Abstract
Background
The biological function of squalene epoxidase (SQLE), an important rate‐limiting enzyme in downstream cholesterol synthesis, is to convert squalene to 2‐3 oxacin squalene. The expression of SQLE in lung cancer is abnormal. We conducted this study to investigate the effect of SQLE expression on lung squamous cell carcinoma (SCC) proliferation, migration, and invasion and its role in extracellular signal‐regulated kinase (ERK) signaling.
Methods
Cell Counting Kit 8, wound healing, and Transwell assays; Western blotting; and quantitative real‐time PCR were used to investigate the effect of SQLE in a lung SCC H520 cell line. Kaplan–Meier analysis was used to identify the prognostic significance of SQLE.
Results
Overexpression of SQLE promoted lung SCC cell proliferation, migration and invasion, whereas knockdown of SQLE expression showed the opposite effect. SQLE can interact with ERK to enhance its phosphorylation. SQLE may contribute to the pathogenesis of lung cancer by modulating ERK signaling. Further survival analysis indicated that high expression of SQLE indicated poor prognosis in lung SCC.
Conclusion
Our study presents novel evidence of potential biomarkers or therapeutic targets for lung SCC therapy and prognosis.
Keywords: ERK signal, invasion, migration, proliferation, SQLE
Introduction
Lung cancer is the most common malignancy and the leading cause of cancer mortality worldwide. Lacking typical symptoms and effective diagnosis methods in early stages, most lung cancers are discovered at locally advanced or metastatic stages.1, 2 With the advances in next‐generation sequencing (NGS) and other molecular biology technology, research of lung cancer driver genes has made breakthrough progress in the past decade. EGRF‐tyrosine kinase inhibitors, anti‐EGFR monoclonal antibodies, and tumor angiogenesis inhibitors have outstanding efficacy for patients with lung adenocarcinoma.3 However, EGFR‐TKIs are only effective for lung squamous cell carcinoma (SCC) with EGFR mutations, and few SCC patients exhibit such mutations. Recently, immune checkpoint inhibitors have been playing an increasingly important role in lung treatment. However, a series of problems remain in regard to immunotherapy, such as selecting effective patients, improving treatment response rate, and treatment strategies combined with other treatment methods.4 Platinum‐based chemotherapy still is the primary strategy for patients with advanced lung SCC. Although FGFR‐1 amplification mutations, DDR2 mutations, and phosphoinositide 3‐kinase pathway alterations may impact the occurrence and development of lung SCC, there is no doubt that lung SCC has genomic complexity and a high mutation load.5 There are currently no universally accepted driver genes or target drugs; therefore, exploring genomic alterations during the development of lung SCC has distinctive significance for early diagnosis and prognosis.
Metabolic alteration, one of the most important features of cancer cells, and cancer development display mutual causality.6, 7 Rapidly growing cancer cells require large amounts of cholesterol to satisfy cell membrane synthesis and mevalonate biosynthesis. Metabolic reprogramming has recently been considered a significant tumor hallmark that plays an important role in the development of cancer.8, 9 Moreover, abnormal accumulation of cholesterol is associated with tumor growth and patient survival.10, 11 Squalene epoxidase (SQLE), codified by the human SQLE gene, and located on human chromosome 8q24.1, is one of key rate‐limiting enzymes in downstream cholesterol synthesis.12 The biological function of SQLE is to convert squalene to 2‐3 oxacin squalene, which can synthesize more important materials, for example, sterol and cholesterol. Abnormal SQLE expression has been reported in some tumors, including breast, pancreatic, liver, and prostate cancers, and lung SCC.13, 14, 15, 16, 17 SQLE may be involved in the development of cancer; however, the underlying biological mechanisms of SQLE in lung SCC require further exploration. Thus, we investigated the effect of SQLE expression on lung SCC proliferation, migration, and invasion and its role in extracellular signal‐regulated kinase (ERK) signaling.
Methods
Cell culture
H520 cells were cultured in RPMI 1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum and 1% streptomycin and penicillin. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere.
