Abstract
Background
The etiology of thymic epithelial tumors is unknown. Murine polyomavirus strain PTA has been shown to induce thymomas in mice. Recently, using diverse molecular techniques, we reported the presence of human polyomavirus 7 (HPyV7) in thymic epithelial tumors. In the present study, we investigated the prevalence of Merkel cell polyomavirus (MCPyV) in thymic epithelial tumors.
Methods
Thirty‐six thymomas were screened for MCPyV by PCR and subsequently tested by DNA and RNA in situ hybridization and immunohistochemistry. Twenty‐six thymomas were diagnosed with myasthenia gravis (MG).
Results
MCPyV DNA was detected by PCR in 7 (19.4%) of the 36 thymic epithelial tumors and in six of these, the presence of MCPyV was confirmed by fluorescence situ hybridization. Of these, 3 (28.6%) revealed weak MCPyV LT‐antigen protein expression. In addition, one of the MCPyV positive thymomas tested positive for MCPyV LT RNA with RNAscope. Of interest, two out of the three thymomas that previously tested positive for MCPyV by immunohistochemistry also tested positive for HPyV7. One of the 11 MG‐negative and 2 of the 25 MG‐positive were positive for MCPyV.
Conclusions
MCPyV DNA and MCPyV protein expression can be detected in human epithelial thymoma; however, to a far lesser extent than HPyV7. Our data strongly indicate that because of its infrequent detection and weak expression, MCPyV is unlikely to play an important role in the etiopathogenesis of human thymomas.
Keywords: Fluorescence in situ hybridization, immunohistochemistry, Merkel cell polyomavirus, thymoma, tumorigenesis
Introduction
Thymomas are rare thymic epithelial tumors that are frequently associated with autoimmune diseases, such as myasthenia gravis (MG, 24–40%).1 The underlying etiology of thymomas is unknown. Several studies have investigated the role of viruses in thymomagenesis.1, 2, 3 We previously assessed the presence of the novel human polyomaviruses 6 and 7 (HPyV6 and 7) in human thymic epithelial tumors, based on the reported induction of thymomas in mouse strains C3H/BiDa and AKR by the polyomavirus strain PTA, and frequently detected HPyV7 in these tumors.4, 5, 6 The number of human polyomaviruses detected has continually increased in recent years.7, 8 To date, only Merkel cell polyomavirus (MCPyV) has been established as a novel human tumor virus in the majority of Merkel cell carcinomas (MCC).8, 9 In MCC, MCPyV is clonally integrated in the tumor DNA and harbors tumor specific truncating oncogenic mutations within the large T (LT) antigen.9, 10, 11 Truncated LT antigen and small T antigen (sTAg) of MCPyV have been shown to inhibit the tumor suppressor protein retinoblastoma (RB) and p53.12, 13, 14 Nicol et al. recently reported the seroprevalence of five novel human polyomaviruses (MCPyV, HPyV6, HPyV7, HPyV9,TSPyV [trichodysplasia spinulosa‐associated polyomavirus]) is increased in an age‐dependent manner.15 In other studies, 86.4% and 56.8–63.6% of adult blood sera (≥ 20 years) were positive for MCPyV‐ and HPyV7‐DNA, respectively.16, 17 In two independent studies using clinicopathologically not further classified thymomas (n = 10), no MCPyV DNA could be detected by PCR.18, 19 Because of these reports and our recent finding that 62.2% of thymomas are HPyV7 positive,6 we aimed to comprehensively assess the prevalence of MCPyV in this clinicopathologically well‐classified cohort of human thymic epithelial tumors by diverse molecular techniques.
Testing the prevalence of MCPyV in formalin‐fixed and paraffin‐embedded (FFPE) tissues of MCC has generated conflicting results in previous studies.20 According to Moshiri et al., the use of a PCR‐based approach in combination with the CM2B4 anti LT‐antigen antibody has the highest specificity and sensitivity.20 Herein, we used a PCR based approach in combination with MCPyV fluorescence situ hybridization (FISH) and immunohistochemistry (IHC) to elucidate the presence of MCPyV in thymic epithelial tumors.
