Summary
Objective
Early onset drug‐resistant epilepsy is a neurologic disorder in which 2 antiepileptic drugs fail to maintain the seizure‐free status of the patient. Heterogeneous clinical presentations make the diagnosis challenging. We aim to identify the underlying genetic causes of a pediatric cohort with drug‐resistant epilepsy and evaluate whether the findings can provide information on patient management.
Methods
We include patients with drug‐resistant epilepsy onset before 18 years of age. Singleton clinical chromosomal microarray (CMA) followed by whole exome sequencing (WES) was performed using genomic DNA. In the first‐tier analysis of the exome data, we aimed to identify disease‐causing mutations in 546 genes known to cause, or to be associated with, epilepsy. For negative cases, we proceeded to exome‐wide analysis. Rare coding variants were interrogated for pathogenicity based on the American College of Medical Genetics and Genomics (ACMG) guidelines.
Results
We recruited 50 patients. We identified 6 pathogenic or likely pathogenic mutations, giving a diagnostic yield of 12%. Mutations were found in 6 different genes: SCN8A, SCN1A, MECP2, CDKL5, DEPDC5, and CHD2. The CDKL5 variant was found to be mosaic. One variant of unknown significance (VUS) in KCNT1 was found in a patient with compatible clinical features. Of note, a reported pathogenic SCN5A mutation known to contribute to Brugada syndrome, was also found in the patient with an SCN1A mutation.
Significance
Our study suggests that singleton WES is an effective diagnostic tool for drug‐resistant epilepsy. Genetic diagnosis can help to consolidate the clinical diagnosis, to facilitate phenotypic expansion, and to influence treatment and management options for seizure control in our patients. In our study, a significant portion of the genetic findings are known to be associated with an increased risk of sudden unexpected death in epilepsy (SUDEP). These findings could assist with more appropriate management in patients with epilepsy.
Keywords: chromosomal microarray, Pediatric‐onset drug‐resistant epilepsy, whole exome sequencing
1.
Key Points.
Whole exome sequencing is an effective diagnostic tool for patients with drug‐resistant epilepsy
Genetic diagnosis can help to identify the SUDEP risk in our cohort
These findings can also help in epilepsy patient management.
2. INTRODUCTION
Since the first epilepsy gene, CHRNA4, was discovered in 1994,1 there has been a paradigm shift in the understanding of epilepsy etiology away from the label of idiopathic and toward a focus on epilepsy genetics. In 2011, this shift became official with the International League Against Epilepsy (ILAE) stating that genetics played a significant role in epilepsy from both clinical and research perspectives.2 By 2013, it was suggested that more than 70% of epilepsies are associated with genetic factors,3 among which monogenic epilepsy accounts for 1%‐2%.4 IZn 2017, ILAE further emphasized the role of genetic factors in the framework of epilepsy classification5 thereby further highlighting the importance of identifying the genetic causes of epilepsies.
The genetic alterations of pediatric‐onset epilepsies range from large size chromosomal copy number variations (CNVs), to small insertions or deletions (indels) and single nucleotide variations (SNVs). In 2014, Olson et al6 demonstrated the importance of CNV detection by chromosomal microarray (CMA) in pediatric patients with epilepsy by establishing that 5% of these patients had phenotypes explainable by CNVs. This diagnostic yield is comparable with autism spectrum disorders and multiple congenital anomalies. Therefore, CMA was recommended as one of the diagnostic tools for patients with early onset epilepsy in clinical settings.
Beyond CNVs, there has been a significant increase in the number of known causative genes for epilepsy in recent years. Next‐generation sequencing, in particular, whole exome sequencing (WES), has become a promising and cost‐effective tool for genetic diagnosis in patients with epilepsy. WES is proving to be especially helpful in the diagnosis of epilepsy, with a diagnostic rate 1.5 times higher in an epilepsy cohort than that seen in nonepilepsy cases.7 In 2013, the Epi4K Consortium had a diagnostic yield of 11% in 264 patients with epileptic encephalopathies using trio WES.8 Further exemplifying the role of WES in difficult to diagnose patients, Dyment et al9 found the genetic diagnosis of 8 patients among 11 patients with intractable epileptic encephalopathy, giving a diagnostic yield of 72% using this method.
