Abstract
Purpose
Fluoroquinolone-resistant (FQR) Escherichia coli causes transrectal prostate biopsy infections. In order to reduce colonization of these bacteria in carriers, we would like to understand the surrounding microbiome to determine targets for decolonization.
Materials and Methods
We perform an observational study to investigate the microbiome differences in men with and without FQR organisms found on rectal culture. A rectal swab with two culturettes was performed on men before an upcoming prostate biopsy procedure as standard of care to perform “targeted prophylaxis.” Detection of FQR was performed by the standard microbiology lab inoculates the swab onto MacConkey agar containing ciprofloxacin. The extra swab was sent for 16S rRNA amplicon sequencing (MiSeq paired-end) using the V1V2 primer. Alpha and beta-diversity analysis were performed using QIIME. We used PERMANOVA to evaluate the statistical significance of beta-diversity distances within and between groups of interest.
Results
We collected 116 rectal swab samples before biopsy for 16S rRNA amplicon sequencing. We identified 18 isolates (15.5%, 18/116) that were positive and had relative reduced diversity profiles (p<0.05). Enterobacteriaceae were significantly over-represented in the FQR subjects (adjusted p=0.03).
Conclusions
Microbiome analysis determined that men colonized with FQR bacteria have less diverse bacterial communities (dysbiosis), higher levels of Enterobacteriaceae and reduced levels of Prevotella disiens. These results may have implications in pre/probiotic intervention studies.
Keywords: Anti-bacterial agents, Escherichia coli, Infection, Microbiota, Prostate
INTRODUCTION
Transrectal ultrasound-guided prostate needle biopsy (TPB) under local anesthetic is the most common technique used to assess for prostate cancer. Studies estimate that nearly 1 million are performed in the United States each year [1]. Historically, TPB has a low infection risk (less than 1%) when done using recommended fluoroquinolone prophylaxis. Rates of post-TPB infection have increased to 6% with more men requiring hospitalization due to related sepsis over the last decade [2].
The cause of the rising infection rates is due to the rapid increase in fluoroquinolone-resistant (FQR) Escherichia coli. These resistant bacteria are colonized in the rectum in 15% to 22% of men at the time of prostate biopsy [3]. The rising incidence of infections has led urologists to either switch to non-fluoroquinolone antibiotic prophylaxis or add more antibiotics, despite an absence of supporting evidence. The concern is that multiple broad-spectrum antibiotics used for a single outpatient procedure may worsen the current critical issue of antibiotic resistance. We utilize a “targeted prophylaxis” approach by obtaining a rectal culture 1 to 2 weeks before the biopsy to direct alternative prophylaxis [4]. We now can detect those men colonized with FQR E. coli before a prostate biopsy.
Our objective was to evaluate the gut microbiome of individuals positive or negative for FQR E. coli to identify bacterial members associated with resistance to FQR colonization. The results could have future implications for reducing FQR before prostate biopsy and reducing infections by non-antibiotic means.
MATERIALS AND METHODS
1. Study design
The study is an observational study to investigate the microbiome differences in men with and without FQR organisms found on rectal culture. A rectal swab was performed on men before an upcoming prostate biopsy procedure as standard of care to perform “targeted prophylaxis.” The culturette had two cotton swabs per sample. We sent one swab to microbiology as standard protocol. The second swab was being stored in a 15 mL conical tube with phosphate-buffered saline (PBS) and transferred to our lab for microbiome processing in our genomics core. Rectal swabs have been determined equivalent to stool culture and are easier to obtain in the office, which improves protocol compliance [5]. Moreover, rectal swabs will be able to be extrapolated to clinical practice. We queried patients and their charts regarding antibiotic exposure in the past year.
