al., 2007).
Table 1.
Xylan as a source of recalcitrance to woody biomass processing and improvements being made to reduce recalcitrance.
| Saccharification and Fermentation (S&F) | Dissolving pulp production (DPP) | |
|---|---|---|
| Main product(s) | • Ethanol [1,2,3]. | • Pure cellulose: nanocellulose viscose (textiles), rayon (tire strings), cellulose acetate films, methylcellulose, nanopaper, surgical stitches [2,4,5,6]. |
| Value added Products from “waste” | • Bioplastics, fermentable lignin, pharmaceuticals, flavourants [2]. | • Xylitol, lignosulphonates, bioinks, nanoparticles, pharmaceuticals, bioplastics [5,7,8,9]. |
| Xylans Impact on the industrial process | • Blockage of glucanase access to cellulose as a result of xylan major domain’s tight association with cellulose reduces saccharification efficiency [10]. • Pre-treatment causes xylan present on the hydrophilic face of cellulose to slide to the hydrophobic face through the action of GlucA .groups, which facilitates additional chemical treatment to remove [12]. • Lignin carbohydrate complexes formed during pre-treatment block xylanases [13, 14, 15, 16] • Pre-treatment causes undesirable and toxic breakdown products which inhibit fermentation into ethanol [17,18]. • Acetyl groups blocking CWDE [19, 20, 21]. • Released acetyl groups alter pH of fermentation fluid thereby inhibiting fermentation [17,18]. • GlucA methylation affects xylose release [27]. • Yeast does not ferment xylose efficiently [17,18]. |
• Xylan major domain’s tight association with cellulose reduces separation and purity of biopolymers [11,12]. • Pre-treatment causes xylan present on the hydrophilic face of cellulose to slide to the hydrophobic face through the action of GlucA groups, which facilitates additional chemical treatment to remove [12]. • Lignin carbohydrate complexes formed during pre-treatment block efficient chemical removal [13,14,15,16] • Pre-treatment causes undesirable and toxic breakdown products which decrease the purity of the separated biopolymers [4] • Strong acid treatment decreases strength of cellulose fibers due to cellulose autohydrolysis [4,22,23]. • Released acetyl groups alter pH of alkaline pulping liquors [24,25,26]. • Calcium bridges and crystal formation around adjacent GlucA groups lead to the use of strong acids for xvlan removal [12]. • Stacking of multiple xylan chain increase stability xylans association with cellulose [12]. |
| Improvements to techniques | • Ionic liquid and microwave assisted heating used during pretreatment increase biomass separation while decreasing toxin and inhibitor production [28,29,30,31]. • Multifunctional enzymes [32.33,34]. • Genetically engineered yeasts [38,39,40,41]. |
• Ionic liquids allow for improved biopolymer separation [28,29,30,31]. • Improved cellulose fibrillation with chemical treatment with 2,2,6,6- tetramethylpiperidine-l-oxyl (TEMPO) [35,36,37]. • WET spinning small cellulose fragments into large [42,43]. |
| Plant biotechnology approaches | Down-regulation of recalcitrance associated genes [44,45,46,47]. Upregulation of recalcitrance reducing genes [48,49]. Knock-out mutagenesis of recalcitrance associated genes [27,50,51]. Vessel complementation of knock-out mutants [52]. Ectopic xylan modifications [53,54,55]. Metabolic engineering [56,57,58,59,60,61] CRISPR:CAS9,dCAS9, or activator and DNA methyltransferase fusion dCAS9 [62,63,64]. Endogenous expression of processing enzymes which can also become active only under specific conditions | 65,66.67,68,69]. Promoter feedback loops [70]. Gene stacking [71]. |
|
References pertaining to numbered items can be found in Supplementary File S2.
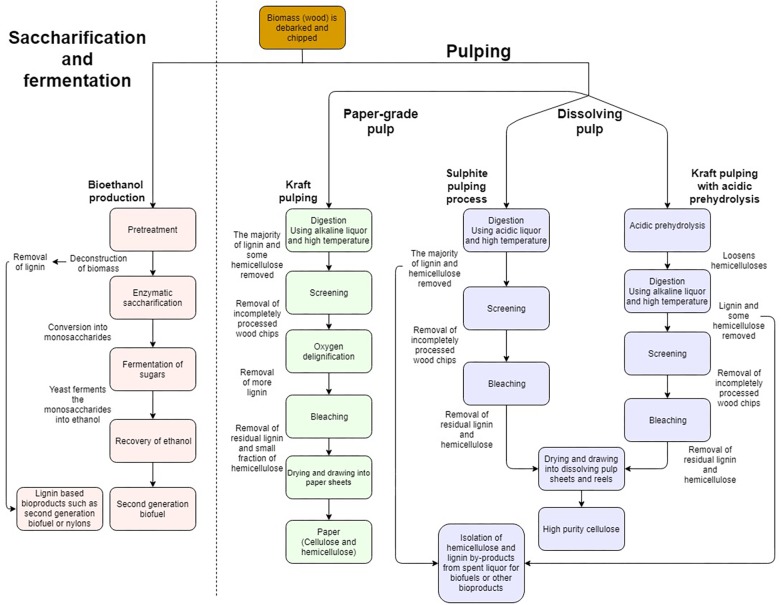
The industrial processing of wood into value added products involves two main approaches summarized in Figure 2. The process of saccharification and fermentation (S&F) refers to the separation of SCW polymers in lignocellulosic biomass through pretreatment, subsequent digestion of the polymers by cell wall degrading enzymes (CWDE) to liberate monomeric cell wall sugars and, ultimately, fermentation of the sugars by microbes to produce bioethanol (Doran-Peterson et al., 2009; Ragauskas et al., 2014; Cheng et al., 2017; Figure 2). In contrast, dissolving pulp processing (DPP) is purely chemical in nature where either highly acidic (sulphite pulping) or basic liquors (Kraft pulping) are used to separate the xylan and lignin from the cellulose at high temperatures yielding only cellulose of high quality and purity (Figure 2). The main aim of S&F industries is to produce the highest possible yield of ethanol at the lowest possible price, whereas chemical cellulose of large degree of polymerization (DP) with high purity and quality is required for cellulose derived products from DPP. These products include multiple new applications for cellulose, such as aerogels, resin impregnated fibers as well as macrofibers which display optical, magnetic and electrical properties. These products are derived from nanofibrillated cellulose (NFC), short cellulose chains which are chemically treated to gain new properties (Walther et al., 2011; Zhu et al., 2016). To accomplish these aims, xylan, lignin, contaminants and inhibitors need to be separated into distinct processing streams for their eventual use as biorefinery feedstocks (Xavier et al., 2010; Vardon et al., 2016). During polymer separation, lignin can also be liberated for applications other than fuels (Table 1). Unfortunately, the separation in the different product components for both S&F and DPP has faced many challenges on the path to commercial viability.
FIGURE 2.
Simplified comparison of industrial processes such as pulping or saccharification and fermentation, which produce renewable bioproducts from lignocellulosic biomass. Comparison of the processing steps where woody biomass is converted into bioproducts. Pulping results in either paper-grade pulp which comprises both cellulose and hemicellulose with lignin removed, or dissolving pulp where both lignin and hemicelluloses are removed to yield only high purity cellulose. Due to the simplified nature of the illustration, several processing steps may be represented as a single process.
Xylan’s Impact on Industrial Processing
Native xylan properties play a significant role in recalcitrance. Efficient separation of xylan from cellulose is a major obstacle for both S&F and DPP as the association is stabilized on multiple levels namely, the backbone itself (Pereira et al., 2017), the spacing of modifications on the backbone (Simmons et al., 2016; Grantham et al., 2017; Pereira et al., 2017) as well as the stabilizing effect of adjacent GlcA modifications that associate via chelating Ca2+ ions in muro (Pereira et al., 2017; Table 1). Interactions between xylan and lignin are less understood, but the quantity of acetyl modifications as well as methylation of GlcA influences lignin composition and solubility (Urbanowicz et al., 2012; Giummarella and Lawoko, 2016; Johnson et al., 2017; Pawar et al., 2017a,b), whereas lignin composition affects release of xylan side groups (Van Acker et al., 2017; Table 1). Modifications on the xylan backbone block access of cell wall degrading enzymes to the biopolymer substrate, which affects the polysaccharide monomerisation in S&F and reduces the efficiency of residual xylan removal by xylanases used by DPP industries as an approach to upgrade chemical cellulose quality (Helle et al., 2003; Qing and Wyman, 2011; Jönsson et al., 2013; Chong et al., 2015; Table 1). Deconstruction and monomerisation of xylan also has a role in recalcitrance as the yeast used (in S&F) to ferment sugars into ethanol do not ferment xylose efficiently and the released acetyl groups alter pH of the fermentation medium in S&F (Rastogi and Shrivastava, 2017; Yusuf and Gaur, 2017) and pulping liquor in alkaline based DPP (Kobayashi et al., 1960; Lacerda et al., 2013; Dong et al., 2016; Table 1). The abovementioned recalcitrance mechanisms require S&F and DPP industries to perform pretreatment, perform additional chemical treatments while constantly replenishing CWDE and yeast to maintain monomerisation and fermentation efficiency.
Pretreatment of biomass is required for biopolymer separation, however, the effect of industrial processing on xylan contributes to a separate set of recalcitrance associated factors. During the heating phase of pretreatment, xylan “slides” from the hydrophilic to the hydrophobic face of cellulose as a result of the GlucA modifications rather than being released (Busse-Wicher et al., 2016b; Pereira et al., 2017), resulting in incomplete separation, thus requiring additional treatment to remove the residual xylan. Adding strong sulphuric acid to dissolving pulps is a strategy to remove residual xylan, but this causes cellulose autohydrolysis; leading to a reduction in cellulose quantity and DP as well as a subsequent reduction in the tensile strength of the resulting product (Sixta, 2006; Arola et al., 2013; Stepan et al., 2016). An alternative measure to remove residual xylan is to treat the extracted cellulose with xylanases, however, xylanase activity is inhibited by the presence of lignin-carbohydrate complexes (LCC), physical (covalent) linkages between xylan and lignin, created either through the harsh chemical treatments or occurring natively in biomass (Gierer and Wännström, 1986; Iversen and Wännström, 1986; Gübitz et al., 1998; Giummarella and Lawoko, 2016; Nishimura et al., 2018). Furthermore, positive metal ions in ionic buffers used during pretreatment react with the calcium which bridge two GlucAs to form a crystal which further limits biopolymer separation (Pereira et al., 2017). Heat and pretreatment create by-products which are fermentation inhibitors and contribute to loss in total yield for biorefinery-centered DPP (Sixta, 2006; Rastogi and Shrivastava, 2017; Yusuf and Gaur, 2017 which facilitates the need to improve both the industrial techniques as well as plant biotechnology targeting xylan biosynthesis.
Improvements to the Industrial Process Methodology
One main improvement that has benefited both S&F and DPP is the use of several ionic liquid (IL) solvents that function to improve dissolution and the separation properties of the lignocellulosic biomass (Roselli et al., 2014; Chen J. et al., 2017). Pulps produced using IL solvents display improved digestion by xylanases which results in higher cellulose quality by reducing residual xylan contamination (Roselli et al., 2014), while enzyme cocktails repertoires for S&F have expanded to contain CWDE which are thermally stable, inhibitor insensitive and multifunctional (Bibi et al., 2014; Nordwald et al., 2014; Shahid et al., 2017); (Table 1). Furthermore, S&F has greatly benefitted from genetically engineered yeasts, which ferment pentoses, break down phenolic inhibitors by secreting laccases, as well as being tolerant to changing pH and growth inhibitors (Larsson et al., 2001; Jin and Jeffries, 2003; Jin et al., 2003; Wei et al., 2013; Table 1). Moreover, the issue with the creation of shorter DP cellulose can potentially be circumvented by using the reducing agent 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO) to create nanofibrillated cellulose (NFC), small fragments of cellulose that maintain its mechanical properties (Saito and Isogai, 2004; Saito et al., 2007; Isogai et al., 2011; Table 1). These NFC fragments can be chemically combined to reconstitute large macrofibers via wet-spinning using a syringe and coagulation bath, which is beneficial as mechanical strength of the chemical cellulose is retained and novel properties can be achieved by impregnation with various chemicals and resins (Iwamoto et al., 2011; Walther et al., 2011; Table 1). Improvements to the techniques have largely benefitted production, however, additional yield improvements and price reduction may be attained by making use of plants which produce biomass with reduced recalcitrance and beneficial processing traits.
Biotechnology Approaches to Improve Industrial Processing
Plant biotechnology approaches aimed at altering endogenous xylan biosynthesis genes (i.e., cisgenic approaches) have shown that targeting xylan properties can result in favorable phenotypes related to plant growth, SCW composition and industrial processing efficiency. Altering expression of genes coding for proteins related to xylan biosynthesis has proven to be an effective strategy. Favorable phenotypes have been observed in Populus as a result of downregulation targeting backbone and RES genes (Biswal et al., 2015; Ratke et al., 2018), as well as overexpression and downregulation of xylan modification genes (Song et al., 2015; Pawar et al., 2017b; Yang et al., 2017; Table 1). Studies in Arabidopsis based on knock-out mutagenesis has yielded positive industrial processing traits when targeting genes associated with GlcA addition as well as GlcA methylation (Mortimer et al., 2010; Urbanowicz et al., 2012; Lyczakowski et al., 2017), whereas irx phenotypes were obtained in knock-outs of backbone, RES and some acetylation related genes (Brown et al., 2007; Peña et al., 2007; Lee et al., 2012c; Manabe et al., 2013; Xiong et al., 2013; Yuan et al., 2013; Schultink et al., 2015; Table 1). Growth of Arabidopsis xylan related irx mutant plants has been restored, while retaining positive processing traits by using a VND7 promoter that complements the expression of the knocked-out gene in vessel cells (Petersen et al., 2012; Table 1). Efficacy of this approach in woody plants remains to be seen. Inducing increased SCW formation in general through positive feedback loops (Yang et al., 2013) as well as altering metabolic flux to SCW wall biopolymer biosynthesis (Tauberger et al., 1999; Fan et al., 2017) has yielded SCW properties that are beneficial to industrial processing, and can be achieved using a wild type or mutant background (Yang et al., 2013; Table 1). The progress currently made in cisgenics now opens the door for the use of novel techniques such as CRISPR, where different CAS9 enzymes can be used to achieve various effects, such as knock-out mutations using standard CAS9 (Fan et al., 2015; Zhou et al., 2015; Park et al., 2017) or altering expression using either dCAS9 or modified dCAS9 fusion proteins (Jinek et al., 2012; Gilbert et al., 2013; Qi et al., 2013; Vojta et al., 2016; Xiong et al., 2017; Table 1).
Progress has also been made in plant engineering efforts relying on heterologous expression of xylan biosynthesis and modification genes. Such transgenic approaches have not only produced favorable phenotypes for industrial processing, but have resulted in plants with novel SCW properties as well as additional traits such as increased stress tolerance. Self-processing plants with increased industrial processing efficiency and biotic defense responses are generated by in planta expression of xylan targeting CWDE from wood rotting fungi that can be active under normal plant growth (Gandla et al., 2014; Pawar et al., 2017b), or only become active upon application of external stimulus (Shen et al., 2012; Mir et al., 2017; Xiao et al., 2018; Table 1). Various studies have shown that xylan modifications differ between plant species and lineages (Allerdings et al., 2006; Rennie and Scheller, 2014; Peña et al., 2016; Smith et al., 2017; de Souza et al., 2018) and modification genes from one lineage can add modifications to the xylan backbone of another (Anders et al., 2012; Xiong et al., 2015; Zhong et al., 2018; Table 1). This field has not been thoroughly explored, but may be a novel manner in which to alter SCW traits. The ability to alter SCW composition and several SCW traits simultaneously has emerged due to recent breakthrough in gene stacking, where various genetic background can be used to not only achieved designer biomass (Gondolf et al., 2014; Eudes et al., 2015, 2016; Smith et al., 2017; Aznar et al., 2018; Table 1).
The above genetic engineering efforts have primarily been tested in Arabidopsis, but several studies have shown that SCW engineering approaches have scaled successfully to Populus (Supplementary File S3). The availability of reference genomes and the possibility of genetic transformation means that similar approaches can be applied and validated on woody biomass crops such as Populus (Song et al., 2006; Tuskan et al., 2006; Maheshwari and Kovalchuk, 2016) and Eucalyptus (Mullins et al., 1997; Myburg et al., 2013, 2014; Klocko et al., 2016). Engineering of SCW traits has been focused on reducing recalcitrance to optimize ethanol yield for S&F industries. Biomass from these modified plants has rarely been used for pulping or DPP applications (Zhou et al., 2017). However, reductions in residual xylan in dissolving pulps may decrease processing costs, thereby increasing the feasibility of replacing many petroleum derived products with alternatives derived from woody biomass.
Novel Biotechnology Strategies
Strategies That Target Xylan Biosynthesis Genes
Unlike cellulose, xylan is synthesized in the Golgi with other hemicelluloses (xyloglucan and glucomannan) and pectins (RGI and RGII), with biosynthesis thought to occur in the medial Golgi network (Scheller and Ulvskov, 2010; Kim and Brandizzi, 2016; Meents et al., 2018). Xylan is synthesized by membrane bound proteins which are responsible for forming the backbone, RES and backbone modifications, and rely on nucleotide sugar, acetyl-CoA as well as S-adenosylmethionine (SAM) precursors that are transported into the Golgi from the cytosol (Pauly and Scheller, 2000; Manabe et al., 2013; Rennie and Scheller, 2014; Ebert et al., 2015). The modifications on the dicot xylan backbone can occur as six possible structural configurations: 2-O monoacetylation, 3-O monoacetylation, 2,3-O diacetylation, α1,2-linked D-GlcA, α1,2-linked 4-O-MeGlcA and 3-O acetylation with α1,2-(Me)GlcA (Ebringerová and Heinze, 2000; Yuan et al., 2013). It is currently accepted that, when biosynthesis is completed, the xylan is packaged in to vesicles in the trans-Golgi and transported to the cell wall. Whether any additional modification to xylan occurs while being transported is unknown. Upon arrival at the cell wall, xylan coats and crosslinks the cellulose microfibrils (Kabel et al., 2007; Busse-Wicher et al., 2014; Simmons et al., 2016). Modification of xylan structure occurs through the action of cell wall bound xylanases and transglycosylases in response to stress (abiotic, biotic and tension) and this can happen throughout SCW formation (Minic et al., 2004; Derba-Maceluch et al., 2015). The modification pattern on xylan has also been shown to differ spatially and temporally throughout stem development (Peralta et al., 2017), but it is not clear whether any of these patterns are altered after deposition into the cell wall. Xylan biosynthesis has been thoroughly reviewed in the past (Rennie and Scheller, 2014; Marriott et al., 2016; Smith et al., 2017), and we therefore only focus on some potential implications of recent studies and how these could lead to novel biotechnology applications. We aim to stimulate hypothesis testing in the xylan research community with some of the ideas we put forward in the subsequent sections.
Xylan Backbone Biosynthesis
Elongation of the xylan backbone is performed by the xylan synthase complex (XSC). XSC related genes have been implicated as recalcitrance factors and recent studies related to the expression of XSC genes point to interesting avenues for further investigation. The XSC is comprised of IRX9, IRX10, and IRX14, these proteins are likely interacting with one another physically, have non-redundant roles and the genes encoding the XSC proteins are preferentially expressed in SCW forming tissues (Wu et al., 2010; Zeng et al., 2010, 2016; Ren et al., 2014; Jiang et al., 2016; Ratke et al., 2018; Figure 1). Additionally, three homologs of the XSC proteins (IRX9L, IRX10L, and IRX14L) have been identified and shown to be involved in the synthesis of PCW xylan indicating two different sets of XSCs for PCW and SCW formation (Mortimer et al., 2015). It has, however, been noted that a SCW XSC mutant can be partially complemented by its PCW XSC homolog when expression is driven by the promoter of the SCW homolog and vice versa (Wu et al., 2010; Mortimer et al., 2015). As expected, the expression of the PCW XSC genes differs from the genes associated with the SCW XSC, but SCW XSC genes also have slightly different expression patterns from each other during SCW formation (Li Z. et al., 2016; Sundell et al., 2017). Expression of these genes is essential for normal plant development, as knocking out a XSC gene leads to dwarfing and irx phenotypes (Brown et al., 2007; Peña et al., 2007; Lee et al., 2012c), but more interestingly, reducing the expression of XSC genes beneficially alters plant growth as well as xylem and SCW properties (Ratke et al., 2018). Therefore, altering XSC expression, rather than completely knocking out the XSC genes, could be a productive biotechnology approach.