Plasmids and transfection
The cells were grown on six‐well plates for 18–24 hours. Transfections were performed using DNAfectin 2100 (Applied Biological Materials Inc., Richmond, BC, Canada). SQLE overexpression plasmids (SQLE) and negative control plasmids (CON083) were obtained from Genechem Co. Ltd (Shanghai, China).
Small interfering RNA and transfection
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used as a reagent to perform transient transfection of small interfering RNAs (siRNAs). SQLE1640 siRNA and control SQLE1853‐siRNA were obtained from GenePharma (Shanghai China).
Reverse transcriptase‐PCR and real‐time PCR
TRIzol Reagent (Invitrogen) was used to isolate total RNA and then reverse transcribed into complementary (c)DNA using the Reverse Transcription System (Promega, Madison, WI, USA); 1 μg RNA was then reversed into cDNA using a ReverTra Ace qPCR Kit (FSQ‐101, Toyobo Co. Ltd., Osaka, Japan). Quantitative real‐time (qRT)‐PCR was carried out using a Bio‐Rad Real‐Time PCR system (Hercules, CA, USA). The sequences of the real‐time primers for SQLE were as follows: forward sequence: 5′‐CTCCAAGTTCAGGAAAAGCCTGG‐3′, reverse sequence: 5′‐GAGAACTGGACTCGGGTTAGCT‐3′. Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as a reference control. The GADPH sequences were as follows: forward sequence: 5′‐CATCACCATCTTCCAGGAGCG‐3′, reverse sequence:5′‐CATCACCATCTTCCAGGAGCG‐3′.
Western blot analysis
Radioimmunoprecipitation assay reagent (Solarbio, Beijing, China) was used to extract protein from different cell groups. Aliquots containing 50 μg of total protein were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membrane, and then locked via non‐specific binding with 5% non‐fat milk. Antibodies against SQLE (1:1000) and p‐ERK (1:1000; Santa Cruz, Biotechnology Inc., Santa Cruz, CA, USA), and GADPH (1:5000, Abcam, Cambridge, MA, USA), respectively, were added and the cells were incubated at 4°C overnight. A secondary antibody was added and cells were incubated at 37°C for two hours. GAPDH was used as the endogenous control. Protein bands were analyzed using enhanced chemiluminescence (ECL) and the signal intensity was measured using Image Lab Software version 5.2.1 (Bio‐Rad) to calculate protein levels.
Cell proliferation assay
Different groups of cells were plated in 96‐well plates (2 × 103 cell/well) after 18–24 hours of transfection. Cell proliferation was assessed every 24 hours using Cell Counting Kit 8 (CCK‐8; Boster, Biological Technology Co. Ltd., Pleasanton, CA, USA). Each experiment was repeated three times.
Cell migration assay
Cell migration was performed by wound healing assay. After 18–24 hours of transfection, cells were plated in six plates (5 × 103 cell/well). When cells were overgrown with a single layer, a 0.10 uL pipetting gun was used to draw a horizontal line in the middle. The floating cells were washed with phosphate buffered saline and serum‐free RPMI 1640 medium was added and the cells were then left to culture for 48 hours. Image J version 1.8.0 (NIH, Bethesda, MD, USA) was used to measure the results of the scratch test. Each experiment was repeated three times.
Cell invasion assay
Cell invasion assays were performed in 24‐well chambers (Corning, Christiansburg, VA, USA) coated with matrigel and serum‐free RPMI 1640 medium. Cells (3–4 × 104 cells/well) in 180 μL serum‐free RPMI 1640 were seeded into the upper chamber, and 600 μL RPMI 1640 with 10% fetal bovine serum was added into the lower chamber. Cells in the upper chamber were removed using a cotton swab after culturing at 37°C in a humidified 5% CO2 atmosphere for 48 hours. Cells attached to the bottom of the membranes were fixed with paraformaldehyde and stained with 1% crystal violet. Each experiment was repeated three times.