2. Population
After institutional review board (IRB) approval (approval number: 05-234H) to obtained rectal swabs stored in our genitourinary biorepository for microbiome collection. Men presented to the South Texas Veterans Health Care System Audie Murphy Division Hospital for evaluation for a “for cause” biopsy usually consisting of elevated PSA or abnormal prostate exam. After shared decision making, the patient and physician determined the indication for a biopsy and the patient would undergo a rectal swab collection to investigate for colonization of FQR E. coli as the standard of care. Physicians chose antibiotic prophylaxis that included ciprofloxacin as baseline and targeted augmented prophylaxis to all patients with fluoroquinolone resistance. We requested banked genitourinary biorepository rectal swabs for research with IRB approval to access the specimens with associated demographics and particularly any antibiotic use in the last year. Those men currently using antibiotics were excluded.
3. Rectal culture
The Liquid Stuart Medium swab and transport system (Copan Diagnostics, Murrieta, CA, USA) was obtained at the time of standard digital rectal examination of the prostate and sent to microbiology. The microbiology lab inoculates the swab onto MacConkey agar containing 10 µg/mL of ciprofloxacin (Hardy Diagnostics, Santa Maria, CA, USA). As a control, the sample was also inoculated onto regular MacConkey agar to ensure that enteric bacteria were indeed on the swab. If after the 24-hour incubation there was no growth on the ciprofloxacin-infused MacConkey agar and there was a growth of normal flora on the other agars, the rectal flora was assumed to be ciprofloxacin-susceptible. Conversely, any Gram-negative (GN) rods growing on the ciprofloxacin-infused MacConkey agar were presumed to be ciprofloxacin-resistant. We analyzed a representative of each distinct colony morphotype using the Vitek 2 analyzer (BioMerieux, Durham, NC, USA) for identification by GN cards and sensitivity testing by antibiotic susceptibility testing cards using Clinical and Laboratory Standards Institute Interpretative Criteria [6].
4. DNA isolation
After collection, the rectal swab is placed in 500 microliters of PBS in a sterile 15 mL conical tube to rinse off the particulate stool. The stool slurry is then transferred to a 1.7 mL microcentrifuge tube. The stool slurry is centrifuged at 12,000g for 5 minutes and the volume of PBS is reduced to 200 microliters. The DNA is subsequently isolated from the 200 microliters of stool specimen using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, USA).
5. Microbiome
Genomic DNA was quantified and the V1–V2 variable region of the 16S rRNA genes amplified with custom-designed primers (F27/R355). The forward Bosshard sequence was AGAGTTTGATCMTGGCTCAG (27F) and the reverse Bosshard sequence was GCTGCCTCCCGTAGGAGT (355R) with the amplicon size of V1–V2 about 320 bp (355-27). Libraries for all samples were prepared and sequenced by Paired-end sequencing (2×300 bp) using the Illumina MiSeq platform. Raw sequences were quality trimmed (Q20) by sickle, and reads shorter than 200 bases were removed. Therefore, only the forward read was used for this analysis. The trimmed sequences were exported as FASTA files.
1) Sample size calculation
Power calculations for 16S amplicon and whole-genome shotgun microbiome studies are particularly challenging, as there is limited uniform data to analyze and uncertainty in the distribution of bacterial taxa throughout the human population [7]. The Human Microbiome Project (HMP) has released a large uniform 16S amplicon survey of gut-associated samples from healthy individuals [8,9,10]. This publicly available data serves as a potential resource for power-analyses by allowing for empirical calculation of the expected variability of taxa in the healthy distal gut microbiome. We obtained taxonomic annotations from 16S profiles of 146 HMP fecal samples and examined the variability of genera among individuals. After subsampling read sets to achieve equivalent sequencing depth across all sample, we identified 27 genus-level groups that on average each represented at least 0.5% of 16S sequences from the gut microbiome. We then computed the associated variance in relative abundance for each of these 27 genera, and used the empirical values to perform a comprehensive power-analysis. For each genus, we estimated the power for a range of proportional shifts in average relative abundance. As expected, increased statistical power was found with higher abundance genera, so here we focus on power results for organisms reflecting an average of 0.5% to 1.0% of 16S sequences in the gut microbiome because detecting differences in these rare taxa may be critical to our study. Among the 10 genera found at this low level in the HMP data, if we assume equal variance in two normally-distributed treatment populations, and require a significance level of α=0.05, we expect that with at least 15 samples per treatment group, we will be able to detect a relative increase of a factor of 4 (e.g., 0.5% mean abundance increased to 2.5%, or 1% mean abundance increased to 5%), with a statistical power of 93.8%. Considering the genera with average frequency of 1% to 10%, we estimate that with at least 15 individuals per treatment group, we will be able to detect a relative increase of a factor of 3 (e.g., 1% mean abundance increased to 4%, or 5% mean abundance increased to 20%), with an expected statistical power of 96.9%.