Methods
Patients and tissues
Thirty‐six FFPE thymoma resection specimens were included in this study (from 19 women and 17 men; mean age 58.3 years, range 34–82 years). Thirty‐five of these had previously been tested for the presence of HPyV7.6 Twenty‐six thymoma patients were known to have a history of MG, of which 24 were anti‐acetylcholine receptor (AChR) antibody positive and two were negative. Twelve of the 26 MG‐positive thymoma patients were immunosuppressed. All clinical pathological data are summarized in Table 1. In addition, non‐neoplastic thymic hyperplasia (n = 20) and fetal thymic (n = 20) tissues were included in this study (summarized in Table S1). All use of this tissue and patient data was in agreement with the Dutch Code of Conduct for Observational Research with Personal Data (2004) and Tissue (2011, www.federa.org/sites/default/files/digital_ version_first_part_code_of_conduct_in_uk_2011_12092012.pdf). Diagnoses were previously defined by histology in routine diagnostics and were reviewed by two experienced pathologists.
Table 1.
Clinicopathological data and MCPyV DNA PCR, FISH, IHC, and RISH results with HPyV7 positivity of the tissues
| Clinicopathological data | MCPyV | HPyV7 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | G | Age | MG | Thymtype | Masaoka‐Koga | Anti AChR | IS/ster. | M1/M2 (178 bp) | VP1 (351 bp) | LT3 (308 bp) | MCPyV FISH | MCPyV RISH | MCPyV IHC CM2B4 | sTAg‐PCR | LTAg‐PCR | FISH | IHC 2t10 |
| 1–1 | F | 73 | + | B1/B2 | I | + | + | − | − | NA | + | + | + | − | |||
| 1–2 | F | 75 | + | B1/B2 | I | + | + | − | − | NA | + | + | + | − | |||
| 1–11 | F | 34 | + | B2 | I | + | − | − | − | − | − | − | ++ | − | |||
| 1–12 | F | 36 | + | A | I | − | + | + | (+) | − | + | n.d. | − | − | − | − | − |
| 1–16 | F | 34 | + | A | I | + | + | + | (+) | − | + | n.d. | − | − | − | − | − |
| 1–17 | M | 69 | + | AB | I | + | + | − | − | − | − | + | ++ | − | |||
| 1–18 | F | 47 | + | AB | I | + | + | − | − | − | + | + | − | − | |||
| 1–19 | M | 68 | − | AB | I | NA | NA | − | − | − | + | + | ++ | + | |||
| 1–21 | F | 38 | + | AB | I | + | − | − | − | − | − | + | − | − | |||
| 1–22 | M | 65 | + | AB/B2 | I | + | − | + | − | (+) | NA | n.d. | − | − | − | − | − |
| 1–28 | M | 38 | − | B1 | I | NA | NA | − | − | − | − | − | − | − | |||
| 1–31 | F | 82 | + | A | I | + | + | + | − | − | + | + | + | + | |||
| 1–32 | M | 47 | + | B2 | I | + | − | + | − | − | n.d. | − | n.d. | − | − | − | − |
| 1–34 | M | 59 | − | AB | I | NA | NA | − | − | − | + | − | ++ | + | |||
| 1–36 | F | 82 | − | AB | I | NA | NA | − | − | − | − | (+) | ++ | + | |||
| 1–39 | F | 78 | − | B1 | I | NA | NA | − | − | − | − | − | − | − | |||
| 1–43 | F | 78 | − | AB | I | NA | NA | + | + | − | + | n.d. | + | − | − | ++ | (+) |
| 1–23 | M | 37 | + | AB | IIA | + | − | + | − | − | + | − | + | + | |||
| 1–24 | M | 68 | + | B2 | IIA | + | − | − | − | − | + | + | ++ | + | |||
| 1–33 | F | 43 | + | B2 | IIA | + | − | − | − | − | − | − | ++ | + | |||
| 1–35 | M | 45 | + | AB | IIA | + | − | − | − | − | + | + | ++ | + | |||
| 1–37 | M | 77 | + | AB | IIA | + | − | − | − | − | − | − | +++ | (+) | |||
| 1–38 | F | 80 | + | AB | IIA | + | + | − | − | + | − | − | − | − | |||
| 1–3 | F | 57 | + | B2/B3 | IIB | + | − | + | − | − | + | + | + | + | |||
| 1–5 | M | 37 | + | B3 | IIB | + | + | + | − | − | + | + | ++ | + | |||
| 1–7 | F | 79 | + | A | IIB | − | − | + | − | − | + | + | ++ | (+) | |||
| 1–8 | M | 58 | + | A/B2 | IIB | + | − | (+) | (+) | + | ++ | (+) | (+) | + | + | +++ | (+) |
| 1–9 | M | 64 | − | B2 | IIB | NA | NA | − | − | − | + | + | ++ | (+) | |||
