Abstract
Macrophages provide key elements of the host response to influenza A virus (IAV) infection, including expression of type I IFN and inflammatory cytokines and chemokines. TBK1 (TNF receptor–associated factor family member–associated NF-κB activator–binding kinase 1) contributes to IFN expression and antiviral responses in some cell types, but its role in the innate response to IAV in vivo is unknown. We hypothesized that macrophage TBK1 contributes to both IFN and non-IFN components of host defense and IAV pathology. We generated myeloid-conditional TBK1 knockout mice and assessed the in vitro and in vivo consequences of IAV infection. Myeloid-specific loss of TBK1 in vivo resulted in less severe host response to IAV, as assessed by decreased mortality, weight loss, and hypoxia and less inflammatory changes in BAL fluid relative to wild-type mice despite no differences in viral load. Mice lacking myeloid TBK1 showed less recruitment of CD64+SiglecF−Ly6Chi inflammatory macrophages, less expression of inflammatory cytokines in the BAL fluid, and less expression of both IFN regulatory factor and NF-κB target genes in the lung. Analysis of sorted alveolar macrophages, inflammatory macrophages, and lung interstitial macrophages revealed that each subpopulation requires TBK1 for distinct components of the response to IAV infection. Our findings define roles for myeloid TBK1 in IAV-induced lung inflammation apart from IFN type I expression and point to myeloid TBK1 as a central and cell type–specific regulator of virus-induced lung damage.
Keywords: influenza, macrophage, TBK1, cytokine, IFN
Clinical Relevance
Innate immune responses during influenza infection can contribute to host tissue damage. The signaling pathways are often amenable to modulation in therapeutically useful ways. Macrophages are a major source of type I IFNs in viral infections, and the kinase TBK1 (TNF receptor–associated factor family member–associated NF-κB activator–binding kinase 1) is critical in most cell types for expression of type I IFN. This study defines additional, discrete, and unexpected roles for TBK1 in the pathogenesis of influenza infection. We show that mice lacking myeloid TBK1 have decreased expression of inflammatory genes and impaired recruitment of inflammatory macrophages to the lung in influenza infection, leading to milder disease.
Influenza A virus (IAV) infection causes significant mortality and morbidity worldwide annually. IAV-induced lung injury results from both viral killing of infected cells and exuberant host immune responses. The host response to IAV has several components, including expression of type I IFNs to restrict viral replication, activation, and recruitment of local and circulating immune cells; killing and clearance of infected or dead cells; and regeneration of injured epithelium. Antiviral therapies for IAV improve clinical outcomes (1), but they must typically be given early in infection. Although IAV strains vary in pathogenicity, multiple lines of evidence suggest that severity of illness corresponds with the degree of host response (2, 3). Thus, there remains a need for host-targeted therapies that prevent tissue damage and can be given after infection is established.
IAV infection stimulates multiple pattern recognition receptors (PRRs) in both infected epithelium and responding immune cells, including alveolar macrophages (AMs) and recruited, bone marrow–derived exudative, or inflammatory macrophages (InfMs) (4–6). Mouse models show that IAV components stimulate Toll-like receptors (TLRs), including TLR3 and TLR7/8; cytosolic nucleic acid sensors retinoic acid inducible gene I (RIG-I), mitochondrial antiviral signaling (MAVS), and DNA-dependent activator of IFN regulatory factors (DAI); and the inflammasome protein NLRP3 (NACHT, LRR, and PYD domains-containing protein 3) (7). PRR stimulation leads to activation of the IKK kinases and the related TNF receptor–associated factor family member–associated NF-κB activator–binding kinase 1 (TBK1), leading to phosphorylation of transcription factors that include members of the NF-κB and IFN regulatory factor (IRF) families (reviewed in Reference 8). Genetic deletion of PRRs or downstream signaling genes in mice reveals surprising and sometimes paradoxical phenotypes in virus- or IAV-infected animals that do not correspond with in vitro cell phenotypes (reviewed in Reference 7). Although some innate immune sensors are required, some are redundant and contribute either to inflammation or to feedback inhibition of other pathways. TBK1 is a convergence point of multiple IAV-driven processes, and thus understanding its function in IAV pathogenesis is useful for developing host-directed therapies.
TBK1 was identified as a binding protein for TNF receptor–associated factor family member–associated NF-κB activator (TANK) (9). In many cell types, TBK1 serves as the major kinase phosphorylating IRF3 and IRF7 and thus is required for expression of type I IFN (10, 11) and other IRF3/7-dependent genes, such as IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), MX dynamin-like GTPase 1 (Mx1), and CXCL10. In some contexts, TBK contributes to activation of NF-κB p65/RelA and promotes expression of primarily p65-dependent genes, such as IL-6 and RANTES (regulated upon activation, normal T cell expressed and secreted) (12, 13), or genes that require a combination of IRF and NF-κB. Conventional deletion of TBK1 in C57BL/6 mice leads to embryonic lethality that is TNF receptor (TNFR) dependent and can be rescued by crossing into either TNFR-null mice or the TNFR2-null Sv129 background (13–15). Cell type–specific deletion in T cells, B cells, and dendritic cells has shown that TBK1 also regulates T-cell migration, NF-κB–dependent immunoglobulin class switching, and gene expression downstream of the type I IFN receptor (16–18).
The in vivo functions of myeloid TBK1 remain undefined. TBK1-deficient bone marrow macrophages (BMMs) from TBK1-knockout (TBK1-KO) Sv129 mice have impaired expression of IFN-β and CCL5/RANTES when stimulated in vitro, but these mice are paradoxically hyperresponsive to intraperitoneal LPS (13). Thus, TBK1 likely contributes to host defense and IAV pathology in a cell type–specific manner.