2) Statistical analysis
Raw paired-end 16S rRNA reads (V1V2 region) were merged into consensus fragments by FLASH [11] and subsequently filtered for quality (target error rate <0.5%) and length (minimum 200 bp) using Trimmomatic and QIIME [12]. Spurious hits to the PhiX control genome were identified using BLASTN and removed. Passing sequences were trimmed of primers, evaluated for chimeras with UCLUST (de novo mode), and screened for human-associated contaminant using Bowtie2 followed by a more sensitive BLASTN search against the GreenGenes 16S rRNA database [13]. Chloroplast and mitochondrial contaminants were detected and filtered using the RDP classifier with a confidence threshold of 80%. High-quality 16S rRNA sequences were assigned to a high-resolution taxonomic lineage using Resphera Insight. Differential abundance analysis utilized the nonparametric permutation difference test for alpha diversity or the negative binomial test (DESeq) for taxonomic count data. The p-values were adjusted using the False Discovery Rate (FDR) [14]. Alpha and beta-diversity analyses were performed using QIIME. The Mann-Whitney U test was employed to evaluate statistical significance of beta-diversity distances within and between FQR and FQS groups.
RESULTS
1. Fluoroquinolone resistance
We collected 131 rectal swab samples from men scheduled to undergo a prostate biopsy, of which 13 (9.9%) were not evaluable. Eight patients did record having antibiotics in the last 6 months and were investigated in the context of those without previous antibiotic use, which did not show a difference (p>0.05). Of the clinical cultures, we identified 18 isolates (15.5%, 18/116) that were FQR E. coli positive (Fig. 1). All bacteria were E. coli except one Klebsiella pneumoniae. Two of the E. coli which were extended beta lactamase producers. Demographics are displayed in Table 1. We did note that obesity by measure of body mass index was more prominent in the fluoroquinolone resistant group (p=0.04). No prostate biopsy infections were recorded.
Fig. 1. Consort flow diagram. FQR, fluoroquinolone-resistant.
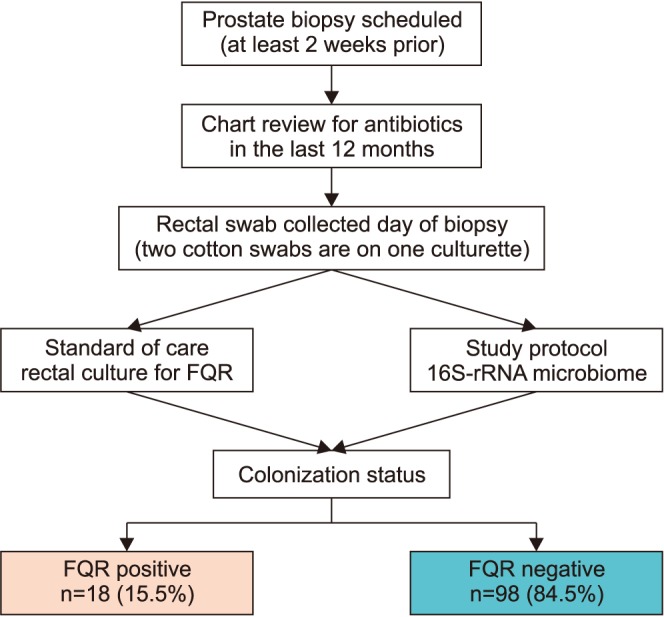
Table 1. Demographics.