The majority of publications on epilepsy genetics focus on epileptic encephalopathy, as this is classified as the most severe type of epilepsy. However, it is also important to recognize the genetic cause of drug‐resistant epilepsy in children. Pediatric‐onset drug‐resistant epilepsy, also known as intractable or refractory epilepsy, is defined by the ILAE as onset of epilepsy before 18‐years‐old with “failure of adequate trials of two tolerated and appropriately chosen and used antiepileptic drug (AED) schedules (whether as monotherapies or in combination) to achieve sustained seizure freedom.”10 It was estimated that approximately 30% of patients with epilepsy fall into this category.11 However, the clinical heterogeneity and diverse disease etiologies make an accurate and specific diagnosis challenging. Patients with difficult‐to‐treat epileptic conditions have a poor quality of life and higher mortality rate when compared to healthy subjects.12A proper molecular diagnosis can aid prognosis, treatment, and patient care. Further understanding of the pathomechanisms could also aid the development of targeted therapy for these patients.
In this study, we aim to identify the disease‐causing mutations in a patient cohort with pediatric‐onset drug‐resistant epilepsy using WES and/or clinical CMA. We also aim to explore the clinical utility of the molecular findings in our patient cohort.
3. METHODS
3.1. Subject recruitment
The study was approved by the institutional review board of the University of Hong Kong/Hospital Authority Hong Kong West Cluster (UW12‐211). Written informed consent was obtained from subjects or their parents. Subjects with neonatal, infantile, or childhood‐onset epilepsy were recruited from Queen Mary Hospital or the Duchess of Kent Children's Hospital, Hong Kong. All of the patients had drug‐resistant epilepsy according to the ILAE definitions.10 Neuroimaging and previous genetic testing (ie, single‐gene testing) had not revealed any underlying epilepsy etiology. Clinical assessment was performed by pediatric neurologists and clinical geneticists.
Genomic DNA was obtained from the peripheral blood using Qiagen Blood Mini Kit (Qiagen). If the clinical geneticist was suspicious of somatic mosaicism in the subject,13, 14 buccal mucosa was also obtained as a germline source of DNA.
3.2. Chromosomal microarray
Clinical CMA was performed using Perkin Elmer‐CGX V2.0 60k oligonucleotide array in Tsan Yuk Hospital (TYH), Hong Kong. The average probe distance was 190 kb with around 28 kb in the targeted regions. Data were analyzed by Genoglyphix software Perkin Elmer (Signature Genomics). Genomic coordinates were based on genome build hg19. Genetic variants were classified based on the American College of Medical Genetics and Genomics (ACMG) practice guidelines. Details of variant interpretation were reported previously.15
3.3. Whole exome sequencing
An exome library was prepared by either TruSeqExome Library Prep Kit (Illumina; n = 26), TruSeq Rapid Exome Library Prep Kit (Illumina; n = 9), or SeqCap EZ Exome + UTR Kit (Roche NimbleGen; n = 15), depending on the availability of the library preparation kit. All preparations and DNA library quality controls were performed according to manufacturer instructions. The DNA libraries were sequenced using Illumina NextSeq500 or HiSeq1500 sequencing platform, with a targeted sequencing coverage of 100×. Details of the library preparation method, sequencer used, and average depth after sequence alignment of each individual is presented in Table S1.
3.4. Data analysis
We used an in‐house developed bioinformatics pipeline for data analysis. In brief, the filtered raw reads were mapped to reference human genome [GRCh37/hg19] by the Burrows‐Wheeler Aligner (BWA) 0.7.10.16 Variant calling was performed using the Genome Analysis Toolkit (GATK) best practices v3.4‐46. The called variants were annotated by Annotate Variation (ANNOVAR).
Variant prioritization was targeted primarily on a customized gene panel with 546 epilepsy‐associated genes (Table S2), integrating 10 epilepsy panels from diagnostic laboratory companies (Baylor Genetics, Ambry Genetics, Emory Genetics Laboratory, Fulgent Genetics. GeneDx, Greenwood Genetic Center, Transgenomic), and a search of the term “epilepsy” in the Online Mendelian Inheritance in Man (OMIM)17 and the National Institutes of Health (NIH) epilepsy genetics initiative databases.18 Coding and canonical splice‐site variants were identified and filtered based on a population frequency of <1% as annotated in the 1000 genome project, the NHLBI GO Exome Sequencing Project (ESP6500), the Exome Aggregation Consortium (ExAC), and the Genome Aggregation Database (gnomAD).19 Rare variants were interrogated for pathogenicity based on the ACMG guideline.20 In silico prediction was performed using Mutation Taster, The Sorting Intolerant from Tolerant (SIFT), Functional Analysis through Hidden Markov Models (FATHMM), and Protein Variation Effect Analyzer (PROVEAN). All detected mutations were validated, and segregation analysis was subjected to the availability of parental samples by Sanger sequencing.