In this study, we examined the role of myeloid TBK1 in influenza infection. Conditional gene deletion demonstrates that myeloid TBK1 is dispensable for viral control but drives the recruitment of InfMs to the infected lung. Mice lacking myeloid TBK1 have less severe host response to influenza. In vitro influenza infection elicits different patterns of gene expression from AMs compared with BMMs. In vivo alveolar, interstitial, or monocyte-derived inflammatory subpopulations of lung macrophages have differing requirements for TBK1 in expression of antiviral genes, inflammatory cytokines, or chemokines. We conclude that myeloid TBK1 contributes to the inflammatory response to influenza apart from its defined role in type I IFN expression, pointing to a role for TBK1 in multiple phases of the host response to influenza.
Methods
Mice
C57BL/6 TBK1lox/lox mice were a kind gift from Dr. Katherine Fitzgerald (University of Massachusetts) and were crossed with LysM-Cre mice from The Jackson Laboratory (stock no. 004781). Colonies were maintained at the University of North Carolina at Chapel Hill. All mouse lines were bred and housed in ventilated cages in pathogen-free facilities. Experiments involving animals were conducted in accordance with the recommendations made by the American Association for Laboratory Animal Science. Procedures were carried out using protocols approved by the University of North Carolina at Chapel Hill School of Medicine Institutional Animal Care and Use Committee.
Influenza Infections
Influenza A/PR/8/34 H1N1 from Charles River (10100374) was titered on MDCK (Madin-Darby canine kidney) cells (19). Eight- to 16-week-old female mice were anesthetized with tribromoethanol. Virus diluted in PBS was administered intranasally as a 5 × 105 egg-infective dose in 40 μl. For studies of intratracheal infections, mice were given a 3,000 egg-infective dose in 40 μl (50% tissue culture–infective dose [TCID50], 22). BMMs or AMs were infected in vitro at multiplicity of infection 1 in Dulbecco’s modified Eagle medium supplemented with 1% BSA and 5 μg/ml TPCK (tosylamide-L-phenylethyl-chloromethyl ketone) trypsin (MilliporeSigma). After 1 hour, the medium was replaced with infection medium consisting of Dulbecco’s modified Eagle medium with 0.15% BSA, 1% FCS, penicillin-streptomycin, and 5 μg/ml TPCK trypsin.
BAL Fluid Collection and Analysis
The lungs of killed mice were lavaged with 1 ml of PBS/2 mM EDTA. BAL cytokines were measured by using the Bio-Plex Pro Mouse Cytokine Group 1 Panel 23-Plex on a Bio-Plex MagPix imager (both from Bio-Rad Laboratories). Mouse IFN-β and IFN-α ELISAs were obtained from InvivoGen. BAL cell differentials and protein concentrations were measured as described elsewhere (20). Lactate dehydrogenase assays were performed by the University of North Carolina Animal Histopathology and Lab Medicine Core.
Flow Cytometry and Cell Sorting
Lungs were digested to single-cell suspensions by intratracheal instillation of 5 mg/ml collagenase I (Worthington Biochemical Corp.) and 0.25 mg/ml DNase I (MilliporeSigma) at 37°C for 30 minutes, followed by mechanical disruption, as previously described (21). Details of antibody staining are provided in the data supplement. Cell sorting was performed using a BD FACSAria II flow cytometer (BD Biosciences).
Statistical Analysis and Graphics
Error bars represent the SEM. For RT-qPCR, all samples were run in technical (assay) triplicate or duplicate. For in vitro ELISAs, all samples were run in assay triplicate. All figures depict experiments that have been replicated a minimum of two times. For Kaplan-Meier survival curves, significance was computed by Gehan-Breslow-Wilcoxon test. ANOVA, two-tailed unpaired t tests, and the Mann-Whitney U tests were performed using Prism software (GraphPad Software), and the level of significance is indicated by *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001. Heatmaps were generated with Heatmapper (22).
Results
Myeloid-Specific Deletion of TBK1 Impairs Macrophage Gene Expression in a Stimulus-Specific Manner
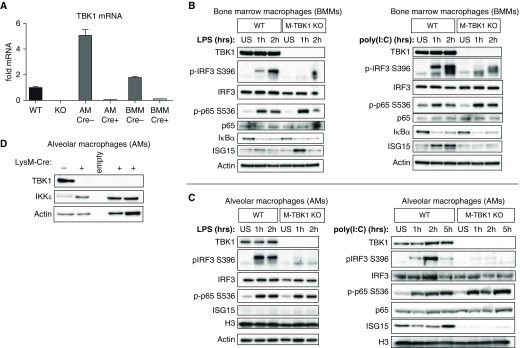
We generated myeloid-deficient TBK1 mice by crossing TBK1fl/fl mice with LysM-Cre mice that express Cre recombinase driven by the lysozyme 2 promoter (16, 23). RT-PCR of TBK1 mRNA shows that Cre expression causes loss of TBK1 expression from both lavageable AMs and BMMs differentiated from LysM-Cre–expressing TBK1fl/fl (M-TBK1-KO) mice that is comparable to that in the well-characterized TBK1-KO mouse embryonic fibroblast line (Figure 1A). BMMs had undetectable expression of TBK1 protein relative to littermate TBK1fl/fl mice that lacked LysM-Cre (wild type [WT]) (Figure 1B), indicating that LysM-Cre can inactivate floxed TBK1 alleles. In contrast, neutrophils had no defect in Tbk1 mRNA expression (data not shown). When stimulated with LPS or polyinosinic:polycytidylic acid [poly(I:C)], BMMs from M-TBK1-KO mice had no defect in degradation of the NF-κB inhibitor IκBα or phosphorylation of p65 S536, but they did show impaired phosphorylation of IRF3 S396 (Figure 1B). Poly(I:C)-stimulated BMMs did show impaired upregulation of ISG15 (IFN-stimulated gene 15), which is driven by both IRF3 and IFN-β. Similar to BMMs, AMs from M-TBK1-KO mice showed loss of TBK1 expression by Western blot analysis (Figure 1D) and loss of IRF3 phosphorylation but intact p65 phosphorylation when stimulated with LPS or poly(I:C). TBK1 was required for ISG15 expression in poly(I:C)-stimulated AMs. TBK1-deficient AMs showed upregulation of IKKε (Figure 1D), perhaps reflecting a compensatory pathway.