| Demographic | FQR positive | FQR negative | p-value |
|---|---|---|---|
| Sample size | 18 | 98 | |
| Race/ethnicity | 0.08 | ||
| White | 9 (50.0) | 50 (51.0) | |
| African-American | 1 (5.6) | 29 (29.6) | |
| Hispanic | 3 (16.7) | 17 (17.3) | |
| Unknown | 3 (16.7) | 5 (5.1) | |
| Age | 65 (61–69) | 68 (63–70) | 0.2 |
| Body mass index | 30 (27–33) | 28 (25–29) | 0.04 |
| Diabetes | 3 (16.7) | 32 (32.7) | 0.3 |
| BPH | 8 (44.4) | 37 (37.8) | 0.3 |
| Previous BPH surgery | 0 (0.0) | 2 (2.0) | |
| Prostate size on ultrasound | 43 (30–63) | 37 (29–45) | 0.8 |
| Number of previous biopsies | 0.4 | ||
| 1 | 6 (33.3) | 21 (21.4) | |
| 2 | 3 (16.7) | 15 (15.3) | |
| 3+ | 1 (5.6) | 4 (4.1) | |
| Family history of prostate cancer | 5 (27.8) | 20 (20.4) | 0.5 |
| Diagnosed with prostate cancer | 8 (44.4) | 55 (56.1) | 0.7 |
| Antibiotic use within last 6 months | 2 (11.1) | 6 (6.1) | 0.6 |
Values are presented as number only, number (%), or median (interquartile range).
FQR, fluoroquinolone-resistant; BPH, benign prostate hypertrophy.
2. Microbiome diversity in FQR+ carriers
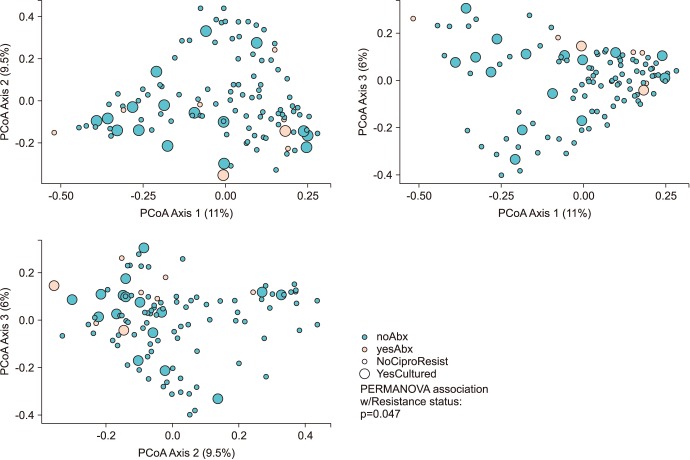
Men positive for FQR E. coli maintained a relatively reduced alpha diversity compared to non-FQR subjects (inverse Simpson; p=0.05). The association of microbial community membership with FQR status was found to be significant for Bray-Curtis (p=0.047) and weighted-UniFrac measures (p=0.01, Fig. 2).
Fig. 2. Beta diversity principal components analysis.