Statistical analysis
One‐way analysis of variance was used to compare cell proliferation, migration, and invasion after transfection. ONCOMINE (https://www.oncomine.org) was used to analyze the expression of SQLE in different cancers. To identify the prognostic value of SQLE, Kaplan–Meier survival curves and log‐rank analysis were conducted. Statistical analysis was conducted using Kaplan–Meier plotter databases. All statistical analyses were performed using SPSS version 24.0 (IBM Corp., Armonk, NY, USA). P < 0.05 was considered statistically significant.
Results
Squalene epoxidase (SQLE) expression is upregulated in cancer tissue
Bioinformatic analyses of gene expression data in the ONCOMINE database revealed that SQLE was significantly highly expressed in a variety of tumor tissues, including breast, pancreas, liver, prostate, and lung SCC (Fig 1). SQLE was significantly elevated compared to normal tissue in lung SCC. This prompted us to further examine SQLE activity in lung cancer development.
Figure 1.
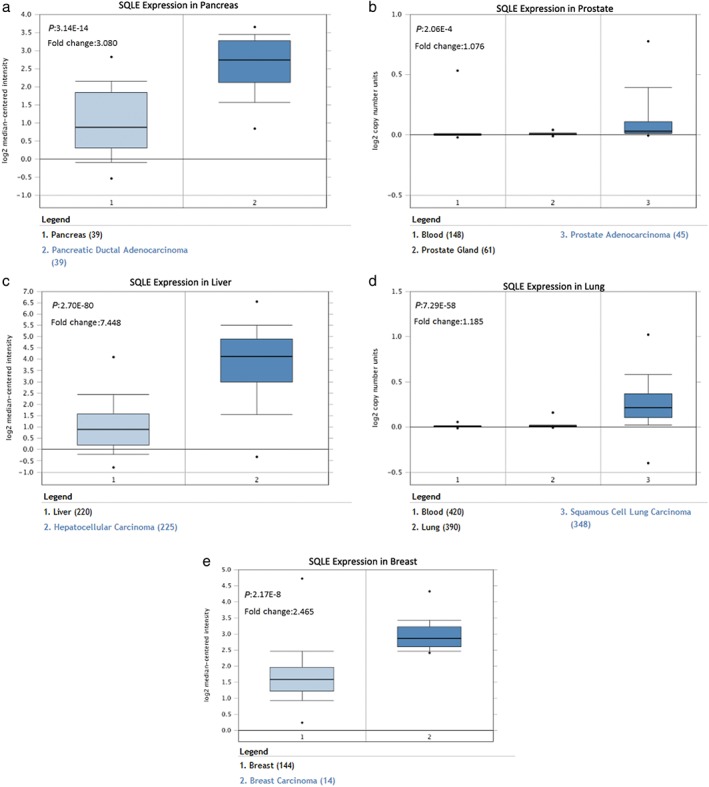
Squalene epoxidase (SQLE) expression levels in pancreatic ductal adenocarcinoma (a), prostate adenocarcinoma (b), hepatocellular carcinoma (c), lung SCC (d), breast carcinoma (e) based on data from the ONCOMINE database.
Overexpression of SQLE promoted proliferation, migration, and invasion of lung SCC H520 cell
To evaluate the functional consequence of SQLE expression in lung SCC, we monitored changes in lung SCC H520 cell behavior following alteration of expression levels. qRT‐PCR and Western blot from RNA and protein levels, respectively, showed SQLE overexpression in cancer cell lines after transfection (Fig 2a–c). Overexpression of SQLE promoted lung squamous cell H520 proliferation, migration, and invasion (Fig 2d–f).
Figure 2.