Figure 1.
Myeloid-specific deletion of TNF receptor–associated factor family member–associated NF-κB activator–binding kinase 1 (TBK1) impairs IFN regulatory factor 3 (IRF3) phosphorylation in macrophages. (A) RT-qPCR of TBK1 mRNA from mouse embryonic fibroblasts, bone marrow macrophages (BMMs), and BAL macrophages. Error bars represent technical triplicates, and data are representative of two independent experiments. (B) Immunoblots of BMMs from wild-type (WT; LysM-Cre−, TBK1flox/flox) or TBK1-knockout (M-TBK1-KO; LysM-Cre+, TBK1flox/flox) mice stimulated in vitro with 100 ng/ml LPS or 25 μg/ml polyinosinic:polycytidylic acid [poly(I:C)] for the indicated times. (C) Immunoblots of alveolar macrophages from WT or M-TBK1-KO mice stimulated in vitro with the same concentrations of LPS or poly(I:C). (D) Immunoblots of alveolar macrophages from WT or multiple M-TBK1-KO mice. Immunoblots are representative of at least two independent experiments. AM = alveolar macrophage; H3 = histone H3; ISG15 = IFN-stimulated gene 15; US = unstimulated.
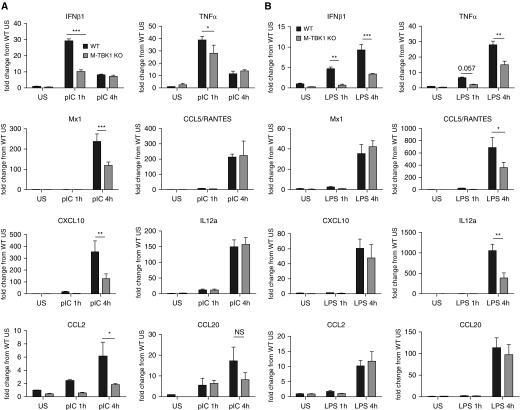
When stimulated with poly(I:C), M-TBK1-KO BMMs showed decreased expression of the mRNAs coding for IFN-β1, CXCL10, Mx1, and CCL2 but largely intact expression of TNF-α, RANTES, IL-12A, and CCL20 mRNA relative to WT controls (Figure 2A). LPS-stimulated M-TBK1-KO BMMs showed less expression of IFN-β1, TNF-α, RANTES, and IL-12A mRNAs but unimpaired mRNA expression of CCL2, CXCL10, Mx1, and CCL20 (Figure 2B). Mx1 is a commonly examined ISG. Although many mouse strains, including C57BL/6, carry a two-exon deletion that renders the Mx1 protein inactive and alters susceptibility to influenza, transcription of Mx1 mRNA is preserved and has been shown to reflect IRF3 and NF-κB activation (24, 25). Similar results were seen with IFIT1, another commonly examined ISG (data not shown). These results indicate that BMMs require TBK1 for activation of IRF and NF-κB targets in a stimulus-dependent manner, which may reflect the relative contribution of upstream MyD88 or TRIF (Toll/IL-1 receptor domain–containing adapter-inducing IFN-β) to activation by each stimulus.
Figure 2.
Myeloid deletion of TBK1 impairs expression of IRF and NF-κB target genes. RT-qPCR was used to assess mRNA of indicated genes from BMMs stimulated with (A) LPS or (B) polyinosinic:polycytidylic acid (pIC) for the indicated times. Data are normalized to the 18S mRNA, expressed as fold change compared with uninfected WT mice by comparative cycle threshold method, and compared by two-tailed unpaired t test. n = 3 per group and least two independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.005. NS = not significant; RANTES = regulated upon activation, normal T cell expressed and secreted.
M-TBK1-KO Mice Have a Less Severe Host Response to Influenza
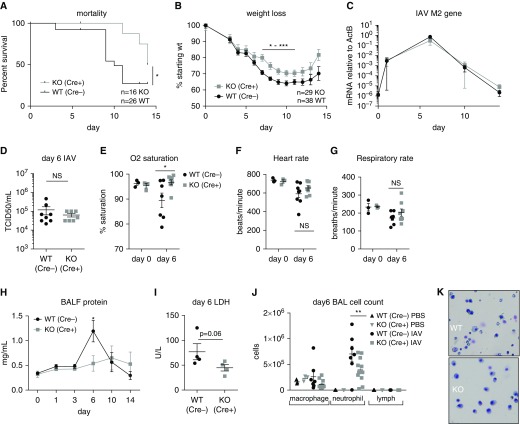
To examine the involvement of myeloid TBK1 in the response to influenza, we challenged M-TBK1-KO mice or their Cre-WT littermates with IAV strain PR8 (TCID50, 366/mouse). Strikingly, M-TBK1-KO mice had decreased and delayed mortality after intranasal infection with IAV (Figure 3A). M-TBK1-KO mice initially lost weight similarly to WT littermates, but from approximately Day 7 after infection, they exhibited less severe weight loss and began to recover earlier (Figure 3B). M-TBK1-KO mice also exhibited decreased mortality and weight loss when PR8 was administered intratracheally (Figures E1A and E1B in the data supplement). When viral load was assessed by RT-qPCR of the IAV M2 gene from lung homogenates, no difference was observed between Days 2 and 14 after either intranasal or intratracheal inoculation (Figures 3C, E1C, and E1D). Viral load assessed by TCID50 of lung homogenates at Day 6 likewise did not differ between WT and M-TBK1-KO mice (Figure 3D). At 6 days after intranasal infection, M-TBK1-KO mice had better oxygenation and no differences in heart rate or respiratory rate (Figures 3E–3G). These results suggest that M-TBK1-KO mice have a less clinically severe response to influenza infection that is significant by Day 6.
Figure 3.