The expression of XSC genes has biotechnology applications by using promoters from SCW XSC genes to express genes of interest or RNA interference (RNAi) fragments specifically in SCW forming tissues (Ratke et al., 2015, 2018), but altering expression of XSC genes may also be a promising biotechnology route to pursue. RNAi mediated downregulation of IRX9 and IRX14 homologs in Populus using an IRX9 promoter lead to an upregulation in cell cycle genes that resulted in taller plants with an increased stem diameter and volume (Ratke et al., 2018). SCW formation was downregulated in the transgenic plants, resulting in xylem cells which had thinner and less recalcitrant SCWs, but no significant reduction in xylose content, perhaps indicating that altering XSC related expression may be a manner in which SCW traits can be changed (Ratke et al., 2018). It remains to be seen whether the abovementioned phenotype would be replicated if a promoter was used from either another XSC related gene or a gene related to another SCW process such as cellulose or lignin biosynthesis. It is also unclear whether the abovementioned phenotype would differ if IRX10 was downregulated. Despite being a contributor to recalcitrance, xylan is a valuable bioproduct of cellulose pulping that can be used for various applications such as hydrogels, nanoparticles and 3D printing bioinks (Gericke et al., 2018; Li and Su, 2018; Xu et al., 2018). It also has dietary value (Huang et al., 2018) as well as being a source of value added products such as acetone, xylitol, xylooligosaccharides, furfural, hydroxymethylfurfural, levulinic acid, polylactic acid and ethanol (Parajó et al., 1997; Zhao et al., 2007; Binder et al., 2010; Deutschmann and Dekker, 2012; Gürbüz et al., 2012; Li et al., 2013; Zhao et al., 2013; Lama et al., 2014; Figure 1). The overexpression of endogenous XSC related genes to increase xylose content has not been attempted previously and there is a need to address the possibility of a metabolic penalty. Recently it has been shown that overexpression of a single PCW associated CesA6-like gene can increase cellulose content (Hu et al., 2017). The type (PCW vs. SCW) of XSC gene that is overexpressed might therefore be a factor to consider when trying to increase xylose content. Additionally, there is a need to investigate whether overexpression of a single XSC gene would be sufficient to increase xylose content, whether co-suppression would occur and whether UDP-xylose flux have to be increased toward xylan biosynthesis?
XSC composition (XSC stoichiometry and XSC interaction with other proteins) is a recent addition to xylan biosynthesis research. Similar to the cellulose synthase (CesA) complexes, XSC stoichiometry differs between plant species (Jiang et al., 2016; Li S. et al., 2016; Zeng et al., 2016; Zhang et al., 2018). Additionally, CesA stoichiometry also differs between tissues (Zhang et al., 2018), xylosyltransferase (XylT) activity is influenced by XSC composition (Zeng et al., 2016), therefore it would be tempting to propose that tissue and species-specific differences in XylT activity (Song et al., 2015; Jiang et al., 2016) could be due to differences in XSC composition. Overexpression of IRX9 and IRX14 (GT43) homologs from cotton in Arabidopsis increased xylose yield, most probably due to upregulation of other xylose biosynthesis related genes (Li et al., 2014). Replacing endogenous Arabidopsis XSC genes with homologs from rice alters XylT activity and chain length (Chiniquy et al., 2013). XSC composition may also influence xylan modification. In wheat it has been shown that XSC interacting proteins such as UDP-arabinopyranose (Arap) mutase (UAM), which reversibly converts UDP-Arap into UDP-arabinofuranose (Araf), produce the substrate required by xylanarabinotransferases (XAT) to add arabinose substitutions to the xylan backbone (Konishi et al., 2007; Anders et al., 2012). Additionally, two proteins annotated as vernalization-related gene 2 (VER2) and germin-like protein (GLP; Zeng et al., 2010; Jiang et al., 2016) also interact with the XSC, and are thought to be associated with the hormones jasmonic acid and auxin, respectively. The XSC physically interacting with these proteins either aids in targeting the XSC correctly to the trans-Golgi (GLP) or to unknown compartments (VER2), which suggests (at least in wheat) that hormone interaction with the XSC regulates SCW xylan biosynthesis (Jiang et al., 2016). Together, these results suggest that XSC composition affects xylan content, modification and transport and it would therefore be desirable to better understand the biotechnology potential of altering XSC related genes.
The results from the preliminary findings on XSC composition point to a number of novel xylan biotechnology approaches. Heterologous expression of XSC genes from other plant species seems to affect xylan properties. Additionally, introducing monocot XSC genes, UAM and XAT into a dicot system could facilitate increased incorporation of ectopic O-2 and/or O-3 arabinose modifications compared to what was previously possible (Anders et al., 2012; Xiong et al., 2013). By putting VER2 expression under the control of different SCW promoters (Tokunaga et al., 2009; Smith et al., 2013; Ratke et al., 2015), or even an inducible promoter (Gatz, 1996; Caddick et al., 1998), one could potentially engineer xylan biosynthesis by altering the transport of the XSC (Jiang et al., 2016). XSC related biotechnology approaches could allow us to have greater control over xylan biosynthesis in general from timing of biosynthesis to altering xylan properties and modifications.
Reducing End Sequence
The RES is a tetrasaccharide comprising of β-Xyl-(1,3)-α-Rha-(1,2)-α-GalA-(1,4)-Xyl, the function of which is currently unknown and the main discussion being whether the RES acts as a primer or terminator. We want to discuss how the RES effects xylan biosynthesis (York and O’Neill, 2008; Smith et al., 2017). The RES has been proposed to act as a primer since in vitro expressed IRX10 was observed to elongate the xylan backbone from the reducing end (Urbanowicz et al., 2014; Smith et al., 2017). This is rather interesting since mutation in the RES related genes, IRX7, IRX7L, IRX8 and PARVUS (Figure 1), lead to variable chain lengths and XylT activity (Brown et al., 2005, 2007; Peña et al., 2007; Lee et al., 2007, 2009; Wu et al., 2010). Plants with mutations in RES genes tend to be severely dwarfed (Rennie and Scheller, 2014), thus complicating the study of RES related genes, but a greater understanding of this process could yield biotechnology tools which could be used to alter xylan content.
Besides the dwarfing effect of RES mutants, more moderate changes in expression patterns of RES genes also affect plant development. It was observed that overexpression of IRX8 in Populus resulted in decreased growth and higher recalcitrance, whereas downregulation had the opposite effect (Biswal et al., 2015, 2018). It remains to be seen whether overexpression and downregulation of other RES genes result in these two opposite phenotypes. If the RES does indeed act as a primer for xylan biosynthesis (Smith et al., 2017), overexpression of the RES genes could also be used to the increase total xylose content (Biswal et al., 2018), by potentially promoting higher rates of xylan biosynthesis initiation.
The biosynthesis of xylan and the heparan sulfate (HS) proteoglycan have often drawn parallels (Smith et al., 2017), as both these polysaccharides are biosynthesised in the Golgi and possess an RES (Kreuger and Kjellén, 2012; Rennie and Scheller, 2014). The RES in HS attaches to a protein required for transport to the cell wall (Kreuger and Kjellén, 2012), so it would be tempting to suggest that perhaps xylan is also attached to a protein required for transport? Some support for this hypothesis exists as arabinogalactan proteins (AGPs) called ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (APAP1) have been found to be attached to PCW xylans (Tan et al., 2013). If the attachment of xylan to an AGP is required for transport, a lack of transport to the cell wall could be the reason for fewer xylan chains being detected in the SCW in RES mutants (Peña et al., 2007; Persson et al., 2007). In the PCW, xylan aids in the interaction of RGI with cellulose (Ralet et al., 2016), the same biopolymers which are affected in RES mutants (Zhong et al., 2005; Brown et al., 2007; Lee et al., 2007; Persson et al., 2007; Figure 1). So does the RES have a role in SCW biopolymer interaction as well? Based on the glycome profiling results obtained from the biomass of plants where IRX8 was either overexpressed or downregulated (Biswal et al., 2015, 2018), it has been suggested that that the RES associates with a homogalacturonan molecule and which may have a direct or indirect effect on guaiacyl lignin polymerization (Persson et al., 2007; Hao et al., 2014; Biswal et al., 2015, 2018; Figure 1). Taking the potential interaction of xylan with other molecules into account could be a manner in which to alter multiple traits simultaneously.
If xylan, like HS (Kreuger and Kjellén, 2012), relies on one or more proteins for transport to the cell wall, the expression of the gene(s) corresponding to the transport proteins could be put under the control of various other SCW promoters to specify the stages of SCW development at which xylan should be transported to the cell wall. Tan et al. (2013) provided the most comprehensive illustration of PCW xylan attached to an AGP. The study also illustrated that xylan was attached to the pectin molecules homogalacturonan and rhamnogalacturonan I. Keeping this structure in mind, it perhaps makes sense that galacturonic acid content was altered in response to IRX8 overexpression and downregulation (Biswal et al., 2015, 2018). Similarly, irx7 mutants may display compromised seed coat mucilage anchoring (Hu et al., 2016) and reduced xylan content at the SCW (Brown et al., 2007) due to lack of attachments to an AGP. Effective transport of xylan may be hindered in both tissues due to a lack of an RES, but before this hypothesis can be tested, we need to determine whether SCW xylan relies on an AGP for transport the cell wall. Fasciclin-like arabinogalactan (FLA) are AGPs expressed during SCW biosynthesis and FLA mutants display differences in xylose content as well as microfibril angle (MacMillan et al., 2010, 2015). A change in microfibril angle has been noted in a few xylan mutants (Derba-Maceluch et al., 2015; Ratke et al., 2018), xylan has previously been suggested to play a role in microfibril angle of cellulose and has been suggested as a reason to why SCW patterning is maintained even when microtubule formation is disrupted (Reis and Vian, 2004; MacMillan et al., 2010; Schneider et al., 2017). Therefore, it could be possible to engineer microfibril angles required by specific industries or biorefinery applications by altering FLA expression patterns. Pectin and xylan can be converted into various value added products through biorefining (Deutschmann and Dekker, 2012; Gürbüz et al., 2012; Xiao and Anderson, 2013; Gericke et al., 2018; Figure 1), overexpressing IRX8 lead to increase in both of these biopolymers (Biswal et al., 2018). However, this plant also displayed increased biomass recalcitrance and decreased growth, if these undesirable traits could be alleviated, the IRX8 overexpressor may have biorefinery applications. The evidence we have provided suggests that there targeting RES synthesis is a viable biotechnology strategy.
Xylan Acetylation
Xylan acetylation is a key factor in recalcitrance. Earlier biotechnology approaches aimed to reduce acetyl content by either targeting the genes involved in acetylation, or by removing the acetyl using ectopically expressed esterases (Manabe et al., 2013; Pogorelko et al., 2013; Xiong et al., 2013). Lately interest has shifted to understanding the effect of acetyl content on SCW traits as well as on plant physiology. The current hypothesized mechanism of xylan acetylation entails cytosolic acetyl-CoA being transported into the Golgi by a REDUCED WALL ACETYLATION (RWA) transporter, where it is sequestered by ALTERED XYLOGLUCAN 9 (AXY9) for polysaccharide acetylation, and subsequently used by xylan acetyltransferases in the TRICHOME BIREFRINGENCE-LIKE (TBL) family to acetylate the xylan backbone (Manabe et al., 2013; Xiong et al., 2013; Yuan et al., 2013; Schultink et al., 2015; Figure 1, 3). Moderate reductions in xylan acetylation (13–20%) through RWA downregulation or the introduction of an acetyl xylan esterase (AnAXE1) from Aspergillus niger has proven to reduce recalcitrance and alter SCW traits in an industrially beneficial way without impacting plant growth (Pawar et al., 2017a,b). Interestingly, plants with increased acetyl content also display reduced recalcitrance in addition to increased growth and stem volume (Yang et al., 2017). Therefore, increasing xylan acetyl content may be an approach to alter SCW traits, increase growth and reduce recalcitrance factors such as LCCs (Giummarella and Lawoko, 2016; Martínez-Abad et al., 2018). Biorefinery applications centered on acetic acid production from biomass may also benefit from biomass with higher acetyl content (Patil et al., 2013; Audu et al., 2018; Figure 1). Furthermore, acidic dissolving pulp production (DPP) benefits from the additional pH decrease caused by released acetyl groups (Chen et al., 2010; Xiao et al., 2015). It has been found that excess acetylation is processed by an esterase in monocots and would be valuable to determine whether this function is shared with dicots (Scheller, 2017; Zhang et al., 2017). Together these findings suggest that total xylan acetyl content is an important target for DPP and biorefinery applications in particular, but xylan acetylation patterns may be equally important and may explain some of the apparently discordant findings from up and down-regulation of xylan acetylation genes.
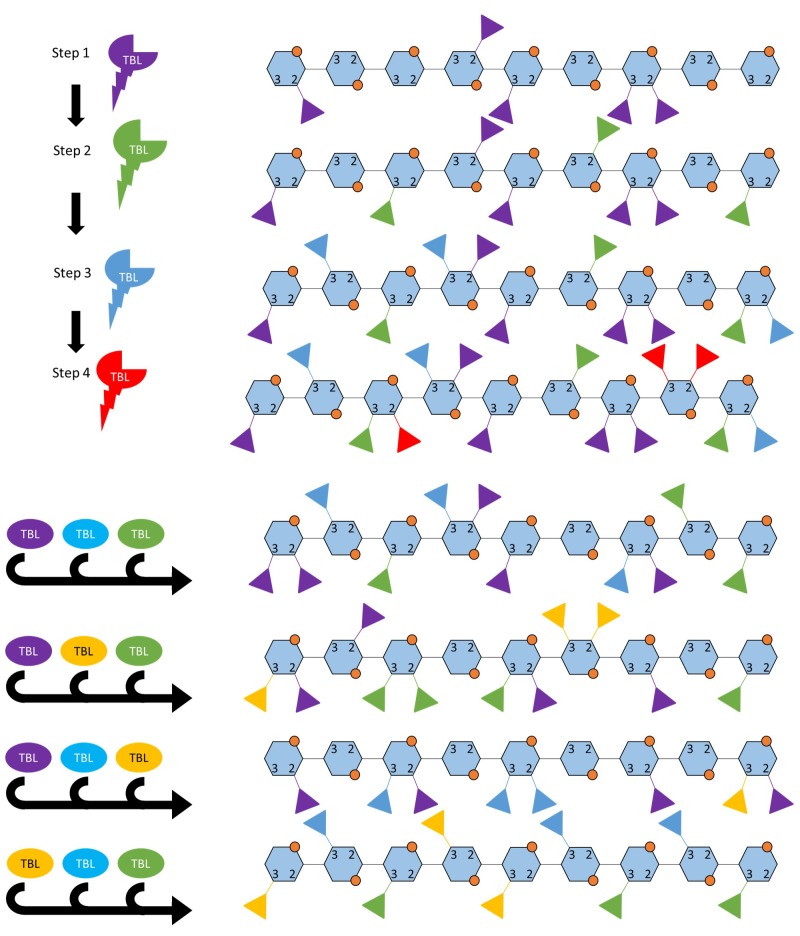
FIGURE 3.
Two possible ways in which TBLs may establish the acetylation pattern. A highly simplified hypothesis of how acetylation patterns may be established by TBL proteins, this model does not incorporate interactions with glucuronic acids or post-synthesis modification by acetyl esterases. (A) Acetylation pattern may occur in a hierarchical or sequential manner where TBL genes are expressed in a certain order for the final pattern to be obtained or the enzyme kinetics determines the order in which the TBL proteins modify the backbone. (B) Certain patterns of acetylation occur when a combination of TBL proteins are present, these TBLs either interact with each other or share transcriptional regulation. The acetylation patterns shown here are hypothetical in order to illustrate the concept. Each TBL of a different color refers to a different TBL.
The nine functionally validated acetyltransferases associated with xylan acetylation all belong to a single clade in the TBL protein family. They all can add acetyl groups to O-2 and O-3 positions on the xylose residue, but differ in enzyme kinetic rate and display differing “preference” for the type of acetylation pattern that is produced suggesting that they differ in regiospecificity (Xiong et al., 2013; Yuan et al., 2013, 2016a,b,c). How the acetylation pattern is established is not fully understood and may be a complex process involving acetyl-CoA precursor supply, differential expression and activity of various xylan biosynthetic proteins and, potentially, editing upon delivery to the SCW. However, based on functional studies of TBL proteins, the activity of these proteins seems to have the largest effect of xylan acetylation (Xiong et al., 2013; Yuan et al., 2013, 2016a,b,c). How these proteins could establish a basal acetylation pattern could possibly include either (i) a hierarchical organization where the TBLs modify the xylan backbone in a specific order, or (ii) a combinatorial mechanism where different combinations of TBLs yield different acetylation patterns, or (iii) a combination of these that is accomplished by fine temporal and spatial transcriptional regulation (Figure 3). Some support for these hypotheses exist as only esk1 (TBL29) mutations result in a large reduction in xylan acetylation and an irx phenotype, whereas other TBL mutants primarily exhibit changes in modification patterns. Additionally, TBL32 and TBL33 require a pre-existing α1,2-(Me)GlcA modification to acetylate the backbone (Xiong et al., 2013; Yuan et al., 2016a,b,c) hinting at an ordered process. Crystal structures of modification enzymes related to xyloglucan and alginate have provided valuable insight into possible mechanisms employed to obtain modification patterns (Sychantha et al., 2017; Culbertson et al., 2018). Acetylation patterns can potentially be established when the xylan enters a channel containing multiple TBLs. Alternatively, the TBL with the highest kinetic rate may preferentially perform the first modification and thereby initiate the pattern. As the xylose chain subsequently increases in length, other TBLs may alternatingly add acetyl groups to extend the pattern. Differentially regulating the TBL genes could yield different combinations of TBL proteins producing different modification patterns, a possibility that could be leveraged in biotechnology approaches to achieve desired acetylation patterns and biomass processing traits.
Transcriptional regulation of xylan acetylation related genes affects total acetyl content, the tissue in which acetylation occurs and the developmental stage at which acetylation occurs. Expression profiles of RWAs seem to determine acetyl-CoA supply for polysaccharide acetylation, as constitutively expressed RWAs from the AB clade supply acetyl-CoA to xyloglucan whereas the SCW expressed RWAs of the CD clade supply acetyl-CoA to xylan (Pawar et al., 2017b). Downregulation of RWA genes was shown to have no effect on plant growth while reducing recalcitrance and altering SCW composition (Pawar et al., 2017b). Overexpression of RWA was specified as having increased sugar release and yield (Macaya-Sanz et al., 2017). It is not currently known whether all TBLs which facilitate SCW xylan acetylation have been identified, but it is known that other TBL clades acetylate different polysaccharides such as xyloglucan (Vogel et al., 2004; Bischoff et al., 2010; Gille et al., 2011b). In the case of xyloglucan, two proteins (TBL22 and TBL27) are involved that perform the same function, but are expressed in different tissues (Gille et al., 2011b). The Populus homolog of TBL45 may be an example of this phenomenon, as it seems to add acetyl to xylan but is highly expressed in phloem and young leaf as opposed to xylem, thereby potentially adding an additional layer of complexity to how acetylation patterns emerge (Yang et al., 2017). In theory, an even large variety of acetyl patterns could be achieved by altering the expression of various combinations of TBLs and overexpressing RWA to provide sufficient acetyl-CoA precursor.
Methylglucuronic Acid Modifications
α1,2-linked D-GlcA modifications that have also been methylated at the O-4 position (MeGlcA) are the only xylan modification currently known to discriminate between the major and minor domains of xylan, PCW and SCW, as well as being associated with recalcitrance and SCW traits. The xylan backbone is modified with GlcA groups exclusively at the O-2 positions by GUX proteins which use UDP-GlcA as precursor, where GUX1 modifies the backbone in an even manner (major domain), GUX2 modifies in a closely and unevenly spaced manner (minor domain) while GUX3 modifies PCW xylan (Mortimer et al., 2010, 2015; Lee et al., 2012a; Bromley et al., 2013). Methylation of GlcA is carried out by GXM proteins from the DUF579 family by using the SAM precursor (Lee et al., 2012b; Urbanowicz et al., 2012; Song et al., 2016; Figure 1), with this type of methylation being exclusive to SCW xylan (Mortimer et al., 2015; Ishii et al., 2017). The ratio of methylated GlcA to unmethylated GlcA is dependent on the kinetic rate of GXM and differs between species (Yuan et al., 2014). What is less clear is the timing of GXM expression. It was found that 35S overexpression of GXM did not have an effect, but lignin linkages and LCCs were not investigated. The biological function of GlcA methylation is currently unclear, but it is known that its presence results in the xylan being more hydrophobic and has been suggested to be related to lignin composition and non-covalent interactions between xylan and lignin (Busse-Wicher et al., 2016b). However, reductions in MeGlcA groups lead to increased xylose release and a higher proportion of S-lignin (Urbanowicz et al., 2012). These groups could be taken advantage of for altering xylan’s association with cellulose through its two domains as well as its influence on lignin composition. This could potentially be achieved by using different SCW promoters rather than general overexpression with 35S to have a more specific effect during wood formation.