4. RESULTS
Fifty patients (male n = 28, female n = 22, the age at epilepsy onset range from 1 day to 9.3 years old, and the median age of onset 7 months) were recruited to our study. Blood was obtained from 47 patients, and buccal swab was obtained from 9 patients. Brain tissue was obtained from one subject with cortical dysplasia. For WES, an average coverage of 61× was achieved. The percentage of coding bases covered more than 10× for 546 gene panel ranged from 86× to 99×, with a median of 95%. We did not identify any positive findings by CMA. However, we identified 6 pathogenic or likely pathogenic variants, and one VUS by WES.
4.1. Patient 1
Results of WES showed a de novo germline variant NM_014191.3(SCN8A):c.2548C>G p.(R850G). SCN8A encodes for the alpha subunits of a sodium channel. Either germline or somatic mutations in this gene cause early infantile epileptic encephalopathy 13 (OMIM #614558). The variant p.(R850G) was absent in the ExAC database, and the constraint metrics of SCN8A missense variant had a z‐score of 7.71. This positive z‐score indicates that this gene is intolerant to a missense variation. Multiple in silico prediction algorithms suggested that this variant is disease‐causing or damaging. In addition, a mutation located in the same amino acid position, p.R850Q, was reported as disease‐causing in Chinese children with epilepsy.21 According to the ACMG guideline,20 the variant was classified as pathogenic and explained the epileptic phenotype well. However, the sensorineural hearing loss has not been reported previously to be associated with the SCN8A mutation. Whole exome analysis did not reveal any causative mutation related to hearing loss.
4.2. Patient 2
We identified a germline DEPDC5 splice‐site variant, NM_014662.4(DEPDC5):c.4427‐2A>G, from the subject's buccal mucosa using WES. The variant was inherited from his asymptomatic father. DEPDC5 is associated with familial focal epilepsy with variable foci (OMIM #604364), and focal cortical dysplasia.22 Around two‐thirds of reported DEPDC5 disease‐causing mutations are loss‐of‐function mutations.23 The c.4427‐2A>G variant was not reported in the gnomAD database. The variant was classified as pathogenic according to the ACMG guideline.20 Adulthood onset up to the age of 50 and incomplete penetrance has been reported previously24 potentially explaining his father's asymptomatic presentation despite being a variant carrier. His father was advised to have a brain magnetic resonance imaging (MRI) scan to check for malformations of cortical development.
4.3. Patient 3
A germline missense disease‐causing variant, NM_004992.3(MECP2):c.473C>T p.(T158M) was found using WES. MECP2 is located in the X chromosome and is required for neuronal maturation. Rett syndrome (OMIM #312750), which is usually found in females due to its lethality in males, is associated with this variant. Indeed, the mutation found in our subject was one of the most common mutations in Rett syndrome in Chinese patients.25
4.4. Patient 4
We found a splice‐site variant, NM_003159.2(CDKL5):c.2277‐2A>C, suggestive of mosaicism by using WES with 28% (19/67) mutant allele by depth. The variant is absent in the mother. The father's DNA was not available. However, given that the mutation is mosaic, it is likely de novo. Examination of the reads alignment and Sanger chromatogram results confirmed that this variant is mosaic. CDKL5 is located on the X chromosome. Loss of function variants, including splicing variants, were one of the pathogenic causes of CDKL5 epilepsy (OMIM #300672). Although most of the CDKL5‐related epilepsy was found in girls, the mosaic CDKL5 mutation has been reported in male patients with epilepsy as well.26, 27
4.5. Patient 5
We identified a germline novel variant, NM_001271.3(CHD2):c.1618G>Ap.(V540I), by using WES. CHD2 is associated with myoclonic encephalopathy. The variant was predicted as disease‐causing and damaging by multiple in silico tools. Furthermore, the variant is located at the N‐terminal domain of the SNF2 family, and it was clustered with other reported pathogenic mutations.28 The variant is absent in the mother. Although the paternal sample is not available for a complete segregation analysis, the variant was classified as likely pathogenic using the ACMG guideline20 and correlated well with the patient's phenotype.
4.6. Patient 6
We found a germline disease‐causing mutation, NM_001202435.2(SCN1A):c.4507G>A p.(E1503K), by using WES. Of interest, a likely pathogenic germline missense variant, NM_000335.4(SCN5A):c.1066G>A p.(D356N), was also identified. The SCN5A variant is related to Brugada syndrome (OMIM #601144) and has been associated with sudden death due to cardiac arrhythmia. In 2010, SCN5A:c.1066G>A p.(D356N) was reported as one of the most common mutations found in patients with Brugada syndrome.29 Because this variant was inherited from the father, we recommended electrocardiography (ECG) surveillance for the patient and her father, and we offered genetic tests to extended family members.