M-TBK1 KO mice have a less severe host response to influenza than WT mice. (A) Survival and (B) weight curves for mice infected intranasally with 5 × 104 egg-infective dose (tissue culture–infective dose [TCID50], 366) of influenza A virus (IAV) strain PR8. Mortality was assessed by Gehan-Breslow-Wilcoxon test and weight by two-way ANOVA with multiple comparisons (Šidák correction). (C) Viral infection level as measured by RT-qPCR of IAV M2 gene from lung homogenates at Days 1–14. (D) Viral infection level as measured by TCID50 from lung homogenates at Day 6. (E) Arterial oxygen saturation, (F) heart rate, and (G) respiratory rate at Days 0 and 6. n = 3–8 mice per group in two independent experiments. (H) BAL fluid (BALF) protein measurements from mice at Days 0–14 after infection. (I) Lactate dehydrogenase (LDH) measurements in BALF from mice at Day 6. (J) Leukocyte counts in BALF assessed using cell counts and cytospins on Day 6. (K) Representative cytospin images from BAL at Day 6. Panels H–J depict six to eight mice per group and two or three independent experiments. Panels D–J reflect analysis by two-tailed unpaired t test. *P < 0.05, **P < 0.01, and ***P < 0.005. ActB = β-actin; U = units.
M-TBK1-KO Have Less Inflammatory Changes in BAL Fluid after Influenza Infection
We also examined markers of lung inflammation within the alveolar space, including BAL fluid protein and leukocytes. Although WT mice accumulated BAL fluid protein that peaked around Day 6, indicative of lung injury, M-TBK1-KO mice had a slower and smaller accumulation of BAL fluid protein (Figure 3H). There was a strong trend toward less lactate dehydrogenase in the BAL fluid (P = 0.06) Figure 3I). Cytospin counts of BAL fluid at Day 6 showed fewer lavageable neutrophils and no difference in macrophage or lymphocyte counts in M-TBK1-KO compared with WT mice (Figures 3J and 3K). Taken together, these data suggest that intraalveolar and luminal airway inflammation is less at the peak of lung inflammation in M-TBK1-KO mice than in WT mice.
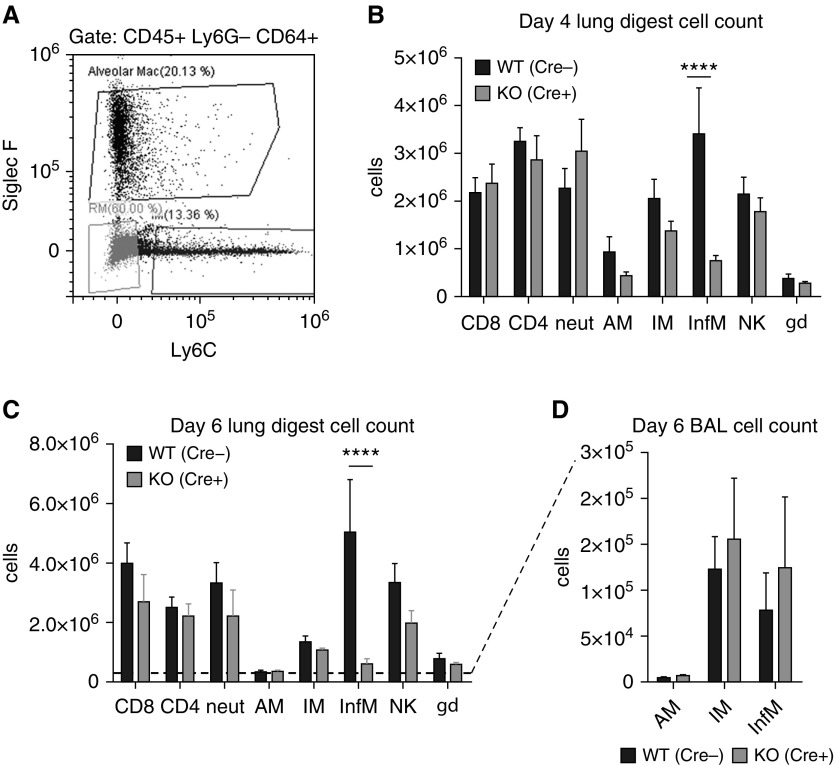
M-TBK1-KO Mice Have Fewer Recruited Inflammatory Lung Macrophages during Influenza Infection
To examine the abundance of immune cells in the lung at after influenza infection, influenza-infected mouse lungs were digested to single-cell suspensions and analyzed by flow cytometry. The leukocytes were identified as CD45+ cells, and markers for macrophages, neutrophils, natural killer (NK) cells, and T cells were included in the immunopanel (gating strategy in Figure E2). The CD64+Ly6G− lung macrophage population was gated to identify macrophage subpopulations of AMs (SiglecF+Ly6C−/low), interstitial macrophages (IMs) (SiglecF−Ly6C−), and recruited InfMs (SiglecF−Ly6C+) (Figure 4A). At Days 4 and 6 after infection, the number of CD64+Ly6G− SiglecF−Ly6C+ InfMs was strikingly less in M-TBK1-KO mice (Figures 4B and 4C). The numbers of AMs and IMs were similar in WT and M-TBK1-KO mice, suggesting that loss of TBK1 does not alter survival or apoptosis of these macrophage subpopulations during influenza. M-TBK1-KO mice showed no change in the numbers of CD4+, CD8+, and γΔT cells and no significant difference in the number of neutrophils or NK cells in the lungs (Figures 4B and 4C). Likewise, immunophenotyping of circulating and splenic cells from uninfected WT and M-TBK1-KO mice showed no differences (Figure E3). When BAL populations were examined, no difference in lavageable AM, IM, or InfM numbers was observed at Day 6 (Figure 4D); of note, we observed far fewer InfMs in BAL fluid than in lung digests during pneumonia. These results suggest that myeloid TBK1 is required for the recruitment of InfMs from the blood and bone marrow to the lung parenchyma but not for the recruitment, proliferation, or survival of other macrophage subpopulations, T cells, or NK cells.