Some unknowns still exist in terms of these modifications namely the transporters needed for SAM and GlcA import into the Golgi during xylogenesis as well as the function of a DUF579 member IRX15. A uronic acid transporter which functions in seed mucilage has recently been identified (Saez-Aguayo et al., 2017). SAM transport into the Golgi has been detected and SAM transporters to the mitochondrion have been found (Palmieri et al., 2006; Ibar and Orellana, 2007). IRX15 and IRX15L are members of the DUF579 protein family, but their functions are not currently known, however, mutagenesis of a single gene leads to improved biomass processing (Brown et al., 2011; Jensen et al., 2011). Mutagenesis of both genes leads to an irx phenotype, shorter xylan chains, a corrugated SCW and an S3 layer which detaches from the S2 layer (Brown et al., 2011; Jensen et al., 2011). It is not currently known whether any editing of the (Me)GlcA group occurs in a manner similar to acetylation in monocots, but it is possible that this may relate to the function of IRX15/IRX15L, or have to do with the transport of xylan from the Golgi to the cell wall (Brown et al., 2011; Jensen et al., 2011). Identification of transporters could allow us to alter precursor supply for GlcA and its methylation through regulating the expression of these genes, while understanding the function of IRX15 may allow us to exploit its function in strategies that target specific SCW layers.
Interaction Between Modifications
Several studies have shown that there is competition and interaction between the xylan machinery predominantly for the modification of the O-2 position of xylose residues. Mutations in several genes related to xylan acetylation result in increased (Me)GlcA content (tbl32 tbl33 is the exception) and vice versa for gux mutants (Busse-Wicher et al., 2014; Lee et al., 2014). The fact that most TBLs can modify the O-2 position is the reason why GlcA increases when these TBLs are mutated, albeit, not as drastically as in the esk1 (tbl29) mutant (Xiong et al., 2013; Yuan et al., 2013, 2016b,c). The interaction between GlcA addition and acetylation is evident from the existence of xylose residues with 3-O acetylation and α1,2-(Me)GlcA. This pattern is produced by the action of TBL32 and TBL33 that add 3-O acetyl groups only to residues where the O-2 position was already occupied by(Me)GlcA (Yuan et al., 2016a; Zhong et al., 2017). The prevalence of these 3-O acetyl modifications increases in several tbl mutants in proportion to the increase of (Me)GlcA at O-2 positions (Yuan et al., 2013, 2016b,c). This residue is found to be the most recalcitrant to esterase treatment and it was found that a separate esterase from Flavobacterium johnsoniae is required to cleave this 3-O acetyl (Razeq et al., 2018; Puchart et al., 2019), perhaps suggesting a role in defense against certain types of pests which do not possess the specific esterase. These results become increasingly important when considering the processability of Populus and Eucalyptus wood as they both contain high proportions of this recalcitrant modification (Evtuguin et al., 2003; Puchart et al., 2019). The function of the 3-O acetylation/α1,2-(Me)GlcA modification pattern is unknown, but it may be linked to defense, or, alternatively, its proximity to (Me)GlcA might have a role in xylan’s association with lignin and it could therefore be a valuable biotechnology target.
An interesting interaction has been noted between GUX1 and ESK1 that may have broader implication for xylan modification. This relates to the types of modification patterns that could be achieved, as well as how xylan modification domains are formed. It has been suggested that GUX1 function relies on ESK1 in order to add GlcA modifications in an evenly spaced manner required for formation of the major domain that associates with the hydrophilic face of cellulose (Grantham et al., 2017). However, the esk1 mutant can be complemented by expressing GUX1 under control of the ESK1 promoter (Xiong et al., 2015). This may indicate either that the timing of ESK1 expression is vital for the establishment of the even modification pattern of the major domain, or that the kinetic rate of GUX1 alone is insufficient to establish the major domain pattern in esk1 (Rennie et al., 2012; Zhong et al., 2017). The increased expression under the control of the ESK1 promoter probably resolves this problem by increasing GUX1 protein abundance. Furthermore, it has been noted that O-2 modification, regardless of the type of modification, is vital for xylan function suggesting that these two modifications, at least in the context of xylan’s interaction with cellulose, fulfill equivalent functions (Xiong et al., 2015; Pereira et al., 2017) that could be exploited in biotechnology strategies to produce trees with different xylan properties and interactions.
It appears that ESK1 and GUX1 are required for major domain formation, but it still needs to be established whether other TBLs are essential for major domain formation, and whether any of the TBLs function specifically together with GUX2 to specify the minor domain (Bromley et al., 2013; Zhong et al., 2017). The gux1 gux2 double mutant has xylan that is devoid of GlcA (i.e., only acetylated; Busse-Wicher et al., 2014; Lee et al., 2014). Conversely, knocking out all SCW xylan associated TBL genes, and expressing GUX genes under the appropriate promoters would lead to xylan with only (Me)GlcA modification. It would be interesting to contrast the SCW properties and composition of plants with xylans that exclusively harbor one type of modification, and determine whether such modified xylans are favorable for specific lignocellulosic biomass applications (Giummarella and Lawoko, 2016; Escudero et al., 2017; Pawar et al., 2017a; Figure 1). The stunted growth of the esk1 mutant complicates efforts to study its effect on SCW composition (Xiong et al., 2013; Yuan et al., 2013). These results indicate that ESK1/TBL29 may be the principal TBL for xylan acetylation, whereas the other TBLs may be providing additional complexity and functionality to the acetylation pattern. If that is indeed the case, the esk1 kaktus (Bensussan et al., 2015) as well as esk1 max 4-7 (Ramírez et al., 2018) double mutants, which exhibit wild type growth and restored vessel morphology, while retaining the altered SCW phenotype of esk1, could be used as genetic backgrounds in a pursuit to completely remove acetyl from the xylan backbone. The rest of the xylan associated TBLs could be knocked out and the TBL gene promoters could be used to drive additional GUX gene expression to yield a plant with only (Me)GlcA modifications. These plants may be highly beneficial for biorefinery applications focused on extracting GlcA for the production of value added products such as glucaric acid, other GlcA derivatives and breakdown products (Zhao et al., 2007, 2013; Gürbüz et al., 2012;Salamone et al., 2014; Li et al., 2018; Figure 1). The identification of more mutants that can rescue irx phenotypes has the potential to allow the study and engineering of SCW and processing traits in plants with highly altered SCW compositions.
Metabolic Engineering of Xylan Traits
Modeling the Interaction Between Xylan Biosynthesis and Other Cellular Processes
Xylan biosynthesis and modification relies on metabolic precursors from primary metabolism including nucleotide sugars, SAM and acetyl-CoA. How metabolism is co-regulated with SCW cell wall formation has previously been investigated for lignin and cellulose biosynthesis (Vanholme et al., 2012; Mizrachi et al., 2013; Yen et al., 2013), but this has not been done for xylan biosynthesis. The valuable insight that systems biology approaches provide may be applied to xylan modification potentially providing insight into how regulation differs between major domain formation required for xylan’s association with cellulose, and minor domain formation required for xylan’s association with lignin (Busse-Wicher et al., 2014; Marriott et al., 2016; Simmons et al., 2016; Peralta et al., 2017). Modification genes associated with the spatial and temporal changes in xylan modification throughout SCW development may be identified, and different SCW traits may arise from re-wiring the expression of these genes. It is known that plant metabolism is extensively altered during SCW development reflecting the need to maintain metabolic homeostasis in developing xylem cells (Li Z. et al., 2016; Ohtani et al., 2016). Understanding how xylan modification is coordinated with precursor metabolic pathways in developing xylem cells may highlight which pathways are the main sources of metabolic precursors such as SAM or acetyl-CoA, and whether alternative metabolic sources are used during different stages of SCW development.
Secondary cell wall formation is a very strong carbon sink in xylem cells and it is imperative that metabolic pools are tightly regulated to ensure optimal resource allocation between normal cellular metabolism and SCW biopolymer synthesis (Mizrachi and Myburg, 2016). Systems biology and systems genetics analyses are network based approaches that involve incorporating multiple “omics” data sets such as genomic, transcriptomic, metabolomic and proteomics data in order to model a biological process of interest (Guerriero et al., 2013). Each of these datasets represent variation in the components of the system, and using network based approaches, associations within and between systems components can be identified, providing a more holistic understanding of regulatory interactions and metabolic interdependencies (Kitano, 2002; Jansen, 2003; Kohl and Noble, 2009). Looking at these metabolic and regulatory processes, and their interactions, by comparing multiple systems components allows identification of emergent properties of the system that could not be otherwise observed (Feltus, 2014; Hillmer, 2015). Systems biology approaches rely on multi-omics experiments, typically on the same genotype that is subjected to different conditions (Barah et al., 2013; Rasmussen et al., 2013), sampled during different developmental stages (Li Z. et al., 2016), and/or creating sequential knock-out lines of many genes involved in a process of interest and observing how the system changes in response to each perturbation (Vanholme et al., 2012). This aids in understanding the dynamics of the process and which components are key targets for perturbation (Chen et al., 2014; Wang et al., 2014; Taylor-Teeples et al., 2015).
Systems genetics relies on using hundreds of genotypes in structured populations, where genetic variation acts as the main perturbation on the system (Civelek and Lusis, 2014). Besides systems components such as transcriptomics, metabolomics or proteomics, systems genetics can also link these components to complex trait variation in a population, for example SCW content and composition, industrial processing efficiency, physiological development and ecological adaptations, all in the context of genetic variation (Drost et al., 2010; Mizrachi et al., 2011, 2017; Civelek and Lusis, 2014; Christie et al., 2017). Individuals used for systems genetics analysis are typically phenotypically normal, therefore, the analysis provides valuable insight into the limits of natural perturbation that the system can handle, whereas mutagenesis of genes in systems biology approaches may be too extreme, leading to associations not found to occur normally. Genomic loci (represented by expression QTLs) associated with multiple components and/or traits, usually represent regulation hotspots with variants usually being found in hub genes or genes responsible for the biosynthesis of widely used metabolic precursors (Porth et al., 2013; Civelek and Lusis, 2014; Christie et al., 2017; Mizrachi et al., 2017). Networks generated from systems genetics analysis can be used to understand directionality of the associations in the system, and be used to understand epistatic interactions and pleotropic effects, which is highly beneficial for understanding highly coordinated processes such as SCW formation (Mizrachi and Myburg, 2016). Both approaches are valuable for identifying novel genes associated with a process, understanding interactions among systems components and for predicting the outcomes of genetic- and metabolic engineering strategies (Thumma et al., 2005; Mizrachi et al., 2011; Mottiar et al., 2016). A common finding in systems level analyses is that primary metabolic pathways are tightly coordinated with the various biosynthetic processes for which they provide precursors and that variation within the abundance of these pathways is associated with variation in complex traits (Vanholme et al., 2012; Li Z. et al., 2016; Ohtani et al., 2016; Mizrachi et al., 2017). Systems biology and genetics approaches may therefore be useful to understand how variation in precursor metabolic pathways affect xylan biosynthesis, the modification patterns, and how this variation ultimately impacts SCW and industrial processing traits.
Nucleotide Sugars
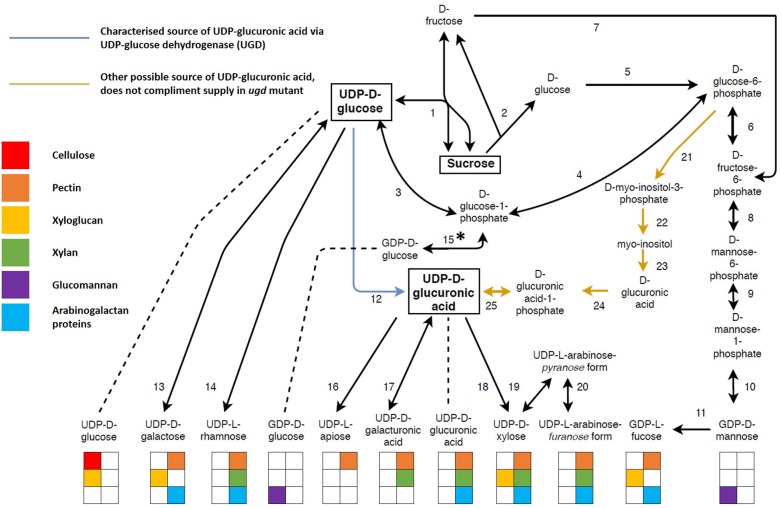
The interconversion of nucleotide sugars is vital for xylan biosynthesis. All nucleotide sugars required for xylan biosynthesis are derived from UDP-Glc (Sharples and Fry, 2007; Bar-Peled and O’Neill, 2011; Reboul et al., 2011), produced together with D-fructose through the reversible action of SUCROSE SYNTHASE (SuSy) (Amor et al., 1995; Salnikov et al., 2001; Coleman et al., 2009; Figure 4 and Supplementary Table S1). In order to effectively partition these sugars between these polysaccharides, some form of co-regulation needs to exist, and it is already known that xylan and cellulose biosynthesis are transcriptionally co-regulated in order to balance UDP-Glc usage for cellulose and xylan via UDP-Xyl (Brown et al., 2005; Ko et al., 2006). Xylan and glucomannan are the two main hemiceluloses present in the SCW (Plomion et al., 2001), but they do not directly compete for substrate. GDP-mannose required for glucomannan biosynthesis is D-fructose derived, while the irreversible sucrose breakdown by INVERTASE (2) produces D-glucose required for the biosynthesis of GDP-glucose, however, this pathway still needs to be characterized in planta (Hinman and Villemez, 1975; Sharples and Fry, 2007; Bar-Peled and O’Neill, 2011). Two strategies for carbon allocation during SCW formation exist, transcriptional co-regulation when the metabolic precursor is shared (e.g., cellulose and xylan), or using precursors that are not derived from competing metabolic pathways (e.g., xylan and glucomannan).
FIGURE 4.
The interconversion of sugar nucleotides derived from sucrose and their use in biopolymer biosynthesis. Sugar nucleotides are the precursors required for the biosynthesis of both primary and secondary cell wall biopolymers with most sugar nucleotides being used for more than one biopolymer. Sucrose is the main source of nucleotide sugars. Its reversible cleavage by sucrose synthase yield D-fructose and UDP-glucose. UDP-glucose serves as a precursor for many other nucleotide sugars, either as a direct source (UDP-galactose, UDP-rhamnose and UDP-glucuronic acid) or an indirect source via from UDP-glucuronic acid (UDP-xylose, UDP-galacturonic acid and UDP-apiose). UDP-glucuronic acid can either be derived from UDP-glucose (blue arrow) or glucose-6-phospate (orange arrows). The former pathway (12) is an almost exclusive source of UDP-glucuronic acid, whereas the latter pathway (21–25) is understudied and is unable to complement an ugd mutant. ∗The enzyme required for this reaction has not been identified in planta. The coloring of the blocks under each nucleotide sugar indicates the biopolymer for which the nucleotide sugar is a precursor. Unidirectional arrows indicate irreversible reactions whereas bidirectional arrows indicate reversible reactions. Key metabolites are highlighted in square boxes. The enzymes (1–25) represented by each number can be found in Supplementary Table S1, along with additional information such as gene ID and cellular localisation.
Apart from xylan biosynthesis, sugar nucleotides are used for the biosynthesis of PCW polysaccharides xyloglucan and the pectins, HG, RGI and RGII (Mølhøj et al., 2003; Mohnen, 2008; Bar-Peled and O’Neill, 2011; Atmodjo et al., 2013; Saez-Aguayo et al., 2017). UDP-sugars may be allocated to xyloglucan, pectin and xylan through spatial and/or temporal regulation, as during the transition from primary to secondary growth in developing xylem, downregulation of genes associated with PCW polysaccharides is observed (Hertzberg et al., 2001). Additionally, transporters can serve as key points of regulation through differential transport activity (Ebert et al., 2015), process specificity (Manabe et al., 2013; Mortimer et al., 2013), alternate functions in different tissues (Saez-Aguayo et al., 2017), or through interactions with the biosynthetic machinery (Gille and Pauly, 2012). In the case of xyloglucan, the cellulose synthase-like C4 (CSLC4) enzyme acts as the biosynthetic machinery and the transporter. Catalysis of the glucan chain begins in the cytosol and the chain is elongated directly into the Golgi lumen by passing through the membrane spanning CSLC4 protein (Davis et al., 2010; Chou et al., 2012). Spatial regulation may be a manner in which to avoid competition for precursors between the synthesis of PCW and SCW polysaccharides, whereas the differential activity of transporters and biosynthetic proteins may be a mechanism by which adequate precursor supply is ensured for Golgi localized polysaccharide synthesis. These mechanisms would be important avenues for metabolic engineering approaches targeted at nucleotide sugar interconversions. More than one polysaccharide could be affected, however, unwanted polysaccharide accumulation or deleterious effects could be avoided by making use of some of the aforementioned regulatory mechanisms.
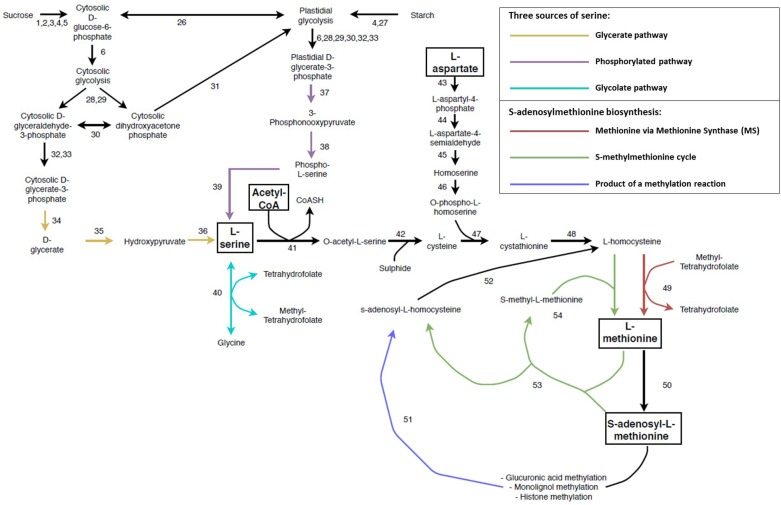
S-Adenosylmethionine
SAM is the universal methyl donor in all forms of life. In SCW forming tissue, it is vital for the biosynthesis of ethylene and polyamines and for the methylation of GlcA modifications on the xylan backbones, monolignols as well as histones and DNA (Inoue et al., 1998; Shen et al., 2002; Roje, 2006; Do et al., 2007; Lee et al., 2012b; Urbanowicz et al., 2012; Sauter et al., 2013; Wang et al., 2016; Hussey et al., 2017). SAM is derived from methionine, which is produced through several conversion steps from the amino acids aspartate and serine, as well as the incorporation of acetyl-CoA (Jander and Joshi, 2009; Sauter et al., 2013; Ros et al., 2014; Figure 5 and Supplementary Table S1). The reliance on multiple key metabolites for its biosynthesis imposes a requirement for tight regulation of SAM biosynthesis. As opposed to bacteria, fungi and animals, there are three sources of serine biosynthesis in plants namely the glycolate pathway, cytosolic glycerate pathway and the plastidial phosphorylated pathway (Ros et al., 2014; Figure 5 and Supplementary Table S1). The glycolate pathway has been suggested as the most important pathway of serine biosynthesis and has been shown to have biotechnology applications. Transgenic Populus overexpressing SERINE HYDROXYMETHYLTRANSFERASE (40) displayed increased growth, stem diameter and sugar release as well as a decrease in lignin content (Zhang et al., 2019). The glycolate pathway is involved in the cycling of folate derivatives (Douce et al., 2001; Loizeau et al., 2007), but is limited to periods of sunlight (Ros et al., 2014). The phosphorylated pathway is the predominant pathway associated with non-green tissues (such as xylem and root; Cascales-Miñana et al., 2013; Toujani et al., 2013) and is also involved in nitrogen metabolism (Anoman et al., 2015). The L-serine from the glycolate pathway may move from the leaves through the phloem to non-photosynthetic tissues (Riens et al., 1991; Lam et al., 1995), thereby supplementing the L-serine derived from the phosphorylated pathway. Although the non-photosynthetic glycerate pathway has been shown to be functional (Kleczkowski and Givan, 1988; Greenler et al., 1989) it has been largely understudied. The 3-PHOSPHOGLYCERATE PHOSPHATASE (34) enzyme has only been partially purified (Randall et al., 1971) and the gene(s) responsible for this function are still to be identified in Arabidopsis (Cascales-Miñana et al., 2013).