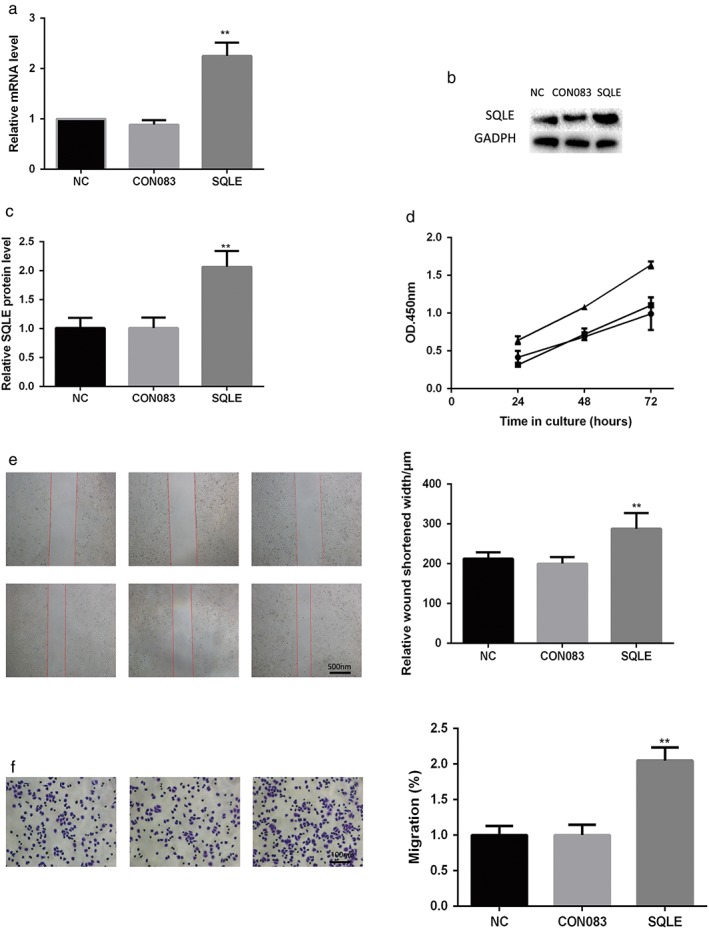
Squalene epoxidase (SQLE) overexpression promoted proliferation, migration, and invasion of lung squamous cell carcinoma (SCC) H520 cells. SQLE overexpression was verified by (a) quantitative real time‐PCR and (b,c) Western blotting. (d) Lung cancer cell proliferation was assayed using Cell Counting Kit 8.  Negative control (NC);
Negative control (NC);  CON083;
CON083;  SQLE. (e) Cell migration was detected by wound healing assay. (f) Cell invasion was detected by transwell assay. Each assay was repeated at least three times. Data from one representative experiment is presented as mean ± standard deviation. **, P < 0.01. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; mRNA, messenger RNA; OD, optical density.
SQLE. (e) Cell migration was detected by wound healing assay. (f) Cell invasion was detected by transwell assay. Each assay was repeated at least three times. Data from one representative experiment is presented as mean ± standard deviation. **, P < 0.01. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; mRNA, messenger RNA; OD, optical density.
Inhibiting SQLE expression reduced proliferation, migration, and invasion of lung SCC H520 cells
To further investigate the effects of SQLE on lung cancer cell growth, migration, and invasion, SQLE expression was knocked down in the H520 lung SCC cell line, which had a relatively high level of expression of endogenous SQLE. Meanwhile, we used qRT‐PCR and Western blot to examine RNA and protein levels, respectively, to detect the efficiency of downregulated SQLE (Fig 3a–c). Knockdown of SQLE cleary suppressed the proliferation, migration, and invasion of lung SCC H520 cells (Fig 3d–f).
Figure 3.