4.7. Patient 7
We identified a missense variant, NM_020822.2(KCNT1):c.1038C>A p.(F346L), via WES. KCNT1 encodes for the sodium‐activated potassium channel and is associated with early infantile epileptic encephalopathy 14 (OMIM #614959). In silico prediction tools were contradictory in their predictions of pathogenicity. The constraint metric in the ExAC database showed a positive z‐score of 3.52 for missense variant. A segregation study by Sanger sequencing found that his father is a mosaic carrier of this variant. Although the pathogenicity of this variant is not as strong as the variants found in other patients and was classified as VUS, the phenotype correlated very well with the reported literature—for example, flushing of the face. In addition to epilepsy, KCNT1 is also related to cardiac problems and leads to sudden unexpected death in epilepsy (SUDEP).30
The clinical features and variants of the subjects reported in this cohort are listed in Tables 1 and 2. Detailed clinical presentations are listed in Data S1. Figure 1 summarizes the exome and Sanger findings of each subject.
Table 1.
Clinical presentations of the patients reported in this cohort
| Patient No. | Sex | Age at onset (mo) | Epilepsy type | AEDs used | EEG findings | MRI findings |
|---|---|---|---|---|---|---|
| 1 | M | 3 | Focal seizure | *Topiramate, levetiracetam | Slow background, multifocal epileptic discharges | Cerebral atrophy |
| 2 | M | 9 | Infantile spasms evolving into focal seizure | *Carbamazepine, levetiracetam, phenobarbitone, phenytoin, steroid, clobazam | Right‐sided hypsarrhythmia evolving into right frontopolar epileptic discharges | Right frontal focal cortical dysplasia |
| 3 | F | 18 | Absence seizure | *Valproate, *phenobarbitone, *clobazam | Slow background, multifocal epileptic discharges | Cerebral atrophy |
| 4 | M | 2 | Infantile spasms evolving into tonic seizure and epileptic spasm | *Valproate, *lamotrigine, *levetiracetam, *ketogenic diet, clobazam, vigabatrin | Hypsarrhythmia evolving into slow background, generalized epileptic discharges | Cerebral atrophy |
| 5 | M | 20 | Myoclonic seizure | *clobazam, valproate, clonazepam | Generalized and focal epileptic discharges | Not performed |
| 6 | F | 13 | Generalized tonic‐clonic seizure, Focal seizure | *Valproate, *topiramate, clobazam, carbamazepine, gabapentin | Unremarkable | Unremarkable |
| 7 | M | 3 | Autonomic seizure with or without generalized tonic‐clonic seizure | *Carbamazepine, *phenobarbitone, *phenytoin, *valproate, *lamotrigine, *clonazepam, *ketogenic diet, topiramate | Slow background, generalized epileptic discharges | Cerebral atrophy and delayed myelination |
AED, anti‐epileptic drug; F, female; M, male.
*Drug administrated during recruitment.
Table 2.
Table summarized the epilepsy‐related variants reported in our cohort
| Patient No. | Source of DNA | Gene | Variant | Amino acid change | Population frequency in ExAC | Report(s) in literature | Inheritance | ACMG Classificationb, 17 | Increased risk of SUDEP? |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Blood | SCN8A | c.2548C>G | p.(R850G) | 0 | Novel | de novo | Pathogenic | Yes39 |
| 2 | Buccal | DEPDC5 | c.4427‐2A>G | / | 0 | Novel | Father | Pathogenic | Yes40 |
| 3 | Blood | MECP2 | c.473C>T | p.(T158M) | 0 | Reported40 | de novo | Likely pathogenic | No |
| 4 | Blood | CDKL5 | c.2277‐2A>C (Mosaic with ~30% variant in blood) | / | 0 | Novel | Not determined | Likely pathogenic | No |
| 5 | Blood | CHD2 | c.1618G>A | p.(V540I) | 0 | Novel | Not determined | Likely pathogenic | No |
| 6 | Blood | SCN1A | c.4507G>A | p.(E1503K) | 0 | Reported41 | de novo | Likely pathogenic | Yes40 |
| SCN5A a | c.1066G>A | p.(D356N) | 0 | Reported26 | Father | Likely pathogenic | Yes29 | ||
| 7 | Blood | KCNT1 | c.1038C>A | p.(F346L) | 0 | Novel | Father (mosaic carrier) | VUS | Yes30, 42 |
Finding related to Brugada syndrome.
Detailed classification was listed in Table S3.
Figure 1.