| 1–15 | F | 53 | + | B3 | IIB | + | + | − | − | − | − | − | + | − | |||
| 1–26 | M | 65 | − | AB | IIB | NA | NA | − | − | (+) | − | − | − | − | − | − | − |
| 1–42 | F | 57 | + | AB | IIB | + | − | − | − | − | − | − | − | − | |||
| 1–10 | F | 73 | + | B3 | III | + | + | − | − | − | − | − | − | − | |||
| 1–13 | F | 54 | − | B2 | III | NA | NA | − | − | − | − | + | + | (+) | |||
| 1–20 | M | 64 | − | B3 | III | NA | NA | ++ | ++ | ++ | + | n.d. | − | − | − | − | − |
| 1–41 | M | 65 | − | B3 | III | NA | NA | − | − | − | + | (+) | +++ | + | |||
| 1–25 | M | 37 | + | B2 | IVA | + | + | + | + | NA | + | n.d. | + | n.d. | n.d. | n.d. | n.d. |
| 25/36 | 24/25 | 12/25 | 13/36 | 8/36 | 4/33 | 6/7 | 1/3 | 3/7 | 15/35 | 17/35 | 22/35 | 15/35 | |||||
| 69.4% | 96.0% | 48.0% | 36.1% | 22.2% | 12.1% | 85.7% | 33.3% | 37.5% | 42.9% | 48.6% | 62.9% | 42.9% | |||||
Clinicopathological data and the results of Merkel cell polyomavirus (MCPyV) DNA PCR, fluorescence in situ hybridization (FISH), immunohistochemistry (IHC) and RNA in situ hybridization (RISH) in relation to the recently reported HPyV7 positivity in these thymomas as published by Rennspiess et al., 2017. MCPyV‐negative ID‐32 and ID‐26 were used as negative controls for IHC and RISH.
Anti‐AChR, anti acetylcholine receptor antibodies; bp, base pairs; F, female; G, gender; HPyV7, human polyomavirus 7; ID, laboratory identification; IS/Ster., immunosuppression/steroids; LTAg, large T‐antigen; M, male; MG, myasthenia gravis; sTAg, small T‐antigen; Thym. type, thymoma type; ++, strong positive; +, positive; (+), weak positive; −, negative; NA, not applicable; n.d., not done.
Merkel cell polyomavirus (MCPyV) detection
PCR was performed with 250 ng of genomic DNA using the AmpliTaq Gold DNA Polymerase (Thermo Fisher Scientific, Naarden, The Netherlands) in a final volume of 50 μL, as recently described.10 DNA quality and integrity were assessed by specimen control size ladder (SCS) as previously described and published for the DNA used.6, 21 All gained PCR fragments were confirmed by sequencing. In addition, Merkel cell carcinoma cell lines MS‐1 and MCC26 were used as positive and negative controls, respectively, to detect MCPyV.
MCPyV detection by fluorescence in situ hybridization (FISH)
MCPyV FISH was performed as previously described.22, 23 In brief, deparaffinized 3 μm thick sections were pretreated with 0.2 M hydrochloric acid, incubated with 1 M NaSCN and digested with 1 mg/mL pepsin (2500–3500 U/mg, Sigma Chemical, St. Louis, MO, USA). The biotin labeled “specific” MCPyV DNA probe was added to the samples at a concentration of 5 ng/μL, followed by denaturation of DNA (five minutes, 80°C) and hybridization overnight (37°C, humid chamber; ThermoBrite System, Abbott Molecular, Abbot Park, IL, USA). Unbound MCPyV DNA probe was stringently washed away. Bound probe was detected by sequential incubation in a combination of secondary antibodies: fluorescein isothiocyanate (FITC) avidin secondary antibody (1:500) and biotin conjugated goat anti‐avidin (1:100; Vector, Brunschwig Chemie, Amsterdam, The Netherlands). Prior to incubation, aspecific binding sites were blocked with Boehringer Blocking reagent (Roche,
Molecular Diagnostics Inc., South Branchburg, NJ, USA). Cell nuclei were counterstained, and cover slipped with 4,6‐diamidino‐2‐phenylindole dihydrochloride (DAPI; 0.2 μg/mL; Vectashield, Vector Laboratories, Burlingame, CA, USA). Samples were visualized using a DM 5000B fluorescence microscope (Leica, Wetzlar, Germany) coupled to an online digital camera (Leica DC 300 Fx) for independent evaluation of FISH signals by two investigators.