3. Microbiome species analysis
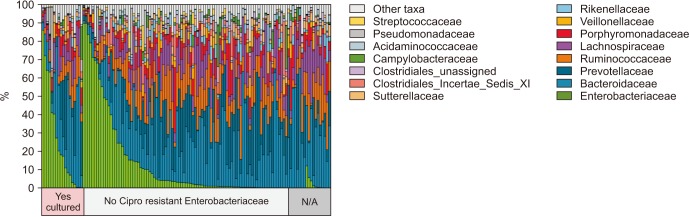
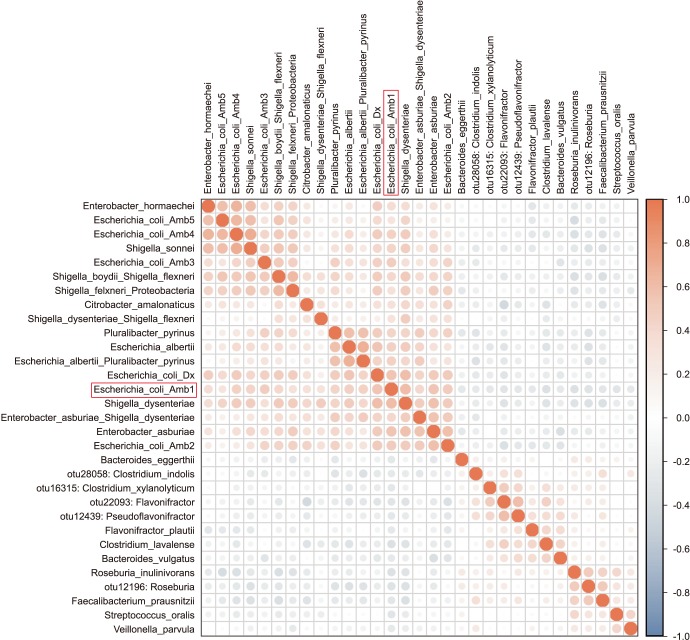
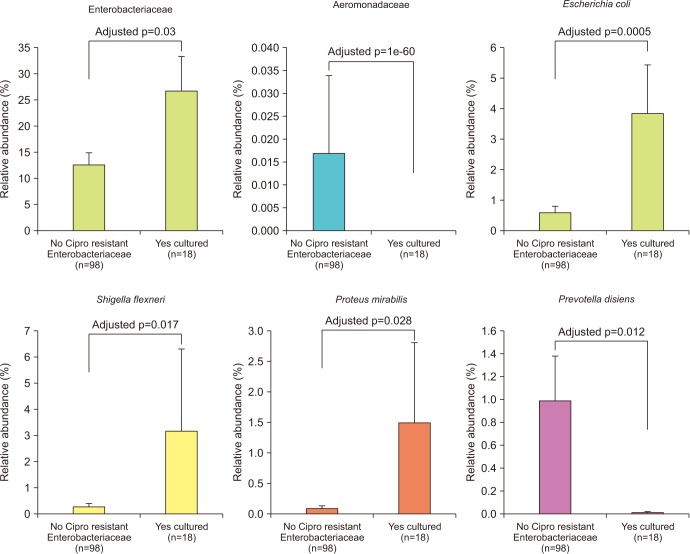
Enterobacteriaceae isolates were significantly overrepresented in the FQR subjects (adjusted p=0.03, Fig. 3). In correlation analysis, we investigate the tope 30 most statistically significant correlations between the dominant E. coli signal (E. coli Amb1) and other well represented species using non-parametric Spearman correlations (all taxa shows p<0.004, Fig. 4). We note consistent positive correlations with other Enterobacteriaceae and negative correlations with Clostridia and Bacteroides members. The bacterial family Aeromonadaceae was absent in the positive FQR culture group despite low relative abundance in non-FQR subjects (adjusted p<0.001). At the species level, Prevotella disiens was significantly enriched in FQR negative subjects (adjusted p=0.012), while Shigella flexneri and Proteus mirabilis were considerably higher in the FQR positive group (adjusted p=0.017 and adjusted p=0.028, respectively, Fig. 5).
Fig. 3. Waterfall plot demonstrating the variability of bacterial taxa associated with or without fluoroquinolone resistant colonization.
Fig. 4. Correlation analysis map.
Fig. 5. Differentially abundant taxa associated with resistance status.
DISCUSSION
Our findings indicate substantial differences in the microbiome comparing those men who are carriers of FQR organisms compared to those who are not. The overgrowth of bacteria, particularly Enterobacteriaceae in those patients colonized with FQR organisms indicate targeting this group of bacteria could be a potential decolonization strategy. A major purpose for our study is to identify the resident microbiome profiles prior to design clinical trials of repurposed pharmaceuticals, prebiotics, or probiotics in this patient population. Considering the only demographic factor significant between the FQR and FQN subjects is body mass index and the link between obesity and the microbiome is well established.