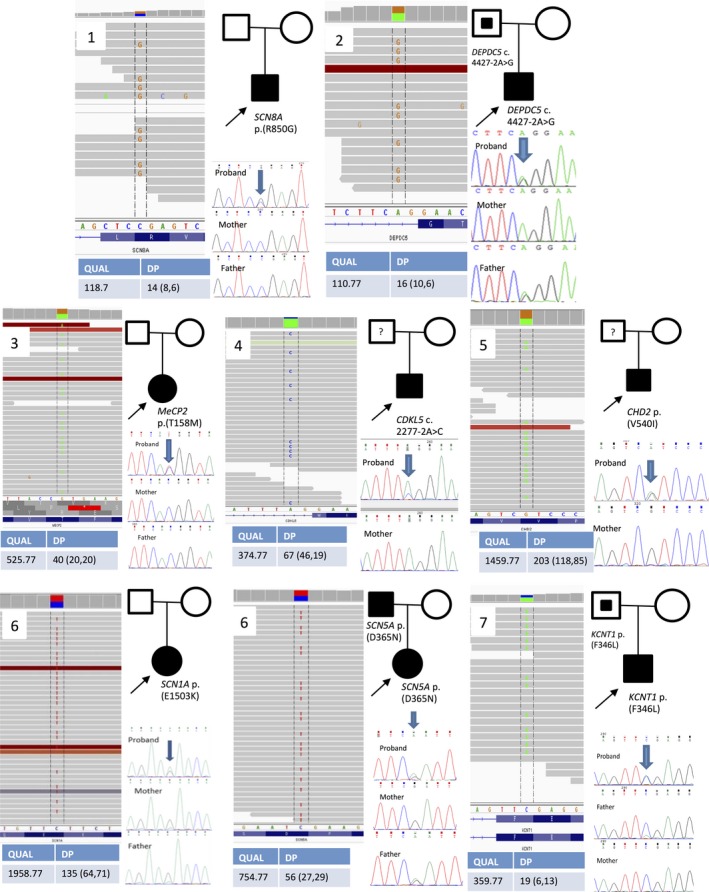
The Integrative Genomics Viewer (IGV) view of the variant from the WES data, pedigree and segregation analysis by Sanger sequencing. For patient 4, the estimated percentage of mosaicism using Depth (DP) was around 30% (20/70). The Sanger chromatogram indicated the variant by the arrow. For the proband, the indicated base has a small blue peak representing the alternated “C” base. The peak height is not identical, suggesting its mosaic nature. At the same variant location, the mother has a green peak only. For patient 7, Sanger chromatogram of the father showed a small green peak indicating that a mosaic variant was found
In summary, the overall diagnostic yield in our study is 12% (6/50). There was one VUS found (1/50, 2%). All patients’ phenotypes correlated well with the clinical features reported for their genetic mutation. Of note, a reported pathogenic SCN5A mutation related to Brugada syndrome was noted in a patient with an SCN1A mutation.
5. DISCUSSION
The diagnostic yield of our study was 12% (6/50). This yield was lower than expected given that the other 2 drug‐resistant epilepsies studies had a diagnostic yield of approximately 20% to 30%.31, 32 It should be noted that our study design was different from that of the previous studies. The study of Ream and Mikati was retrospective, with drug‐resistant epilepsy patients presenting with developmental delay, epileptic encephalopathy, and generalized epilepsy.31 Our recruitment criterion is pediatric‐onset drug‐resistant epilepsy subjects only. This may have decreased the percentage of patients with an epilepsy‐associated variant. The study of Parrini et al32 has a large cohort (n = 349), with age at onset in the first years of life, whereas ours is a small cohort with later age at onset. The other possible explanation is that we have excluded the 15 patients with SMEI because SCN1A Sanger sequencing is available in our center.33 Patient 6 was recruited into our present study because her phenotype was less typical of Severe myoclonic epilepsy of infants (SMEI) (seizure onset after 1 year of age with normal early neurodevelopmental status before and absence of myoclonus), and thus Sanger sequencing for SCN1A was not initiated.
In addition, we did not have any positive results by CMA where a reported diagnostic figure was 5%.6 This could be explained by the reported cohort being recruited using International Classification of Diseases, Ninth Revision (ICD‐9) codes for epilepsy or seizures, while our inclusion criteria were more stringent involving subjects with drug‐resistant epilepsy specifically.