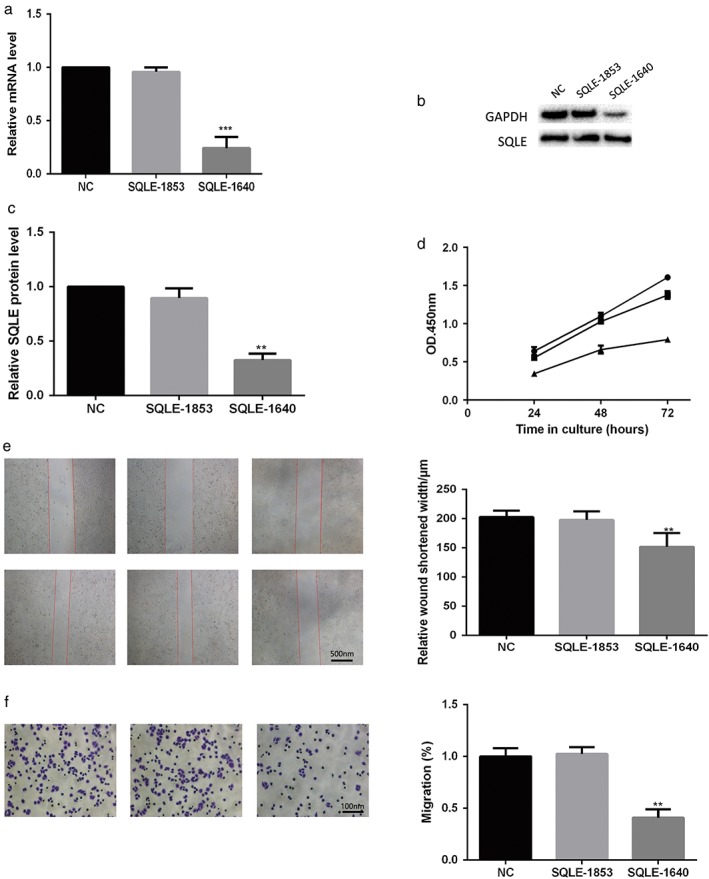
Inhibition of squalene epoxidase (SQLE) expression reduced proliferation, migration, and invasion in lung squamous cell carcinoma (SCC) H520 cells and was verified by (a) quantitative real time‐PCR and (b,c) Western blotting. (d) Lung cancer cell proliferation was assayed using Cell Counting Kit 8. Negative control (NC) ;
;  SQLE‐1853;
SQLE‐1853;  SQLE‐1640. (e) Cell migration was detected by wound healing assay. (f) Cell invasion was detected by transwell assay. Each assay was repeated at least three times. Data from one representative experiment is presented as mean ± standard deviation. **, P < 0.01 ***, P < 0.001. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; mRNA, messenger RNA; OD, optical density.
SQLE‐1640. (e) Cell migration was detected by wound healing assay. (f) Cell invasion was detected by transwell assay. Each assay was repeated at least three times. Data from one representative experiment is presented as mean ± standard deviation. **, P < 0.01 ***, P < 0.001. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase; mRNA, messenger RNA; OD, optical density.
Alteration of SQLE expression regulated extracellular signal‐regulated kinase phosphorylation levels
According to reports, the derivates of mevalonate play a crucial role in the activation of ERK signaling.18 It is well known that ERK signaling is critically involved in cancer development. In order to investigate the possible molecular mechanism of SQLE in cancer development, we examined the ERK signaling level when SQLE expression was altered. SQLE overexpression in H520 cells significantly stimulated ERK phosphorylation, indicating activation of the ERK signaling pathway (Fig 4a,b). Consistent with these observations, inhibiting the expression of SQLE in H520 cells effectively downregulated the phosphorylation level of ERK (Fig 4c,d). In summary, these results revealed that alteration of SQLE expression regulated ERK phosphorylation levels. SQLE may promote the malignant behavior of lung SCC by activating the ERK singling pathway.
Figure 4.
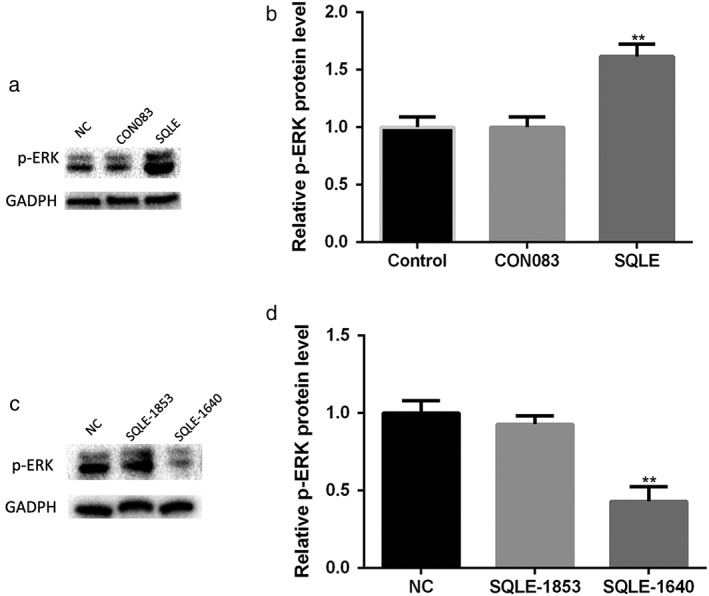
Alteration of squalene epoxidase (SQLE) expression regulated extracellular signal‐regulated kinase (ERK) phosphorylation levels. (a,b) Overexpression of SQLE stimulated the phosphorylation of ERK in lung squamous cell carcinoma (SCC) H520 cells. (c,d) SQLE knockdown inhibited ERK phosphorylation in lung SCC H520 cells. **, P < 0.01. GAPDH, glyceraldehyde 3‐phosphate dehydrogenase.