RNA‐in situ hybridization
RNAscope 2.5 HD RED assay (cat. no. 322350; Advanced Cell Diagnostics, Newark, CA, USA) was performed using target probes to MCPyV on tissue sections according to the manufacturer’s instructions and methods followed by Wang et al.24 Human peptidylprolyl isomerase B (Hs‐PPIB) and bacterial dihydrodipicolinate reductase (DapB) were used as positive and negative controls, respectively. FFPE HeLa cells (Advanced Cell Diagnostics) were also used as negative and positive controls, as suggested by Advanced Cell Diagnostics. In addition, we used a FFPE MCC tissue block that previously tested MCPyV‐positive by DNA‐PCR and immunohistochemistry as a positive control. Four investigators independently evaluated the slides.
Immunohistochemistry (IHC) and double staining procedure
We used an anti LT‐antigen MCPyV antibody (clone: CM2B4, dilution 1:50; Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA). Immunohistochemical staining was conducted on a Dako Autostainer Link 48 using the EnVision FLEX Visualization Kit K8008 (Dako, Carpinteria, CA, USA) according to standard diagnostic routine protocols and manufacturer’s instructions.
Results
MCPyV detection by DNA PCR and FISH
A total of 13 of 36 (36.1%) thymomas tested positive using M1/2, 8 (22.2%) tested positive using VP1, and 4 tested positive using LT3 primers (Table 1). Sequence analyses of the PCR amplicons identified all PCR products as MCPyV DNA sequences. In total, 7 (19.4%) of 36 thymomas tested positive for two out of three different MCPyV DNA PCRs (Table 1, Fig 1) and thus were considered MCPyV‐positive. Of these seven MCPyV‐positive thymomas, five had a history of MG. In these five, the presence of MCPyV was confirmed by MCPyV FISH (Fig 2a). FISH analysis revealed relative weak punctuate hybridization signals, which were mostly heterogeneous patterns in all cases tested. None of the follicular hyperplasia or fetal thymus tissues were positive for MCPyV‐DNA.
Figure 1.
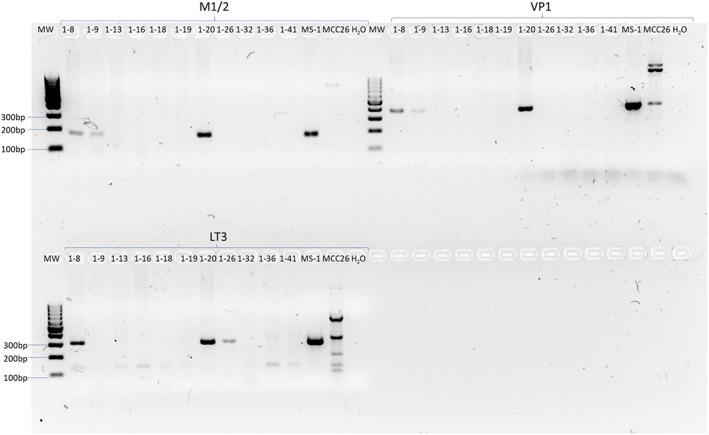
Representative PCR of 11 thymic tissues. Representative results of Merkel cell polyomavirus (MCPyV) M1/2 (178 base pairs [bp]), VP1 (351 bp), and LT3 (308 bp) PCR primer set of 11 human epithelial thymic DNA and two Merkel cell carcinoma cell lines. MS‐1 was used as a positive control and MCC26 as a negative control. Amplification could be observed for VP1 and M1/2 primer sets for ID 1‐8, ID 1‐9 and ID 1‐20. For the LT3 primer set, ID 1‐8, ID 1‐20, and ID‐26, a PCR product could be detected. free, free space; H2O, water control; MW, molecular weight marker.
Figure 2.