Our findings suggest that the application of genetic testing can benefit our patients in several ways:
6. CONSOLIDATION OF CLINICAL DIAGNOSIS
Genetic diagnosis can help to consolidate clinical diagnosis. Patient 3 first presented with a congenital malformation, that is, sacrococcygeal myelomeningocele with essentially normal neurodevelopment except for mild gross motor delay, until regression began between 1 and 2 years of age without progressive microcephaly. This clinical picture is not particularly suggestive of Rett syndrome, and this patient's eventual diagnosis was via WES. By using WES, we were able to identify the reported de novo MECP2 missense disease‐causing variant and help the patient avoid further investigation. The same principle can also be exemplified by patients 1, 4, and 5, when the intractable epilepsy phenotype is mixed with a variable combination of severe global developmental delay, movement disorder, upper motor neuron syndrome, oromotor dysfunction, cortical visual impairment, and sensorineural deafness. Without a proper molecular diagnosis, clinicians might further proceed to subsequent neurometabolic testing. These metabolic tests can be time consuming, expensive, and invasive (such as muscle biopsy). Genetic counseling based on the de novo or mosaic finding is also useful, as the psychological pressure of having another affected child can be relieved.
In our study, we have also extracted DNA from buccal mucosa if the patients presented with a phenotype suspicious of either tissue overgrowth or cortical dysplasia syndrome. The primary aim is to identify somatic mutations for genetic overgrowth syndrome.14 This kind of mutation would probably be missed by sequencing of blood DNA. Indeed, although no overgrowth syndrome was identified, we have made the genetic diagnosis in one patient with focal epilepsy and cortical dysplasia as evidenced by their MRI findings.
7. PROVIDING TREATMENT RECOMMENDATION
In addition to providing an accurate diagnosis, elucidating the genetic cause of pediatric‐onset drug‐resistant epilepsy can also guide clinical management. For example, patient 1 has a disease‐causing variant in the sodium channel gene SCN8A. A sodium channel blocker, oxcarbazepine, has been found to be beneficial in SCN8A‐related epilepsy in a Chinese patient.21 For patient 7 with a KCNT1 variant, quinidine may be effective in controlling the epilepsy. It should be noted that although quinidine may be beneficial to this patient from an epilepsy standpoint, extra care and monitoring is required as the drug is an antiarrhythmic agent that may induce cardiac arrythmias.34 For Patient 2 with a DEPDC5 disease‐causing variant, epilepsy surgery may cure the disease if the epileptogenic area can be completely removed.22
Beyond treatment suggestions, a verified genetic diagnosis can warn clinicians of treatment choices that should be avoided. For patients 4 and 5 who have CDKL5 and CHD2 variants, respectively, a ketogenic diet is proven not to have long‐term efficacy35 and has not been shown to be effective in seizure management.28 Careful consideration and close monitoring should be taken before starting and throughout a ketogenic diet in these 2 patients as significant side effects may be introduced. In the case of patient 6 with coexistence of SCN1A and SCN5A pathogenic variants, the commonly used antiepileptic drug carbamazepine is not suitable as it has been shown to worsen the seizure status of patients with SCN1A variants.36
8. IDENTIFICATION OF SUDEP RISK IN PATIENTS WITH EPILEPSY
From our cohort, we identified 4 patients with increased SUDEP risk.
Various risk factors for SUDEP have been proposed—for example, sex, seizure frequency, and types and dosages of AEDs used. However, no concrete conclusions have been drawn.37 In 2017, Bagnall et al38 reported respiratory dysfunction, cardiac arrhythmia, and electroencephalography (EEG) suppression as the major mechanism of death in SUDEP patients. A higher genetic risk was found in 8 genes, including genes encoded for ion channels such as KCNA1, KCNQ1, KCNH2, SCN1A, SCN2A, SCN5A, SCN8A, and DEPDC5, a regulatory gene in the mammalian target of rapamycin (mTOR) pathway.
Patient 1 is a boy with a novel SCN8A disease‐causing variant, one of the 8 genes associated with a higher SUDEP risk. Furthermore, mutations in SCN8A have been reported to cause cardiac arrhythmia by hyperexcitability of sodium channel NAv1.6 in mice.39 This evidence indicates that regular ECG surveillance might be of benefit.
For patient 2 with a DEPDC5 splicing variant, it was reported that nonsense variants in this gene cause a higher SUDEP risk. However, the causal relationship and the molecular/pathogenic mechanism have not yet been elucidated.40 Close monitoring with proper surveillance, for example, serial EEG studies and MRI of the brain are beneficial to this patient's management.
The SCN5A variant in patient 6 is both a known contributor to Brugada syndrome and a risk factor for SUDEP. This indicates that this patient has a multitude of risk factors for cardiac arrythmias and requires regular ECG surveillance. The patient has demonstrated the scenario of “one man two diseases,” which was introduced since the application of genomic testing in disease studies.41 The variant will probably be missed if only targeted SCN1A sequencing was performed.