Figure 4.
M-TBK1-KO mice have fewer recruited inflammatory lung macrophages during influenza infection. Mice were infected intranasally with 5 × 104 egg-infective dose and lungs were harvested and digested to single-cell suspensions. (A) Representative flow cytometric plot from CD45+/Ly6G−/CD64+ gate showing definition of resident alveolar (AM; Siglec F+/Ly6Clow), resident interstitial (IM; Siglec F−/Ly6C−), and recruited inflammatory (InfM; Siglec F−/Ly6C+) macrophages. Total number of relevant cell populations identified by flow cytometry at (B) Day 4 and (C) Day 6 after infection. (D) Macrophage subpopulations recovered from BAL at Day 6 after infection. n = 6–8 mice per genotype per group and two independent experiments analyzed by Mann-Whitney U test. NK = natural killer cells. ****P < 0.001. gd = γδ.
M-TBK1-KO Mice Have Less Lung Inflammatory Gene Expression and Lower BAL Cytokine Levels
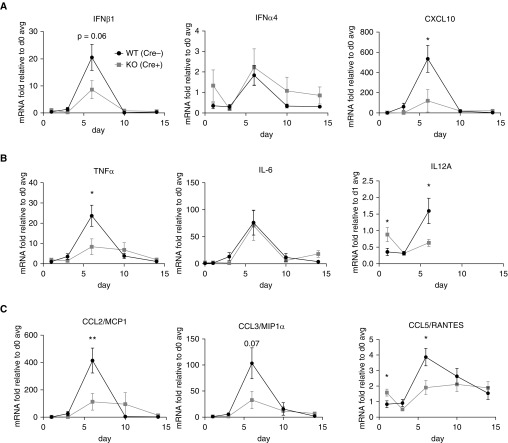
To examine lung inflammation, we measured gene expression by RT-qPCR of whole-lung homogenates from influenza-infected mice starting at Day 2 and extending through Day 14 after infection, at which time the mice have cleared the infection and are in the process of resolving the lung injury. M-TBK1-KO mice had lower levels of CXCL10 mRNA and a trend toward less IFN-β1 mRNA at Day 6, whereas IFN-α4 was only weakly induced and not impaired in M-TBK1-KO mice (Figure 5A). M-TBK1-KO mice had significantly impaired expression of TNF-α and IL-12A mRNA at Day 6, whereas IL-6 mRNA expression was unimpaired in KO mice (Figure 5B). M-TBK1-KO mice had less CCL2/monocyte chemoattractant protein 1 (MCP1) and CCL5/RANTES mRNA and a trend toward less CCL3/macrophage inflammatory protein-1α (MIP-1α) at Day 6 (Figure 5C).
Figure 5.
M-TBK1-KO mice have less lung inflammatory gene expression. RT-qPCR of indicated mRNAs from lung homogenates. Commonly examined (A) IRF3 targets, (B) NF-κB targets, and (C) chemokines are shown. Lungs from intranasally infected mice were snap frozen, and mRNA and cDNA were prepared. Data are normalized to 18S mRNA and expressed as fold change compared with an average of Day 0 mice. n = 5–8 mice per point and two independent experiments analyzed by Mann-Whitney U test. *P < 0.05 and **P < 0.01. MCP1 = monocyte chemoattractant protein 1; MIP1α = macrophage inflammatory protein 1α.
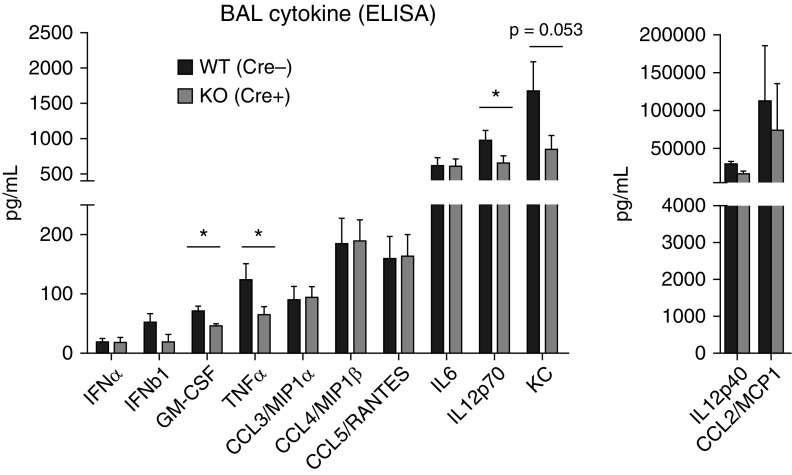
To examine lung inflammation, type I IFN and cytokine expression in BAL fluid from M-TBK1-KO and WT mice at 6 days after infection was measured using multiplex ELISAs. Type I IFNs were expressed at low levels in BAL fluid, and neither IFN-α nor IFN-β was significantly less at Day 3 or Day 6 after infection in M-TBK1-KO mice (Figures 6 and E5), suggesting that myeloid TBK1 is not required for secretion of type I IFN into the airspace at these times, in agreement with the mRNAs. In comparison, significant differences in multiple BAL cytokines were identified between genotypes. M-TBK1-KO mice expressed lower levels of TNF-α, GM-CSF (granulocyte–macrophage colony–stimulating factor), and IL-12 p70 (Figure 6) with trends toward less expression of KC (keratinocyte chemoattractant). BAL levels of RANTES, IL-6, and MIP-1α/β were largely intact. Taken together, these results suggest that myeloid TBK1 contributes to expression of certain inflammatory cytokines in the lung, either directly through gene expression or indirectly by altering the abundance of inflammatory cells in the lung and airspace.
Figure 6.
M-TBK1-KO mice have less BAL cytokine expression. ELISA of indicated cytokines and chemokines from BAL fluid of infected mouse lungs harvested at Day 6 after infection. n = 3–8 mice per point and two separate experiments analyzed by Mann-Whitney U test. *P < 0.05. GM-CSF = granulocyte–macrophage colony–stimulating factor; KC = keratinocyte chemoattractant.