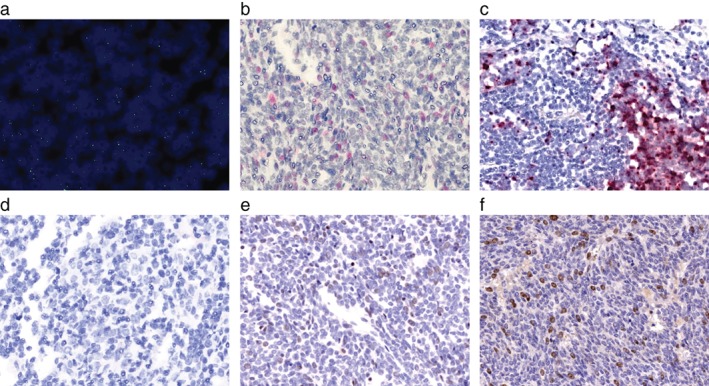
Detection of Merkel cell polyomavirus (MCPyV) and human polyomavirus 7 (HPyV7) in thymoma ID 1‐8. (a) Magnification (63x) an overlay of nuclei 4,6‐diamidino‐2‐phenylindole dihydrochloride staining and MCPyV fluorescence in situ hybridization of thymoma ID 1‐8 shows that MCPyV DNA is present in thymoma nuclei. Single dots per nuclei were detected. (b) RNAscope RNA in situ hybridization (10x) was performed on the thymoma ID 1‐8, (c) MCPyV‐positive Merkel cell carcinoma (MCC) tissue, and (d) and thymoma ID 1‐32. MCPyV LT RNA are detected in (b) thymoma cells, which is comparable to the dot‐like signals in (c) MCPyV‐positive MCC cells. (d) In thymoma ID‐32, no MCPyV RNA is present. Immunohistochemistry (10x) for (e) MPCPyV with the CM2B4 antibody and (f) HPyV7 with the 2t10 antibody reveal MCPyV and HPyV7 expressing thymoma cells. MCPyV expression is weaker than HPyV7.
MCPyV LT IHC and MCPyV LT RNA‐ISH
All seven positive MCPyV thymomas were assessed for MCPyV LT‐antigen expression by IHC. Three out of seven (42.9%) revealed the presence of LT‐antigen in thymoma tissues (Fig 2e, Table 1). The expression of MCPyV LT in these tissues was weak. In addition, three thymoma tissues, two MCPyV‐negative (ID 1–26 and ID 1–32) and one MCPyV‐positive (ID 1–8); and one MCPyV‐positive MCC tissue were analyzed for MCPyV LT‐antigen RNA (Fig 2b,d, Table 1). The MCC cells revealed strong MCPyV RNA detection in the cytoplasm (Fig 2c). ID 1–8 showed weak detection of MCPyV RNA. ID 1–26 and ID 1–32 showed no RNA amplification (Fig 2c, Table 1).
Of interest, thymoma tissue ID 1–8 was positive for both polyomaviruses: HPyV7 and MCPyV (Fig 2f, Table 1), whereas the MCPyV expression was weaker compared to HPyV7.
Discussion
We recently reported the presence of HPyV7 in 46.0% (protein level) to 62.2% (DNA level) of thymomas.6 Herein, we assessed the presence of MCPyV on DNA level by PCR and FISH, on RNA level by RNA‐ISH (RISH), and on protein level by IHC. In order to reliably detect MCPyV in FFPE tissues, Moshiri et al. recently reported that the use of a PCR‐based approach in combination with the CM2B4 anti LT‐antigen antibody yielded the highest specificity and sensitivity.20 In the present study, we used a PCR based approach in combination with MCPyV FISH, RISH, and IHC in order to comprehensively elucidate the presence of MCPyV in the same clinicopathologically well‐classified thymoma cohort in which we recently assessed the presence of HPyV7. None of thymic hyperplasia (n = 20) or fetal thymic tissues were positive for MCPyV assessed by PCR. Overall 19.4% (n = 7) thymomas tested positive for the presence of MCPyV DNA (VP1, M1/2, and LT3). The presence of MCPyV DNA was confirmed in six of seven PCR positive thymomas by FISH analysis. The bands and signals obtained by PCR and FISH were weak. Correspondingly, weak MCPyV LT‐antigen expression on protein level was found in three of seven thymomas. The prevalence of MCPyV was far lower than the prevalence of HPyV7 in these thymomas.