The KCNT1 variant found in patient 7 was also associated with SUDEP. Previously, KCNT1 was reported to cause cardiac arrhythmia.30 Recent study, however, demonstrated that respiratory failure could contribute to the sudden death in KCNT1‐mutated patients.42
These findings are essential in the medical management of the patients. Because SUDEP usually occurs in patients from the 30s to the 40s,40 long‐term follow‐up to adulthood will be beneficial to these patients. Furthermore, the risk of sudden death may be monitored by simple clinical tests, for example, ECG. This shows how CMA and WES are useful in this cohort even beyond their epilepsy status.
One of the limitations of our study is a lack of longitudinal follow‐up of the patients. This is required to prove the findings on increased risk of SUDEP. Further study of the risk of an individual gene in causing SUDEP would also be interesting but was beyond the scope of this particular study.
To conclude, the diagnostic yield of CMA and WES in our cohort is 12%. Our finding demonstrated that a proper genetic diagnosis is crucial for patients with pediatric‐onset drug‐resistant epilepsy.
DISCLOSURE
None of the authors has any conflict of interest to disclose.
9.
We confirm that we have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Supporting information
ACKNOWLEDGMENTS
We would like to thank The Society for the Relief of Disabled Children and The Edward and Yolanda Wong Fund for their support.
Tsang MH‐Y, Leung GK‐C, Ho AC‐C, et al. Exome sequencing identifies molecular diagnosis in children with drug‐resistant epilepsy. Epilepsia Open. 2019;4:63–72. 10.1002/epi4.12282
Contributor Information
Ada Wing‐Yan Yung, Email: ayung@hku.hk.
Cheuk‐Wing Fung, Email: fcw1209m@hku.hk.
Brian Hon‐Yin Chung, Email: bhychung@hku.hk.
REFERENCES
- 1. Beck C, Moulard B, Steinlein O, et al. A nonsense mutation in the alpha4 subunit of the nicotinic acetylcholine receptor (CHRNA4) cosegregates with 20q‐linked benign neonatal familial convulsions (EBNI). Neurobiol Dis 1994;1:95–9. [DOI] [PubMed] [Google Scholar]
- 2. Ottman R, Hirose S, Jain S, et al. Genetic testing in the epilepsies—Report of the ILAE genetics commission. Epilepsia 2010;51:655–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hildebrand MS, Dahl HH, Damiano JA, et al. Recent advances in the molecular genetics of epilepsy. J Med Genet 2013;50:271–9. [DOI] [PubMed] [Google Scholar]
- 4. Weber YG, Lerche H. Genetic mechanisms in idiopathic epilepsies. Dev Med Child Neurol 2008;50:648–54. [DOI] [PubMed] [Google Scholar]
- 5. Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia 2017;58:512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol 2014;75:943–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helbig KL, Farwell Hagman KD, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med 2016;18:898–905. [DOI] [PubMed] [Google Scholar]
- 8. Epi4K Consortium , Epilepsy Phenome/Genome Project , Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyment DA, Tetreault M, Beaulieu CL, et al. Whole‐exome sequencing broadens the phenotypic spectrum of rare pediatric epilepsy: a retrospective study. Clin Genet 2015;88:34–40. [DOI] [PubMed] [Google Scholar]
- 10. Kwan P, Arzimanoglou A, Berg AT, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia 2010;51:1069–77. [DOI] [PubMed] [Google Scholar]
- 11. Kwan P, Brodie MJ. Refractory epilepsy: mechanisms and solutions. Expert Rev Neurother 2006;6:397–406. [DOI] [PubMed] [Google Scholar]
- 12. Laxer KD, Trinka E, Hirsch LJ, et al. The consequences of refractory epilepsy and its treatment. Epilepsy Behav 2014;37:59–70. [DOI] [PubMed] [Google Scholar]
- 13. Lee JH, Huynh M, Silhavy JL, et al. De novo somatic mutations in components of the PI3K‐AKT3‐mTOR pathway cause hemimegalencephaly. Nat Genet 2012;44:941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeung KS, Tso WWY, Ip JJK, et al. Identification of mutations in the PI3K‐AKT‐mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Mol Autism 2017;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tao VQ, Chan KYK, Chu YWY, et al. The clinical impact of chromosomal microarray on paediatric care in Hong Kong. PLoS ONE 2014;9:e109629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and accurate short read alignment with Burrows‐Wheeler Transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Online Mendelian Inheritance in Man, OMIM® . McKusick‐Nathans Institute of Genetic Medicine, Johns Hopkins University; (Baltimore, MD: ). World Wide Web https://omim.org/ [Google Scholar]