TBK1 Deletion Impairs BMM and AM Transcriptional Responses Differently during Influenza Infection In Vitro
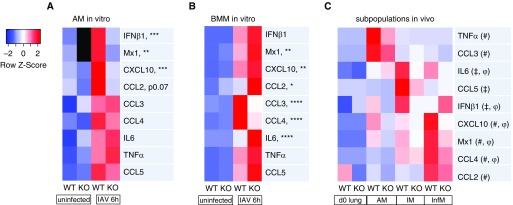
Our in vivo studies show that M-TBK1-KO mice show no change in AM number but a large defect in recruitment of InfMs, which are derived from BMMs. Furthermore, AMs and BM-derived monocytes and macrophages are noted to play different roles in influenza infection (5, 26). We assessed the role of TBK1 in their transcriptional response to influenza infection using the H1N1 PR8 strain in vitro. In response to IAV infection for 6 hours, lavageable AMs from M-TBK1-KO mice showed a pronounced defect in expression of mRNAs coding for IFN-β1, Mx1, and CXCL10 mRNA and a trend toward decreased CCL2 (Figures 7A and E6) but largely intact expression of CCL3, CCL4, CCL5, and TNF-α. In contrast, BMMs from M-TBK1-KO mice had intact induction of IFN-β1, Mx1, and CXCL10 mRNA but impaired expression of CCL3 and CCL4 and elevated expression of IL-6 (Figures 7B and E6). Complementary results were seen by ELISA (Figure E4A). Notably, BMMs transcriptionally upregulate TBK1 at least twofold when infected with IAV, whereas AMs do not (Figure E4B). These results indicate that AMs and BMMs respond differently upon encountering influenza virus.
Figure 7.
TBK1 deletion impairs BMM and AM transcriptional responses differently during influenza virus infection in vitro and has subtype-specific effects on lung macrophages in vivo. RT-qPCR of (A) AMs or (B) BMMs infected with multiplicity of infection 1 of IAV strain PR8 in vitro for 6 hours. Data were normalized to the 18S mRNA and expressed as fold change compared with uninfected WT by the comparative cycle threshold method. Heatmap depicts z-score of mRNA transcript expression level scaled to row mean. n = 3 per group and two independent experiments. Asterisks depict significance of P values for infected WT versus TBK1-KO samples (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001). More detailed graphs for each gene are shown in Figure E6 in the data supplement. (C) Macrophage subpopulations were isolated from lung digests using FACS at Day 4 after infection and subjected to RT-qPCR of indicated genes. mRNA levels were normalized to ActB or to 18S by the comparative cycle threshold method. Data reflect three pooled mice analyzed in technical triplicate. All rows attain statistical significance by two-way ANOVA. Symbols depict significance (P < 0.05) by Šidák’s multiple comparisons method between WT and KO for AMs#, IMs‡, or InfMsφ. Detailed graphs for each gene are shown in Figure E7.
To determine how macrophage subpopulations contribute to TBK1-dependent gene expression in vivo, we isolated AMs, IMs, and InfMs from lung digests using FACS at Day 4 after infection Although AMs are numerically depleted at this time, they appear to be the major macrophage producers of TNF-α and CCL3 (Figures 7C and E7), which both require TBK1 for full expression. In contrast, IMs are major producers of TBK1-dependent IL-6 and IFN-β1 at this time, whereas InfMs are major producers of TBK1-dependent CXCL10 and Mx1. These data show that that these macrophage subpopulations have differing requirements for TBK1 in antiviral and inflammatory gene expression. They also demonstrate that the in vivo gene expression patterns of macrophage subpopulations are not well reflected by either AMs or BMMs infected in vitro.
Discussion
In the present study, we examined the role of myeloid TBK1 in influenza infection. We observed that myeloid deletion of TBK1 results in decreased mortality and weight loss in influenza-infected mice without altering the level of viral infection. These results implicate myeloid TBK1 in the inflammatory lung injury that can accompany severe influenza infection. We observed no difference in the level of virus or viral genome between WT and M-TBK1-KO mice, suggesting that infectivity, viral replication, and viral clearance are not obviously dependent on myeloid TBK1. These data also suggest that viral load is not the primary driver of the morbidity or mortality. Although the decreased disease severity in M-TBK1-KO mice is surprising, other mouse models support the idea that targeting innate immune pathways can decrease host injury without compromising viral control. Mice with conventional deletion of TLR7, TLR3, and MyD88 have decreased influenza-related mortality and pathology (reviewed in Reference 7). In comparison, deletion of other components, such as IFNAR, IKKε, or PKR, or combined deletion of MyD88/TRIF or TLR3/TLR7 increases mortality and morbidity. This may reflect either protein-specific functions or the fact that inflammation and injury represent the summation of many pathways acting simultaneously.
Most strikingly, we noted a marked decrease in the number of recruited CD64+SiglecF−Ly6C+ InfMs in the lungs of M-TBK1-KO mice (Figure 4) during the peak of lung injury, despite no significant alteration in expression of monocyte-attracting chemokines such as MCP1/CCL2. InfMs are known to be major producers of CXCL10 (27), and consistent with this, we observed decreased lung CXCL10 in M-TBK1-KO mice. Multiple lines of evidence implicate recruited InfMs in the severity of influenza lung injury in both humans and mice (26, 28–30). Recruited mouse InfMs may derive from one or more monocyte pools, including the CCR2+, CX3CR1+, or intermediate monocyte pools (31). Further studies will be required to delineate whether TBK1 is required in some or all monocyte populations. Taken together, these observations point to a role for myeloid TBK1 in promoting the recruitment of bone marrow–derived InfMs into the lung during influenza infection.