Of all MG‐positive thymomas tested for MCPyV, five were positive on DNA level and three on protein level. Based on this small number, no conclusion can be drawn concerning an association between MCPyV and MG in thymoma. In addition, the weak DNA and protein detection correlates with the weak prevalence of MCPyV RNA compared to the RNA detection of the MCPyV‐positive MCC tissue assessed by RISH in ID 1–8. In contrast to Torrachio et al. and Hashida et al. we used a larger number of MCPyV detection techniques in a larger thymoma cohort, which very likely explains why we were able to identify some MCPyV positive thymoma cases.18,19 However, given the weak DNA amplification and the low protein expression in these few, it can be concluded that MCPyV is weakly present in three thymomas, thus MCPyV is very unlikely to play a role in human thymomagenesis. In the context of our previously reported frequent finding of HPyV7 within the same thymoma cohort, the low prevalence of MCPyV indirectly supports a role of HPyV7 in human thymomagenesis.
Remarkably, most thymoma cases that tested positive for HPyV7 or MCPyV in this cohort were of stage II or III according to the Masaoka classification (Table 1). Eleven of 14 stage II thymomas were positive for one of these human polyomaviruses. In addition, one of the three MCPyV positive tissues was classified as stage IIIB (Table 1). The higher thymoma stages are considered to be more invasive.25, 26 The sTAg of MCPyV was recently observed to provide cells the ability to migrate;27 however, in as much this applies to HPyV7 and thymic epithelial cells currently remains speculation.
Of interest, to the best of our knowledge, we show for the first time the concomitant detection of HPyV7 and MCPyV in the same tumor tissue using diverse molecular techniques. Interestingly, MCPyV and HPyV7 are also present with a high seroprevalence in the same blood.16, 17 Detection of the MCPyV genome by PCR and FISH reveals that MCPyV can infect thymic epithelial cells. In the present study we show that thymic epithelial tumors can harbor both HPyV7 and MCPyV, although the prevalence of MCPyV in human epithelial tumors is far lower than reported for HPyV7. Our data strongly indicate that MCPyV is very unlikely to play an important role in the etiopathogenesis of human thymomas.
Disclosure
No authors report any conflict of interest.
Supporting information
Table S1. Clinicopathological Data of follicular hyperplasia tissue results with human polyomavirus 7 (HPyV7) positivity, as published by Rennspiess et al., 2017. Anti‐AChR, anti‐acetylcholine receptor antibodies; F, female; G., gender; ID, laboratory identification; IS/Ster., immunosuppression/ steroids; M, male; MG, myasthenia gravis; NA, not applicable; Thym. type, thymoma type.
Acknowledgments
This research was supported by RWTH Aachen University through Graduiertenförderung nach Richtlinien zur Förderung des wissenschaftlichen Nachwuchses (RFwN).
References
- 1. Cavalcante P, Barberis M, Cannone M et al Detection of poliovirus‐infected macrophages in thymus of patients with myasthenia gravis. Neurology 2010; 74: 1118–26. [DOI] [PubMed] [Google Scholar]
- 2. McGuire LJ, Huang DP, Teoh R, Arnold M, Wong K, Lee JC. Epstein‐Barr virus genome in thymoma and thymic lymphoid hyperplasia. Am J Pathol 1988; 131: 385–90. [PMC free article] [PubMed] [Google Scholar]
- 3. Inghirami G, Chilosi M, Knowles DM. Western thymomas lack Epstein‐Barr virus by Southern blotting analysis and by polymerase chain reaction. Am J Pathol 1990; 136: 1429–36. [PMC free article] [PubMed] [Google Scholar]
- 4. Sanjuan N, Porrás A, Otero J, Perazzo S. Expression of major capsid protein VP‐1 in the absence of viral particles in Thymomas induced by murine polyomavirus. J Virol 2001; 75: 2891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wirth JJ, Fluck MM. Immunological elimination of infected cells as the candidate mechanism for tumor protection in polyomavirus‐infected mice. J Virol 1991; 65: 6985–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rennspiess D, Pujari S, Keijzers M et al Detection of Human Polyomavirus 7 in human thymic epithelial tumors. J Thorac Oncol 2015; 10: 360–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gheit T, Dutta S, Oliver J et al Isolation and characterization of a novel putative human polyomavirus. Virology 2017; 506: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schadendorf D, Lebbé C, zur Hausen A et al Merkel cell carcinoma: Epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer 2017; 71: 53–69. [DOI] [PubMed] [Google Scholar]