- 18. CURE'S Epilepsy Genetics Initiative. (n.d.). Retrieved from https://www.cureepilepsy.org/egi/
- 19. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein‐coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kong W, Zhang Y, Gao Y, et al. SCN8A mutations in Chinese children with early onset epilepsy and intellectual disability. Epilepsia 2015;56:431–8. [DOI] [PubMed] [Google Scholar]
- 22. Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol 2015;77:675–83. [DOI] [PubMed] [Google Scholar]
- 23. Baulac S. Genetics advances in autosomal dominant focal epilepsies: focus on DEPDC5. Prog Brain Res 2014;213:123–39. [DOI] [PubMed] [Google Scholar]
- 24. Dibbens LM, de Vries B, Donatello S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet 2013;45:546–51. [DOI] [PubMed] [Google Scholar]
- 25. Li MR, Pan H, Bao XH, et al. MECP2 and CDKL5 gene mutation analysis in Chinese patients with Rett syndrome. J Hum Genet 2007;52:38–47. [DOI] [PubMed] [Google Scholar]
- 26. Mei D, Darra F, Barba C, et al. Optimizing the molecular diagnosis of CDKL5 gene‐related epileptic encephalopathy in boys. Epilepsia 2014;55:1748–53. [DOI] [PubMed] [Google Scholar]
- 27. Stosser MB, Lindy AS, Butler E, et al. High frequency of mosaic pathogenic variants in genes causing epilepsy‐related neurodevelopmental disorders. Genet Med 2018;20:403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas RH, Zhang LM, Carvill GL, et al. CHD2 myoclonic encephalopathy is frequently associated with self‐induced seizures. Neurology 2015;84:951–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A‐encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm 2010;7:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moller RS, Heron SE, Larsen LH, et al. Mutations in KCNT1 cause a spectrum of focal epilepsies. Epilepsia 2015;56:e114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ream MA, Mikati MA. Clinical utility of genetic testing in pediatric drug‐resistant epilepsy: a pilot study. Epilepsy Behav 2014;37:241–8. [DOI] [PubMed] [Google Scholar]
- 32. Parrini E, Marini C, Mei D, et al. Diagnostic targeted resequencing in 349 patients with drug‐resistant pediatric epilepsies identifies causative mutations in 30 different genes. Hum Mutat 2017;38:216–25. [DOI] [PubMed] [Google Scholar]
- 33. Kwong AK, Fung CW, Chan SY, et al. Identification of SCN1A and PCDH19 mutations in Chinese children with Dravet syndrome. PLoS ONE 2012;7:e41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mikati MA, Jiang YH, Carboni M, et al. Quinidine in the treatment of KCNT1‐positive epilepsies. Ann Neurol 2015;78:995–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lim Z, Wong K, Olson HE, et al. Use of the ketogenic diet to manage refractory epilepsy in CDKL5 disorder: experience of >100 patients. Epilepsia 2017;58:1415–22. [DOI] [PubMed] [Google Scholar]
- 36. Ceulemans B, Boel M, Claes L, et al. Severe myoclonic epilepsy in infancy: toward an optimal treatment. J Child Neurol 2004;19:516–21. [DOI] [PubMed] [Google Scholar]
- 37. Tomson T, Nashef L, Ryvlin P. Sudden unexpected death in epilepsy: current knowledge and future directions. Lancet Neurol 2008;7:1021–31. [DOI] [PubMed] [Google Scholar]
- 38. Bagnall RD, Crompton DE, Semsarian C. Genetic basis of sudden unexpected death in epilepsy. Front Neurol. 2017;8:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frasier CR, Wagnon JL, Bao YO, et al. Cardiac arrhythmia in a mouse model of sodium channel SCN8A epileptic encephalopathy. Proc Natl Acad Sci U S A. 2016;113(45):12838–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bagnall RD, Crompton DE, Petrovski S, et al. Exome‐based analysis of cardiac arrhythmia, respiratory control, and epilepsy genes in sudden unexpected death in epilepsy. Ann Neurol 2016;79:522–34. [DOI] [PubMed] [Google Scholar]
- 41. Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole‐exome sequencing. JAMA 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuchenbuch M, Kaminska A, Chemaly N, et al. Long term outcome of patients with Epilepsy with migrating focal seizure in infancy (EMFSI) due to KCNT1 mutation. Eur J Paediatr Neurol 2017;21:e189. [Google Scholar]
- 43. Adkins NL, Georgel PT. MeCP2: structure and function. Biochem Cell Biol 2011;89:1–11. [DOI] [PubMed] [Google Scholar]
- 44. Wang JW, Shi XY, Kurahashi H, et al. Prevalence of SCN1A mutations in children with suspected Dravet syndrome and intractable childhood epilepsy. Epilepsy Res 2012;102:195–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