Survival analysis
To further explore the impact of SQLE expression levels on lung SCC patient survival, we performed survival analysis using the Kaplan–Meier plotter databases. Information of lung SCC patients was obtained from The Cancer Genome Atlas database. Samples were divided into high and low expression groups and survival analysis was performed. The results indicated that SQLE was obviously associated with the survival of lung SCC patients. Higher SQLE indicated shorter overall survival (Fig 5), suggesting that high expression of SQLE is not conducive to prognosis in patients with lung SCC.
Figure 5.
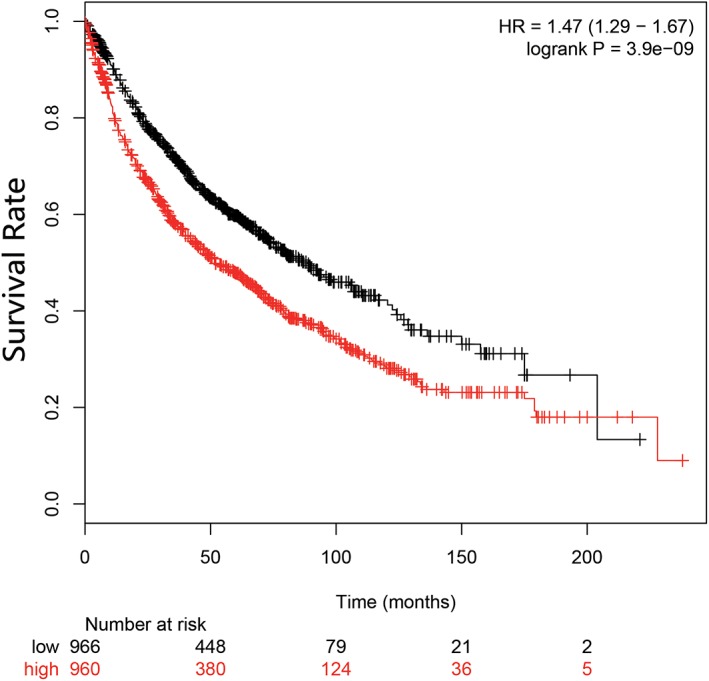
Kaplan–Meier plots for squalene epoxidase (SQLE) in lung squamous cell carcinoma (SCC).  low;
low;  high. HR, hazard ratio.
high. HR, hazard ratio.
Discussion
The occurrence and development of cancer is extremely complicated. Although therapeutic strategies for early‐stage SCC are similar to other histologic subtypes of NSCLC, there are limited therapeutic options for advanced lung SCC in comparison to lung adenocarcinoma. This is in part because of the complexity of lung SCC driver genes. There is still no universally accepted targeted therapy for advanced lung SCC, in which a biomarker is used to select patients most likely to benefit. With the vigorous development of bioinformatic and gene test technologies, increasing research on tumor driver genes is being conducted.