FIGURE 5.
Biosynthesis and cycling of s-adenosylmethionine. For s-adenosylmethionine (SAM) to be produced, methionine needs to be biosynthesised first which requires sulfide, the cofactor acetyl-CoA as well as the two amino acids aspartate and serine. There are three possible sources from which serine can be derived, with the glycolate pathway (light blue) being predominant in autotrophic tissues and the phosphorylated pathway (purple) predominating in heterotrophic tissues, whereas the metabolic context of the glycerate pathway (orange) is still unknown. SAM SYNTHETASE (50) is the enzyme responsible for converting methionine into SAM (Peleman et al., 1989). SAM can then be used by countless methyltransferases, which in turn add a methyl to their target molecule, releasing S-adenosylhomocysteine (SAH) as a by-product (Roje, 2006; dark blue). The SAH released from these reactions is recycled through the action of SAH HYDROLASE (SAHH; 52; Rocha et al., 2005; Pereira et al., 2007) which yields adenosine and homocysteine that is subsequently converted to methionine by METHIONINE SYNTHASE (MS; 49; red) using a folate cofactor (Ravanel et al., 2004; Loizeau et al., 2007). Due to the importance of SAM, homeostasis needs to be maintained, which occurs through the action of SAM:METHIONINE S-METHYLTRANSFERASE (53) which converts SAM and methionine into S-methylmethionine (SMM) and SAH (green). SMM produced in the leaves, can be moved into the phloem and transported to different tissues. The accumulation of SMM and methionine in the seeds is done in both a spatial and temporal manner (Frank et al., 2015), but whether the SMM cycle is used in other non-reproductive tissues such as xylem during SCW formation is still unclear. This pathway is the most common source of SAM in seeds. SMM produced in the leaves arrives at the seeds via the phloem, and HOMOCYSTEINE S-METHYLTRANSFERASE (54) subsequently uses the SMM and homocysteine to produce two methionine molecules (Cohen et al., 2017). The accumulation of SMM and methionine in the seeds is done in both a spatial and temporal manner (Frank et al., 2015), but whether the SMM cycle is used in other non-reproductive tissues such as xylem during SCW formation is still unclear. Unidirectional arrows indicate irreversible reactions, bidirectional arrows indicate reversible reactions, whereas lines with multiple numbers next to it indicates multiple enzymatic reactions. Key metabolites are highlighted in square boxes. The enzymes (26–54) represented by each number can be found in Supplementary Table S1, along with additional information such as gene ID and cellular localisation. Several enzymatic steps are repeated from Figure 4.
SAM is used as a cofactor for countless methylation reactions. S-adenosylhomocysteine (SAH; Rocha et al., 2005; Pereira et al., 2007) is produced as a by-product but is reconstituted to SAM through the action of SAH hydrolase (52), methionine synthase (Ravanel et al., 2004; Loizeau et al., 2007) and SAM synthetase (SAMS; 50; Peleman et al., 1989). Previous studies have noted that 80% of methionine produced by the plant is used for SAM biosynthesis (Giovanelli et al., 1985) and the role of SAM is especially highlighted during germination, where methionine was observed to accumulate before being converted to SAM the following day (Gallardo et al., 2002). A similar process may occur during xylogenesis (Ohtani et al., 2016). SAM homeostasis as well as transport to reproductive tissues is through the action of the s-methylmethionine (SMM) cycle. SMM produced in the leaves, can be moved into the phloem and transported to different tissues to act as a source of SAM (Cohen et al., 2017). The accumulation of SMM and methionine in the seeds occurs in both a spatial and temporal manner (Frank et al., 2015), but whether the SMM cycle is used in other non-reproductive tissues such as xylem during SCW formation is still unclear. Homeostasis has been noted between monolignol methylation and xylan GlcA methylation as gxm mutants exhibit an increase in monolignol methylation, whereas reductions in both forms of methylation were observed when genes of the folate biosynthesis pathway were mutated (Urbanowicz et al., 2012; Srivastava et al., 2015). Homeostasis of SAM may be determined by the action of a yet undiscovered Golgi localized SAM transporter, which may be a reason why overexpression of GXM2 and GXM3 did not display an altered lignin composition opposite to that of the gxm3 mutant (Urbanowicz et al., 2012; Yuan et al., 2014). Additionally, polysaccharide biosynthesis and methylation may influence SAM transporter activity. Golgi microsomes have been used to study SAM intake into the Golgi. Intake increased when a combination of either UDP-GlcA and acetyl-CoA or UDP-GalA and acetyl-CoA was present whereas intake of SAM decreased when SAH was present (Ibar and Orellana, 2007).
Acetyl-CoA
Acetyl-CoA is a vital metabolite which acts as a universal acetyl donor for multiple processes such as fatty acid biosynthesis and the acetylation of molecules such as polysaccharides (xylan, xyloglucan, pectins and glucomannan), metabolites (phenolics and isoprenoids) as well as proteins (e.g., histones); (Pauly and Scheller, 2000; Fatland et al., 2005; Oliver et al., 2009; Chen C. et al., 2017). Compartmentalisation of acetyl-CoA between the plastid (fatty acids) and cytosol (latter mentioned processes) is a mechanism that is used to ensure that different competing processes receive adequate amounts of cofactor (Schwender et al., 2006; Faria-Blanc et al., 2018). The cytosolic ATP-CITRATE LYASE (ACL) (67) enzyme that converts citrate into oxaloacetate and acetyl-CoA is vital for cellular function as acl mutants are embryo lethal, whereas downregulation yields plants with “bonsai” phenotypes (Fatland et al., 2002, 2005; Figure 6 and Supplementary Table S1). The citrate required for this reaction is derived primarily from the mitochondrial tricarboxylic acid (TCA) cycle that obtains carbon skeletons via pyruvate from cytosolic glycolysis (Picault et al., 2002; Giegé et al., 2003). Plants have two glycolytic pathways (cytosolic and plastidial), each with its own distinct enzymes, and the pyruvate produced from glycolysis is essential for acetyl-CoA generation (Plaxton, 1996; Givan, 1999; Giegé et al., 2003; Schwender et al., 2006). Cytosolic glycolysis derives pyruvate from sugars such as UDP-Glc, glucose-6-phosphate (G-6-P) or fructose-6-phosphate (F-6-P) from sucrose breakdown (Plaxton, 1996; Givan, 1999; Giegé et al., 2003; Stein et al., 2016). Plastidial glycolysis uses cytosolic G-6-P, triose phosphates and phosphoenolpyruvate which are transported into the plastid by various transporters (Fischer and Weber, 2002; Schwender et al., 2003, 2006; Flores-Tornero et al., 2017). Alternatively starch breakdown can supply glucose-1-phosphate (Plaxton, 1996; Givan, 1999; Ho et al., 2017; Pick and Avidan, 2017). Although mitochondrial and plastidial glycolysis are the predominant sources of acetyl-CoA, other pathways have been found to contribute to both acetyl-CoA pools.
FIGURE 6.
Multiple metabolic sources of plastidial and cytosolic acetyl-CoA. Plastidial glycolysis contributes pyruvate which is converted to acetyl-CoA by the plastidial Pyruvate Dehydrogenase (PDH) (58) complex for fatty acid biosynthesis (Johnston et al., 1997; Lin et al., 2003; green arrows) whereas cytosolic glycolysis is the main metabolic route which contributes pyruvate toward the TCA cycle for respiration in the mitochondrion (Luethy et al., 1995). In the mitochondrion, the pyruvate is broken down to acetyl-CoA which forms part of the TCA cycle through the action of CITRATE SYNTHASE (66) which produces mitochondrial citrate (red) from acetyl-CoA and oxaloacetate. Mitochondrial citrate (red) is transported to the cytosol (purple) and converted by heteromeric ATP-CITRATE LYASE (67) into acetyl-CoA and oxaloacetate. This is the main source of cytosolic acetyl-CoA pool whereas the resulting oxaloacetate is transported back into the mitochondrion in exchange for more citrate (orange). Acetaldehyde derived either from fermentation or from ethanol from other tissues can either enter the plastid where it contributes to fatty acid biosynthesis (green arrows), or be converted to acetate in the cytosol and enter the peroxisome. Acetyl-CoA can be generated in the peroxisome from acetate (79), β-oxidation of fatty acids (80–82), or breakdown of isoleucine (82, 86, 87, 88, 90, 92). The peroxisomal acetyl-CoA produced from the three aforementioned pathways can contribute to the cytosolic acetyl-CoA through the TCA either as citrate (blue) or succinate from the glyoxylate cyle (85). The breakdown of amino acids in the mitochondrion supplies various TCA cycle intermediates (gold) which can either contribute to respiration or the cytosolic acetyl-CoA pool. Unidirectional arrows indicate irreversible reactions, bidirectional arrows indicate reversible reactions whereas lines with multiple numbers next to it indicates multiple enzymatic reactions. Key metabolites are highlighted in square boxes, whereas acetyl-CoA is highlighted by ovals. The enzyme represented by each number can be found in Supplementary Table S1, along with additional information such as gene ID and cellular localization with steps 55–98 being displayed above. Several enzymatic steps are repeated from and Figure 4, 5.
Under hypoxic and/or energy deficient conditions, fermentation pathways become active in order to keep glycolysis functioning by maintaining NAD+ generation which is accomplished by converting pyruvate to ethanol (Tadege et al., 1999; Zabalza et al., 2009). Two enzymes are involved in this conversion namely: PYRUVATE DECARBOXYLASE (76) which converts pyruvate to a toxic intermediate, acetaldehyde, and subsequently ALCOHOL DEHYDROGENASE (77) converts the acetaldehyde to ethanol, effectively regenerating NAD+ from NADH (Bucher et al., 1995; Tadege and Kuhlemeier, 1997; Tadege et al., 1999; Figure 6 and Supplementary Table S1). The ethanol is hypothesized to move through the vasculature from roots (hypoxic conditions) to the upper organs to be converted back into acetaldehyde and then into acetate by ALDEHYDE DEHYDROGENASE (78); (Kreuzwieser et al., 1999; Oliver et al., 2009). This acetate can enter fatty acid biosynthesis in the plastid (Mellema et al., 2002), but has been shown to not greatly contribute to this process (Lin and Oliver, 2008), or can enter the peroxisome and potentially contribute to the cytosolic acetyl-CoA pool through TCA (Turner et al., 2005; Allen et al., 2011; Figure 6). Acetyl-CoA produced in the peroxisome can exit in two ways, (i) the citrate enters the glyoxylate pathway by which succinate is produced and is able to rejoin the TCA or (ii) the citrate enters the TCA directly (Mettler and Beevers, 1980; Salon et al., 1988; Eastmond et al., 2000). It is, however, unknown whether citrate exiting the peroxisome can contribute directly to the cytosolic acetyl-CoA pool as a substrate for ACL. Besides acetate, β-oxidation and the degradation of isoleucine use this pathway in order to provide carbon skeletons to the TCA cycle (Baker et al., 2006; Hildebrandt et al., 2015; Figure 6 and Supplementary Table S1).
The breakdown of amino acids (especially BCAA and lysine) is an understudied source of acetyl-CoA in plants. The enzymes responsible still need to be identified as several of the degradation steps are currently inferred by homology to yeast, bacteria and mammals (Anderson et al., 1998; Schuster and Binder, 2005; Araújo et al., 2010; Hildebrandt et al., 2015; Figure 6 and Supplementary Table S1). The acetyl-CoA produced by these pathways has been shown to be required by oligeneous diatoms for the biosynthesis of fatty acids and the related genes become upregulated during late SCW formation in inducible VND7 Arabidopsis (Ge et al., 2014; Li Z. et al., 2016). Due to the importance of acetyl-CoA, the existence of so many biosynthetic pathways may provide redundancy in order to maintain the cytosolic acetyl-CoA pool under various developmental stages and environmental conditions. Alternatively, these pathways may be required to maintain TCA cycle activity when the flux of carbon in glycolysis is redirected to other processes such as phenylalanine biosynthesis (Ohtani et al., 2016). Xylan acetylation and the production of SAM for monolignol and GlcA methylation both use acetyl-CoA, which suggests that competition for acetyl-CoA occurs and needs to be regulated in some manner. It is possible that cytosolic acetyl-CoA pools are maintained by multiple pathways in order to reduce competition for the cofactor among xylan acetylation and the methylation of GlcA and monolignols. If true, identifying underlying regulatory mechanisms responsible for reducing cofactor competition could provide novel avenues to alter SCW traits through metabolic engineering.
Conclusion
Xylan biosynthesis is a process that affects SCW formation, the composition and interaction of SCW components, as well as the efficiency of the deconstruction of lignocellulosic biomass for the production of various bioproducts. How perturbing xylan traits and modification patterns may affect SCW composition and processing still needs further investigation. Additional unknowns include the genes which code for the Golgi localized GlcA and SAM transporters, the exact functions of IRX15/IRX15L, the full complement of XSC and RES synthesizing proteins, and whether there are any more xylan associated proteins. Work in monocots suggest that several other proteins are involved in xylan biosynthesis and interact with the biosynthetic machinery (Zeng et al., 2010; Jiang et al., 2016). It still remains to be seen whether the interacting partners are conserved in other plant lineages. The presence of a UAM in the XSC may provide adequate UDP-Araf, which could explain why rice XAT fails to add sufficient amounts of arabinose modifications to xylan in Arabidopsis (Zeng et al., 2010; Xiong et al., 2015; Jiang et al., 2016). The fact that hormone binding proteins were found to interact with the XSC may indicate that hormones dictate SCW composition through xylan biosynthesis. Furthermore, the finding that xylan mutants are often more tolerant to stresses may be linked to altered interactions between the XSC and interacting proteins, which means that stress tolerant plants could be generated by manipulating xylan properties (Xin and Browse, 1998; Keppler and Showalter, 2010; Jiang et al., 2016; Escudero et al., 2017). In mutants such as esk1 that exhibits altered metabolism, carbon flux may be redirected to upregulated defense pathways as opposed to SCW, affecting not only xylan but also other polysaccharides. This scenario may be occurring in other xylan mutants as well, which explains the reduction in the content of polysaccharides such as glucomannan and pectin.
The multiple proteins involved in xylan biosynthesis and modification, the requirement of different xylan conformations needed to associate with cellulose and lignin, as well as spatial and temporal changes in xylan modification during SCW development suggest that different combinations of enzymes may be needed during different SCW formation stages. Since xylan shares metabolic precursors with cellulose and lignin, co-regulation of not only structural genes, but also primary metabolism needs to occur for SCW formation to take place correctly. Systems biology and systems genetics approaches could identify which parts of the biosynthetic machinery are responsible for xylan-cellulose and xylan-lignin interactions, as well as the ultimate composition and structure of the two biopolymers. Such approaches will also aid in identifying the pathways that are vital for supplying precursors and that, when perturbed, might lead to changes in xylan modification patterns. The mechanisms behind the formation of the two xylan domains still needs to be understood, as well as what effect supply of metabolic precursors has on establishing these domains. Such understanding would inform metabolic engineering efforts aimed at altering SCW properties. The only currently known effect of metabolism on xylan biosynthesis is that a stronger flux of sucrose increases total xylan content, and that xylan biosynthetic rates are temporally linked to the sucrose produced in the leaves and supplied to the xylem via the phloem (Ishihara et al., 2015).
Metabolic engineering approaches have the potential to target multiple processes at once, using genetic modifications that can be applied in a tissue-specific manner, to yield the trait of interest without causing undesirable effects on plant growth and defense. This approach has been applied successfully to increase SCW biomass, increase lignin content, alter lignin composition and reduce methyl content in the SCW, all of which displayed beneficial properties for the process of interest (Huntley et al., 2003; Srivastava et al., 2015; Eloy et al., 2017; Fan et al., 2017; Zhang et al., 2019). CRISPR gene editing could prove to be a useful tool for metabolic engineering, as it has been successfully applied to alter SCW traits (Fan et al., 2015; Zhou et al., 2015; Park et al., 2017) and provides the option to either knock-out or alternatively regulate genes involved in interdependent metabolic processes (Jinek et al., 2012; Qi et al., 2013; Vojta et al., 2016; Xiong et al., 2017). Additionally, knocked-out or downregulated pathway steps could be replaced with alternative enzymes to “rewire” metabolism in ways that would achieve desirable industrial processing traits without disrupting plant growth and defenses (Aznar et al., 2018). Finally, greater understanding of xylan biosynthesis in diverse plant lineages could drive successful introduction of novel modifications to fundamentally alter SCW composition in woody biomass. Producing a tree with an ideal biomass composition for all industrial applications might not be possible. However, xylan bioengineering may be an approach whereby woody biomass with customized composition can be created for different biorefinery application.
Author Contributions
MW conceived, wrote and made figures for the review. VM, EM, and AM contributed to the outline of the review, and provided extensive edits to both the text and figures.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded in part by the National Research Foundation (NRF) of South Africa – Bioinformatics and Functional Genomics Programme (BFG Grant UID 86936 and 97911), the Technology and Human Resources for Industry Programme (THRIP Grant UID 96413), the Department of Science and Technology (DST, Strategic Grant for the Eucalyptus Genomics Platform) and by Sappi Forest Research through the Forest Molecular Genetics (FMG) Programme at the University of Pretoria (UP). MW acknowledges postgraduate scholarship support from the NRF. VM acknowledges a postdoctoral fellowship support from UP.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00176/full#supplementary-material
List of enzymes which catalyze the biosynthesis of sugar nucleotides, s-adenosyl methionine and acetyl-CoA. (a) The numbering of the enzymes corresponds to Figure 4–6. (b) Name of the enzyme. (c) Not all accessions were provided for each reaction, just one representative from each cellular compartment. Accessions and cellular localisation were obtained from TAIR and SUBA3, respectively.
References pertaining to numbered items in Table 1.
Biotechnology approaches that have scaled from Arabidopsis to Populus.