- 9. Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human merkel cell carcinoma. Science (New York, NY) 2008; 319: 1096–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kassem A, Schöpflin A, Diaz C et al Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res 2008; 68: 5009–13. [DOI] [PubMed] [Google Scholar]
- 11. Shuda M, Feng H, Kwun HJ et al T antigen mutations are a human tumor‐specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci 2008; 105: 16272–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Houben R, Adam C, Baeurle A et al An intact retinoblastoma protein‐binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer 2012; 130: 847–56. [DOI] [PubMed] [Google Scholar]
- 13. Pipas JM, Levine AJ. Role of T antigen interactions with p53 in tumorigenesis. Semin Cancer Biol 2001; 11: 23–30. [DOI] [PubMed] [Google Scholar]
- 14. Harms PW, Patel RM, Verhaegen ME et al Distinct gene expression profiles of viral‐ and non‐viral associated Merkel cell carcinoma revealed by transcriptome analysis. J Invest Dermatol 2013; 133: 936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nicol JTJ, Robinot R, Carpentier A et al Age‐specific seroprevalences of Merkel cell polyomavirus, human polyomaviruses 6, 7, and 9, and Trichodysplasia Spinulosa‐associated polyomavirus. Clin Vaccine Immunol: CVI 2013; 20: 363–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Šroller V, Hamšíková E, Ludvíková V, Musil J, Němečková Š, Saláková M. Seroprevalence rates of HPyV6, HPyV7, TSPyV, HPyV9, MWPyV and KIPyV polyomaviruses among the healthy blood donors. J Med Virol 2016; 88: 1254–61. [DOI] [PubMed] [Google Scholar]
- 17. Kamminga S, van der Meijden E, Feltkamp MCW, Zaaijer HL. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS One 2018; 13: e0206273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toracchio S, Foyle A, Sroller V et al Lymphotropism of Merkel cell polyomavirus infection, Nova Scotia, Canada. Emerg Infect Dis 2010; 16: 1702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hashida Y, Imajoh M, Nemoto Y et al Detection of Merkel cell polyomavirus with a tumour‐specific signature in non‐small cell lung cancer. Br J Cancer 2013; 108: 629–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moshiri AS, Doumani R, Yelistratova L et al Polyomavirus‐negative Merkel cell carcinoma: A more aggressive subtype based on analysis of 282 cases using multimodal tumor virus detection. J Invest Dermatol 2017; 137: 819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Dongen JJM, Langerak AW, Brüggemann M et al Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED‐2 Concerted Action BMH4‐CT98‐3936. Leukemia 2003; 17: 2257–317. [DOI] [PubMed] [Google Scholar]
- 22. Haugg AM, Speel E‐JM, Pantulu ND et al Fluorescence in situ hybridization confirms the presence of Merkel cell polyomavirus in chronic lymphocytic leukemia cells. Blood 2011; 117: 5776–7. [DOI] [PubMed] [Google Scholar]
- 23. Haugg AM, Rennspiess D, Hausen AZ et al Fluorescence in situ hybridization and qPCR to detect Merkel cell polyomavirus physical status and load in Merkel cell carcinomas. Int J Cancer 2014; 135: 2804–15. [DOI] [PubMed] [Google Scholar]
- 24. Wang L, Harms PW, Palanisamy N et al Age and gender associations of virus positivity in Merkel cell carcinoma characterized using a novel RNA in situ hybridization assay. Clin Cancer Res 2017; 23: 5622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kolen KV, Pierrache L, Heyman S, Pauwels P, Schil PV. Prognostic factors and genetic markers in thymoma. Thoracic Cancer 2010; 1: 133–40. [DOI] [PubMed] [Google Scholar]
- 26. Huang J, Detterbeck FC, Wang Z, Loehrer PJ. Standard outcome measures for thymic malignancies. J Thorac Oncol 2010; 5: 2017–23. [DOI] [PubMed] [Google Scholar]
- 27. Knight LM, Stakaityte G, Wood JJ et al Merkel cell polyomavirus small T antigen mediates microtubule destabilization to promote cell motility and migration. J Virol 2015; 89: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Clinicopathological Data of follicular hyperplasia tissue results with human polyomavirus 7 (HPyV7) positivity, as published by Rennspiess et al., 2017. Anti‐AChR, anti‐acetylcholine receptor antibodies; F, female; G., gender; ID, laboratory identification; IS/Ster., immunosuppression/ steroids; M, male; MG, myasthenia gravis; NA, not applicable; Thym. type, thymoma type.