Measurement of gene expression from lungs of infected WT and M-TBK1-KO mice showed alterations in multiple genes, including TNFα, CCL2, CCL5, and CXCL10, with trends toward impaired expression of IFN-β1 and CCL3. When we examined sorted macrophage subpopulations from infected WT and M-TBK1-KO mice, we saw that TBK1 functioned differently in different lung macrophages. AMs appeared to be a major source of TNF-α and CCL3 at Day 4 after infection and required TBK1 for full expression, whereas IMs were a major source of IL-6, IFN-β1, and CCL5, for which they required TBK1. In comparison, InfMs produced abundant CXCL10 and CCL4 in a TBK1-dependent manner. These data suggest that the M-TBK1-KO phenotype of decreased lung inflammatory gene expression includes specific contributions of TBK1 in multiple macrophage subpopulations. In addition, some TBK1-dependent gene and cytokine expression in IAV infection may derive from non-IAV danger signals that activate TBK1, such as hyaluronan and self-DNA from dying cells (32, 33), and these signals may contribute to differences in mRNAs that require TBK1 only under certain circumstances, such as TNF-α, IL-12A, and CCL5 (Figures 2 and 5–7). M-TBK1-KO mice have preserved numbers of lung neutrophils in lung digests but fewer lavageable neutrophils in the alveolar space. The production of lower levels of cytokines and neutrophil chemokines, including TNF-α and KC, suggests that this is the reason for decreased recruitment to the alveolar space. Although LysM-Cre targets all myeloid cells, we observed intact expression of TBK1 mRNA in the lavageable neutrophils of M-TBK1-KO mice (data not shown), suggesting that the defect in recruitment is likely due to less production of inflammatory mediators rather than to a defect in recognition, adhesion, and migration by neutrophils.
Of note, we observed only a modest reduction in the level of type I IFN protein and mRNA in the lung across the course of infection (Figures 5 and 6A), suggesting that myeloid TBK1 is largely dispensable for type I IFN in this setting. This may reflect compensation by IKKε, which we observed to be upregulated in TBK1-KO AMs (Figure 1C) or by other kinases, such as IKKβ (34), or that nonmyeloid cells such as respiratory epithelium contribute significantly to type I IFN expression.
These studies show that LysM-Cre–induced deletion of Tbk1 results in complete loss of TBK1 protein in both BMMs and AMs. This depletion effectively mimics changes seen in macrophages from conventional TBK1/Tnfsfr1a-KO mice (14) and Sv129 TBK1-KO mice that lack Tnfrsf1b (TNF receptor superfamily member 1B) (13). M-TBK1-KO AMs and BMMs infected with IAV in vitro displayed complementary transcriptional defects; TBK1-null AMs are strongly impaired in expression of IFN-β1 and ISGs such as Mx1 and CXCL10, whereas TBK1-null BMMs express IFN-β1, Mx1, and CXCL10 in the setting of IAV but have impaired expression of genes such as CCL3 and CCL4 (Figure 7). We propose that TBK1 has macrophage subpopulation–specific contributions to IRF and NF-κB activation. This may be mediated either by cell type–specific expression of TBK1 adaptor proteins (TANK, OPTN [optineurin], Azi2 [5-azacytidine–induced protein 2]), alterations in TBK1-activating scaffolds such as TRAF (TNF receptor–associated factor) family members and STING (stimulator of IFN genes), or some other mechanism. Activation of TBK1 by hyperactivating STING mutations results in elevated serum levels of multiple NF-κB–driven cytokines, including TNF-α, MCP1, MIP-1α/β, and G-CSF (granulocyte colony-stimulating factor) (35), and this effect persists when IRF3 is deleted, further implicating TBK1 as a driver in inflammation independent of the IRF axis. We observed that TBK1 contributed to the expression of classical IRF targets (such as IFN-β, Mx1, and IFIT1), classical NF-κB targets (such as TNF-α), and genes that are driven by a combination of transcription factors. The detailed TBK1-dependent signaling pathways that each macrophage subpopulation uses to perform diverse functions remain to be defined.
In summary, we demonstrate that myeloid TBK1 contributes to morbidity and mortality in influenza-infected mice and is largely dispensable for viral control. Mice lacking myeloid TBK1 have decreased numbers of InfMs at the peak of infection and injury, pointing to a role for myeloid TBK1 in promoting the generation and recruitment of bone marrow–derived InfMs. TBK1 promotes differing patterns of inflammatory gene expression in lung macrophage subtypes. TBK1 inhibition by small-molecule inhibitors may be a point of intervention in late or established severe influenza infection.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Chirag H. Patel, Samuel L. Collins, and Jonathan D. Powell of The Johns Hopkins University School of Medicine for sharing reagents and techniques and for critical review of the manuscript. The authors also thank Jessica R. Martin, John C. Gomez, and Jason R. Mock for discussion and sharing techniques.
Footnotes
Supported by National Institutes of Health grants 5K12HL119998 (C.M.D.) and R01HL114388 (C.M.D.) and American Heart Association grant 12POST12030011 (R.S.H.). The content is solely the view of the authors and does not necessarily represent the official views of the funding organizations.