References
- Allen E., Moing A., Wattis J. A. D., Larson T., Maucourt M., Graham I. A., et al. (2011). Evidence that ACN1 (ACETATE NON-UTILIZING 1) prevents carbon leakage from peroxisomes during lipid mobilization in Arabidopsis seedlings. Biochem. J. 437 505–513. 10.1042/BJ20101764 [DOI] [PubMed] [Google Scholar]
- Allerdings E., Ralph J., Steinhart H., Bunzel M. (2006). Isolation and structural identification of complex feruloylated heteroxylan side-chains from maize bran. Phytochemistry 67 1276–1286. 10.1016/j.phytochem.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Amor Y., Haigler C. H., Johnson S., Wainscott M., Delmer D. P. (1995). A membrane-associated form of SUCROSE SYNTHASE and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. U.S.A. 92 9353–9357. 10.1073/pnas.92.20.9353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N., Wilkinson M. D., Lovegrove A., Freeman J., Tryfona T., Pellny T. K., et al. (2012). Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc. Natl. Acad. Sci. U.S.A. 109 989–993. 10.1073/pnas.1115858109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. D., Che P., Song J., Nikolau B. J., Wurtele E. S. (1998). 3-METHYLCROTONYL-COENZYME A CARBOXYLASE is a component of the mitochondrial leucine catabolic pathway in plants. Plant Physiol. 118 1127–1138. 10.1104/pp.118.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anoman A. D., Muñoz-Bertomeu J., Rosa-Téllez S., Flores-Tornero M., Serrano R., Bueso E., et al. (2015). Plastidial glycolytic GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE is an important determinant in the carbon and nitrogen metabolism of heterotrophic cells in Arabidopsis. Plant Physiol. 169 1619–1637. 10.1104/pp.15.00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W. L., Ishizaki K., Nunes-Nesi A., Larson T. R., Tohge T., Krahnert I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22 1549–1563. 10.1105/tpc.110.075630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arola S., Malho J.-M., Laaksonen P., Lille M., Linder M. B. (2013). The role of hemicellulose in nanofibrillated cellulose networks. Soft Matter 9 1319–1326. 10.1039/C2SM26932E [DOI] [Google Scholar]
- Atmodjo M. A., Hao Z., Mohnen D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64 747–779. 10.1146/annurev-arplant-042811-105534 [DOI] [PubMed] [Google Scholar]
- Audu I., Brosse N., Winter H., Hoffmann A., Bremer M., Fischer S., et al. (2018). Acetyl groups in Typha capensis: fate of acetates during organosolv and ionosolv pulping. Polymers 10:619 10.3390/polym10060619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar A., Chalvin C., Shih P. M., Maimann M., Ebert B., Birdseye D. S., et al. (2018). Gene stacking of multiple traits for high yield of fermentable sugars in plant biomass. Biotechnol. Biofuels 11:2. 10.1186/s13068-017-1007-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A., Graham I. A., Holdsworth M., Smith S. M., Theodoulou F. L. (2006). Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 11 124–132. 10.1016/j.tplants.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Barah P., Winge P., Kusnierczyk A., Tran D. H., Bones A. M. (2013). Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PLoS One 8:e58987. 10.1371/journal.pone.0058987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M., O’Neill M. A. (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 62 127–155. 10.1146/annurev-arplant-042110-103918 [DOI] [PubMed] [Google Scholar]
- Bensussan M., Lefebvre V., Ducamp A., Trouverie J., Emilie G., Fortabat M.-N., et al. (2015). Suppression of dwarf and irregular xylem phenotype generates low-acetylated biomass lines in Arabidopsis thaliana. Plant Physiol. 168 452–463. 10.1104/pp.15.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibi Z., Ansari A., Zohra R. R., Aman A., Ul Qader S. A. (2014). Production of xylan degrading endo-1, 4-β-xylanase from thermophilic Geobacillus stearothermophilus KIBGE-IB29. J. Radiat. Res. Appl. Sci. 7 478–485. 10.1016/j.jrras.2014.08.001 [DOI] [Google Scholar]
- Binder J. B., Blank J. J., Cefali A. V., Raines R. T. (2010). Synthesis of furfural from xylose and xylan. Chemsuschem 3 1268–1272. 10.1002/cssc.201000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V., Nita S., Neumetzler L., Schindelasch D., Urbain A., Eshed R., et al. (2010). TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 153 590–602. 10.1104/pp.110.153320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal A. K., Atmodjo M. A., Pattathil S., Amos R. A., Yang X., Winkeler K., et al. (2018). Working towards recalcitrance mechanisms: increased xylan and homogalacturonan production by overexpression of GAlactUronosylTransferase12 (GAUT12) causes increased recalcitrance and decreased growth in Populus. Biotechnol. Biofuels 11:9. 10.1186/s13068-017-1002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal A. K., Hao Z., Pattathil S., Yang X., Winkeler K., Collins C., et al. (2015). Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol. Biofuels 8:41. 10.1186/s13068-015-0218-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley J. R., Busse-Wicher M., Tryfona T., Mortimer J. C., Zhang Z., Brown D. M., et al. (2013). GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J. 74 423–434. 10.1111/tpj.12135 [DOI] [PubMed] [Google Scholar]
- Brown D., Wightman R., Zhang Z., Gomez L. D., Atanassov I., Bukowski J.-P., et al. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66 401–413. 10.1111/j.1365-313X.2011.04501.x [DOI] [PubMed] [Google Scholar]
- Brown D. M., Goubet F., Wong V. W., Goodacre R., Stephens E., Dupree P., et al. (2007). Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. Plant J. 52 1154–1168. 10.1111/j.1365-313X.2007.03307.x [DOI] [PubMed] [Google Scholar]
- Brown D. M., Zeef L. A. H., Ellis J., Goodacre R., Turner S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17 2281–2295. 10.1105/tpc.105.031542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher M., Brander K. A., Sbicego S., Mandel T., Kuhlemeier C. (1995). Aerobic fermentation in tobacco pollen. Plant Mol. Biol. 28 739–750. 10.1007/BF00021197 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M., Gomes T. C. F., Tryfona T., Nikolovski N., Stott K., Grantham N. J., et al. (2014). The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 79 492–506. 10.1111/tpj.12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse-Wicher M., Grantham N. J., Lyczakowski J. J., Nikolovski N., Dupree P. (2016a). Xylan decoration patterns and the plant secondary cell wall molecular architecture. Biochem. Soc. Trans. 44 74–78. 10.1042/BST20150183 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M., Li A., Silveira R. L., Pereira C. S., Tryfona T., Gomes T. C. F., et al. (2016b). Evolution of xylan substitution patterns in gymnosperms and angiosperms: implications for xylan interaction with cellulose. Plant Physiol. 171 2418–2431. 10.1104/pp.16.00539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caddick M. X., Greenland A. J., Jepson L., Krause K.-P., Qu N., Riddell K. V., et al. (1998). An ethanol inducible gene switch for plants used to manipulate carbon metabolism. Nat. Biotechnol. 16 177–180. 10.1038/nbt0298-177 [DOI] [PubMed] [Google Scholar]
- Cascales-Miñana B., Muñoz-Bertomeu J., Flores-Tornero M., Anoman A. D., Pertusa J., Alaiz M., et al. (2013). The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25 2084–2101. 10.1105/tpc.113.112359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Li C., Wang Y., Renaud J., Tian G., Kambhampati S., et al. (2017). Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants 3 814–824. 10.1038/s41477-017-0023-7 [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Song J., Wang J. P., Lin Y.-C., Ducoste J., Shuford C. M., et al. (2014). Systems biology of lignin biosynthesis in Populus trichocarpa: heteromeric 4-COUMARIC ACID:COENZYME A LIGASE protein complex formation, regulation, and numerical modeling. Plant Cell 26 876–893. 10.1105/tpc.113.119685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Jiang Q., Yang G., Wang Q., Fatehi P. (2017). Ultrasonic-assisted ionic liquid treatment of chemithermomechanical pulp fibers. Cellulose 24 1483–1491. 10.1007/s10570-016-1180-y [DOI] [Google Scholar]
- Chen X., Lawoko M., Heiningen A. (2010). Kinetics and mechanism of autohydrolysis of hardwoods. Bioresour. Technol. 101 7812–7819. 10.1016/j.biortech.2010.05.006 [DOI] [PubMed] [Google Scholar]
- Cheng N., Koda K., Tamai Y., Yamamoto Y., Takasuka T. E., Uraki Y. (2017). Optimization of simultaneous saccharification and fermentation conditions with amphipathic lignin derivatives for concentrated bioethanol production. Bioresour. Technol. 232 126–132. 10.1016/j.biortech.2017.02.018 [DOI] [PubMed] [Google Scholar]
- Chiniquy D., Varanasi P., Oh T., Harholt J., Katnelson J., Singh S., et al. (2013). Three novel rice genes closely related to the Arabidopsis IRX9, IRX9L, and IRX14 genes and their roles in xylan biosynthesis. Front. Plant Sci. 4:83. 10.3389/fpls.2013.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.-L., Derba-Maceluch M., Koutaniemi S., Gómez L. D., McQueen-Mason S. J., Tenkanen M., et al. (2015). Active fungal GH115 α-glucuronidase produced in Arabidopsis thaliana affects only the UX1-reactive glucuronate decorations on native glucuronoxylans. BMC Biotechnol. 15:56. 10.1186/s12896-015-0154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.-H., Pogorelko G., Zabotina O. A. (2012). Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. 159 1355–1366. 10.1104/pp.112.199356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie N., Myburg A. A., Joubert F., Murray S. L., Carstens M., Lin Y.-C., et al. (2017). Systems genetics reveals a transcriptional network associated with susceptibility in the maize-grey leaf spot pathosystem. Plant J. 89 746–763. 10.1111/tpj.13419 [DOI] [PubMed] [Google Scholar]
- Civelek M., Lusis A. J. (2014). Systems genetics approaches to understand complex traits. Nat. Rev. Genet. 15 34–48. 10.1038/nrg3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen H., Salmon A., Tietel Z., Hacham Y., Amir R. (2017). The relative contribution of genes operating in the S-methylmethionine cycle to methionine metabolism in Arabidopsis seeds. Plant Cell Rep. 36 731–743. 10.1007/s00299-017-2124-1 [DOI] [PubMed] [Google Scholar]
- Coleman H. D., Yan J., Mansfield S. D. (2009). SUCROSE SYNTHASE affects carbon partitioning to increase cellulose production and altered cell wall ultrastructure. Proc. Natl. Acad. Sci. U.S.A. 106 13118–13123. 10.1073/pnas.0900188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson A. T., Ehrlich J. J., Choe J-Y., Honzatko R. B., Zabotina O. A. (2018). Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers. Proc. Natl. Acad. Sci. U.S.A. 115 6064–6069. 10.1073/pnas.1801105115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J., Brandizzi F., Liepman A. H., Keegstra K. (2010). Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. Plant J. 64 1028–1037. 10.1111/j.1365-313X.2010.04392.x [DOI] [PubMed] [Google Scholar]
- de Souza W. R., Martins P. K., Freeman J., Pellny T. K., Michaelson L. V., Sampaio B. L., et al. (2018). Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytol. 218 81–93. 10.1111/nph.14970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartini J. D., Pattathil S., Miller J. S., Li H., Hahn M. G., Wyman C. E. (2013). Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energy Environ. Sci. 6 898–909. 10.1039/c3ee23801f [DOI] [Google Scholar]
- Derba-Maceluch M., Awano T., Takahashi J., Lucenius J., Ratke C., Kontro I., et al. (2015). Suppression of xylan endotransglycosylase PtxtXyn10A affects cellulose microfibril angle in secondary wall in aspen wood. New Phytol. 205 666–681. 10.1111/nph.13099 [DOI] [PubMed] [Google Scholar]
- Deutschmann R., Dekker R. F. H. (2012). From plant biomass to bio-based chemicals: latest developments in xylan research. Biotechnol. Adv. 30 1627–1640. 10.1016/j.biotechadv.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Diaz A. B., Moretti M. M. S., Bezerra-Bussoli C., Carreira Nunes C. C., Blandino A., da Silva R., et al. (2015). Evaluation of microwave-assisted pretreatment of lignocellulosic biomass immersed in alkaline glycerol for fermentable sugars production. Bioresour. Technol. 185 316–323. 10.1016/j.biortech.2015.02.112 [DOI] [PubMed] [Google Scholar]
- Do C.-T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., et al. (2007). Both CAFFEOYL COENZYME A 3-O-METHYLTRANSFERASE 1 and CAFFEIC ACID O-METHYLTRANSFERASE 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226 1117–1129. 10.1007/s00425-007-0558-3 [DOI] [PubMed] [Google Scholar]
- Dong S., Bortner M. J., Roman M. (2016). Analysis of the sulfuric acid hydrolysis of wood pulp for cellulose nanocrystal production: a central composite design study. Ind. Crops Prod. 93 76–87. 10.1016/j.indcrop.2016.01.048 [DOI] [Google Scholar]
- Doran-Peterson J., Jangid A., Brandon S. K., DeCrescenzo-Henriksen E., Dien B., Ingram L. O. (2009). Simultaneous saccharification and fermentation and partial saccharification and co-fermentation of lignocellulosic biomass for ethanol production. Methods Mol. Biol. 581 263–280. 10.1007/978-1-60761-214-8_17 [DOI] [PubMed] [Google Scholar]
- Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001). The GLYCINE DECARBOXYLASE system: a fascinating complex. Trends Plant Sci. 6 167–176. 10.1016/S1360-1385(01)01892-1 [DOI] [PubMed] [Google Scholar]
- Drost D. R., Benedict C. I., Berg A., Novaes E., Novaes C. R. D. B., Yu Q., et al. (2010). Diversification in the genetic architecture of gene expression and transcriptional networks in organ differentiation of Populus. Proc. Natl. Acad. Sci. U.S.A. 107 8492–8497. 10.1073/pnas.0914709107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P. J., Germain V., Lange P. R., Bryce J. H., Smith S. M., Graham I. A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. U.S.A. 97 5669–5674. 10.1073/pnas.97.10.5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B., Rautengarten C., Guo X., Xiong G., Stonebloom S., Smith-Moritz A. M., et al. (2015). Identification and characterization of a Golgi-localized UDP-XYLOSE TRANSPORTER family from Arabidopsis. Plant Cell 27 1218–1227. 10.1105/tpc.114.133827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebringerová A., Heinze T. (2000). Xylan and xylan derivatives – biopolymers with valuable properties. Naturally occurring xylans structures, isolation procedures and properties. Macromol. Rapid Commun. 21 542–556. [DOI] [Google Scholar]
- Eloy N. B., Voorend W., Lan W., Saleme M. L., Cesarino I., Vanholme R., et al. (2017). Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiol. 173 998–1016. 10.1104/pp.16.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero V., Jordá L., Sopeña-Torres S., Mélida H., Miedes E., Muñoz-Barrios A., et al. (2017). Alteration of cell wall xylan acetylation triggers defense responses that counterbalance the immune deficiencies of plants impaired in the β-subunit of the heterotrimeric G-protein. Plant J. 92 386–399. 10.1111/tpj.13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Pereira J. H., Yogiswara S., Wang G., Teixeira Benites V., Baidoo E. E. K., et al. (2016). Exploiting the substrate promiscuity of hydroxycinnamoyl-CoA:shikimate hydroxycinnamoyl transferase to reduce lignin. Plant Cell Physiol. 57 568–579. 10.1093/pcp/pcw016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Sathitsuksanoh N., Baidoo E. E. K., George A., Liang Y., Yang F., et al. (2015). Expression of a bacterial 3-dehydroshikimate dehydratase reduces lignin content and improves biomass saccharification efficiency. Plant Biotechnol. J. 13 1241–1250. 10.1111/pbi.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evtuguin D. V., Tomás J. L., Silva A. M. S., Neto C. P. (2003). Characterization of an acetylated heteroxylan from Eucalyptus globulus Labill. Carbohydr. Res. 338 597–604. 10.1016/S0008-6215(02)00529-3 [DOI] [PubMed] [Google Scholar]
- Fan C., Feng S., Huang J., Wang Y., Wu L., Li X., et al. (2017). AtCesA8-driven OsSUS3 expression leads to largely enhanced biomass saccharification and lodging resistance by distinctively altering lignocellulose features in rice. Biotechnol. Biofuels 10:221. 10.1186/s13068-017-0911-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Liu T., Li C., Jiao B., Li S., Hou Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5:12217. 10.1038/srep12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria-Blanc N., Mortimer J., Dupree P. (2018). A transcriptomic analysis of xylan mutants does not support the existence of a secondary cell wall integrity system in Arabidopsis. Front. Plant Sci. 9:384. 10.3389/fpls.2018.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland B. L., Ke J., Anderson M. D., Mentzen W. I., Cui L. W., Allred C. C., et al. (2002). Molecular characterization of a heteromeric ATP-CITRATE LYASE that generates cytosolic acetyl-Coenzyme A in Arabidopsis. Plant Physiol. 130 740–756. 10.1104/pp.008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland B. L., Nikolau B. J., Wurtele E. S. (2005). Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-CITRATE LYASE in Arabidopsis. Plant Cell 17 182–203. 10.1105/tpc.104.026211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus F. A. (2014). Systems genetics: a paradigm to improve discovery of candidate genes and mechanisms underlying complex traits. Plant Sci. 223 45–48. 10.1016/j.plantsci.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Fischer K., Weber A. (2002). Transport of carbon in non-green plastids. Trends Plant Sci. 7 345–351. 10.1016/S1360-1385(02)02291-4 [DOI] [PubMed] [Google Scholar]
- Flores-Tornero M., Anoman A. D., Rosa-Téllez S., Toujani W., Weber A. P. M., Eisenhut M., et al. (2017). Overexpression of the TRIOSE PHOSPHATE TRANSLOCATOR (TPT) complements the abnormal metabolism and development of plastidial glycolytic GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE mutants. Plant J. 89 1146–1158. 10.1111/tpj.13452 [DOI] [PubMed] [Google Scholar]
- Frank A., Cohen H., Hoffman D., Amir R. (2015). Methionine and S-methylmethionine exhibit temporal and spatial accumulation patterns during the Arabidopsis life cycle. Amino Acids 47 497–510. 10.1007/s00726-014-1881-1 [DOI] [PubMed] [Google Scholar]
- Gallardo K., Job C., Groot S. P. C., Puype M., Demol H., Vandekerckhove J., et al. (2002). Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol. Plant. 116 238–247. 10.1034/j.1399-3054.2002.1160214.x [DOI] [PubMed] [Google Scholar]
- Gandla M. L., Derba-Maceluch M., Liu X., Gerber L., Master E. R., Mellerowicz E. J., et al. (2014). Expression of a fungal glucuronoyl esterase in Populus: effects on wood properties and saccharification efficiency. Phytochemistry 112 210–220. 10.1016/j.phytochem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Gatz C. (1996). Chemically inducible promoters in transgenic plants. Curr. Opin. Biotechnol. 7 168–172. 10.1016/S0958-1669(96)80008-5 [DOI] [Google Scholar]
- Ge F., Huang W., Chen Z., Zhang C., Xiong Q., Bowler C., et al. (2014). METHYLCROTONYL-CoA CARBOXYLASE regulates triacylglycerol accumulation in the model diatom Phaeodactylum tricornutum. Plant Cell 26 1681–1697. 10.1105/tpc.114.124982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gericke M., Gabriel L., Geitel K., Benndorf S., Trivedi P., Fardim P., et al. (2018). Synthesis of xylan carbonates – An approach towards reactive polysaccharide derivatives showing self-assembling into nanoparticles. Carbohydr. Polym. 193 45–53. 10.1016/j.carbpol.2018.03.083 [DOI] [PubMed] [Google Scholar]
- Giegé P., Heazlewood J. L., Roessner-Tunali U., Millar A. H., Fernie A. R., Leaver C. J., et al. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15 2140–2151. 10.1105/tpc.012500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer J., Wännström S. (1986). Formation of ether bonds between lignins and carbohydrates during alkaline pulping processes. Holzforschung 40 347–352. 10.1515/hfsg.1986.40.6.347 [DOI] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Cheng K., Skinner M. E., Liepman A. H., Wilkerson C. G., Pauly M. (2011a). Deep sequencing of voodoo lily (Amorphophallus konjac): an approach to identify relevant genes involved in the synthesis of the hemicellulose glucomannan. Planta 234 515–526. 10.1007/s00425-011-1422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., de Souza A., Xiong G., Benz M., Cheng K., Schultink A., et al. (2011b). O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, Proteins with a TBL and DUF231 domain. Plant Cell 23 4041–4053. 10.1105/tpc.111.091728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S., Pauly M. (2012). O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3:12. 10.3389/fpls.2012.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanelli J., Mudd S. H., Datko A. H. (1985). Quantitative analysis of pathways of methionine metabolism and their regulation in Lemna. Plant Physiol. 78 555–560. 10.1104/pp.78.3.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giummarella N., Lawoko M. (2016). Structural basis for the formation and regulation of lignin-xylan bonds in birch. ACS Sustain. Chem. Eng. 4 5319–5326. 10.1021/acssuschemeng.6b00911 [DOI] [Google Scholar]