SQLE, a monooxygenase, is one of the key rate‐limiting enzymes of cholesterol synthesis. It is involved in cholesterol biosynthesis, cellular metabolism, and the regulation of organ specificity. The results of our bioinformatic analyses and the findings of previous studies show that SQLE is abnormally expressed in various types of cancer, including breast, pancreatic, liver, and prostate cancers, and lung SCC.13, 14, 15, 16, 17 SQLE is significantly elevated in lung SCC compared to normal tissue. It has been reported that high expression of SQLE can increase the risk of early metastasis in estrogen receptor‐positive breast cancer.13 SQLE, frequent Myc gene amplification, and DNA hypomethylation, have been identified as a genuine metabolic oncogenes in breast cancer.14 In pancreatic cancer, SQLE expression is associated with radioresistance in vitro.15 In colorectal cancer, SQLE expression is associated with patient prognosis.16 Previous studies have shown that SQLE expression in lung SCC is significantly higher than matched paracancerous tissue and is related to prognosis.17 These results suggest that SQLE is involved in the occurrence and development of lung SCC.
We explored the role of SQLE by regulating the expression of SQLE in a lung SCC H520 cell line. CCK‐8, wound healing, and Transwell assays were used to investigate the effect of SQLE on proliferation, migration, and invasion, respectively. The results revealed that enforced expression of SQLE caused accelerated lung SCC cell proliferation, migration, and invasion, while inhibition of SQLE expression reduced the malignant behavior of lung SCC cells. These results suggest that SQLE plays a role in the occurrence and development of lung SCC and may have oncogenic potential.
At the molecular level, SQLE regulated ERK signaling and promoted ERK‐phosphorylation, which in turn appears to be closely associated with its tumor‐promoting activity. However, the specific details of the mechanisms of SQLE activation of the ERK signaling pathway require further exploration. Hydroxymethylglutaryl coenzyme A reductase, another rate‐limiting enzyme in the cholesterol synthesis pathway, could regulate ERK signals in the development of esophageal squamous cell carcinoma (ESCC),19 suggesting that it may promote cancer development via the ERK signal pathway. The mevalonate metabolic pathway is upstream of the cholesterol synthesis pathway. In addition to providing downstream products for cholesterol synthesis, the mevalonate metabolic pathway also produces intermediates, such as farnesyl pyrophosphate and geranylgeranyl pyrophosphate, which are responsible for modifying Ras, Rho, Cdc42, and so on. Ras could activate the ERK signaling pathway and cause rapid cell proliferation.20 Rho and Cdc42 could enhance cell migration and invasion by regulating cytoskeletal rearrangement.21 Moreover, in our study, SQLE was observed to activate ERK signaling, which has been reported as an effector of the mevalonate pathway.
Extracellular signal regulated kinase (ERK) is an important part of mitogen‐activated protein kinase (MAPK), which plays an important role in regulating various physiological processes, such as cell growth, reproduction, and apoptosis. ERK protein is transferred to the nucleus to regulate cell transcription and phosphorylation and then participates in the activation of various enzymes, promoting cell proliferation and differentiation after the activation of the Ras‐Raf‐MEK pathway.22 Activation of ERK can also promote EGFR ligands and forms the key link for tumor growth. Therefore, EGFR/Ras/Raf/MEK/ERK is an important pathway for tumor growth, and the key molecule in this signaling pathway is expected to become a new strategy for targeted cancer treatment.
Furthermore, the results of survival analysis indicate that SQLE is associated with survival of patients with lung SCC. High expression of SQLE was significantly associated with shorter overall survival (Fig 5). This suggests that high expression of SQLE is not conducive to the prognosis of patients with lung SCC. SQLE expression might be used as an index for the prognosis of lung SCC.
In conclusion, we have provided evidence of the effect of SQLE on lung SCC occurrence and development. SQLE promotes lung SCC cell proliferation, migration, and invasion by interacting with the ERK signaling pathway. Our findings present novel evidence of potential biomarkers or targets for lung SCC therapy and prognosis.
Disclosure
No authors report any conflict of interest.
Acknowledgment
The authors wish to thank the Department of Oncology, Affiliated Hospital of Qingdao University for their support of this study.