Author Contributions: R.S.H.: conceived of the study and oversaw all experimentation and interpretation of results; R.S.H. and J.T.-C.: conducted the majority of the experiments, analyzed the results, and wrote the paper with contributions from C.M.D.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as 10.1165/rcmb.2018-0122OC on October 5, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hsu J, Santesso N, Mustafa R, Brozek J, Chen YL, Hopkins JP, et al. Antivirals for treatment of influenza: a systematic review and meta-analysis of observational studies. Ann Intern Med. 2012;156:512–524. doi: 10.7326/0003-4819-156-7-201204030-00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aevermann BD, Pickett BE, Kumar S, Klem EB, Agnihothram S, Askovich PS, et al. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci Data. 2014;1:140033. doi: 10.1038/sdata.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandes M, Klauschen F, Kuchen S, Germain RN. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu YR, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, et al. Flow cytometric analysis of myeloid cells in human blood, bronchoalveolar lavage, and lung tissues. Am J Respir Cell Mol Biol. 2016;54:13–24. doi: 10.1165/rcmb.2015-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse Mx1 to human IRF7. Curr Opin Immunol. 2016;38:109–120. doi: 10.1016/j.coi.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helgason E, Phung QT, Dueber EC. Recent insights into the complexity of Tank-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 2013;587:1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 9.Pomerantz JL, Baltimore D. NF-κB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999;18:6694–6704. doi: 10.1093/emboj/18.23.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 11.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 12.Delhase M, Kim SY, Lee H, Naiki-Ito A, Chen Y, Ahn ER, et al. TANK-binding kinase 1 (TBK1) controls cell survival through PAI-2/serpinB2 and transglutaminase 2. Proc Natl Acad Sci USA. 2012;109:E177–E186. doi: 10.1073/pnas.1119296109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marchlik E, Thakker P, Carlson T, Jiang Z, Ryan M, Marusic S, et al. Mice lacking Tbk1 activity exhibit immune cell infiltrates in multiple tissues and increased susceptibility to LPS-induced lethality. J Leukoc Biol. 2010;88:1171–1180. doi: 10.1189/jlb.0210071. [DOI] [PubMed] [Google Scholar]
- 14.Bonnard M, Mirtsos C, Suzuki S, Graham K, Huang J, Ng M, et al. Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-κB-dependent gene transcription. EMBO J. 2000;19:4976–4985. doi: 10.1093/emboj/19.18.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmermans S, Van Montagu M, Libert C. Complete overview of protein-inactivating sequence variations in 36 sequenced mouse inbred strains. Proc Natl Acad Sci USA. 2017;114:9158–9163. doi: 10.1073/pnas.1706168114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin J, Xiao Y, Chang JH, Yu J, Hu H, Starr R, et al. The kinase TBK1 controls IgA class switching by negatively regulating noncanonical NF-κB signaling. Nat Immunol. 2012;13:1101–1109. doi: 10.1038/ni.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao Y, Zou Q, Xie X, Liu T, Li HS, Jie Z, et al. The kinase TBK1 functions in dendritic cells to regulate T cell homeostasis, autoimmunity, and antitumor immunity. J Exp Med. 2017;214:1493–1507. doi: 10.1084/jem.20161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Zhou X, Chang M, Nakaya M, Chang JH, Xiao Y, et al. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun. 2015;6:6074. doi: 10.1038/ncomms7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisfeld AJ, Neumann G, Kawaoka Y. Influenza A virus isolation, culture and identification. Nat Protoc. 2014;9:2663–2681. doi: 10.1038/nprot.2014.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada M, Gomez JC, Chugh PE, Lowell CA, Dinauer MC, Dittmer DP, et al. Interferon-γ production by neutrophils during bacterial pneumonia in mice. Am J Respir Crit Care Med. 2011;183:1391–1401. doi: 10.1164/rccm.201004-0592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dial CF, Tune MK, Doerschuk CM, Mock JR. Foxp3+ regulatory T cell expression of keratinocyte growth factor enhances lung epithelial proliferation. Am J Respir Cell Mol Biol. 2017;57:162–173. doi: 10.1165/rcmb.2017-0019OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 24.Roberts ZJ, Goutagny N, Perera PY, Kato H, Kumar H, Kawai T, et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol Cell Biol. 1988;8:4518–4523. doi: 10.1128/mcb.8.10.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole SL, Dunning J, Kok WL, Benam KH, Benlahrech A, Repapi E, et al. M1-like monocytes are a major immunological determinant of severity in previously healthy adults with life-threatening influenza. JCI Insight. 2017;2:e91868. doi: 10.1172/jci.insight.91868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tighe RM, Liang J, Liu N, Jung Y, Jiang D, Gunn MD, et al. Recruited exudative macrophages selectively produce CXCL10 after noninfectious lung injury. Am J Respir Cell Mol Biol. 2011;45:781–788. doi: 10.1165/rcmb.2010-0471OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J Immunol. 2008;180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- 29.Lin KL, Sweeney S, Kang BD, Ramsburg E, Gunn MD. CCR2-antagonist prophylaxis reduces pulmonary immune pathology and markedly improves survival during influenza infection. J Immunol. 2011;186:508–515. doi: 10.4049/jimmunol.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coates BM, Staricha KL, Koch CM, Cheng Y, Shumaker DK, Budinger GRS, et al. Inflammatory monocytes drive influenza A virus–mediated lung injury in juvenile mice. J Immunol. 2018;200:2391–2404. doi: 10.4049/jimmunol.1701543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi C, Pamer EG. Monocyte recruitment during infection and inflammation. Nat Rev Immunol. 2011;11:762–774. doi: 10.1038/nri3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Black KE, Collins SL, Hagan RS, Hamblin MJ, Chan-Li Y, Hallowell RW, et al. Hyaluronan fragments induce IFNβ via a novel TLR4-TRIF-TBK1-IRF3-dependent pathway. J Inflamm. 2013;10:23. doi: 10.1186/1476-9255-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schattgen SA, Gao G, Kurt-Jones EA, Fitzgerald KA. Cutting edge: DNA in the lung microenvironment during influenza virus infection tempers inflammation by engaging the DNA sensor AIM2. J Immunol. 2016;196:29–33. doi: 10.4049/jimmunol.1501048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez-Pelaez M, Lamont DJ, Peggie M, Shpiro N, Gray NS, Cohen P. Protein kinase IKKβ-catalyzed phosphorylation of IRF5 at Ser462 induces its dimerization and nuclear translocation in myeloid cells. Proc Natl Acad Sci USA. 2014;111:17432–17437. doi: 10.1073/pnas.1418399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warner JD, Irizarry-Caro RA, Bennion BG, Ai TL, Smith AM, Miner CA, et al. STING-associated vasculopathy develops independently of IRF3 in mice. J Exp Med. 2017;214:3279–3292. doi: 10.1084/jem.20171351. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.