- Givan C. V. (1999). Evolving concepts in plant glycolysis: two centuries of progress. Biol. Rev. 74 277–309. 10.1017/S0006323199005344 [DOI] [Google Scholar]
- Gondolf V. M., Stoppel R., Ebert B., Rautengarten C., Liwanag A. J. M., Loqué D., et al. (2014). A gene stacking approach leads to engineered plants with highly increased galactan levels in Arabidopsis. BMC Plant Biol. 14:344. 10.1186/s12870-014-0344-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami T., Saikia C. N., Baruah R. K., Sarma C. M. (1996). Characterization of pulp obtained from Populus deltoides plants of different ages using, I. R., XRD and SEM. Bioresour. Technol. 57 209–214. 10.1016/0960-8524(96)00069-7 [DOI] [Google Scholar]
- Grantham N. J., Wurman-Rodrich J., Terrett O. M., Lyczakowski J. J., Stott K., Iuga D., et al. (2017). An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 3 859–865. 10.1038/s41477-017-0030-8 [DOI] [PubMed] [Google Scholar]
- Greenler J. M., Sloan J. S., Schwartz B. W., Becker W. M. (1989). Isolation, characterization and sequence analysis of a full-length cDNA clone encoding NADH-dependent HYDROXYPYRUVATE REDUCTASE from cucumber. Plant Mol. Biol. 13 139–150. 10.1007/BF00016133 [DOI] [PubMed] [Google Scholar]
- Gübitz G. M., Stebbing D. W., Johansson C. I., Saddler J. N. (1998). Lignin-hemicellulose complexes restrict enzymatic solubilization of mannan and xylan from dissolving pulp. Appl. Microbiol. Biotechnol. 50 390–395. 10.1007/s002530051310 [DOI] [Google Scholar]
- Guerriero G., Sergeant K., Hausman J.-F. (2013). Integrated -omics: a powerful approach to understanding the heterogeneous lignification of fibre crops. Int. J. Mol. Sci. 14 10958–10978. 10.3390/ijms140610958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürbüz E. I., Wettstein S. G., Dumesic J. A. (2012). Conversion of hemicellulose to furfural and levulinic acid using biphasic reactors with alkylphenol solvents. ChemSusChem 5 383–387. 10.1002/cssc.201100608 [DOI] [PubMed] [Google Scholar]
- Hao Z., Avci U., Tan L., Zhu X., Glushka J., Pattathil S., et al. (2014). Loss of Arabidopsis GAUT12/IRX8 causes anther indehiscence and leads to reduced G lignin associated with altered matrix polysaccharide deposition. Front. Plant Sci. 5:357. 10.3389/fpls.2014.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle S., Cameron D., Lam J., White B., Duff S. (2003). Effect of inhibitory compounds found in biomass hydrolysates on growth and xylose fermentation by a genetically engineered strain of S-cerevisiae. Enzyme Microb. Technol. 33 786–792. 10.1016/S0141-0229(03)00214-X [DOI] [Google Scholar]
- Hertzberg M., Aspeborg H., Schrader J., Andersson A., Erlandsson R., Blomqvist K., et al. (2001). A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. U.S.A. 98 14732–14737. 10.1073/pnas.261293398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt T. M., Nunes Nesi A., Araújo W. L., Braun H.-P. (2015). Amino acid catabolism in plants. Mol. Plant 8 1563–1579. 10.1016/j.molp.2015.09.005 [DOI] [PubMed] [Google Scholar]
- Hillmer R. A. (2015). Systems biology for biologists. PLoS Pathog. 11:e1004786. 10.1371/journal.ppat.1004786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel M. E., Ding S.-Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., et al. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315 804–807. 10.1126/science.1137016 [DOI] [PubMed] [Google Scholar]
- Hinman M. B., Villemez C. L. (1975). Glucomannan biosynthesis catalyzed by Pisum sativum enzymes. Plant Physiol. 56 608–612. 10.1104/pp.56.5.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.-H., Nakanishi A., Kato Y., Yamasaki H., Chang J.-S., Misawa N., et al. (2017). Dynamic metabolic profiling together with transcription analysis reveals salinity-induced starch-to-lipid biosynthesis in alga Chlamydomonas sp. JSC4. Sci. Rep. 7:45471. 10.1038/srep45471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking M. B. (1997). Vanillin: synthetic flavoring from spent sulfite liquor. J. Chem. Educ. 74 1055–1059. 10.1021/ed074p1055 [DOI] [Google Scholar]
- Hu H., Zhang R., Feng S., Wang Y., Wang Y., Fan C., et al. (2017). Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol. J. 16 976–988. 10.1111/pbi.12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Li J., Yang X., Zhao X., Wang X., Tang Q., et al. (2016). Irregular xylem 7 (IRX7) is required for anchoring seed coat mucilage in Arabidopsis. Plant Mol. Biol. 92 25–38. 10.1007/s11103-016-0493-4 [DOI] [PubMed] [Google Scholar]
- Huang J., Xu Q., Zheng Y., Qian L., Guan X., Lin B. (2018). In vitro digestibility and fermentability of naturally acetylated xylans from bamboo shavings. Cogent Biol. 4 1–8. 10.1080/23312025.2018.1491087 [DOI] [Google Scholar]
- Hunter D., De la Mare A. C. (1978). Papermaking : the History and Technique of an Ancient Craft. New York, NY: Dover Publications. [Google Scholar]
- Huntley S. K., Ellis D., Gilbert M., Chapple C., Mansfield S. D. (2003). Significant increases in pulping efficiency in C4H-F5H-transformed poplars: improved chemical savings and reduced environmental toxins. J. Agric. Food Chem. 51 6178–6183. 10.1021/jf034320o [DOI] [PubMed] [Google Scholar]
- Hussey S. G., Loots M. T., van der Merwe K., Mizrachi E., Myburg A. A. (2017). Integrated analysis and transcript abundance modelling of H3K4me3 and H3K27me3 in developing secondary xylem. Sci. Rep. 7:3370. 10.1038/s41598-017-03665-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibar C., Orellana A. (2007). The import of S-adenosylmethionine into the Golgi apparatus is required for the methylation of homogalacturonan. Plant Physiol. 145 504–512. 10.1104/pp.107.104679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Sewalt V. J., Murray G. B., Ni W., Stürzer C., Dixon R. A. (1998). Developmental expression and substrate specificities of alfalfa CAFFEIC ACID 3-O-METHYLTRANSFERASE and CAFFEOYL COENZYME A 3-O- METHYLTRANSFERASE in relation to lignification. Plant Physiol. 117 761–770. 10.1104/pp.117.3.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Obata T., Sulpice R., Fernie A. R., Stitt M. (2015). Quantifying protein synthesis and degradation in Arabidopsis by dynamic 13CO2 labeling and analysis of enrichment in individual amino acids in their free pools and in protein. Plant Physiol. 168 74–93. 10.1104/pp.15.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T., Matsuoka K., Ono H., Ohnishi-Kameyama M., Yaoi K., Nakano Y., et al. (2017). Characterization of xylan in the early stages of secondary cell wall formation in tobacco bright yellow-2 cells. Carbohydr. Polym. 176 381–391. 10.1016/j.carbpol.2017.08.108 [DOI] [PubMed] [Google Scholar]
- Isogai A., Saito T., Fukuzumi H. (2011). TEMPO-oxidized cellulose nanofibers. Nanoscale 3 71–85. 10.1039/c0nr00583e [DOI] [PubMed] [Google Scholar]
- Iversen T., Wännström S. (1986). Lignin-carbohydrate bonds in a residual lignin isolated from pine kraft pulp. Holzforschung 40 19–22. 10.1515/hfsg.1986.40.1.19 [DOI] [Google Scholar]
- Iwamoto S., Isogai A., Iwata T. (2011). Structure and mechanical properties of wet-spun fibers made from natural cellulose nanofibers. Biomacromolecules 12 831–836. 10.1021/bm101510r [DOI] [PubMed] [Google Scholar]
- Jander G., Joshi V. (2009). Aspartate-derived amino acid biosynthesis in Arabidopsis thaliana. Arabidopsis Book 7:e0121. 10.1199/tab.0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. C. (2003). Studying complex biological systems using multifactorial perturbation. Nat. Rev. Genet. 4 145–151. 10.1038/nrg996 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Johnson N. R., Wilkerson C. G. (2014). Arabidopsis thaliana IRX10 and two related proteins from psyllium and Physcomitrella patens are xylan xylosyltransferases. Plant J. 80 207–215. 10.1111/tpj.12641 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Kim H., Cocuron J.-C., Orler R., Ralph J., Wilkerson C. G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66 387–400. 10.1111/j.1365-313X.2010.04475.x [DOI] [PubMed] [Google Scholar]
- Jiang N., Wiemels R. E., Soya A., Whitley R., Held M., Faik A. (2016). Composition, assembly, and trafficking of a wheat xylan synthase complex. Plant Physiol. 170 1999–2023. 10.1104/pp.15.01777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S., Zhang G., Zhang P., Li F., Wang S., Fan S., et al. (2016). Microwave assisted alkaline pretreatment to enhance enzymatic saccharification of catalpa sawdust. Bioresour. Technol. 221 26–30. 10.1016/j.biortech.2016.09.033 [DOI] [PubMed] [Google Scholar]
- Jin Y.-S., Jeffries T. W. (2003). Changing flux of xylose metabolites by altering expression of XYLOSE REDUCTASE and XYLITOL DEHYDROGENASE in recombinant Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 105–108, 277–286. 10.1385/ABAB:106:1-3:277 [DOI] [PubMed] [Google Scholar]
- Jin Y.-S., Ni H., Laplaza J. M., Jeffries T. W. (2003). Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-XYLULOKINASE activity. Appl. Environ. Microbiol. 69 495–503. 10.1128/AEM.69.1.495-503.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. M., Kim H., Ralph J., Mansfield S. D. (2017). Natural acetylation impacts carbohydrate recovery during deconstruction of Populus trichocarpa wood. Biotechnol. Biofuels 10:48. 10.1186/s13068-017-0734-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. L., Luethy M. H., Miernyk J. A., Randall D. D. (1997). Cloning and molecular analyses of the Arabidopsis thaliana plastid PYRUVATE DEHYDROGENASE subunits. Biochim. Biophys. Acta 1321 200–206. 10.1016/S0005-2728(97)00059-5 [DOI] [PubMed] [Google Scholar]
- Jönsson L. J., Alriksson B., Nilvebrant N.-O. (2013). Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol. Biofuels 6:16. 10.1186/1754-6834-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabel M. A., van den Borne H., Vincken J.-P., Voragen A. G. J., Schols H. A. (2007). Structural differences of xylans affect their interaction with cellulose. Carbohydr. Polym. 69 94–105. 10.1016/j.carbpol.2006.09.006 [DOI] [Google Scholar]
- Keppler B. D., Showalter A. M. (2010). IRX14 and IRX14-LIKE, two glycosyl transferases involved in glucuronoxylan biosynthesis and drought tolerance in Arabidopsis. Mol. Plant 3 834–841. 10.1093/mp/ssq028 [DOI] [PubMed] [Google Scholar]
- Kim S.-J., Brandizzi F. (2016). The plant secretory pathway for the trafficking of cell wall polysaccharides and glycoproteins. Glycobiology 26 940–949. 10.1093/glycob/cww044 [DOI] [PubMed] [Google Scholar]
- Kitano H. (2002). Computational systems biology. Nature 420 206–210. 10.1038/nature01254 [DOI] [PubMed] [Google Scholar]
- Kleczkowski L. A., Givan C. V. (1988). Serine formation in leaves by mechanisms other than the glycolate pathway. J. Plant Physiol. 132 641–652. 10.1016/S0176-1617(88)80223-2 [DOI] [Google Scholar]
- Klein A., Grabner M. (2015). Analysis of construction timber in rural Austria: wooden log walls. Int. J. Archit. Herit. 9 553–563. 10.1080/15583058.2013.804608 [DOI] [Google Scholar]
- Klemm D., Heublein B., Fink H.-P., Bohn A. (2005). Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. 44 3358–3393. 10.1002/anie.200460587 [DOI] [PubMed] [Google Scholar]
- Klocko A. L., Ma C., Robertson S., Esfandiari E., Nilsson O., Strauss S. H. (2016). FT overexpression induces precocious flowering and normal reproductive development in Eucalyptus. Plant Biotechnol. J. 14 808–819. 10.1111/pbi.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.-H., Beers E. P., Han K.-H. (2006). Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol. Genet. Genomics 276 517–531. 10.1007/s00438-006-0157-1 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Sakai Y., Iizuka K. (1960). Hydrolysis of cellulose in a small amount of concentrated sulfuric acid. Bull. Agric. Chem. Soc. Japan 24 443–449. 10.1080/03758397.1960.10857689 [DOI] [Google Scholar]
- Kohl P., Noble D. (2009). Systems biology and the virtual physiological human. Mol. Syst. Biol. 5:292. 10.1038/msb.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Takeda T., Miyazaki Y., Ohnishi-Kameyama M., Hayashi T., O’Neill M. A., et al. (2007). A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17 345–354. 10.1093/glycob/cwl081 [DOI] [PubMed] [Google Scholar]
- Kreuger J., Kjellén L. (2012). Heparan sulfate biosynthesis: regulation and variability. J. Histochem. Cytochem. 60 898–907. 10.1369/0022155412464972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzwieser J., Scheerer U., Rennenberg H. (1999). Metabolic origin of acetaldehyde emitted by poplar (Populus tremula ×P. alba) trees. J. Exp. Bot. 50 757–765. [Google Scholar]
- Lacerda T. M., Zambon M. D., Frollini E. (2013). Effect of acid concentration and pulp properties on hydrolysis reactions of mercerized sisal. Carbohydr. Polym. 93 347–356. 10.1016/j.carbpol.2012.10.039 [DOI] [PubMed] [Google Scholar]
- Lairez D., Cathala B., Monties B., Bedos-Belval F., Duran H., Gorrichon L. (2005). Aggregation during coniferyl alcohol polymerization in pectin solution: a biomimetic approach of the first steps of lignification. Biomacromolecules 6 763–774. 10.1021/bm049390y [DOI] [PubMed] [Google Scholar]
- Lam H. M., Coschigano K., Schultz C., Melo-Oliveira R., Tjaden G., Oliveira I., et al. (1995). Use of Arabidopsis mutants and genes to study amide amino acid biosynthesis. Plant Cell 7 887–898. 10.1105/tpc.7.7.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lama L., Tramice A., Finore I., Anzelmo G., Calandrelli V., Pagnotta E., et al. (2014). Degradative actions of microbial xylanolytic activities on hemicelluloses from rhizome of Arundo donax. AMB Express 4:55. 10.1186/s13568-014-0055-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson S., Cassland P., Jönsson L. J. (2001). Development of a Saccharomyces cerevisiae strain with enhanced resistance to phenolic fermentation inhibitors in lignocellulose hydrolysates by heterologous expression of LACCASE. Appl. Environ. Microbiol. 67 1163–1170. 10.1128/AEM.67.3.1163-1170.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Teng Q., Huang W., Zhong R., Ye Z.-H. (2009). The F8H glycosyltransferase is a functional paralog of FRA8 involved in glucuronoxylan biosynthesis in Arabidopsis. Plant Cell Physiol. 50 812–827. 10.1093/pcp/pcp025 [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R., Ye Z.-H. (2012a). Arabidopsis GUX proteins are glucuronyltransferases responsible for the addition of glucuronic acid side chains onto xylan. Plant Cell Physiol. 53 1204–1216. 10.1093/pcp/pcs064 [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R., Yuan Y. X., Haghighat M., Ye Z.-H. (2012b). Three Arabidopsis DUF579 domain-containing GXM proteins are methyltransferases catalyzing 4-O-methylation of glucuronic acid on xylan. Plant Cell Physiol. 53 1934–1949. 10.1093/pcp/pcs138 [DOI] [PubMed] [Google Scholar]
- Lee C., Zhong R., Ye Z.-H. (2012c). Arabidopsis family GT43 members are xylan xylosyltransferases required for the elongation of the xylan backbone. Plant Cell Physiol. 53 135–143. 10.1093/pcp/pcr158 [DOI] [PubMed] [Google Scholar]
- Lee C., Teng Q., Zhong R., Ye Z.-H. (2014). Alterations of the degree of xylan acetylation in Arabidopsis xylan mutants. Plant Signal. Behav. 9:e27797. 10.4161/psb.27797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Zhong R., Richardson E. A., Himmelsbach D. S., McPhail B. T., Ye Z.-H. (2007). The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 48 1659–1672. 10.1093/pcp/pcm155 [DOI] [PubMed] [Google Scholar]
- Li L., Huang J., Qin L., Huang Y., Zeng W., Rao Y., et al. (2014). Two cotton fiber-associated glycosyltransferases, GhGT43A1 and GhGT43C1, function in hemicellulose glucuronoxylan biosynthesis during plant development. Physiol. Plant. 152 367–379. 10.1111/ppl.12190 [DOI] [PubMed] [Google Scholar]
- Li Q., Koda K., Yoshinaga A., Takabe K., Shimomura M., Hirai Y., et al. (2015). Dehydrogenative polymerization of coniferyl alcohol in artificial polysaccharides matrices: effects of xylan on the polymerization. J. Agric. Food Chem. 63 4613–4620. 10.1021/acs.jafc.5b01070 [DOI] [PubMed] [Google Scholar]
- Li S., Bashline L., Zheng Y., Xin X., Huang S., Kong Z., et al. (2016). Cellulose synthase complexes act in a concerted fashion to synthesize highly aggregated cellulose in secondary cell walls of plants. Proc. Natl. Acad. Sci. U.S.A. 113 11348–11353. 10.1073/pnas.1613273113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Su X. (2018). Multifunctional smart hydrogels: potential in tissue engineering and cancer therapy. J. Mater. Chem. B 6 4714–4730. 10.1039/C8TB01078A [DOI] [PubMed] [Google Scholar]
- Li Y., Xue Y., Cao Z., Zhou T., Alnadari F. (2018). Characterization of a uronate dehydrogenase from Thermobispora bispora for production of glucaric acid from hemicellulose substrate. World J. Microbiol. Biotechnol. 34:102. 10.1007/s11274-018-2486-8 [DOI] [PubMed] [Google Scholar]
- Li Z., Omranian N., Neumetzler L., Wang T., Herter T., Usadel B., et al. (2016). A transcriptional and metabolic framework for secondary wall formation in Arabidopsis. Plant Physiol. 172 1334–1351. 10.1104/pp.16.01100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Qu H., Li C., Zhou X. (2013). Direct and efficient xylitol production from xylan by Saccharomyces cerevisiae through transcriptional level and fermentation processing optimizations. Bioresour. Technol. 149 413–419. 10.1016/j.biortech.2013.09.101 [DOI] [PubMed] [Google Scholar]
- Lin M., Behal R., Oliver D. J. (2003). Disruption of plE2, the gene for the E2 subunit of the plastid PYRUVATE DEHYDROGENASE complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Mol. Biol. 52 865–872. 10.1023/A:1025076805902 [DOI] [PubMed] [Google Scholar]
- Lin M., Oliver D. J. (2008). The role of ACETYL-COENZYME A SYNTHETASE in Arabidopsis. Plant Physiol. 147 1822–1829. 10.1104/pp.108.121269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizeau K., Gambonnet B., Zhang G.-F., Curien G., Jabrin S., Van Der Straeten D., et al. (2007). Regulation of one-carbon metabolism in Arabidopsis: the N-terminal regulatory domain of CYSTATHIONINE γ-SYNTHASE is cleaved in response to folate starvation. Plant Physiol. 145 491–503. 10.1104/pp.107.105379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy M. H., Miernyk J. A., Randall D. D. (1995). The mitochondrial PYRUVATE DEHYDROGENASE complex: nucleotide and deduced amino-acid sequences of a cDNA encoding the Arabidopsis thaliana E1 α-subunit. Gene 164 251–254. 10.1016/0378-1119(95)00465-I [DOI] [PubMed] [Google Scholar]
- Lund C. H., Bromley J. R., Stenbæk A., Rasmussen R. E., Scheller H. V., Sakuragi Y. (2015). A reversible Renilla luciferase protein complementation assay for rapid identification of protein-protein interactions reveals the existence of an interaction network involved in xyloglucan biosynthesis in the plant Golgi apparatus. J. Exp. Bot. 66 85–97. 10.1093/jxb/eru401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczakowski J. J., Wicher K. B., Terrett O. M., Faria-Blanc N., Yu X., Brown D., et al. (2017). Removal of glucuronic acid from xylan is a strategy to improve the conversion of plant biomass to sugars for bioenergy. Biotechnol. Biofuels 10:224. 10.1186/s13068-017-0902-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaya-Sanz D., Chen J., Kalluri U. C., Muchero W., Tschaplinski T. J., Gunter L. E., et al. (2017). Agronomic performance of Populus deltoides trees engineered for biofuel production. Biotechnol. Biofuels 10:253. 10.1186/s13068-017-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan C. P., Mansfield S. D., Stachurski Z. H., Evans R., Southerton S. G. (2010). FASCICLIN-LIKE ARABINOGALACTAN proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J. 62 689–703. 10.1111/j.1365-313X.2010.04181.x [DOI] [PubMed] [Google Scholar]