References
- 1. Hoffman PC, Mauer AM, Vokes EE. Lung cancer. Lancet 2000; 355: 479–85. [DOI] [PubMed] [Google Scholar]
- 2. Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med 2004; 350: 379–92. [DOI] [PubMed] [Google Scholar]
- 3. Kumar M, Ernani V, Owonikoko TK. Biomarkers and targeted systemic therapies in advanced non‐small cell lung cancer. Mol Aspects Med 2015; 45: 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haanen JB, Robert C. Immune checkpoint inhibitors. Prog Tumor Res 2015; 42: 55–66. [DOI] [PubMed] [Google Scholar]
- 5. Gandara DR, Hammerman PS. Squamous cell lung cancer: From tumor genomics to cancer therapeutics. Clin Cancer Res 2015; 21: 2236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kroemer G, Pouyssegur J. Tumor cell metabolism: Cancer’s Achilles’ heel. Cancer Cell 2008; 13: 472–82. [DOI] [PubMed] [Google Scholar]
- 7. Silvius JR, del Giudice D, Lafleur M. Cholesterol at different bilayer concentrations can promote or antagonize lateral segregation of phospholipids of differing acyl chain length. Biochemistry 1996; 35: 15198–208. [DOI] [PubMed] [Google Scholar]
- 8. Silvente‐Poirot S, Poirot M. Cholesterol metabolism and cancer: The good, the bad and the ugly. Curr Opin Pharmacol 2012; 12: 673–6. [DOI] [PubMed] [Google Scholar]
- 9. Hu J, Locasale JW, Bielas JH et al Heterogeneity of tumor‐induced gene expression changes in the human metabolic network. Nat Biotechnol 2013; 31: 522–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhuang L, Kim J, Adam RM, Solomon KR, Freeman MR. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J Clin Invest 2005; 115: 959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brusselmans K, Timmermans L, Van de Sande T et al Squalene synthase, a determinant of Raft‐associated cholesterol and modulator of cancer cell proliferation. J Biol Chem 2007; 282: 18777–85. [DOI] [PubMed] [Google Scholar]
- 12. Nagai M, Sakakibara J, Wakui K et al Localization of the squalene epoxidase gene (SQLE) to human chromosome region 8q24.1. Genomics 1997; 44: 141–3. [DOI] [PubMed] [Google Scholar]
- 13. Helms MW, Kemming D, Pospisil H et al Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer 2008; 99: 774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brown DN, Caffa I, Cirmena G et al Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep 2016; 18: 619435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Souchek JJ, Baine MJ, Lin C et al Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer 2014; 111: 1139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuen HF, McCrudden CM, Huang YH, Tham JM et al TAZ expression as a prognostic indicator in colorectal cancer. PloS One 2013; 8: e54211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang HY, Li HM, Yu Z, Yu XY, Guo K. Expression and significance of squalene epoxidase in squamous lung cancerous tissues and pericarcinoma tissues. Thorac Cancer 2014; 5: 275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang T, Seah S, Loh X et al Simvastatin‐induced breast cancer cell death and deactivation of PI3K/Akt and MAPK/ERK signalling are reversed by metabolic products of the mevalonate pathway. Oncotarget 2016; 7: 2532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong C, Fan L, Yao F, Shi J, Fang W, Zhao H. HMGCR is necessary for the tumorigenecity of esophageal squamous cell carcinoma and is regulated by Myc. Tumor Biol 2014; 35: 4123–9. [DOI] [PubMed] [Google Scholar]
- 20. Luan Z, He Y, Alattar M, Chen Z, He F. Targeting the prohibitin scaffold‐CRAF kinase interaction in Ras‐ERK‐driven pancreatic ductal adenocarcinoma. Mol Cancer 2014; 13: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luckashenak N, Wähe A, Breit K, Brakebusch C, Brocker T. Rho‐family GTPase Cd42 controls migration of Langerhans cell in vivo. J Immunol 2013; 190: 27–35. [DOI] [PubMed] [Google Scholar]
- 22. You B, Yang YL, Xu Z et al Inhibition of ERK1/2 down‐regulates the Hippo/YAP signaling pathway in human NSCLC cells. Oncotarget 2015; 6: 4357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]