The microbiome has been implicated as a reservoir for antimicrobial resistance and termed the “resistome” [15]. The high density of bacteria allow for communication and sharing of genetic material creating increased likelihood of transfer of microbial resistance. The international metagenomics of human intestinal tract project has identified nearly some 3.3 million different bacterial genes among human microbiomes [16]. The detection of individual groups of gut microbial communities characterized by presence of similar antibiotic resistance profiles has been termed “Resistotypes” similar to the term “Enterotypes” usually describing the dominant microbial bacteria in the gut [17]. One of the particular resistome gene clusters was detected specifically within the European population and contained the mexb and smed genes imparting resistance to specifically fluoroquinolones [18].
Physicians have had significant concerns regarding the rising fluoroquinolone resistance patterns, specifically in urologic surgical patients. “Targeted prophylaxis” focuses on the detection of FQR organisms before a transrectal prostate biopsy in order to alter antibiotic prophylaxis regimens [4,19]. We see this opportunity to also assess for microbiome evaluation and determination of decolonization with various drugs or vaccines could then render the patient free from FQR organisms prior to prostate biopsy. The technique could also be applied to preoperative preparation and recurrent infections. Rather than using more antibiotics, we propose a process in which we could manipulate the microbiome before the prostate biopsy to reduce infection rates. Given our findings, men colonized with FQR E. coli have an abundant overgrowth of Enterobacteriaceae and potentially could be targeted utilizing new pharmaceuticals targeting FimH receptors to deplete uropathogenic bacteria and reduce antibiotic-resistant bacteria colonization [20]. In addition to microbiome manipulation with prebiotics or probiotics, vaccines have been previously used against FimH as well [21,22]. Vaccines are unlikely to be the future to prevent these infections because newer data have shown that E. coli can shed the FimH protein and alter its pathogenic capabilities [23]. With our current data, we are poised to test various subtle microbiome manipulations to attempt to “decolonize” men with fluoroquinolone resistant organisms. In turn, this could lower the hospitalization rate from infection associated with prostate biopsy by four-fold [24]. We anticipate this research would also have implications regarding research in other urologic (prostatitis and recurrent urinary tract infections) and non-urologic disease profiles [25].
Two profiles that favored the absence of FQR organisms were at the family level Aeromonadaceae and P. disiens. Prevotella has been considered the more favorable or less inflammatory enterotype [17]. Individuals with an enterotype characterized by enriched proportions of Prevotella have significantly higher plasma concentration of trimethylamine-N-oxide (TMAO) [26,27,28,29]. The biogenic reduction of trimethylamine oxide (TMAO) leads to the metabolite trimethylamine (TMA) to modify the antibiotic resistance profiles of bacteria [30]. Therefore, increasing provetella and/or TMAO may assist with the reduction of antibiotic resistant organisms.
Limitations of the study include a small number of fluoroquinolone resistant subjects and large sample size could provide more robust data for sub-analysis. We also cannot account for the bias from an individual's diet, which can alter microbiome composition. We attempt to identify those patients with previous antibiotic use within 6 months because this may alter microbiome results. We did not document probiotic or supplemental use and is rarely documented in the medical record. Probiotic, supplement, and antibiotic use all play important roles in microbiome research and could alter results.
CONCLUSIONS
Microbiome analysis determined that men colonized with FQR bacteria have less diverse bacterial communities, higher levels of Enterobacteriaceae and reduced levels of P. disiens. These results may have implications in pre/probiotic or new pharmaceuticals to target Enterobacteriaceae overgrowth with intervention studies.
ACKNOWLEDGMENTS
We thank all the patients, physicians, residents and research coordinators who worked together to accomplish this project.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI104681. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All materials will be available upon request through the Antibiotic Leadership Group (ALRG) lead by Duke University.
Footnotes
CONFLICTS OF INTEREST: Resphera Biosciences was paid to perform the microbiome analysis, and Dr. White is the owner of the company.