- MacMillan C. P., Taylor L., Bi Y., Southerton S. G., Evans R., Spokevicius A. (2015). The FASCICLIN-LIKE ARABINOGALACTAN protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol. 206 1314–1327. 10.1111/nph.13320 [DOI] [PubMed] [Google Scholar]
- Maheshwari P., Kovalchuk I. (2016). Agrobacterium-mediated stable genetic transformation of Populus angustifolia and Populus balsamifera. Front. Plant Sci. 7:296. 10.3389/fpls.2016.00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe Y., Verhertbruggen Y., Gille S., Harholt J., Chong S.-L., Pawar P. M., et al. (2013). REDUCED WALL ACETYLATION proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol. 163 1107–1117. 10.1104/pp.113.225193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott P. E., Gómez L. D., McQueen-Mason S. J. (2016). Unlocking the potential of lignocellulosic biomass through plant science. New Phytol. 209 1366–1381. 10.1111/nph.13684 [DOI] [PubMed] [Google Scholar]
- Martínez-Abad A., Giummarella N., Lawoko M., Vilaplana F. (2018). Differences in extractability under subcritical water reveal interconnected hemicellulose and lignin recalcitrance in birch hardwoods. Green Chem. 20 2534–2546. 10.1039/C8GC00385H [DOI] [Google Scholar]
- McCann M. C., Carpita N. C. (2015). Biomass recalcitrance: a multi-scale, multi-factor, and conversion-specific property. J. Exp. Bot. 66 4109–4118. 10.1093/jxb/erv267 [DOI] [PubMed] [Google Scholar]
- Meents M. J., Watanabe Y., Samuels A. L. (2018). The cell biology of secondary cell wall biosynthesis. Ann. Bot. 121 1107–1125. 10.1093/aob/mcy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellema S., Eichenberger W., Rawyler A., Suter M., Tadege M., Kuhlemeier C. (2002). The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J. 30 329–336. 10.1046/j.1365-313X.2002.01293.x [DOI] [PubMed] [Google Scholar]
- Mettler I. J., Beevers H. (1980). Oxidation of NADH in glyoxysomes by a malate-aspartate shuttle. Plant Physiol. 66 555–560. 10.1104/pp.66.4.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z., Rihouey C., Do C. T., Lerouge P., Jouanin L. (2004). Purification and characterization of enzymes exhibiting β-D-xylosidase activities in stem tissues of Arabidopsis. Plant Physiol. 135 867–878. 10.1104/pp.104.041269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir B. A., Myburg A. A., Mizrachi E., Cowan D. A. (2017). In planta expression of hyperthermophilic enzymes as a strategy for accelerated lignocellulosic digestion. Sci. Rep. 7:11462. 10.1038/s41598-017-11026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrachi E., Berger D. K., Mansfield S. D., Myburg A. A. (2013). Functional Genomics and Systems Genetics of Cellulose Biosynthesis in Eucalyptus. Ph.D. thesis, University of Pretoria, Pretoria. [Google Scholar]
- Mizrachi E., Mansfield S. D., Myburg A. A. (2011). Cellulose factories: advancing bioenergy production from forest trees. New Phytol. 194 54–62. 10.1111/j.1469-8137.2011.03971.x [DOI] [PubMed] [Google Scholar]
- Mizrachi E., Myburg A. A. (2016). Systems genetics of wood formation. Curr. Opin. Plant Biol. 30 94–100. 10.1016/j.pbi.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Mizrachi E., Verbeke L., Christie N., Fierro A. C., Mansfield S. D., Davis M. F., et al. (2017). Network-based integration of systems genetics data reveals pathways associated with lignocellulosic biomass accumulation and processing. Proc. Natl. Acad. Sci. U.S.A. 114 1195–1200. 10.1073/pnas.1620119114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11 266–277. 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Mølhøj M., Verma R., Reiter W.-D. (2003). The biosynthesis of the branched-chain sugar D-apiose in plants: functional cloning and characterization of a UDP-D-APIOSE/UDP-D-XYLOSE SYNTHASE from Arabidopsis. Plant J. 35 693–703. 10.1046/j.1365-313X.2003.01841.x [DOI] [PubMed] [Google Scholar]
- Mortimer J. C., Faria-Blanc N., Yu X., Tryfona T., Sorieul M., Ng Y. Z., et al. (2015). An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14. Plant J. 83 413–426. 10.1111/tpj.12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J. C., Miles G. P., Brown D. M., Zhang Z., Segura M. P., Weimar T., et al. (2010). Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc. Natl. Acad. Sci. U.S.A. 107 17409–17414. 10.1073/pnas.1005456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer J. C., Yu X., Albrecht S., Sicilia F., Huichalaf M., Ampuero D., et al. (2013). Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25 1881–1894. 10.1105/tpc.113.111500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S. D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37 190–200. 10.1016/j.copbio.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Mullins K. V., Llewellyn D. J., Hartney V. J., Strauss S., Dennis E. S. (1997). Regeneration and transformation of Eucalyptus camaldulensis. Plant Cell Rep. 16 787–791. 10.1007/s002990050321 [DOI] [PubMed] [Google Scholar]
- Mutwil M., Ruprecht C., Giorgi F. M., Bringmann M., Usadel B., Persson S. (2009). Transcriptional wiring of cell wall-related genes in Arabidopsis. Mol. Plant 2 1015–1024. 10.1093/mp/ssp055 [DOI] [PubMed] [Google Scholar]
- Myburg A. A., Grattapaglia D., Tuskan G. A., Hellsten U., Hayes R. D., Grimwood J., et al. (2014). The genome of Eucalyptus grandis. Nature 510 356–362. 10.1038/nature13308 [DOI] [PubMed] [Google Scholar]
- Myburg A. A., Lev-Yadun S., Sederoff R. R. (eds). (2013). “Xylem structure and function,” in eLS (Chichester: John Wiley & Sons, Ltd; ). [Google Scholar]
- Nasatto L. P., Pignon F., Silveira L. J., Duarte E. M., Noseda D. M., Rinaudo M. (2015). Methylcellulose, a cellulose derivative with original physical properties and extended applications. Polymers 7 777–803. 10.3390/polym7050777 [DOI] [Google Scholar]
- Nishimura H., Kamiya A., Nagata T., Katahira M., Watanabe T. (2018). Direct evidence for α ether linkage between lignin and carbohydrates in wood cell walls. Sci. Rep. 8:6538. 10.1038/s41598-018-24328-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordwald E. M., Brunecky R., Himmel M. E., Beckham G. T., Kaar J. L. (2014). Charge engineering of cellulases improves ionic liquid tolerance and reduces lignin inhibition. Biotechnol. Bioeng. 111 1541–1549. 10.1002/bit.25216 [DOI] [PubMed] [Google Scholar]
- Ohtani M., Morisaki K., Sawada Y., Sano R., Uy A. L. T., Yamamoto A., et al. (2016). Primary metabolism during biosynthesis of secondary wall polymers of protoxylem vessel elements. Plant Physiol. 172 1612–1624. 10.1104/pp.16.01230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A., Joshi H. J., Rennie E. A., Ebert B., Manisseri C., Heazlewood J. L., et al. (2010). An integrative approach to the identification of Arabidopsis and rice genes involved in xylan and secondary wall development. PLoS One 5:e15481. 10.1371/journal.pone.0015481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. J., Nikolau B. J., Wurtele E. S. (2009). Acetyl-CoA—Life at the metabolic nexus. Plant Sci. 176 597–601. 10.1016/j.plantsci.2009.02.005 [DOI] [Google Scholar]
- Palmieri L., Arrigoni R., Blanco E., Carrari F., Zanor M. I., Studart-Guimaraes C., et al. (2006). Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 142 855–865. 10.1104/pp.106.086975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajó J. C., Domínguez H., Domínguez J. M. (1997). Xylitol production from Eucalyptus wood hydrolysates extracted with organic solvents. Process Biochem. 32 599–604. 10.1016/S0032-9592(97)00016-2 [DOI] [Google Scholar]
- Paredez A. R., Somerville C. R., Ehrhardt D. W. (2006). Visualization of CELLULOSE SYNTHASE demonstrates functional association with microtubules. Science 312 1491–1495. 10.1126/science.1126551 [DOI] [PubMed] [Google Scholar]
- Park J.-J., Yoo C. G., Flanagan A., Pu Y., Debnath S., Ge Y., et al. (2017). Defined tetra-allelic gene disruption of the 4-COUMARATE:COENZYME A LIGASE 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol. Biofuels 10:284. 10.1186/s13068-017-0972-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil R. M., Genco J., Pendse H., van Heiningen A. (2013). Cleavage of acetyl groups from northeast hardwood for acetic acid production in kraft pulp mills. Tappi J. 12 57–67. [Google Scholar]
- Pauly M., Gille S., Liu L., Mansoori N., de Souza A., Schultink A., et al. (2013). Hemicellulose biosynthesis. Planta 238 627–642. 10.1007/s00425-013-1921-1 [DOI] [PubMed] [Google Scholar]
- Pauly M., Scheller H. V. (2000). O-Acetylation of plant cell wall polysaccharides: identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 210 659–667. 10.1007/s004250050057 [DOI] [PubMed] [Google Scholar]
- Pawar P. M., Derba-Maceluch M., Chong S.-L., Gandla M. L., Bashar S. S., Sparrman T., et al. (2017a). In muro deacetylation of xylan affects lignin properties and improves saccharification of aspen wood. Biotechnol. Biofuels 10:98. 10.1186/s13068-017-0782-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar P. M., Ratke C., Balasubramanian V. K., Chong S.-L., Gandla M. L., Adriasola M., et al. (2017b). Downregulation of RWA genes in hybrid aspen affects xylan acetylation and wood saccharification. New Phytol. 214 1491–1505. 10.1111/nph.14489 [DOI] [PubMed] [Google Scholar]
- Pawar P. M., Koutaniemi S., Tenkanen M., Mellerowicz E. J. (2013). Acetylation of woody lignocellulose: significance and regulation. Front. Plant Sci. 4:118. 10.3389/fpls.2013.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S., van de Lindt J. W., Popovski M., Berman J. W., Dolan J. D., Ricles J., et al. (2016). Cross-laminated timber for seismic regions: progress and challenges for research and implementation. J. Struct. Eng. 142:e2514001 10.1061/(ASCE)ST.1943-541X.0001192 [DOI] [Google Scholar]
- Peleman J., Boerjan W., Engler G., Seurinck J., Botterman J., Alliotte T., et al. (1989). Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-ADENOSYLMETHIONINE SYNTHETASE. Plant Cell 1 81–93. 10.1105/tpc.1.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M. J., Kulkarni A. R., Backe J., Boyd M., O’Neill M. A., York W. S. (2016). Structural diversity of xylans in the cell walls of monocots. Planta 244 589–606. 10.1007/s00425-016-2527-1 [DOI] [PubMed] [Google Scholar]
- Peña M. J., Zhong R., Zhou G.-K., Richardson E. A., O’Neill M. A., Darvill A. G., et al. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19 549–563. 10.1105/tpc.106.049320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta A. G., Venkatachalam S., Stone S. C., Pattathil S. (2017). Xylan epitope profiling: an enhanced approach to study organ development-dependent changes in xylan structure, biosynthesis, and deposition in plant cell walls. Biotechnol. Biofuels 10:245. 10.1186/s13068-017-0935-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C. S., Silveira R. L., Dupree P., Skaf M. S. (2017). Effects of xylan side-chain substitutions on xylan-cellulose interactions and implications for thermal pretreatment of cellulosic biomass. Biomacromolecules 18 1311–1321. 10.1021/acs.biomac.7b00067 [DOI] [PubMed] [Google Scholar]
- Pereira L. A. R., Todorova M., Cai X., Makaroff C. A., Emery R. J. N., Moffatt B. A. (2007). Methyl recycling activities are co-ordinately regulated during plant development. J. Exp. Bot. 58 1083–1098. 10.1093/jxb/erl275 [DOI] [PubMed] [Google Scholar]
- Persson S., Caffall K. H., Freshour G., Hilley M. T., Bauer S., Poindexter P., et al. (2007). The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19 237–255. 10.1105/tpc.106.047720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G. P., Somerville C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102 8633–8638. 10.1073/pnas.0503392102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen P. D., Lau J., Ebert B., Yang F., Verhertbruggen Y., Kim J. S., et al. (2012). Engineering of plants with improved properties as biofuels feedstocks by vessel-specific complementation of xylan biosynthesis mutants. Biotechnol. Biofuels 5:84. 10.1186/1754-6834-5-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N., Palmieri L., Pisano I., Hodges M., Palmieri F. (2002). Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: bacterial expression, reconstitution, functional characterization, and tissue distribution. J. Biol. Chem. 277 24204–24211. 10.1074/jbc.M202702200 [DOI] [PubMed] [Google Scholar]
- Pick U., Avidan O. (2017). Triacylglycerol is produced from starch and polar lipids in the green alga Dunaliella tertiolecta. J. Exp. Bot. 68 4939–4950. 10.1093/jxb/erx280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton W. C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 185–214. 10.1146/annurev.arplant.47.1.185 [DOI] [PubMed] [Google Scholar]
- Plomion C., Leprovost G., Stokes A. (2001). Wood formation in trees. Plant Physiol. 127 1513–1523. 10.1104/pp.010816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko G. V., Lionetti V., Fursova O. V., Sundaram R. M., Qi M., Whitham S. A., et al. (2013). Arabidopsis and Brachypodium transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 162 9–23. 10.1104/pp.113.214460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porth I., Klapšte J., Skyba O., Hannemann J., McKown A. D., Guy R. D., et al. (2013). Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 200 710–726. 10.1111/nph.12422 [DOI] [PubMed] [Google Scholar]
- Puchart V., Mørkeberg Krogh K. B. R., Biely P. (2019). Glucuronoxylan 3-O-acetylated on uronic acid-substituted xylopyranosyl residues and its hydrolysis by GH10, GH11 and GH30 endoxylanases. Carbohydr. Polym. 205 217–224. 10.1016/j.carbpol.2018.10.043 [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Q., Wyman C. E. (2011). Supplementation with xylanase and β-xylosidase to reduce xylo-oligomer and xylan inhibition of enzymatic hydrolysis of cellulose and pretreated corn stover. Biotechnol. Biofuels 4:18. 10.1186/1754-6834-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas A. J., Beckham G. T., Biddy M. J., Chandra R., Chen F., Davis M. F., et al. (2014). Lignin valorization: improving lignin processing in the biorefinery. Science 344:1246843. 10.1126/science.1246843 [DOI] [PubMed] [Google Scholar]
- Ralet M.-C., Crépeau M.-J., Vigouroux J., Tran J., Berger A., Sallé C., et al. (2016). Xylans provide the structural driving force for mucilage adhesion to the Arabidopsis seed coat. Plant Physiol. 171 165–178. 10.1104/pp.16.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez V., Xiong G., Mashiguchi K., Yamaguchi S., Pauly M. (2018). Growth- and stress-related defects associated with wall hypoacetylation are strigolactone-dependent. Plant Direct 2:e00062 10.1002/pld3.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall D. D., Tolbert N. E., Gremel D. (1971). 3-PHOSPHOGLYCERATE PHOSPHATASE in plants: II. Distribution, physiological considerations, and comparison with P-GLYCOLATE PHOSPHATASE. Plant Physiol. 48 480–487. 10.1104/pp.48.4.480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Barah P., Suarez-Rodriguez M. C., Bressendorff S., Friis P., Costantino P., et al. (2013). Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 161 1783–1794. 10.1104/pp.112.210773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi M., Shrivastava S. (2017). Recent advances in second generation bioethanol production: an insight to pretreatment, saccharification and fermentation processes. Renew. Sustain. Energy Rev. 80 330–340. 10.1016/j.rser.2017.05.225 [DOI] [Google Scholar]
- Ratke C., Pawar P. M.-A., Balasubramanian V. K., Naumann M., Duncranz M. L., Derba-Maceluch M., et al. (2015). Populus GT43 family members group into distinct sets required for primary and secondary wall xylan biosynthesis and include useful promoters for wood modification. Plant Biotechnol. J. 13 26–37. 10.1111/pbi.12232 [DOI] [PubMed] [Google Scholar]
- Ratke C., Terebieniec B. K., Winestrand S., Derba-Maceluch M., Grahn T., Schiffthaler B., et al. (2018). Downregulating aspen xylan biosynthetic GT43 genes in developing wood stimulates growth via reprograming of the transcriptome. New Phytol. 219 230–245. 10.1111/nph.15160 [DOI] [PubMed] [Google Scholar]
- Ravanel S., Block M. A., Rippert P., Jabrin S., Curien G., Rébeillé F., et al. (2004). Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279 22548–22557. 10.1074/jbc.M313250200 [DOI] [PubMed] [Google Scholar]
- Razeq F. M., Jurak E., Stogios P. J., Yan R., Tenkanen M., Kabel M. A., et al. (2018). A novel acetyl xylan esterase enabling complete deacetylation of substituted xylans. Biotechnol. Biofuels 11:74. 10.1186/s13068-018-1074-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul R., Geserick C., Pabst M., Frey B., Wittmann D., Lütz-Meindl U., et al. (2011). Down regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 286 39982–39992. 10.1074/jbc.M111.255695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis D., Vian B. (2004). Helicoidal pattern in secondary cell walls and possible role of xylans in their construction. C. R. Biol. 327 785–790. 10.1016/j.crvi.2004.04.008 [DOI] [PubMed] [Google Scholar]
- Ren Y., Hansen S. F., Ebert B., Lau J., Scheller H. V. (2014). Site-directed mutagenesis of IRX9, IRX9L and IRX14 proteins involved in xylan biosynthesis: glycosyltransferase activity is not required for IRX9 function in Arabidopsis. PLoS One 9:e105014. 10.1371/journal.pone.0105014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E. A., Hansen S. F., Baidoo E. E. K., Hadi M. Z., Keasling J. D., Scheller H. V. (2012). Three members of the Arabidopsis glycosyltransferase 8 family are xylan glucuronosyltransferases. Plant Physiol. 159 1408–1417. 10.1104/pp.112.200964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie E. A., Scheller H. V. (2014). Xylan biosynthesis. Curr. Opin. Biotechnol. 26 100–107. 10.1016/j.copbio.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Restolho J. A., Prates A., de Pinho M. N., Afonso M. D. (2009). Sugars and lignosulphonates recovery from Eucalyptus spent sulphite liquor by membrane processes. Biomass Bioenergy 33 1558–1566. 10.1016/j.biombioe.2009.07.022 [DOI] [Google Scholar]
- Riens B., Lohaus G., Heineke D., Heldt H. W. (1991). Amino acid and sucrose content determined in the cytosolic, chloroplastic, and vacuolar compartments and in the phloem sap of spinach leaves. Plant Physiol. 97 227–233. 10.1104/pp.97.1.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha P. S. C. F., Sheikh M., Melchiorre R., Fagard M., Boutet S., Loach R., et al. (2005). The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING 1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 17 404–417. 10.1105/tpc.104.028332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje S. (2006). S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67 1686–1698. 10.1016/j.phytochem.2006.04.019 [DOI] [PubMed] [Google Scholar]
- Ros R., Muñoz-Bertomeu J., Krueger S. (2014). Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci. 19 564–569. 10.1016/j.tplants.2014.06.003 [DOI] [PubMed] [Google Scholar]
- Roselli A., Hummel M., Monshizadeh A., Maloney T., Sixta H. (2014). Ionic liquid extraction method for upgrading Eucalyptus kraft pulp to high purity dissolving pulp. Cellulose 21 3655–3666. 10.1007/s10570-014-0344-x [DOI] [Google Scholar]
- Ruprecht C., Mutwil M., Saxe F., Eder M., Nikoloski Z., Persson S. (2011). Large-scale co-expression approach to dissect secondary cell wall formation across plant species. Plant Physiol. 2:23. 10.3389/fpls.2011.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Aguayo S., Rautengarten C., Temple H., Sanhueza D., Ejsmentewicz T., Sandoval-Ibañez O., et al. (2017). UUAT1 is a Golgi-localized UDP-uronic acid transporter that modulates the polysaccharide composition of Arabidopsis seed mucilage. Plant Cell 29 129–143. 10.1105/tpc.16.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Isogai A. (2004). TEMPO-mediated oxidation of native cellulose. The effect of oxidation conditions on chemical and crystal structures of the water-insoluble fractions. Biomacromolecules 5 1983–1989. 10.1021/bm0497769 [DOI] [PubMed] [Google Scholar]
- Saito T., Kimura S., Nishiyama Y., Isogai A. (2007). Cellulose nanofibers prepared by TEMPO-mediated oxidation of native cellulose. Biomacromolecules 8 2485–2491. 10.1021/bm0703970 [DOI] [PubMed] [Google Scholar]