References
- 1.Loeb S, Carter HB, Berndt SI, Ricker W, Schaeffer EM. Complications after prostate biopsy: data from SEER-Medicare. J Urol. 2011;186:1830–1834. doi: 10.1016/j.juro.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson DA, Barrett LK, Rogers BA, Freeman JT, Hadway P, Paterson DL. Infectious complications following transrectal ultrasound-guided prostate biopsy: new challenges in the era of multidrug-resistant Escherichia coli. Clin Infect Dis. 2013;57:267–274. doi: 10.1093/cid/cit193. [DOI] [PubMed] [Google Scholar]
- 3.Roberts MJ, Williamson DA, Hadway P, Doi SA, Gardiner RA, Paterson DL. Baseline prevalence of antimicrobial resistance and subsequent infection following prostate biopsy using empirical or altered prophylaxis: a bias-adjusted meta-analysis. Int J Antimicrob Agents. 2014;43:301–309. doi: 10.1016/j.ijantimicag.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Taylor AK, Zembower TR, Nadler RB, Scheetz MH, Cashy JP, Bowen D, et al. Targeted antimicrobial prophylaxis using rectal swab cultures in men undergoing transrectal ultrasound guided prostate biopsy is associated with reduced incidence of postoperative infectious complications and cost of care. J Urol. 2012;187:1275–1279. doi: 10.1016/j.juro.2011.11.115. [DOI] [PubMed] [Google Scholar]
- 5.Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9:e101344. doi: 10.1371/journal.pone.0101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CLSI. Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. CLSI document M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. [Google Scholar]
- 7.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 15.Penders J, Stobberingh EE, Savelkoul PH, Wolffs PF. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh TS, Gupta SS, Nair GB, Mande SS. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS One. 2013;8:e83823. doi: 10.1371/journal.pone.0083823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liss MA, Johnson JR, Porter SB, Johnston B, Clabots C, Gillis K, et al. Clinical and microbiological determinants of infection after transrectal prostate biopsy. Clin Infect Dis. 2015;60:979–987. doi: 10.1093/cid/ciu1129. [DOI] [PubMed] [Google Scholar]
- 20.Spaulding CN, Klein RD, Ruer S, Kau AL, Schreiber HL, Cusumano ZT, et al. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature. 2017;546:528–532. doi: 10.1038/nature22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CH, Pinkner JS, Roth R, Heuser J, Nicholes AV, Abraham SN, et al. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci U S A. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, et al. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A. 1998;95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liss MA, Taylor SA, Batura D, Steensels D, Chayakulkeeree M, Soenens C, et al. Fluoroquinolone resistant rectal colonization predicts risk of infectious complications after transrectal prostate biopsy. J Urol. 2014;192:1673–1678. doi: 10.1016/j.juro.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Ericsson AC, Franklin CL. Manipulating the gut microbiota: methods and challenges. ILAR J. 2015;56:205–217. doi: 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stremmel W, Schmidt KV, Schuhmann V, Kratzer F, Garbade SF, Langhans CD, et al. Blood trimethylamine-N-oxide originates from microbiota mediated breakdown of phosphatidylcholine and absorption from small intestine. PLoS One. 2017;12:e0170742. doi: 10.1371/journal.pone.0170742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoyles L, Jiménez-Pranteda ML, Chilloux J, Brial F, Myridakis A, Aranias T, et al. Metabolic retroconversion of trimethylamine N-oxide and the gut microbiota. Microbiome. 2018;6:73. doi: 10.1186/s40168-018-0461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu CN, Lu PC, Lo MH, Lin IC, Chang-Chien GP, Lin S, et al. Gut microbiota-dependent trimethylamine N-oxide pathway associated with cardiovascular risk in children with early-stage chronic kidney disease. Int J Mol Sci. 2018;19:E3699. doi: 10.3390/ijms19123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Létoffé S, Audrain B, Bernier SP, Delepierre M, Ghigo JM. Aerial exposure to the bacterial volatile compound trimethylamine modifies antibiotic resistance of physically separated bacteria by raising culture medium pH. MBio. 2014;5:e00944–e00913. doi: 10.1128/mBio.00944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]