- Salamone S., Boisbrun M., Didierjean C., Chapleur Y. (2014). From d-glucuronic acid to l-iduronic acid derivatives via a radical tandem decarboxylation–cyclization. Carbohydr. Res. 386 99–105. 10.1016/j.carres.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Salnikov V. V., Grimson M. J., Delmer D. P., Haigler C. H. (2001). SUCROSE SYNTHASE localizes to cellulose synthesis sites in tracheary elements. Phytochemistry 57 823–833. 10.1016/S0031-9422(01)00045-0 [DOI] [PubMed] [Google Scholar]
- Salon C., Raymond P., Pradet A. (1988). Quantification of carbon fluxes through the tricarboxylic acid cycle in early germinating lettuce embryos. J. Biol. Chem. 263 12278–12287. [PubMed] [Google Scholar]
- Sauter M., Moffatt B., Saechao M. C., Hell R., Wirtz M. (2013). Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 451 145–154. 10.1042/BJ20121744 [DOI] [PubMed] [Google Scholar]
- Scheller H. V. (2017). Plant cell wall: never too much acetate. Nat. Plants 3:17024. 10.1038/nplants.2017.24 [DOI] [PubMed] [Google Scholar]
- Scheller H. V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61 263–289. 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- Schneider R., Tang L., Lampugnani E. R., Barkwill S., Lathe R., Zhang Y., et al. (2017). Two complementary mechanisms underpin cell wall patterning during xylem vessel development. Plant Cell 29 2433–2449. 10.1105/tpc.17.00309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A., Naylor D., Dama M., Pauly M. (2015). The role of the plant-specific ALTERED XYLOGLUCAN9 protein in Arabidopsis cell wall polysaccharide O-acetylation. Plant Physiol. 167 1271–1283. 10.1104/pp.114.256479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster J., Binder S. (2005). The mitochondrial BRANCHED-CHAIN AMINOTRANSFERASE (AtBCAT-1) is capable to initiate degradation of leucine, isoleucine and valine in almost all tissues in Arabidopsis thaliana. Plant Mol. Biol. 57 241–254. 10.1007/s11103-004-7533-1 [DOI] [PubMed] [Google Scholar]
- Schwender J., Ohlrogge J. B., Shachar-Hill Y. (2003). A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278 29442–29453. 10.1074/jbc.M303432200 [DOI] [PubMed] [Google Scholar]
- Schwender J., Shachar-Hill Y., Ohlrogge J. B. (2006). Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 281 34040–34047. 10.1074/jbc.M606266200 [DOI] [PubMed] [Google Scholar]
- Shahid S., Tajwar R., Akhtar M. W. (2017). A novel trifunctional, family GH10 enzyme from Acidothermus cellulolyticus 11B, exhibiting endo-xylanase, arabinofuranosidase and acetyl xylan esterase activities. Extremophiles 22 109–119. 10.1007/s00792-017-0981-8 [DOI] [PubMed] [Google Scholar]
- Sharples S. C., Fry S. C. (2007). Radioisotope ratios discriminate between competing pathways of cell wall polysaccharide and RNA biosynthesis in living plant cells. Plant J. 52 252–262. 10.1111/j.1365-313X.2007.03225.x [DOI] [PubMed] [Google Scholar]
- Shen B., Li C., Tarczynski M. C. (2002). High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-ADENOSYL-L-METHIONINE SYNTHETASE 3 gene. Plant J. 29 371–380. 10.1046/j.1365-313X.2002.01221.x [DOI] [PubMed] [Google Scholar]
- Shen B., Sun X., Zuo X., Shilling T., Apgar J., Ross M., et al. (2012). Engineering a thermoregulated intein-modified xylanase into maize for consolidated lignocellulosic biomass processing. Nat. Biotechnol. 30 1131–1136. 10.1038/nbt.2402 [DOI] [PubMed] [Google Scholar]
- Simmons T. J., Mortimer J. C., Bernardinelli O. D., Pöppler A.-C., Brown S. P., deAzevedo E. R., et al. (2016). Folding of xylan onto cellulose fibrils in plant cell walls revealed by solid-state NMR. Nat. Commun. 7:13902. 10.1038/ncomms13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixta H. (2006). Handbook of Pulp. Weinheim: Wiley-VCH; 1009–1067. 10.1002/9783527619887 [DOI] [Google Scholar]
- Sixta H., Iakovlev M., Testova L., Roselli A., Hummel M., Borrega M., et al. (2013). Novel concepts of dissolving pulp production. Cellulose 20 1547–1561. 10.1016/j.biortech.2011.01.073 [DOI] [PubMed] [Google Scholar]
- Smith P. J., Wang H.-T., York W. S., Peña M. J., Urbanowicz B. R. (2017). Designer biomass for next-generation biorefineries: leveraging recent insights into xylan structure and biosynthesis. Biotechnol. Biofuels 10:286. 10.1186/s13068-017-0973-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. A., Schuetz M., Roach M., Mansfield S.D., Ellis B., Samuels L. (2013). Neighboring parenchyma cells contribute to Arabidopsis Xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25 3988–3999. 10.1105/tpc.113.117176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Gui J., Liu C., Sun J., Li L. (2016). Suppression of PtrDUF579-3 expression causes structural changes of the glucuronoxylan in Populus. Front. Plant Sci. 7:493. 10.3389/fpls.2016.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Lu S., Chen Z.-Z., Lourenco R., Chiang V. L. (2006). Genetic transformation of Populus trichocarpa genotype Nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol. 47 1582–1589. 10.1093/pcp/pcl018 [DOI] [PubMed] [Google Scholar]
- Song L., Zeng W., Wu A., Picard K., Lampugnani E. R., Cheetamun R., et al. (2015). Asparagus spears as a model to study heteroxylan biosynthesis during secondary wall development. PLoS One 10:e0123878. 10.1371/journal.pone.0123878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A. C., Chen F., Ray T., Pattathil S., Peña M. J., Avci U., et al. (2015). Loss of function of FOLYLPOLYGLUTAMATE SYNTHETASE 1 reduces lignin content and improves cell wall digestibility in Arabidopsis. Biotechnol. Biofuels 8:224. 10.1186/s13068-015-0403-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein O., Avin-Wittenberg T., Krahnert I., Zemach H., Bogol V., Daron O., et al. (2016). Arabidopsis FRUCTOKINASES are important for seed oil accumulation and vascular development. Front. Plant Sci. 7:2047. 10.3389/fpls.2016.02047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepan A. M., Michud A., Hellstén S., Hummel M., Sixta H. (2016). IONCELL-P&F: pulp fractionation and fiber spinning with ionic liquids. Ind. Eng. Chem. Res. 55 8225–8233. 10.1021/acs.iecr.6b00071 [DOI] [Google Scholar]
- Sundell D., Street N. R., Kumar M., Mellerowicz E. J., Kucukoglu M., Johnsson C., et al. (2017). AspWood: high-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29 1585–1604. 10.1105/tpc.17.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sychantha D., Jones C. S., Little D. J., Moynihan P. J., Robinson H., Galley N. F., et al. (2017). In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA). PLoS Pathog. 13:e1006667. 10.1371/journal.ppat.1006667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Dupuis I., Kuhlemeier C. (1999). Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci. 4 320–325. 10.1016/S1360-1385(99)01450-8 [DOI] [PubMed] [Google Scholar]
- Tadege M., Kuhlemeier C. (1997). Aerobic fermentation during tobacco pollen development. Plant Mol. Biol. 35 343–354. 10.1023/A:1005837112653 [DOI] [PubMed] [Google Scholar]
- Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25 270–287. 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauberger E., Hoffmann-Benning S., Fleischer-Notter H., Willmitzer L., Fisahn J. (1999). Impact of INVERTASE overexpression on cell size, starch granule formation and cell wall properties during tuber development in potatoes with modified carbon allocation patterns. J. Exp. Bot. 50 477–486. 10.1093/jxb/50.333.477 [DOI] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T. W., Gaudinier A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517 571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumma B. R., Nolan M. F., Evans R., Moran G. F. (2005). Polymorphisms in CINNAMOYL-COA REDUCTASE (CCR) are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171 1257–1265. 10.1534/genetics.105.042028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga N., Kaneta T., Sato S., Sato Y. (2009). Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiol. Plant. 136 237–249. 10.1111/j.1399-3054.2009.01233.x [DOI] [PubMed] [Google Scholar]
- Toujani W., Muñoz-Bertomeu J., Flores-Tornero M., Rosa-Téllez S., Anoman A. D., Alseekh S., et al. (2013). Functional characterization of the plastidial 3-PHOSPHOGLYCERATE DEHYDROGENASE family in Arabidopsis. Plant Physiol. 163 1164–1178. 10.1104/pp.113.226720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. E., Greville K., Murphy E. C., Hooks M. A. (2005). Characterization of Arabidopsis fluoroacetate-resistant mutants reveals the principal mechanism of acetate activation for entry into the glyoxylate cycle. J. Biol. Chem. 280 2780–2787. 10.1074/jbc.M407291200 [DOI] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Urbanowicz B. R., Peña M. J., Moniz H. A., Moremen K. W., York W. S. (2014). Two Arabidopsis proteins synthesize acetylated xylan in vitro. Plant J. 80 197–206. 10.1111/tpj.12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz B. R., Peña M. J., Ratnaparkhe S., Avci U., Backe J., Steet H. F., et al. (2012). 4-O-methylation of glucuronic acid in Arabidopsis glucuronoxylan is catalyzed by a Domain of Unknown Function family 579 protein. Proc. Natl. Acad. Sci. U.S.A. 109 14253–14258. 10.1073/pnas.1208097109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Acker R., Déjardin A., Desmet S., Hoengenaert L., Vanholme R., Morreel K., et al. (2017). Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol. 175 1018–1039. 10.1104/pp.17.00834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R., Storme V., Vanholme B., Sundin L., Christensen J. H., Goeminne G., et al. (2012). A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 24 3506–3529. 10.1105/tpc.112.102574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardon D. R., Rorrer N. A., Salvachúa D., Settle A. E., Johnson C. W., Menart M. J., et al. (2016). cis,cis-Muconic acid: separation and catalysis to bio-adipic acid for nylon-6,6 polymerization. Green Chem. 18 3397–3413. 10.1039/C5GC02844B [DOI] [Google Scholar]
- Vogel J. P., Raab T. K., Somerville C. R., Somerville S. C. (2004). Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40 968–978. 10.1111/j.1365-313X.2004.02264.x [DOI] [PubMed] [Google Scholar]
- Vojta A., Dobrinić P., Tadić V., Boćkor L., Korać P., Julg B., et al. (2016). Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44 5615–5628. 10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A., Timonen J. V. I., Díez I., Laukkanen A., Ikkala O. (2011). Multifunctional high-performance biofibers based on wet-extrusion of renewable native cellulose nanofibrils. Adv. Mater. 23 2924–2928. 10.1002/adma.201100580 [DOI] [PubMed] [Google Scholar]
- Wang J. P., Naik P. P., Chen H.-C., Shi R., Lin C.-Y., Liu J., et al. (2014). Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26 894–914. 10.1105/tpc.113.120881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ci D., Li T., Li P., Song Y., Chen J., et al. (2016). The role of DNA methylation in xylogenesis in different tissues of poplar. Front. Plant Sci. 7:1003. 10.3389/fpls.2016.01003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardrop A. B., Preston R. D. (1947). Organization of the cell walls of tracheids and wood fibres. Nature 160 911–913. 10.1038/160911a0 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Meents M. J., McDonnell L. M., Barkwill S., Sampathkumar A., Cartwright H. N., et al. (2015). Visualization of cellulose synthases in Arabidopsis secondary cell walls. Science 350 198–203. 10.1126/science.aac7446 [DOI] [PubMed] [Google Scholar]
- Wei N., Quarterman J., Kim S. R., Cate J. H. D., Jin Y.-S. (2013). Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat. Commun. 4:2580. 10.1038/ncomms3580 [DOI] [PubMed] [Google Scholar]
- Wu A.-M., Hörnblad E., Voxeur A., Gerber L., Rihouey C., Lerouge P., et al. (2010). Analysis of the Arabidopsis IRX9/IRX9-L and IRX14/IRX14-L pairs of glycosyltransferase genes reveals critical contributions to biosynthesis of the hemicellulose glucuronoxylan. Plant Physiol. 153 542–554. 10.1104/pp.110.154971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A. M., Correia M. F., Pereira S. R., Evtuguin D. V. (2010). Second-generation bioethanol from eucalypt sulphite spent liquor. Bioresour. Technol. 101 2755–2761. 10.1016/j.biortech.2009.11.092 [DOI] [PubMed] [Google Scholar]
- Xiao C., Anderson C. (2013). Roles of pectin in biomass yield and processing for biofuels. Front. Plant Sci. 4:67. 10.3389/fpls.2013.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Wang C.-Z., Bian J., Sun R.-C. (2015). Optimization of bamboo autohydrolysis for the production of xylo-oligosaccharides using response surface methodology. RSC Adv. 5 106219–106226. 10.1039/C5RA18508D [DOI] [Google Scholar]
- Xiao Y., He X., Ojeda-Lassalle Y., Poovaiah C., Coleman H. D. (2018). Expression of a hyperthermophilic endoglucanase in hybrid poplar modifies the plant cell wall and enhances digestibility. Biotechnol. Biofuels 11:225. 10.1186/s13068-018-1224-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z., Browse J. (1998). eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proc. Natl. Acad. Sci. U.S.A. 95 7799–7804. 10.1073/pnas.95.13.7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Cheng K., Pauly M. (2013). Xylan O-acetylation impacts xylem development and enzymatic recalcitrance as indicated by the Arabidopsis mutant tbl29. Mol. Plant 6 1373–1375. 10.1093/mp/sst014 [DOI] [PubMed] [Google Scholar]
- Xiong G., Dama M., Pauly M. (2015). Glucuronic acid moieties on xylan are functionally equivalent to O-acetyl-substituents. Mol. Plant 8 1119–1121. 10.1016/j.molp.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Xiong T., Meister G. E., Workman R. E., Kato N. C., Spellberg M. J., Turker F., et al. (2017). Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci. Rep. 7:6732. 10.1038/s41598-017-06757-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Wang X., Sandler N., Willför S., Xu C. (2018). Three-dimensional printing of wood-derived biopolymers: a review focused on biomedical applications. ACS Sustain. Chem. Eng. 6 5663–5680. 10.1021/acssuschemeng.7b03924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Mitra P., Zhang L., Prak L., Verhertbruggen Y., Kim J.-S., et al. (2013). Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 11 325–335. 10.1111/pbi.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yoo C. G., Winkeler K. A., Collins C. M., Hinchee M. A. W., Jawdy S. S., et al. (2017). Overexpression of a Domain of Unknown Function 231-containing protein increases O-xylan acetylation and cellulose biosynthesis in Populus. Biotechnol. Biofuels 10:311. 10.1186/s13068-017-0998-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J. Y., Senger R. S., Gillaspy G. E. (2013). Systems Metabolic Engineering of Arabidopsis for Increased Cellulose Production. MSc dissertation, Virginia Polytechnic Institute and State University, Blacksburg, VA. [Google Scholar]
- York W. S., O’Neill M. A. (2008). Biochemical control of xylan biosynthesis — which end is up? Curr. Opin. Plant Biol. 11 258–265. 10.1016/j.pbi.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Teng Q., Lee C., Zhong R., Ye Z.-H. (2014). Modification of the degree of 4-O-methylation of secondary wall glucuronoxylan. Plant Sci. 219–220, 42–50. 10.1016/j.plantsci.2014.01.005 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Teng Q., Zhong R., Ye Z.-H. (2013). The Arabidopsis DUF231 domain-containing protein ESK1 mediates 2-O- and 3-O-acetylation of xylosyl residues in xylan. Plant Cell Physiol. 54 1186–1199. 10.1093/pcp/pct070 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Teng Q., Zhong R., Haghighat M., Richardson E. A., Ye Z.-H. (2016a). Mutations of Arabidopsis TBL32 and TBL33 affect xylan acetylation and secondary wall deposition. PLoS One 11:e0146460. 10.1371/journal.pone.0146460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Teng Q., Zhong R., Ye Z.-H. (2016b). TBL3 and TBL31, two Arabidopsis DUF231 domain proteins, are required for 3-O-monoacetylation of xylan. Plant Cell Physiol. 57 35–45. 10.1093/pcp/pcv172 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Teng Q., Zhong R., Ye Z.-H. (2016c). Roles of Arabidopsis TBL34 and TBL35 in xylan acetylation and plant growth. Plant Sci. 243 120–130. 10.1016/j.plantsci.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Zhong R., Cui D., Phillips D. R., Ye Z.-H. (2018). A novel rice xylosyltransferase catalyzes the addition of 2-O-xylosyl side chains onto the xylan backbone. Plant Cell Physiol. 59 554–565. 10.1093/pcp/pcy003 [DOI] [PubMed] [Google Scholar]
- Yusuf F., Gaur N. A. (2017). “Engineering Saccharomyces cerevisiae for C5 fermentation: a step towards second-generation biofuel production,” in Metabolic Engineering for Bioactive Compounds, eds Kalia V., Saini A. (Singapore: Springer; ), 157–172. [Google Scholar]
- Zabalza A., van Dongen J. T., Froehlich A., Oliver S. N., Faix B., Gupta K. J., et al. (2009). Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol. 149 1087–1098. 10.1104/pp.108.129288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Jiang N., Nadella R., Killen T. L., Nadella V., Faik A. (2010). A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiol. 154 78–97. 10.1104/pp.110.159749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Lampugnani E. R., Picard K. L., Song L., Wu A.-M., Farion I. M., et al. (2016). Asparagus IRX9, IRX10, and IRX14A are components of an active xylan backbone synthase complex that forms in the Golgi apparatus. Plant Physiol. 171 93–109. 10.1104/pp.15.01919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang L., Li F., Zhang D., Liu X., Wang H., et al. (2017). Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat. Plants 3:17017. 10.1038/nplants.2017.17 [DOI] [PubMed] [Google Scholar]
- Zhang J., Li M., Bryan A. C., Yoo C. G., Rottmann W., Winkeler K. A., et al. (2019). Overexpression of a serine hydroxymethyltransferase increases biomass production and reduces recalcitrance in the bioenergy crop Populus. Sustain. Energy Fuels 3 195–207. 10.1039/C8SE00471D [DOI] [Google Scholar]
- Zhang X., Dominguez P. G., Kumar M., Bygdell J., Miroshnichenko S., Sundberg B., et al. (2018). Cellulose synthase stoichiometry in aspen differs from Arabidopsis and Norway spruce. Plant Physiol. 117 1096–1107. 10.1104/pp.18.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Holladay J. E., Brown H., Zhang Z. C. (2007). Metal chlorides in ionic liquid solvents convert sugars to 5-hydroxymethylfurfural. Science 316 1597–1600. 10.1126/science.1141199 [DOI] [PubMed] [Google Scholar]
- Zhao J., Xu L., Wang Y., Zhao X., Wang J., Garza E., et al. (2013). Homofermentative production of optically pure L-lactic acid from xylose by genetically engineered Escherichia coli B. Microb. Cell Factor. 12:57. 10.1186/1475-2859-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Cui D., Ye Z.-H. (2017). Regiospecific acetylation of xylan is mediated by a group of DUF231-containing O-acetyltransferases. Plant Cell Physiol. 58 2126–2138. 10.1093/pcp/pcx147 [DOI] [PubMed] [Google Scholar]
- Zhong R., Peña M. J., Zhou G.-K., Nairn C. J., Wood-Jones A., Richardson E. A., et al. (2005). Arabidopsis FRAGILE FIBER8, which encodes a putative glucuronyltransferase, is essential for normal secondary wall synthesis. Plant Cell 17 3390–3408. 10.1105/tpc.105.035501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Runge T., Karlen S. D., Ralph J., Gonzales-Vigil E., Mansfield S. D. (2017). Chemical pulping advantages of zip-lignin hybrid poplar. ChemSusChem 10 3565–3573. 10.1002/cssc.201701317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Jacobs T. B., Xue L.-J., Harding S. A., Tsai C.-J. (2015). Exploiting SNPs for biallelic CRISPR mutations in the outcrossing woody perennial Populus reveals 4-COUMARATE:COA LIGASE specificity and redundancy. New Phytol. 208 298–301. 10.1111/nph.13470 [DOI] [PubMed] [Google Scholar]
- Zhu H., Luo W., Ciesielski P. N., Fang Z., Zhu J. Y., Henriksson G., et al. (2016). Wood-derived materials for green electronics, biological devices, and energy applications. Chem. Rev. 116 9305–9374. 10.1021/acs.chemrev.6b00225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of enzymes which catalyze the biosynthesis of sugar nucleotides, s-adenosyl methionine and acetyl-CoA. (a) The numbering of the enzymes corresponds to Figure 4–6. (b) Name of the enzyme. (c) Not all accessions were provided for each reaction, just one representative from each cellular compartment. Accessions and cellular localisation were obtained from TAIR and SUBA3, respectively.
References pertaining to numbered items in Table 1.
Biotechnology approaches that have scaled from Arabidopsis to Populus.